Abstract
Background
Integrative Conjugative Elements (ICEs) are important factors in the plasticity of microbial genomes. An element related to the ICE Tn4371 was discovered during a bioinformatic search of the Ralstonia pickettii 12J genome. This element was analysed and further searches carried out for additional elements.
A PCR method was designed to detect and characterise new elements of this type based on this scaffold and a culture collection of fifty-eight Ralstonia pickettii and Ralstonia insidiosa strains were analysed for the presence of the element.
Results
Comparative sequence analysis of bacterial genomes has revealed the presence of a number of uncharacterised Tn4371-like ICEs in the genomes of several β and γ- Proteobacteria. These elements vary in size, GC content, putative function and have a mosaic-like structure of plasmid- and phage-like sequences which is typical of Tn4371-like ICEs. These elements were found after a through search of the GenBank database. The elements, which are found in Ralstonia, Delftia, Acidovorax, Bordetella, Comamonas, Acidovorax, Congregibacter, Shewanella, Pseudomonas Stenotrophomonas, Thioalkalivibrio sp. HL-EbGR7, Polaromonas, Burkholderia and Diaphorobacter sp. share a common scaffold. A PCR method was designed (based on the Tn4371- like element detected in the Ralstonia pickettii 12J genome) to detect and characterise new elements of this type.
Conclusion
All elements found in this study possess a common scaffold of core genes but contain different accessory genes. A new uniform nomenclature is suggested for ICEs of the Tn4371 family. Two novel Tn4371-like ICE were discovered and characterised, using the novel PCR method described in two different isolates of Ralstonia pickettii from laboratory purified water.
Background
Integrative Conjugative Elements (ICEs) carry functional modules involved in their conjugative transfer, chromosomal integration and for control of expression of ICE genes [1]. ICEs are maintained in their host via site-specific integration and establishment at a unique site or sites in their host [2-7]. ICEs have been discovered in the genomes of various low G+C Gram-positive bacteria, various α, β- and γ-Proteobacteria, and Bacteroides species [8]. The first ICE found was Tn916 from Bacteroides species [8].
One of the best models of ICEs is a family of elements called the R391\SXT family that are found in γ-Proteobacteria. These are interesting elements as over 25 have been found to date in organisms spread across the world. They share a common core scaffold of genes related to integration, excision, transfer and regulation. Different elements can possess different fitness determinants such as antibiotic resistances, heavy metal resistances, and error-prone DNA repair systems [9].
Tn4371 is a 55-kb ICE, which allows its host to degrade biphenyl and 4-chlorobiphenyl. It was isolated after mating between Cupriavidus oxalaticus (Ralstonia oxalatica) A5 carrying the broad-host-range conjugative plasmid RP4 and Cupriavidus metallidurans (Ralstonia metallidurans) CH34. Selection was applied for transconjugants that expressed the heavy metal resistances from CH34 and grew with biphenyl as a sole source of carbon and energy [10]. The transconjugants carried an RP4 plasmid with a 55-kb insert near its tetracycline resistance operon. The insert was shown to transpose to other locations and hence was called Tn4371 [10-12]. Tn4371 has been sequenced [13] and closely related elements have been found in the genome sequences of a number of bacteria including Ralstonia solanacearum GMI1000, a phytopathogen from French Guyana [14], Cupriavidus metallidurans CH34, a heavy metal resistant bacteria from Belgium [15], Erwinia chrysanthemi 3937, aphytopathogen [16] and Azotobacter vinelandii AvOP, a nitrogen-fixing bacterium isolated from soil in the USA [13,17]. None of these other elements possessed the biphenyl and 4-chlorobiphenyl degradation genes.
The Tn4371-like ICEs characterised to date are mosaic in structure consisting of Ti-RP4-like transfer systems, an integrase region, plasmid maintenance genes and accessory genes [13]. All the characterised elements integrate into sites on the bacterial genomes with a conserved 5'-TTTTTCAT-3' sequence, termed the attB site [11]. Tn4371 transposition most likely involves a site-specific integration/excision process, since the ends of the element can be detected covalently linked as a transfer intermediate [11,13]. Integration is catalysed by a tyrosine based site specific recombinase related to bacteriophage and ICE family integrases [18].
A small number of putative ICEs have been discovered following sequence analyses of genomes of various low G+C Gram-positive bacteria [19], various α, β- and γ-Proteobacteria [20-22], and Bacteroides species [23].
We now report the discovery and comparative analysis of a number of novel uncharacterised Tn4371-like ICEs from several different bacterial species. These elements are also mosaics of plasmid and other genes and posses a common scaffold with apparent hotspots containing insertions of different presumably adaptive genes. Using sequences from the common scaffold a PCR method was developed to discover and characterise new Tn4371-like ICEs in different bacteria. Here we report on the use of this method to discover and characterise two new Tn4371-like ICEs in Ralstonia pickettii strains isolated from a purified water system. Furthermore we propose a uniform nomenclature for newly discovered ICEs of the Tn4371 family
Results and Discussion
Bioinformatic analysis of Tn4371-like ICEs
Using bioinformatic analysis tools, searches of the genome databases for elements similar to the Tn4371 element were carried out using the original Tn4371 sequence as a probe. The method used was similar to that used to detect novel members of the R391/SXT family of ICEs in Enterobacteriaceae [22]. In this study novel unreported ICEs closely related to Tn4371 were discovered in the genome sequences of several different bacteria including the β-proteobacteria, two elements in Delftia acidovorans SPH-1, and a single element Comamonas testosteroni KF-1, Acidovorax avenae subsp. citrulli AAC00-1, Bordetella petrii DSM12804, Acidovorax sp. JS42, Polaromonas naphthalenivorans CJ2 plasmid pPNAP01, Burkholderia pseudomallei MSHR346 and Diaphorobacter sp. TPSY [Table 1]. Novel elements were also found in the γ-proteobacteria Congregibacter litoralis KT71, Shewanella sp. ANA-3, Pseudomonas aeruginosa 2192, Pseudomonas aeruginosa PA7, Pseudomonas aeruginosa PACS171b, Pseudomonas aeruginosa UCBPP-PA14, Stenotrophomonas maltophilia K279a, Thioalkalivibrio sp. HL-EbGR7 [Table 2]. The element in Bordetella petrii DSM12804 was previously identified but not analyzed in a paper by Lechner et al., [24]. The elements found in Delftia acidovorans SPH-1, Comamonas testosteroni KF-1 and Bordetella petrii DSM12804 were also partially characterised along with further information on the elements in Cupriavidus metallidurans CH34 in a paper by Van Houdt et al., [25]. Geographically all these bacteria were found in different locations in both Europe and the Americas and were isolated from many different environments including activated sludge, polluted water and clinical situations [Table 1 and 2]. All elements contained different inserts [containing accessory genes] in the core backbone except for those found in Delftia acidovorans SPH-1 and Comamonas testosteroni KF-1. The size of the newly discovered elements varied from 42 to 70 Kb and the GC content from 59 to 65% [Table 1 and 2].
Table 1.
Size and %GC Content, accessory Genes contained in and the location and environment of isolated strains containing Tn4371-like ICEs fromβ-Proteobacteria
| Tn4371-like Elements | Size | %GC Content | Location | Environment | Accessory Genes | Reference | Name | Accession Number |
|---|---|---|---|---|---|---|---|---|
| Ralstonia pickettii 12J | 54121 bp | 64.63 | USA | Copper-contaminated sediment from a lake | Lipid metabolism | [74] | ICETn43716033 | CP001068 |
| Acidovorax avenae subsp. citrulli AAC00-1 | 59844 bp | 63.12 | USA | Watermelon | Insertion Sequences metabolism |
[75] | ICETn43716036 | NC_008752 |
| Delftia acidovorans SPH-1 | 57901 bp | 63.66 | Germany | Activated sludge | czc metal resistance pumps | [76] | ICETn43716037 | NC_010002 |
| Comamonas testosteroni KF-1 | 52455 bp | 63.77 | Switzerland | Activated sludge | czc metal resistance pumps | [76] | ICETn43716038 | NZ_AAUJ0100000 |
| Acidovorax sp. JS42 | 53489 bp | 62.88 | USA | Groundwater | Multidrug resistance pump Insertion Sequences |
[77] | ICETn43716039 | NC_008782 |
|
Bordetella petrii DSM12804 |
47191 bp | 63.73 | Germany | River sediment | Aromatic compounds metabolism | [78] | ICETn43716040 | NC_010170 |
| Burkholderia pseudomallei MSHR346 | 49278 bp | 62.21 | Australia | Melioidosis patient | metabolism | N/A | ICETn43716064 | CP001408 |
| Polaromonas naphthalenivorans CJ2 plasmid pPNAP01 | 70106 bp | 62.89 | USA | Coal-tar-waste contaminated site | Biphenyl degradation | [79] | ICETn43716065 | CP000530 |
| Diaphorobacter sp. TPSY | 49020 bp | 65.30 | USA | Soil | czc metal resistance pumps | [80] | ICETn43716066 | CP001392 |
| Delftia acidovorans SPH-1 | 66755 bp | 64.94 | Germany | Activated sludge | Various types of metal resistance pumps | [76] | ICETn43716067 | NC_010002 |
Table 2.
Size and %GC Content, accessory Genes contained in and the location and environment of isolated strains containing Tn4371-like ICEs from γ-Proteobacteria
| Tn4371-like Elements | Size | %GC Content | Location | Environment | Accessory Genes | Reference | Name | Accession Number |
|---|---|---|---|---|---|---|---|---|
| Shewanella sp. ANA-3 | 45233 bp | 59.43 | USA | Arsenate treated wood pier | Multidrug resistance pump | [81] | ICETn43716034 | NC_008577 |
| Congregibacter litoralis KT71 | 50661 bp | 59.52 | North Sea | Ocean-surface water | RND type multidrug efflux pump | [82] | ICETn43716035 | NZ_AAOA01000008 |
| Pseudomonas aeruginosa 2192 | 48538 bp | 62.62 | USA | Cystic fibrosis patient | RND type multidrug efflux pump | [83] | ICETn43716041 | NZ_AAKW01000024 |
| Pseudomonas aeruginosa PA7 | 55287 bp | 52.38 | Argentina | Clinical wound isolate | Multiple antibiotic resistance genes Potassium transporter system |
[84] | ICETn43716042 | NC_009656 |
| Stenotrophomonas maltophilia K279a | 43509 bp | 62.76 | UK | Blood infection | Multidrug resistance pump | [85] | ICETn43716068 | AM743169 |
| Pseudomonas aeruginosa UCBPP-PA14 | 43172 bp | 65.55 | USA | Burn patient | czc metal resistance pumps | [86] | ICETn43716069 | CP000438 |
| Pseudomonas aeruginosa PACS171b | 42156 bp | 64.12 | USA | Cystic fibrosis patient | Arsenate resistance pumps | [87] | ICETn43716070 | EU595746 |
| Thioalkalivibrio sp. HL-EbGR7 | 42540 bp | 64.95 | Unknown | bioreactor removing sulfide from biogas | Potassium transporter system | [88] | ICETn43716071 | CP001392 |
Characterisation of Tn4371-like ICEs in whole genome sequences
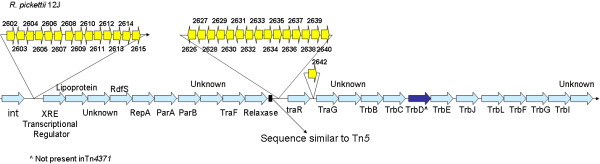
The core structure conserved amongst known Tn4371-like ICEs is presented in Fig. 1. At the attR end of the elements a putative int gene [that bears similarities to tyrosine based site-specific recombinases historically called phage-like integrases [20], possessing the R-H-R-Y tetrad] is found [Additional file 1]. A phylogenetic study was carried out on all available Tn4371-like int genes and tyrosine recombinases from phages and other ICEs. The phylogenetic tree can be seen in Additional file 2. These Tn4371-like int genes grouped with the int genes of ICE Hin1056, an ICE from Haemophilus influenzae and from phages related to the P22 phage. The int gene was found in all characterised elements and was followed by nonconserved ORFs which differed from element to element. These ORFs include putative DNA helicases and nucleases, proteins with β-lactamase domains, similar to RadC DNA repair proteins, putative reductases, transposases of insertion sequences, putative ubiquitin-activating enzymes, putative transcriptional regulators and many different hypothetical proteins whose functions are unknown [Fig. 1, Additional file 3]. These ORF's were found in differing arrangements in each of the different elements. Polaromonas naphthalenivorans CJ2 plasmid pPNAP01 contained biphenyl degradation genes in this area of the element and these genes are similar to those found in the original Tn4371 element but are found in a different part of the element. Pseudomonas aeruginosa PACS171b and the second Delftia acidovorans SPH-1 element have an arsenate resistance system located in this region. This system is related to the ars system, and has the genes arsH, arsC, arsB and arsA in the operon in this bacterium. The function of arsH is unknown; however it is necessary for resistance to arsenic in the Yersinia enterocolitica virulence plasmid pYV [27]. The arsC gene encodes a soluble arsenate reductase which reduces intracellular arsenate to arsenite for efflux from the cell [28]. The arsA gene codes for a unique ATPase which binds to the ArsB membrane protein forming an anion transporting arsenite pump [28]. The arsD gene encodes an inducer independent regulatory protein which controls the upper level of operon expression [29]. The second Delftia acidovorans SPH-1 element has genes related to the Mer (Mercury Resistance) operon: merR, merT, merP and merA. The merR gene controls regulation of the operon, merT and merP transport of the mercury ions and merA reduction of the mercury ions [30]. This region also contains a predicted czc [Cd/Zn/Co] efflux system [31,32]. Czc mediates the inducible resistance to Co2+, Zn2+ and Cd2+, the protein products of gens czcA, czcB and czc form a membrane-bound protein complex catalysing an energy dependant efflux of these three metal ions [33].
Figure 1.
Common core scaffold of Tn4371-like ICEs (in blue) and above inserted genes present in R. pickettii ICETn43716033 (in yellow).
Following the integrase gene two conserved genes [ORF00013 and ORF00014 in Tn4371] were present in most elements except those in C. litoralis KT71 and Shewanella sp. ANA-3. These ORF's are related to proteins encoded by genes located near the transfer origin of Escherichia coli F plasmid [Q9WTE4 and Q9S4W2]. Although the function of the first protein is unknown, the second shows similarity to ParB-like nucleases initially identified as a critical element in the faithful partitioning of plasmid DNA during cell division in the absence of selection pressure [34,35]. Subsequently, a number of similar proteins have been identified in prokaryotes and archea which carry out the function of segregation of genomic DNA during cell division. ParB homologs are present in almost all eubacteria chromosomes [36].
The next region on all elements contains proteins similar of the XRE [Xenobiotic Responsive Element] family of transcriptional regulators, a putative lipoprotein with a DNA binding domain and a protein of unknown function. The XRE family behave as lambda repressor-like proteins associated with different phages, including Staphylococcus aureus phage phi 11 [37] and the Bacillus subtilis defective prophage PBSX [[38], Fig. 1]. Two different homologues of the XRE were found in different elements one related to that found in the original Tn4371 element (R. pickettii 12J, D. acidovorans SPH-1, A. avenae subsp. citrulli AAC00-1, C. testosteroni KF-1 and Acidovorax sp. JS42, C. litoralis KT71, Shewanella sp. ANA-3, P. aeruginosa 2192 and P. aeruginosa PA7, P. aeruginosa PACS171b, Thioalkalivibrio sp. HL-EbGR7 and B. pseudomallei MSHR346). A different XRE was found in the remaining elements: B. petrii DSM 12804, S. maltophilia K279a, P. aeruginosa UCBPP-PA14, Diaphorobacter sp. TPSY, P. naphthalenivorans CJ2 plasmid pPNAP01 and the second element of Delftia acidovorans SPH-1.
Following on from the XRE transcriptional regulators, a protein [ORF00035 of Tn4371] was found with similarity to the RdfS excisionase [CAD31514] of ICEMlSymR7A, the symbiosis island of Mesorhizobium loti R7A [39]. Most excisionases, also called recombination directionality factors [RDF's], share a number of conserved features: they are small [usually <100 amino acids] DNA-binding proteins, that are typically basic with the majority of known RDFs having isoelectric points in the range of pH 8-10 [40]. The size of the ORF00035 protein homologues found in this comparative analysis ranged from 89-98 aa [amino acid] and had pI's ranging from 8.14 to 9.59. BlastP scores showed approximately 50% aa identity with the ICEMlSymR7A RdfS, over approximately 55 aa for all of the putative RdfSs discovered in this study [Fig. 1]. No excisionase was found in the second Delftia acidovorans SPH-1 element. The location of this ORF is also of interest as usually excisionases are found close to the integrase gene in most ICEs particularly the SXT/R391 family [41].
The next sequence region of the elements contained plasmid-related genes whose predicated products were related to the RepA [replication] protein of Pseudomonas plasmid pVS1 [BAA96327, 42], plasmid pMLb of M. loti MAFF303099 [NP_109574, 43] and plasmid pEMT8 [CAC94910, 44]; this gene maybe involved in replication of the element [13]. The ParA partition protein of the type Ib family [45] and its associated ParB protein was also found but in all cases the ParB was truncated. Rep and Par proteins have been proposed to act as a stabilisation system for the maintenance of mobile elements in bacterial genomes [19,36], similar to the toxin-anti-toxin system encoded by ORFs s044 and s045 of the SXT-ICE [46]. Qui et al. found that the P. aeruginosa ICE PAPI-1 contains a homologue of the plasmid and chromosome partitioning gene soj (parA). They demonstrated that deletion of the soj homologue from PAPI-1 resulted in complete loss of PAPI-1 from P. aeruginosa. The mechanism by which the Soj protein promotes PAPI-1 maintenance remains to be elucidated [47]. Similar genes to soj have been found in ICE Hin1056 and ICEA [20,48]. This region was followed by an ORF encoding a conserved hypothetical protein [ORF00040 in Tn4371] whose function is unknown [Fig. 1].
This sequence is followed by a region containing transfer like proteins, the first being a putative conjugation protein TraF related to the pilus assembly proteins of IncP plasmids. This TraF protein is a protease that acts upon the pilus assembly protein TrbC [49]. The second is a putative relaxase-like protein [ORF00041 in Tn4371] that has similarity to the VirD2 protein of Ti plasmids and to the RlxS [relaxase, CAD31511] of ICEMlSymR7A. Transfer and maintenance of ICEMlSymR7A in cells has been shown to be dependent on the relaxase protein RlxS [[39], Fig. 1]. A relaxase, usually encoded by the plasmid, recognizes oriT, makes a single-strand DNA break (a nick) in oriT, and covalently attaches to the 5' end of the nicked DNA strand via a phosphotyrosyl linkage. No helicase domain was found in examining the protein so this indicates that the element may use leading-strand DNA synthesis (rolling-circle replication) from the nicked 3' end to promote strand displacement and single-strand DNA transfer [50,51].
Following the putative relaxase-like protein is a variable region encoding a number of different ORFs, which vary from element to element; these genes encode putative antibiotic genes, heavy metal resistance pumps and degradative and metabolic enzymes which may have originated by transposition into the element. The sequence between the putative relaxase gene and the first gene of the variable region, in all elements, is similar to the sequence of an area of Tn5 [U00004] indicating that the diversity in this region maybe due to one or a number of Tn5 mediated insertion events. This variable region in the novel ICE in R. pickettii 12J encodes a putative set of lipid metabolising genes [Fig. 1]. These are closely related to genes from Pseudomonas putida W619 [NZ_AAVY01000010.1] and from the pREC1 plasmid from Rhodococcus erythropolis PR4 [NC_007486] [52]. A. avenae subsp. citrulli AAC00-1 contained insertion sequences and homologues to general metabolism proteins whose exact functions are unknown. D. acidovorans SPH-1 and C. testosteroni KF-1 contain a predicted czc [Cd/Zn/Co] efflux system [31,32] in their variable regions. The novel element in Acidovorax sp. JS42 contains genes that show similarity to a multidrug resistance pump and insertion sequences [InterPro Scan] in this region. In the variable region in B. petrii DSM 12804 there are various proteins that are putatively involved in degradation, however their exact function is unknown. Burkholderia pseudomallei MSHR346 has genes that are putatively involved in xenobiotic metabolism; however again their exact function is unknown. Polaromonas naphthalenivorans CJ2 plasmid pPNAP01 contains a putative antibiotic resistance pump and metabolism proteins whose role have not been identified. Diaphorobacter sp. TPSY contains a predicted czc [Cd/Zn/Co] efflux system similar to those in D. acidovorans SPH-1 and C. testosteroni KF-1. The second D. acidovorans SPH-1 contains a copper resistance system Cop related to that of Pseudomonas syringae. The genes in this system are laid out in the following order copSR copABFCD. copSR is a two-component signal transduction system, which is required for the copper-inducible expression of copper resistance [53]. CopA and CopC are abundant periplasmic copper binding proteins, and CopB is associated with copper accumulation in the outer membrane. No specific function for CopD has been determined yet [54]. CopF is involved in the cytoplasmic detoxification of copper ions [55]. In the novel element associated with Shewanella sp. ANA-3 the variable region encodes genes that shares similarities with a chloramphenicol efflux pump [InterPro Scan]. C. litoralis KT71 and P. aeruginosa 2192 have a putative resistance nodulation division [RND] type multidrug efflux pump related to the mex system of P. aeruginosa [56] and the oqx system of E. coli plasmid pOLA52 [57] encoded. Apart from antibiotics, the broad substrate range of the Mex efflux systems of P. aeruginosa also includes organic solvents, biocides, dyes, and cell signalling molecules [58]. In the ICE of P. aeruginosa PA7 this variable region encodes homologs of genes for antibiotic resistance including neomycin/kanamycin resistance, bleomycin resistance, and streptomycin resistance related to the antibiotic resistance genes from Tn5 [U00004]. There are also a set of genes with similarity to the kdpFABC system. The KdpFABC complex acts as a high affinity K+ uptake system. In E. coli, the complex is synthesized when the constitutively expressed low affinity K+ uptake systems Trk and Kup can no longer meet the cell's demand for potassium due to external K+ limitation Altendorf et al., 1992 K. Altendorf, A. Siebers and W. Epstein, The KDP ATPase of Escherichia coli, Ann. NY Acad. Sci. 671 (1992), pp. 228-243. Full Textvia CrossRef|View Record in Scopus Cited By in Scopus (22)[59]. Stenotrophomonas maltophilia K279a had a putative Major Facilitator Superfamily (MFS) efflux pump that usually function as specific exporters for certain classes of antimicrobial agents. This is related to the emrAB system from E. coli [60]. P. aeruginosa UCBPP-PA14 has a predicted czc [Cd/Zn/Co] efflux system similar to those in D. acidovorans SPH-1 and C. testosteroni KF-1. P. aeruginosa PACS171b contains a homolog of UspA- the Universal Stress Protein. The UspA protein is important for survival during cellular growth arrest in E. coli, but the exact physiological role of the protein is unknown [61]. Thioalkalivibrio sp. HL-EbGR7 has a set of genes with approximately 88% aa identity to the putative KdpFABC system in P. aeruginosa PA7. This variability is suggestive that this region may be a hotspot for insertion or recombination where insertion clearly does not disrupt or affect the expression of neighbouring genes. The variation in predicted gene function, size and lack of homology between elements is suggestive of this region contributing a number of different adaptive traits to hosts containing these ICEs.
Following this variable region is encoded a putative transcriptional regulator protein TraR and a homologue of the type IV coupling protein TraG [similar to those in IncP plasmids]. TraG is responsible for DNA transfer during conjugation and is a putative DNA binding protein [62]. Interestingly the gene order of this region and the order of genes preceding it are also suggestive of an insertion [of the variable region just discussed] into a primordial transfer module.
The putative DNA binding gene traG is followed by a group of genes encoding proteins [TrbBCDEJLFGI] with similarity to the mating-pair formation [mpf] apparatus or type IV secretion system closely related to IncP and Ti plasmids. This system presumably mediates the DNA transfer of the ICE to recipient cells [63,64]. These genes show similarity to those required for conjugative transfer of the Agrobacterium Ti plasmid, pNGR234a and RP4, except that two genes, trbK and trbH, found on these plasmids are missing [65]. In the Tn4371-like elements the gene order was trb BCDEJLFGI in all the characterised elements found in this study and similar to the molecular organisation in ICEMlSymR7A [[19], Fig. 1]. The TrbB, TrbC, TrbE, TrbG, and TrbL proteins are involved in the creation of the mpf apparatus, TrbC is involved in pilus formation and TrbE displays ATPase activity [65].
The novel ICEs detected in this study are integrated into various locations in the genomes of the host bacteria where they were discovered. In Acidovorax sp. JS42 other partial copies of Tn4371-like elements were also found in addition to the full element reported here. Two elements were discovered and characterised in D. acidovorans SPH-1. A further partial element was found in B. petrii this however lacked the intTn4371 gene. This situation is similar to that found in R. metallidurans CH34 and indicates that duplication or multiple insertions of the elements occur in bacteria. Near complete copies of Tn4371-like elements were also found in Burkholderia ambifaria AMMD and Burkholderia multivorans ATCC17616, where both were found to lack the Tn4371-like integrase gene suggesting that the elements may no longer be mobile. New elements were also found in Ralstonia solanacearum MolK2 and a second element in Diaphorobacter sp. TPSY, these share similarities in the stabilisation and transfer regions of the element to Tn4371-like elements but they have a different integrase region not related to the intTn4371 gene.
All of the elements reported here [Table 1 and 2] appear to share a common scaffold or backbone that is approximately 24 kb in size containing a 1.5 kb integrase gene; an 8.5 kb replication/stability gene cluster and a 14 kb conjugal transfer/mating pair formation cluster [Fig. 1]. A visual representation of this can be seen in Figs. 2, 3, 4 and 5 where the various sequences were aligned for comparison, the core scaffold identified and 'adaptive' genes highlighted which vary from element to element.
Figure 2.
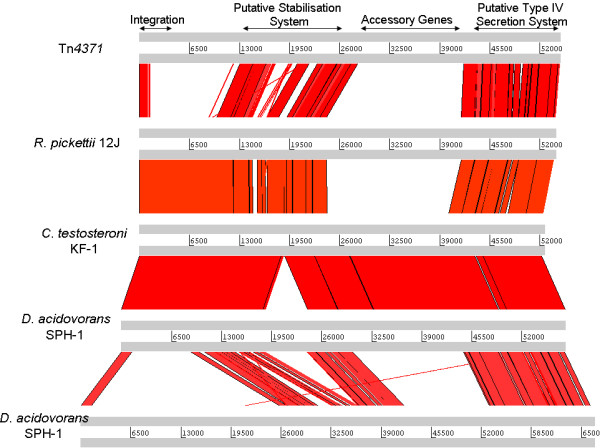
Use of the Artemis comparison tool to analysis Tn4371-like ICE sequences of Tn4371, R. pickettii 12J, both elements from D. acidovorans SPH-1 and C. testosteroni KF-1. All ICEs analysed shared extensive sequence homology, and general gene order. Arrows on top delimit the functional regions whose order is well conserved in all Tn4371-like ICEs.
Figure 3.
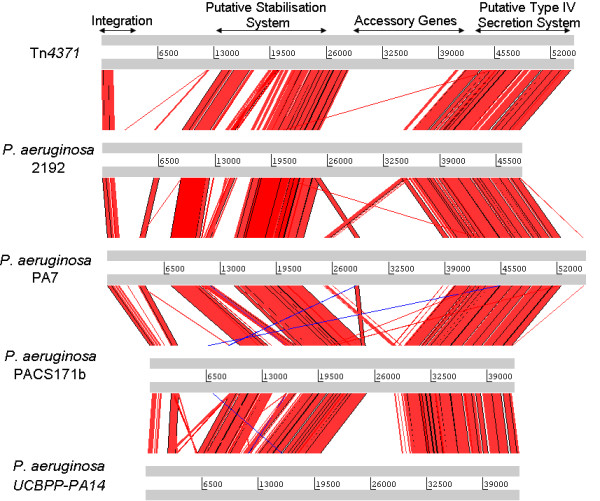
Use of the Artemis comparison tool to analysis Tn4371-like ICE sequences of Tn4371, P. aeruginosa 2192, P. aeruginosa PA7, P. aeruginosa UCBPP-PA14 and P. aeruginosa PACS171b. All ICEs analysed shared extensive sequence homology, and general gene order. Arrows on top delimit the functional regions whose order is well conserved in all Tn4371-like ICEs.
Figure 4.
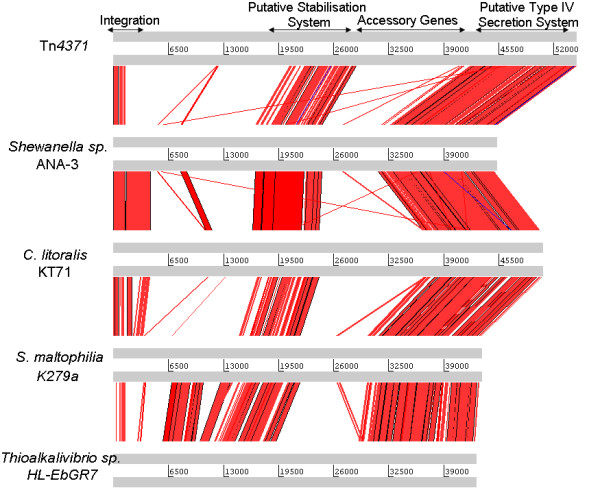
Use of the Artemis comparison tool to analysis Tn4371-like ICE sequences of Tn4371, Shewanella sp. ANA-3, C. litoralis KT71, S. maltophilia K279a and Thioalkalivibrio sp. HL-EbGR7. All ICEs analysed shared extensive sequence homology, and general gene order. Arrows on top delimit the functional regions whose order is well conserved in all Tn4371-like ICEs.
Figure 5.
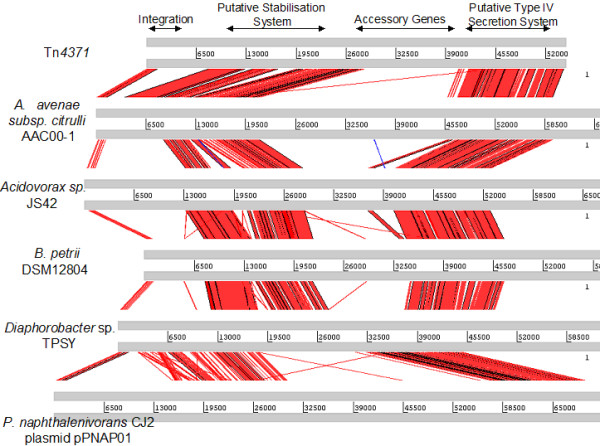
Use of the Artemis comparison tool to analysis Tn4371-like ICE sequences of Tn4371, A. avenae subsp. citrulli AAC00-1, Acidovorax sp. JS42, B. petrii DSM12804, Diaphorobacter sp. TPSY and P. naphthalenivorans CJ2 plasmid pPNAP01. All ICEs analysed shared extensive sequence homology, and general gene order. Arrows on top delimit the functional regions whose order is well conserved in all Tn4371-like ICEs.
Bioinformatic comparisons were performed between the genes that make up the core scaffold region of the ICE and these ranged from the highly conserved traG gene, with 84 to 96% aa identity, trbE gene, with 76 to 94% aa identity, and the parA gene, with 90 to 97% aa identity, to the less-conserved traR gene, with 53 to 84% aa identity. On average the genes that we ascribed to the core showed > 75% aa identity and were also related by gene order. All gene numbers and a basic description of the genes are included in Additional file 3.
Defining the Tn4371 family of ICEs and nomenclature
These elements have been classed as ICEs as we believe at this moment in time this is the best terminology currently available. They follow all the criteria of ICEs having integration and transfer modules, possessing an excisionase gene and having genes and gene layout (rdfS, rlxS and the trb genes) similar to other ICEs namely ICEMlSymR7A. The original element can also excise from bacterial chromosome and form a circular intermediate [9], however the element has not been shown to transfer between different bacteria, and this could be due to the original element lacking the trbD gene [13].
Although the elements identified in this study are not identical, they share a similar core backbone that, in our view, warrants their inclusion into the Tn4371 ICE family. All encode a related integrase, related maintenance and transfer genes and the gene order of homologous genes are similar, if one were to remove variable inserted regions which differ from element to element. We propose that any ICE that encodes an integrase gene closely related to intTn4371, defined as over 70% protein homology and that has similar maintenance and transfer genes be considered part of the Tn4371 family of ICEs.
Given the number of Tn4371-like elements discovered in this study, it seems sensible to name newly described ICEs of the Tn4371 family with a uniform nomenclature. We propose adapting the system used for naming transposons described by Roberts et al., [66]. This system is a website http://www.ucl.ac.uk/eastman/tn/ based system which assigns Tn numbers in sequence e.g. Tn6033, Tn6034, etc and the elements were then called ICETn43716033, ICETn43716034, etc to distinguish that they are ICEs of the Tn4371 family. The names assigned to the elements discovered in this study are listed in Table 1 and 2. This system was chosen as other systems such as that used by Burrus et al., [8] for naming members of the SXT\R391 family of ICEs are not regulated and can differ between laboratories leading to confusion.
Tn4371-like ICE detection and molecular characterisation
Following the discovery of the widespread nature of Tn4371-like ICEs in the genomes of many new organisms, PCR primers were designed to amplify important genes of the core scaffold to aid in the rapid identification of new Tn4371-like elements. We tested this on a culture collection of fifty-eight Ralstonia pickettii and Ralstonia insidiosa strains from various environments and geographic locations. The PCR primers were based on conserved consensus sequences of core genes identified from all the elements identified in this study and those reported previously.
The results in Fig. 6 show the genes encoding a putative integrase [int], the putative stabilisation system [repA, parA, parB], a homologue to the DNA transfer protein [traG] and a putative pilus assembly and synthesis protein [trbI]. DNA sequencing of the four amplicons in the tester strains demonstrated that Tn4371-like sequences exist in the genome of R. pickettii ULM001. While this data clearly demonstrates the presence of Tn4371-like elements in tester strains the possibility of multiple elements in such strains cannot be excluded, although out sequencing of resulting amplicons is suggestive of only one element.
Figure 6.
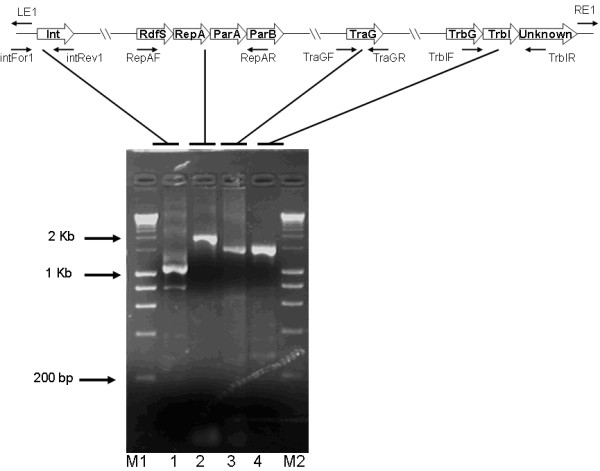
Amplification of genes of the putative Tn4371-like ICE ICETn43716043 in Ralstonia pickettii strain ULM001 (a laboratory purified water isolate). A scheme of the amplified genes is shown above the 0.7% agarose gel of the PCR products generated with the primers listed in Table 2. Open white arrows denote ORFs of the Ralstonia pickettii ICE, and small black arrows represent the relative location of primers. Lanes M1 and M2 contain 200-10000 bp molecular size markers (Bioline Hyperladder I), respectively. The lanes and the product sizes are as follows: Lane 1, int gene and flanking bases (1035 bp); Lane 2 RepA gene (1657 bp), Lane 3 traG gene (1483 bp); Lane 4 trbI gene (1597 bp).
Three of the fifty-eight Ralstonia isolates, ULM001, ULM003 and ULM006 [which were laboratory purified water isolates from different locations in France] showed positive amplification for intTn4371 integrase gene when tested with the intFor1 and intRev1 primer pair in PCR amplification [Table 3]. Sequencing revealed that the ULM001 int gene showed 85% and 99% nucleotide identity to the Tn4371 int gene and ICETn43716033 int gene, respectively. The RepAF and RepAR primers also amplified the repA gene and the parA gene in ULM001, ULM003 and ULM006. Sequencing these amplicons revealed that in ULM001 the repA and parB genes were present and showed 88% and 99% nucleotide identity to the RepA and ParA genes from Tn4371 and ICETn43716033 respectively. A traGTn4371 homolog was also detected in ULM001, ULM003 and ULM006 following PCR amplification. Sequencing revealed that the ULM001 traGTn4371 gene showed 91% and 89% nucleotide identity to traG from Tn4371 and ICETn43716033 respectively. TrbIF and TrbIR primers were used to amplify the trbI gene in ULM001 and ULM003 while no amplification occurred in ULM006. Sequencing showed that the ULM001 amplicon was a homolog, which had 88% and 99% nucleotide identity to the trbI gene from Tn4371 and ICETn43716033 respectively. The absence of a trbI gene amplicon in ULM006 may indicate a deleted gene or truncated element in this strain. The use of these primer sets has thus revealed the presence of two new elements, which can then be further characterised. The ICEs detected in this study from Ralstonia pickettii were named ICETn43716043 and ICETn43716044 using the nomenclature system described above, a general map of the elements can be seen in Fig. 6.
Table 3.
Primers used to map Tn4371-like ICEs
| Genes | Size (bp) | Primers | Tm(°C) |
R. pickettii 12J Position |
Accession no. | |
|---|---|---|---|---|---|---|
| Start | Stop | |||||
| CirIm | ~220 | RE1 GCATGGAAGACTTGACAG LE1 GAGCTTGAGTTTTGCCACG |
54 | N\A | N\A | FM244490 |
| int | 1035 | intFor1 TTTCATTTCACCATGACTCCAG intRev1 GAGAGCAGTCGATAGGCTTCC |
61.7 | 2715201 | 2716235 | FM244486 |
| RepA, ParA ParB |
1657 | RepAF GAGACTACCAGCGCCTCAAG RepAR ACGTGTTCATGAGGACTTCTCC |
55 | 2734598 | 2736255 | FM244487 |
| traG | 1483 | traGF GTTCGAGTGGTGGTTCTTCTTC traGR GAAATTGCTGTCCGCGTAGTAG |
61 | 2757179 | 2758661 | FM244488 |
| trbI | 1597 | trbIF AACTGACCATGAGCCAGGAC trbIR AAAGCTCCTCAAAAGCGAAAG |
62 | 2767516 | 2769113 | FM244489 |
The attL and attR region of Tn4371 ICEs
Analysis of hosts harbouring Tn4371-like elements indicated that integration occurred at an 8-bp attB site generating attL and attR element chromosomal junctions [[11], Fig. 7a]. An alignment of the first and last 200 bp of the elements analysed in this study with Tn4371-like element from previous studies showed the attL site had a sequence of TTTTC/TA/GT and attR had a sequence of TTTTC/TA/GT for some bacteria, while others had no direct repeats. These alignments can be seen in Additional file 4. The exact sequence of the direct repeat for each element is presented in Table 4. The absence of direct repeats in some of these elements may mean that they are no longer mobile. Tn4371 has been shown to excise from the RP4 plasmid in Ralstonia eutropha forming a circular extrachromosomal intermediate [[10], Fig. 7a] as a transfer intermediate. The strains in which we detected Tn4371-like elements were examined to see if they also excised forming extrachromosomal intermediates [CirIm] using a PCR assay that allowed amplification across the circular junction but which would not amplify if the element were integrated. Primer LE1 is specific to integrated Tn4371-like ICE DNA at the attL left-end where as primer RE1 is specific to integrated Tn4371-like ICE at the attR right-end [Fig. 7a, Table 3]. Both primers are oriented towards the Tn4371- like ICE junctions, and PCR product will be generated only if the respective left and right ends [attL and attR sites] excise from the chromosome and circularise [CirIm], reconstituting attP [attachment locus on the element]. A model of integration and excision of the ICE can be seen in Fig. 7a. PCR products of ~220-bp were obtained from ICETn43716043 [ULM001] and ICETn43716044 [ULM003] [Fig. 7b.], indicating that a circular extrachromosomal form of the element is present in these cells, while no PCR product was obtained from ULM006 [Fig. 7b]. The sequencing of the attP region of ICETn43716043 gave an attL region of TTTTTCAT and an attR region of TACTTTTT. This rapid amplification across the circular attP junction can also be utilised for the rapid identification of Tn4371-like elements. It is possible that the PCR may have picked up tandems of the element if those happened to be intermediates in "transposition".
Figure 7.
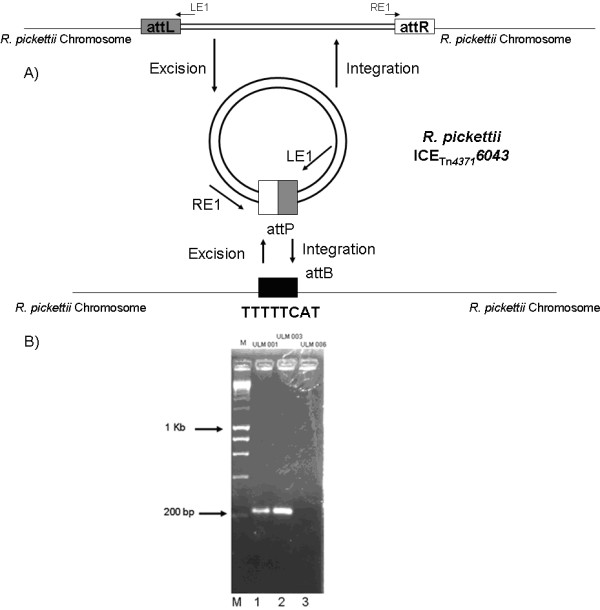
A) Schematic representation of Tn4371 excision and insertion into the R. pickettii chromosome. Primer LE1 and RE1 are the primers for detection of the circular form of the element. B) Agarose gel of attP of ICETn43716043 and ICETn43716044. Lanes M contains 200-10000 bp molecular size markers (Bioline Hyperladder I), Lane 1 ULM001, Lane2 ULM002, Lane 3 ULM006.
Table 4.
Direct repeats at the ends of each element
| Tn4371-like Elements | Direct repeats |
|---|---|
| Ralstonia pickettii 12J | TTTTTCAT |
| Shewanella sp. ANA-3 | TTTTTTAT |
| Congregibacter litoralis KT71 | TTTTTTAT |
| Acidovorax avenae subsp. citrulli AAC00-1 | TTTTTCAT |
| Delftia acidovorans SPH-1 | TTTTTCAT |
| Comamonas testosteroni KF-1 | TTTTTCAT |
| Pseudomonas aeruginosa 2192 | TTTTTTAT |
| Pseudomonas aeruginosa PA7 | TTTTTTGT |
| Stenotrophomonas maltophilia K279a | TTTTTTGT |
| Pseudomonas aeruginosa PACS171b | TTTTTTAT |
| Diaphorobacter sp. TPSY | TTTTTCAT |
| Delftia acidovorans SPH-1 | TTTTTCAT |
| Acidovorax sp. JS42 | NP |
| Bordetella petrii DSM12804 | NP |
| Thioalkalivibrio sp. HL-EbGR7 | NP |
| Burkholderia pseudomallei MSHR346 | NP |
| Polaromonas naphthalenivorans CJ2 plasmid pPNAP01 | NP |
| Pseudomonas aeruginosa PA14 | NP |
NP, Not Present
Conclusion
Tn4371-like ICEs are found in a wide range of γ-proteobacteria and β-proteobacteria from both clinical and environmental sources. These types of bacteria are known for their large metabolic repertoires and these elements could potentially be a source of acquisition of adaptive functions for these organisms. The discovery of the Tn4371-like ICEs in the P. aeruginosa strains, S. maltophilia K279a and B. pseudomallei MSHR346 are the first reports of these elements found in human pathogens. This along with the discovery of putative antibiotic resistance genes in their genomes indicates that these elements may have an impact in clinical situations. The discovery and characterisation of novel Tn4371-like elements as reported here adds significantly to the repertoire of such elements and helps define the core scaffold of such elements. It is clear that these elements are highly adaptable and may contribute significantly to the metabolic capabilities of their host. This study increases the knowledge available about these elements adding data on eighteen new elements to the five already known. A new nomenclature system for Tn4371-like elements was designed to avoid confusion in the naming of these elements. The primer system used to detect and characterise the Tn4371-like ICEs in Ralstonia pickettii ULM001 and ULM003 could be adapted and used for other bacterial species for the rapid screening of such elements.
Methods
Bacterial strains and growth conditions
The strains used in this study are shown in Table 5. All the strains were stored at -20°C in Nutrient Broth [Biolab, Budapest, Hungary] with 50% glycerol. Isolates were grown aerobically on Nutrient Agar [Biolab, Budapest, Hungary] and incubated overnight at 30°C.
Table 5.
Ralstonia Strains used in this work
| Strain | Source |
|---|---|
|
R. pickettii JCM5969, NCTC11149, DSM6297, CIP73.23 CCUG3318 |
Culture Collections |
|
R. pickettii CCM2846 CCUG18841 |
Culture Collections |
|
R.insidiosa ATCC4199 |
Culture Collection |
|
R.insidiosa LMG21421 |
Culture Collection |
|
R. pickettii ULC193, ULC194, ULC277, ULC297, ULC298, ULC224, ULC421 |
Microbiology laboratory of Limerick Regional Hospital (Cystic Fibrosis Patients) |
|
R. pickettii ULI785, ULI788, ULI790, ULI791, ULI796, ULI798, ULI800, ULI801, ULI804, ULI806, ULI807, ULI818, ULI159, ULI162, ULI165, ULI167, ULI169, ULI171, ULI174, ULI181, ULI187, ULI188, ULI193 |
Isolated from various Millipore Purified water systems (Ireland) |
|
R.insidiosa ULI821, ULI797, ULI785, ULI181, ULI794, ULI185, ULI166, ULI819, ULI784, ULI163, ULI795 |
Isolated from various Millipore Purified water systems (Ireland) |
|
R. pickettii ULM001, ULM002, ULM003, ULM004, ULM005, ULM006 |
Isolated from various Millipore Purified water systems (France) |
|
R. pickettii ULM007, ULM008, ULM009, ULM010, ULM011 |
Isolated from various Millipore Purified water systems (Ireland) |
|
R.insidiosa ULM008, ULM009 |
Isolated from various Millipore Purified water systems (Ireland) |
Molecular analysis of genes of Tn4371-like ICEs
PCR primers were designed based on the conserved aligned scaffold common to all ICEs characterised in this study and from the consensus sequence of the Ralstonia pickettii 12J Tn4371 ICE using the Primer 3 program [[67], http://frodo.wi.mit.edu/]. All primers are listed in Table 5. The cycling conditions were as follows: initial denaturation (98°C, 2 min); 35 cycles consisting of denaturation [98°C for 15 s], primer annealing [TA [estimated primer annealing temperature], 1 min], and extension [72°C, 1 min/kb]; followed by a final extension step [72°C, 10 min]. Amplification was carried out with a GC buffer [in a total reaction of 100 μL containing 0.2 mM deoxynucleoside triphosphates, 100 pmol of each primer, 8 μL of genomic template DNA, and 3 units of Phusion polymerase [New England Biolabs, UK]. Amplification was carried out using a GeneAmp 2400 Thermocycler. Bacterial DNA for PCR amplification was extracted according to Ausubel et al. [68]. Amplicons to be sequenced were directly purified from the PCR reaction by the NucleoSpin Extract II kit [Macherey-Nagel, Düren] according to the manufacturer's instructions. Sequence analysis was performed by Euorfins-MWG [Germany] using both the forward and reverse primers listed in Table 3.
Bioinformatic Analysis of the Tn4371-like ICEs in genomes
All analysed DNA sequences were retrieved from the GenBank database http://www.ncbi.nlm.nih.gov.
DNA and protein sequences similar to Tn4371 [[13], AJ536756] were detected within the NCBI nonredundant nucleotide and protein databases http://www.ncbi.nlm.nih.gov via blastp and blastn analysis using the original Tn4371 sequence as a probe [69]. Assembly and comparison with other Tn4371-like sequences was performed with the Artemis Comparison Tool [ACT] [[70], http://www.sanger.ac.uk/Software/ACT]. The complete DNA sequences were also manually annotated to verify the deposited sequence. The similarity of proteins encoded by the element was determined as % aa identities over the entire protein to its Tn4371 equivalent via blastp. Unknown ORFs were analysed using InterProScan http://www.ebi.ac.uk/InterProScan/, [71]] to locate motifs or domains where similarity with known proteins was low or absent. Size and total % GC content was determined using the GC-Profile program [[72], http://tubic.tju.edu.cn/GC-Profile/]. Phylogenetic and molecular evolutionary analyses were conducted using genetic-distance-based neighbour-joining algorithms within MEGA version 4.0 [[73], http://www.megasoftware.net/]
Nucleotide sequence accession numbers
The DNA sequences described in this article have been assigned the accession numbers listed in Table 3.
Authors' contributions
MRP was responsible for conception of the study, experimental design, data collection, and analysis and preparation of the manuscript. JTP and CCA participated in experimental design, data analysis and preparation of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Alignment of the conserved domains among the site-specific recombinases of the tyrosine integrase family. Alignment of the conserved domains among the site-specific recombinases of the tyrosine integrase family from phages, conjugative transposons, plasmids and other sources. R (Arginine) being in Domain I and H (Histidine)-R-Y (Tyrosine) in Domain II.
Phylogenetic tree of the Integrase proteins from Tn4371-like integrases available on the GenBank database and other Phage and ICE integrases. Phylogenetic tree of the Integrase proteins from available Tn4371-like integrases available on the GenBank database and other Phage and ICE integrases. Cluster analysis was based upon the neighbour joining method. Numbers at branch-points are percentages of 1000 bootstrap resamplings that support the topology of the tree. The scale bar represents 0.2 substitutions per nucleotide position.
Gene numbers for genes in the elements discovered in this study. The gene numbering for genes of the elements discovered in this study. Genes with yellow background are the scaffold genes of the element.
Alignment of the first/last 200 bp of Tn4371-like ICEs using ClustalW. Fig S1a: Alignment of the first 200 bp of Tn4371-like ICEs using ClustalW. Fig S1b: Alignment of the last 200 bp of I Tn4371-like ICEs using ClustalW.
Contributor Information
Michael P Ryan, Email: Michael.P.Ryan@ul.ie.
J Tony Pembroke, Email: Tony.Pembroke@ul.ie.
Catherine C Adley, Email: Catherine.Adley@ul.ie.
Acknowledgements
MPR was funded was provided by a Postgraduate bursary from the Chemical and Environmental Science Department, Faculty of Science and Engineering, University of Limerick. We would like to thank the reviewers for their useful suggestions.
References
- Toussaint A, Merlin C. Mobile elements as a combination of functional modules. Plasmid. 2002;47:26–35. doi: 10.1006/plas.2001.1552. [DOI] [PubMed] [Google Scholar]
- Burrus V, Pavlovic G, Decaris B, Guédon G. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- Churchward G. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz ML, editor. Washington DC: American Society for Microbiology; 2002. Conjugative transposons and related mobile elements; pp. 177–191. [Google Scholar]
- Merlin C, Mahillon J, Nesvera J, Toussaint A. In: The Horizontal Gene Pool: Bacterial Plasmids and Gene Spread. Thomas CM, editor. Newark: Hardwood Academic Publishers; 2000. Gene recruiters and transporters: the modular structure of bacterial elements; pp. 363–408. [Google Scholar]
- Smith CJ, Tribble GD, Bayley DP. Genetic elements of Bacteroides species: a moving story. Plasmid. 1998;40:12–29. doi: 10.1006/plas.1998.1347. [DOI] [PubMed] [Google Scholar]
- Hochhut B, Waldor MK. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol Microbiol. 1999;32:99–110. doi: 10.1046/j.1365-2958.1999.01330.x. [DOI] [PubMed] [Google Scholar]
- Osborn MA, Boltner D. When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid. 2002;48:202–212. doi: 10.1016/S0147-619X(02)00117-8. [DOI] [PubMed] [Google Scholar]
- Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. 2004;155:376–386. doi: 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Burrus V, Marrero J, Waldor MK. The current ICE age: Biology and evolution of SXT-related integrating conjugative elements. Plasmid. 2006;55:173–183. doi: 10.1016/j.plasmid.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Springael D, Kreps S, Mergeay M. Identification of a catabolic transposon, Tn4371, carrying biphenyl and 4-chlorobiphenyl degradation genes in Alcaligenes eutrophus A5. J Bacteriol. 1993;175:1674–1681. doi: 10.1128/jb.175.6.1674-1681.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlin C, Springael D, Toussaint A. Tn4371: a modular structure encoding a phage-like integrase, a Pseudomonas-like catabolic pathway and RP4/Ti-like transfer. Plasmid. 1999;41:40–54. doi: 10.1006/plas.1998.1375. [DOI] [PubMed] [Google Scholar]
- Springael D, Diels L, Mergeay M. Transfer and expression of PCB-degradative genes into heavy metal resistant Alcaligenes eutrophus strains. Biodegradation. 1994;5:343–357. doi: 10.1007/BF00696469. [DOI] [PubMed] [Google Scholar]
- Toussaint A, Merlin C, Monchy S, Benotmane MA, Leplae R, Mergeay M, Springael D. The biphenyl- and 4-chlorobiphenyl-catabolic transposon Tn4371, a member of a new family of genomic islands related to IncP and Ti plasmids. Appl Environ Microbiol. 2003;69:4837–4845. doi: 10.1128/AEM.69.8.4837-4845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher CA, Barberis PA, Trigalet AP, Demery DA. Transposon mutagenesis of Pseudomonas solanacearum: Isolation of Tn5-induced avirulent mutants. J Gen Microbiol. 1985;131:2449–2457. [Google Scholar]
- Mergeay M, Houba C, Gerits J. Extrachromosomal inheritance controlling resistance to cadmium, cobalt and zinc ions: evidence from curving in a Pseudomonas. Arch Int Physiol Biochim. 1978;86:440–442. [PubMed] [Google Scholar]
- Kotoujansky A, Lemattre M, Boitard P. Utilization of a thermosensitive episome bearing transposon Tn10 to isolate Hfr donor strains of Erwinia carotovora subsp. chrysanthemi. J Bacteriol. 1982;150:122–131. doi: 10.1128/jb.150.1.122-131.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess BK, Jacobs DB, Stiefel EI. Large-scale purification of high activity Azotobacter vinelandii nitrogenase. Biochim Biophys Acta. 1980;614:196–209. doi: 10.1016/0005-2744(80)90180-1. [DOI] [PubMed] [Google Scholar]
- McGrath BM, O'Halloran JA, Piterina AV, Pembroke JT. Molecular tools to detect the IncJ elements: a family of integrating, antibiotic resistant mobile genetic elements. J Microbiol Meth. 2006;66:32–42. doi: 10.1016/j.mimet.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Burrus V, Pavlovic G, Decaris B, Guédon G. The ICESt 1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid. 2002;48:77–97. doi: 10.1016/S0147-619X(02)00102-6. [DOI] [PubMed] [Google Scholar]
- Sullivan JT, Trzebiatowski JR, Cruickshank RW, Gouzy J, Brown SD, Elliot RM, Fleetwood DJ, McCallum NG, Rossbach U, Stuart GS, Weaver JE, Webby RJ, de Bruijn FJ, Ronson CW. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J Bacteriol. 2002;184:3086–3095. doi: 10.1128/JB.184.11.3086-3095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Zain Z, Turner SL, Cerdeño-Tárraga AM, Lilley AK, Inzana TJ, Duncan AJ, Harding RM, Hood DW, Peto TE, Crook DW. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J Bacteriol. 2004;186:8114–8122. doi: 10.1128/JB.186.23.8114-8122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pembroke JT, Piterina AV. A novel ICE in the genome of Shewanella putrefaciens W3-18-1: comparison with the SXT/R391 ICE-like elements. FEMS Microbiol Lett. 2006;264:80–88. doi: 10.1111/j.1574-6968.2006.00452.x. [DOI] [PubMed] [Google Scholar]
- Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, Cordum H, Van Brunt A, Kim K, Fulton RS, Fulton LA, Clifton SW, Wilson RK, Knight RD, Gordon JI. Evolution of Symbiotic Bacteria in the Distal Human Intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M, Schmitt K, Bauer S, Hot D, Hubans C, Levillain E, Locht C, Lemoine Y, Gross R. Genomic island excisions in Bordetella petrii. BMC Microbiol. 2009;9:141. doi: 10.1186/1471-2180-9-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt R, Monchy S, Leys N, Mergeay M. New mobile genetic elements in Cupriavidus metallidurans CH34, their possible roles and occurrence in other bacteria. Antonie Van Leeuwenhoek. 2009;96:205–226. doi: 10.1007/s10482-009-9345-4. [DOI] [PubMed] [Google Scholar]
- Nunes-Düby SE, Kwon HJ, Tirumalai RS, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan D, Colleran E. Arsenical resistance in the IncHI2 plasmids. Plasmid. 2002;47:234–240. doi: 10.1016/S0147-619X(02)00012-4. [DOI] [PubMed] [Google Scholar]
- Ji G, Silver S. Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid pI258. Proc Natl Acad Sci USA. 1992;89:9474–9478. doi: 10.1073/pnas.89.20.9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Rosen BP. The arsD gene encodes a second trans-acting regulatory protein of the plasmid-encoded arsenical resistance operon. Mol Microbiol. 1993;8:615–623. doi: 10.1111/j.1365-2958.1993.tb01605.x. [DOI] [PubMed] [Google Scholar]
- Nascimento AM, Chartone-Souza E. Operon mer: bacterial resistance to mercury and potential for bioremediation of contaminated environments. Genet Mol Res. 2003;2:92–101. [PubMed] [Google Scholar]
- Lelie D van der, Schwuchow T, Schwidetzky U, Wuertz S, Baeyens W, Mergeay M, Nies DH. Two-component regulatory system involved in transcriptional control of heavy-metal homoeostasis in Alcaligenes eutrophus. Mol Microbiol. 1997;23:493–503. doi: 10.1046/j.1365-2958.1997.d01-1866.x. [DOI] [PubMed] [Google Scholar]
- Grosse C, Grass G, Anton A, Franke S, Santos AN, Lawley B, Brown NL, Nies DH. Transcriptional organization of the czc heavy-metal homeostasis determinant from Alcaligenes eutrophus. J Bacteriol. 1999;181:2385–2393. doi: 10.1128/jb.181.8.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies DH, Nies A, Chu L, Silver S. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci USA. 1989;86:7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S, Ziese M, Sternberg N. A novel role for site-specific recombination in maintenance of bacterial replicons. Cell. 1981;25:729–736. doi: 10.1016/0092-8674(81)90180-X. [DOI] [PubMed] [Google Scholar]
- Gerlitz M, Hrabak O, Schwab H. Partitioning of broad host range Plasmid RP4 is a complex system involving site-specific recombination. J Bacteriol. 1990;172:6194–6203. doi: 10.1128/jb.172.11.6194-6203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell C, Thomas CM. The bacterial ParA-ParB partitioning proteins. J Biotechnol. 2001;91:1–34. doi: 10.1016/S0168-1656(01)00293-0. [DOI] [PubMed] [Google Scholar]
- Das M, Ganguly T, Chattoraj P, Chanda PK, Bandhu A, Lee CY, Sau S. Purification and characterization of repressor of temperate S. aureus phage phi11. J Biochem Mol Biol. 2007;40:740–748. doi: 10.5483/bmbrep.2007.40.5.740. [DOI] [PubMed] [Google Scholar]
- McDonnell GE, McConnell DJ. Overproduction, isolation, and DNA-binding characteristics of Xre, the repressor protein from the Bacillus subtilis defective prophage PBSX. J Bacteriol. 1994;176:5831–5834. doi: 10.1128/jb.176.18.5831-5834.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay JP, Sullivan JT, Stuart GS, Lamont IL, Ronson CW. Excision and transfer of the Mesorhizobium loti R7A symbiosis island requires an integrase IntS, a novel recombination directionality factor RdfS, and a putative relaxase RlxS. Mol Microbiol. 2006;62:723–734. doi: 10.1111/j.1365-2958.2006.05396.x. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Hatfull GF. Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res. 2001;29:2205–2216. doi: 10.1093/nar/29.11.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Halloran JA, McGrath BM, Pembroke JT. The orf4 gene of the enterobacterial ICE, R391, encodes a novel UV-inducible recombination directionality factor, Jef, involved in excision and transfer of the ICE. FEMS Microbiol Lett. 2007;272:99–105. doi: 10.1111/j.1574-6968.2007.00747.x. [DOI] [PubMed] [Google Scholar]
- Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, Wade J, Walsh U, O'Gara F, Haas D. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol Plant Microbe Interact. 2000;13:232–237. doi: 10.1094/MPMI.2000.13.2.232. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kiyokawa C, Kohara M, Matsumoto M, Matsuno A, Mochizuki Y, Nakayama S, Nakazaki N, Shimpo S, Sugimoto M, Takeuchi C, Yamada M, Tabata S. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 2000;7:331–338. doi: 10.1093/dnares/7.6.331. [DOI] [PubMed] [Google Scholar]
- Gstalder ME, Faelen M, Mine N, Top EM, Mergeay M, Couturier M. Replication functions of new broad host range plasmids isolated from polluted soils. Res Microbiol. 2003;154:499–509. doi: 10.1016/S0923-2508(03)00143-8. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Moller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- Dziewit L, Jazurek M, Drewniak L, Baj J, Bartosik D. The SXT conjugative element and linear prophage N15 encode toxin-antitoxin-stabilizing systems homologous to the tad-ata module of the Paracoccus aminophilus plasmid pAMI2. J Bacteriol. 2007;189:1983–1997. doi: 10.1128/JB.01610-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Gurkar AU, Lory S. Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2006;103:19830–19835. doi: 10.1073/pnas.0606810104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenda M, Barbe V, Gourgues G, Mangenot S, Sagne E, Citti C. A new integrative conjugative element occurs in Mycoplasma agalactiae as chromosomal and free circular forms. J Bacteriol. 2006;188:4137–4141. doi: 10.1128/JB.00114-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase J, Lanka E. A specific protease encoded by the conjugative DNA transfer systems of IncP and Ti plasmids is essential for pilus synthesis. J Bacteriol. 1997;179:5728–5735. doi: 10.1128/jb.179.18.5728-5735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd DR, Matson SW. Nicking by transesterification: the reaction catalysed by a relaxase. Mol Microbiol. 1997;25:1011–1022. doi: 10.1046/j.1365-2958.1997.5241885.x. [DOI] [PubMed] [Google Scholar]
- Lanka E, Wilkins BM. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- Sekine M, Tanikawa S, Omata S, Saito M, Fujisawa T, Tsukatani N, Tajima T, Sekigawa T, Kosugi H, Matsuo Y, Nishiko R, Imamura K, Ito M, Narita H, Tago S, Fujita N, Harayama S. Sequence analysis of three plasmids harboured in Rhodococcus erythropolis strain PR4. Environ Microbiol. 2006;8:334–346. doi: 10.1111/j.1462-2920.2005.00899.x. [DOI] [PubMed] [Google Scholar]
- Mills SD, Jasalavich CA, Cooksey DA. A two-component regulatory system required for copper-inducible expression of the copper resistance operon of Pseudomonas syringae. J Bacteriol. 1993;175:1656–1664. doi: 10.1128/jb.175.6.1656-1664.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JS, Cooksey DA. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc Natl Acad Sci USA. 1991;88:8915–8919. doi: 10.1073/pnas.88.20.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergeay M, Monchy S, Vallaeys T, Auquier V, Benotmane A, Bertin P, Taghavi S, Dunn J, Lelie D van der, Wattiez R. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol Rev. 2003;27:385–410. doi: 10.1016/S0168-6445(03)00045-7. [DOI] [PubMed] [Google Scholar]
- Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty LK, Pechere JC. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- Hansen LH, Johannesen E, Burmolle M, Sorensen AH, Sorensen SJ. Plasmid-encoded multidrug efflux-pump conferring resistance to olaquindox in Escherichia coli. Antimicrob Agents Chemother. 2004;48:3332–3337. doi: 10.1128/AAC.48.9.3332-3337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos CJ, Carson CF, Chang BJ, Riley TV. Role of the MexAB-OprM Efflux Pump of Pseudomonas aeruginosa in Tolerance to Tea Tree (Melaleuca alternifolia) Oil and Its Monoterpene Components Terpinen-4-ol, 1,8-Cineole, and α-Terpineol. Appl Environ Microbiol. 2008;74:1932–1935. doi: 10.1128/AEM.02334-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebers A, Altendorf K. In: Alkali cation transport systems in prokaryotes. Bakker EP, editor. CRC Press, Boca Raton, Florida; 1993. K+-translocating Kdp-ATPases and other bacterial P-type ATPases; pp. 225–252. [Google Scholar]
- Furukawa H, Tsay JT, Jackowski S, Takamura Y, Rock CO. Thiolactomycin resistance in Escherichia coli is associated with the multidrug resistance efflux pump encoded by emrAB. J Bacteriol. 1993;175:3723–3729. doi: 10.1128/jb.175.12.3723-3729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachin L, Nannmark U, Nyström T. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J Bacteriol. 2005;187:6265–6272. doi: 10.1128/JB.187.18.6265-6272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Ruth FX, de la Cruz F, Coll M. Structure and role of coupling proteins in conjugal DNA transfer. Res Microbiol. 2002;153:199–204. doi: 10.1016/S0923-2508(02)01313-X. [DOI] [PubMed] [Google Scholar]
- Schroder G, Lanka E. The mating pair formation system of conjugative plasmids-A versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54:1–25. doi: 10.1016/j.plasmid.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Lawley D, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- Li PL, Everhart DM, Farrand SK. Genetic and sequence analysis of the pTiC58 trb locus, encoding a mating-pair formation system related to members of the type IV secretion family. J Bacteriol. 1998;180:6164–6172. doi: 10.1128/jb.180.23.6164-6172.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AP, Chandler M, Courvalin P, Guédon G, Mullany P, Pembroke T, Rood JI, Smith CJ, Summers AO, Tsuda M, Berg DE. Revised Nomenclature for Transposable Genetic Elements. Plasmid. 2008;60:167–173. doi: 10.1016/j.plasmid.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S, Misener S, editor. Totowa, NJ: Humana Press; 2000. Primer3 on the WWW for general users and for biologist programmers; pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. John Wiley & Sons, New York; 1997. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan-an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Gao F, Zhang CT. GC-Profile: a web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. 2006;34:W686–W691. doi: 10.1093/nar/gkl040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Isaacs N, Fett J, Simpson S, Long DT, Marsh TL. Microbial diversity and resistance to copper in metal-contaminated lake sediment. Microb Ecol. 2003;45:191–202. doi: 10.1007/s00248-002-1035-y. [DOI] [PubMed] [Google Scholar]
- Walcott RR, Fessehaie A, Castro AC. Differences in pathogenicity between two genetically distinct groups of Acidovorax avenae subsp. citrulli on cucurbit hosts. J Phytopathol. 2004;152:277–285. doi: 10.1111/j.1439-0434.2004.00841.x. [DOI] [Google Scholar]
- Schleheck D, Knepper TP, Fischer K, Cook AM. Mineralization of individual congeners of linear alkylbenzenesulfonate by defined pairs of heterotrophic bacteria. Appl Environ Microbiol. 2004;70:4053–4063. doi: 10.1128/AEM.70.7.4053-4063.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler BE, Wallace WH, Spain JC. Biodegradation of 2-nitrotoluene by Pseudomonas sp. strain JS42. Appl Environ Microbiol. 1994;60:3466–3469. doi: 10.1128/aem.60.9.3466-3469.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wintzingerode F, Schattke A, Siddiqui RA, Rösick U, Göbel UB, Gross R. Bordetella petrii sp. nov., isolated from an anaerobic bioreactor, and emended description of the genus Bordetella. Int J Sys Evol Microbiol. 2001;51:1257–1265. doi: 10.1099/00207713-51-4-1257. [DOI] [PubMed] [Google Scholar]
- Jeon CO, Park W, Ghiorse WC, Madsen EL. Polaromonas naphthalenivorans sp. nov., a naphthalene-degrading bacterium from naphthalene-contaminated sediment. Int J Syst Evol Microbiol. 2004;54:93–97. doi: 10.1099/ijs.0.02636-0. [DOI] [PubMed] [Google Scholar]
- Coates JD, Weber KA, Scherer M, Achenbach LA. The Diverse Microbiology of Anaerobic Fe(II) Oxidation. American Geophysical Union, Fall Meeting; 2007. abstract #B22D-01. [Google Scholar]
- Saltikov CW, Cifuentes A, Venkateswaran K, Newman DK. The ars detoxification system is advantageous but not required for As (V) respiration by the genetically tractable Shewanella species strain ANA-3. Appl Environ Microbiol. 2003;69:2800–2809. doi: 10.1128/AEM.69.5.2800-2809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs BM, Spring S, Teeling H, Quast C, Wulf J, Schattenhofer M, Yan S, Ferriera S, Johnson J, Glöckner FO, Amann R. Characterization of a marine gammaproteobacterium capable of aerobic anoxygenic photosynthesis. Proc Natl Acad Sci USA. 2007;104:2891–2896. doi: 10.1073/pnas.0608046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna SL, Sherman NE, Kinter MT, Goldberg JB. Comparison of proteins expressed by Pseudomonas aeruginosa strains representing initial and chronic isolates from a cystic fibrosis patient: an analysis by 2-D gel electrophoresis and capillary column liquid chromatography-tandem mass spectrometry. Microbiology. 2000;146:2495–2508. doi: 10.1099/00221287-146-10-2495. [DOI] [PubMed] [Google Scholar]
- Brodinova NS, Baskakova NV, Moroz AF, Vertiev IuV, Mokrievich NM. [Exotoxin A production during Pseudomonas aeruginosa PA-7 cultivation in Martin's broth] Zh Mikrobiol Epidemiol Immunobiol. 1984;4:22–26. Article in Russian. [PubMed] [Google Scholar]
- Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, Saunders D, Arrowsmith C, Carver T, Peters N, Adlem E, Kerhornou A, Lord A, Murphy L, Seeger K, Squares R, Rutter S, Quail MA, Rajandream MA, Harris D, Churcher C, Bentley SD, Parkhill J, Thomson NR, Avison MB. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 2008;9:R74. doi: 10.1186/gb-2008-9-4-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, Diggins LT, He J, Saucier M, Déziel E, Friedman L, Li L, Grills G, Montgomery K, Kucherlapati R, Rahme LG, Ausubel FM. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden HS, Gillett W, Saenphimmachak C, Lim R, Zhou Y, Jacobs MA, Chang J, Rohmer L, D'Argenio DA, Palmieri A, Levy R, Haugen E, Wong GK, Brittnacher MJ, Burns JL, Miller SI, Olson MV, Kaul R. Large-insert genome analysis technology detects structural variation in Pseudomonas aeruginosa clinical strains from cystic fibrosis patients. Genomics. 2008;91:530–537. doi: 10.1016/j.ygeno.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin DY, Bosch PL van den, Abbas B, Janssen AJ, Muyzer G. Microbiological analysis of the population of extremely haloalkaliphilic sulfur-oxidizing bacteria dominating in lab-scale sulfide-removing bioreactors. Appl Microbiol Biotechnol. 2008;80:965–975. doi: 10.1007/s00253-008-1598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the conserved domains among the site-specific recombinases of the tyrosine integrase family. Alignment of the conserved domains among the site-specific recombinases of the tyrosine integrase family from phages, conjugative transposons, plasmids and other sources. R (Arginine) being in Domain I and H (Histidine)-R-Y (Tyrosine) in Domain II.
Phylogenetic tree of the Integrase proteins from Tn4371-like integrases available on the GenBank database and other Phage and ICE integrases. Phylogenetic tree of the Integrase proteins from available Tn4371-like integrases available on the GenBank database and other Phage and ICE integrases. Cluster analysis was based upon the neighbour joining method. Numbers at branch-points are percentages of 1000 bootstrap resamplings that support the topology of the tree. The scale bar represents 0.2 substitutions per nucleotide position.
Gene numbers for genes in the elements discovered in this study. The gene numbering for genes of the elements discovered in this study. Genes with yellow background are the scaffold genes of the element.
Alignment of the first/last 200 bp of Tn4371-like ICEs using ClustalW. Fig S1a: Alignment of the first 200 bp of Tn4371-like ICEs using ClustalW. Fig S1b: Alignment of the last 200 bp of I Tn4371-like ICEs using ClustalW.