Abstract
Phosphorylation is a dynamic post-translational protein modification that is the basis of a general mechanism for maintaining and regulating protein structure and function, and of course underpins key cellular processes through signal transduction. In the last several years, many studies of large-scale profiling of phosphoproteins and mapping phosphorylation sites from cultured human cells or tissues by mass spectrometry technique have been published; however, there is little information on general (or global) phosphoproteomic characterization and description of the content of phosphoprotein analytes within the circulation. Circulating phosphoproteins and phosphopeptides could represent important disease biomarkers because of their well-known importance in cellular function, and these analytes frequently are mutated and activated in human diseases such as cancer. Here we report an initial attempt to characterize the phosphoprotein content of serum. To accomplish this, we developed a method in which phosphopeptides are enriched from digested serum proteins and analyzed by LC-MS/MS using LTQ-Orbitrap (CID) and LTQ-ETD mass spectrometers. Using this approach we identified ~100 unique phosphopeptides with stringent filtering criteria and a lower than 1% false discovery rate.
Keywords: Titanium dioxide, phosphopeptide, serum, LTQ-Orbitrap, LTQ-ETD, mass spectrometry
1. Introduction
Post-translational phosphorylation is a common and important mechanism of acute and reversible regulation of protein function in mammalian cells. Dynamic phosphorylation of proteins on serine, threonine and tyrosine residues is recognized as a key mode of regulating cell cycle, cell growth, cell differentiation and metabolism.1-4 To understand exactly why a particular protein becomes phosphorylated, it may be necessary to identify precisely which amino acid residues are phosphorylated. These residues then can be changed by site-directed mutagenesis, and the mutated protein can be examined for changes in activity, intracellular localization, and association with other proteins in the cell. In addition, identification of phosphorylation sites could reveal which protein kinase regulates the protein and thereby help to understand the biological function and significance of signal transduction pathway. Today, mass spectrometry (MS)-based methods are used routinely for protein identification, and recent mass spectrometry instrumental advancements enable a highly efficient method for global profiling of phosphoproteins and phosphorylation sites.5,6 The MS strategy consists of enzymatic digestion of proteins to yield peptides, which are analyzed by MS either directly or after enrichment. When compared to the mass predicted from the amino acid sequence, a mass increment of 80 Da indicates the presence of a phosphate group (HPO3) on a peptide. The phosphorylation site can be mapped to a specific residue by tandem mass spectrometry (MS/MS) whereby a peptide is activated in the gas phase and fragmented to produce structurally-informative fragment ions. However, the stoichiometry of most phosphorylation is relatively low – only a small fraction of the available intracellular pool of a protein is phosphorylated, and phosphorylated peptides are notoriously difficult to analyze in the presence of a much greater abundance of non-phosphorylated peptides, especially when analyzing a highly complex biological sample. Therefore, in most cases, enrichment of phosphopeptides by tyrosine-specific antibodies, immobilized metal ion affinity chromatography (IMAC), or titanium dioxide (TiO2) chromatography, is advantageous or required before MS analysis.7-9
Collision-induced dissociation (CID) is the most widely used technique to fragment peptide ions in a mass spectrometer and has proven to be extremely useful for amino acid sequence assignment and for post-translational modification studies. However, low-energy collisional activation of phosphorylated peptides typically results in the predominant loss of phosphoric acid with inadequate fragmentation of the peptide backbone, whereby only limited sequence information is obtained, and large peptides (>2500 Da) are difficult to fragment by CID. Electron-transfer dissociation (ETD) is a newly developed fragmentation method, with which it has been shown that phosphorylation is preserved while the backbone of the peptide is fragmented to yield c and z product ions.10,11 The resulting ETD spectra typically exhibit an informative ion series indicating both the amino acid sequence and the phosphorylation site. ETD also has the advantage of enabling efficient fragmentation of larger peptides with charge states of 3+ or higher, but can yield poorer fragmentation efficiency for smaller peptides with lower (1+, 2+) charge states.12
In the last several years, several reports of large-scale profiling of phosphoproteins and mapping phosphorylation sites from bacteria, yeast, mammalian cells have been published,13-17 however, there is little information on the characterization of the phosphoproteome of serum or plasma. The serum proteome, containing more than 9000 proteins based on Plasma Proteome Project (http://www.hupo.org/research/hppp/), is a very complex matrix in which protein concentration can range over more than ten orders of magnitude, and 90% of the total protein composition is constituted by ~20 abundant proteins, such as albumin, IgGs, complement components, and apolipoproteins.18-20 The complexity of serum protein mixture and the high background level of the most abundant proteins make MS-based phosphoprotein identification and localization of phosphorylation sites a challenge.
Recently Hu et al reported the profiling of endogenous serum phosphorylated peptides by TiO2-enrichment and MALDI-TOF MS detection.21 However, the characterization and description of the content of phosphoprotein analytes in serum were limited. In this present work, we report an approach to identify a large number of phosphopeptides and their phosphorylation sites using TiO2-based phosphopeptide enrichment followed by LC-MS/MS analysis using high sensitivity nano-electrospray and an LTQ-Orbitrap instrument. The latter provides high accuracy mass measurement that is essential for the validation of modified peptide identifications and the reduction of false positive identifications. We also explored the use of ETD, the newly emerged MS method for the study of protein post-translational modifications. Comparison of immunoaffinity (anti-phosphotyrosine antibody) versus chemical (TiO2) enrichment methods, as well as single versus dual enrichment strategies, yielded an initial assessment of the phosphoprotein/peptide content of human serum, which can serve as a launch-point for further exploration and analysis.
2. Materials and methods
2.1. Materials
Dithiothreitol (DTT), iodoacetamide, urea, ammonium bicarbonate, 2,5-dihydroxybenzoic acid (DHB), trifluoroacetic acid (TFA), and phosphoprotein bovine beta-casein were from Sigma-Aldrich; protease inhibitor and phosphatase inhibitor cocktail tablets were from Roche; Coomassie (Bradford) protein assay kit was from Pierce; anti-phosphotyrosine (4G10) agarose conjugate was from Upstate; trypsin was from Promega; LysC was from Wako Chemicals. Human angiotensin I (AngI) peptide DRVYIHPFHL, and human tyrosine phosphorylated Angiotensin II (AngII-P) DRVpYIHPF were from Calbiochem; phosphopeptides ETTCSKEpSNEELTESCETK and AIPVAQDLNAPSDWDpSR were synthesized from Peptide 2.0 Inc.. The single clinical specimen chosen for analysis, a pre-operative, pre-treatment serum sample procured from a male subject with prostate cancer undergoing radical prostatectomy, was obtained under IRB approval (University of California-Irvine) and patient consent.
2.2. Albumin/IgG depletion
Qproteome Albumin/IgG Depletion Kit (Qiagen, Catalogue number 37521) was used for depletion of albumin and IgG from 40 μL (~2 mg) serum sample. The kit contains spin columns and immobilized monoclonal antibodies for depletion of albumin and IgG. Diluted serum samples with added protease and phosphatase inhibitors were applied to the resin in the spin column, which was then sealed and incubated on a shaker. Serum proteins, depleted of most albumin and IgG, were recovered by centrifugation. The albumin and IgG in the sample, which were bound by antibodies immobilized on a solid support in the spin column, were mixed with 8 M urea and then recovered by centrifugation.
2.3. Trypsin/LysC digestion and desalting
U266 cells were cultured to semi-confluency, treated with 10 mM pervanadate for 30 minutes, stripped from the flask, and collected by centrifugation. The cells were washed by phosphate buffered saline (PBS) three times to remove culture media. The pelleted cells (~10 million) were lysed by mixing with 200 μL of 8 M urea, sonicated for 30 seconds, and centrifuged at 16000 g for 10 minutes. The supernatant was transferred to another centrifugation tube, and the protein concentration was measured by Bradford Assay.
Proteins from cell lysate, raw serum, eluted serum proteins that were depleted of albumin and IgG, and eluted albumin and IgG from resin, were spiked with the phosphoprotein bovine beta-casein as an internal standard, reduced by 10 mM DTT in the presence of 8 M urea for 30 minutes at 37 °C, and then alkylated by 50 mM iodoacetamide at room temperature. The concentrated urea in the sample was diluted to a final concentration of 2 M, and the proteins were digested by trypsin or LysC at 37 °C for 6 hours in a buffer containing ammonium bicarbonate (50 mM, pH 9). The digestion mixture was then acidified by adding glacial acetic acid to a final concentration of 2% and desalted by SepPak column (Waters).
2.4. 4G-10 enrichment of phosphopeptides from digested serum
Acetonitrile in the tryptic serum peptide solution that was eluted from SepPak column was removed by SpeedVac, and the pH of the sample was adjusted to 7.5 with a 0.1 N NaOH solution. The sample was then mixed with 50 μL of phosphotyrosine-specific antibody 4G-10 immobilized on agarose in Tris buffered saline (TBS) buffer and incubated in a shaker for overnight at 4 °C. After centrifugation and removal of supernatant, the resin was washed with TBS buffer three times. The resin then was mixed with 8 M urea, and the bound phosphopeptides were harvested from the supernatant. The sample then was desalted by ZipTip (Millipore).
2.5. TiO2 enrichment of phosphopeptides
Phosphopeptides were enriched from the mixture of all tryptic peptides using TiO2 as described by Thingholm22 with modification. In brief, a 30-cm piece of fused silica capillary tubing (360 μm OD, 200 μm ID, Polymicro Technologies, Phoenix, AZ) was attached to the frit end of Inline MicroFilter Assembly (Upchurch Scientific), and TiO2 loose media (GL Sciences, Inc) were slurry-packed into the tubing using a Pressure Cell (Brechbühler Inc.) to form a 200 μm × 2 cm TiO2 column. 1 pmol of standard phosphopeptide angiotensin II phosphate was added to the SepPak- or ZipTip-cleaned sample. The sample then was mixed with an equal volume of Loading Buffer (200 mg/mL DHB, 5% TFA, 80% acetonitrile), and loaded into the TiO2 column using the Pressure Cell with flow rate of 3 μL per minute. The column was washed by 200 μL of Wash Buffer 1 (40 mg/mL DHB, 2% TFA, 80% acetonitrile) and 2 × 200 μL of a second Wash Buffer 2 (2% TFA, 50% acetonitrile) to remove non-phosphopeptides. Phosphopeptides were eluted from the column with the Elution Buffer (5% ammonia solution). Ammonia in the eluate was removed by lyophilization (~3 minutes), and the sample was acidified by adding glacial acetic acid to a final concentration of 2%, and desalted by ZipTip. 100fmol of standard peptide angiotensin I were spiked into the enriched sample as an internal standard. For two rounds of TiO2 enrichment, after removal of ammonia in the elution fraction from the first round of TiO2 purification, the sample was mixed with an equal volume of Loading Buffer, loaded onto another TiO2 column, washed, and eluted as in the first enrichment.
2.6. Mass spectrometry for phosphopeptide identification
The purified phosphopeptides were analyzed by reversed-phase liquid chromatography nanospray tandem mass spectrometry (LC-MS/MS) using an LTQ-Orbitrap mass spectrometer (Thermo Fisher).23 The reversed-phase LC column was slurry-packed in-house with 5 μm, 200 Å pore size C18 resin (Michrom BioResources, CA) in a 100 μm i.d. × 10 cm long piece of fused silica capillary (Polymicro Technologies, Phoenix, AZ) with a laser-pulled tip. After sample injection, the column was washed for 5 minutes with mobile phase A (0.1% formic acid), and peptides were eluted using a linear gradient of 0% mobile phase B (0.1% formic acid, 80% acetonitrile) to 45% mobile phase B in 120 minutes at 200 nL/minute, then to 100% B in an additional 5 minutes. The LTQ-Orbitrap mass spectrometer was operated in a data-dependent mode in which each full MS scan (60,000 resolving power) was followed by eight MS/MS scans where the eight most abundant molecular ions were dynamically selected and fragmented by collision-induced dissociation (CID) using a normalized collision energy of 35%. Tandem mass spectra were searched against the NCBI human protein database using SEQUEST (Bioworks software, ThermoFisher) with full tryptic cleavage constraints, static cysteine alkylation by iodoacetamide, and variable phosphorylation of Ser/Thr/Tyr and methionine oxidation. Confident phosphopeptide identifications were determined using stringent filter criteria for database match scoring followed by manual evaluation of the results. The purified phosphopeptides were analyzed also by LC-MS/MS using an LTQ-ETD mass spectrometer (ThermoFisher). The LTQ-ETD mass spectrometer was operated in a data-dependent mode in which each full MS scan was followed by five MS/MS scans with supplemental activation. Tandem mass spectra were searched against the NCBI human protein database using EQUEST (Bioworks software, ThermoFisher) with full LysC cleavage constraints, static cysteine alkylation by iodoacetamide, and variable phosphorylation of Ser/Thr/Tyr. Confident phosphopeptide identifications were determined using stringent filter criteria for database match scoring.
3. Results
3.1. Method Optimization
Because we used a modified method to pack the TiO2 column for phosphopeptide enrichment, we evaluated this method using human cell lysate as a test experiment. The TiO2 enriched phosphopeptides from trypsin-digested 200 μg cell lysate with spiked 200 ng (10 pmol) beta casein were analyzed by LC-MS/MS using LTQ-Orbitrap mass spectrometer. The phosphopeptide FQpSEEQQQTEDELQDK (2+ ion m/z 1031.4178 and 3+ ion m/z 687.9477) from beta casein and the standard phosphopeptide AngII-P (2+ ion m/z 563.7577), which was added into the trypsin-digested sample, were detected with high S/N and identified from the raw MS data based on the peptide m/z value and MS2 spectra. The search results were filtered using the following criteria: “ranked top #1; Xcorr versus charge 1.8, 2.5 for 2+, 3+ ions; mass accuracy 3 ppm; probability of randomized identification of peptide < 0.05”, which yielded 486 matched MS2 spectra. Among these, 62 (12.8%) spectra were matched to non-phosphopeptides, and 424 (87.2%) spectra were matched to phosphopeptides. A total of 228 unique phosphopeptides was identified from this 200 μg cell lysate sample. The estimated “false discovery rate (FDR)” is lower than 1% by searching a combined forward-reversed database as described by Elias.24 These results demonstrate that phosphopeptides from crude cell lysate were highly enriched, and this method can be applied to more complexed serum samples.
3.2. Sensitivity Assessment
Our high performance liquid chromatography (HPLC)-coupled LTQ-Orbitrap system was tested and found capable of allowing detection of 1 fmol of the phosphopeptide standard AngII-P (data not shown). In order to estimate the detection limit for phosphopeptides enriched from serum, a sensitivity test experiment was performed. In this experiment, phosphopeptides were enriched from trypsin-digested 40 μL (2 mg) serum samples that had been spiked with 1 μg (50 pmol), 100 ng (5 pmol), 10 ng (500 fmol), and/or 1 ng (50 fmol) beta-casein and then analyzed by LC-MS/MS. The beta casein phosphopeptide FQpSEEQQQTEDELQDK was identified from serum samples that contained 1 μg, 100 ng, and 10 ng beta casein with relatively high MS peak intensity/area, and the 1 ng beta-casein-spiked sample yielded low MS peak intensity/area (Figure 1S). These results indicate that our method could allow detection of serum phosphoproteins/phosphopeptides with amounts greater than approximately 10-100 fmol. Identification of lower abundance serum phosphoproteins/phosphopeptides could be a challenge because a fraction of these peptides could be lost during desalting and/or TiO2 enrichment steps, and the amount of certain purified phosphopeptides could be too low to be detected. We also noticed that most of the abundant serum phosphopeptides were repeatedly identified from serum samples that had been spiked with various amount of beta casein, indicating the method was reproducible.
3.3. Comparison of phosphoprotein enrichment methodologies
We then carried out several approaches to characterize serum phosphoproteins, as shown in the Figure 1 flowchart. From a single TiO2 enrichment of phosphopeptides from 40 μL (~ 2 mg) serum 66.2% of matched MS2 spectra correspond to phosphopeptides (Table 1), and this percentage is lower than that of cell lysate. A total of 48 unique phosphopeptides were identified from this sample. This number is much lower than that of from even 200 μg cell lysate, indicating that the abundance of phosphorylated proteins/peptides in serum is low compared with cells. Most of these identified phosphopeptides have the modification at serine and threonine, and only two have modification at tyrosine. It is apparent that serine and threonine residues undergo phosphorylation more often than tyrosine residues in serum, similar to that of cells whose phospho-amino acid content ratio (pSer:pThr:pTyr) is 1800:200:1.25 We then tested if a higher purity of phosphopeptides could be obtained by performing an additional TiO2 enrichment of the eluted fraction from the first TiO2 enrichment. The result shows that, with two rounds of TiO2 enrichment of phosphopeptides from 40 μL (~ 2 mg) serum, 94.2% of matched MS2 spectra correspond to phosphopeptides, indicating that the purity of phosphopeptides was significantly increased. The number of identified unique phosphopeptides moderately increased to 60 (Table 1). As shown in Table 2, which lists the identified phosphopeptides from two steps TiO2 enrichment, many of these phosphopeptides are from abundant serum proteins such as apolipoproteins, kininogen 1, serine (or cysteine) proteinase inhibitor, alpha-2-HS-glycoprotein and complement components. Based on semi-quantitative MS2 spectra counts, the most abundant phosphopeptides are ETTCSKEpSNEELTESCETK from kininogen 1, ATEDEGpSEQKIPEATNR from serine (or cysteine) proteinase inhibitor clade C, and HTFMGVVSLGSPpSGEVSHPR from alpha-2-HS-glycoprotein.
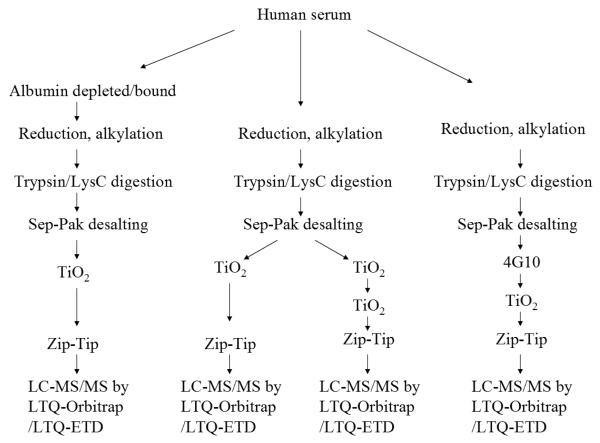
Figure 1.
Flowchart of the experimental procedure. Groups of serum samples were digested by trypsin or LysC, and phosphopeptides were purified by single/dual TiO2-enrichment strategies as well as immunoaffinity for LC-MS/MS analyses using LTQ-Orbitrap and/or LTQ-ETD, followed by phosphopeptide identification by database search and manual data evaluation.
Table 1.
Summary of serum phosphopeptide identification by LTQ-Orbitrap a
| Total matched spectra |
Total non- phosphopeptide spectra (% total) |
Total phosphopeptide spectra (% total) |
Total unique phosphopeptides |
|
|---|---|---|---|---|
| One round TiO2 enrichment |
526 | 178 (33.8%) | 348 (66.2%) | 48 |
| Two rounds TiO2 enrichment |
275 | 16 (5.8%) | 259 (94.2%) | 60 |
| Albumin/IgG depletion fraction |
357 | 86 (24.1%) | 271 (75.9%) | 23 |
| Albumin/IgG bound fraction |
320 | 238 (74.4%) | 82 (25.6%) | 27 |
| Cell lysate | 486 | 62 (12.8%) | 424 (87.2%) | 228 |
All results were obtained using the following criteria to filter the SEQUEST search results: “ranked top #1; Xcorr versus charge 1.8, 2.5 for 2+, 3+ ions; mass accuracy 3 ppm; probability of randomized identification of peptide < 0.05”. The estimated false discovery rate by searching combined forward and reversed database is lower than 1%.
Table 2.
Serum phosphopeptides identified from two steps TiO2 enrichment and LTQ-Orbitrap analysis
| Protein description | Phosphopeptide sequence | MS2 spectra count |
|---|---|---|
| kininogen 1 | ETTCSKEpSNEELTESCETK | 124 |
| ETTCpSKESNEELTESCETK | 7 | |
| ETTCpSKEpSNEELTESCETK | 1 | |
| EpTTCSKESNEELTESCETK | 8 | |
| ETpTCSKESNEELTESCETK | 6 | |
| EpTpTCSKESNEELTESCETK | 2 | |
| serine (or cysteine) proteinase inhibitor, clade C (antithrombin), member 1 |
ATEDEGpSEQKIPEATNR | 16 |
| serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
VVQAPKEEEEDEQEApSEEK | 2 |
| VVQAPKEEEEDEQEApSEEKApSEEEK | 2 | |
| alpha-2-HS-glycoprotein | HTFMGVVSLGSPpSGEVSHPR | 10 |
| HTFMGVVSLGSPSGEVpSHPR | 1 | |
| HTFMGVVSLGpSPpSGEVSHPR | 1 | |
| apolipoprotein L1 isoform a precursor | VTEPISAEpSGEQVER | 4 |
| VTEPIpSAESGEQVER | 4 | |
| VTEPIpSAEpSGEQVER | 3 | |
| apolipoprotein A-II preproprotein | pSKEQLTPLIK | 2 |
| EPCVEpSLVSQYFQTVTDYGK | 1 | |
| EPCVESLVpSQYFQTVTDYGK | 1 | |
| apolipoprotein A-IV precursor | ISApSAEELR | 3 |
| apolipoprotein AV | EPApTLKDSLEQDLNNMNK | 1 |
| EPATLKDpSLEQDLNNMNK | 1 | |
| apolipoprotein A-I preproprotein | THLAPYpSDELR | 1 |
| apolipoprotein E precursor | GEVQAMLGQpSTEELR | 2 |
| inter-alpha globulin inhibitor H2 polypeptide |
SLPGEpSEEMMEEVDQVTLYSYK | 3 |
| secreted phosphoprotein 1 isoform b | ISHELDSApSSEVN | 2 |
| ISHELDpSASSEVN | 1 | |
| FRIpSHELDpSASSEVN | 1 | |
| AIPVAQDLNAPSDWDpSR | 1 | |
| secreted phosphoprotein 2, 24kDa | KDpSGEDPATCAFQR | 1 |
| NNYLFGLIpSDEpSIpSEQFYDR | 1 | |
| insulin-like growth factor binding protein 3 isoform b precursor |
YKVDYESQSTDTQNFpSSESKR | 2 |
| clusterin isoform 1 | VTTVASHTSDpSDVPSGVTEVVVK | 1 |
| VTTVASHpTpSDSDVPSGVTEVVVK | 1 | |
| fibrinogen, alpha polypeptide isoform alpha-E preproprotein |
PGSTGTWNPGpSSER | 2 |
| complement component 3 precursor | IPIEDGpSGEVVLSR | 2 |
| SGpSDEVQVGQQR | 1 | |
| pSGSDEVQVGQQR | 1 | |
| complement component 9 | AEQCCEETASpSISLHGK | 2 |
| albumin precursor | YICENQDSISpSK | 2 |
| YICENQDpSISSK | 1 | |
| heparin cofactor II precursor | GGETAQpSADPQWEQLNNK | 2 |
| coagulation factor V precursor | CIPDDDEDSpYEIFEPPESTVMATR | 1 |
| CIPDDDEDpSYEIFEPPESTVMATR | 1 | |
| YKVDYESQSpTDTQNFpSSESKR | 1 | |
| golgi phosphoprotein 2 | RQVEKEEpTNEIQVVNEEPQR | 1 |
| TFPGFFSPMLGEFVSETEpSR | 1 | |
| chordin-like 2 | PHSQNLPLDSDQEpSQEAR | 1 |
| selenoprotein P precursor | DMPApSEDLQDLQKK | 2 |
| plasminogen-related protein B2 | AFQYHpSKEQQCVIMAENR | 1 |
| hypothetical protein LOC54964 | SSAINEEDGSpSEEGVVINAGK | 2 |
| FIANpSQEPEIR | 1 | |
| vitronectin precursor | DpSWEDIFELLFWGR | 2 |
| matrix, extracellular | INQELSpSKENIVQER | 1 |
| phosphoglycoprotein with ASARM motif (bone) |
||
| angiopoietin-like 3 precursor | IDQDNSpSFDSLSPEPK | 1 |
| fibronectin 1 isoform 2 preproprotein | RPGGEPSPEGTTGQpSYNQYSQR | 1 |
| RPGGEPpSPEGTTGQSYNQYSQR | 1 | |
| chondroitin sulfate proteoglycan 2 (versican) |
QEIESETTpSEEQIQEEK | 1 |
| QEIEpSETTSEEQIQEEK | 1 | |
| SPARC-like 1 | HIQETEWQpSQEGK | 1 |
| bovine beta casein (spiked in standard) |
FQpSEEQQQTEDELQDK | 8 |
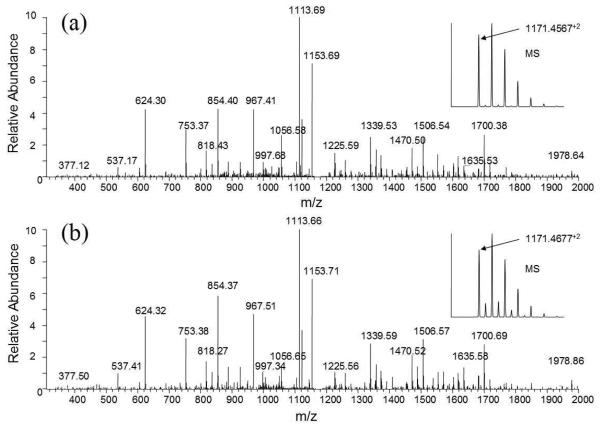
Two of these identified phosphopeptides, ETTCSKEpSNEELTESCETK from kininogen-1 and AIPVAQDLNAPSDWDpSR from secreted phosphoprotein 1, were obtained as synthetic peptides from a company, and their MS2 spectra (the former peptide was reduced and alkylated prior to LC-MS) were acquired using the LTQ-Orbitrap. The MS2 spectra of these phosphopeptides obtained from TiO2 enriched serum were almost identical to the MS2 spectra of synthesized peptides (Figures 2 and 2S), indicating accurate identifications.
Figure 2.
Verification of identified phosphopeptide ETTCSKEpSNEELTESCETK from kininogen by MS2 spectra comparison. (a) CID spectrum of the phosphopeptide (2+ ion m/z 1171.4567) obtained from TiO2 enriched serum phosphopeptides; (b) CID spectrum of synthetic phosphopeptide ETTCSKEpSNEELTESCETK.
We further tested the effect of depletion of abundant serum proteins on phosphopeptides identification. Albumin/IgG depletion was performed with 40 μL (2 mg) serum, and phosphopeptides from both the depletion fraction and the albumin/IgG bound fraction were TiO2-enriched and analyzed using the LTQ-Orbitrap. It was found that 75.9% of the MS2 spectra were matched to phosphopeptides enriched from the depletion fraction, which is higher compared with that without depletion but lower than that for the cell lysate. In addition, 25.6% of the MS2 matched to phosphopeptide enriched from the 2 mg albumin/IgG bound fraction (Table 1). There were 23 unique phosphopeptides identified in the depletion fraction, which is lower than that without depletion and indicates that depletion of abundant serum proteins did not improve serum phosphopeptide identification. Interestingly, 27 unique phosphopeptides were identified from albumin/IgG bound fraction, indicating that a significant amount of serum phosphoproteins/peptides are tightly bound with the carrier proteins albumin and IgGs.
We also tried to use phosphotyrosine-specific antibody to immunoprecipitate tyrosinephosphorylated peptides from digested serum, and only one tyrosine-phosphorylated peptide was identified. This result is similar to that of one step of TiO2 enrichment, therefore, the enrichment using a phosphotyrosine-specific antibody did not improve the identification of tyrosine phosphorylated peptides from serum.
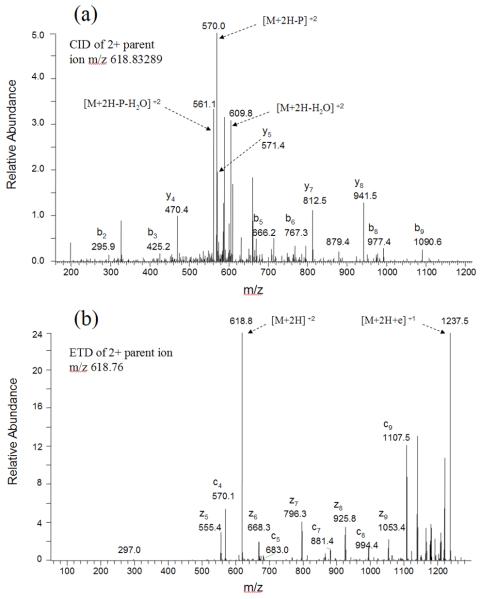
We next characterized TiO2-enriched serum phosphopeptides from LysC-digested samples using a different mass spectrometer and fragmentation method, LTQ-ETD, and we obtained similar results. Using one step of TiO2 enrichment of phosphopeptides from LysC-digested 40 μL (~ 2 mg) serum, 45.7% of matched MS2 spectra were matched to phosphopeptides (Table 3), and 15 unique phosphopeptides were identified after the EQUEST search results were filtered using the stringent criteria of “ranked top #1; Probability (EQUEST) > 7”. With two rounds of TiO2 enrichment of phosphopeptides from 40 μL (~ 2 mg) serum, 82.0% of matched MS2 spectra correspond to phosphopeptides, and 13 unique phosphopeptides were identified. Again, several phosphopeptides were identified from both depletion fraction and albumin/IgG bound fraction obtained from albumin/IgG depletion. Some of the phosphopeptides identified by LTQ-ETD, such as pSKEQLTPLIK from Apolipoprotein A-II and HIQETEWQpSQEGK from SPARC-like 1 protein, were identified also using the LTQ-Orbitrap. As shown in Figures 3 and 3S, the CID and ETD spectra obtained enabled the confident identification of these two peptides. Several multiply phosphorylated peptides, such as QVSSLpSpSGVIQEALATNMK from microfilament and actin filament cross-linker protein, SKEEpSHEQpSAEQGK from SPARC-like 1 protein, were identified by LTQ-ETD but not by LTQ-Orbitrap.
Table 3.
Summary of serum phosphopeptide identification by LTQ-ETD a
| Total matched spectra |
Total non- phosphopeptide spectra (% total) |
Total phosphopeptide spectra (% total) |
Total unique phosphopeptides |
|
|---|---|---|---|---|
| One round TiO2 enrichment |
46 | 25 (54.3%) | 21 (45.7%) | 15 |
| Two rounds TiO2 enrichment |
50 | 9 (18.0%) | 41 (82.0%) | 13 |
| Albumin/IgG depletion fraction |
33 | 11 (33.3%) | 22 (66.7%) | 13 |
| Albumin/IgG bound fraction |
12 | 6 (50.0%) | 6 (50.0 %) | 4 |
All results were obtained using the following criteria to filter the EQUEST search results: “ranked top #1; Probability (EQUEST) > 7”.
Figure 3.
Example CID and ETD spectra of the doubly-charged phosphopeptide pSKEQLTPLIK from Apolipoprotein A-II. (a) CID spectrum labeled to show singly-charged b and y ions, as well as ions corresponding to neutral losses of the phosphate group and water. (b) ETD spectrum labeled to show singly-charged c and z ions, as well as ions corresponding to unfragmented and charge-reduced parent ion.
From this first global phosphoproteomic analysis of serum, using several strategies, approximately 100 serum phosphopeptides were identified with high confidence.
4. Discussion
While the characterization and description of tissue phosphoproteins is the source of intense investigation by many scientists throughout the world, similar analysis of the phosphoprotein content of serum and plasma remains largely non-existent. The paucity of information is largely due to a) the low abundance of the circulating phosphoanalytes due to enzymatic degradation once entering into the circulation and b) the fact that most phosphoprotein/peptides that are found in the circulation arise from a tissue source and are immediately diluted into the circulation thus dramatically diluting the endogenous concentration. Of course, these analytical issues are further compounded by the high concentrations of resident proteins such as albumin and immunoglobulins. Our efforts centered on the development of an approach that provided the most comprehensive view of the serum phosphoproteome, that yielded the highest number of individual analytes. This effort was not meant to serve as a complete description of the serum phosphoproteome nor identify disease-specific biomarker candidates, but as an initial effort to begin the description of this potentially important information archive. We chose a single serum sample, obtained from a patient with prostate cancer prior to prostatectomy, to analyze with different technological approaches so that we could methodically understand the effect of the phosphoprotein enrichment procedures and MS techniques on analyte yield and complexity.
Recently, Thingholm reported a new, improved procedure for the purification of phosphorylated peptides using TiO2 column,22 which substantially enhances the enrichment efficiency for phosphopeptides without any prior chemical modification. With this modified TiO2 column packing technique, we here show that phosphopeptides from serum samples were highly enriched and ~100 unique serum phosphopeptides were identified by tandem mass spectrometry using high performance LTQ-Orbitrap and LTQ-ETD instruments. However, the actual number of serum phosphopeptides could be higher because: 1) the current SEQUEST search was performed using indexed peptide database with full tryptic cleavage constraints. We noticed that a large number of MS2 spectra in the raw data contain apparent neutral loss peak from phosphate group but the spectra did not confidently match with phosphopeptide in the full tryptic peptide database. It is very likely that these MS2 spectra correspond to partial tryptic phosphopeptides or non-tryptic phosphopeptides generated by other proteases; 2) the current SEQUEST search result was filtered by stringent criteria to reach ~ 1% false discovery rate. It is well known that MS2 spectrum quality of many phosphopeptides is poor due to neutral loss of phosphate group, and the SEQUEST score of some phosphopeptides is low and could be filtered out by the stringent criteria. These low scored phosphopeptides could be identified by relaxing the SEQUEST filter criteria and performing subsequent manual verification; 3) the current ETD raw data was acquired from low mass accuracy LTQ-ETD mass spectrometer. EQUEST was used to search database and the result was filtered by stringent criteria, however, for the low mass accuracy MS data, it is difficult to find optimum filter criteria to yield a large number of identified phosphopeptides with low a false discovery rate such as 1%. It is likely that more serum phosphopeptides could be confidently identified by using the most updated high mass accuracy hybrid mass spectrometer such as the LTQ-ETD-Orbitrap.
The serum proteome is a complex mixture predominated by high-abundance resident proteins, such as albumin and other carrier proteins, together with proteins that originate from circulating blood cells and tissues.26 Some of the identified phosphoproteins are from abundant serum proteins, such as kininogen 1, alpha-2-HS-glycoprotein, apolipoproteins. Based on MS2 spectra counts, peptide ETTCSKEpSNEELTESCETK from kininogen 1 is the most abundant serum phosphopeptide. The plasma and tissue kininogen-kallikrein-kinn system has been reported to play important roles in the cardiovascular system, intestinal inflammatory disease and arthritis.27-30 It remains unclear what kinds of kinases are responsible for the phosphorylation of these proteins and what is the function of these serum phosphoproteins. We also identified some cellular phosphopeptides that presumably entered the blood from the surrounding tissue, such as VTTVASHTSDpSDVPSGVTEVVVK from clusterin, AIPVAQDLNAPSDWDpSR from secreted phosphoprotein 1, HIQETEWQpSQEGK from SPARC-like 1 protein. The glycoprotein clusterin is a key player in apoptosis, cell cycle control, as well as many diseases including cancer.31,32 The secreted phosphoprotein 1, also named osteopontin, is expressed and secreted by numerous human cancers. It has pivotal role in cellular signaling and tumor metastasis.33,34 The SPARC-like 1 protein showed tumor-suppressor function in cultured pancreatic cancer cells and has been reported to be down regulated in cancers.35-37 These modified forms of proteins are a reflection of ongoing physiological and pathological events, and could be a treasure trove of biomarkers for early cancer diagnosis.
Supplementary Material
Acknowledgement
We thank Dr. David Ornstein from University of California for providing the human pre-operation prostate cancer serum that was used for this study. The research is supported by funding from the College of Science at George Mason University.
5. References
- 1.Venter JC. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. co-workers. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Signaling—2000 and beyond. Cell. 2000;100(1):113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 3.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278(5346):2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 4.Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr. Opin. Cell Biol. 2009;21(2):140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yates JR., 3rd. Database searching using mass spectrometry data. Electrophoresis. 1998;19(6):893–900. doi: 10.1002/elps.1150190604. [DOI] [PubMed] [Google Scholar]
- 6.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 7.Stensballe A, Andersen S, Jensen ON. Characterization of phosphoproteins from electrophoretic gels by nanoscale Fe(III) affinity chromatography with off-line mass spectrometry analysis. Proteomics. 2001;1(2):207–222. doi: 10.1002/1615-9861(200102)1:2<207::AID-PROT207>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 8.Mann M, Ong SE, Gronborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20(6):261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 9.Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 2004;76(14):3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- 10.Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Analysis of phosphorylation site on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2007;104(7):2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiesner J, Premsler T, Sickmann A. Application of electron transfer dissociation (ETD) for the analysis of posttranslational modifications. Proteomics. 2008;8(21):4466–4483. doi: 10.1002/pmic.200800329. [DOI] [PubMed] [Google Scholar]
- 12.Molina H, Matthiesen R, Kandasamy K, Pandey A. Comprehensive comparison of collision induced dissociation and electron transfer dissociation. Anal. Chem. 2008;80(13):4825–4835. doi: 10.1021/ac8007785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macek B, Gnad F, Soufi B, Kumar C, Olsen JV, Mijakovic I, Mann M. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics. 2008;7(2):299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villén J, Elias JE, Gygi SP. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 2007;6(3):1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 15.Cantin GT, Yi W, Lu B, Park SK, Xu T, Lee JD, Yates JR., 3rd. Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J. Proteome Res. 2008;7(3):1346–1351. doi: 10.1021/pr0705441. [DOI] [PubMed] [Google Scholar]
- 16.Pan C, Gnad F, Olsen JV, Mann M. Quantitative phosphoproteome analysis of a mouse liver cell line reveals specificity of phosphatase inhibitors. Proteomics. 2008;8(21):4534–4546. doi: 10.1002/pmic.200800105. [DOI] [PubMed] [Google Scholar]
- 17.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 2007;104(5):1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 19.Deutsch EW, Eng JK, Zhang H, King NL, Nesvizhskii AI, Lin B, Lee H, Yi EC, Ossola R, Aebersold R. Human plasma peptide atlas. Proteomics. 2005;5(13):3497–3500. doi: 10.1002/pmic.200500160. [DOI] [PubMed] [Google Scholar]
- 20.Thadikkaran L, Siegenthaler MA, Crettaz D, Queloz PA, Schneider P, Tissot JD. Recent advances in blood-related proteomics. Proteomics. 2005;5(12):3019–3034. doi: 10.1002/pmic.200402053. [DOI] [PubMed] [Google Scholar]
- 21.Hu L, Zhou H, Li Y, Sun S, Guo L, Ye M, Tian X, Gu J, Yang S, Zou H. Profiling of endogenous serum phosphorylated peptides by titanium (IV) immobilized mesoporous silica particles enrichment and MALDI-TOF MS detection. Anal. Chem. 2009;81(1):94–104. doi: 10.1021/ac801974f. [DOI] [PubMed] [Google Scholar]
- 22.Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nature Protocols. 2006;1(4):1929–1935. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- 23.Luchini A, Geho DH, Bishop B, Tran D, Xia C, Dufour RL, Jones CD, Espina V, Patanarut A, Zhou W, Ross MM, Tessitore A, Petricoin EF, Liotta LA. Smart hydrogel particles: Biomarker harvesting: One-step affinity purification, size exclusion, and protection against degradation. Nano Lett. 2008;8(1):350–361. doi: 10.1021/nl072174l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elias JE, Hass W, Faherty BK, Gygi SP. Nature Methods. 2005;2(9):667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 25.Hunter T, Sefton BM. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc. Natl. Acad. Sci. U.S.A. 1980;77(3):1311–1315. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liotta LA, Ferrari M, Petricoin E. Clinical Proteomics: written in blood. Nature. 2003;425(6961):905. doi: 10.1038/425905a. [DOI] [PubMed] [Google Scholar]
- 27.Marcondes S, Antunes W. The plasma tissue kininogen-kallikrein-kinin system: role in the cardiovascular system. Curr. Med. Chem. Cardiovasc. Hemotol. Agents. 2005;3(1):33–44. doi: 10.2174/1568016052773351. [DOI] [PubMed] [Google Scholar]
- 28.Isordia-Salas I, Pixley RA, Sainz IM, Martinez-Murillo C, Colman RW. The role of plasma high molecular weight kininogen in experimental intestinal and systemic inflammation. Arch. Med. Res. 2005;36(1):87–95. doi: 10.1016/j.arcmed.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Moreau ME, Garbacki N, Molinaro G, Brown NJ, Marceau F, Adam A. The kallikrein-kinin system: current and future pharmacological targets. J. Pharmacol. Sci. 2005;99(1):6–38. doi: 10.1254/jphs.srj05001x. [DOI] [PubMed] [Google Scholar]
- 30.Sainz IM, Pixley RA, Colman RW. Fifty years of research on the plasma kallikreinkinin system: from protein structure and function to cell biology and in-vivo pathophysiology. J. Thromb. Haemost. 2007;98(1):77–83. [PubMed] [Google Scholar]
- 31.Shannan B, Seifert M, Boothman DA, Tilgen W, Reichrath J. Clusterin and DNA repair: a new function in cancer for a key player in apoptosis and cell cycle control. J. Mol. Histol. 2006;37(57):183–188. doi: 10.1007/s10735-006-9052-7. [DOI] [PubMed] [Google Scholar]
- 32.Rizzi F, Bettuzzi S. Targeting Clusterin in prostate cancer. J. Physiol. Pharmacol. 2008;59(Suppl 9):265–274. [PubMed] [Google Scholar]
- 33.El-Tanani MK. Role of osteopontin in cellular signaling and metastatic phenotype. Front Biosci. 2008;13:4276–4284. doi: 10.2741/3004. [DOI] [PubMed] [Google Scholar]
- 34.Wai PY, Kuo PC. Osteopontin: regulation in tumor metastasis. Cancer Metastasis Rev. 2008;27(1):103–118. doi: 10.1007/s10555-007-9104-9. [DOI] [PubMed] [Google Scholar]
- 35.Isler SG, Schenk S, Bendik I, Schraml P, Novotna H, Moch H, Sauter G, Ludwig CU. Genomic organization and chromosomal mapping of SPARC-like 1, a gene down regulated in cancers. Int. J. Oncol. 2001;18(3):521–526. doi: 10.3892/ijo.18.3.521. [DOI] [PubMed] [Google Scholar]
- 36.Esposito I, Kayed H, Keleg S, Giese T, Sage EH, Schirmacher P, Friess H, Kleeff J. Tumor-suppressor function of SPARC-like protein 1/Hevin in pancreatic cancer. Neoplasia. 2007;9(1):8–17. doi: 10.1593/neo.06646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isler SG, Ludwig CU, Chiquet-Ehrismann R, Schenk S. Evidence for transcriptional repression of SPARC-like 1, a gene down regulated in human lung tumors. Int. J. Oncol. 2004;25(4):1073–1079. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.