Abstract
Most Ewing's sarcomas harbor chromosomal translocations that encode fusions between EWS and ETS family members. The most common fusion, EWS/FLI, consists of an EWSR1-derived strong transcriptional activation domain fused, in frame, to the DNA binding domain-containing portion of FLI1. EWS/FLI functions as an aberrant transcription factor to regulate genes that mediate the oncogenic phenotype of Ewing's sarcoma. One of these regulated genes, NR0B1, encodes a co-repressor protein, and likely plays a transcriptional role in tumorigenesis. However, the genes that NR0B1 regulates and the transcription factors it interacts with in Ewing's sarcoma are largely unknown. We used transcriptional profiling and chromatin immunoprecipitation to identify genes that are regulated by NR0B1, and compared these data to similar data for EWS/FLI. While the transcriptional profile overlapped as expected, we also found that the genome-wide localization of NR0B1and EWS/FLI overlapped as well, suggesting that they regulate some genes coordinately. Further analysis revealed that NR0B1 and EWS/FLI physically interact. This protein-protein interaction is likely to be relevant for Ewing's sarcoma development because mutations in NR0B1 that disrupt the interaction have transcriptional consequences and also abrogate oncogenic transformation. Taken together, these data suggest that EWS/FLI and NR0B1 physically interact, coordinately modulate gene expression, and mediate the transformed phenotype of Ewing's sarcoma.
Keywords: EWS/FLI, NR0B1, DAX1, Ewing's sarcoma
Introduction
Ewing's sarcoma is an aggressive bone-associated tumor that affects the pediatric population. The majority of Ewing's sarcomas harbor a reciprocal translocation, t(11;22)(q24;q12), which links a strong transcriptional activation domain from EWSR1 to the ETS DNA-binding portion of FLI1 (1). The EWS/FLI fusion is required for Ewing's sarcoma oncogenesis, as inhibition of fusion function or expression results in the loss of transformation of Ewing's sarcoma cells (2-4). Thus, understanding the function of EWS/FLI is critical to understanding Ewing's sarcoma development.
EWS/FLI is thought to function as a transcriptional activator (5-7). However, in Ewing's sarcoma cells, three times as many genes are down-regulated by EWS/FLI than are up-regulated (4, 8, 9). One hypothesis for this observation is that some EWS/FLI up-regulated gene targets function as transcriptional repressors. Indeed, this was supported by the demonstration that one EWS/FLI up-regulated gene product, NKX2.2, functions as a transcriptional repressor in Ewing's sarcoma. However, NKX2.2-mediated gene repression accounts for a small portion of the EWS/FLI down-regulated gene expression signature, suggesting that other targets may also function as repressors (10).
A second critical target, NR0B1 (DAX1), is an attractive candidate to mediate gene repression downstream of EWS/FLI. We recently demonstrated that NR0B1 is directly regulated by EWS/FLI and that it is required for the transformed phenotype of Ewing's sarcoma cells (8, 11). NR0B1 is an orphan member of the nuclear hormone receptor superfamily. NR0B1 is unusual because it lacks a conventional DNA binding domain, hence it is not thought to directly interact with DNA like other family members (12). While the molecular function of NR0B1 in Ewing's sarcoma is unknown, it appears to function primarily as a transcriptional co-repressor during the development and function of the hypothalamic-pituitary-adrenal-gonadal axis (13). To better understand the role of NR0B1 in Ewing's sarcoma, we tested the hypothesis that it functions as a transcriptional co-regulator during oncogenesis.
Materials and Methods
Constructs and Retroviruses
For “knockdown” experiments, previously described NR0B1-RNAi, luc-RNAi, and EF-2-RNAi constructs were utilized (4, 8). For over-expression experiments, a 3x-FLAG-tag was introduced onto the amino-terminus of NR0B1 and its mutants, EWS/FLI and its mutants, and wild-type FLI1 in the pMSCV-Neo retroviral vector (Clontech). For yeast-two-hybrid experiments, wild-type NR0B1 (14), NR0B1 mutants, wild-type EWS/FLI, and EWS/FLI mutants were cloned into pGBKT7 and pGADT7 (Clontech). For luciferase assays, approximately 700 bp of the NR0B1 intron (Supplementary Data 1) was cloned upstream of the SV40 promoter in the pGL3-Promoter luciferase reporter vector (Promega).
Cell Culture
Ewing's sarcoma cell lines A673, TC71, and EWS502, and the human embryonic kidney cell line 293EBNA, were grown as previously described (8, 15). Following retroviral infection, polyclonal cell populations were prepared by growth in selection media (2 mg/ml puromycin, 300 mg/ml G418). Soft-agar transformation assays and 3T5 growth assays were performed as previously described (8).
Microarray Analysis
RNA preparation, microarray hybridization, normalization, and analysis were performed as previously described (4, 10). Expression data were filtered for a 5-fold change across samples, with a minimal “delta” value of 50. Overlaps between different gene sets were performed using the VennMaster program and Chi square analysis (4, 10).
Chromatin immunoprecipitation (ChIP), Sequential ChIP, and Whole genome localization studies (ChIP-chip)
ChIP and ChIP-chip were performed as previously described (10, 11), by using M2-anti-FLAG (F1804; Sigma), anti-FLI-1-X, anti-cMyc (sc-356X or sc-764 respectively; Santa Cruz Biotechnology), or anti-α-tubulin (CP06; Calbiochem). Quantitative PCR was performed with NR0B1 or ALB primers (Supplementary Table S1). Sequential ChIP was performed with the Re-ChIP-IT kit (Active Motif) and the above listed antibodies.
Luciferase Assays
cDNA constructs (described in Constructs and Retroviruses above) and 293EBNA were utilized as previously described (11). Two-tailed Student's t- tests were used for statistical comparisons.
Yeast-Two-Hybrid Assays
AH109 yeast cells were transformed with indicated combinations of plasmids using the Matchmaker kit (Clontech). Yeast were then plated on SD/-Leu/-Trp plates or SD/-Leu/-Trp/-His plates. Plates were incubated for 4 days and colony growth counted. Three-aminotriazole (Sigma) was included in some instances to minimize autoactivation effects.
Co-Immunoprecipitation Assays
293EBNA were co-transfected with the indicated constructs, protein was extracted using NP40 lysis buffer (0.05 M Tris-HCl pH 7.4, 0.15 M NaCl, 1% NP40, 1 mM EDTA, protease inhibitor [Roche]). Co-immunoprecipitation experiments were conducted using FLAG-M2-Agarose beads (Sigma) or Dynabeads M-280 (Dynal) per manufacturer's directions.
Immunodetection
Western blots were performed with the indicated antibodies: M2-anti-FLAG, anti-FLI-X, anti-α-tubulin, or anti-mSin3A (gift from D. Ayer, ref. 16). Immunofluorescence experiments were performed using M2-anti-FLAG and anti-FLI-X primary antibodies per manufacturer's instructions (Sigma). Alexafluor 488 goat anti-mouse and Alexafluor 568 goat anti-rabbit (Invitrogen) were utilized as secondary antibodies.
Subcellular Fractionization
Transfected 293EBNA cells were collected and resuspended in hypotonic buffer (20 mM Hepes pH 7.5, 5 mM NaF, 0.1 mM EDTA, 10 μM Na2MoO4) and incubated on ice. 0.5% NP40 was then added and the homogenate was centrifuged. The cytoplasmic fraction was collected, while the nuclear pellet was resuspended in complete lysis buffer (400 mM NaCl, 20 mM Hepes pH 7.5, 10 mM NaF, 10 mM p-nitrophenyl phosphate, 1 mM NaVO3, 0.1 mM EDTA, 10 μM Na2MoO4, 10 mM ß-glycerophosphate, 20% Glycerol, 0.1 mM DTT, protease inhibitor [Roche]) and incubated with rocking at 4°C. After centrifugation the nuclear cell extract was then collected.
Results and Discussion
NR0B1 both up- and down-regulates gene expression in Ewing's sarcoma
To determine NR0B1's effect on gene expression in Ewing's sarcoma, we utilized our previously described RNAi-based loss-of-function approach in the patient-derived Ewing's sarcoma TC71 cell line (8, 10). We chose TC71 cells for this assay because “knockdown” of NR0B1 in the TC71 cell line results in a loss of transformation without affecting growth in tissue culture, thus the confounding transcriptional effects seen with other cell lines (e.g., A673 and EWS502) could be avoided (8). The transcriptional signature was determined using Affymetrix U133plus2 microarrays. The signal-to-noise metric was used to rank-order genes whose expression correlated with NR0B1 expression, and permutation testing at the 99% confidence level defined the cohort of NR0B1-regulated genes. As would be expected, based on prior assertions that NR0B1 functions as a transcriptional co-repressor, we found 846 genes that were down-regulated by the protein. Surprisingly, we found that NR0B1 also up-regulated 1131 genes (Supplementary Table S2).
To define NR0B1's contribution to the EWS/FLI transcriptional profile, we compared the gene signatures of NR0B1 to that previously reported for EWS/FLI (10). We found 300 NR0B1 regulated genes overlapped with the EWS/FLI signature (p = 1.05 × 10−57; Fig. 1A). When the data were segregated into up- and down-regulated gene sets, we found 159 of the NR0B1 down-regulated genes were also down-regulated by EWS/FLI (p = 1.05 × 10−63; Fig. 1B), and that 18 of the NR0B1 up-regulated genes were also up-regulated by EWS/FLI (p = 0.006; Fig. 1C). These data confirm that NR0B1 contributes to EWS/FLI's transcriptional signature, and support a hierarchical relationship where EWS/FLI regulates NR0B1 (among other genes), which then transcriptionally influences other downstream targets. Given the higher level of statistical significance of the overlap between down-regulated genes as compared to up-regulated genes, we speculate that NR0B1 has a more prominent function as a transcriptional repressor in Ewing's sarcoma.
Figure 1.
NR0B1 target genes contribute to the EWS/FLI gene expression signature. A, B, C, Venn diagrams representing the overlap between the overall NR0B1 and EWS/FLI gene expression signatures, and down- and up-regulated genes, respectively. The Chi square-determined p-value is indicated.
Identification of direct NR0B1 targets
To define direct NR0B1 targets, we used our previously described “knockdown/rescue” approach to replace endogenous NR0B1 with a 3x-FLAG-tagged version (8). We next performed genome-wide localization studies, using a chromatin immunoprecipitation (ChIP) approach followed by promoter microarray analysis (ChIP-chip). These microarrays interrogate ~17,000 human promoters, and include regions from −5.5 kb to +2.5 kb relative to the transcriptional start site. We found that 250 genes were directly occupied by NR0B1 (Supplementary Table S3).
Analysis revealed a small but insignificant overlap between the NR0B1 ChIP-chip and NR0B1 microarray datasets. We believe this result may be due to a combination of factors. First, gene expression profiles are comprised of both direct and indirect transcriptional targets. Therefore, it is possible that only a small portion of NR0B1-regulated genes are direct targets. Second, a portion of directly occupied NR0B1 sites may be transcriptionally inert and these are not reflected in the NR0B1 gene signature. Third, the ChIP microarray only evaluates promoter region occupancy. If NR0B1 has many functional binding sites outside of the interrogated region, then the sensitivity of our data overlap analysis is reduced. Lastly, while every effort is taken to identify genes that are statistically significantly different from background levels, the amount of “noise” present in each gene list is unknown. Such noise will reduce the ability to observe statistically significant similarities between datasets. Similar findings were also observed between the EWS/FLI gene expression profile and ChIP-chip datasets (K.G. and S.S. unpublished observations, and ref. 17).
Some genes are bound by both NR0B1 and EWS/FLI
We recently reported that EWS/FLI bound to ~900 genes, including NR0B1 (11). In addition to promoter binding, we also noted strong binding to the NR0B1 intron (K.G. and S.S., unpublished observations). Interestingly, our ChIP-chip analysis demonstrated that NR0B1 bound this same region. Indeed, the pattern of NR0B1 binding mirrored the EWS/FLI binding pattern (Fig. 2A) and directed ChIP assays confirmed these results (Fig. 2B).
Figure 2.
Coordinate occupancy of NR0B1 and EWS/FLI at specific genomic loci. A, Probe enrichment pattern for NR0B1 and EWS/FLI ChIP-chip derived from two separate biological samples per experimental condition. Relative genomic positioning of probes was determined by the Integrated Genome Browser software program (Affymetrix). Probe p-values were determined by the Agilent ChIP Analytics program. NR0B1, KCNN2, and HSPA4L are examples of overlapping probe patterns, while GPR101 represents distinct probe distribution. B, ChIP of the NR0B1 intronic region using the indicated antibodies. Data are plotted as fold enrichment of the intron compared to the enrichment of a negative control gene (ALB). Error bars indicate standard deviation of three independent experiments. Asterisks signify p < 0.05. C, Venn diagram representing the overlap between ChIP-chip identified bound NR0B1 and EWS/FLI gene targets. The Chi square-determined p-value is indicated.
To determine if additional loci demonstrated a similar binding pattern, we identified 20 genes bound by both proteins in the two ChIP-chip datasets (p = 0.0003; Fig. 2C). Evaluation of these dual-bound genes revealed that 90% displayed the same overlapping probe enrichment pattern for both NR0B1 and EWS/FLI (e.g., Fig. 2A). Importantly, our negative control ChIP-chip experiments did not show similar findings, demonstrating that the enrichment pattern is specific to NR0B1 and EWS/FLI (data not shown). Furthermore, a few genes had distinct probe binding patterns for EWS/FLI and NR0B1 suggesting that such similarities were not simply technical artifacts (e.g., Fig. 2A). These data therefore demonstrate that NR0B1 and EWS/FLI occupy the same regions of genomic DNA at a subset of loci.
NR0B1 and EWS/FLI directly interact
NR0B1 is not believed to bind directly to DNA, rather it is thought to predominantly function as a transcriptional co-regulator by interacting with transcription factors to modulate gene expression (12). The identity of these transcription factors in Ewing's sarcoma is unknown. We have demonstrated that both EWS/FLI and NR0B1 are bound to the same genomic loci (Fig. 2C). In addition, recent observations from various proteomic assays suggest that wild-type EWSR1 and NR0B1 may exist in a large protein complex (18, 19). We therefore speculated that NR0B1 and EWS/FLI directly interact. To test this hypothesis we first performed directed yeast two-hybrid assays (Y2H). We fused NR0B1 to the GAL4 DNA binding domain as bait, and EWS/FLI to the GAL4 activation domain as prey. Interactions were assessed through expression of a HIS3 reporter gene placed downstream of GAL4 DNA binding sites. Thus, bait-prey interactions enabled growth of yeast on histidine deficient plates. We found that when the two proteins were co-expressed as bait and prey, the HIS3 reporter was activated, implying a direct protein-protein interaction between NR0B1 and EWS/FLI. Importantly, the HIS3 reporter was also activated when EWS/FLI was used as bait and NR0B1 as prey (Supplementary Table S4).
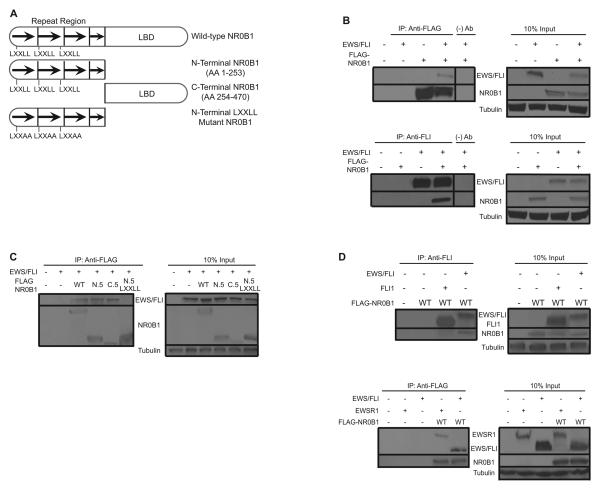
To begin to identify the interacting regions of NR0B1 and EWS/FLI, mutant NR0B1 constructs were generated and tested using the directed Y2H approach (Fig. 3A). We found that both the amino-and carboxyl-termini of NR0B1 independently interacted with EWS/FLI, suggesting that both the amino- and carboxyl-domains have EWS/FLI binding sites (Supplementary Table S4).
Figure 3.
NR0B1 and EWS/FLI directly interact. A, Schematic of wild-type and mutant NR0B1 constructs. NR0B1 consists of an amino-terminal domain (amino acids 1 – 253) that contains 3½ alanine-glycine rich repeating units harboring three LXXLL motifs, and a carboxyl-terminal domain (amino acids 253 – 470) that is homologous to other nuclear hormone receptors ligand binding domains. B, Western blot analysis of input and co-immunoprecipitation samples of 293EBNA cells transfected with wild-type EWS/FLI and NR0B1. Immunoprecipitation experiments and western blots were performed with the indicated antibodies. C, Co-immunoprecipitation of 293EBNA transfected with EWS/FLI and various NR0B1 mutant constructs, using the indicated antibodies. WT indicates the NR0B1 wild-type allele. N.5 and C.5 represent the amino- and carboxy-terminal domains of NR0B1, respectively. N.5 LXXLL indicates amino-terminal NR0B1 with all three LXXLL motifs mutated. All NR0B1 constructs are 3x-FLAG-tagged. Input samples were utilized to ensure appropriate expression of all constructs tested. D, Co-immunoprecipitation of 293EBNA transfected with NR0B1 and EWS/FLI parental proteins EWSR1 or FLI1, using the indicated antibodies.
NR0B1 is known to interact with some of its protein-binding partners (e.g., SF1, ER, AR) through the three LXXLL motifs found in its amino-terminal half (20, 21). We therefore tested whether these contributed to the NR0B1-EWS/FLI interaction. We mutated each LXXLL motif to LXXAA, both singly and in combination, and analyzed these constructs using directed Y2H assays. The results using single and double LXXLL motif mutations were variable and not reproducible. However, when all three LXXLL motifs were mutated, NR0B1 was unable to interact with EWS/FLI (Supplementary Table S4).
To validate the Y2H data using a different experimental approach, we performed co-immunoprecipitation experiments following transfection of EWS/FLI and 3x-FLAG-tagged NR0B1. We found that EWS/FLI co-immunoprecipitated (Fig. 3B) and co-localized (Fig. 4A) with NR0B1. Furthermore, consistent with the Y2H data, both the amino- and carboxyl-termini of NR0B1 also immunoprecipitated EWS/FLI (Fig. 3C). Mutation of the NR0B1 LXXLL motifs again abolished this interaction without affecting its ability to localize to the nucleus (Fig. 3C and Fig. 4B). Introduction of mutations in the carboxyl-terminus of NR0B1 rendered the protein relatively unstable, and thus the EWS/FLI-interacting domain in this region could not be mapped. In addition, co-localization studies indicate the carboxyl-terminal half of NR0B1 resides predominantly in the cytoplasm (Fig. 4B). Therefore, mapping studies using this mutant may not accurately reflect the native interaction between NR0B1 and EWS/FLI.
Figure 4.
NR0B1 and EWS/FLI co-localize to the nucleus. A, Immunofluorescence of EWS502 Ewing's sarcoma cells infected with the indicated cDNA constructs and detected with the indicated antibody. Nuclei are shown by DAPI staining and 293EBNA cells are shown as a negative control. B, C, A673 cells infected or 293EBNA transfected, respectively, with the indicated cDNA constructs and subfractionated. Western blots were performed using the designated antibodies. Tubulin is utilized as a control for the cytoplasmic fraction, while mSin3A is used as a control for the nuclear fraction.
We then repeated our Y2H assays using only the EWS- or FLI-domain of EWS/FLI to begin to identify the NR0B1 interacting site(s) on EWS/FLI. Neither construct activated the HIS3 reporter. These data imply the isolated domains are unable to interact with NR0B1 and suggest both domains are required for the NR0B1-EWS/FLI interaction (Supplementary Table S4).
To determine if the protein interaction is indeed unique to the EWS/FLI fusion product, we performed co-immunoprecipitation experiments with the full-length EWS/FLI parental proteins EWSR1 and FLI1. Although, FLI1 was predominantly nuclear, it did not interact with NR0B1. In contrast, EWSR1 co-localized and co-immunoprecipitated with NR0B1 (Fig. 4C and 3D). The EWSR1 result is unexpected because the Y2H data suggest that both the EWS- and FLI-domain are required for the NR0B1-EWS/FLI interaction. This discordance may be due to improper folding of the isolated portions of the EWS- or FLI-domain mutant constructs or may reflect a lack of necessary cofactors in the yeast system. The EWSR1 data therefore suggest that a NR0B1 binding site is most likely present in the EWS domain of EWS/FLI. Indeed, the EWS amino-terminal domain is believed to lack native structure making the region a highly amenable binding site for a multitude of protein interaction partners (22). Taken together, the co-immunoprecipitation and Y2H experiments demonstrate that EWS/FLI and NR0B1 participate in a protein-protein interaction, most likely mediated through the NR0B1 LXXLL motifs and the EWS domain of EWS/FLI.
NR0B1 and EWS/FLI interact on chromatin
Because NR0B1 and EWS/FLI directly interact and are coordinately present at a subset of promoters, we next sought to determine whether NR0B1 and EWS/FLI interact on chromatin. We performed a sequential ChIP assay with the same NR0B1 “knockdown/rescue” A673 Ewing's cells we utilized for our ChIP-chip and directed ChIP studies. We first precipitated and eluted 3x-FLAG-NR0B1 bound chromatin to isolate genomic loci directly affiliated with NR0B1. Following chromatin resuspension, we performed a second immunoprecipitation with anti-FLI antibody to isolate genomic regions also bound by EWS/FLI. We then assessed occupancy at the NR0B1 intronic region. We chose this region because our ChIP-chip experiments demonstrated NR0B1 and EWS/FLI enriched several probes within the NR0B1 intron and independent directed ChIP assays confirmed binding by both NR0B1 and EWS/FLI (Fig. 2A and 2B). Our sequential ChIP experiments demonstrated enrichment of the NR0B1 intronic region relative to a negative control region (Fig. 5A). Importantly, the intronic region was not enriched when the same technique was applied to A673 cells not expressing the 3x-FLAG-NR0B1 construct, nor was it enriched with control antibodies (Figure 5A). To further validate these results the sequential ChIP experiments were repeated using a different Ewing's sarcoma cell line (TC71) and a similar trend was observed (Fig. 5A). These results, taken together with the Y2H and immunoprecipitation data, support the hypothesis that NR0B1 and EWS/FLI physically interact and concurrently occupy the same genomic region.
Figure 5.
NR0B1 and EWS/FLI interact on chromatin and their co-expression influences transcription. A, Sequential ChIP of NR0B1 and EWS/FLI in A673 and TC71 Ewing's sarcoma cells. Using our “knockdown/rescue” approach, endogenous NR0B1 was first replaced with a 3x-FLAG-tagged-NR0B1 allele. NR0B1 was immunoprecipitated first with anti-FLAG antibody, the isolated NR0B1-associated chromatin mixture was then subject to anti-FLI immunoprecipitation. A673 and TC71 cells (lacking any FLAG construct) and non-specific antibodies were used as controls. Data are plotted as fold enrichment for the NR0B1 intronic region compared to the average enrichment of a negative control gene (ALB). Bars are representative of four of six independent experiments. B, Luciferase assays in 293EBNA co-transfected with the ~700bp NR0B1 intronic response element upstream of a minimal promoter and the indicated cDNA constructs. Relative luciferase activity is the ratio of firefly luciferase activity to Renilla luciferase activity (to control for transfection efficiency). Error bars indicate standard deviations, and the asterisk indicates p < 0.05.
Transcriptional consequences of EWS/FLI and NR0B1 co-expression
Because both EWS/FLI and NR0B1 have transcriptional function, we next sought to determine if there was a transcriptional effect due to the NR0B1-EWS/FLI interaction. We cloned ~700bp of the NR0B1 intronic region upstream of a luciferase reporter construct containing a minimal promoter derived from SV40. The NR0B1 intronic region was chosen for these assays because it was identified as a mutual binding site for NR0B1 and EWS/FLI by our multiple ChIP studies. This construct was co-transfected into 293EBNA cells with NR0B1 and/or EWS/FLI, and luciferase activity determined. We found that the intron was not responsive to EWS/FLI by itself (Fig. 5B). In contrast, luciferase activity was increased ~3 fold with NR0B1 (Fig. 5B). When both proteins were co-expressed, luciferase activity was reduced to basal levels (Fig. 5B). In this setting, then, EWS/FLI inhibits NR0B1-mediated transcriptional activity. Because a full-length triple LXXLL mutant form of NR0B1 did not stimulate activity from this reporter, the contribution of NR0B1's LXXLL motifs to the inhibitory effect of EWS/FLI could not be assessed. These results suggest that at the NR0B1 intron, NR0B1 binds an unidentified transcription factor through its LXXLL motifs to enable transcriptional activation, and that EWS/FLI abrogates this effect through direct interaction with NR0B1.
The EWS/FLI-interacting region of NR0B1 is required for oncogenic transformation
We previously demonstrated that NR0B1 expression is critical to the Ewing's sarcoma transformed phenotype (8). To assess the biological significance of the NR0B1-EWS/FLI protein interaction to oncogenesis we performed “knockdown/rescue” soft-agar colony formation experiments using mutant forms of NR0B1 in two different Ewing's sarcoma cell lines (A673 and TC71). “Knockdown” of endogenous NR0B1 abrogated colony growth and re-expression of NR0B1 fully rescued transformation, as previously reported (8). In contrast, neither amino-terminal nor carboxyl-terminal NR0B1 mutants were capable of rescuing transformation (data not shown). These data suggest that both domains are necessary for NR0B1's function in Ewing's sarcoma. One limitation of this interpretation, however, is that it is dependent on data derived from large structural protein alterations. Indeed, relatively little is known about how the entire amino- or carboxyl- domains of NR0B1 function, rather most work has focused on sub-domains within the protein (e.g., the LXXLL motifs). Therefore, we generated a full-length triple LXXLL NR0B1 mutant allele to test a construct with domain mutations incapable of interacting with EWS/FLI and to minimize the effect of large structural deletions on NR0B1 function. We found this mutant was also unable to rescue transformation (Fig. 6A and 6B). The LXXLL mutant protein had similar expression levels, subcellular localization, and tissue-culture growth patterns as wild-type NR0B1 (data not shown). These data demonstrate that intact LXXLL motifs are required for NR0B1's participation in the oncogenic phenotype of Ewing's sarcoma and suggest our experimental system may be a useful approach towards enhancing our understanding about the function of NR0B1's structural domains in tumorigenesis.
Figure 6.
The NR0B1-EWS/FLI interacting domain is critical for transformation. A, Soft agar colony formation in A673 and TC71 Ewing's sarcoma cells infected with the indicated constructs. NR0B1 LXXLL Mutant cDNA represents the full-length triple LXXLL NR0B1 mutant allele. B, Quantification of colonies formed in soft agar. Error bars indicate standard deviations of duplicate assays.
Conclusions
NR0B1 is an enigmatic protein, and this is particularly true in Ewing's sarcoma, where a role for this protein has only recently been discovered (8, 23). The data in this report provide new understanding of the mechanisms by which NR0B1 functions in cancer. We demonstrated that NR0B1 influences both transcriptional repression and activation during Ewing's sarcoma oncogenesis. We have also shown that NR0B1and EWS/FLI are coordinately present at a subset of promoters and display a direct protein-protein interaction. In addition, we demonstrated that the regions of NR0B1 required for the EWS/FLI interaction are also required for its transcriptional and tumorigenic functions. Taken together, our data suggest that NR0B1 and EWS/FLI physically interact to influence gene expression and mediate the transformed phenotype of Ewing's sarcoma.
EWS/FLI is the principle oncoprotein in Ewing's sarcoma. Most prior data suggest that the fusion functions as a transcriptional activator (24-26). However, recent studies (using RNAi-based approaches) demonstrated that EWS/FLI down-regulates more genes than it up-regulates (4, 8, 9). Some of this down-regulated signature appears to be mediated by EWS/FLI target genes, such as NKX2.2 (10), and as shown in this report, NR0B1.
In contrast to its function as a transcriptional co-repressor, emerging evidence suggests that NR0B1 may also have transcriptional activating functions in some settings (27, 28). Indeed, our own data supports this dual activity of NR0B1. We demonstrated that the NR0B1 transcriptional profile consists of both down-regulated and up-regulated genes, and we found that NR0B1 serves as an activator at the NR0B1 intron. In addition, the inhibition of NR0B1's activating function by interaction with EWS/FLI may be a method for modulating the transcriptional influence of both proteins and may dynamically affect gene target expression at specific loci. For example, the interaction at the NR0B1 intronic region could be a means to fine tune NR0B1 expression levels in Ewing's sarcoma cells.
Our data provide important insights into how the critical orphan nuclear hormone receptor, NR0B1, contributes to Ewing's sarcoma tumorigenesis, and sets the stage for future work focused on understanding the biochemical mechanisms underlying these functions. Such an understanding may allow for the development of antagonists and/or synthetic ligands that modulate NR0B1 activity. Indeed, elucidating the role of other nuclear hormone family members (e.g., estrogen receptor, androgen receptor, and the retinoic acid receptor) in a variety of cancers has lead to the development of more directed and effective therapies (29-31). Such an approach focusing on NR0B1 in Ewing's sarcoma may result in novel therapeutic options for patients affected by this devastating pediatric cancer.
Supplementary Material
Acknowledgements
We gratefully acknowledge D. Nix for technical assistance, D. Ayer, and members of the Lessnick and Bild laboratories for in depth discussions. M.K. was supported by T32-CA093247-07, S.L.L. was supported by the Liddy Shriver Sarcoma Initiative, the Terri Anna Perine Sarcoma Fund, the Sunbeam Foundation, the Department of Pediatrics at University of Utah, and Huntsman Cancer Institute/Huntsman Cancer Foundation. We also acknowledge NIH support to the Huntsman Cancer Institute (P30-CA042014). This work was also supported by United States Public Health Service National Research Service Award GM07104 (A.K.I.) and NIH Grant R01-HD39322 (E.R.B.M.).
References
- 1.Turc-Carel C, Aurias A, Mugneret F, et al. Chromosomes in Ewing's sarcoma. I. An evaluation of 85 cases of remarkable consistency of t(11;22)(q24;q12) Cancer Genet Cytogenet. 1988;32(2):229–38. doi: 10.1016/0165-4608(88)90285-3. [DOI] [PubMed] [Google Scholar]
- 2.Kovar H, Aryee DN, Jug G, et al. EWS/FLI-1 antagonists induce growth inhibition of Ewing tumor cells in vitro. Cell Growth Differ. 1996;7(4):429–37. [PubMed] [Google Scholar]
- 3.Ouchida M, Ohno T, Fujimura Y, Rao VN, Reddy ES. Loss of tumorigenicity of Ewing's sarcoma cells expressing antisense RNA to EWS-fusion transcripts. Oncogene. 1995;11(6):1049–54. [PubMed] [Google Scholar]
- 4.Smith R, Owen LA, Trem DJ, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell. 2006;9(5):405–16. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 5.May WA, Gishizky ML, Lessnick SL, et al. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc Natl Acad Sci U S A. 1993;90(12):5752–6. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.May WA, Lessnick SL, Braun BS, et al. The Ewing's sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol. 1993;13(12):7393–8. doi: 10.1128/mcb.13.12.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lessnick SL, Braun BS, Denny CT, May WA. Multiple domains mediate transformation by the Ewing's sarcoma EWS/FLI- 1 fusion gene. Oncogene. 1995;10(3):423–31. [PubMed] [Google Scholar]
- 8.Kinsey M, Smith R, Lessnick SL. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing's sarcoma. Mol Cancer Res. 2006;4(11):851–9. doi: 10.1158/1541-7786.MCR-06-0090. [DOI] [PubMed] [Google Scholar]
- 9.Prieur A, Tirode F, Cohen P, Delattre O. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol Cell Biol. 2004;24(16):7275–83. doi: 10.1128/MCB.24.16.7275-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Owen LA, Kowalewski AA, Lessnick SL. EWS/FLI mediates transcriptional repression via NKX2.2 during oncogenic transformation in Ewing's sarcoma. PLoS ONE. 2008;3(4):e1965. doi: 10.1371/journal.pone.0001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gangwal K, Sankar S, Hollenhorst PC, et al. Microsatellites as EWS/FLI response elements in Ewing's sarcoma. Proc Natl Acad Sci U S A. 2008;105(29):10149–54. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lalli E, Sassone-Corsi P. DAX-1, an unusual orphan receptor at the crossroads of steroidogenic function and sexual differentiation. Mol Endocrinol. 2003;17(8):1445–53. doi: 10.1210/me.2003-0159. [DOI] [PubMed] [Google Scholar]
- 13.Iyer AK, McCabe ER. Molecular mechanisms of DAX1 action. Mol Genet Metab. 2004;83(12):60–73. doi: 10.1016/j.ymgme.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 14.Iyer AK, Zhang YH, McCabe ER. Dosage-sensitive sex reversal adrenal hypoplasia congenita critical region on the X chromosome, gene 1 (DAX1) (NR0B1) and small heterodimer partner (SHP) (NR0B2) form homodimers individually, as well as DAX1-SHP heterodimers. Mol Endocrinol. 2006;20(10):2326–42. doi: 10.1210/me.2005-0383. [DOI] [PubMed] [Google Scholar]
- 15.Lessnick SL, Dacwag CS, Golub TR. The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell. 2002;1(4):393–401. doi: 10.1016/s1535-6108(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 16.Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89(3):341–7. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 17.Guillon N, Tirode F, Boeva V, Zynovyev A, Barillot E, Delattre O. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS One. 2009;4(3):e4932. doi: 10.1371/journal.pone.0004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun C, Nakatake Y, Akagi T, et al. Dax1 binds to Oct3/4 and inhibits its transcriptional activity in embryonic stem cells. Mol Cell Biol. 2009;29(16):4574–83. doi: 10.1128/MCB.01863-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Rao S, Chu J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444(7117):364–8. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki T, Kasahara M, Yoshioka H, Morohashi K, Umesono K. LXXLL-related motifs in Dax-1 have target specificity for the orphan nuclear receptors Ad4BP/SF-1 and LRH-1. Mol Cell Biol. 2003;23(1):238–49. doi: 10.1128/MCB.23.1.238-249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, Thomsen JS, Johansson L, Gustafsson JA, Treuter E. DAX-1 functions as an LXXLL-containing corepressor for activated estrogen receptors. J Biol Chem. 2000;275(51):39855–9. doi: 10.1074/jbc.C000567200. [DOI] [PubMed] [Google Scholar]
- 22.Lee KA. Ewings family oncoproteins: drunk, disorderly and in search of partners. Cell Res. 2007;17(4):286–8. doi: 10.1038/cr.2007.22. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Aragoncillo E, Carrillo J, Lalli E, et al. DAX1, a direct target of EWS/FLI1 oncoprotein, is a principal regulator of cell-cycle progression in Ewing's tumor cells. Oncogene. 2008;27(46):6034–43. doi: 10.1038/onc.2008.203. [DOI] [PubMed] [Google Scholar]
- 24.Lessnick SL, Braun BS, Denny CT, May WA. Multiple domains mediate transformation by the Ewing's sarcoma EWS/FLI-1 fusion gene. Oncogene. 1995;10(3):423–31. [PubMed] [Google Scholar]
- 25.May WA, Gishizky ML, Lessnick SL, et al. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc Natl Acad Sci U S A. 1993;90(12):5752–6. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.May WA, Lessnick SL, Braun BS, et al. The Ewing's sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol. 1993;13(12):7393–8. doi: 10.1128/mcb.13.12.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–61. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu B, Yang WH, Gerin I, Hu CD, Hammer GD, Koenig RJ. Dax-1 and steroid receptor RNA activator (SRA) function as transcriptional coactivators for steroidogenic factor 1 in steroidogenesis. Mol Cell Biol. 2009;29(7):1719–34. doi: 10.1128/MCB.01010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baselga J, Norton L. Focus on breast cancer. Cancer Cell. 2002;1(4):319–22. doi: 10.1016/s1535-6108(02)00066-1. [DOI] [PubMed] [Google Scholar]
- 30.Isaacs W, De Marzo A, Nelson WG. Focus on prostate cancer. Cancer Cell. 2002;2(2):113–6. doi: 10.1016/s1535-6108(02)00103-4. [DOI] [PubMed] [Google Scholar]
- 31.Parmar S, Tallman MS. Acute promyelocytic leukaemia:a review. Expert Opin Pharmacother. 2003;4(8):1379–92. doi: 10.1517/14656566.4.8.1379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.