Abstract
Cell signaling plays a central role in the etiology of cancer. Numerous therapeutics in use or under development target signaling proteins, however off-target effects often limit assignment of positive clinical response to the intended target. As direct measurements of signaling protein activity are not generally feasible during treatment, there is a need for more powerful methods to determine if therapeutics inhibit their targets and when off-target effects occur.
We have used the Bayesian Decomposition algorithm and data on transcriptional regulation to create a novel methodology, DESIDE (Differential Expression for SIgnaling DEtermination), for inferring signaling activity from microarray measurements. We applied DESIDE to deduce signaling activity in gastrointestinal stromal tumor cell lines treated with the targeted therapeutic imatinib mesylate (Gleevec). We detected the expected reduced activity in the KIT pathway, as well as unexpected changes in the P53 pathway. Pursuing these findings, we have determined that imatinib-induced DNA damage is responsible for the increased activity of P53, identifying a novel off-target activity for this drug.
We then used DESIDE on data from resected, post-imatinib treatment tumor samples and identified a pattern in these tumors similar to that at late time points in the cell lines, and this pattern correlated with initial clinical response. The pattern showed increased activity of ELK1 and STAT3 transcription factors, which are associated with the growth of side population cells.
DESIDE infers the global reprogramming of signaling networks during treatment, permitting treatment modification that leverages ongoing drug development efforts, which is crucial for personalized medicine.
Introduction
Our present understanding of cancer has demonstrated the importance of cell signaling processes in etiology and treatment response. As signaling proteins, including those with gain- and loss-of-function mutations, provide a logical target for therapeutic intervention, numerous compounds targeting signaling proteins are under development (1). The success of imatinib mesylate (IM) in treating chronic myelogenous leukemia has greatly increased the hope for targeted therapy and spurred further development of BCR-ABL kinase inhibitors (2), however the varying specificity of the therapeutics and the potential for reprogramming of cell signaling in response to treatment make the discovery of predictive biomarkers essential for drug development and patient treatment.
Gastrointestinal stromal tumors (GIST) are mesenchymal digestive tract tumors (3). The most common primary sites are the stomach (60-70%) (4), small intestine (25-35%) (5), and colon and rectum (10%) (6). These tumors contain smooth muscle and neural elements and are thought to arise from the interstitial cells of Cajal (7, 8). GISTs are clinically diagnosed by immunohistochemical KIT staining by the CD117 antibody. KIT is a receptor tyrosine kinase (RTK) that can initiate RAS and PI3K signaling. The majority (~70%) of GISTs possess gain-of-function mutations in KIT, while ~10% possess gain-of-function mutations in the RTK PDGFRA, with the rest having no such mutations (wild-type) (9, 10). The primary treatment for GIST is surgical resection, however high-risk GIST has a high incidence of recurrence (11). Since 2002, IM has become a standard treatment for patients with metastatic and/or unresectable GIST, with objective responses or stable disease obtained in > 80% of patients (12, 13).
The expected specific targeting of IM to KIT in GIST makes it an ideal system for developing methods to understand therapeutics that target cell signaling processes. The desired effect of a signaling inhibitor, such as IM, is the loss of propagation of an aberrant signal through a pathway. The logical method to measure the effect of this inhibitor is to look for changes in protein post-translational modifications upon treatment, since most signal propagation involves protein phosphorylation. The ability to make these measurements is presently limited in vivo during treatment. In addition, such measurements target specific proteins, thus losing the ability to identify off-target effects and unexpected activation of other signaling processes.
An alternative approach is to use a mature technology capable of global measurements, such as microarrays, to obtain a full picture of the cellular response. Since transcriptional changes resulting from activation or suppression of transcriptional regulators are primary endpoints for many signaling processes, microarrays provide a potential tool for exploring signaling globally. There are three primary impediments to this approach. First, the “wiring diagrams” for cell signaling processes are not fully determined and vary between cell types. Second, the transcriptional response to a signal is convoluted with the responses to other biological drivers of transcription, with most if not all genes under the control of multiple transcriptional regulators. Third, a good statistic for measuring the probability of activity of a regulator (or transcription factor, TF) that accounts for the multiple regulation of genes has not been developed.
Although the wiring diagrams defining cell signaling relationships (protein-protein interactions) are far from complete, several core pathways affecting disease have been detailed, especially in cancer studies (1, 14). The determination that these pathways play critical roles in embryogenesis has led to a substantial knowledge-base for understanding basic signaling (15), and these can be specialized to cells of interest by manual review of the literature (16). In this way a core signaling network can be created for a system of interest, with the critical pathways linked to TFs.
There has been intensive development of analysis methods to address multiple regulation. Our own work on Bayesian Decomposition (BD) for spectroscopic imaging (17) and later for microarrays (18), was followed by non-negative matrix factorization (NMF) for imaging (19) and later for microarrays (20, 21). Similar approaches were developed in statistics (22) and machine learning (23). These methods provide a set of tools for isolating transcriptional responses related to individual TFs, or more accurately, to sets of TFs that are active in the same samples in the data, since these approaches are data driven.
Statistical methods have been developed to look at enhancement of gene sets in microarray data. Initially, hypergeometric tests were used to determine if a gene set (e.g., a gene ontology category) was overrepresented in a sample (24). The development of gene set enrichment analysis (25) allowed the overall ranks of genes within a set to play a role in the statistic. However, these methods do not handle multiple regulation or inclusion of uncertainty. As genes vary strongly in the degree of transcriptional regulation, there are order of magnitude differences in the variances that transcript levels show related to phenotypic response (26). By including uncertainty levels, this issue can be addressed, as we demonstrate in this work.
We present here the results of the novel technique, DESIDE, for identifying changes in signaling activity during intervention with targeted therapeutics. Other modeling approaches to signaling are in use. These include ordinary differential equation (ODE) models (27) and reconstruction of small networks from protein measurements (28) or prior data (29). In contrast, we wish to estimate changes in signaling on a larger network and confine our method by assuming the connectivity is known, while minimizing the need for a large number of biochemical constants. In addition, by focusing on downstream effectors of pathways and genome-wide microarray measurements, we can discover novel signaling activity that is not initially in the model and is beyond the capability of ODE and proteomics-based methods to discover due to limited coverage. Recent developments in generation of mRNA from frozen core needle biopsies also suggests that routine clinical pathology can provide our method with the data needed to move into clinical practice (30).
Materials and Methods
GIST-T1 Cell Culture and Microarray Hybridization
GIST-T1 cells (31) were grown in triplicate cultures and treated with 10 μM IM. Cells were harvested at nine time points during treatment (details in supplemental file 1), and total RNA was isolated from the samples using TRIzol reagent (Invitrogen). Microarrays were processed using standard Agilent protocols and using RNeasy from Qiagen.
Agilent Microarray Scanning and Data Preprocessing
Microarrays were scanned using an Agilent G2505B Scanner, and Agilent summary files were preprocessed with R/Bioconductor (version 2.4.1) using the limma package (version 2.9.13) (32). Standard limma normalization procedures were performed, providing relative transcript levels for 44,000 probes across 26 conditions, comprising triplicate measurements at all time points except 24 hr, where only duplicate measurements were made.
Probes were assigned to Unigene clusters (33), using the ASAP annotation tool (34). At each time point, we combined all probes across the replicate arrays measuring the same Unigene gene, obtaining mean values and standard deviations for 20,656 genes. We annotated all genes using TRANSFAC (35) version 2008.4 based on physical evidence of regulation, providing 1363 genes linked to TFs. Raw data is provided in the Gene Expression Omnibus (GSE17018), and the processed data used for analysis is provided in supplementary file 2.
Estimation of Signaling Activity
We estimated signaling activity by use of BD (see (36) for a statistical or (18) for a biology-focused description of the algorithm), production of a p-value from a Z-score for each TF, and linking of signaling pathway activity to TF activity.
We applied BD to identify transcriptional signatures within the data. Our data matrix, D, comprised 1363 rows (genes) by 9 columns (time-points) and was factored into two lower dimensional matrices, a P matrix whose rows provide P patterns and an A matrix that apportions the behavior of the genes to these patterns. Mathematically, BD performs a decomposition using Markov chain Monte Carlo (37) as
| (1) |
where the genes are indexed by i, the conditions by j, and the patterns by p. As for PCA (38), issues remain in choosing P, however we used our previously described approach to identify five patterns as suitable (39). We performed three separate runs of BD to address variability and potential lack of convergence criteria.
For estimating TF activity, we introduced a Z-score based statistic that, unlike a hypergeometric test, is independent of threshold and utilizes variance measures.. For each transcriptional regulator, we defined the gene set Gt with gi ∈Gt if and only if gi is regulated by the TF t. We then obtain a statistic for the TF with R known targets,
| (2) |
where r indexes the target gene, t the TF, p the pattern, A is the mean in the A matrix, and σ the uncertainty on the mean. This provides a statistic for the overrepresentation of target genes of a transcriptional regulator in a pattern, which we converted to a p-value through random sample tests for each transcriptional regulator with more than five known targets. This provided probability estimates of regulatory activity for 230 regulators in each pattern.
To link the probability of TF activity to signaling, we created a simplified model of KIT signaling in GIST, including IGF1R based on its role in GIST (40, 41). As multiple regulators lie downstream in these pathways, we interpreted the results across the three varying patterns by visualizing p-values. For display, we rescaled the values such that p < 0.5 → 0 – 1 for overrepresentation and p < 0.5 → −1 – 0 for underrepresentation of TF targets.
COMET Assays, qRT-PCR, and Western Blots
For the COMET assay, GIST-T1 cells were treated with either 10 μM of IM for 48 hours, 4 Gy of gamma-irradiation followed by a 2 hour recovery, or with IM followed by irradiation. Standard COMET assay protocols were followed to obtain CometSlides (Trevigen). The DNA was stained with SYBR fluorescent dye and visualized with an immunofluorescent microscope. At least 75 comets were counted for each treatment, and at least three independent comets assays were carried out. For qRT-PCR, RNA was isolated from GIST-T1 cell lines at six time points, reverse transcribed to cDNA, and measured by real-time PCR for three target genes and the endogenous control gene, HPRT. All data were normalized to HPRT expression. For immunoblotting, whole cell extracts were prepared and the proteins evaluated as previously described (42).
Tumor Samples and Analysis
63 patients with primary or recurrent operable GIST were enrolled onto the Radiation Therapy Oncology Group (RTOG) 0132 trial from 18 institutions (43). All patients signed informed consent following IRB approval for this study. Tumor samples were obtained from the surgical specimens obtained at the time of resection following neoadjuvant/preoperative IM. Fresh-frozen samples were collected from all participating institutions and shipped to the RTOG tissue bank prior to evaluation. Total RNA was isolated from frozen tissue samples using TRIzol reagent according to standard protocols. Due to patients leaving the study and poor RNA quality, only 22 samples were analyzed by microarray with 21 having initial tumor response measurements.
BD analysis was performed on the microarray data from the 22 samples. Preprocessing as described above for the cell line data was used to obtain data for the same 1363 genes. Using the methodology described above, we estimated 11 dimensions to fully describe the data. Analysis of the patterns revealed three with significant correlation with volume of initial tumor shrinkage.. Significant correlation was defined as greater than one standard deviation away from mean correlation with response across all 11 patterns (mean = -1.4 × 10-3, standard deviation = 0.28). To compare to clinical response, logistic regression was performed on these patterns in terms of initial clinical response, as defined previously in (44). These patterns were compared for transcription factor activity estimates with patterns identified in the cell line data. Raw data are provided in the Gene Expression Omnibus (GSE15966), and the processed data used for analysis is provided in supplementary file 3.
Results
Five transcriptional patterns capture all time-dependent behavior in imatinib treated GIST-T1 cells
We created a time series cell line data set for analysis using the normalized Agilent data for the IM treatment of GIST-T1 cells. Probes were combined by Unigene ID, and we retained genes with a known TF in TRANSFAC Professional 2008.4. We analyzed this data of transcript levels for 1363 genes across 9 time points using BD, positing three to seven patterns. We chose five patterns based on consistency, using methods developed previously (45). We then applied BD three times to this data generating 500,000 MCMC samples each time, obtaining normalized χ2 fits of 19246, 19267, and 19272.
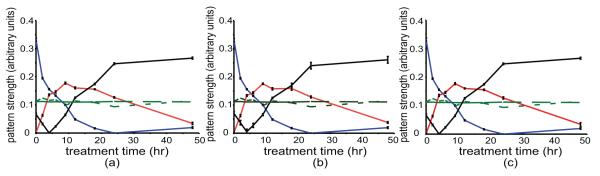
The mean values for each of the three applications of BD for the five patterns are pictured in Figure 1. Error bars represent the standard deviation from the Markov chain sampling for the patterns varying during IM treatment. Two patterns are basically constant across the time points. The other three patterns varied with treatment: pattern 1) falling with IM treatment and reaching a low at 24 hours, pattern 2) transiently rising with a peak between 9 and 18 hours, and pattern 3) rising continuously after 6 hours. For each pattern, there is a strength of assignment of each gene, together with a standard deviation on that strength. Although we do not focus on behavior of individual genes, we validated our microarray measurements by qRT-PCR measurements at six time points on three well-studied targets of the TFs of interest (CDC25A, JAK3, and SOCS3) in IM-treated GIST-T1 cell culture, verifying that the arrays were correctly estimating relative expression levels (supplementary file 4).
Figure 1.
The five patterns identified by BD in the cell line data. The curves show the rows of the P matrix for five patterns, for each of the three analyses, a) χ2 = 19267, b) χ2 = 19246, c) χ2 = 19272. Error bars (three varying patterns) represent standard deviations from the Markov chain sampling. The patterns that vary significantly with IM treatment show a decline with treatment, a transient peak at ~9 - 18 hr, and an increase with treatment beginning at ~6 hr.
Statistical analysis identifies significant changes in transcription factor activities
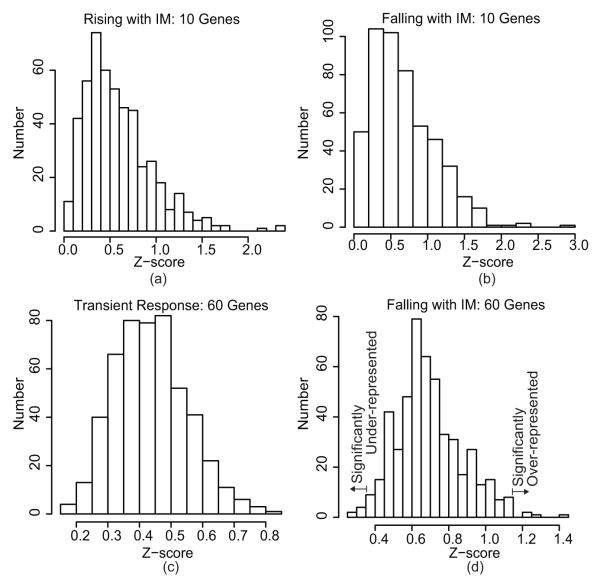
In order to infer the activity of transcriptional regulators, we generated a Z-score for each regulator in TRANSFAC that had more than five known targets (230 transcriptional regulators). We modified the TRANSFAC list for two TFs of special interest in our network model. For the FOXO family we combined all members, since there is strong overlap in potential targets (46). For CREB1, we combined the lists for CREB and CREB1, as these refer to the same factor. First, a Z-score for each gene in each pattern was generated. Then, the Z-score for a regulator was defined as the average Z-score of all targets of the regulator (Equation 2), and statistical significance was determined by generating Z-scores for random sets of genes from the associated pattern. We performed 500 random draws, thus permutation testing sets a limit of p ≥ 0.002. Since the random samples provide a limit for both chance overrepresentation and underrepresentation of targets, comparison of the Z-score to the distribution from random draws provided an indication of increased or decreased activity (Figure 2).
Figure 2.
The permutations for Z-score significance testing, produced by random draws from each pattern showing variation with IM. The top panel (a - b) provides sample histograms of the Z-score for random draws of 10 genes, and the bottom panel (c - d) provides those for 50 genes. Panel (d) shows where significant Z-scores would fall.
The results for TFs rated significantly greater than expected by this procedure in two of the three BD analyses for a threshold α = 0.05 are presented in Table 1. The full list for each of the three runs is presented in supplementary file 5. We discuss the link to the signaling network in detail in the next section, however we note that a number of key transcription factors related to signaling processes affected in cancer are present. ELK1, the downstream effector of the MAPK pathway, shows an initial decline and then recovery. The oncogene MYC appears in the pattern declining with IM, and it is not significantly represented in the pattern that rises later. The DNA damage response regulator P53 is the strongest detection in the transient response, although it also shows up in the pattern that declines initially with IM treatment. Although not included initially due to having too few known targets, we analyzed P63 and P73 by the same method. Both were significantly upregulated in the transient pattern, but only P63 was significantly upregulated in the decreasing pattern.
Table 1.
The transcription factors with significant activity in the three patterns. Complete details for all 230 transcriptional regulators are presented in supplemental file 5.
| Falling with IM | Transient Rise with IM | Rising with IM |
|---|---|---|
| Elk-1 Elk-1-isoform1 SRF PEA3 RPB-Jκ SRF-L c-Myc Egr-1 NF-E2p45 Clock-BMAL1 Smad3 c-Fos:c-Jun E2F:DP p53-isoform1 HSF-1 Nrf-2 c-Jun HIF-1 ATF2 |
p53 p53-isoform1 PEA3 Erm c-Fos:c-Jun δCREB ATF3 FOSB NF-YA NF-Y E2F:DP NF-YB Smad3 Sp3 GKLF GLI HES-1 Egr-2 |
Elk-1 RPB-Jκ SRF-L HIF-1α STAT1α NERF-1a ATF-2 RelA:p65 Egr-2 NFκB SRF ELF-1 |
The transcriptional effectors of the signaling network respond initially as expected to imatinib treatment
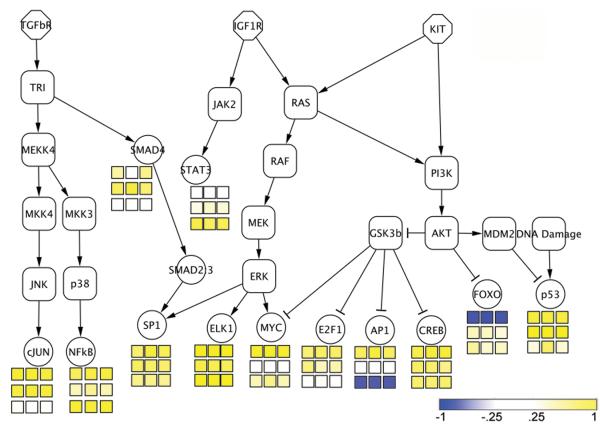
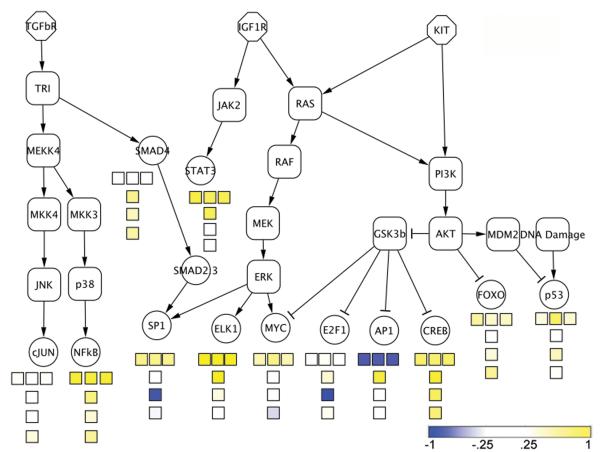
We estimate activity of signaling pathways by looking at the overall changes of known downstream effectors of pathway activity, as in Figure 3, produced with Cytoscape (47). In the square boxes in this figure, yellow represents TFs with a p-value close to 0 when testing for overrepresentation of targets in the pattern, while blue represents TFs with a p-value close to 0 when testing for underrepresentation of targets in the pattern. We have rescaled the values such that p < 0.5 → 0 – 1 for overrepresentation and p < 0.5 → —1 – 0 for underrepresentation. For each of the three patterns varying with IM treatment, we show the values obtained from each of the three BD analyses. The top three squares under each regulator are for pattern 1: declining with IM treatment, the middle three are for pattern 2: rising transiently with IM treatment, and the bottom three are for pattern 3: increasing with IM treatment.
Figure 3.
Transcription factor (TF) responses. The square boxes provide a color representation of activity of the TF, with deeper yellow indicating high activity and deeper blue lack of activity and potential suppression. The top boxes are for the pattern declining during IM treatment, the middle boxes are for the transient pattern peaking at ~9 - 18 hours, and the bottom boxes are for the pattern rising with treatment. The first column is associated with Figure 1(a), the second with 1(b), and the third with 1(c).
In pattern 1, which declines upon treatment with IM (top box), the effectors of RAS-RAF signaling, ELK1 and MYC, show high activity. FOXO, the directly repressed target of AKT1, shows low activity, while the targets effectively upregulated by AKT1 (through repression of the repressor GSK3B), E2F1, AP-1, and CREB1, show high activity. Taken as a whole, this is consistent with identifying pattern 1 as associated with active KIT signaling. Since pattern 1 declines during IM treatment, this verifies that the downstream effectors of constituitively activated KIT show declining activity with IM treatment, as expected.
The p53 transcription factor is highly active at 9 - 18 hours and is responding to unexpected DNA damage
In pattern 2, the transient response pattern (middle box), P53 shows a strong signal for activity. The increased activity in P53 suggested a DNA damage response, and we predicted therefore that IM unexpectedly damages DNA.
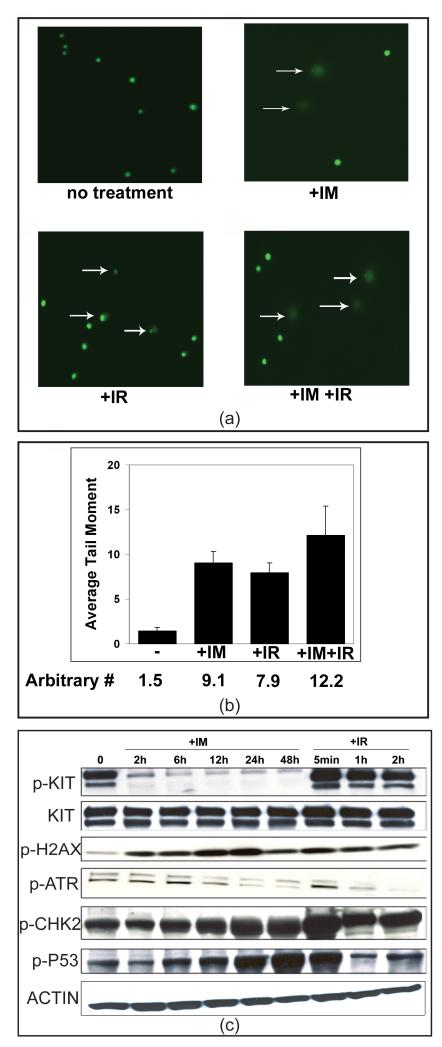
In order to validate our prediction that IM treatment of GIST-T1 cells leads to DNA damage, we used COMET assays on GIST-T1 cells treated with control, IM, radiation, and IM together with radiation, as shown in Figure 4a&b. The radiation is known to induce DNA damage, so it functions as a positive control. The IM treatment causes a somewhat larger though statistically similar amount of damage as the radiation treatment, and there is some indication of increased damage if both IM and radiation are used together, suggesting possible sensitization of the cells by IM. It is clear that IM induces DNA damage, as predicted by our analysis from the microarray data.
Figure 4.
The results of IM treatment on DNA damage and repair pathways response in GIST cells. Panel (a) shows a COMET assay to assess DNA damage to GIST-T1 cells untreated, treated with 10 μM IM for 48 hr, treated with 4 Gy of gamma radiation followed by recovery, or the combination of both. The data across replicate measurements is summarized in (b) and confirms the prediction based on P53 activity that DNA is damaged during IM treatment. To determine whether DNA damage repair pathways are activated in response to IM-induced DNA damage, immunoblotting of various pathway components was performed after either IM treatment or IR exposure followed by recovery (positive control) for the times indicated (c).
To verify that P53 response was generated via DNA damage, we performed immunoblotting of p-H2AX, p-ATR, p-CHK2, and p-P53 together with KIT and p-KIT. As shown in Figure 4c, the DNA damage response proteins rise following IM treatment as p-KIT declines, with p-ATR peaking at 6 hr, immediately before our measured P53 transcriptional response. Other components remain phosphorylated, although the transcriptional response declines.
The GIST-T1 cell line shows increased activity of ELK1 and STAT3 within 24 hours of imatinib treatment
In pattern 3, which increases after ~6 hr (bottom box), we see a return to high activity in ELK1, though not in MYC nor in coordinated changes in targets of AKT1 signaling. In addition, STAT3 shows increased activity. Interestingly, activity of MEK (and thus ELK1) and STAT3, coupled to mTOR (FRAP) activation, which cannot be determined by this technique, have been identified as hallmarks of side population cells (48), which are considered highly enriched for stem cells. To verify the activation, we performed immunoblotting of p-ELK1 and p-STAT3, which showed increased phosphorylation beginning at 6 – 12 hours (see supplemental file 6).
The strength of a pattern in tumors similar to the late time point pattern in GIST-T1 cells correlates with initial tumor response
Using BD and positing 11 patterns based on consistency, we analyzed the data from resected IM-treated patient tumors collected in the RTOG 0132 trial. Three patterns showed significant correlation with initial tumor shrinkage, defined as at least one standard deviation of correlation away from the mean for all 11 patterns. In order to determine clinical significance, we used logistic regression in terms of clinical initial response as defined in (44). In Figure 5, the results for these three patterns are shown together with pattern 3 from the cell line data (i.e., increasing with IM treatment, rising with IM line in Figure 1, bottom row in Figure 3).
Figure 5.
The transcription factor responses for tumors compared to the increasing pattern in the cell line. The three boxes on top give the activity estimates for the cell line, while the individual boxes give the estimates for three of the patterns in tumors. The top box has a pattern similar to that of the cell line, except for SP-1 activity. This pattern correlates with initial clinical response with p = 0.06. The second and third pattern do not correspond to cell line patterns, and are anticorrelated and correlated respectively with initial tumor response.
The first pattern from the tumor samples is positively correlated with initial response (r = 0.34, logistic p = 0.06) and is highly similar to the pattern from the cell line data. The main differences are the high activity of AP-1 in the tumors compared to the cell line, and slightly weaker activity in SP1 and MYC. Importantly this pattern shows high activity in ELK1 and STAT3, as would be expected if side population cells were forming a significant percentage of harvested cells.
Other patterns for tumors showing correlation with initial response are not similar to cell line patterns
The second pattern from the tumors shows negative correlation (r = −0.36, logistic p-value = 0.17) with initial response, while the third pattern shows positive correlation (r = 0.52, logistic p-value = 0.09). Neither shows strong consistency with any cell line pattern. The second pattern shows significantly lower activity in SP1 and E2F1, with no heightened activity from ELK1 or STAT3. The third pattern shows little activity of any of the transcriptional effectors of the network, and it likely represents a behavior unrelated to these signaling processes. Plots of pattern strength against clinical response are shown in supplemental file 7.
Discussion
Cell signaling processes have been implicated in human disease etiology, and signaling proteins and their interactions have become targets of therapeutic development. Presently, there are hundreds of compounds in development, and high-throughput searches for inhibitors of kinases, which often function in signaling pathways, are being undertaken (49). In vivo these compounds often hit multiple targets, and sometimes do not affect the intended target, either due to mutations in those targets or delivery and competitive binding issues. This makes development of methods to identify targets of new molecular medicines essential for drug development, personalized treatment, and development of combination therapies that can hit multiple pathways simultaneously.
We have demonstrated here the ability to use Bayesian Decomposition coupled to a novel metric of enrichment of known transcriptional targets to infer pathway activity that drives changes in the activity levels of transcriptional regulators. As these are often the primary effectors of signaling processes, this approach allows us to infer when inhibitors have had the desired effect, turning off a pathway. With this approach, we have shown that imatinib mesylate (IM) downregulates constituitively activated KIT in GIST-T1 cells, including its downstream transcriptional effectors. Importantly, since we have used a global measure of transcriptional response, we have also identified unexpected changes in signaling, including increased activity of P53 after 9 - 18 hr of IM treatment. We predicted that this increased activity was due to unexpected DNA damage from IM, and we confirmed this by comet assay and immunoblotting.
In the late time points, increased activity of STAT3 and ELK1 appears, two hallmarks of side population cells enriched for cancer stem cells. The third hallmark, upregulation of mTOR, cannot be detected by our method. The MYC transcription factor does not show a return to high activity at late time points, which may be due to continued suppression by the ongoing IM treatment. We also analyzed resected tumors from patients receiving presurgical IM treatment to reduce tumor volume. We identified within these patients a pattern that is similar to this cell line pattern and that correlates with initial clinical response during IM treatment. Although the analysis did not reach standard statistical significance, this likely reflects the very small sample size of post surgical specimens. Importantly the pattern includes increased activity of STAT3 and ELK1. This suggests that tumors may respond well to initial treatment, but that a reserve population of stem-like cells may then become active. This could explain the lack of correlation between initial tumor response and long-term progression free survival (PFS) observed in RTOG 0132. We identified additional patterns that showed correlation with initial tumor shrinkage though poorer correlation with initial clinical response, including one pattern that showed very little increase or decrease in transcription factor activity related to the core signaling pathways.
The lack of an ability to determine mTOR activity here highlights the desirability of having multiple types of measurements available in systems under study. While we could look for side population cells using targeted proteomics, that would only give a partial picture. Measurements on the transcriptional regulators through their targets by microarrays can provide information on whether signals have been modified downstream of the protein whose state is measured directly. Overall, this suggests an ideal study might be a mixture of targeted experiments to test specific hypotheses and global measurements to identify unexpected behaviors.
While we have used information from TRANSFAC to drive our statistical analysis, this approach is also promising for inferring regulation that is not yet identified in TRANSFAC or which contradicts such information. As transcriptional regulation of any gene is likely to be context specific (e.g., cell line, present state of a cell, etc.) (50), we can use inconsistencies in prior information from TRANSFAC to refine our knowledge of transcriptional regulation in specific cases. This should lead directly to an ability to tailor treatments to specific individuals based on the signaling activity in individual tumors.
Supplementary Material
Acknowledgements
Grant support: NLM LM009382 (mfo), Maryland CRF (mfo), NCI Hopkins CCSG (mfo), NCI CA106588 (akg), NCI U10-RTOG supplement CA21661 (akg), NIH Training Grant CA009035 (lr).
We thank the JH Kimmel microarray core for assistance, and the FCCC Biosample Repository Core and Clinical Molecular Genetics Laboratory for support of the tissue studies. We thank Robert Clarke of Georgetown University for helpful suggestions on the manuscript and the anonymous RTOG reviewers for helpful comments. We acknowledge the support of Ms. Tania Stutman and the GIST Cancer Research Fund for a fellowship (lr).
Footnotes
Bibliography
- 1.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 2.O’Hare T, Eide CA, Deininger MW. New Bcr-Abl inhibitors in chronic myeloid leukemia: keeping resistance in check. Expert Opin Investig Drugs. 2008;17:865–78. doi: 10.1517/13543784.17.6.865. [DOI] [PubMed] [Google Scholar]
- 3.Corless CL, Heinrich MC. Molecular pathobiology of gastrointestinal stromal sarcomas. Annu Rev Pathol. 2008;3:557–86. doi: 10.1146/annurev.pathmechdis.3.121806.151538. [DOI] [PubMed] [Google Scholar]
- 4.Miettinen M, Lasota J. Gastrointestinal stromal tumors: review on morphology, molecular pathology, prognosis, and differential diagnosis. Arch Pathol Lab Med. 2006;130:1466–78. doi: 10.5858/2006-130-1466-GSTROM. [DOI] [PubMed] [Google Scholar]
- 5.Miettinen M, Kopczynski J, Makhlouf HR, et al. Gastrointestinal stromal tumors, intramural leiomyomas, and leiomyosarcomas in the duodenum: a clinicopathologic, immunohistochemical, and molecular genetic study of 167 cases. Am J Surg Pathol. 2003;27:625–41. doi: 10.1097/00000478-200305000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Tworek JA, Goldblum JR, Weiss SW, Greenson JK, Appelman HD. Stromal tumors of the anorectum: a clinicopathologic study of 22 cases. Am J Surg Pathol. 1999;23:946–54. doi: 10.1097/00000478-199908000-00013. [DOI] [PubMed] [Google Scholar]
- 7.van Roggen JF Graadt, van Velthuysen ML, Hogendoorn PC. The histopathological differential diagnosis of gastrointestinal stromal tumours. J Clin Pathol. 2001;54:96–102. doi: 10.1136/jcp.54.2.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazur MT, Clark HB. Gastric stromal tumors. Reappraisal of histogenesis. Am J Surg Pathol. 1983;7:507–19. doi: 10.1097/00000478-198309000-00001. [DOI] [PubMed] [Google Scholar]
- 9.Debiec-Rychter M, Sciot R, Le Cesne A, et al. KIT mutations and dose selection for imatinib in patients with advanced gastrointestinal stromal tumours. Eur J Cancer. 2006;42:1093–103. doi: 10.1016/j.ejca.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 10.Tarn C, Merkel E, Canutescu AA, et al. Analysis of KIT mutations in sporadic and familial gastrointestinal stromal tumors: therapeutic implications through protein modeling. Clin Cancer Res. 2005;11:3668–77. doi: 10.1158/1078-0432.CCR-04-2515. [DOI] [PubMed] [Google Scholar]
- 11.Eisenberg BL, Judson I. Surgery and imatinib in the management of GIST: emerging approaches to adjuvant and neoadjuvant therapy. Ann Surg Oncol. 2004;11:465–75. doi: 10.1245/ASO.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. The New England journal of medicine. 2002;347:472–80. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 13.Verweij J, Casali PG, Zalcberg J, et al. Progression-free survival in gastrointestinal stromal tumours with high-dose imatinib: randomised trial. Lancet. 2004;364:1127–34. doi: 10.1016/S0140-6736(04)17098-0. [DOI] [PubMed] [Google Scholar]
- 14.Yeh JJ, Der CJ. Targeting signal transduction in pancreatic cancer treatment. Expert Opin Ther Targets. 2007;11:673–94. doi: 10.1517/14728222.11.5.673. [DOI] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–6. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W, Bagaitkar J, Watabe K. Roles of AKT signal in breast cancer. Front Biosci. 2007;12:4011–9. doi: 10.2741/2367. [DOI] [PubMed] [Google Scholar]
- 17.Ochs MF, Stoyanova RS, Arias-Mendoza F, Brown TR. A new method for spectral decomposition using a bilinear Bayesian approach. J Magn Reson. 1999;137:161–76. doi: 10.1006/jmre.1998.1639. [DOI] [PubMed] [Google Scholar]
- 18.Moloshok TD, Klevecz RR, Grant JD, Manion FJ, Speier WFt, Ochs MF. Application of Bayesian Decomposition for analysing microarray data. Bioinformatics. 2002;18:566–75. doi: 10.1093/bioinformatics/18.4.566. [DOI] [PubMed] [Google Scholar]
- 19.Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–91. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- 20.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101:4164–9. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim PM, Tidor B. Subsystem identification through dimensionality reduction of large-scale gene expression data. Genome Res. 2003;13:1706–18. doi: 10.1101/gr.903503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West M. Bayesian factor regression models in the “large p, small n” paradigm. In: Bernardo JM, Bayarri MJ, Berger JO, Dawid AP, editors. Bayesian Statistics 7. Oxford University Press; Oxford: 2003. [Google Scholar]
- 23.Liao JC, Boscolo R, Yang YL, Tran LM, Sabatti C, Roychowdhury VP. Network component analysis: reconstruction of regulatory signals in biological systems. Proc Natl Acad Sci U S A. 2003;100:15522–7. doi: 10.1073/pnas.2136632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Draghici S, Khatri P, Bhavsar P, Shah A, Krawetz SA, Tainsky MA. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 2003;31:3775–81. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes JD, Estep PW, Tavazoie S, Church GM. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol. 2000;296:1205–14. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- 27.Birtwistle MR, Hatakeyama M, Yumoto N, Ogunnaike BA, Hoek JB, Kholodenko BN. Ligand-dependent responses of the ErbB signaling network: experimental and modeling analyses. Molecular systems biology. 2007;3:144. doi: 10.1038/msb4100188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sachs K, Itani S, Carlisle J, Nolan GP, Pe’er D, Lauffenburger DA. Learning signaling network structures with sparsely distributed data. J Comput Biol. 2009;16:201–12. doi: 10.1089/cmb.2008.07TT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee S, Speed TP. Network inference using informative priors. Proc Natl Acad Sci U S A. 2008;105:14313–8. doi: 10.1073/pnas.0802272105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tebbit C, Zhai J, Untch B, et al. Novel tumor sampling strategies to enable microarray gene expression signatures in breast cancer: a study to determine feasibility and reproducibility in the context of clinical care. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-008-0301-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taguchi T, Sonobe H, Toyonaga S, et al. Conventional and molecular cytogenetic characterization of a new human cell line, GIST-T1, established from gastrointestinal stromal tumor. Lab Invest. 2002;82:663–5. doi: 10.1038/labinvest.3780461. [DOI] [PubMed] [Google Scholar]
- 32.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wheeler DL, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kossenkov A, Manion FJ, Korotkov E, Moloshok TD, Ochs MF. ASAP: automated sequence annotation pipeline for web-based updating of sequence information with a local dynamic database. Bioinformatics. 2003;19:675–6. doi: 10.1093/bioinformatics/btg056. [DOI] [PubMed] [Google Scholar]
- 35.Matys V, Kel-Margoulis OV, Fricke E, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–10. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ochs MF. Bayesian Decomposition. In: Parmigiani G, Garrett E, Irizarry R, Zeger S, editors. The Analysis of Gene Expression Data: Methods and Software. Springer Verlag; New York: 2003. [Google Scholar]
- 37.Gilks WR, Richardson S, Spiegelhalter DJ. Markov chain Monte Carlo in practice. Chapman & Hall; London: 1996. [Google Scholar]
- 38.Cangelosi R, Goriely A. Component retention in principal component analysis with application to cDNA microarray data. Biol Direct. 2007;2:2. doi: 10.1186/1745-6150-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bidaut G, Manion FJ, Garcia C, Ochs MF. WaveRead: automatic measurement of relative gene expression levels from microarrays using wavelet analysis. J Biomed Inform. 2006;39:379–88. doi: 10.1016/j.jbi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Braconi C, Bracci R, Bearzi I, et al. Insulin-like growth factor (IGF) 1 and 2 help to predict disease outcome in GIST patients. Ann Oncol. 2008;19:1293–8. doi: 10.1093/annonc/mdn040. [DOI] [PubMed] [Google Scholar]
- 41.Tarn C, Rink L, Merkel E, et al. Insulin-like growth factor 1 receptor is a potential therapeutic target for gastrointestinal stromal tumors. Proc Natl Acad Sci U S A. 2008;105:8387–92. doi: 10.1073/pnas.0803383105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarn C, Godwin AK. The molecular pathogenesis of gastrointestinal stromal tumors. Clinical colorectal cancer. 2006;6(Suppl 1):S7–17. doi: 10.3816/ccc.2006.s.002. [DOI] [PubMed] [Google Scholar]
- 43.Eisenberg BL, Harris J, Blanke CD, et al. Phase II trial of neoadjuvant/adjuvant imatinib mesylate (IM) for advanced primary and metastatic/recurrent operable gastrointestinal stromal tumor (GIST): early results of RTOG 0132/ACRIN 6665. J Surg Oncol. 2009;99:42–7. doi: 10.1002/jso.21160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rink L, Skorobogatko Y, Kossenkov A, et al. Genetic Signatures Indicate Predictable Rapid Response to Imatinib Mesylate Treatment in Gastrointestinal Stromal Tumors. Mol Cancer Therapeutics. doi: 10.1158/1535-7163.MCT-09-0193. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bidaut G, Suhre K, Claverie JM, Ochs MF. Determination of strongly overlapping signaling activity from microarray data. BMC Bioinformatics. 2006;7:99. doi: 10.1186/1471-2105-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgering BMT. A brief introduction to FOXOlogy. Oncogene. 2008;27:2258. doi: 10.1038/onc.2008.29. [DOI] [PubMed] [Google Scholar]
- 47.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou J, Wulfkuhle J, Zhang H, et al. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci U S A. 2007;104:16158–63. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldstein DM, Gray NS, Zarrinkar PP. High-throughput kinase profiling as a platform for drug discovery. Nat Rev Drug Discov. 2008;7:391–7. doi: 10.1038/nrd2541. [DOI] [PubMed] [Google Scholar]
- 50.Hallstrom TC, Mori S, Nevins JR. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell. 2008;13:11–22. doi: 10.1016/j.ccr.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.