Abstract
MicroRNAs are a class of small non-coding RNAs that are found in plants, animals, and some viruses. They modulate the gene function at the post-transcriptional level and act as a fine tuner of various processes, such as development, proliferation, cell signaling, and apoptosis. They are associated with different types and stages of cancer. Recent studies have shown the involvement of microRNAs in liver diseases caused by various factors, such as Hepatitis C, Hepatitis B, metabolic disorders, and by drug abuse. This review highlights the role of microRNAs in liver diseases and their potential use as therapeutic molecules.
Keywords: MicroRNA, Hepatitis, Fatty liver, Fibrosis, Cirrhosis
INTRODUCTION
The recent discovery of several types of non-coding RNAs revealed that the transcriptomes of higher eukaryotes are much more complex than originally anticipated. The non-coding RNAs, including microRNAs (miRNA), piwi interacting RNAs, small nucleolar RNAs, small interfering RNAs, long non-coding RNAs and antisense RNAs, have newly-discovered roles in the biology of health and diseases. These RNAs serve as modulators of genes involved in various biological pathways, such as development, cell differentiation, cell proliferation, cell death, chromosome modifications, virus pathogenesis, and oncogenesis[1-3]. Among these RNAs, miRNAs are small RNA molecules of approximate 21 nucleotides that are present in most eukaryotes[4]. Since the discovery of the first miRNA, lin-4, in 1993[5], more than 500 miRNAs have been reported in the mammalian genome[6].
It is postulated that about 1%-5% of genes in animals encode miRNAs, while 10%-30% of protein-coding genes are predicted miRNA targets[7,8], and have a specific miRNA signature in normal or cancer cells[9,10]. Although our understanding of the specific roles of miRNAs in cellular functions is only beginning, several studies have shown that miRNAs play a pivotal role in the most critical biological events, including development, proliferation, differentiation, cell fate determination, apoptosis, signal transduction, organ development, hematopoietic lineage differentiation, host-viral interactions, and carcinogenesis[4,11-13].
Overview of miRNAs
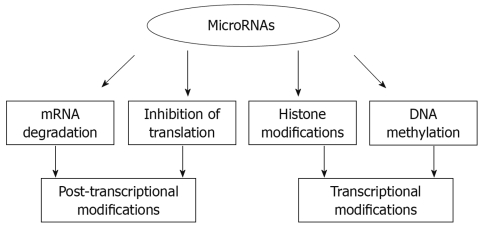
Despite the emerging critical role of miRNAs, the mechanism of their action is yet to be fully understood. The widely-known mode of gene regulation by miRNAs occurs at the post-transcriptional level by either specific inhibition of translation or induction of mRNA cleavage[14]. The essential role of miRNAs in cellular physiology was elegantly shown in mice lacking the Dicer enzyme that is required for the processing of the precursor miRNAs into the mature form[15].
Until now, the major focus of miRNA research was on their role in the cytoplasm, however recent reports indicate a diverse role of miRNAs, such as regulation of target genes by acting at 5’ UTR[16], regulation of DNA methylation[17,18] and import of the mature miRNA into the nucleus, suggesting other functional modes[19]. It is becoming clear that miRNAs not only regulate gene expression at the post-transcriptional level, but they are also capable of modifying chromatin (Figure 1).
Figure 1.
Various modes of gene regulation by miRNAs.
In this review, we discuss the role of the newly emerging class of RNA, miRNAs, in different types of liver diseases.
MiRNAs IN LIVER DISEASES
Improvements in the characterization and functional analysis techniques for miRNAs has not only uncovered their role in various cellular processes, but also revealed abnormal patterns of miRNA expression in various diseases, such as cancer[20], viral infection[21], inflammation[22], diabetes[23], cardiovascular[24], and Alzheimer[25]. In this review, we focus on the involvement of miRNAs in liver diseases caused by various factors, such as viral hepatitis, fatty liver due to alcohol abuse or metabolic syndrome, drug-induced liver disease, and by autoimmune processes (summarized in Table 1).
Table 1.
Expression of miRNAs in liver diseases
| Liver | Upregulated | Downregulated | Ref. |
| HCV | miR-215, miR-16, miR-199, miR-155, miR-146 | miR-122, miR-320, miR-191 | [53] |
| HBV | miR-181a, miR-200b and miR-146a | miR-15a | [56] |
| Drug overdose | miR-711 (liver), | miR-29b(liver), | [62] |
| Plasma: miR-122 miR-15a, miR-21, miR-101b, miR-148a and miR-192 | miR-710 (plasma) | ||
| NAFLD | miR-122, miR-34a, miR-31, miR-103, miR-107, miR-194, miR-335-5p, miR-221 and miR-200a | miR-29c, miR-451, miR-21 | [69,70] |
| ALD | miR-212, miR-320, miR-486, miR-705, and miR-1224 | miR-27b, miR-214, miR-199a-3p, miR-182, miR-183, miR-200a, and miR-322 | [71,72] |
| PBC | miR-328 and miR-299 | miR-122a and miR-26a | [73] |
| Liver fibrosis | miR-27a, miR-27b,miR-30, | miR-9,miR-721 and miR-301 | [80] |
| HCC | miR-18, miR-21, miR-221, miR-222, miR-224, miR-373 and miR-301 | miR-122, miR-125, miR-130a, miR-150, miR-199, miR-200 and let-7 family members | [81,90-95] |
HCV: Hepatitis C virus; HBV: Hepatitis B virus; NAFLD: Non-alcoholic fatty liver disease; ALD: Alcoholic liver disease; PBC: Primary biliary cirrhosis; HCC: Hepatocellular carcinoma.
MiRNAs in host-viral response and viral hepatitis
Viruses are equipped with complex machinery to exploit the host biosynthetic pathways and to defend against host cellular responses[26]. Recent studies have revealed the involvement of miRNA-mediated RNA-silencing pathways during viral-host cell interactions[27]. However, little is known about the role of cellular miRNAs against viral infection in eukaryotic organisms. Interestingly, viruses not only exploit the cellular miRNAs, but also encode their own miRNAs, adding another layer of complexity[28,29]. Therefore, it is important to study the abundance and distribution of miRNAs within the host cell during viral replication and latency, which will help us to understand the molecular regulation by the host cellular system as well as the simultaneous attempts by viruses to overcome the host defense.
Role of miRNAs in immune response: The interaction of parenchymal and immune cells plays a unique role in the liver in response to liver insults by viruses, bacteria, toxins, or antigens. There is compelling evidence that characterizes miRNAs as key regulators of innate and adaptive immune responses[22,30] and, therefore, they might play a role in inflammatory, autoimmune, or viral diseases of the liver. The importance of miRNAs in the liver immune system is highlighted by the fact that mice lacking Dicer 1 function in the liver were unable to produce mature miRNA and showed progressive hepatocyte damage, apoptosis, and portal inflammation[31].
The innate immune response is the first line of defense against noxious agents and is mediated by adaptors, such as the toll like receptors (TLRs), which recognize specific molecular patterns. Signals triggered by TLRs are involved in most liver diseases (from viral to inflammatory conditions) as well as in liver regeneration[32]. Several miRNAs (such as miR-155 or miR-146a/b) have been implicated in the regulation of TLR-induced signaling and might associated with liver pathophysiology. In particular, miR-155 was upregulated in monocytes and macrophages both upon TLR2, TLR3, TLR4, and TLR9 stimulation and after exposure to cytokines such as tumor necrosis factor (TNF)-α and interferons (IFN)[33-35]. The exact function of this miRNA in macrophages is not completely clear, but it might exert a positive regulation on TNF-α release, through targeting genes involved in nuclear factor-κB (NF-κB) signaling or enhancing TNF-α translation[34]. miR-146 is also upregulated in response to TLR2, TLR4, and TLR5 stimulation in monocytes[33]. This miRNA seems to decrease the release of inflammatory mediators, such as interleukin (IL)-1β or IL-8[36], possibly by downregulating IL-1 receptor-associated kinase 1 and TNF receptor-associated factor 6, and act as a negative regulator of TLR signaling[33]. Most of the studies describing the importance of these miRNAs in the innate immune response have been performed in macrophages or monocytes in vitro, and therefore the elucidation of specific miRNA functions in vivo in Kupffer cells, the resident liver macrophage, still awaits confirmation.
Neutrophil leukocyte infiltration in the liver is also a part of the innate immune response, which is common to liver injury, hepatic stress, or systemic inflammation signals[37]. Regulators of granulocyte function and activation, such as miR-223, might also be able to play a role in liver disease. Several reports have shown that the transcription factor C/EBPα binds to the miR-223 promoter and enhances its expression[38-40]. Given the importance of antigen presentation in the liver, the involvement of miRNAs in this function could be also of interest. In line with this, Hashimi et al[41] have recently shown that inhibition of both miR-21 and miR-34a in monocyte-derived dendritic cells led to a decrease in endocytic capacity and altered differentiation.
Apart from their role in the innate immune responses, miRNAs are associated with many aspects of both normal and abnormal immune adaptive responses, making them likely actors in autoimmune liver diseases or, particularly, viral hepatitis[22,30]. Virtually every T and B cell type seems to be associated with a different miRNA expression profile, and therefore an interesting avenue of research is to identify the miRNAs signature associated with specific liver diseases. Regarding specific miRNA functions, miR-181 has been described as a key quantitative regulator of T cell response to antigens[42]. MiR-150 plays a crucial role in B cell differentiation because its over-expression results in a selective defect in B cell development that blocks the transition from pro-B cell to pre-B cell[43]. Of note, miR-155 is implicated in both T and B cell function and differentiation promoting T helper 1 vs type 2 differentiation[44,45] and is essential in the production of antigen-specific antibodies[46]. These data highlight the importance of this miRNA as a central actor in immune regulation.
Hepatitis C virus (HCV) infection: HCV is an enveloped RNA virus of the Flavivirus family, and is capable of causing both acute and chronic hepatitis in humans by infecting liver cells. It is a major cause of chronic liver disease, with about 170 million people infected worldwide[47]. Up to 70% of patients will have persistent infection after inoculation, making it a fatal disease. The disease varies widely, from asymptomatic chronic infection to cirrhosis and hepatocellular carcinoma (HCC)[47]. Since the discovery of HCV, the treatment of hepatitis C has considerably improved by the use of pegylated interferons with ribavirin; however, more than 50% of people still do not respond to current medication and thus there is need for better drug therapy.
Recent studies have shown the involvement of miRNAs in the regulation of HCV infection. MiR-122 is first identified liver-specific cellular miRNA, which has been shown to enhance the replication of HCV by targeting the viral 5’ non-coding region[48]. It appears that HCV replication is associated with an increase in expression of cholesterol biosynthesis genes that are regulated by miR-122 and hence is considered as a potential target for antiviral intervention[49].
To date, there are some controversies around the role of miR-122 in HCV infection. Recently, Henke et al[50] showed that miR-122 stimulates HCV translation by enhancing the association of ribosomes with the viral RNA. The findings describing the role of miR-122 in HCV replication are of special interest, not only because of its novel mode of action, but because they also provide the first evidence of miRNAs linked to infectious disease. The development of therapeutic products to inhibit the miR-122 is believed to be an attractive approach to treat HCV patients.
By contrast, Sarasin-Filipowicz et al[51] found that liver biopsies from chronic hepatitis C patients undergoing IFN therapy revealed no correlation between miR-122 expression and viral load. Moreover, they found markedly decreased miR-122 levels in people who did not have any virological response during later IFN therapy. These reports indicate that the role of miR-122 in HCV viral replication is still controversial and awaits future research.
While some host miRNAs are beneficial for the virus, others inhibit viral replication. Studies by Pedersen et al[52] demonstrated that IFN-β stimulation, an antiviral cytokine, induces numerous cellular miRNAs, specifically eight miRNAs (miR-1, miR-30, miR-128, miR-196, miR-296, miR-351, miR-431 and miR-448) that have sequence-predicted targets within the HCV genomic RNA. Overexpression of these miRNAs in infected liver cells considerably attenuated viral replication. From this report it seems that host miRNAs have evolved to target viral genes and inhibit their replication, and thus might represent part of the host antiviral immune response. Very recently, Peng et al[53] carried out a computational study of hepatitis C virus associated miRNAs-mRNA regulatory modules in human livers. They found differential profiles of cellular miRNAs that target the genes involved in chemokine (16 genes such as CXCL12 etc.), B cell receptor, PTEN (13 genes), IL-6 ERK/ MAPK (18 genes; Ras, Erk3 and STAT3 etc.) and JAK/STAT signaling pathways, suggesting a critical role of miRNAs in the replication, propagation, and latency of virus in the host cell. Specifically, they found that miR-122, miR-320, and miR-191 were downregulated, whereas miR-215, miR-16, miR-26, miR-130, miR-199 and miR-155 were upregulated. These findings suggest that miRNAs have the potential to become novel drug targets in virally induced infectious or malignant diseases.
Hepatitis B virus (HBV) infection: HBV infection is a global problem and it can cause acute or chronic hepatitis B, liver cirrhosis, and, in some instances HCC[54]. The available therapy is only partially effective against the virus and development of better therapy remains an important issue. Little is known about the involvement of miRs during HBV infection. Considering the fact that cellular miRNAs play an important role in viral pathogenesis, it is likely that they have a role in HBV infection.
A differential miRNAs expression pattern was found in the livers of HBV and HCV infected individuals with hepatocellular cancer[55]. A total of nineteen miRNAs were clearly differentiated between HBV and HCV groups, out of which thirteen miRNAs were downregulated in the HCV group, whereas six showed a decreased expression in the HBV group. Some of the differentially regulated miRNAs between the HCV and HBV groups were miR-190, miR-134, miR-151, miR-193, miR-211, and miR-20. Interestingly, in the same study, it was shown that pathway analysis of targeted genes using infection-associated miRNAs could differentiate the genes into two groups. For instance, in HBV-infected livers, pathways related to cell death, DNA damage, recombination, and signal transduction were activated, and those related to immune response, antigen presentation, cell cycle, proteasome, and lipid metabolism were activated in HCV-infected livers[55].
In a second study, the profiling of cellular miRNAs of a stable HBV expressing cell line HepG2.2.15 and its parent cell line HepG2, showed that eighteen miRNAs were differentially expressed between the two cell lines[56]. Of them, eleven were upregulated and seven were downregulated. For instance, miR-181a, miR-200b, and miR-146a were found to be upregulated and miR-15a was downregulated. It remains to be evaluated if differentially regulated miRNAs could be exploited as potential biomarkers to differentiate between HCV and HBV in the initial stages of pathogenesis.
MiRNAs in drug-induced liver injury
Drug-induced liver injury is a serious clinical health problem and is the leading cause of drugs being removed from the market[57]. Acetaminophen overdose is the most common cause of fulminant liver failure. Many studies have been carried out to identify reliable and sensitive early blood markers for liver injury using high throughput technologies[58]. However, these efforts have so far failed to yield markers that are superior compared to the existing aminotransferase-based markers, and the need for better and stable biomarkers persists. Increasing attention is being paid to circulating miRNAs as potential biomarkers because of their stability over enzymes or proteins[59-61]. However, not much is known about the roles of miRNAs in drug-induced liver injury. A study by Wang et al[62] showed differential miRNAs profiling in a mouse model of acetaminophen overdose. Significant differences were found in the levels of miRNAs in both liver tissues and plasma between control and acetaminophen overdosed animals. Using miRPortal (www.miRportal.net), pathways involving antigen processing and presentation, apoptosis, B cell receptor signaling pathways, cytokine-cytokine receptor interaction, and cell cycles were found to be potential targets of miRNAs that showed decreased levels in the liver after acetaminophen exposure. In contrast, pathways involved in various signaling transduction and cell-cell interactions, such as gap-junctions, focal adhesion, and MAPK signaling pathways were potentially affected by miRNAs with increased levels in the liver[62].
Interestingly, it was observed that most miRNAs that were found to decrease in the liver exhibited increased levels in the plasma in overdosed mice. For instance, miR-122 and miR-192 showed higher levels in plasma, but they were decreased in tissue, whereas miR-710 and miR-711 were elevated in liver tissues; however, they displayed lower levels in plasma after acetaminophen overdose. Moreover, liver specific miRNAs, such as miR-122 and miR-192, showed dose and time-dependent changes in the plasma that paralleled serum aminotransferase levels and liver injury on histopathology. These findings suggest the potential of using specific circulating miRNAs as sensitive and informative biomarkers for drug-induced liver injury.
MiRNAs in non-alcoholic fatty liver disease (NAFLD)
NAFLD is the most common form of chronic liver disease worldwide and a major public health concern in the modern society[63]. It is characterized by excess fat accumulation in liver, which ranges from simple steatosis to steatohepatitis, and cirrhosis in the absence of heavy alcohol consumption[64]. The major risk factors for the development of NAFLD include metabolic syndrome, such as obesity, type 2 diabetes mellitus, and dyslipidemia[65]. Although progress has been made in understanding the factors causing NAFLD, more needs to be done to dissect the underlying regulatory networks. Compelling evidences indicate the role of miRNAs in energy metabolism and liver functions[66,67]. For instance, miR-143 has been shown to play a role in adipose differentiation[68].
MiR-122, which was found to be upregulated in NAFLD, is known to regulate genes involved in fatty acid biosynthesis[69], and administration of this miRNA antagonist into mice resulted in reduced levels of plasma cholesterol, increased hepatic fatty acid oxidation, and decreased synthesis of hepatic fatty acid and cholesterol.
Li et al[70] reported that eight miRNAs (miR-34a, miR-31, miR-103, miR-107, miR-194, miR-335-5p, miR-221 and miR-200a) were upregulated and three miRNAs (miR-29c, miR-451, miR-21) were downregulated in the livers of ob/ob mice. These findings suggested a connection between miRNAs and metabolic disorders, and the role of miRNA in NAFLD deserves further investigation.
Role of miRNAs in alcoholic liver disease (ALD)
ALD, alcoholic hepatitis, and cirrhosis represent a major segment of liver diseases worldwide. A recent report by Dolganiuc et al[71] analyzed the miRNA expression profile in a murine model of ALD. Liver samples from mice fed an ethanol-containing diet (Lieber-DeCarli) showed features of alcoholic steatohepatitis and had an increased expression of miR-320, miR-486, miR-705, and miR-1224. On the other hand, decreased expression was found for several miRNAs, including miR-27b, miR-214, miR-199a-3p, miR-182, miR-183, miR-200a, and miR-322. Furthermore, livers from mice with alcoholic and with non-alcoholic fatty liver shared common alterations in their miRNA profiles (such as upregulation of miR-705 and miR-1224). However, the functions and physiological roles of these miRNAs in ALD are yet to be determined. Tang et al[72] have suggested a novel mechanism for ethanol-induced intestinal permeability. They found that ethanol intake resulted in the induction of miR-212, which in turn downregulated the Zonula occludens (ZOP) protein. The decrease in ZOP protein levels led to intestinal barrier disruption and thus increased intestinal permeability. The interest of this finding lies in the association of intestinal permeability with the development of ALD, therefore linking miR-212 with the pathogenesis of this disease.
Primary biliary cirrhosis (PBC)
Recent studies indicate that the role of miRNAs is yet to be explored in liver diseases associated with autoimmune aggression. In PBC, altered hepatic miRNA expression was found[73]. miRNA profiling of explanted livers from subjects with PBC revealed differential expression of thirty-five independent miRNAs compared to controls. Downregulation of miR-122a and miR-26a, and increased expression of miR-328 and miR-299, were confirmed by PCR in the PBC livers. The role of miRNAs in autoimmune hepatitis is yet to be studied.
Liver fibrosis
Chronic hepatitis contributes to liver fibrosis, which has been linked to fibrosis due to fibrin deposition by hepatic stellate cells (HSC)[74]. The relationship between miRNAs and fibrosis has been shown in the context of specific diseases, such as diabetic kidney sclerosis[75] or myocardial fibrosis[76], and some of the mechanisms described could be involved in common pathways of fibrosis in other diseases. A recent report by Kato et al[77] has described that transforming growth factor (TGF)-β activates Akt kinase by enhancing the expression of both miR-216a and miR-217. These miRNAs, in turn, target PTEN, an inhibitor of Akt activation that has also been described to play a role in hepatic stellate cell activation[78]. Hepatic stellate cell activation has been identified as the major driver of liver fibrosis. Activation of HSC is associated with upregulation of various signaling pathways. In cirrhotic rat livers, HSC activation was associated with mitochondrial damage indicated by increased Bcl-2 and downregulated caspase-9 levels. The miRNA profile of the same rat HSC indicated upregulation of thirteen and downregulation of twenty-two, signaling pathways by miRNAs[79]. In another in vitro study, overexpression of miR-27a and miR-27b resulted in reversal of the activated phenotype of stellate cells to a more quiescent phenotype with increased fat accumulation and decreased proliferation[80]. The effects of miR-27a and miR-27b were mediated by regulation of the retinoid X receptor in the hepatic stellate cells.
Although there are only a few studies in the literature on miRNA genes and HCV-induced liver fibrosis, given the critical role of miRNA, its possible that they play a critical role in HCV infection, in the progression of fibrosis, and in the prediction of treatment response. Jiang et al[81] showed increased miRNA expression in cirrhotic and hepatitis-positive liver samples and suggested that important changes in miRNA expression occur during the development of chronic viral hepatitis and cirrhosis.
HCC
HCC is the third-leading cause of death from cancer and the fifth most common malignancy worldwide[82]. Liver cancer is a complex disease with exceptional heterogeneity in cause and outcome, involving epigenetic and chromosomal instability[83] and abnormalities in the expression of both coding and non-coding genes, including miRNAs[84,85]. The major etiologies of HCC include chronic liver disease due to chronic hepatitis B or hepatitis C virus infection, alcoholic steatohepatitis, metabolic disorders such as nonalcoholic steatohepatitis or insulin resistance, hereditary hemochromatosis, and immune-related diseases such as PBC and autoimmune hepatitis[86].
There are several studies in the literature showing specific miRNA signatures in HCC formation[87-89] and progression that could be exploited as potential cancer biomarkers. Briefly, miR-18, miR-21, miR-221, miR-222, miR-224, miR-373, and miR-301 were reported to be upregulated[81,90,91] whereas miR-122, miR-125, miR-130a, miR-150, miR-199, miR-200, and let-7 family members[92-95] were found to be downregulated in HCC by most studies. These miRNAs target the genes involved in cell cycle and cell death regulation, including cyclin-dependent inhibitors p27/CDKN1B and p57/CDKN1C, or the PI3K antagonist phosphatase and tensin homolog (PTEN). Budhu et al[96] defined twenty miRNAs, as a HCC metastasis signature, and showed that the twenty-miRNA-based signature was capable of predicting survival and recurrence of HCC in patients with multinodular or solitary tumors, including those with early-stage disease. miR-219-1, miR-207, and miR-338 were the most highly upregulated, whereas miR-34a, miR-30c-1, and miR-148a were highly downregulated in metastasis cases. Target Scan analysis revealed that SPTBN2, GTF2H1, PSCD3, VAMP3, and SLC20A2 are the target genes which were affected by multiple miRNAs related to metastasis and might be the part of signaling pathways that significantly contribute to this phenotype. Moreover, this miRNA signature was an independent and significant predictor of patient prognosis when compared to other available clinical parameters[96]. This study suggested that these twenty miRNAs can assist in HCC prognosis and might have clinical relevance. Further functional studies of these miRNAs could help to elucidate the mechanism leading to HCC metastasis. Deregulation of these twenty miRNAs in metastatic HCC implies that altered expression of their target genes might contribute to the development or recurrence of metastasis, and miR-122 is one example of this metastasis signature[97].
THE POTENTIAL OF MiRNAs IN DIAGNOSTICS, DISEASE PROGNOSIS, AND THERAPY
Increasing evidence indicates that miRNAs play an important role in a wide range of liver diseases, ranging from cancers and viral hepatitis, to metabolic diseases. The unique expression profile of miRNAs in different types and at different stages of cancer, and in other diseases, suggest that these small molecules can be exploited as novel biomarkers for disease diagnostics and might present a new strategy for miRNA gene therapy.
Recently, Kota et al[98] showed that administration of miR-26a, (which is under-expressed in HCC cells and downregulates cyclins D2 and E2) using adeno-associated virus (AAV) resulted in the inhibition of cancer cell proliferation, induction of tumor-specific apoptosis, and dramatic protection from disease progression without toxicity in a mouse model of HCC, suggesting the potential therapeutic use of miRNAs. This study showed that anti-miRNA compounds could be delivered in vivo safely and with efficacy; thus opening the way for translation of these basic research findings to clinical applications. It is also important to consider other factors, because some miRNA genes such as miR-1 undergo methylation-mediated regulation in HCC cell lines[99], suggesting a strong link between the DNA methylome and the miRNAome. In particular, there are some reports showing that the expression profiles of miRNA differ between malignant hepatocytes, malignant cholangiocytes, and benign liver cancer[90], suggesting that miRNA profiling could be used as molecular diagnostic markers in liver disease.
CONCLUSION
It is increasingly evident that miRNAs play a novel and important role in regulation of gene expression. In recent years, research focusing on small molecules such as miRNAs has intensified to understand their role in health and disease. The excitement of exploring their regulatory potential will remain for years to come, given the observation that miRNAs could be ideal therapeutic targets for many diseases. We are at the beginning of understanding of the diverse roles of miRNAs in fine-tuning biological pathways, and in future years we will have a better picture of this complex regulatory mechanism. Presently, there is need for miRNAs-based diagnostics and gene therapy; however, these techniques are in their infancy.
Footnotes
Supported by NIH Grant AA011576-10A1 to Gyongyi Szabo and an Alfonso Martin Escudero Foundation Scholarship to Miguel Marcos
Peer reviewer: Dr. T Choli-Papadopoulou, Associate Professor, Aristotle University of Thessaloniki, School of Chemistry, Department of Biochmistry, Thessaloniki 55124, Greece
S- Editor Wang YR L- Editor Stewart GJ E- Editor Ma WH
References
- 1.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 2.Voinnet O. Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- 3.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 6.Barringhaus KG, Zamore PD. MicroRNAs: regulating a change of heart. Circulation. 2009;119:2217–2224. doi: 10.1161/CIRCULATIONAHA.107.715839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 8.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One. 2009;4:e6229. doi: 10.1371/journal.pone.0006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Fontana L, Sorrentino A, Condorelli G, Peschle C. Role of microRNAs in haemopoiesis, heart hypertrophy and cancer. Biochem Soc Trans. 2008;36:1206–1210. doi: 10.1042/BST0361206. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Kim DH, Saetrom P, Snove O Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 19.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 20.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 21.Houzet L, Yeung ML, de Lame V, Desai D, Smith SM, Jeang KT. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology. 2008;5:118. doi: 10.1186/1742-4690-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 23.Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 24.Latronico MV, Catalucci D, Condorelli G. Emerging role of microRNAs in cardiovascular biology. Circ Res. 2007;101:1225–1236. doi: 10.1161/CIRCRESAHA.107.163147. [DOI] [PubMed] [Google Scholar]
- 25.Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci Lett. 2009;459:100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 26.Sullivan CS. New roles for large and small viral RNAs in evading host defences. Nat Rev Genet. 2008;9:503–507. doi: 10.1038/nrg2349. [DOI] [PubMed] [Google Scholar]
- 27.Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nair V, Zavolan M. Virus-encoded microRNAs: novel regulators of gene expression. Trends Microbiol. 2006;14:169–175. doi: 10.1016/j.tim.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bi Y, Liu G, Yang R. MicroRNAs: novel regulators during the immune response. J Cell Physiol. 2009;218:467–472. doi: 10.1002/jcp.21639. [DOI] [PubMed] [Google Scholar]
- 31.Hand NJ, Master ZR, Le Lay J, Friedman JR. Hepatic function is preserved in the absence of mature microRNAs. Hepatology. 2009;49:618–626. doi: 10.1002/hep.22656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seki E, Brenner DA. Toll-like receptors and adaptor molecules in liver disease: update. Hepatology. 2008;48:322–335. doi: 10.1002/hep.22306. [DOI] [PubMed] [Google Scholar]
- 33.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 35.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perry MM, Moschos SA, Williams AE, Shepherd NJ, Larner-Svensson HM, Lindsay MA. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol. 2008;180:5689–5698. doi: 10.4049/jimmunol.180.8.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramaiah SK, Jaeschke H. Role of neutrophils in the pathogenesis of acute inflammatory liver injury. Toxicol Pathol. 2007;35:757–766. doi: 10.1080/01926230701584163. [DOI] [PubMed] [Google Scholar]
- 38.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 39.Fukao T, Fukuda Y, Kiga K, Sharif J, Hino K, Enomoto Y, Kawamura A, Nakamura K, Takeuchi T, Tanabe M. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129:617–631. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- 40.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 41.Hashimi ST, Fulcher JA, Chang MH, Gov L, Wang S, Lee B. MicroRNA profiling identifies miR-34a and miR-21 and their target genes JAG1 and WNT1 in the coordinate regulation of dendritic cell differentiation. Blood. 2009;114:404–414. doi: 10.1182/blood-2008-09-179150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 43.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci USA. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 46.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 48.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 49.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarasin-Filipowicz M, Krol J, Markiewicz I, Heim MH, Filipowicz W. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat Med. 2009;15:31–33. doi: 10.1038/nm.1902. [DOI] [PubMed] [Google Scholar]
- 52.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng X, Li Y, Walters KA, Rosenzweig ER, Lederer SL, Aicher LD, Proll S, Katze MG. Computational identification of hepatitis C virus associated microRNA-mRNA regulatory modules in human livers. BMC Genomics. 2009;10:373. doi: 10.1186/1471-2164-10-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kiyosawa K, Sodeyama T, Tanaka E, Gibo Y, Yoshizawa K, Nakano Y, Furuta S, Akahane Y, Nishioka K, Purcell RH. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology. 1990;12:671–675. doi: 10.1002/hep.1840120409. [DOI] [PubMed] [Google Scholar]
- 55.Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K, Kaneko S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–1112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- 56.Liu Y, Zhao JJ, Wang CM, Li MY, Han P, Wang L, Cheng YQ, Zoulim F, Ma X, Xu DP. Altered expression profiles of microRNAs in a stable hepatitis B virus-expressing cell line. Chin Med J (Engl) 2009;122:10–14. doi: 10.3901/jme.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 57.Halegoua-De Marzio D, Navarro VJ. Drug-induced hepatotoxicity in humans. Curr Opin Drug Discov Devel. 2008;11:53–59. [PubMed] [Google Scholar]
- 58.Ozer J, Ratner M, Shaw M, Bailey W, Schomaker S. The current state of serum biomarkers of hepatotoxicity. Toxicology. 2008;245:194–205. doi: 10.1016/j.tox.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 59.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 61.Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One. 2009;4:e6229. doi: 10.1371/journal.pone.0006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clark JM, Brancati FL, Diehl AM. Nonalcoholic fatty liver disease. Gastroenterology. 2002;122:1649–1657. doi: 10.1053/gast.2002.33573. [DOI] [PubMed] [Google Scholar]
- 64.Farrell GC, Larter CZ. Nonalcoholic fatty liver disease: from steatosis to cirrhosis. Hepatology. 2006;43:S99–S112. doi: 10.1002/hep.20973. [DOI] [PubMed] [Google Scholar]
- 65.Marchesini G, Brizi M, Bianchi G, Tomassetti S, Bugianesi E, Lenzi M, McCullough AJ, Natale S, Forlani G, Melchionda N. Nonalcoholic fatty liver disease: a feature of the metabolic syndrome. Diabetes. 2001;50:1844–1850. doi: 10.2337/diabetes.50.8.1844. [DOI] [PubMed] [Google Scholar]
- 66.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 67.Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, Zavolan M, Stoffel M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci USA. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 69.Jin X, Ye YF, Chen SH, Yu CH, Liu J, Li YM. MicroRNA expression pattern in different stages of nonalcoholic fatty liver disease. Dig Liver Dis. 2009;41:289–297. doi: 10.1016/j.dld.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 70.Li S, Chen X, Zhang H, Liang X, Xiang Y, Yu C, Zen K, Li Y, Zhang CY. Differential expression of MicroRNAs in mouse liver under aberrant energy metabolic status. J Lipid Res. 2009:Epub ahead of print. doi: 10.1194/jlr.M800509-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dolganiuc A, Petrasek J, Kodys K, Catalano D, Mandrekar P, Velayudham A, Szabo G. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol Clin Exp Res. 2009;33:1704–1710. doi: 10.1111/j.1530-0277.2009.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang Y, Banan A, Forsyth CB, Fields JZ, Lau CK, Zhang LJ, Keshavarzian A. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin Exp Res. 2008;32:355–364. doi: 10.1111/j.1530-0277.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 73.Padgett KA, Lan RY, Leung PC, Lleo A, Dawson K, Pfeiff J, Mao TK, Coppel RL, Ansari AA, Gershwin ME. Primary biliary cirrhosis is associated with altered hepatic microRNA expression. J Autoimmun. 2009;32:246–253. doi: 10.1016/j.jaut.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qian Y, Feldman E, Pennathur S, Kretzler M, Brosius FC 3rd. From fibrosis to sclerosis: mechanisms of glomerulosclerosis in diabetic nephropathy. Diabetes. 2008;57:1439–1445. doi: 10.2337/db08-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Diez J. Do microRNAs regulate myocardial fibrosis? Nat Clin Pract Cardiovasc Med. 2009;6:88–89. doi: 10.1038/ncpcardio1415. [DOI] [PubMed] [Google Scholar]
- 77.Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11:881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takashima M, Parsons CJ, Ikejima K, Watanabe S, White ES, Rippe RA. The tumor suppressor protein PTEN inhibits rat hepatic stellate cell activation. J Gastroenterol. 2009;44:847–855. doi: 10.1007/s00535-009-0073-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo CJ, Pan Q, Cheng T, Jiang B, Chen GY, Li DG. Changes in microRNAs associated with hepatic stellate cell activation status identify signaling pathways. FEBS J. 2009;276:5163–5176. doi: 10.1111/j.1742-4658.2009.07213.x. [DOI] [PubMed] [Google Scholar]
- 80.Ji J, Zhang J, Huang G, Qian J, Wang X, Mei S. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 2009;583:759–766. doi: 10.1016/j.febslet.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 81.Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 83.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 84.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Varnholt H. The role of microRNAs in primary liver cancer. Ann Hepatol. 2008;7:104–113. [PubMed] [Google Scholar]
- 86.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 87.Braconi C, Patel T. MicroRNA expression profiling: a molecular tool for defining the phenotype of hepatocellular tumors. Hepatology. 2008;47:1807–1809. doi: 10.1002/hep.22326. [DOI] [PubMed] [Google Scholar]
- 88.Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, Odenthal M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 89.Wang Y, Lee AT, Ma JZ, Wang J, Ren J, Yang Y, Tantoso E, Li KB, Ooi LL, Tan P, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283:13205–13215. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- 90.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 91.Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 92.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 93.Gramantieri L, Fornari F, Callegari E, Sabbioni S, Lanza G, Croce CM, Bolondi L, Negrini M. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12:2189–2204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Croce CM, Bolondi L, Negrini M. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res. 2009;15:5073–5081. doi: 10.1158/1078-0432.CCR-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fornari F, Gramantieri L, Giovannini C, Veronese A, Ferracin M, Sabbioni S, Calin GA, Grazi GL, Croce CM, Tavolari S, et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009;69:5761–5767. doi: 10.1158/0008-5472.CAN-08-4797. [DOI] [PubMed] [Google Scholar]
- 96.Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 97.Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 98.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]