Abstract
Genome-wide association studies are now widely used tools to identify genes and/or regions which may contribute to the development of various diseases. With case-control data a 2×3 contingency table can be constructed for each SNP to perform genotype-based tests of association. An increasingly common technique to increase the power to detect an association is to collapse each 2×3 table into a table assuming either a dominant or recessive mode of inheritance (2×2 table). We consider three different methods of determining which genetic model to choose and show that each of these methods of collapsing genotypes increases the type I error rate (i.e., the rate of false positives). However, one of these methods does lead to an increase in power compared with the usual genotype- and allele-based tests for most genetic models.
Keywords: genome-wide association, type I error rate
1 Introduction
Genome-wide association studies (GWAS) are invaluable tools in the search for genes involved with complex diseases. Recent technology allows for the examination of hundreds of thousands of markers at a time located throughout the genome. For example, Affymetrix has a chip that features over 900K SNPs (Affymetrix Genome-Wide Human SNP Array 6.0) and Illumina has developed a chip with over one million loci (Illumina human1M-duo). In the context of complex diseases this vast coverage of the human genome is particularly important since there may be many genes involved in the disease process. We consider a case-control approach to genome-wide association studies of single nucleotide polymorphisms (SNPs).
In very broad terms there are two approaches to the analysis of case-control genome-wide association studies: one is genotype-based and the other allele-based. The genotype-based tests can involve analysis of a 2 × contingency table where no assumptions are made regarding modes of inheritance, or 2 × 2 tables where some genetic model is considered. The allele-based test also uses a 2 × 2 contingency table but simply counts alleles. The latter test is more sensitive to additive effects, while the former tests are more powerful if the assumed mode of inheritance is correct. Sasieni (1997) showed that the 2 × 3 genotype-based test is more powerful than the allele-based unless both cases and controls are in Hardy-Weinberg equilibrium (HWE). To cover all bases, some investigators opt to perform several tests of association for each SNP in the study (e.g., Kato et al., 2008; Wellcome Trust Case Control Consortium, 2007; Rudd et al., 2006; Smyth et al., 2006; Hu et al., 2005). Obviously this further compounds the problem of multiple testing when analyzing hundreds of thousands of SNPs, as acknowledged by Hu et al. As an alternative, some researchers select to perform only one test for each SNP, then follow up only significant results with a different test (e.g., DeWan et al., 2006; Hamada et al., 2005; Klein et al., 2005). Such an approach decreases the number of tests performed relative to using two tests per SNP. Other investigators choose to only perform one test, for example, Hampe et al. (2007) and De Ferrari et al. (2007) use only allele-based tests to detect associations. Alternatively, Yeager et al. (2007) opted to use only genotype-based tests in order to detect effects which may or may not be additive.
There have also been studies which select one result from multiple tests to represent each SNP, such that there is only one p-value reported per marker. Li et al. (2008) performed the 2 × 3 genotype-based χ2 test, and evaluated three genetic models via logistic regression: dominant, recessive and additive. For the regression analyses, one model was chosen to represent each SNP using the Bayes Information Criterion. Bonferroni corrections were made to account for the fact that several genetic models were fit. Lencz et al. (2007) used a similar approach but selected either a dominant or recessive model based on likelihood ratio tests. Zheng et al. (2006) go a step further and propose several composite statistics which are functions of the optimal test statistics over several genetic models. Advantages of this approach are that it allows the model-specific tests to be correlated, and that it would be robust to misspecification of the underlying genetic model because it simultaneously considers a range of possibilities. On the other hand, it may be computationally intense since each genetic model must be fit for each SNP, the correlations among these model-specific tests must be estimated, and permutation tests employed to evaluate significance. Clearly, how to select a test/model to represent each SNP in testing procedures has been of interest and a simple solution remains unclear. Therefore we undertook a study to evaluate and compare several intuitive approaches to the selection of one test/model for each SNP. Our focus was on increasing power by decreasing the degrees of freedom for a contingency table analysis.
For contingency table analysis of case-control GWAS, there are essentially two general approaches. The full genotype-based analysis constructs a 2 × 3 contingency table for each SNP where the two rows are disease status, and the three possible genotypes form the columns. A simple χ2 test with two degrees of freedom is performed to determine whether the frequencies of any of the genotypes are associated with the affected phenotype. For the allele-based approach, a 2 × 2 contingency table is constructed for each SNP where the rows are disease status and the columns are alleles. Again, a χ2 test of association is used, however, there is only one degree of freedom. This difference in the number of degrees of freedom is important since power is often greater for tests with smaller degrees of freedom. For the case where the minor allele frequency (MAF) is small, or heterozygosity is associated with disease, the genotype-based approach may be more powerful than the allele-based one despite the fact that the tests of association have more degrees of freedom.
Alternative forms of the genotype-based test of association seek to decrease the number of degrees of freedom by collapsing the full contingency table to a 2 × 2 one that is still genotype-based but assumes either a dominant or recessive genetic model. This collapsing is the contingency table analog of the approaches employed by Lencz et al., Hu et al., and Li et al. Assuming a dominant model for the A allele, the first genotype column would be the number of subjects with the aa genotype and the second would be the number of subjects with either the aA or AA genotypes. Similarly, if a recessive model is assumed for the A allele then the first genotype column would be the number with either aa or aA genotypes and the second would be those with AA genotypes. Consider the following 2 × 3 contingency table:
| aa | aA | AA | |
|---|---|---|---|
| Case | x1 | x2 | x3 |
| Control | z1 | z2 | z3 |
where xg and zg are the number of cases and controls with genotype g (g = aa, aA, AA), respectively. The corresponding table assuming a dominant mode of inheritance for the A allele is:
| aa | aA or AA | |
|---|---|---|
| Case | x1 | x2 + x3 |
| Control | z1 | z2 + z3 |
and the recessive model would be:
| aa or aA | AA | |
|---|---|---|
| Case | x1 + x2 | x3 |
| Control | z1 + z2 | z3 |
The choice on how to collapse the full 2 × 3 contingency table can be made using several different criteria. For each SNP in the genome-wide association analyses, the 2 × 2 contingency tables are constructed using some criterion and tests are performed using the corresponding one degree of freedom χ2 test of association.
Because the collapsing approaches consider multiple genetic models, there is a possibility that they have inflated type I error rates. In addition, if the collapsing criteria induce a bias towards the alternative hypothesis, the type I error rate will be further inflated. Lencz et al. allude to the problem in regards to their approach which selects the mode of inheritance based on a likelihood ratio test. In this situation, it is clear that the collapsing criterion induces a bias away from the null hypothesis. The inflated type I error rates must be accounted for in any meaningful comparison of these approaches. Of interest is the relative power of these approaches. Via simulation, we confirm the intuition that these collapsing approaches are indeed associated with an increased type I error rate.
2 Materials and Methods
For all simulation studies we considered three different criteria for selecting how to collapse the full 2 × 3 contingency table into a dominant or recessive one. The first selects the 2 × 2 table with the largest odds-ratio (OR). For example, if the dominant 2 × 2 table has an OR of 2 while the recessive table has an OR of 1.5, then the analysis for that SNP assumes a dominant mode of inheritance. We refer to this as the OR criterion approach. An alternative criterion is to select the 2 × 2 contingency table with the largest χ2 test statistic, which is equivalent to choosing the model with the smallest p-value. This is referred to as the χ2 approach. Choosing the most significant model has also been used in the context of likelihood ratio tests (Lencz et al.) and generalized linear models (Wellcome Trust Case Control Consortium; Hu et al.). These OR and χ2 criteria may induce a bias toward the alternative, so we additionally consider a third approach which should not introduce any bias. This method combines the rarest homozygotic genotype with the heterozygotes. For example, if the frequency of the aa genotype (obtained by pooling cases and controls) is less than that of the AA genotype, then a recessive model is chosen. This is referred to as the rare homozygotes criterion.
Initially we examined the type I error rates for five analytic methods: the allele-based, the full 2 × 3 contingency table, and the three collapsing approaches: OR, χ2 and rare homozygotes. Since it is common to analyze each SNP independently, the simulations considered only one SNP. The genotypes for each individual were generated from the frequencies p2, 2pq and q2 (where p is the frequency of the a allele) assuming HWE in the general population and no association with disease. Without loss of generality we assumed an equal number of cases and controls. Each simulation consisted of 1000000 iterations and considered several sample sizes and minor allele frequencies (MAFs). P-values were computed assuming the test statistics are χ2 distributed with either 1 or 2 degrees of freedom, as appropriate. The type I error rate for each approach was then estimated as the proportion of iterations for which that approach yielded a p-value less than 0.05.
To compare the power for the five aforementioned analytic methods, the genotypic data were generated assuming an underlying genetic model and HWE in the general population. Let fg denote the penetrance associated with genotype g. Thus, the population prevalence, denoted by κ, is given by faap2 + 2faApq + fAAq2 where q = 1 − p. It can be shown that the distribution of genotypes conditional on disease status is given by
The first three probabilities above were used to generate N cases, and the latter three probabilities to generate N controls.
As before, each simulation consisted of 1000000 iterations where several sample sizes and MAFs were considered, however, only results for N = 250 are presented here. For each of these sample size and frequency combinations four genetic models were considered: dominant, recessive, additive and a scenario where heterozygosity is associated with disease. The details of these models are given in Table 1. The power was estimated as the proportion of iterations for each approach that rejects the null hypothesis, that is, the proportion of iterations in which the test statistic exceeds a critical limit. For each analysis method, its own critical value was chosen such that 5% of iterations generated under no association furnished test statistics exceeding this limit. In this manner, all power evaluations were based on a type I error rate of 5% and thus can be directly compared.
Table 1.
Penetrance definitions for the power simulations.
| Model | faa | faA | fAA |
|---|---|---|---|
| Dominant | 0.05 | 0.09 | 0.09 |
| Recessive | 0.05 | 0.05 | 0.09 |
| Additive | 0.05 | 0.07 | 0.09 |
| Heterozygosity | 0.05 | 0.09 | 0.05 |
Also of interest is the accuracy of the collapsing criteria, that is, how often do they select the 2 × 2 table corresponding to the true underlying genetic model. Only the OR and χ2 criteria are considered because the rare homozygotes approach does not explicitly evaluate any evidence for a particular mode of inheritance. Within these power simulations, the accuracies of these two genotype-based collapsing approaches were estimated as the proportion of iterations for which each approach selects the genetic model used to generate the data.
3 Results
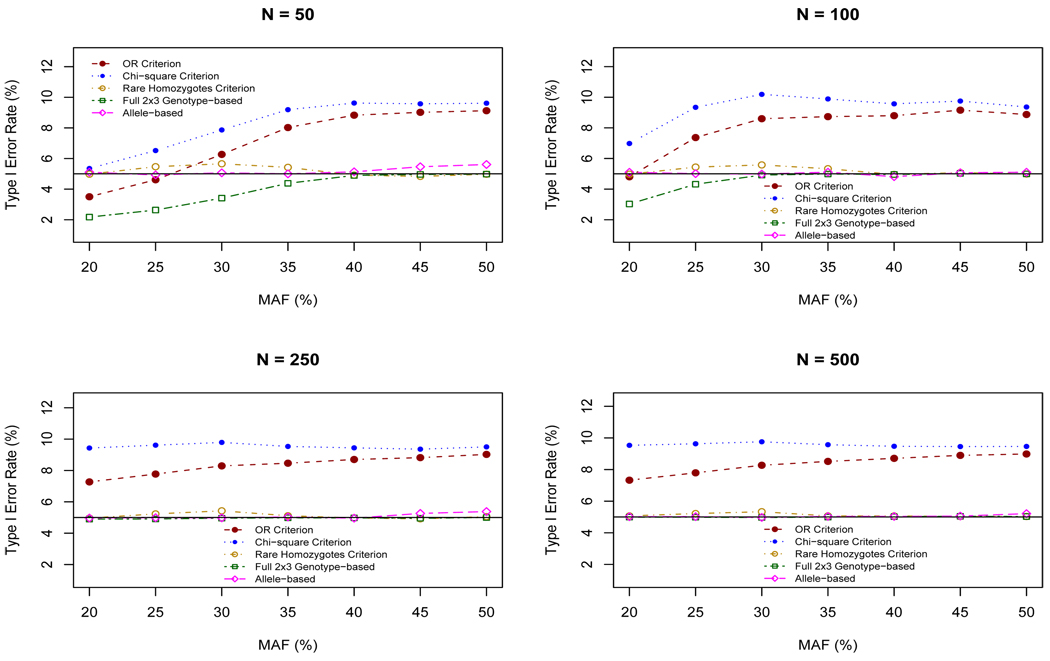
The type I error rates for the five analytic approaches are given in Figure 1. As expected, the type I error rates for the full 2 × 3 contigency table, the allele-based and rare homozygotes approaches hover around 5%. It is also clear that there is a substantial increase in the type I error rate for the OR and χ2 collapsing approaches. The latter approach has a type I error rate which varies from 5% to 10%. The OR criterion approach yields type I error rates that mimic the χ2 criterion but are slightly less for all sample sizes and MAFs.
Figure 1.
Type I error rates for various sample sizes and MAFs.
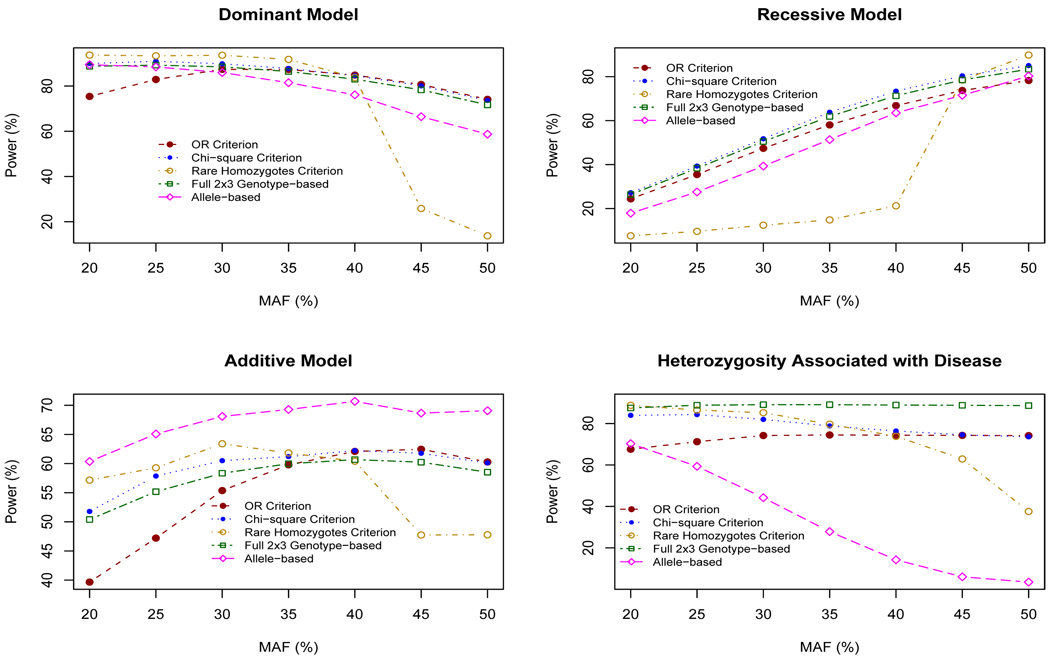
Figure 2 gives the power estimates for four genetic models with 250 cases and 250 controls. Recall that these power estimates correspond to 5% type I error rates. There is a marked increase in the power of the OR and χ2 collapsing approaches over the allele-based test for all but the additive model. In all the genetic models the power of the χ2 approach equaled or exceeded that of the OR approach. For an MAF approaching 50%, the rare homozygotes criterion has very poor power assuming a dominant, additive or heterozygotic susceptibility model. As expected, the allele-based approach has greater power than the other tests for the additive genetic model. If the disease is associated with heterozygosity, the allele-based approach is very poorly powered, particularly as the MAF increases. These patterns are also observed with sample sizes of 50, 100 and 500 (results not presented here). The 2 × 3 genotype-based test is approximately as powerful as the OR and χ2 approaches for all but the heterozygotic susceptibility model.
Figure 2.
Power estimates for four genetic models with N = 250.
The results of how well the OR and χ2 collapsing approaches perform with regard to selecting the correct model are given in Table 2. The χ2 approach generally outperforms the OR approach. For larger sample sizes both selection criteria are highly accurate, often over 90%, and, as expected, the accuracy increases with sample size.
Table 2.
Comparison of the accuracy of the OR and χ2 approaches.
| Dominant Modela |
Recessive Modelb |
||||
|---|---|---|---|---|---|
| MAF | N | OR | χ2 | OR | χ2 |
| 0.50 | 50 | 73% | 70% | 54% | 72% |
| 100 | 83% | 81% | 65% | 84% | |
| 250 | 96% | 94% | 84% | 96% | |
| 500 | 99% | 99% | 95% | 100% | |
| 0.45 | 50 | 70% | 71% | 56% | 70% |
| 100 | 82% | 83% | 67% | 82% | |
| 250 | 95% | 96% | 84% | 95% | |
| 500 | 99% | 99% | 94% | 99% | |
| 0.40 | 50 | 67% | 73% | 57% | 69% |
| 100 | 79% | 85% | 66% | 80% | |
| 250 | 93% | 97% | 83% | 94% | |
| 500 | 98% | 100% | 93% | 99% | |
| 0.35 | 50 | 62% | 76% | 58% | 65% |
| 100 | 74% | 86% | 65% | 76% | |
| 250 | 90% | 97% | 80% | 91% | |
| 500 | 97% | 100% | 90% | 98% | |
| 0.30 | 50 | 57% | 80% | 58% | 59% |
| 100 | 69% | 88% | 64% | 72% | |
| 250 | 85% | 98% | 76% | 87% | |
| 500 | 94% | 100% | 87% | 96% | |
| 0.25 | 50 | 50% | 83% | 58% | 51% |
| 100 | 62% | 89% | 62% | 67% | |
| 250 | 78% | 98% | 72% | 82% | |
| 500 | 88% | 100% | 82% | 93% | |
| 0.20 | 50 | 45% | 85% | 58% | 43% |
| 100 | 53% | 91% | 61% | 57% | |
| 250 | 69% | 98% | 67% | 75% | |
| 500 | 79% | 100% | 75% | 87% | |
faa = 0.05, faA = 0.09, fAA = 0.09
faa = 0.05, faA = 0.05, fAA = 0.09
4 Discussion
Recent analyses of genome-wide association studies for a wide range of diseases have employed several different tests of association for each SNP (e.g., Kato et al.; Rudd et al.; Wellcome Trust Case Control Consortium). Given the problem of multiple comparisons in GWAS, it may not be desirable to further compound it by performing multiple tests per SNP. To protect against missing any associations by not choosing the proper underlying genetic model, Zheng et al. proposed an approach which uses composite statistics that consider several modes of inheritance simulatenously. As stated previously, this approach can potentially be computationally intense. The objective of this paper was to develop simple and intuitive ways of selecting one genetic model to represent each SNP based on decreasing the degrees of freedom for a contingency table analysis, and evaluating them with respect to their type I error rates and power.
Our approach to increasing the power to detect an association in genome-wide case-control studies is to collapse the full 2 × 3 contingency tables using some criterion into a 2 × 2 table which assumes either a dominant or recessive genetic model. The decrease in the dimensions of the contingency table decreases the number of degrees of freedom for the χ2 test of association, thus potentially increasing power. Collapsing tables into dominant or recessive models may be informative if the causal SNP is being examined, but not if the SNP is in incomplete linkage disequilibrium with the causal one. However, the objective here is to maximize power via decreasing the dimensions of the contingency table, not to make inferences regarding the mode of inheritance.
The results of the first set of simulations (Figure 1) show that the OR and χ2 collapsing approaches have inflated type I error rates, as expected. Both of these criteria induce a bias away from the null hypothesis by selecting the model indicating a greater “effect”. This bias is clearly greater for the χ2 criterion and is reflected in the fact that it has a greater type I error rate than the OR approach for all sample sizes and MAFs. Further, the χ2 approach is, in essence, utilizing an order statistic (maximum) which most likely does not follow a χ2 distribution. Incorrectly assuming such a distribution may also alter the type I error rate. Since the rare homozygotes criterion does not select a 2 × 2 table based on any evidence of association there is no inflation of the rate of false positives.
The power simulations yielded several interesting results. Firstly, in all scenarios considered, using the χ2 criterion was more powerful than use of the OR. If the OR is small, then this approach may not detect the true underlying model and thus power is reduced relative to the χ2 which can detect even small effect sizes. For all combinations of sample size, MAF and most of the underlying genetic models considered, these two collapsing approaches are as or more powerful than the genotype-based test, but the difference decreases as the sample size increases since the power for all approaches nears 100%. Secondly, the rare homozygotes criterion is poorly powered for most genetic models as the MAF approaches 50%. This boundary value of the MAF suggests that on average half the time the criterion will select a dominant table and the other half a recessive table, regardless of the true genetic model. For this rare homozygotes approach there was no increase in power relative to the full 2 × 3 genotype-based test despite the fewer degrees of freedom because it incorrectly selects the 2 × 2 table. Thirdly, when heterozygosity is associated with disease there are no additive effects and thus the allele-based approach is extremely poorly powered. Lastly, the major result of the power simulations is that the χ2 collapsing approach is more powerful than the allele-based tests for all but the additive model.
Permutation methods should be used to determine the significance threshold corresponding to a 5% type I error rate because the null distribution is generally unknown. In the GWA paradigm, case status would be randomly permuted for, say, K iterations while the genotypes remain fixed and the selected collapsing approach is applied to each permuted sampled. There are several possible ways to evaluate significance. First, consider application of the Westfall and Young (1989) approach to GWAS and let pmin,k denote the smallest p-value, over all SNPs, at iteration k (k = 1,…,K). The adjusted p-value for a particular SNP is then the percent of the K iterations for which pmin,k is greater than the observed p-value of that SNP. This adjusted p-value can then be compared to a pre-determined significance threshold, such as 5%. Alternatively, one may use the fifth percentile of the K values of pmin,k as a significance threshold for the observed p-value, as suggested by Dudbridge and Gusnanto (2007). Note that both approaches to computing thresholds simulatenously account for the correlation between SNPs and the multiple testing issue. In addition, such permutation testing also accounts for the fact that in the χ2 approach multiple models are being considered.
In summary, collapsing the full 2 × 3 contingency tables into either dominant or recessive tables using the one with the largest χ2 test statistic is as or more powerful than the genotype- and allele-based tests for most genetic models.
Footnotes
We would like to thank Dr. Josephine Hoh and Dr. Andrew DeWan for their invaluable input in the early conceptual development of this project. This work was supported by grant MH44292 from the National Institute of Mental Health and by an NSFC grant from the Chinese Government (project number 30730057).
References
- De Ferrari G, Papassotiropoulos A, Biechele T, Wavrant De-Vrieze F, Avila M, Major M, Myers A, Sáez K, Henríquez J, Zhao A, Wollmer M, Nitsch R, Hock C, Morris C, Hardy J, Moon R. Common genetic variation within the low-density lipoprotein receptor-related protein 6 and late-onset Alzheimer’s disease. Proc Natl Acad Sci, USA. 2007;104(22):9434–9439. doi: 10.1073/pnas.0603523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWan A, Liu M, Hartman S, Zhang S, Liu D, Zhao C, Tam P, Chan W, Lam D, Snyder M, Barnstable C, Pang C, Hoh J. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314(5801):989–992. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- Dudbridge F, Gusnanto A. Estimating signficance thresholds for genome-wide association scans; Presented at the annual meeting of the American Society of Human Genetics; San Deigo, CA. 2007. [October 25 2007]. Abstract from www.ashg.org/genetics/ashg07s/index.shtml. [Google Scholar]
- Hamada D, Takata Y, Osabe D, Nomura K, Shinohara S, Egawa H, Nakano S, Shinomiya F, Scafe C, Reeve V, Miyamoto T, Moritani M, Kunika K, Inoue H, Yasui N, Itakura M. Association between single-nucleotide polymorphisms in the SEC8L1 gene, which encodes a subunit of the exocyst complex, and rheumatoid arthritis in a Japanese population. Arthritis Rheum. 2005;52(5):1371–1380. doi: 10.1002/art.21013. [DOI] [PubMed] [Google Scholar]
- Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega F, Briggs J, Günther S, Prescott N, Onnie C, Häsler R, Sipos B, Fölsch U, Lengauer T, Platzer M, Mathew C, Krawczak M, Schreiber S. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39(2):207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- Hu N, Wang C, Hu Y, Yang H, Giffen C, Tang Z, Han X, Goldstein A, Emmert-Buck M, Buetow K, Taylor P, Lee M. Genome-wide association study in esophageal cancer using GeneChip mapping 10K array. Cancer Res. 2005;65(7):2542–2546. doi: 10.1158/0008-5472.CAN-04-3247. [DOI] [PubMed] [Google Scholar]
- Kato N, Miyata T, Tabara Y, Katsuya T, Yanai K, Hanada H, Kamide K, Nakura J, Kohara K, Takeuchi F, Mano H, Yasunami M, Kimura A, Kita Y, Ueshima H, Nakayama T, Soma M, Hata A, Fujioka A, Kawano Y, Nakao K, Sekine A, Yoshida T, Nakamura Y, Saruta T, Ogihara T, Sugano S, Miki T, Tomoike H. High-density association study and nomination of susceptibility genes for hypertension in the Japanese National Project. Hum Mol Genet. 2008;17(4):617–627. doi: 10.1093/hmg/ddm335. [DOI] [PubMed] [Google Scholar]
- Klein R, Zeiss C, Chew E, Tsai J, Sackler R, Haynes C, Henning A, San-Giovanni J, Mane S, Mayne S, Bracken M, Ferris F, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencz T, Morgan T, Athanasiou M, Dain B, Reed C, Kane J, Kucherlapati R, Malhotra A. Converging evidence for a pseudoautsomal cytokine receptor gene locus in schizophrenia. Mol Psychiatry. 2007;12(6):572–580. doi: 10.1038/sj.mp.4001983. [DOI] [PubMed] [Google Scholar]
- Li H, Wetten S, Li L, St. Jean P, Upmanyu R, Surh L, Hosford D, Barnes M, Briley J, Borrie M, Coletta N, Delisle R, Dhalla D, Ehm M, Feldman H, Fornazzari L, Gauthier S, Goodgame N, Guzman D, Hammond S, Hollingworth P, Hsiung G, Johnson J, Kelly D, Keren R, Kertesz A, King K, Lovestone S, Loy-English I, Matthews P, Owen M, Plumpton M, Pryse-Phillips W, Prinjha R, Richardson J, Saunders A, Slater A, St George-Hyslop P, Stinnett S, Swartz J, Taylor R, Wherrett J, Williams J, Yarnall D, Gibson R, Irizarry M, Middleton L, Roses A. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65(1):45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- Rudd M, Webb E, Matakidou A, Sellick G, Williams R, Bridle H, Eisen T, Houlston R GELCAPS Consortium. Variants in the GH-IGF axis confer susceptibility to lung cancer. Genome Res. 2006;16(6):693–701. doi: 10.1101/gr.5120106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasieni P. From genotypes to genes: doubling the same size. Biometrics. 1997;53(4):1253–1261. [PubMed] [Google Scholar]
- Smyth D, Cooper J, Bailey R, Field S, Burren O, Smink L, Guja C, Ionescu-Tirgoviste C, Widmer B, Dunger B, Savage D, Walker N, Clayton D, Todd J. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38(6):617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall P, Young S. p value adjustment for multiple tests in multivariate binomial models. J Am Stat Soc. 1989;84(407):780–786. [Google Scholar]
- Yeager M, Orr N, Hayes R, Jacobs K, Kraft P, Wacholder S, Minichiello M, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats B, Calle E, Feigelson H, Thun M, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher F, Giovannucci E, Willett W, Cancel-Tassin G, Cussenot O, Valeri A, Andriole G, Gelmann E, Tucker M, Gerhard D, Fraumeni J, Jr., Hoover R, Hunter D, Chanock S, Thomas G. Genome-wide association study of prostate cancer identifies a second risk lcosu at 8q24. Nat Genet. 2007;39(5):645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- Zheng G, Friedlin B, Gastwirth J. Comparison of robust tests for genetic association using case-control studies. IMS Lecture Notes - Monograph Series, 2nd Lehmann Symposium - Optimality. 2006;49:253–265. [Google Scholar]