Abstract
Green tea polyphenols (GTPs) have been reported to inhibit DNA methylation in cultured cells. Here we tested whether oral consumption of GTPs affects normal or cancer specific DNA methylation in vivo, using mice. Wildtype (WT) and Transgenic Adenocarcinoma of Mouse Prostate (TRAMP) mice were administered 0.3% GTPs in drinking water beginning at 4 weeks of age. To monitor DNA methylation, we measured 5-methyl-deoxycytidine (5mdC) levels, methylation of the B1 repetitive element, and methylation of the Mage-a8 gene. Each of these parameters were unchanged in prostate, gut, and liver from WT mice at both 12 and 24 weeks of age, with the single exception of a decrease of 5mdC in the liver at 12 weeks. In GTP-treated TRAMP mice, 5mdC levels and the methylation status of four loci hypermethylated during tumor progression were unaltered in TRAMP prostates at 12 or 24 weeks. Quite surprisingly, GTP treatment did not inhibit tumor progression in TRAMP mice, although known pharmacodynamic markers of GTPs were altered in both WT and TRAMP prostates. We also administered 0.1%, 0.3%, or 0.6% GTPs to TRAMP mice for 12 weeks and measured 5mdC levels and methylation of B1 and Mage-a8 in prostate, gut, and liver tissues. No dose-dependent alterations in DNA methylation status were observed. Genome-wide DNA methylation profiling using the HELP assay also revealed no significant hypomethylating effect of GTP. These data indicate that oral administration of GTPs does not affect normal or cancer-specific DNA methylation in the murine prostate.
Keywords: green tea polyphenols, TRAMP, prostate cancer, DNA methylation, HELP assay
Introduction
Prostate cancer is the most commonly diagnosed cancer and the second most common cause of cancer death in United States men (1). The majority of prostate cancer patients are diagnosed after the age of 65 and prostate cancer tends to be slow growing during the early stages of the disease (1, 2). These characteristics suggest that chemopreventive treatments for men at high risk for prostate cancer may be especially effective at reducing the number of deaths due to this disease.
DNA methylation patterns are altered in many cancers, including prostate cancer (3). A common alteration is de novo hypermethylation of gene promoters, leading to transcriptional silencing (4). Because hypermethylated genes are often tumor suppressors, hypomethylating agents may be viable options for cancer treatment or prevention (4). The most commonly studied hypomethylating agent is the classical DNA methyltransferase (Dnmt) inhibitor 5-aza-2′-deoxycytidine (decitabine) (5). Decitabine is a cytosine analog that is incorporated into DNA and covalently binds to cytosine DNA methyltransferases, depleting the cell of these enzymes and resulting in DNA hypomethylation upon cellular replication (5). However, decitabine treatment also results in DNA damage as it leads to covalent adduct formation from the bound Dnmt proteins (6). This DNA damage causes cytotoxicity in both tumor cells and normal proliferating cells. Although decitabine is approved for the treatment of Myelodysplastic Syndrome and is currently under investigation for the treatment of several types of cancer, it is unlikely to be suitable for chemoprevention. Global DNA hypomethylation is associated with chromosomal instability (7, 8) and for this reason, long-term treatments with potent hypomethylating agents could be problematic. However, it was recently reported that prolonged treatment of Apcmin/+ mice with zebularine, an orally available hypomethylating agent, resulted in minimal side effects and prevented intestinal tumor formation (9).
Studies searching for novel non-toxic DNA methylation inhibitors have resulted in the identification of several naturally occurring compounds that may act through this mechanism (10-14). One such potential DNA methylation inhibitor is green tea (15). In silico studies show that epigallocatechin-3-gallate (EGCG), the main polyphenolic component of green tea, can fit into the binding pocket of DNA methyltransferase 1 (Dnmt1) (11). In addition, several green tea catechins, including catechin and epicatechin, are targets of catechol O-methyltransferase metabolism (13). Therefore, treatment with Green Tea Polyphenols (GTPs) results in decreased S-adenosyl methionine (SAM), the essential cofactor for Dnmts, and increased S-adenosyl homocysteine, a noncompetitive inhibitor of Dnmts (13). Reversal of locus-specific DNA hypermethylation in cancer cell lines after EGCG or GTP treatment has been attributed to each of these mechanisms (11, 13). Further studies demonstrate that although EGCG is the main component of green tea, other GTPs, including catechin and epicatechin, have greater oral bioavailability (16). In addition, data from numerous studies indicate that many of the pharmacological effects of GTPs, which includes multiple polyphenolic components, are not recapitulated by using EGCG alone (17). It is therefore likely that a formulation containing all GTPs would be more effective at inhibiting DNA methylation in vivo than EGCG alone.
Despite the above mentioned studies, the ability of EGCG to inhibit DNA methylation remains controversial, as two published studies have not found evidence for hypomethylating activity using this agent (18, 19). Chuang et al. reported that purified EGCG did not inhibit DNA methylation at single copy loci or repetitive DNA elements in three different human cancer cell lines (18). Similarly, Streseman et al. reported that EGCG treatment did not produce a significant effect on DNA methylation in HCT116 human colorectal cancer cells (19). While these reports clearly show that EGCG is not a potent DNA hypomethylating agent in cancer cell lines, they did not address the potential effects of green tea extracts (GTPs) on DNA methylation, nor did they assess the potential effects of GTPs or EGCG in vivo. Thus, it remains possible that GTPs could affect DNA methylation in a physiological setting.
It has been reported that GTP treatment of TRansgenic Adenocarcinoma of Mouse Prostate (TRAMP) mice inhibits prostate tumorigenesis and metastasis (20, 21). In addition, our previous studies demonstrated that DNA methylation patterns are altered during TRAMP tumor progression, with global DNA hypomethylation beginning at early stages and locus-specific DNA hypermethylation occurring primarily in late stage disease (22-24). Based on these findings and the contrasting in vitro results regarding the hypomethylating activity of GTPs, we sought to test whether oral consumption of green tea extract affects normal or cancer specific DNA methylation in WT or TRAMP mice. We hypothesized that long term consumption of a mixture containing all GTPs could result in DNA hypomethylation in vivo and this may be particularly evident in an environment wherein DNA methylation is already disrupted, as it is in TRAMP tumors. In the present study, we provided GTPs in the drinking water of both wild type (WT) and TRAMP mice and examined normal and cancer specific DNA methylation patterns. We measured 5-methyl-deoxycytidine (5mdC) levels and locus-specific DNA methylation in several normal tissues, including prostate, and in TRAMP prostate tissues. Overall, our data suggest that GTPs do not inhibit DNA methylation in vivo.
Materials and Methods
Animals and Tissues
WT and TRAMP strain matched 50:50 C57Bl/6:FVB mice were bred in the RPCI animal housing facility in accordance with an IACUC approved protocol. Mice were contained in microisolation cages and provided food and control or GTP water ad libitum. Body, prostate and urogenital tract (UG) weights were measured upon sacrifice. Tissue samples were obtained as previously described (23). Tumor and metastatic incidence were determined using a dissecting microscope and palpable tumor incidence was determined by palpating animals directly before sacrifice. Dnmt1 hypomorphic mice (25) (C57Bl/6) were used as a control for DNA hypomethylation and were kindly provided by Dr. Peter Laird (USC). Dnmt1 hypomorph prostate, gut, and liver tissues were microdissected at necropsy. Samples were flash frozen in liquid nitrogen, and stored at −80°C until use.
Green Tea Treatment
Green tea extract was purchased from LKT Laboratories, Inc. (St. Paul, MN). Extracts contained 95% tea polyphenols, including 70% catechins and 35% EGCG. Small amounts of caffeine (3%) were also contained within the mixture. At 0.1% concentration in mouse drinking water this formulation of GTPs approximates human consumption of three cups of green tea per day (21). Freshly prepared green tea solution was provided to animals in their drinking water as the only source of liquid (changed 3 times per week), as previously described (21). The extract powder was kept in an amber bottle and green tea liquid was provided to mice in dark bottles to reduce light exposure. Treatment was started at either 4 or 6 weeks of age and animals were sacrificed at 12, 18, or 24 weeks of age. Animal appearance throughout treatment and final body weights of the mice were examined to confirm that the concentrations used were non-toxic (Supplementary Fig. S1A-B & Supplementary Fig. S2A), and liquid volumes were measured to confirm that mice consumed at least as much green tea as water (Supplementary Table S1). The total volume consumed per cage was measured 3 times a week, consumption per week was divided by the number of animals in each cage to obtain ml liquid consumed/mouse/week (Supplementary Table S1).
Determination of 5-methyl-deoxycytidine (5mdC) levels
5mdC levels were determined using liquid chromatography-electrospray ionization quadrapole mass spectrometry (LC-MS) as described previously (26). Genomic DNAs were isolated using the Puregene DNA isolation kit (Qiagen, Valencia, CA) and 1 μg genomic DNA samples were digested using 4 units of Nuclease S1 (Fermentas, Glen Burnie, MD).
Bisulfite Pyrosequencing
Genomic DNAs were isolated as above and sodium bisulfite conversion was performed using the EZ DNA Methylation Kit (Zymo Research, Orange, CA). Control methylated DNA was produced by M. SssI (New England Biolabs, Ipswich, MA) conversion of mouse genomic DNA. Control unmethylated DNA was produced by two consecutive whole genome amplifications of mouse genomic DNA (Illustra GenomiPhi V2 DNA Amplification Kit, GE Healthcare, Pittsburgh, PA). DNA from prostate, gut, or liver from WT and Dnmt1 hypomorphic mice were also used as positive and negative controls. A bisulfite pyrosequencing assay for the murine B1 element was performed as described previously (23). A pyrosequencing assay for Mage-a8 was performed as described previously (27). Pyrosequencing of the purified single-stranded PCR product was accomplished using the PSQ HS96 Pyrosequencing System (Biotage AB, Uppsala, Sweden). The sequence analyzed for B1 repetitive element contains 2 CpG sites and the sequence analyzed for Mage-a8 contains 6 CpG sites. All samples were analyzed in duplicate and the mean methylation value of all sites were averaged for each sample.
Mass Array Quantitative Methylation Analysis (MAQMA)
Sodium bisulfite converted genomic DNAs were obtained as described above. Primers to analyze methylation of Irx3, Cacna1a, Cdkn2a, and Nrxn2 were designed using Primer3 design software and PCR product fragments were checked for MAQMA compatibility using R-seqmeth as previously described (28). Primer sequences were either published previously or are available upon request (22). MAQMA was performed using the MassARRAY Compact system as described previously (22).
HpaII Tiny Fragment Enrichment by Ligation-Mediated PCR (HELP) assay
We used the high-resolution HELP assay to assess genome-wide changes in DNA methylation due to the administration of GTPs (29). In total, we analyzed four biological samples: 1) a WT normal prostate at 24 weeks of age, following 18 weeks of treatment with 0 % GTP, 2) a WT normal prostate at 24 weeks of age, following 18 weeks of treatment with 0.3 % GTP, 3) a TRAMP prostate tumor at 24 weeks of age, following 18 weeks of treatment with 0 % GTP, and 4) a TRAMP prostate tumor at 24 weeks of age, following 18 weeks of treatment with 0.3 % GTP. The WT samples selected for HELP assay had 5mdC values close to the mean of their respective experimental groups. The TRAMP samples selected for the HELP assay also had 5mdC values close to the mean of their respective experimental groups, and in addition both contained a 60-100 mg primary tumor in the dorsal prostate lobe. After DNA isolation, two separate digestion reactions for each sample, using either HpaII or MspI enzymes, were performed. The resulting products were ligated to a two adaptor set mixture that provided priming sites for ligation-mediated PCR. The samples were then labeled using 9-mer random primers, and conjugated with Cy5 for HpaII and Cy3 for MspI as previously described (30). Both HpaII and MspI representations for each sample were co-hybridized to a custom 2.1 million feature Roche Nimblegen microarray at the Roche-NimbleGen Service Laboratory (Reykjavik, Iceland). Approximately 1 million loci were interrogated by the assay, which provided coverage of >90% of all murine refSeq genes, using the criteria of at least one HpaII fragment within 1kb of the refSeq promoter (29). We assessed hybridization quality and performed data pre-processing, including quantile normalization, as previously described (31). Changes in methylation state were defined using a HpaII/MspI ratio threshold of zero, where hypomethylated loci had a positive ratio reflecting HpaII enrichment, whereas hypermethylated loci had a negative value.
Quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
RNA samples were extracted from mouse tissues and converted to cDNA as described previously (24). PCR reactions were conducted using qPCR SYBR MasterMix (Eurogentec, San Diego, CA) and the 7300 Real Time PCR System (Applied Biosystems, Foster City, CA). Primer sequences for analysis of Clusterin, Ssat, and 18s rRNA were either designed using the Primer3 design software or were previously reported (32). SYBR green absolute quantification analysis was used to determine target gene copy number, which was normalized to 18s rRNA.
Hematoxylin and Eosin (H&E) and Immunohistochemistry Staining
Tissue staining was completed as described previously (23). Briefly, five micron thick tissue sections were cut from paraffin embedded blocks and mounted on slides. Slides were de-parafinized and rehydrated with Xylene and graded alcohol and equilibrated with Tris-phosphate buffer. Samples were stained with H&E or incubated with SV40 T antigen antibody (BD PharMingen, San Diego, CA) for immunohistochemistry with diaminobenzidene staining, dehydrated through alcohol into xylene, and mounted with glass coverslips. Tissue sections were scored for tumor grade using a compound Olympus XI-50 microscope. One slide was analyzed for each mouse in the study as a representative section of prostatic tissue.
Statistical Analyses
All statistical analyses were completed using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA). Significant differences between sample groups for DNA methylation, mRNA expression, and body, prostate and UG weights were determined with a two tailed non-parametric Mann-Whitney test. To test for differences in tumor, palpable tumor, and metastatic incidence, we used the Fisher's Exact Test.
For comparison of pathological grade, a Disease Index was calculated from the percent of each pathological stage determined for each lobe, which was multiplied by a linearly increasing number to represent disease progression. Percent of normal tissue was multiplied by 0, percent Prostatic Intraepithelial Neoplasia (PIN) was multiplied by 1, percent Well Differentiated Tumor (WD) was multiplied by 2, percent Moderately Differentiated Tumor (MD) was multiplied by 3, and percent Poorly Differentiated Tumor (PD) was multiplied by 4 for each lobe of each prostate sample. This resulted in single values for each prostatic lobe for each sample, which were then compared using the Mann-Whitney test (Supplementary Fig. S3).
Results
Effect of GTP consumption on DNA methylation in WT mice
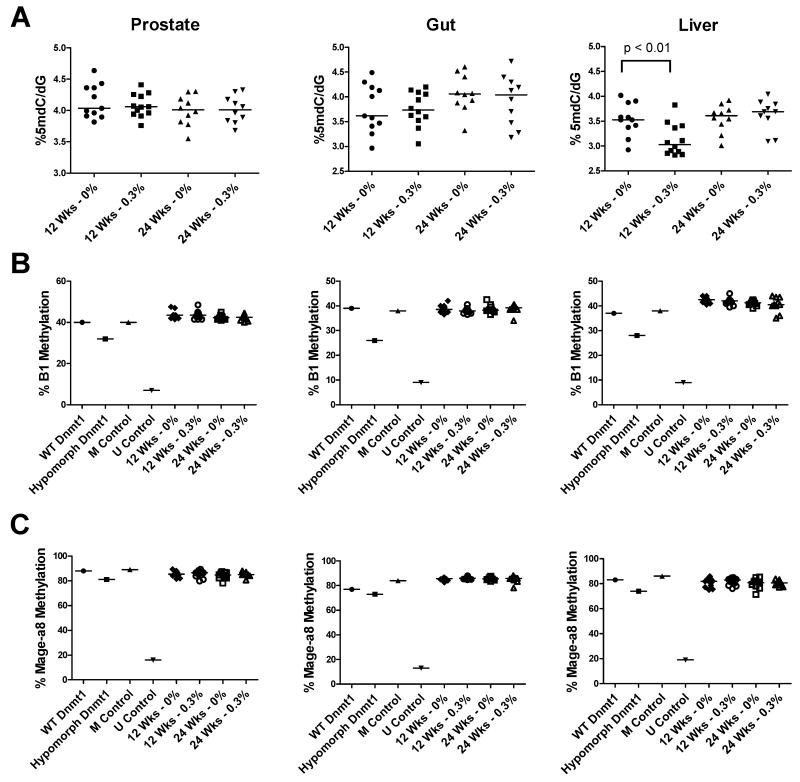
To determine if GTPs alter normal DNA methylation, we treated 10-20 WT mice with either 0% or 0.3% GTPs from 4 weeks to 12 or 24 weeks of age and obtained prostate, gut, and liver tissues for analyses. This concentration of green tea has been used in previous mouse studies and is achievable in the human diet (21). We examined prostate tissue as a control for comparison to the TRAMP tumor model, small intestine (gut) tissue as a normal tissue that has direct exposure to the green tea and high cellular turnover, and normal liver tissue that may also have high exposure as a main site of metabolism of these compounds (16). First, we measured 5mdC levels by LC-MS and found that these were unchanged in prostate or gut tissue following GTP treatment at either time point (Fig. 1A). However, we did observe a significant decrease in 5mdC levels in liver samples from 12 week old GTP-treated mice compared to control, but this effect was not observed at 24 weeks (Fig. 1A).
Fig. 1. Effect of GTP consumption on DNA methylation in wild-type (WT) mice.
A) 5mdC levels in prostate, gut, and liver tissue in control or GTP-treated WT mice at 12 and 24 weeks of age was measured by LC-MS as described in Materials and Methods. B) B1 repetitive element methylation in prostate, gut, and liver tissue in control or GTP-treated WT mice at 12 and 24 weeks of age was measured by bisulfite pyrosequencing as described in Materials and Methods. C) Mage-a8 methylation in prostate, gut, and liver tissue in control or GTP-treated WT mice at 12 and 24 weeks of age by bisulfite pyrosequencing as described in Materials and Methods. Mice were administered GTPs beginning at 4 weeks of age. Organ matched DNA was used from WT or Dnmt1 hypomorphic mice in addition to in vitro methylated (M Control) and unmethylated (U Control) DNA as controls. Each symbol represents an individual sample and the bar indicates the median of each group. Mann-Whitney Test was used to determine significant differences.
We next used bisulfite pyrosequencing to determine whether DNA methylation of two normally methylated loci, the B1 repetitive element and Mage-a8 gene, was altered with GTP treatment. Matched tissue DNA from WT or Dnmt1 hypomorphic mice in addition to in vitro methylated and unmethylated DNA were included as positive and negative assay controls. In spite of being able to clearly measure changes in DNA methylation of Mage-a8 and B1 in the controls, there was no change in methylation of these loci in prostate, gut or liver from WT mice treated with 0.3% GTPs as compared to control mice (Fig. 1B-C). These results suggest that GTP consumption does not alter DNA methylation at these loci in various normal mouse tissues in WT mice.
Effect of GTP consumption on DNA methylation in TRAMP mice
We treated 20-40 TRAMP mice with either 0% or 0.3% GTPs from the age of 4 weeks to 12 or 24 weeks. We chose the time points of 12 and 24 weeks based on a previous study demonstrating the development of early stage disease at 12 weeks and late stage disease at 24 weeks, and our knowledge that DNA methylation is altered in prostates from TRAMP mice at these ages (23, 33).
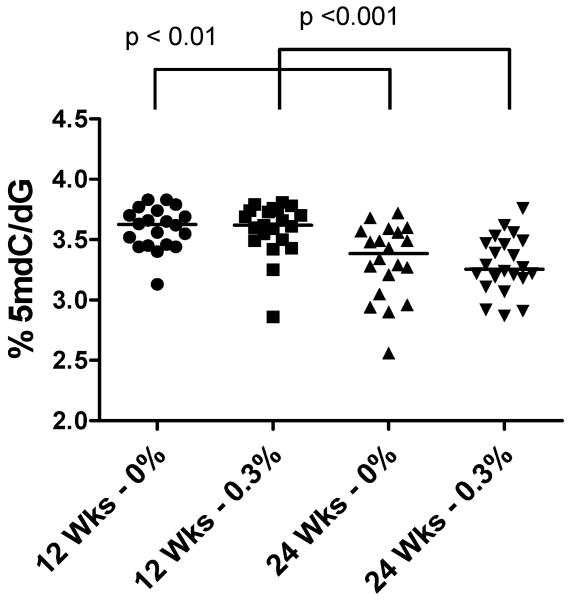
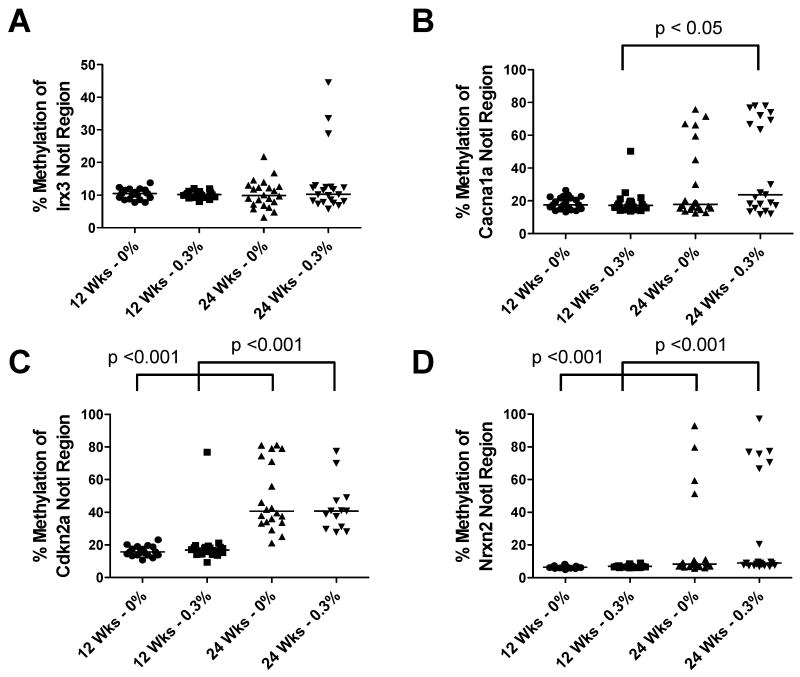
In both experimental groups, 5mdC levels were decreased at 24 weeks of age compared to 12 weeks of age, confirming our previous study (Fig. 2) (23). Notably, 5mdC levels were unchanged in 0.3% GTP-treated TRAMP mice compared to control mice at either 12 or 24 weeks of age (Fig. 2). The possibility remained that GTPs could prevent or reverse aberrant locus-specific hypermethylation, as has been shown in vitro (11, 13). Thus, we next analyzed DNA methylation at four genes (Irx3, Cacna1a, Cdkn2a, and Nrxn2) that we had previously shown become hypermethylated during TRAMP tumor progression (22-24). In TRAMP tumors, Irx3 is hypermethylated near its promoter coinciding with decreased expression, while Cacna1a, Cdkn2a, and Nrxn2 are all hypermethylated downstream of their promoters correlating with increased mRNA expression (22-24). As expected, there was an overall trend of increased DNA methylation in TRAMP prostate samples from 24 week old versus 12 week old mice with several statistically significant changes (Fig. 3A-D). However, there was no difference between samples from control or GTP-treated mice at all four loci (Fig. 3A-D), indicating that GTP consumption does not inhibit cancer-specific hypermethylation of these loci in TRAMP tumors.
Fig. 2. Effect of GTP consumption on 5mdC levels in TRAMP mice.
5mdC levels in prostate from control or GTP-treated TRAMP mice at 12 and 24 weeks of age was determined by LC-MS analysis as described in Materials and Methods. Mice were administered GTPs beginning at 4 weeks of age. Each symbol represents an individual sample and the bar indicates the median of each group. Mann-Whitney Test was used to determine significant differences.
Fig. 3. Effect of GTP consumption on locus-specific DNA hypermethylation in TRAMP mice.
Methylation of Irx3 (A), Cacna1a (B), Cdkn2a (C), and Nrxn2 (D) in prostate tissue from control or GTP-treated TRAMP mice at 12 and 24 weeks of age was determined by MAQMA analysis as described in Materials and Methods. Mice were administered GTPs beginning at 4 weeks of age. Each symbol represents an individual sample and the bar indicates the median of each group. Mann-Whitney Test was used to determine significant differences.
Effect of GTP consumption on prostate cancer in TRAMP mice
GTP treatment of TRAMP mice has previously been reported to inhibit tumor progression (20, 21, 34). We next examined this effect in the same group of TRAMP mice treated with 0.3% GTPs described above. We measured pathological stage, quantified tumor and metastatic incidence, and measured prostate and urogenital tract (UG) weights (normalized to body weight) as indicators of tumor size. To rule out potential artifacts, we also confirmed that GTP treatment did not alter SV40 T antigen (Tag) expression in prostate tissues from TRAMP mice (data not shown).
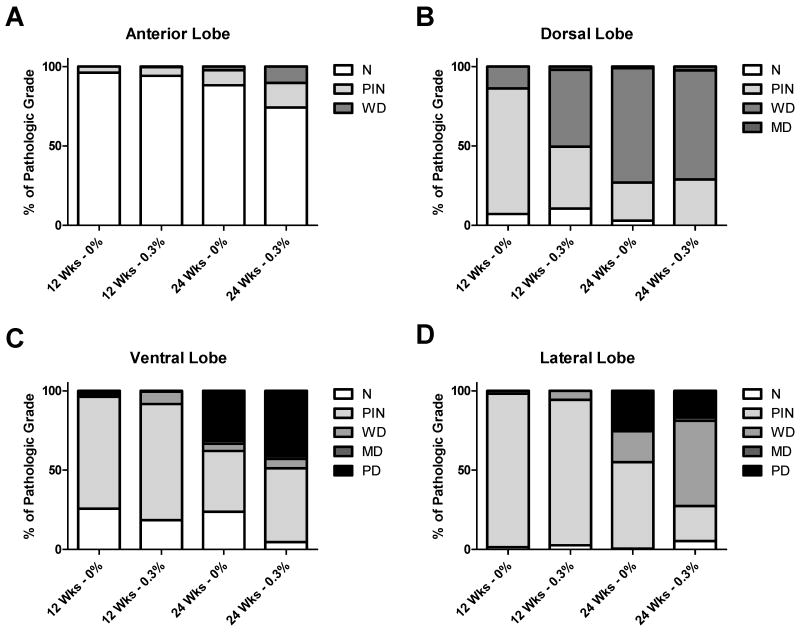
The percent of tissue in each prostatic lobe that was normal (N), prostatic intraepithelial neoplasia (PIN), well differentiated tumor (WD), moderately differentiated tumor (MD), or poorly differentiated tumor (PD) was determined from Hematoxylin and Eosin (H&E) stained slides, as described previously (33). Unexpectedly, compared to control mice, we observed no inhibition of tumor progression with GTP treatment in any of the four prostatic lobes (Fig. 4A-D). In fact, after calculating a Disease Index based on the histological analysis, we observed modest but statistically significant increases in disease progression in prostates from GTP-treated animals 24 weeks of age in the anterior lobe, at 12 weeks of age in the dorsal lobe, with a trend towards significance at 12 weeks of age in the ventral lobe, and no change in the lateral lobe (Supplementary Fig. S3). Furthermore, tumor incidence increased by 12%, palpable tumor incidence increased by 18% and metastatic incidence increased by 10% in 24 week old 0.3% GTP-treated mice compared to control mice, although these differences were not statistically significant (Table 1).
Fig. 4. Effect of GTP consumption on prostate cancer in TRAMP mice.
Microscopic analysis of hematoxylin and eosin staining of prostate tissue from control and GTP-treated TRAMP mice at 12 and 24 weeks of age. Percent of each pathological grade (N-Normal, PIN-Prostatic Intraepithelial Neoplasia, WD-Well Differentiated, MD-Moderately Differentiated, PD-Poorly Differentiated) was determined and averaged for all of the animals in each group for the four lobes of mouse prostate: A) Anterior, B) Dorsal, C) Ventral, and D) Lateral. One section from the prostate of each mouse was analyzed at 40× magnification to score pathological grade.
Table 1. Primary tumor and metastatic incidence in TRAMP mice treated with 0% or 0.3% Green Tea Polyphenols (GTP).
| Treatment Group | Age at sacrifice | Primary tumor incidence† | Palpable tumor incidence † | Metastatic incidence † |
|---|---|---|---|---|
| 0% GTP | 12 weeks | 0/42 (0%) | 0/42 (0%) | 0/42 (0%) |
| 0.3% GTP | 12 weeks | 1/40 (2.5%) | 0/40 (0%) | 0/40 (0%) |
| 0% GTP | 24 weeks | 16/22 (72.7%) | 6/22 (27.3%) | 2/22 (9.1%) |
| 0.3% GTP | 24 weeks | 17/20 (85%) | 9/20 (45%) | 4/20 (20%) |
Fisher's exact test insignificant for differences in tumors, palpable tumors, and metastases between control and treated mice
Analysis of prostate and UG weights relative to body weight in GTP-treated TRAMP mice revealed that both parameters were increased at 12 weeks, but unchanged at 24 weeks, compared to control mice (Supplementary Fig. S1C-D). We also observed an increase in body, prostate, and UG weight in WT mice treated with 0.3% GTPs compared to control (Supplementary Fig. S2A-C). When prostate and UG weights were normalized to body weight, UG weight in 12 week old treated WT mice remained elevated (Supplementary Fig. S2D-E). These data suggest that the increased prostate and UG weights we observed in TRAMP mice treated with GTPs may not be entirely related to disease progression. Nonetheless, green tea consumption clearly did not inhibit TRAMP tumor progression in our experiments.
Mice consume 0.3% green tea extract and known pharmacodynamic markers are altered in the murine prostate after treatment
As part of the above described studies of GTP treatment in WT and TRAMP mice we measured the approximate amount of green tea liquid or water that was consumed/mouse/week to establish that the mice were consuming green tea. Our findings confirmed this and interestingly, both TRAMP and WT mice consumed greater amounts of green tea as compared to water control (an average of 12 ml/mouse/week more) (Supplementary Table S1). Increased consumption of GTP relative to water controls could be related to the increased body weight observed in both TRAMP and WT mice treated with GTP (Supplementary Fig. S1A and S2A).
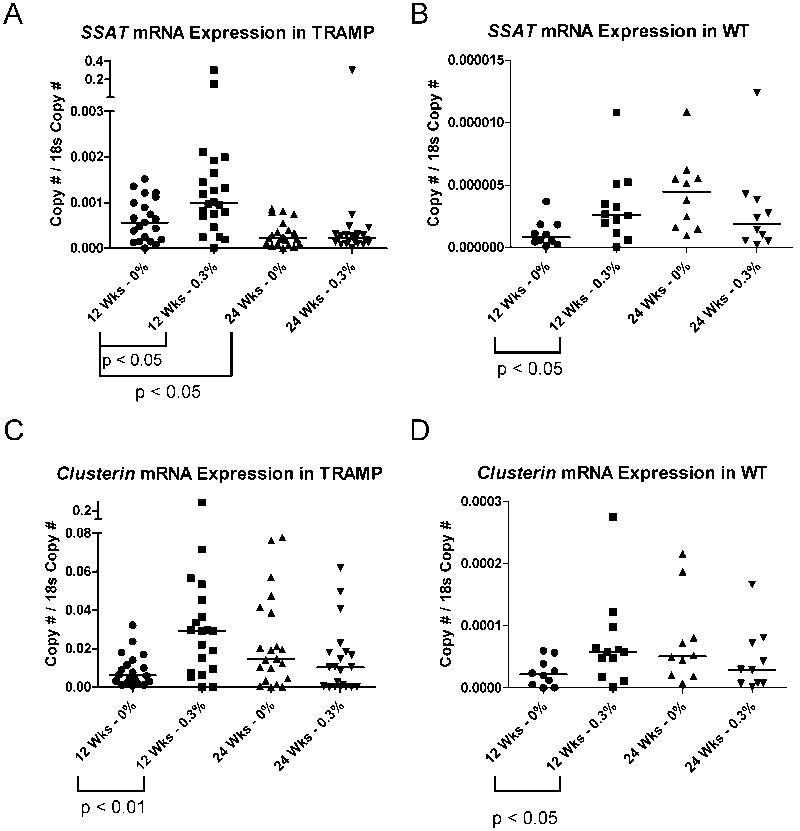
As a positive control for GTP action in the prostate, we measured the expression of two genes that have been shown to be altered with GTP treatment. Previous studies indicate that Ssat mRNA expression decreases during TRAMP tumor progression, but increases upon GTP treatment (32), and Clusterin mRNA levels increase following GTP treatment in TRAMP mice (20, 32). Confirming these data, we found that Ssat expression is decreased in TRAMP prostate tissue from 24 week old mice compared to samples from 12 week old mice (Fig. 5A). Because Ssat has a 5′ CpG island, we investigated the possibility that Ssat downregulation at 24 weeks in TRAMP mice may correlate with promoter DNA hypermethylation. Using MSP assays, we did not observe evidence for hypermethylation of the Ssat promoter in TRAMP (data not shown).
Fig. 5. Ssat and Clusterin are upregulated in TRAMP and WT prostates following 0.3% GTP treatment.
A) Ssat mRNA expression in prostates from either control or GTP-treated TRAMP mice at 12 and 24 weeks of age was measured by qRT-PCR as described in Materials and Methods. B) Ssat mRNA expression in prostates from control or GTP-treated WT mice at 12 and 24 weeks of age was measured by qRT-PCR as described in Materials and Methods. C) Clusterin mRNA expression in prostates from control or GTP-treated TRAMP mice at 12 and 24 weeks of age was measured by qRT-PCR as described in Materials and Methods. D) Clusterin mRNA expression in prostates from control or GTP-treated WT mice at 12 and 24 weeks of age was measured by qRT-PCR as described in Materials and Methods. Mice were administered GTPs beginning at 4 weeks of age. Each symbol represents an individual sample and the bar indicates the median of each group. Mann-Whitney Test was used to determine significant differences.
Notably, Ssat expression was increased in prostate tissue from both TRAMP and WT mice treated with 0.3% GTPs at 12 weeks (Fig. 5A-B). This change was not observed at 24 weeks of age in either TRAMP or WT prostates (Fig. 5A-B). Similar to the response observed with Ssat, Clusterin mRNA expression was increased in prostate tissue from GTP-treated TRAMP and WT mice at 12 weeks, but not at 24 weeks of age (Fig. 5C-D). Overall these data demonstrate that the mice in our studies both consumed the green tea extracts and that GTP consumption altered known pharmacodynamic markers in the prostates of both WT and TRAMP mice at 12 weeks of age.
Consumption of GTPs does not inhibit DNA methylation in normal or tumor tissues in TRAMP mice over a range of GTP concentrations
Because we did not observe significant inhibition of DNA methylation in vivo at a concentration of 0.3% GTPs, we completed an additional small scale study (5-10 mice/treatment group) in which we treated TRAMP mice with 0.1%, 0.3%, and 0.6% GTPs from 6 to 18 weeks of age. Our purpose was to confirm data from the first experiment using 0.3% GTPs, as well as to examine a range of concentrations, including a higher dose of 0.6% GTPs, to thoroughly test whether green tea consumption might inhibit DNA methylation in vivo.
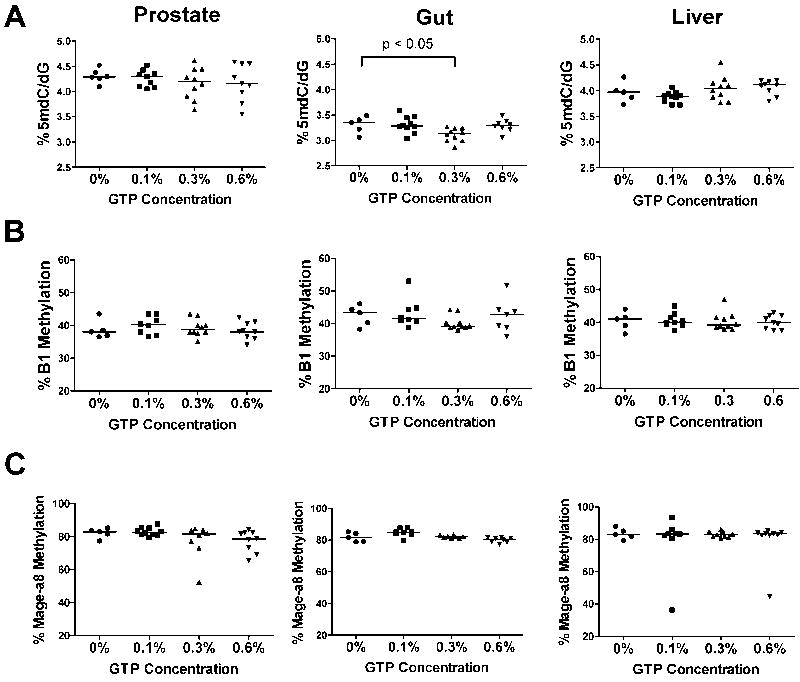
We examined global 5mdC levels and B1 repetitive element and Mage-a8 methylation in prostate, gut, and liver tissues from treated TRAMP mice at 18 weeks of age. Prostate samples displayed increasing variability but no significant directional change of 5mdC levels with increasing GTP dose (Fig. 6A). Gut samples from TRAMP mice treated with GTPs had modestly reduced 5mdC at 0.3% but not at 0.1% or 0.6% concentrations (Fig. 6A). There was no significant change in 5mdC levels in liver samples from GTP-treated mice (Fig. 6A). Some variability was detected in Mage-a8 and B1 repetitive element methylation amongst the three tissues and treatment concentrations examined, but none of these differences were statistically significant (Fig. 6B-C). In addition, we examined whether methylation of CDKN2a in the TRAMP prostate was altered by treatment with the highest concentration of GTPs (0.6%). There was significant hypermethylation of CDKN2a in control-treated mice, and this was unchanged in the 0.6% GTP group (data not shown). In summary, we observed several instances of increased or decreased methylation levels, but no consistent dose-dependent decrease of 5mdC levels or locus-specific DNA methylation with GTP treatment, in any of the examined tissues.
Fig. 6. Effect of GTP consumption on DNA methylation in TRAMP mice over a range of concentrations.
A) 5mdC levels in prostate, gut, and liver tissue from control or GTP-treated TRAMP mice at 18 weeks of age was determined by LC-MS analysis as described in Materials and Methods. B) B1 repetitive element methylation in prostate, gut, and liver tissue from control or GTP-treated TRAMP mice at 18 weeks of age was determined by bisulfite pyrosequencing as described in Materials and Methods. C) Mage-a8 methylation in prostate, gut, and liver tissue from control or GTP-treated TRAMP mice at 18 weeks of age was determined by bisulfite pyrosequencing as described in Materials and Methods. Mice were administered GTPs beginning at 6 weeks of age. Each symbol represents an individual sample and the bar indicates the median of each group. Mann-Whitney Test was used to determine significant differences.
Genome-wide DNA methylation analysis using the HELP assay confirms a lack of DNA hypomethylation in GTP-treated murine prostate
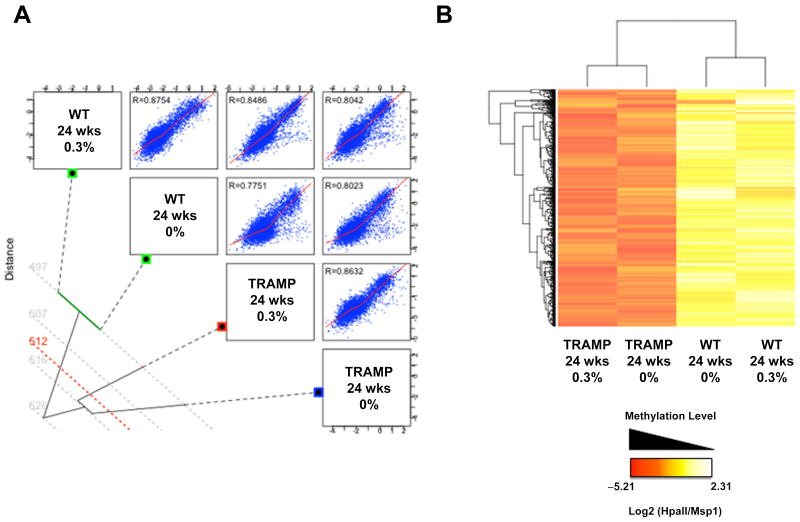
As a final test of the potential in vivo DNA hypomethylating activity of GTP, we utilized the high resolution HELP assay, an epigenomic approach, to measure DNA methylation genome-wide (29, 30). Using HELP, we profiled the methylation status of approximately one million loci in the mouse genome, using custom-made genomic microarrays, as described in Materials and Methods. In total we analyzed four samples (due to the high cost of this approach) including one WT and TRAMP sample each, from both the 0% and 0.3% GTP treatment groups. The criteria for the selection of specific samples are described in Materials and Methods. We initially analyzed HELP data using unsupervised clustering followed by Pearson Correlation testing. We observed that WT samples (either control or GTP-treated) clustered together, as did the TRAMP samples (Fig. 7A). In contrast, WT samples were much less correlated with TRAMP samples, regardless of GTP treatment status (Fig. 7A). We further analyzed HELP data by selecting loci that were hypermethylated in TRAMP samples relative to WT samples for specific analysis (Fig. 7B). The data illustrate that GTP treatment has a negligible global effect on DNA methylation status in either TRAMP or WT prostates (Fig. 7B). While sporadic methylation changes in both directions were apparent, there was not a tendency towards hypomethylation effects in the GTP-treated samples, as would be anticipated for a DNA methylation inhibitor (Fig. 7B).
Fig 7. Genome-wide DNA methylation profiling of GTP-treated normal murine prostate and TRAMP prostate using HELP assays.
WT and TRAMP prostate tissues were collected at 24 weeks of age, following 18 weeks of treatment with either 0% or 0.3% GTPs. Genomic DNAs were harvested and utilized for global DNA methylation analysis at HpaII sites using the high-resolution HELP assay, as described in the Materials and Methods. A) Unsupervised clustering of HELP and global Pairwise (Pearson) correlations is shown. R values indicate the Pearson correlation for each pair. The data illustrate a consistent pattern of methylation between the two WT samples (R=0.8754), as well as the two TRAMP samples (R=0.8632). In contrast, there was divergence in the methylation pattern of the WT samples versus the TRAMP samples (mean R value=0.8075). B) Two-dimensional hierarchical clustering of genes differentially methylated between TRAMP and WT, illustrated by a heatmap. Supervised analysis identified 2035 HpaII-amplifiable fragments with a log2 ratio (HpaII/MspI) of >1.3 between TRAMP and WT. The heat map illustrates 1000 random points selected from this group. Cases are represented in the columns; probe sets are represented in the rows. The data illustrates a high similarity of the DNA methylation pattern in the 0% and 0.3% GTP treatment groups in either sample type (WT or TRAMP), for the loci that are hypermethylated in TRAMP.
Discussion
Recent in vitro studies have examined the separate components of green tea for their ability to inhibit DNA methylation (11, 13, 18, 19), but to our knowledge there have been no previous reports on the effects of GTPs on DNA methylation in vivo. Prior data suggest that distinct polyphenolic components in green tea may each be important for inhibition of DNA methylation and the natural state of green tea may thus be the most effective at hypomethylating DNA (13, 15). In addition, many in vivo mouse studies and all human epidemiological studies are based on consumption of green tea, not the isolated polyphenolic components (21, 35, 36). Therefore, to investigate whether green tea inhibits DNA methylation in vivo as a potential mechanism of action, we used whole green tea extracts administered to mice in drinking water. First, we tested whether consumption of green tea could alter normal patterns of DNA methylation by examining prostate, gut, and liver from WT mice treated with 0.3% GTPs, and also gut and liver from TRAMP mice treated with several concentrations of GTPs. We did find two instances (WT liver at 12 weeks and TRAMP gut at 18 weeks) where 5mdC levels were modestly decreased after 0.3% GTP treatment. However, no DNA hypomethylation was observed in 13 other instances, suggesting that GTPs do not have a significant effect on normal DNA methylation in vivo.
The ability of green tea and its components to reverse aberrant locus-specific DNA hypermethylation in cancer cell lines is controversial (11, 13, 18, 19). Here we examined whether GTP treatment alters either 5mdC levels or tumor specific DNA hypermethylation in a murine tumor model. We did not observe any significant changes in 5mdC levels in prostate tissues from TRAMP mice treated with GTPs. Additionally, there were no instances of hypomethylation of the B1 repetitive element, Mage-a8 gene promoter, or at four specific loci that are commonly hypermethylated in TRAMP. Finally, genome-wide DNA methylation profiling using HELP assays did not reveal significant effects of GTP treatment on DNA methylation; in contrast, dramatic DNA methylation differences were observed between normal and tumor prostate. Overall these data strongly suggest that GTPs do not inhibit DNA methylation in vivo, and particularly in the murine prostate. This supports two previous studies that found negligible DNA hypomethylation effects following treatment of cancer cell lines with EGCG (18, 19). A possible explanation for the contrasting results from some in vitro and our in vivo analyses of the hypomethylating activity of GTPs is that insufficient amounts of polyphenols are retained in prostate, gut, or liver tissues to lead to DNA hypomethylation. This emphasizes the issues of stability and bioavailability of GTPs that limit the use of these compounds as therapeutic agents (37). It remains possible that the DNA hypomethylating activity of GTPs in vivo may be highly tissue- or locus-specific, involving tissues or loci outside of the scope of the current study.
The most unanticipated result of our study was that, contrary to previous reports (20, 21, 34), GTP treatment did not inhibit tumor progression in TRAMP mice, as measured by tumor pathology, incidence, or size. Confirming that GTP was delivered effectively to the prostate in our experiments, we observed increased expression of Ssat and Clusterin in the prostates of GTP treated TRAMP mice, as expected (20, 32). Interestingly, GTP treatment induced these genes at 12, but not 24 weeks of age. This could be due to age-related insensitivity to GTP action in the murine prostate.
Of the seven prior reports characterizing the effects of GTPs in TRAMP (20, 21, 32, 34, 38-40), only three reported effects on tumor incidence or progression relative to untreated mice (20, 21, 34). The other studies focused on molecular parameters and did not report antitumor activity compared to untreated TRAMP mice (32, 38-40). The range of GTP concentrations, and EGCG, used in our study are consistent with the concentrations used in the previous studies (20, 21, 34). However, some differences exist between our studies, the first being the mouse strain. Both our study and Gupta et al. utilized 50:50 C57/Bl6:FVB TRAMP mice, while Caporali et al. and Adami et al. used pure C57/Bl6 TRAMP mice (20, 21, 34). Mouse strain is an important factor in TRAMP tumor development since tumors in the pure C57 background frequently invade into the seminal vesicles, whereas in the 50:50 background the tumors are usually contained within the prostate (41). This may effect the palpability of tumors, which was one major parameter quantified by Caporali et al. (20). A second difference in these studies is variations in the age of the mice at the beginning and end of GTP treatment. In an attempt to maximize potential effects on DNA methylation, we began GTP treatment at 4 or 6 weeks of age when the SV40 transgene starts to be expressed, whereas two other studies began treatment at 8 weeks, several weeks after the transgene has been activated (20, 21). Earlier treatment of TRAMP mice could potentially enhance the selective outgrowth of GTP-resistant tumors. However, the recent report from Adami et al. indicated that the chemopreventive effect of GTP in TRAMP is only fully revealed with treatment beginning at early ages, e.g. 6 weeks (34), supporting the design of our study. Finally, Gupta et al. sacrificed mice at 32 weeks of age (21), while we sacrificed mice at 24 weeks. It is possible that the effects of green tea on prostate tumor progression are manifested most significantly between the ages of 24 and 32 weeks in the C57/Bl6:FVB strain of TRAMP mice. Slight variations in the formulation of GTPs and TRAMP mice colonies may also have affected the outcome on tumor progression. Despite the caveats mentioned above, the data clearly show that the chemopreventive activity of GTPs in the TRAMP model is inconsistent.
It is essential for potential DNA methylation inhibitors to be active in vivo in order to use them to prevent or treat human disease. Here we have examined the effects of green tea polyphenols on DNA methylation using mice. We utilized both WT and prostate tumor model mice to test whether GTP consumption alters normal and/or cancer-specific DNA methylation. We find that GTP consumption does not alter DNA methylation in either setting, or inhibit TRAMP tumor progression, even at high concentrations. Thus, our study does not support the contention that green tea is a DNA methylation inhibitor, and highlights the importance of combining in vitro analyses with in vivo validation studies.
Supplementary Material
A) Body weight in control and 0.3% GTP-treated TRAMP mice at 12 and 24 weeks of age. B) Body weight in control and 0.1%, 0.3%, and 0.6% GTP-treated TRAMP mice at 18 weeks of age. C) Prostate weight normalized to body weight in control and 0.3% GTP-treated TRAMP mice at 12 and 24 weeks of age. D) UG weight normalized to body weight in control and 0.3% GTP-treated TRAMP mice at 12 and 24 weeks of age. Each symbol represents an individual sample and the bar indicates the median of each group. Mann-Whitney Test was used to determine significant differences.
A) Body weight in control and GTP-treated WT mice at 12 and 24 weeks of age. B) Prostate weight in control and GTP-treated WT mice at 12 and 24 weeks of age. C) UG weight in control and GTP-treated WT mice at 12 and 24 weeks of age. D) Prostate weight normalized to body weight in control and GTP-treated WT mice at 12 and 24 weeks of age. E) UG weight normalized to body weight in control and GTP-treated WT mice at 12 and 24 weeks of age. Each symbol represents an individual sample and the bar indicates the median of each group. Mann-Whitney Test was used to determine significant differences.
A) Disease Index values based on pathological grading of the anterior lobe in control and GTP-treated TRAMP mice at 12 and 24 weeks of age. B) Disease Index values based on pathological grading of the dorsal lobe in control and GTP-treated TRAMP mice at 12 and 24 weeks of age. A) Disease Index values based on pathological grading of the ventral lobe in control and GTP-treated TRAMP mice at 12 and 24 weeks of age. A) Disease Index values based on pathological grading of the lateral lobe in control and GTP-treated TRAMP mice at 12 and 24 weeks of age. Disease Index was calculated as described in the Materials and Methods (Statistical Analyses section). Each symbol represents an individual sample and the bar indicates the median of each group. Mann-Whitney Test was used to determine significant differences.
Acknowledgments
We thank Dr. Margot Ip for critical review of the manuscript, Dr. Dominic Smiraglia for assistance with MAQMA primer design, and Dr. Peter Laird (USC Norris Comprehensive Cancer Center) for providing Dnmt1 hypomorphic mice. We thank Dr. Michael Moser for helpful input on mouse treatments, Petra Link for assistance with MSP, and Dr. Masako Suzuki (Albert Einstein College of Medicine) for assistance with HELP assays. This work was supported in part, by the NCI Cancer Center Support Grant to the Roswell Park Cancer Institute CA016056, NIH R21CA128062 (ARK), NIH 5T32CA009072 (SRMK), and DOD PC060354 (SRMK).
References
- 1.NCI. SEER Cancer Statistics Review 1975-2005. 2007 [Google Scholar]
- 2.Nelson PS, Montgomery B. Unconventional therapy for prostate cancer: good, bad or questionable? Nat Rev Cancer. 2003;3:845–58. doi: 10.1038/nrc1210. [DOI] [PubMed] [Google Scholar]
- 3.Nelson WG, Yegnasubramanian S, Agoston AT, et al. Abnormal DNA methylation, epigenetics, and prostate cancer. Front Biosci. 2007;12:4254–66. doi: 10.2741/2385. [DOI] [PubMed] [Google Scholar]
- 4.Gronbaek K, Hother C, Jones PA. Epigenetic changes in cancer. APMIS. 2007;115:1039–59. doi: 10.1111/j.1600-0463.2007.apm_636.xml.x. [DOI] [PubMed] [Google Scholar]
- 5.Ghoshal K, Bai S. DNA methyltransferases as targets for cancer therapy. Drugs Today (Barc) 2007;43:395–422. doi: 10.1358/dot.2007.43.6.1062666. [DOI] [PubMed] [Google Scholar]
- 6.Karpf AR, Jones DA. Reactivating the expression of methylation silenced genes in human cancer. Oncogene. 2002;21:5496–503. doi: 10.1038/sj.onc.1205602. [DOI] [PubMed] [Google Scholar]
- 7.Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–92. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 8.Karpf AR, Matsui S. Genetic disruption of cytosine DNA methyltransferase enzymes induces chromosomal instability in human cancer cells. Cancer Res. 2005;65:8635–9. doi: 10.1158/0008-5472.CAN-05-1961. [DOI] [PubMed] [Google Scholar]
- 9.Yoo C, Chuang JC, Byun H, Egger G, Yang AS, Dubeau L, Long T, Laird PW, Marquez VE, Jones PA. Long-term Epigenetic Therapy with Oral Zebularine Has Minimal Side Effects and Prevents Intestinal Tumors in Mice. Cancer Prevention Research. 2008;1:233–40. doi: 10.1158/1940-6207.CAPR-07-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang MZ, Chen D, Sun Y, Jin Z, Christman JK, Yang CS. Reversal of hypermethylation and reactivation of p16INK4a, RARbeta, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res. 2005;11:7033–41. doi: 10.1158/1078-0432.CCR-05-0406. [DOI] [PubMed] [Google Scholar]
- 11.Fang MZ, Wang Y, Ai N, et al. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–70. [PubMed] [Google Scholar]
- 12.King-Batoon A, Leszczynska JM, Klein CB. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ Mol Mutagen. 2008;49:36–45. doi: 10.1002/em.20363. [DOI] [PubMed] [Google Scholar]
- 13.Lee WJ, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005;68:1018–30. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- 14.Lin RK, Hsu CH, Wang YC. Mithramycin A inhibits DNA methyltransferase and metastasis potential of lung cancer cells. Anticancer Drugs. 2007;18:1157–64. doi: 10.1097/CAD.0b013e3282a215e9. [DOI] [PubMed] [Google Scholar]
- 15.Fang M, Chen D, Yang CS. Dietary polyphenols may affect DNA methylation. J Nutr. 2007;137:223S–8S. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- 16.Henning SM, Choo JJ, Heber D. Nongallated compared with gallated flavan-3-ols in green and black tea are more bioavailable. J Nutr. 2008;138:1529S–34S. doi: 10.1093/jn/138.8.1529S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bode AM, Dong Z. Epigallocatechin 3-gallate and green tea catechins: United they work, divided they fail. Cancer prevention research (Philadelphia, Pa. 2009;2:514–7. doi: 10.1158/1940-6207.CAPR-09-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuang JC, Yoo CB, Kwan JM, et al. Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2′-deoxycytidine. Mol Cancer Ther. 2005;4:1515–20. doi: 10.1158/1535-7163.MCT-05-0172. [DOI] [PubMed] [Google Scholar]
- 19.Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006;66:2794–800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- 20.Caporali A, Davalli P, Astancolle S, et al. The chemopreventive action of catechins in the TRAMP mouse model of prostate carcinogenesis is accompanied by clusterin over-expression. Carcinogenesis. 2004;25:2217–24. doi: 10.1093/carcin/bgh235. [DOI] [PubMed] [Google Scholar]
- 21.Gupta S, Hastak K, Ahmad N, Lewin JS, Mukhtar H. Inhibition of prostate carcinogenesis in TRAMP mice by oral infusion of green tea polyphenols. Proc Natl Acad Sci U S A. 2001;98:10350–5. doi: 10.1073/pnas.171326098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camoriano M, Kinney SR, Moser MT, et al. Phenotype-specific CpG island methylation events in a murine model of prostate cancer. Cancer Res. 2008;68:4173–82. doi: 10.1158/0008-5472.CAN-07-6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morey Kinney SR, Smiraglia DJ, James SR, Moser MT, Foster BA, Karpf AR. Stage-specific alterations of DNA methyltransferase expression, DNA hypermethylation, and DNA hypomethylation during prostate cancer progression in the transgenic adenocarcinoma of mouse prostate model. Mol Cancer Res. 2008;6:1365–74. doi: 10.1158/1541-7786.MCR-08-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morey SR, Smiraglia DJ, James SR, et al. DNA methylation pathway alterations in an autochthonous murine model of prostate cancer. Cancer Res. 2006;66:11659–67. doi: 10.1158/0008-5472.CAN-06-1937. [DOI] [PubMed] [Google Scholar]
- 25.Eads CA, Nickel AE, Laird PW. Complete genetic suppression of polyp formation and reduction of CpG-island hypermethylation in Apc(Min/+) Dnmt1-hypomorphic Mice. Cancer Res. 2002;62:1296–9. [PubMed] [Google Scholar]
- 26.Song L, James SR, Kazim L, Karpf AR. Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal Chem. 2005;77:504–10. doi: 10.1021/ac0489420. [DOI] [PubMed] [Google Scholar]
- 27.Link PA, Gangisetty O, James SR, et al. Distinct roles for histone methyltransferases G9a and GLP in cancer germ-line antigen gene regulation in human cancer cells and murine embryonic stem cells. Mol Cancer Res. 2009;7:851–62. doi: 10.1158/1541-7786.MCR-08-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coolen MW, Statham AL, Gardiner-Garden M, Clark SJ. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements. Nucleic Acids Res. 2007;35:e119. doi: 10.1093/nar/gkm662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oda M, Glass JL, Thompson RF, et al. High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res. 2009;37:3829–39. doi: 10.1093/nar/gkp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khulan B, Thompson RF, Ye K, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome research. 2006;16:1046–55. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson RF, Reimers M, Khulan B, et al. An analytical pipeline for genomic representations used for cytosine methylation studies. Bioinformatics (Oxford, England) 2008;24:1161–7. doi: 10.1093/bioinformatics/btn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scaltriti M, Belloni L, Caporali A, et al. Molecular classification of green tea catechin-sensitive and green tea catechin-resistant prostate cancer in the TRAMP mice model by quantitative real-time PCR gene profiling. Carcinogenesis. 2006;27:1047–53. doi: 10.1093/carcin/bgi287. [DOI] [PubMed] [Google Scholar]
- 33.Kaplan-Lefko PJ, Chen TM, Ittmann MM, et al. Pathobiology of autochthonous prostate cancer in a pre-clinical transgenic mouse model. Prostate. 2003;55:219–37. doi: 10.1002/pros.10215. [DOI] [PubMed] [Google Scholar]
- 34.Adhami VM, Siddiqui IA, Sarfaraz S, et al. Effective prostate cancer chemopreventive intervention with green tea polyphenols in the TRAMP model depends on the stage of the disease. Clin Cancer Res. 2009;15:1947–53. doi: 10.1158/1078-0432.CCR-08-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brausi M, Rizzi F, Bettuzzi S. Chemoprevention of Human Prostate Cancer by Green Tea Catechins: Two Years Later. A Follow-up Update. Eur Urol. 2008;54:472–3. doi: 10.1016/j.eururo.2008.03.100. [DOI] [PubMed] [Google Scholar]
- 36.Kurahashi N, Sasazuki S, Iwasaki M, Inoue M, Tsugane S. Green tea consumption and prostate cancer risk in Japanese men: a prospective study. Am J Epidemiol. 2008;167:71–7. doi: 10.1093/aje/kwm249. [DOI] [PubMed] [Google Scholar]
- 37.Huo C, Wan SB, Lam WH, et al. The challenge of developing green tea polyphenols as therapeutic agents. Inflammopharmacology. 2008;16:248–52. doi: 10.1007/s10787-008-8031-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adhami VM, Siddiqui IA, Ahmad N, Gupta S, Mukhtar H. Oral consumption of green tea polyphenols inhibits insulin-like growth factor-I-induced signaling in an autochthonous mouse model of prostate cancer. Cancer Res. 2004;64:8715–22. doi: 10.1158/0008-5472.CAN-04-2840. [DOI] [PubMed] [Google Scholar]
- 39.McCarthy S, Caporali A, Enkemann S, et al. Green tea catechins suppress the DNA synthesis marker MCM7 in the TRAMP model of prostate cancer. Mol Oncol. 2007;1:196–204. doi: 10.1016/j.molonc.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siddiqui IA, Shukla Y, Adhami VM, et al. Suppression of NFkappaB and its regulated gene products by oral administration of green tea polyphenols in an autochthonous mouse prostate cancer model. Pharm Res. 2008;25:2135–42. doi: 10.1007/s11095-008-9553-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenberg NM, DeMayo F, Finegold MJ, et al. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci U S A. 1995;92:3439–43. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Body weight in control and 0.3% GTP-treated TRAMP mice at 12 and 24 weeks of age. B) Body weight in control and 0.1%, 0.3%, and 0.6% GTP-treated TRAMP mice at 18 weeks of age. C) Prostate weight normalized to body weight in control and 0.3% GTP-treated TRAMP mice at 12 and 24 weeks of age. D) UG weight normalized to body weight in control and 0.3% GTP-treated TRAMP mice at 12 and 24 weeks of age. Each symbol represents an individual sample and the bar indicates the median of each group. Mann-Whitney Test was used to determine significant differences.
A) Body weight in control and GTP-treated WT mice at 12 and 24 weeks of age. B) Prostate weight in control and GTP-treated WT mice at 12 and 24 weeks of age. C) UG weight in control and GTP-treated WT mice at 12 and 24 weeks of age. D) Prostate weight normalized to body weight in control and GTP-treated WT mice at 12 and 24 weeks of age. E) UG weight normalized to body weight in control and GTP-treated WT mice at 12 and 24 weeks of age. Each symbol represents an individual sample and the bar indicates the median of each group. Mann-Whitney Test was used to determine significant differences.
A) Disease Index values based on pathological grading of the anterior lobe in control and GTP-treated TRAMP mice at 12 and 24 weeks of age. B) Disease Index values based on pathological grading of the dorsal lobe in control and GTP-treated TRAMP mice at 12 and 24 weeks of age. A) Disease Index values based on pathological grading of the ventral lobe in control and GTP-treated TRAMP mice at 12 and 24 weeks of age. A) Disease Index values based on pathological grading of the lateral lobe in control and GTP-treated TRAMP mice at 12 and 24 weeks of age. Disease Index was calculated as described in the Materials and Methods (Statistical Analyses section). Each symbol represents an individual sample and the bar indicates the median of each group. Mann-Whitney Test was used to determine significant differences.