Abstract
The mite Psoroptes ovis is the causative agent of sheep scab. Although not usually fatal, the disease can spread rapidly and is a serious animal welfare concern. Vaccine development against ectoparasites has primarily focussed on two sources of candidate vaccine antigens – “exposed” antigens that are secreted in saliva during feeding on a host and “concealed” antigens that are usually expressed in the parasite gut and may be involved in digestion. Here, we sought to identify genes encoding proteins important for mite feeding and digestion by a subtractive suppressive hybridisation approach comparing mRNA transcript abundance in “fed” and “starved” mites. The study identified a variety of genes which are up-regulated by feeding mites. These included group 1, 5, 7 and 13 allergens including the previously described cysteine protease Pso o 1. In addition, numerous novel genes were identified here including some encoding potential salivary gland proteins and others encoding proteins which may facilitate feeding such as a serum opacity factor. An olfactory receptor-like protein was identified in the starved mite population which may help the mite to identify a host.
Keywords: Psoroptes, sheep, gene, protein, feeding
1. INTRODUCTION
The mite Psoroptes ovis is the causative agent of sheep scab which is, arguably, the most important ectoparasitic disease of sheep in the United Kingdom. Disease costs in the UK have been estimated at £8 million per annum [14]. The disease presents as a type I hypersensitivity reaction with the release of serous exudate through the skin, which then dries to from the crusty scabs from which the disease takes its name e.g. [26]. The clinical signs of sheep scab include anorexia, weight loss, loss of fleece and pelt condition, and general loss of production. Although not usually fatal, the disease can spread rapidly and is a serious animal welfare concern.
Disease control has been achieved by annual dipping of sheep in the UK but this is no longer compulsory and the available dips have become more restricted due to risks to operator health and environmental concerns. As a result of these changes, the prevalence of sheep scab has escalated and the disease is now endemic in both hill and lowland sheep in all areas of the British Isles, with an estimated 7 000 outbreaks in 2004 [3].
There have been sustained efforts over the last 10–15 years to develop new methods of control, including vaccination. The feasibility of vaccination is supported by the observation that prior infestation with the mite leads to a suppression of lesion development in subsequent infestations, indicative of a protective immune response [26]. Vaccination of sheep with soluble extracts of P. ovis has resulted in 5-fold reductions in mite numbers and more than 50% reduction in lesion area [23]. After further fractionation of this extract, a sub-fraction was identified which stimulated high titres of mite-specific serum antibodies, inhibited lesion growth to less than a third of that seen in controls and reduced mite numbers by more than 13-fold from 28 days after a challenge infection in vaccinated sheep [24].
Vaccine development against ectoparasites has focussed primarily on two sources of candidate vaccine antigens – “exposed” antigens that are secreted in saliva during feeding on a host and “concealed” antigens that are normally hidden from the host. The latter are usually expressed in the parasite gut and may be involved in feeding, food processing and digestion, the best example being the commercial vaccine based on Bm86, a midgut membrane-bound protein of unknown function in the cattle tick, Rhipicephalus (Boophilus) microplus [28].
A “concealed antigen” approach to P. ovis vaccine development is also a possibility given that ovine IgG was detected on the surface of the gut cells of feeding stages of freshly harvested P. ovis [20] and the digestive enzymes of Psoroptes mites have been proposed as potential vaccine candidates [15].
Currently, little is known about the digestive processes in P. ovis although aminopeptidase, aspartyl and cysteinyl protease activities have been detected [11, 16] and a small expressed sequence tag (EST) survey identified several EST encoding proteases which may be required for feeding [12]. Suppressive subtractive hybridisation (SSH) can be utilised to compare gene expression between two similar populations of cDNA [8] and has been applied widely in parasite biology, for example to identify gender-specific [5, 17] and stage-specific [19] gene expression. Here we have used SSH to compare gene expression between actively feeding and starved P. ovis, in order to identify genes up-regulated during feeding. The protein products of these genes may be essential for mite digestion and, hence, be good vaccine candidates.
2. MATERIALS AND METHODS
2.1. Parasite material
Mites, sufficient to fill a 1.5 mL microfuge tube, were collected from infested sheep fleeces immediately after post mortem and divided into two groups. One group (Fed) was immediately snap frozen in liquid nitrogen, and the other (starved) was transferred to a 500 mL capacity, vented-cap tissue culture flask. This flask was kept at high relative humidity, at 25 °C for 4 days, and then live mites were removed from the tissue culture flask and snap frozen.
2.2. Suppressive subtractive hybridisation
Total RNA was extracted from both groups of mites as described previously [18] and cDNA was synthesised from 2 μg of total RNA from either fed or starved mites using the Super SMART™ PCR cDNA Synthesis Kit and amplified by PCR (15 cycles) according to the kit instructions (Clontech, Europe1). The resultant products were purified using Clontech Chroma-spin™-100 columns. The cDNA was then digested with Rsa-I, adaptors were ligated according to the kit instructions and then the fed mite (tester) cDNA was hybridized with an excess of unfed mite cDNA (driver) to remove common cDNA transcripts from the adapter-ligated fed mite cDNA, generating the “fed mite-library”. The reciprocal procedure was performed using unfed-tester cDNA and fed-driver cDNA to construct an “unfed-library”. The hybridised cDNA populations were then diluted in dH2O, amplified by PCR with primers to the adaptor sequences (Clontech) and the resultant products purified using a QIAquick PCR Purification kit (Qiagen, Crawley, UK2). The products were then ligated into the pGEM®-T Easy cloning vector (Promega, Southampton, UK) and the reaction products used to transform competent Escherichia coli (strain JM109) cells as described in the manufacturer’s protocol (Promega). On the basis of blue/white colour selection, 100 white colonies resulting from the forward subtraction (fed), and 50 colonies from the reverse subtraction (unfed) were picked and inserts were confirmed by colony PCR. The corresponding positive colonies were used to grow 5 mL overnight cultures in Luria Bertani medium (LB) for producing plasmid DNA using a Wizard miniprep kit. Plasmid DNA was then sequenced using a Mega BACE capillary sequencer.
2.3. Sequence analysis
Nucleotide sequences were compared with those in public databases including the GenBank non-redundant database, using the Basic Local Alignment Search Tool (BLASTn and BLASTx, [1]) programme from the National Center for Biotechnology Information. Sequence alignments were performed using CLUSTAL W3.
2.4. Semi-quantitative PCR
Confirmation of cDNA subtraction from the SSH analysis was sought using semi-quantitative reverse transcriptase PCR (RT-PCR). Template RNA from fed or starved mites was transcribed to cDNA and then amplified using a Super Script 1 Step RT-PCR kit (Invitrogen, Renfrew, UK4), and gene specific oligonucleotide primers to target genes identified in the subtraction analyses (primer sequences available from authors on request). PCR reaction mixes comprised 1× reaction mix (Invitrogen; containing 1.2 mM MgSO4 and 0.2 mM of each dNTP), 0.5 μg template RNA, 2 μM forward primer, 2 μM reverse primer, and 200 U of Super Script II reverse transcriptase (Invitrogen). Reactions were incubated at 50 °C for 30 min, followed by 94 °C for 2 min, before 15, 20, 25, and 30 cycles of 94 °C for 15 s, 57.5 °C for 30 s, and 72 °C for 1 min. This was followed by a final extension period of 72 °C for 10 min before cooling to 4 °C. PCR products were analysed by separation on 1% agarose gels containing Gel Red (Biotium, USA) to visualise DNA.
3. RESULTS
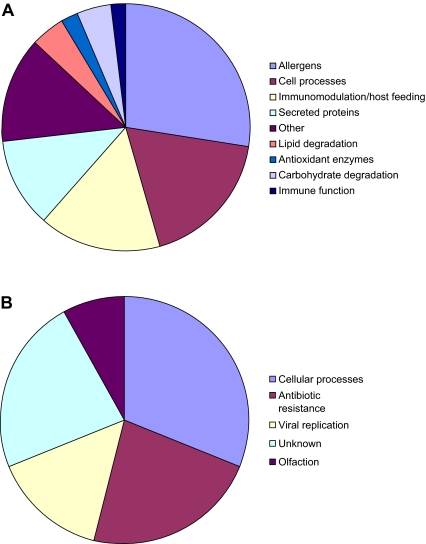
Subtractive hybridisation was applied to identify genes associated with feeding in the sheep scab mite Psoroptes ovis by comparing gene transcription in fed and starved mites. The sequences obtained for each population were submitted to BLAST searches, and the information gathered is summarised in Figures 1A (fed) and 1B (starved); Tables I (fed) and II (starved).
Figure 1.
Classification of proposed functions of genes identified in SSH libraries representing fed (Panel A) and starved (Panel B) sheep scab mites (P. ovis). (A color version of this figure is available at www.vetres.org.)
Table I.
Sequence analysis of clones up-regulated by fed mites. Each sequence was BLAST searched under NCBI and TIGR databases. The E value is a statistical value of the similarity of the two sequences being compared calculated by the BLAST programme [1]. The length of the alignment describes the length (in nucleotides) of the region of similarity between the two sequences.
| Classification | N | Function | Homologue | E value | Identity (%) | Similarity (%) | Length of alignment |
|---|---|---|---|---|---|---|---|
| Allergens | 12 | Pso o I allergen | Psoroptes ovis | 1e-22 | 100 | 100 | 233 |
| Group 5 allergen | Dermatophagoides pteronyssinus | 5e-18 | 61 | 77 | 74 | ||
| Group 7 allergen | Dermatophagoides pteronyssinus | 9e-32 | 61 | 82 | 74 | ||
| Group 13 allergen | Suidasia medanensis | 9e-04 | 47 | 68 | 47 | ||
| Secreted proteins | 5 | Salivary gland protein | Anopheles gambiae | 3e-31 | 92 | 98 | 203 |
| Oesophageal gland protein | Heterodera glycines | 6e-70 | 93 | 93 | 122 | ||
| Salivary gland protein | Ixodes scapularis | 0.012 | 63 | 66 | 35 | ||
| Antioxidant enzymes | 1 | Superoxide dismutase | Taiwanofungus camphorates | 0.004 | 91 | 91 | 24 |
| Carbohydrate metabolism | 2 | Phosphoglycerate kinase | Schistosoma mansoni | 8e-29 | 57 | 74 | 107 |
| Enolase | Glycyphagus domesticus | 6e-45 | 77 | 88 | 131 | ||
| Lipid metabolism | 2 | Phosphatidylethanolamine binding protein | Tribolium castaneum | 1e-38 | 66 | 80 | 109 |
| Fatty acid binding protein | Suidasia medanensis | 9e-04 | 47 | 68 | 47 | ||
| Cell processes | 7 | ATP synthase | Rhipicephalus sanguineus | 9e-22 | 56 | 81 | 47 |
| Methyl transferase | Drosophila pseudo-obscura | 4e-11 | 41 | 61 | 116 | ||
| Cytochrome c oxidase | Archichauliodes sp. | 3e-16 | 45 | 62 | 82 | ||
| Cytochrome monooxygenase | Zea mays | 2e-32 | 45 | 54 | 220 | ||
| Cadherin 1 | Gallus gallus | 8e-05 | 44 | 61 | 58 | ||
| Cadherin-like protein | Rhipicephalus appendiculatus | 4e-50 | 81 | 81 | 128 | ||
| Ribosomal RNA | Ixodes ricinus | 4e-65 | 82 | 82 | 159 | ||
| Other | 4 | Alpha haemolysin homologue | Ixodes scapularis | 1e-40 | 76 | 76 | 213 |
| Serum opacity-like factor | Ixodes scapularis | 2e-20 | 79 | 79 | 100 | ||
| Cell adhesion molecule | Tribolium castaneum | 7e-15 | 45 | 65 | 218 | ||
| Cuticle protein DFP1 | Dermatophagoides pteronyssinus | 2e-11 | 68 | 72 | 86 |
Table II.
Sequence analysis of clones up-regulated in the unfed mite population. Sequences were BLAST searched against the NCBI and TIGR databases. The E value is a statistical value of the similarity of the two sequences being compared calculated by the BLAST programme. The percentage identity is the percentage of identical amino acids between the submitted sequence and the homologue. The percentage similarity is the percentage of similar amino acids shared between the two sequences [1]. The length of the alignment describes the length (in number of amino acids) of the region of similarity between the two sequences.
| Classification | N | Function | Homologue | E value | Identity (%) | Similarity (%) | Length of alignment |
|---|---|---|---|---|---|---|---|
| Cellular processes | 2 | Cadherin 8 | Mus musculus | 6e-04 | 78 | 78 | 66 |
| Signal recognition docking protein | Ixodes scapularis | 2e-06 | 77 | 77 | 103 | ||
| Viral replication | 2 | Matrix protein | Rabies virus | 2e-86 | 98 | 98 | 155 |
| Unknown | 3 | Hypothetical protein | Ixodes scapularis | 2e-74 | 79 | 79 | 189 |
| Hypothetical protein | Plasmodium falciparum | 2e-42 | 89 | 91 | 87 | ||
| Olfaction | 1 | Olfactory receptor-like protein | Ixodes scapularis | 1e-36 | 68 | 68 | 153 |
| Antibiotic resistance | 2 | Beta lactamase | Escherichia coli | 1e-89 | 87 | 22 | 36 |
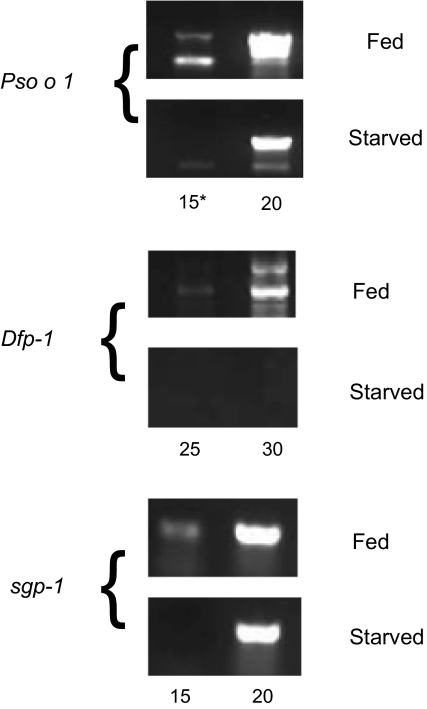
Semi-quantitative RT-PCR (Fig. 2) and dot-blot macroarray analyses (not shown) indicated that the majority of the genes selected were expressed in both populations but at a higher level in the “fed” population. In the fed population, 44 of 100 selected sequences were confirmed, by macroarray, as up-regulated.
Figure 2.
RT-PCR estimates of transcript abundance for 3 genes identified as up-regulated in fed compared to unfed mites by SSH. Dfp-1 was expressed exclusively in fed mites whereas Pso o 1 and the salivary gland protein (sgp-1) were up-regulated in this population compared to the unfed. *Indicates cycle number in semi-quantitative PCR.
A large proportion (30%) of the genes identified in the fed mite dataset were highly homologous to allergens, either from P. ovis itself, or from house dust mites or storage mites. The most abundant transcript in the fed mite dataset was Pso o I, the P. ovis homolog of the house dust mite group 1 allergen, which appeared 8 times. Other allergens identified were homologues of the group 5, group 7, and group 13 allergens.
Several putative secreted proteins were also identified in the fed mite pool, including homologues of proteins localised to the salivary glands of the deer tick, Ixodes scapularis, the mosquito Anopheles gambiae and the oesophageal glands of a nematode Heterodera glycines.
4. DISCUSSION
By using suppressive subtractive hybridisation it has been possible to identify a group of genes that are up-regulated in P. ovis during feeding, and thus which may encode proteins directly involved in digestion. The proposed biological functions of the proteins encoded by these genes expressed by fed mites are illustrated in Figure 1A. Feeding-induced genes included those encoding allergens, secreted proteins, antioxidant enzymes, carbohydrate and lipid degrading enzymes, proteins involved in cellular processes, and also some homologues of blood-meal digesting proteins from ticks and nematodes. The genes up-regulated in starved mites (see Fig. 1B) included one encoding an olfactory receptor protein which may help the mite to locate a host or feeding site or to initiate feeding behaviour.
Some genes were exclusively expressed by fed mites (e.g. a homologue of a cuticle protein Dfp-1 (Fig. 2 and Tab. I) but, for the majority, approximately 5-fold up regulation was evident (e.g. Fig. 2, Pso-1, salivary gland protein). The efficiency of subtractive hybridization depends upon a variety of factors such as driver:tester ratio, or DNA and salt concentrations. Nonetheless, the lack of functional overlap amongst the genes selected in the two populations confirms the utility of SSH for this type of analysis.
A large proportion of the genes identified (30%) in the fed mite population were highly homologous to allergens, either from P. ovis itself, or from house dust mites or storage mites (Fig. 1A and Tab. I). The most abundant EST in the fed mite dataset was Pso o 1, a cysteine proteinase and a homologue of the house dust mite group 1 allergen Der p 1. Pso o 1 is likely to play a part in digestion [11] being expressed in the mite gut and present in mite excretions [18].
Homologues of the group 5 and 7 allergens, two of the most immunoreactive house dust mite allergens [25, 27] were also enriched in fed mites. The functions of these two allergen groups remain undefined, but the group 5 allergen has been localised to the midgut and faecal pellets of the house dust mite Blomia tropicalis [9], possibly indicative of a role in digestion. Moreover, a homologue of the house dust mite group 13 allergen was also enriched in fed mites. Group 13 allergens are fatty acid binding proteins (FABP) [21], which are involved in lipid metabolism and can act as carriers of lipids across membranes. Given that P. ovis mites live in a lipid rich skin environment, the FABP may be required for the supply of essential lipids to the mite. Other parasites, for example nematodes, are unable to synthesise complex lipids and must acquire them from their host [2].
This study identified several transcripts, in the fed mite cDNA pool encoding proteins with a putative glandular origin, implying secretion (Tab. I). These included homologues of a salivary gland protein of the deer tick, Ixodes scapularis [22], a salivary gland protein from Anopheles gambiae and two sequences which were homologous to oesophageal gland proteins from the plant nematode Heterodera glycines. P. ovis does not penetrate the host, and has not been shown conclusively to possess salivary or oesophageal glands, however, a recent report [13], described a putative secreted/excreted peroxiredoxin in P. ovis which was localised to a region immediately posterior to the mouthparts which the authors suggested could be a rudimentary salivary gland.
Several sequences showed high similarity to genes with a possible role in blood meal digestion. One of these, a serum opacity-like factor from the deer tick, I. scapularis, is an orthologue of bacterial apolipoproteinases which makes host blood cells become opaque and binds to mammalian fibronectin and fibrinogen [6, 10]. Scab mites have not been observed to ingest red blood cells, but they have been shown to ingest host eosinophils and immunoglobulins [20]. If these are taken up by the mite and move to its gut then it is possible that other blood cell types are also taken up for digestion and a serum opacity-like factor may play a role in this process.
Alpha-haemolysin-like homologues appeared three times in the fed mite dataset (Tab. I) with two sequences showing homology to sequences from the deer tick I. scapularis and one to a protein from Ascaris suum. Alpha haemolysin is a bacterial virulence factor which lyses red blood cells, and is also cytotoxic to leukocytes and fibroblasts [4]. By analogy to studies in bacteria, these alpha-haemolysins may help the mite to counteract potentially damaging effects which may accrue from the intensive eosinophil infiltration of the infection site in the sheep.
In starved mites, a very limited number of genes were expressed at a sufficiently higher level than in fed. One differentially up-regulated transcript worthy of note was a homologue of an olfactory receptor protein of I. scapularis. Little is known about olfactory receptors in ticks and mites, but in insects they are located on the maxillary palps and antennae [7] and are involved in locating food sources. The mites could use olfactory receptors to help locate the host or a preferred feeding site. This dataset also contained a homologue of a signal peptide docking protein. These proteins act as signal peptide receptors in the rough endoplasmic reticulum and homologues are present in Ixodes spp. EST datasets5. The viral replication proteins show homology to matrix proteins and may play a role in transcription and replication. Alternatively, and perhaps most likely because of the low numbers, these sequences and the antibiotic resistance homologues detected may simply reflect microbial colonisation of mites which is masked in fed mites by up regulation of genes associated with feeding and downstream processes.
To summarise, the genes shown to be up-regulated in actively feeding mites encoded a wide variety of proteins, many of which may potentially be involved in feeding. These included several allergens, secreted proteins (some from salivary glands) and proteins involved in the breakdown of blood cells. On the other hand, genes up-regulated by starved mites encoded mostly housekeeping proteins, with the exception of homologues of an olfactory receptor gene and a viral matrix protein. By using suppressive subtractive hybridisation we have been able to identify several proteins which may be involved in the mite’s digestion, and thus would make interesting candidate vaccine antigens.
Acknowledgments
The authors gratefully acknowledge funding by the Department for Environment Food and Rural Affairs, UK Research Contract OD0544 and by the Scottish Government Rural and Environment Research and Analysis Directorate (RERAD). We are very grateful to John Huntley and David Frew (Moredun Research Institute) for the provision of mites.
Footnotes
References
- 1.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Basic local alignment search tool, J. Mol. Biol. (1990) 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2.Barrett J., Biochemistry of parasitic helminths, University Park Press, Baltimore, MD, 1981 [Google Scholar]
- 3.Bisdorff B., Milnes A., Wall R., Prevalence and regional distribution of scab, lice and blowfly strike in sheep in Great Britain, Vet. Rec. (2006) 158:749–752 [DOI] [PubMed] [Google Scholar]
- 4.Cavalieri S.J., Bohach G.A., Snyder I.S., Escherichia coli alpha-hemolysin: characteristics and probable role in pathogenicity, Microbiol. Rev. (1984) 48:326–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cottee P.A., Nisbet A.J., Abs El-Osta Y.G., Webster T.L., Gasser R.B., Construction of gender-enriched cDNA archives for adult Oesophagostomum dentatum by suppressive subtractive hybridization and microarray analysis of expressed sequence tags, Parasitology (2006) 132:691–708 [DOI] [PubMed] [Google Scholar]
- 6.Courtney H.S., Zhang Y.M., Frank M.W., Rock C.O., Serum opacity factor, a streptococcal virulence factor that binds to apolipoproteins A-I and A-II and disrupts high density lipoprotein structure, J. Biol. Chem. (2006) 281:5515–5521 [DOI] [PubMed] [Google Scholar]
- 7.Dahanukar A., Hallem E.A., Carlson J.R., Insect chemoreception, Curr. Opin. Neurobiol. (2005) 15:423–430 [DOI] [PubMed] [Google Scholar]
- 8.Diatchenko L., Lau Y.F., Campbell A.P., Chenchik A., Moqadam F., Huang B., et al. , Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries, Proc. Natl. Acad. Sci. USA (1996) 93:6025–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y.F., Wang D.Y., Ong T.C., Tay S.L., Yap K.H., Chew F.T., Identification and characterization of a novel allergen from Blomia tropicalis: Blo t 21, J. Allergy Clin. Immunol. (2007) 120:105–112 [DOI] [PubMed] [Google Scholar]
- 10.Jeng A., Sakota V., Li Z., Datta V., Beall B., Nizet V., Molecular genetic analysis of a group A Streptococcus operon encoding serum opacity factor and a novel fibronectin-binding protein, SfbX, J. Bacteriol. (2003) 185:1208–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenyon F., Knox D., The proteinases of Psoroptes ovis, the sheep scab mite -their diversity and substrate specificity, Vet. Parasitol. (2002) 105:317–325 [DOI] [PubMed] [Google Scholar]
- 12.Kenyon F., Welsh M., Parkinson J., Whitton C., Blaxter M.L., Knox D.P., Expressed sequence tag survey of gene expression in the scab mite Psoroptes ovis – allergens, proteases and free-radical scavengers, Parasitology (2003) 126:451–460 [DOI] [PubMed] [Google Scholar]
- 13.McNair C.M., Nisbet A.J., Billingsley P.F., Knox D.P., Molecular characterization, expression and localization of a peroxiredoxin from the sheep scab mite, Psoroptes ovis, Parasitology (2009) 136:453–460 [DOI] [PubMed] [Google Scholar]
- 14.Nieuwhof G.J., Bishop S.C., Costs of the major endemic diseases of sheep in Great Britain and the potential benefits of reduction in disease impact, Anim. Sci. (2005) 81:23–29 [Google Scholar]
- 15.Nisbet A.J., Billingsley P.F., Immunological control of scab mites: digestive enzymes as candidate compounds, Vet. Parasitol. (1999) 83:231–239 [DOI] [PubMed] [Google Scholar]
- 16.Nisbet A.J., Billingsley P.F., A comparative survey of the hydrolytic enzymes of ectoparasitic and free-living mites, Int. J. Parasitol. (2000) 30:19–27 [DOI] [PubMed] [Google Scholar]
- 17.Nisbet A.J., Gasser R.B., Profiling of gender-specific gene expression for Trichostrongylus vitrinus (Nematoda: Strongylida) by microarray analysis of expressed sequence tag libraries constructed by suppression-subtractive hybridisation, Int. J. Parasitol. (2004) 34:633–643 [DOI] [PubMed] [Google Scholar]
- 18.Nisbet A.J., MacKellar A., McLean K., Brennan G.P., Huntley J.F., Eukaryotic expression of recombinant Pso o 1, an allergen from Psoroptes ovis, and its localization, Parasitology (2007) 134:83–89 [DOI] [PubMed] [Google Scholar]
- 19.Nisbet A.J., Redmond D.L., Matthews J.B., Watkins C., Yaga R., Jones J.T., et al. , Stage-specific gene expression in Teladorsagia circumcincta (Nematoda: Strongylida), Int. J. Parasitol. (2008) 38:829–838 [DOI] [PubMed] [Google Scholar]
- 20.Pettit D., Smith W.D., Richardson J., Munn E.A., Localisation and characterisation of ovine immunoglobulin within the sheep scab mite, Psoroptes ovis, Vet. Parasitol. (2000) 89:231–239 [DOI] [PubMed] [Google Scholar]
- 21.Puerta L., Kennedy M.W., Jim N.S., Caraballo L., Structural and ligand binding analysis of recombinant Blo t 13 allergen from Blomia tropicalis mite, a fatty acid binding protein, Int. Arch. Allergy Immunol. (1999) 119:181–184 [DOI] [PubMed] [Google Scholar]
- 22.Ribeiro J.M., Arcon-Chaidez F., Francischetti I.M., Mans B.J., Mather T.N., Valenzuela J.G., Wikel S.K., An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks, Insect. Biochem. Mol. Biol. (2006) 36:111–129 [DOI] [PubMed] [Google Scholar]
- 23.Smith W.D., Bates P., Pettit D.M., van den Broek A., Taylor M.A., Attempts to immunise sheep against the scab mite, Psoroptes ovis, Parasite Immunol. (2002) 24:303–310 [DOI] [PubMed] [Google Scholar]
- 24.Smith W.D., Pettit D.M., Immunisation against sheep scab: preliminary identification of fractions of Psoroptes ovis which confer protective effects, Parasite Immunol. (2004) 26:307–314 [DOI] [PubMed] [Google Scholar]
- 25.Thomas W.R., Smith W., Towards defining the full spectrum of important house dust mite allergens, Clin. Exp. Allergy (1999) 29:1583–1587 [DOI] [PubMed] [Google Scholar]
- 26.Van den Broek A.H.M., Huntley J.F., Machel J., Taylor M.A., Bates P., Miller H.R.P., Cutaneous and systemic responses during primary and challenge infestations of sheep with the sheep scab mite, Psoroptes ovis, Parasite Immunol. (2000) 22:407–414 [DOI] [PubMed] [Google Scholar]
- 27.Weghofer M., Thomas W.R., Pittner G., Horak F., Valenta R., Vrtala S., Comparison of purified Dermatophagoides pteronyssinus allergens and extract by two-dimensional immunoblotting and quantitative immunoglobulin E inhibitions, Clin. Exp. Allergy (2005) 35:1384–1391 [DOI] [PubMed] [Google Scholar]
- 28.Willadsen P., Vaccination against ectoparasites, Parasitology (2006) 133(Suppl.):S9–S25 [DOI] [PubMed] [Google Scholar]