Abstract
CD4+Foxp3+ regulatory T cells (Tregs) maintain peripheral tolerance and influence immune responses to foreign antigens. The thymus is an important source of Tregs, but controversy exists as to whether T cells are selected into the Treg lineage based on signals received through TCRs specific for self-peptides. To examine the specificity of TCRs expressed by Tregs and its effect on CD4+ T cell development, we generated Treg-TCR transgenic mice. Deletion of >90% of CD4+ T cells in RAG sufficient mice, and nearly 100% deletion in RAG−/− mice expressing this TCR indicate that the TCR is specific for an unknown, naturally expressed peptide in the thymus. Deletion occurs late in development, suggesting this peptide is presented by APCs in the thymic medulla. These studies are the first to describe the effects of expressing a Treg-TCR on CD4+ T cell development. The implications of our data for models of Treg selection are discussed.
Keywords: T Cell Receptors, Repertoire Development, Thymus, Tolerance/Suppression/Anergy
Introduction
During the generation of the T cell repertoire, a population of CD4+CD25+Foxp3+ regulatory T cells (Tregs) emerges from the thymus [1]. These cells play a critical role in preventing autoimmunity, and also influence immune responses to foreign antigens. Some of the signals involved in generating and maintaining a functional population of Tregs are now being elucidated, but the role of TCR specificity in Treg development is controversial. One model proposes that Treg development is instructed by TCRs with high avidity for self-peptides presented in the thymus. Studies that report increases in the proportion of Tregs when transgenic T cells are forced to see their cognate antigens in the thymus have been interpreted as evidence for this instructive model [2–7]. The instructive model is also supported by reports of a limited amount of overlap between the TCR sequences expressed within the conventional T cell and Treg repertoire [8–10]. These data have been interpreted as evidence that Treg TCRs are likely to be specific for self-peptides. This model has been challenged by studies that failed to see the induction of CD4+Foxp3+ T cells in TCR/antigen doubly transgenic mice [11, 12], and a recent report that proposes Treg development is initiated in the double negative stage of thymocyte development, well before agonist-ligand induced positive selection occurs [13]. Recent studies also report that a significant amount of overlap does exist between TCR sequences expressed by conventional T cells and Tregs, suggesting that Tregs may also be specific for foreign peptides [10, 14, 15].
In order to gain insight to the specificity of TCRs expressed by Tregs, we isolated and cloned a TCR from a thymic-derived Treg and generated transgenic mice expressing this TCR. Deletion of >90% of CD4+ T cells in RAG sufficient mice, and nearly 100% deletion in RAG1−/− mice expressing this TCR indicates that this TCR is specific for an unknown, naturally expressed self-peptide in the thymus. Transgenic expression of this Treg-TCR led to deletion of CD4+Foxp3− and CD4+Foxp3+ T cells in the CD4SP stage of development. These data are the first to demonstrate that a TCR expressed by a thymic-derived Treg can be specific for an endogenous self-peptides presented in the thymus. How these data relate to current models of Treg development and specificity is discussed.
Results
Identifying and Isolating TCRs expressed by Tregs
It is widely believed that Tregs develop in TCR transgenic RAG1-sufficient mice due to rearrangement of endogenous TCR α–chains that pair with the transgenic TCR β–chain to generate TCRs specific for self-peptides that instruct Treg development. To examine this, we isolated CD4+CD25+ T cells from OTII TCR transgenic mice, and sorted those that express endogenously rearranged TCR Vα3, Vα8, or Vα11 (Fig. 1 A). These particular Vα chains were selected because of the availability of mAbs that allowed us to detect T cells co-expressing these TCR α-chains at their surface.
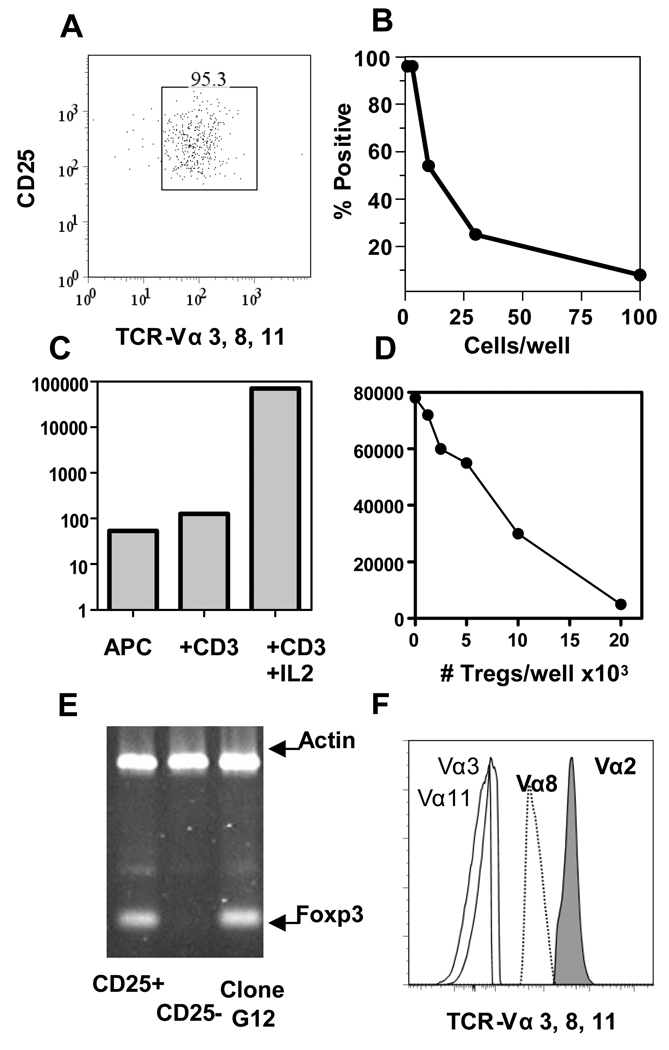
Figure 1. Cloning Tregs and identifying Treg-TCRs.
(A) >95% of the CD4+ T cells express CD25 and TCR Vα3, Vα8, and Vα11 after sorting lymph nodes cells from OTII TCR transgenic mice. (B) Results of plating the sorted cells in limiting dilution and scoring wells for T cell growth. (C) The proliferation of an individual clone, G12, to APCs alone, APCs + anti-CD3, and APCs + anti-CD3 + IL-2. (D) Clone (G12) was tested for the ability to suppress the proliferation of freshly isolated CD4+ T cells responding to anti-CD3. (E) Foxp3 expression by freshly isolated CD4+CD25+ T cells, CD4+CD25− T cells, and the Treg T cell clone G12. (F) The expression of TCR Vα8, and Vα11 was analyzed by flow cytometry on clone G12, along with Vα2 and Vα3 as negative controls.
After sorting, T cells were immediately cloned by limiting dilution. The frequency of sorted T cells that grew under these conditions was between 1 in 20 and 1 in 40 (Fig. 1B). T cell clones were identified in plates in which 37% or less of wells were positive for T cell growth. Individual clones were expanded, and assayed in vitro for a phenotype consistent with Tregs; they were anergic to anti-CD3 stimulation, able to proliferate in response to anti-CD3 with exogenous IL-2, and capable of suppressing the proliferation of freshly isolated CD4+T cells in a dose-dependent manner. Clone G12 was one of many clones that met these criteria (Fig 1C–D). This clone also expressed Foxp3 at a level similar to that of freshly isolated Tregs (Fig. 1E). In addition to expressing the TCR-Vα2 and TCR-Vβ5 encoded by transgenes in OTII mice, this clone also expressed endogenously rearranged TCR-Vα8 (Fig. 1F). We cloned and sequenced the endogenously rearranged TCR Vα8 from cDNA derived from the G12-Treg clone, and used standard techniques to generate TCR transgenic constructs.
Analysis of T Cells in the Periphery of Treg-TCR transgenic mice
TCR transgenic mice that expressed the endogenous TCR Vα8 isolated from the G12 Treg clone along with the TCR Vβ5 from the OTII TCR were generated on the C57BL/6 background (termed Treg-TCR), and backcrossed to RAG1−/− mice (termed Treg-TCRxRAG1−/−) to limit TCR expression to the Treg-TCR. We could then evaluate the effects of expressing a Treg-TCR on CD4+ T cell development.
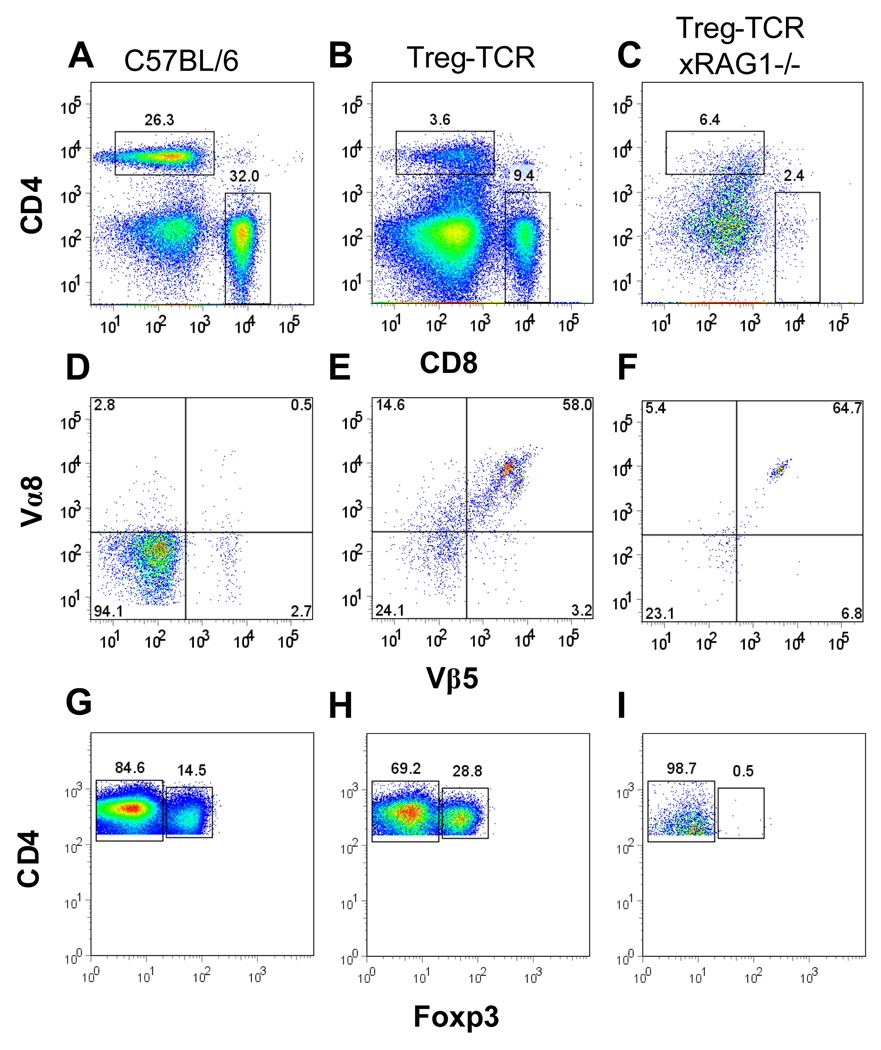
Analysis of spleens from Treg-TCR mice revealed that there are fewer CD4+ T cells present compared to non-transgenic littermate controls (Fig. 2A–B). Furthermore, CD4+ T cells are virtually absent from the spleens of Treg-TCRxRAG1−/− mice (Fig. 2C). CD4+ T cells express the transgenic Treg-TCR Vα8/Vβ5 (Fig 2D–F), and expression of both chains is detectable on Foxp3− and Foxp3+ T cells (data not shown). While the percentage of CD4+ T cells expressing Foxp3 is higher in Treg-TCR mice, Foxp3+ T cells are not detected in Treg-TCRxRAG1−/− mice (Fig. 2G–I). Accounting for cell numbers, CD4+Foxp3− T cells are reduced by 5-fold in Treg-TCR, and >120-fold in Treg-TCRxRAG1−/− mice; CD4+Foxp3+ T cells are reduced by 2-fold in Treg-TCR mice, and >150-fold in Treg-TCRxRAG1−/− mice (Table I). In addition, CD4+ cells from Treg-TCR mice are CD44 high and CD62L low, a phenotype often associated with effector/memory T cells (data not shown). Together these results show that transgenic expression of the TCR isolated from the original Treg-clone leads to a reduction in the numbers of CD4+Foxp3− and CD4+Foxp3+ T cells in the periphery of transgenic mice.
Figure 2. Reduced CD4+Foxp3− and CD4+Foxp3+ T cells in the periphery of Treg-TCR transgenic mice.
(A–C) The proportions of CD4+ and CD8+ T cell in the spleens of C57BL/6, Treg-TCR, and Treg-TCRxRAG1−/− transgenic mice. (D–F) Expression of TCR Vβ5 and TCR Vα8 on CD4+ gated T cells from the spleen. (G–H) Foxp3 expression on CD4+ gated T cells from the spleen. Total numbers of cells are shown in Table I. Results shown are representative of similar analysis performed on 7 to 10 individual mice in each group.
Table I. Reduced numbers of CD4+Foxp−, and CD4+Foxp+ splenocytes and thymocytes in Treg-TCR transgenic mice.
Shown are the mean +/− standard error for the total number of cells in the spleen and thymus of mice from each of the indicated strains. The number of mice in each group is represented by N= in the column title. The total numbers of cells were multiplied by the percentages of CD4+Foxp3− and CD4+Foxp3+ to determine the number of each population in individual mice. N.D. stands for not determined. The cell numbers were compared using a one-way Anova with a Bonferroni post-hoc test at a significance level of 0.05. The Treg-TCR and Treg-TCRxRAG1−/− have a negative effect on the number of CD4+ and Foxp3+ cells compared to the B6 mice. For example, a comparison of the number of thymocytes between the mouse strains suggests that the differences are significant for total thymocytes (F = 5.606, p = 0.019) and highly significant for the CD4+Foxp3− (F = 17.371, p < 0.001) and CD4+Foxp3+ (F = 12.185, p = 0.001). The reduction of CD4+Foxp3− and CD4+Foxp3+ splenocytes of Treg-TCR, Treg-TCRxRAG1−/−, and Treg-TCRxOTII were also highly significant (p <0.05).
| B6 (N=7) |
Treg-TCR (N=7) |
Treg-TCRxRAG1−/− (N=4) |
Treg-TCRxOTII (N=4) |
|
|---|---|---|---|---|
| Total Splenocytes | 63 ×106 +/− 6.1 | 43 ×106 +/− 6.5 | 5.2 ×106 +/− 1.1 | N.D. |
| CD4+Foxp3− | 6.2 ×106 +/− 0.80 | 1.3 ×106 +/− 0.32 | 0.05 ×106 +/− 0.021 | N.D. |
| CD4+Foxp3+ | 7.1 ×105+/− 0.16 | 3.7 ×105 +/− 1.2 | 0.04 ×105+/− 0.0001 | N.D. |
| Total Thymyocytes | 99 ×106 +/− 8.0 | 52 ×106 +/− 7.4 | 78 ×106 +/− 12.5 | 65 ×106 +/− 6.1 |
| CD4+Foxp3− | 61.0 ×105 +/− 12.0 | 2.3 ×105 +/− 0.75 | 1.9 ×105 +/− 0.47 | 3.5 ×105+/− 0.40 |
| CD4+Foxp3+ | 1.9 ×105+/− 0.37 | 0.41 ×105 +/− 0.17 | 0.068 ×105+/− 0.0037 | 0.042 ×105+/− 0.0021 |
Thymocyte Development in Treg-TCR Transgenic Mice
We next examined how the expression of this Treg-TCR influenced T cell development in the thymus. The thymi from Treg-Treg-TCR mice, TCRxRAG1−/−, and negative littermates as controls are similar in size and only modest differences are observed in the cellularity in the three groups of mice (Table I). Despite this similarity, a ~7-fold reduction in the percentage of CD4 single positive thymocytes (CD4SP) is observed in Treg-TCR mice, and ~20-fold reduction is seen in TCRxRAG1−/− mice (Fig. 3A–C). The number of CD4+Foxp3− T cells is reduced by ~25-fold, and although the percentage of CD4+Foxp3+ T cells is increased in Treg-TCR mice (Fig. 3A–B), the absolute number of CD4+Foxp3+ T cells is reduced by ~5-fold and (Table I). A very small percentage of the CD4SP cell express Foxp3 in Treg-TCRxRAG1−/− mice, and the numbers of CD4+Foxp3− and CD4+Foxp3+ T cells are reduced by >25-fold (Table I). Expression of TCR Vα8/Vβ5 was detected on both Foxp3− and Foxp3+ T cells, and a majority of CD4SP thymocytes expressed high levels of CD44 (data not shown). These data demonstrate that this particular TCR recognizes a self-peptide that is naturally expressed and presented in the thymus, and that transgenic expression of this TCR leads to deletion of CD4+Foxp3− and CD4+Foxp3+ thymocytes.
Figure 3. Deletion of CD4+Foxp3− and CD4+Foxp3+ thymocytes in Treg-TCR transgenic mice.
The expression of CD4 and CD8 from (A) negative control littermates (C57BL/6), (B) Treg-TCR transgenic mice, and (C) Treg-TCRxRAG1−/− (D) Treg-TCRxOTII, and (E) OTII TCR transgenic mice. Expression of CD25 and Foxp3 on CD4+CD8− gated thymocytes from (E) negative control littermates, (F) Treg-TCR transgenic mice, (G) Treg-TCRxRAG1−/− mice, (H) Treg-TCRxOTII, and (I) OTII TCR transgenic mice. Total numbers of thymocytes are shown in Table I. Results are representative of similar analysis of between 4 and 10 individual mice in each group.
Since the original Treg clone (G12) also expressed OTII-derived TCR-Vα2 (Fig. 1F), we crossed the Treg-TCR mice to OTII TCR transgenic mice to generate mice expressing both the OTII TCR Vα2/Vβ5 and the Treg-TCR Vα8/Vβ5 TCRs. FACS analysis confirmed that thymocytes from double TCR transgenic mice expressed similar levels of both the OTII-TCR and Treg-TCR (data not shown). Thymocytes from OTII single transgenic mice contain a large percentage of CD4SP thymocytes, as this TCR is known to positively select CD4+ T cells on the C57BL/6 background (Fig. 3E). The total number of thymocytes is slightly increased in Treg-TCRxOTII mice, but the percentage of CD4SP is reduced to 1% compared to 35% in OTII mice (Fig. 3D–E). As the proportion of CD4+Foxp3+ T cells is similar between the OTII and Treg-TCRxOTII mice (Fig. 3H–I), the 35-fold decrease in CD4SP thymocytes leads to similar reductions in the absolute numbers of both CD4+Foxp3− T cells and CD4+Foxp3+ T cells in Treg-TCRxOTII mice compared to OTII TCR transgenic mice (Table I).
TCR and Foxp3 Expression by Treg-TCR Thymocytes
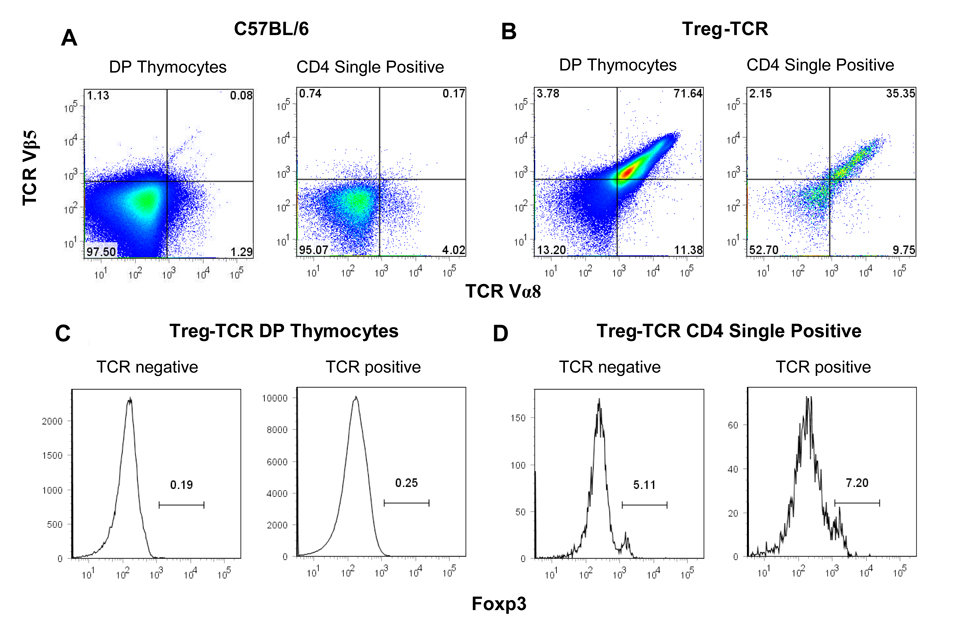
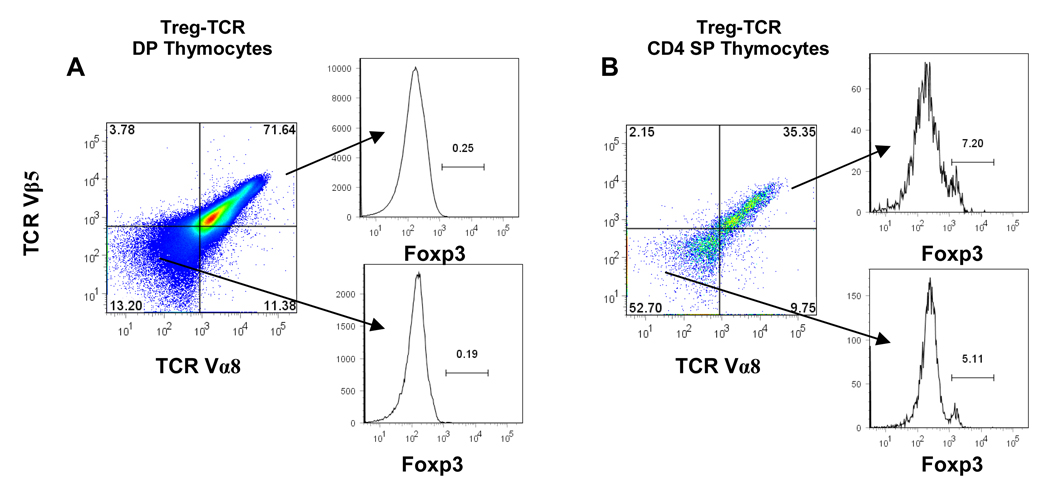
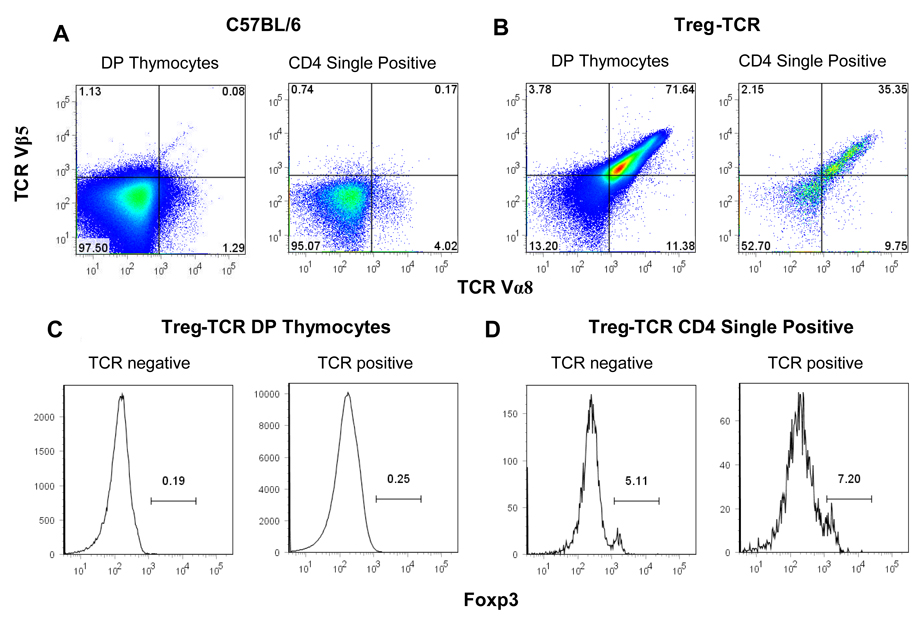
While a large fraction of thymocytes are deleted in Treg-TCR mice, we wanted to determine whether the T cells that escaped negative selection expressing the transgenic TCR preferentially expressed Foxp3. A majority (~70%) of the CD4+CD8+ (DP) thymocytes express both TCR Vα8 and TCR Vβ5 (Figure 4A). As expected, very few of the DP thymocytes express Foxp3. A lower proportion of the CD4 single positive (CD4SP) thymocytes express the transgenic TCR Vα8 and TCR Vβ5. In fact, ~50% of the CD4SP thymocytes to not express either of the transgenic TCRs, although staining for total TCR shows uniform staining with TCR levels only slightly lower than non-transgenic thymocytes (data not shown), indicating that the Treg-TCR intermediate and low thymocytes are also expressing endogenously rearranged TCR chains (Fig. 4B and data not shown). Thus, there is very little correlation between the expression of this autoreactive Treg-TCR, and the expression of Foxp3.
Figure 4. TCR and Foxp3 expression in CD4+CD8+ double positive (DP), and CD4 single positive (SP) thymocytes from Treg-TCR transgenic mice.
The expression of the transgenic TCR Vα8 and TCR Vβ5 on DP gated thymocytes (A), and CD4 SP gated thymocytes (B) from Treg-TCR transgenic mice. Foxp3 expression is shown after gating on the TCR Vα8/Vβ5 high, intermediate, and low DP thymocytes (A) and CD4SP thymocytes (B). Results are represented of 7 individual mice in each group.
Discussion
Expression of this Treg-TCR as a transgene results in deletion of CD4 SP thymocytes, demonstrating that this TCR is specific for a self-peptide that is naturally processed and presented in the thymus. Deletion of Treg-TCR expressing CD4+ T cells appears to occur late in thymic development, as the number of total thymocytes is only modestly reduced in the transgenic mice, and the decrease is most significant for CD4SP thymocytes. In addition, double positive thymocytes expressing the Treg-TCR are not reduced in number. The fact that deletion occurs late in thymic development makes it likely that this Treg-TCR is specific for a peptide presented by an APC residing in the thymic medulla. One possibility is that this TCR recognizes a tissue specific peptide presented by medullary thymic epithelial cells, which have been shown to express tissue restricted antigens in an Aire dependent manner [16–18]. Identifying the thymic APC and the self-antigen recognized by this TCR is the focus of ongoing experiments.
Although we observed negative selection in Treg-TCR transgenic mice, expressing this self-reactive Treg-TCR did not enhance Treg development. Why is it that expressing this Treg TCR did not enhance the development of CD4+Foxp3+ T cells? One possibility is that Treg development is not induced by the signals received through a self-reactive TCR. It has been proposed that self-reactive Tregs are less sensitive to negative selection than self-reactive conventional T cells [11]. If this is the case, it is possible that the original clone was instructed into the Treg lineage by signals unrelated to the TCR, and it survived negative selection with the lower levels of TCR than is expressed in the TCR transgenic T cells. Our data showing a lack of Treg induction, and a lack of correlation between the expression of this autoreactive TCR and Foxp3 demonstrate that this self-reactive Treg-TCR is not sufficient to enhance Treg development when expressed as a transgene in mice. On the other hand, we cannot rule out the possibility that the level of TCR expression in the transgenic mice is higher than in the original clone, and thymocytes from the transgenic mice receive a stronger signal than the original clone, which causes deletion instead of Treg induction. While this is possible, it is interesting that the CD4 SP thymocytes that escaped negative selection expressing low levels of the TCR, did not have a higher frequency of Foxp3+ than the TCR− CD4 SP thymocytes. Experiments are underway to test several more TCRs isolated from Tregs, and examine whether they are specific for self-peptides in the thymus, and whether they induce the development of CD4+Foxp3+ T cells.
At present, the percentage of Tregs that express TCRs specific for self-peptides compared to the percentage of Treg that express TCRs specific for non-self peptides is not known. The instructive models predict that all TCRs examined will manifest signs of autoreactivity (negative selection or Treg induction in TCR transgenic mice); while the non-instructive, stochastic models predict that a proportion of the TCRs specific for foreign peptides, and will not be deleted and will not induce Treg-development. Determining whether specificity of Treg-TCRs is crucial to understanding how they regulate immune responses to foreign and self-antigens. Furthermore, before cellular biotherapy with Tregs can be used as a potential treatment of autoimmune diseases, it will be useful to understand whether self-reactive Tregs are specific for ubiquitous self-antigens, and suppress in a bystander fashion, or whether they are specific for tissue-specific antigens, and suppress in an antigen-specific manner.
Materials and Methods
Mice, antibodies and reagents
OTII TCR transgenic mice have been described previously and were purchased from Taconic [19]. C56BL/6 mice, and C57BL/6xRAG1−/− mice were purchased from the National Cancer Institute animal facility (Frederick, MD). All mice were housed under specific pathogen free conditions in our animal facility and cared for in accordance with NIH institutional guidelines.
Antibodies were purchased from BD Pharmingen (San Diego, CA), with the exception of anti-CD4-TC (Tri-color), which was purchased from Caltag Laboratories (Burlingame, CA), and anti-FoxP3-APC (FJK-16s), which was purchased from eBioscience. Human recombinant IL-2 was purchased from Peprotech (Rockyhill, NJ). DTA-1 (anti-GITR) was purified on a protein G Sepharose column. Flow cytometry was performed on BD FACScan or FACSCaliber and analyzed using CellQuest or FlowJo software (Beckton Dickinson).
Limiting dilution assays and in vitro assays
CD4+CD25+TCR Vα3+α8+α11+ T cells were sorted from OTII TCR transgenic mice. T cells were plated after sorting ranging from 300 to 0.3 cells/well in 96 well U-bottom plates in the presence of 5×105 T-depleted irradiated splenocytes, 0.5 µg/mL anti-CD3, 50 Units/mL rhu-IL-2, and 1 µg/mL of DTA-1. Each well was visually examined and scored for growth on day 7. Growth positive wells were expanded and assayed for proliferation to 5×105 T-depleted irradiated splenocytes with or without 0.5 µg/mL of anti-CD3 and 50 units/mL of rhu-IL-2. In the suppression assays, CD4+ T cells (5×104) from C57BL/6 mice were incubated with irradiated T-depleted spleen cells (5×104) and 0.25 µg/mL of anti-CD3 for 3 days in the presence of varying numbers of T cell clones.
PCR
Expression of Foxp3 mRNA was analyzed by PCR. cDNA was prepared using Superscript III (Invitrogen) using random primers, and 0.5 ug of total RNA from individual clones, and sorted populations of CD4+CD25− and CD4+CD25+ Tregs.
DNA constructs and generation of Treg-TCR transgenic mice
cDNA was prepared from total RNA extracted from 2×106 G12 T cell clones. The Vα segment of the G12 TCR receptor was identified by PCR using a Cα complementary primer and primers complementary to various TCR Vα chains. PCR products were sequenced, and the TCR was identified as TRAV12-1. A CD2 based expression vector was used to express the TRAV12-1 [20]. A construct containing the full length cDNA for the TCR Vβ5 expressed by OTII TCR transgenic mice was a generous gift from Dr. Francis Carbone. The generation and screening of transgenic mice was performed at the National Institute of Allergy and Infectious Diseases transgenic facility (Frederick Cancer Research and Development Center, Frederick, MD).
ACKNOWLEDGEMENTS
This work was supported by intramural NIH funds. We would like to thank Dr. Ronald Schwartz for helpful discussions, Lionell Feigenbaum for generating the TCR transgenic mice, and Kanaan Natarajan, Karen Bijwaard, Mark Ebel, Sherri Koehm, and Sarah Tanksley for technical assistance. We also thank Maureen Donlin for help with statistics, and Dr. Carbone for providing the OTII TCR Vβ5 transgenic construct. This research was supported by the Intramural Research Program of the NIH, NIAID.
Abbreviations
- Tregs
regulatory T cells
- TCR
T cell receptor
- SP
single positive
- DP
double positive
- RAG1−/−
Recombinase-Activating Gene 1 knockout
Footnotes
The authors declare no financial or commercial conflict of interest.
References
- 1.Fontenot JD, Dooley JL, Farr AG, Rudensky AY. Developmental regulation of Foxp3 expression during ontogeny. J Exp Med. 2005;202:901–906. doi: 10.1084/jem.20050784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apostolou I, Sarukhan A, Klein L, von Boehmer H. Origin of regulatory T cells with known specificity for antigen. Nat Immunol. 2002;3:756–763. doi: 10.1038/ni816. [DOI] [PubMed] [Google Scholar]
- 3.Jordan MS, Boesteanu A, Reed AJ, Petrone AL, Holenbeck AE, Lerman MA, Naji A, Caton AJ. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001;2:301–306. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 4.Kawahata K, Misaki Y, Yamauchi M, Tsunekawa S, Setoguchi K, Miyazaki J, Yamamoto K. Generation of CD4(+)CD25(+) regulatory T cells from autoreactive T cells simultaneously with their negative selection in the thymus and from nonautoreactive T cells by endogenous TCR expression. J Immunol. 2002;168:4399–4405. doi: 10.4049/jimmunol.168.9.4399. [DOI] [PubMed] [Google Scholar]
- 5.Larkin J, 3rd, Rankin AL, Picca CC, Riley MP, Jenks SA, Sant AJ, Caton AJ. CD4+CD25+ Regulatory T Cell Repertoire Formation Shaped by Differential Presentation of Peptides from a Self-Antigen. J Immunol. 2008;180:2149–2157. doi: 10.4049/jimmunol.180.4.2149. [DOI] [PubMed] [Google Scholar]
- 6.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burchill MA, Yang J, Vang KB, Moon JJ, Chu HH, Lio CW, Vegoe AL, Hsieh CS, Jenkins MK, Farrar MA. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28:112–121. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsieh CS, Liang Y, Tyznik AJ, Self SG, Liggitt D, Rudensky AY. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21:267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 9.Pacholczyk R, Ignatowicz H, Kraj P, Ignatowicz L. Origin and T cell receptor diversity of Foxp3+CD4+CD25+ T cells. Immunity. 2006;25:249–259. doi: 10.1016/j.immuni.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 10.Wong J, Obst R, Correia-Neves M, Losyev G, Mathis D, Benoist C. Adaptation of TCR repertoires to self-peptides in regulatory and nonregulatory CD4+ T cells. J Immunol. 2007;178:7032–7041. doi: 10.4049/jimmunol.178.11.7032. [DOI] [PubMed] [Google Scholar]
- 11.van Santen HM, Benoist C, Mathis D. Number of T reg cells that differentiate does not increase upon encounter of agonist ligand on thymic epithelial cells. J Exp Med. 2004;200:1221–1230. doi: 10.1084/jem.20041022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shih FF, Mandik-Nayak L, Wipke BT, Allen PM. Massive thymic deletion results in systemic autoimmunity through elimination of CD4+ CD25+ T regulatory cells. J Exp Med. 2004;199:323–335. doi: 10.1084/jem.20031137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pennington DJ, Silva-Santos B, Silberzahn T, Escorcio-Correia M, Woodward MJ, Roberts SJ, Smith AL, Dyson PJ, Hayday AC. Early events in the thymus affect the balance of effector and regulatory T cells. Nature. 2006;444:1073–1077. doi: 10.1038/nature06051. [DOI] [PubMed] [Google Scholar]
- 14.Pacholczyk R, Kern J, Singh N, Iwashima M, Kraj P, Ignatowicz L. Nonself-antigens are the cognate specificities of Foxp3+ regulatory T cells. Immunity. 2007;27:493–504. doi: 10.1016/j.immuni.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fazilleau N, Bachelez H, Gougeon ML, Viguier M. Cutting edge: size and diversity of CD4+CD25high Foxp3+ regulatory T cell repertoire in humans: evidence for similarities and partial overlapping with CD4+CD25− T cells. J Immunol. 2007;179:3412–3416. doi: 10.4049/jimmunol.179.6.3412. [DOI] [PubMed] [Google Scholar]
- 16.Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Aire regulates negative selection of organ-specific T cells. Nat Immunol. 2003;4:350–354. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- 17.Aschenbrenner K, D'Cruz LM, Vollmann EH, Hinterberger M, Emmerich J, Swee LK, Rolink A, Klein L. Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat Immunol. 2007;8:351–358. doi: 10.1038/ni1444. [DOI] [PubMed] [Google Scholar]
- 18.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 19.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhumabekov T, Corbella P, Tolaini M, Kioussis D. Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J Immunol Methods. 1995;185:133–140. doi: 10.1016/0022-1759(95)00124-s. [DOI] [PubMed] [Google Scholar]