Abstract
Microarray data has a high dimension of variables but available datasets usually have only a small number of samples, thereby making the study of such datasets interesting and challenging. In the task of analyzing microarray data for the purpose of, e.g., predicting gene-disease association, feature selection is very important because it provides a way to handle the high dimensionality by exploiting information redundancy induced by associations among genetic markers. Judicious feature selection in microarray data analysis can result in significant reduction of cost while maintaining or improving the classification or prediction accuracy of learning machines that are employed to sort out the datasets. In this paper, we propose a gene selection method called Recursive Feature Addition (RFA), which combines supervised learning and statistical similarity measures. We compare our method with the following gene selection methods:
Support Vector Machine Recursive Feature Elimination (SVMRFE)
Leave-One-Out Calculation Sequential Forward Selection (LOOCSFS)
Gradient based Leave-one-out Gene Selection (GLGS)
To evaluate the performance of these gene selection methods, we employ several popular learning classifiers on the MicroArray Quality Control phase II on predictive modeling (MAQC-II) breast cancer dataset and the MAQC-II multiple myeloma dataset. Experimental results show that gene selection is strictly paired with learning classifier. Overall, our approach outperforms other compared methods. The biological functional analysis based on the MAQC-II breast cancer dataset convinced us to apply our method for phenotype prediction. Additionally, learning classifiers also play important roles in the classification of microarray data and our experimental results indicate that the Nearest Mean Scale Classifier (NMSC) is a good choice due to its prediction reliability and its stability across the three performance measurements: Testing accuracy, MCC values, and AUC errors.
Introduction
Using microarray techniques, researchers can measure the expression levels for tens of thousands of genes in a single experiment. This ability allows scientists to investigate the functional relationship between the cellular and physiological processes of biological organisms and genes at a genome-wide level. The preprocessing procedure for the raw microarray data consists of background correction, normalization, and summarization. After preprocessing, a high level analysis, such as gene selection, classification, or clustering, is applied to profile the gene expression patterns [1]. In the high-level analysis, partitioning genes into closely related groups across time and classifying patients into different health statuses based on selected gene signatures have become two main tracks of microarray data analysis in the past decade [2]–[6]. Various standards related to systems biology are discussed by Brazma et al. [7]. When sample sizes are substantially smaller than the number of features or genes, statistical modeling and inference issues become challenging as the familiar “large p small n problem” arises. Designing feature selection methods that lead to reliable and accurate predictions by learning classifiers, therefore, is an issue of great theoretical as well as practical importance in high dimensional data analysis.
To address the “curse of dimensionality” problem, three basic strategies have been proposed for feature selection: filtering, wrapper, and embedded methods. Filtering methods select subset features independently from the learning classifiers and do not incorporate learning [8]–[11]. One of the weaknesses of filtering methods is that they only consider the individual feature in isolation and ignore the possible interaction among features. Yet the combination of certain features may have a net effect that does not necessarily follow from the individual performance of features in that group [12]. A consequence of the filtering methods is that we may end up with selecting groups of highly correlated features/genes, which present redundant information to the learning classifier to ultimately worsen its performance. Also, if there is a practical limit on the number of features to be chosen, one may not be able to include all informative features.
To avoid the weakness of filtering methods, wrapper methods wrap around a particular learning algorithm that can assess the selected feature subsets in terms of the estimated classification errors and then build the final classifier [13]. Wrapper methods use a learning machine to measure the quality of subsets of features. One recent well-known wrapper method for feature/gene selection is Support Vector Machine Recursive Feature Elimination (SVMRFE) [14], which refines the optimum feature set by using Support Vector Machines (SVM). The idea of SVMRFE is that the orientation of the separating hyper-plane found by the SVM can be used to select informative features; if the plane is orthogonal to a particular feature dimension, then that feature is informative, and vice versa. In addition to microarray data analysis, SVMRFE has been widely used in high-throughput biological data analyses and other areas involving feature selection and pattern classification [15].
Wrapper methods can noticeably reduce the number of features and significantly improve the classification accuracy [16], [17]. However, wrapper methods have the drawback of high computational load, making them less desirable as the dimensionality increases. The embedded methods perform feature selection simultaneously with learning classifiers to achieve better computational efficiency than wrapper methods while maintaining similar performance. LASSO [18], [19], logic regression with the regularized Laplacian prior [20], and Bayesian regularized neural network with automatic relevance determination [21] are examples of embedded methods.
To improve classification of microarray data, Zhou and Mao proposed SFS-LS bound and SFFS-LS bound algorithms for optimal gene selection by combining the sequential forward selection (SFS) and sequential floating forward selection (SFFS) with LS (Least Squares) bound measure [22]. Tang et al. designed two methods of gene selection, leave-one-out calculation sequential forward selection (LOOCSFS) and the gradient based leave-one-out gene selection (GLGS) [23]. Diaz-Uriarte and De Andres [24] presented a method for gene selection by calculating the out of bag errors with random forest [25].
In human genetic research, exploiting information redundancy from highly correlated genes can potentially reduce the cost and simultaneously improve the reliability and accuracy of learning classifiers that are employed in data analysis. To exploit the information redundancy that exists among the huge number of variables and improve classification accuracy of microarray data, we propose a gene selection method, Recursive Feature Addition (RFA), which is based on supervised learning and similarity measures. We compare RFA with SVMRFE, LOOCSFS, and GLGS by using the MAQC-II breast cancer dataset to predict pre-operative treatment response (pCR) and estrogen receptor status (erpos) and compare RFA with SVMRFE and LOOCSFS on the MAQC-II multiple myeloma dataset to predict the overall survival milestone outcome (OSMO, 730-day cutoff) and to predict event-free survival milestone outcome (EFSMO, 730-day cutoff).
Results
Results on MAQC-II Breast Cancer Dataset
We compare MSC-based RFA methods with GLGS, LOOCSFS, and SVMRFE on MAQC-II breast cancer dataset. Tables 1 to 10 list the cancer related genes of the first 100 features selected by these methods. In comparison to GLGS, LOOCSFS, and SVMRFE, RFA is associated with a greater number of currently known cancer related genes for prediction of pCR and a smaller number of currently known cancer related genes for prediction of erpos. Since disease status is not simply related to the number of these cancer related genes, we obtain the prediction performance by running multiple experiments, and compare the average prediction performance using the following measurements: testing accuracy, Matthews Correlation Coefficients (MCC) [26], [27] that has been used in MAQC-II consortium [28], and area under the receiver operating curve (AUC) errors with classifiers NMSC, NBC, SVM and UDC (uncorrelated normal based quadratic Bayes classifier) [29].
Table 1. The 34 cancer related genes of the 100 features selected by NBC-MSC on original training group of MAQC-II breast cancer data for pCR prediction.
| Symbol | Synonym(s) | Entrez Gene Name | Affymetrix |
| ALB | ALB1, ALBUMIN, Albumin 1, Albza, DKFZp779N1935, PRO0883, PRO0903, PRO1341, SA, SERUM ALBUMIN, SERUM ALBUMIN CHAIN A, Serum albumin precursor | albumin | 214837_at |
| C10ORF81 | 9930023K05RIK, bA211N11.2, FLJ23537, HEL185, MGC99964, RGD1559884, RP11-211N11.2 | chromosome 10 open reading frame 81 | 219857_at |
| CDK10 | BC017131, MGC112847, PISSLRE | cyclin-dependent kinase 10 | 210622_x_at |
| CEACAM1 | bb-1, BGP, BGP1, BGPA, Bgpd, BGPI, BGPR, C-CAM, C-CAM1, CCAM105, CD66, CD66A, Cea-1, Cea-7, CEACAM1-4L, ECTO ATPASE, HV2, mCEA1, Mhv-1, MHVR, MHVR1, mmCGM1, mmCGM1a, mmCGM2, Pp120 | carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | 211889_x_at |
| CHRNB4 | Acrb-4, NACHR BETA4 | cholinergic receptor, nicotinic, beta 4 | 207516_at |
| CR1 | C3b/C4b receptor, C3BR, CD35, CD46, Cr1l, Crry, KN, Mcp, mCRY, MGC102484, SCR1 | complement component (3b/4b) receptor 1 (Knops blood group) | 217552_x_at |
| CXCL3 | Cinc-2, CINC-2a, CINC-2b, Cinc3, Cxcl2, Dcip1, Gm1960, GRO ALPHA, GRO BETA, GRO GAMMA, GRO1, Gro2, GRO3, GROA, GROb, GROg, KC, MGSA, Mgsa-b, MIP-2, MIP-2a, MIP-2b, Mip2 alpha, N51, Scyb, Scyb2, SCYB3 | chemokine (C-X-C motif) ligand 3 | 207850_at |
| CXCL13 | ANGIE, ANGIE2, BCA-1, BLC, BLC1, BLR1L, CXC CHEMOKINE, Loc498335, SCYB13 | chemokine (C-X-C motif) ligand 13 | 205242_at |
| DKK1 | Dkk1 predicted, mdkk-1, SK | dickkopf homolog 1 (Xenopus laevis) | 204602_at |
| DRD2 | D2, D2 DOPAMINE RECEPTOR, D2a dopamine receptor, D2DR, D2R, D2S, DOPAMINE D2 RECEPTOR, Dr2 | dopamine receptor D2 | 216924_s_at |
| EED | HEED, l(7)5Rn, l7Rn5, lusk, WAIT-1 | embryonic ectoderm development | 209572_s_at |
| GINS3 | 2700085M18Rik, AI616142, FLJ13912, PSF3, RGD1308153 | GINS complex subunit 3 (Psf3 homolog) | 218719_s_at |
| GPS2 | AI505953, AMF-1, MGC104294, MGC119287, MGC119288, MGC119289 | G protein pathway suppressor 2 | 209350_s_at |
| GRIA2 | GLUR-B, GluR-K2, GLUR2, GLUR2 IONOTROPIC, HBGR2 | glutamate receptor, ionotropic, AMPA 2 | 205358_at |
| GSN | DKFZp313L0718, GELSOLIN, MGC28083, MGC95032 | gelsolin (amyloidosis, Finnish type) | 214040_s_at |
| IFNAR1 | ALPHA CHAIN OF TYPE I IFNR, AVP, BETA R1, CD118, Ifar, IFN RECEPTOR TYPE 1, IFN TYPE 1 RECEPTOR, IFN-alpha-beta-R, IFN-ALPHA-REC, IFNalpha/betaR, IFNAR, IFNBR, IFRC, Infar, INFAR1, Interferon Receptor, LOC284829, Type I infr | interferon (alpha, beta and omega) receptor 1 | 204191_at |
| ITGB4 | AA407042, C230078O20, CD104, INTEGRIN-BETA 4 | integrin, beta 4 | 204989_s_at |
| IVD | 1300016K07Rik, 6720455E18Rik, ACAD2, AI463340, Isovaleryl-Coa Dehydrogenase | isovaleryl Coenzyme A dehydrogenase | 216958_s_at |
| KL | ALPHA KLOTHO, alpha-kl, KLOTHO | klotho | 205978_at |
| N4BP1 | AI481586, C81621, FLJ31821, KIAA0615, MGC176730, MGC7607, RGD1305179 | NEDD4 binding protein 1 | 32069_at |
| NAIP | AV364616, BIRC1, BIRC1A, Birc1b, Birc1e, BIRC1F, D13Lsd1, FLJ18088, FLJ42520, FLJ58811, LGN1, LOC652755, Naip-rs1, Naip-rs3, Naip-rs4, Naip-rs4A, Naip1, Naip2, Naip5, Naip6, NLRB1, psiNAIP, RGD1559914 | NLR family, apoptosis inhibitory protein | 204861_s_at |
| NDST1 | 1200015G06RIK, HSNST, HSST, HSST1, NST1 | N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 | 202608_s_at |
| PAICS | 2610511I09Rik, ADE2, ADE2H1, AIRC, DKFZp781N1372, MGC1343, MGC5024, MGC93240, PAIS | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | 214664_at |
| PAWR | PAR-4 | PRKC, apoptosis, WT1, regulator | 204005_s_at |
| PDPK1 | MGC20087, MGC35290, PDK1, PRO0461 | 3-phosphoinositide dependent protein kinase-1 | 204524_at |
| PHLDA1 | DT1P1B11, MGC131738, PHRIP, PQ-RICH, Proline- and glutamine-rich, TDAG, TDAG51 | pleckstrin homology-like domain, family A, member 1 | 218000_s_at |
| PPP1R15A (includes EG:23645) | 9630030H21, GADD34, MYD116, Myeloid Differentiation, Peg-3, PP1 REGULATORY SUBUNIT, Ppp1r15a | protein phosphatase 1, regulatory (inhibitor) subunit 15A | 202014_at |
| RASSF1 | 123F2, AA536941, AU044980, D4Mgi37, MGC94319, NORE2A, PTS, Rassf1A, Rassf1B, Rassf1C, RDA32, REH3P21 | Ras association (RalGDS/AF-6) domain family member 1 | 204346_s_at |
| SYT1 | AW124717, DKFZp781D2042, G630098F17Rik, P65, SVP65, SYNAPTOTAGMIN 1, SYT | synaptotagmin I | 203999_at |
| TACSTD2 | C80403, EGP-1, GA733, GA733-1, Ly97, M1S1, MGC141612, MGC141613, MGC72570, Prp1, TROP2 | tumor-associated calcium signal transducer 2 | 202286_s_at |
| TEK | AA517024, CD202B, Hyk, MGC139569, TIE-2, VMCM, VMCM1 | TEK tyrosine kinase, endothelial | 206702_at |
| TTF1 | AV245725, RGD1565673, Ttf-I | transcription termination factor, RNA polymerase I | 204772_s_at |
| ZEB1 | 3110032K11Rik, AREB6, BZP, DELTA-EF1, MEB1, MGC133261, NIL-2-A, Nil2, Tcf18, TCF8, TCP8, TF8, TRANSCRIPTION FACTOR 8, ZEB, ZFHEP, Zfhep2, ZFHX1A, Zfx1a, Zfx1ha, [delta]EF1 | zinc finger E-box binding homeobox 1 | 212758_s_at |
| ZNF10 | KOX1 | zinc finger protein 10 | 216350_s_at |
Table 2. The 34 cancer related genes of the 100 features selected by NMSC-MSC on original training group of MAQC-II breast cancer data for pCR prediction.
| Symbol | Synonym(s) | Entrez Gene Name | Affymetrix |
| ALB | ALB1, ALBUMIN, Albumin 1, Albza, DKFZp779N1935, PRO0883, PRO0903, PRO1341, SA, SERUM ALBUMIN, SERUM ALBUMIN CHAIN A, Serum albumin precursor | albumin | 214837_at |
| ARAF | 1200013E08Rik, ARAF1, AW495444, PKS, PKS2, RAFA1 | v-raf murine sarcoma 3611 viral oncogene homolog | 201895_at |
| C10ORF81 | 9930023K05RIK, bA211N11.2, FLJ23537, HEL185, MGC99964, RGD1559884, RP11-211N11.2 | chromosome 10 open reading frame 81 | 219857_at |
| CDH1 | AA960649, Arc-1, Cadherin 1, CD324, CDHE, CSEIL, E-CADHERIN, E-CADHERIN 120 KDA, ECAD, L-CAM, MGC107495, Um, UVO, uvomorulin | cadherin 1, type 1, E-cadherin (epithelial) | 201131_s_at |
| CDK10 | BC017131, MGC112847, PISSLRE | cyclin-dependent kinase 10 | 210622_x_at |
| CEACAM1 | bb-1, BGP, BGP1, BGPA, Bgpd, BGPI, BGPR, C-CAM, C-CAM1, CCAM105, CD66, CD66A, Cea-1, Cea-7, CEACAM1-4L, ECTO ATPASE, HV2, mCEA1, Mhv-1, MHVR, MHVR1, mmCGM1, mmCGM1a, mmCGM2, Pp120 | carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | 211889_x_at |
| CEBPE | C/EBP EPSILON, C/EBPe, C/EPBe, CRP1, Gm294, MGC124002, MGC124003 | CCAAT/enhancer binding protein (C/EBP), epsilon | 214523_at |
| CHRNB4 | Acrb-4, NACHR BETA4 | cholinergic receptor, nicotinic, beta 4 | 207516_at |
| CR1 | C3b/C4b receptor, C3BR, CD35, CD46, Cr1l, Crry, KN, Mcp, mCRY, MGC102484, SCR1 | complement component (3b/4b) receptor 1 (Knops blood group) | 217552_x_at |
| CXCL3 | Cinc-2, CINC-2a, CINC-2b, Cinc3, Cxcl2, Dcip1, Gm1960, GRO ALPHA, GRO BETA, GRO GAMMA, GRO1, Gro2, GRO3, GROA, GROb, GROg, KC, MGSA, Mgsa-b, MIP-2, MIP-2a, MIP-2b, Mip2 alpha, N51, Scyb, Scyb2, SCYB3 | chemokine (C-X-C motif) ligand 3 | 207850_at |
| CXCL13 | ANGIE, ANGIE2, BCA-1, BLC, BLC1, BLR1L, CXC CHEMOKINE, Loc498335, SCYB13 | chemokine (C-X-C motif) ligand 13 | 205242_at |
| DRD2 | D2, D2 DOPAMINE RECEPTOR, D2a dopamine receptor, D2DR, D2R, D2S, DOPAMINE D2 RECEPTOR, Dr2 | dopamine receptor D2 | 216924_s_at |
| DYRK1A | 2310043O08Rik, D16Ertd272e, D16Ertd493e, DUAL-SPECIFICITY TYROSINE-(Y)-PHOSPHORYLATION REGULATED KINASE 1A, DYRK, DYRK1, HP86, MGC150253, MGC150254, mmb, MNB, MNBH, Mp86, PSK47 | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | 211541_s_at |
| EPOR | EP-R, ERYTHROPOIETIN RECEPTOR, MGC108723, MGC138358 | erythropoietin receptor | 215054_at |
| FAM153A | KIAA0752, NY-REN-7 | family with sequence similarity 153, member A | 211166_at |
| GINS3 | 2700085M18Rik, AI616142, FLJ13912, PSF3, RGD1308153 | GINS complex subunit 3 (Psf3 homolog) | 218719_s_at |
| GRIA2 | GLUR-B, GluR-K2, GLUR2, GLUR2 IONOTROPIC, HBGR2 | glutamate receptor, ionotropic, AMPA 2 | 205358_at |
| GSN | DKFZp313L0718, GELSOLIN, MGC28083, MGC95032 | gelsolin (amyloidosis, Finnish type) | 214040_s_at |
| IFNAR1 | ALPHA CHAIN OF TYPE I IFNR, AVP, BETA R1, CD118, Ifar, IFN RECEPTOR TYPE 1, IFN TYPE 1 RECEPTOR, IFN-alpha-beta-R, IFN-ALPHA-REC, IFNalpha/betaR, IFNAR, IFNBR, IFRC, Infar, INFAR1, Interferon Receptor, LOC284829, Type I infr | interferon (alpha, beta and omega) receptor 1 | 204191_at |
| ITGB4 | AA407042, C230078O20, CD104, INTEGRIN-BETA 4 | integrin, beta 4 | 204989_s_at |
| KL | ALPHA KLOTHO, alpha-kl, KLOTHO | klotho | 205978_at |
| LPAR1 | 5031439C20, AI326300, clone 4.9, EDG2, ENDOTHELIAL DIFFERENTIATION LYSOPHOSPHATIDIC ACID G-PROTEIN-COUPLED RECEPTOR 2, Gpcr26, GPR26, Kdt2, LPA receptor 1, LPA1, LPA1 RECEPTOR, LPA2, LYSOPHOSPHATIDIC ACID G-PROTEIN-COUPLED RECEPTOR, MGC105279, MGC29102, Mrec1.3, rec.1.3, vzg-1 | lysophosphatidic acid receptor 1 | 204037_at |
| MCF2 | B230117G22Rik, DBL, MGC159138, RGD1566098 | MCF.2 cell line derived transforming sequence | 208017_s_at |
| MYO10 | AW048724, D15Ertd600e, FLJ10639, FLJ21066, FLJ22268, FLJ43256, KIAA0799, MGC131988, mKIAA0799, Myo10 (predicted), myosin-X | myosin X | 201976_s_at |
| NAIP | AV364616, BIRC1, BIRC1A, Birc1b, Birc1e, BIRC1F, D13Lsd1, FLJ18088, FLJ42520, FLJ58811, LGN1, LOC652755, Naip-rs1, Naip-rs3, Naip-rs4, Naip-rs4A, Naip1, Naip2, Naip5, Naip6, NLRB1, psiNAIP, RGD1559914 | NLR family, apoptosis inhibitory protein | 204860_s_at |
| NDST1 | 1200015G06RIK, HSNST, HSST, HSST1, NST1 | N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 | 202608_s_at |
| PHLDA1 | DT1P1B11, MGC131738, PHRIP, PQ-RICH, Proline- and glutamine-rich, TDAG, TDAG51 | pleckstrin homology-like domain, family A, member 1 | 218000_s_at |
| PPP1R15A (includes EG:23645) | 9630030H21, GADD34, MYD116, Myeloid Differentiation, Peg-3, PP1 REGULATORY SUBUNIT, Ppp1r15a | protein phosphatase 1, regulatory (inhibitor) subunit 15A | 202014_at |
| RASSF1 | 123F2, AA536941, AU044980, D4Mgi37, MGC94319, NORE2A, PTS, Rassf1A, Rassf1B, Rassf1C, RDA32, REH3P21 | Ras association (RalGDS/AF-6) domain family member 1 | 204346_s_at |
| SIAH1 | AA982064, AI853500, D9MGI7, FLJ08065, hSIAH1, HUMSIAH, SIAH, Siah1a, Sinh1a | seven in absentia homolog 1 (Drosophila) | 202981_x_at |
| TTF1 | AV245725, RGD1565673, Ttf-I | transcription termination factor, RNA polymerase I | 204772_s_at |
| VAMP2 | FLJ11460, mVam2, RATVAMPB, RATVAMPIR, SYB, SYB2, SYNAPTOBREVIN 2, Vamp ii | vesicle-associated membrane protein 2 (synaptobrevin 2) | 201557_at |
| ZNF10 | KOX1 | zinc finger protein 10 | 216350_s_at |
| ZNF205 | 4933429B21, AI835008, Krox-8, Zfp13, ZINC FINGER PROTEIN 205, ZNF210 | zinc finger protein 205 | 206416_at |
Table 3. The 26 cancer related genes of the 100 features selected by GLGS on original training group of MAQC-II breast cancer data for pCR prediction.
| Symbol | Synonym(s) | Entrez Gene Name | Affymetrix |
| ADAM15 | MDC15, METARGIDIN, tMDCVI | ADAM metallopeptidase domain 15 | 217007_s_at |
| ARAF | 1200013E08Rik, ARAF1, AW495444, PKS, PKS2, RAFA1 | v-raf murine sarcoma 3611 viral oncogene homolog | 201895_at |
| ASNS | AS, ASPARAGINE SYNTHETASE, MGC93148, TS11 | asparagine synthetase | 205047_s_at |
| B4GALT5 | 9430078I07Rik, AW049941, AW539721, BETA4-GALT-IV, beta4Gal-T5, beta4GalT-V, gt-V, MGC138470 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 | 221485_at |
| CEACAM1 | bb-1, BGP, BGP1, BGPA, Bgpd, BGPI, BGPR, C-CAM, C-CAM1, CCAM105, CD66, CD66A, Cea-1, Cea-7, CEACAM1-4L, ECTO ATPASE, HV2, mCEA1, Mhv-1, MHVR, MHVR1, mmCGM1, mmCGM1a, mmCGM2, Pp120 | carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | 211889_x_at |
| CXCL3 | Cinc-2, CINC-2a, CINC-2b, Cinc3, Cxcl2, Dcip1, Gm1960, GRO ALPHA, GRO BETA, GRO GAMMA, GRO1, Gro2, GRO3, GROA, GROb, GROg, KC, MGSA, Mgsa-b, MIP-2, MIP-2a, MIP-2b, Mip2 alpha, N51, Scyb, Scyb2, SCYB3 | chemokine (C-X-C motif) ligand 3 | 207850_at |
| CYCS (includes EG:54205) | CYC, CYCS, CYCSA, CYCT, CYCTA, CYTC, CYTOCHROME C, HCS, MGC93634, T-Cc | cytochrome c, somatic | 208905_at |
| DRD2 | D2, D2 DOPAMINE RECEPTOR, D2a dopamine receptor, D2DR, D2R, D2S, DOPAMINE D2 RECEPTOR, Dr2 | dopamine receptor D2 | 216924_s_at |
| EGFR | 9030024J15RIK, AI552599, EGF RECEPTOR, EGF-TK, EGFR1, EGFRec, ER2, ERBB, ERBB1, Errp, HER1, MENA, PIG61, wa-2, Wa5 | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 201983_s_at |
| FBLN1 | FBLN, FIBULIN 1 | fibulin 1 | 207834_at |
| FIS1 | 2010003O14Rik, CGI-135, Riken cDNA 2010003o14, TTC11 | fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) | 218034_at |
| FOXC1 | ARA, ch, fkh-1, FKHL7, FLJ11796, FLJ11796 FIS, FREAC-3, frkhda, IGDA, IHG1, IRID1, Mf1, Mf4, rCG 44068, rCG_44068 | forkhead box C1 | 213260_at |
| FTL | FERRITIN LIGHT CHAIN, FTL1, Ftl2, L-FERRITIN, MGC102130, MGC102131, MGC118079, MGC118080, MGC71996, RGD1560687, RGD1561055, RGD1566189, YB24D08 | ferritin, light polypeptide | 201265_at |
| GFRA1 | AU042498, GDNFR, Gdnfr alpha, GDNFRA, GFR-ALPHA-1, GRFA1, MGC23045, Ret, RET1L, RETL1, TRNR1 | GDNF family receptor alpha 1 | 205696_s_at |
| ITGB4 | AA407042, C230078O20, CD104, INTEGRIN-BETA 4 | integrin, beta 4 | 204989_s_at |
| KL | ALPHA KLOTHO, alpha-kl, KLOTHO | klotho | 205978_at |
| LEF1 | 3000002B05, AI451430, DKFZp586H0919, LEF, TCF/LEF, TCF1ALPHA | lymphoid enhancer-binding factor 1 | 221557_s_at |
| LMO4 | A730077C12Rik, Crp3, Etohi4, MGC105593 | LIM domain only 4 | 209205_s_at |
| LPAR2 | EDG-4, FLJ93869, IPA2, LPA receptor 2, LPA2, LPA2 RECEPTOR, Pbx4, RGD1561336 | lysophosphatidic acid receptor 2 | 206723_s_at |
| MKI67 | D630048A14Rik, KI-67, KIA, MIB1, MIKI67A, MKI67A | antigen identified by monoclonal antibody Ki-67 | 212022_s_at |
| NAIP | AV364616, BIRC1, BIRC1A, Birc1b, Birc1e, BIRC1F, D13Lsd1, FLJ18088, FLJ42520, FLJ58811, LGN1, LOC652755, Naip-rs1, Naip-rs3, Naip-rs4, Naip-rs4A, Naip1, Naip2, Naip5, Naip6, NLRB1, psiNAIP, RGD1559914 | NLR family, apoptosis inhibitory protein | 204861_s_at |
| PPP1R15A (includes EG:23645) | 9630030H21, GADD34, MYD116, Myeloid Differentiation, Peg-3, PP1 REGULATORY SUBUNIT, Ppp1r15a | protein phosphatase 1, regulatory (inhibitor) subunit 15A | 202014_at |
| RARB | A830025K23, BETA RAR, HAP, LOC51036, NR1B2, RAR BETA, RAR-EPSILON, RRB2 | retinoic acid receptor, beta | 208530_s_at |
| TEK | AA517024, CD202B, Hyk, MGC139569, TIE-2, VMCM, VMCM1 | TEK tyrosine kinase, endothelial | 206702_at |
| TTF1 | AV245725, RGD1565673, Ttf-I | transcription termination factor, RNA polymerase I | 204772_s_at |
| USF2 | bHLHb12, FIP, MGC91056 | upstream transcription factor 2, c-fos interacting | 202152_x_at |
Table 4. The 25 cancer related genes of the 100 features selected by LOOCSFS on original training group of MAQC-II breast cancer data for pCR prediction.
| Symbol | Synonym(s) | Entrez Gene Name | Affymetrix |
| ADAM17 | Alpha Secretase, CD156b, cSVP, MGC71942, TACE, TNFA CONVERTASE | ADAM metallopeptidase domain 17 | 213532_at |
| APP | A beta 25–35, A-BETA 40, A-BETA 42, AAA, ABETA, ABPP, AD1, Adap, AL024401, AMYLOID BETA, AMYLOID BETA 40, AMYLOID BETA 40 HUMAN, AMYLOID BETA 42, Amyloid beta A4, AMYLOID BETA PEPTIDE 40, Amyloid precursor, Amyloidogenic glycoprotein, App alpha, APPI, appican, BETAAPP, CTFgamma, CVAP, E030013M08RIK, Nexin II, P3, PN2, PreA4, PROTEASE NEXIN2 | amyloid beta (A4) precursor protein | 214953_s_at |
| ARAF | 1200013E08Rik, ARAF1, AW495444, PKS, PKS2, RAFA1 | v-raf murine sarcoma 3611 viral oncogene homolog | 201895_at |
| CEACAM1 | bb-1, BGP, BGP1, BGPA, Bgpd, BGPI, BGPR, C-CAM, C-CAM1, CCAM105, CD66, CD66A, Cea-1, Cea-7, CEACAM1-4L, ECTO ATPASE, HV2, mCEA1, Mhv-1, MHVR, MHVR1, mmCGM1, mmCGM1a, mmCGM2, Pp120 | carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | 211889_x_at |
| CTSL2 | 1190035F06Rik, Cathepsin l, CATHEPSIN V, CATHL, CATL2, Ctsl, Ctsl1, CTSU, CTSV, fs, MEP, MGC125957, nkt | cathepsin L2 | 210074_at |
| CYR61 | AI325051, CCN1, Cysteine-rich protein 61, GIG1, IGFBP10, MGC93040 | cysteine-rich, angiogenic inducer, 61 | 210764_s_at |
| DKK1 | Dkk1 predicted, mdkk-1, SK | dickkopf homolog 1 (Xenopus laevis) | 204602_at |
| DRD2 | D2, D2 DOPAMINE RECEPTOR, D2a dopamine receptor, D2DR, D2R, D2S, DOPAMINE D2 RECEPTOR, Dr2 | dopamine receptor D2 | 216924_s_at |
| ETS2 | AU022856, ETS2IT1 | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | 201329_s_at |
| GRIA2 | GLUR-B, GluR-K2, GLUR2, GLUR2 IONOTROPIC, HBGR2 | glutamate receptor, ionotropic, AMPA 2 | 205358_at |
| ITGB4 | AA407042, C230078O20, CD104, INTEGRIN-BETA 4 | integrin, beta 4 | 204990_s_at |
| KIF3A | 111-11-71, 111-11-86, AF180004, AF180009, AW124694, KIF3, Kifl, Kns3 | kinesin family member 3A | 213623_at |
| KLF6 | Aa1017, AI448727, BCD1, C86813, COPEB, CPBP, DKFZp686N0199, Erythropoietin 1, FM2, FM6, GBF, Ierepo1, IEREPO3, KRUPPEL LIKE ZINC FINGER PROTEIN ZF9, PAC1, PROTO-ONCOGENE BCD, Proto-oncogene BCD1, R75280, ST12, ZF9 | Kruppel-like factor 6 | 211610_at |
| LTBP1 | 9430031G15Rik, 9830146M04, MGC163161, TGFB | latent transforming growth factor beta binding protein 1 | 202729_s_at |
| MCF2 | B230117G22Rik, DBL, MGC159138, RGD1566098 | MCF.2 cell line derived transforming sequence | 208017_s_at |
| REST | 2610008J04RIK, AA407358, D14MGI11, MGC150099, NRSF, XBR | RE1-silencing transcription factor | 204535_s_at |
| SDC1 | AA408134, AA409076, BB4, CD138, HSPG, SDC, SYN1, Synd, SYND1, SYNDECA, Syndecan, SYNDECAN-1 | syndecan 1 | 201287_s_at |
| SEMA3B | AW208495, FLJ34863, LUCA-1, SEMA, SEMA5, SEMAA, semaV | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B | 203071_at |
| SLPI | ALK1, ALP, ANTILEUKOPROTEASE, antileukoproteinase, BLPI, HUSI, HUSI-I, MPI, Secretory Leukoprotease Inhibitor, SLP1, WAP4, WFDC4 | secretory leukocyte peptidase inhibitor | 203021_at |
| SP1 | 1110003E12RIK, AA450830, AI845540, Sp1 (trans spliced isoform), SP1-1, Trans-acting transcription factor 1 | Sp1 transcription factor | 214732_at |
| TCF7L2 (includes EG:6934) | LOC679869, mTcf-4B, mTcf-4E, TCF-4, TCF4B, TCF4E, Tcf7l2 | transcription factor 7-like 2 (T-cell specific, HMG-box) | 216511_s_at |
| TNFAIP3 | A20, MAD6, MGC104522, MGC138687, MGC138688, OTUD7C, TNF ALPHA-INDUCED PROTEIN 3, TNF-inducible early response, TNFA1P2, Tnfip3 | tumor necrosis factor, alpha-induced protein 3 | 202644_s_at |
| TTF1 | AV245725, RGD1565673, Ttf-I | transcription termination factor, RNA polymerase I | 204772_s_at |
| TXN | ADF, AW550880, DKFZp686B1993, EOSINOPHIL CYTOTOXICITY FACTOR, MGC151960, MGC61975, THIOREDOXIN, TRX, TRX1, Txn1 | thioredoxin | 208864_s_at |
Table 5. The 20 cancer related genes of the 100 features selected by SVMRFE on original training group of MAQC-II breast cancer data for pCR prediction.
| Symbol | Synonym(s) | Entrez Gene Name | Affymetrix |
| APP | A beta 25–35, A-BETA 40, A-BETA 42, AAA, ABETA, ABPP, AD1, Adap, AL024401, AMYLOID BETA, AMYLOID BETA 40, AMYLOID BETA 40 HUMAN, AMYLOID BETA 42, Amyloid beta A4, AMYLOID BETA PEPTIDE 40, Amyloid precursor, Amyloidogenic glycoprotein, App alpha, APPI, appican, BETAAPP, CTFgamma, CVAP, E030013M08RIK, Nexin II, P3, PN2, PreA4, PROTEASE NEXIN2 | amyloid beta (A4) precursor protein | 214953_s_at |
| CLEC3B | DKFZp686H17246, TETRANECTIN, TN, TNA, TTN | C-type lectin domain family 3, member B | 205200_at |
| CXCL9 | BB139920, CMK, crg-10, Humig, MGC105312, MIG, SCYB9 | chemokine (C-X-C motif) ligand 9 | 203915_at |
| DKK1 | Dkk1 predicted, mdkk-1, SK | dickkopf homolog 1 (Xenopus laevis) | 204602_at |
| DRD2 | D2, D2 DOPAMINE RECEPTOR, D2a dopamine receptor, D2DR, D2R, D2S, DOPAMINE D2 RECEPTOR, Dr2 | dopamine receptor D2 | 216924_s_at |
| EPOR | EP-R, ERYTHROPOIETIN RECEPTOR, MGC108723, MGC138358 | erythropoietin receptor | 215054_at |
| ESR1 | AA420328, Alpha estrogen receptor, AU041214, DKFZp686N23123, ER, ER ALPHA, Er alpha (46 kDa isoform), ER66, ERA, ER[a], ESR, ESRA, ESTR, ESTRA, ESTROGEN RECEPTOR ALPHA, ESTROGEN RECEPTOR1, NR3A1, RNESTROR, TERP-1 | estrogen receptor 1 | 215552_s_at |
| FAM153A | KIAA0752, NY-REN-7 | family with sequence similarity 153, member A | 211166_at |
| GSN | DKFZp313L0718, GELSOLIN, MGC28083, MGC95032 | gelsolin (amyloidosis, Finnish type) | 214040_s_at |
| IFNAR1 | ALPHA CHAIN OF TYPE I IFNR, AVP, BETA R1, CD118, Ifar, IFN RECEPTOR TYPE 1, IFN TYPE 1 RECEPTOR, IFN-alpha-beta-R, IFN-ALPHA-REC, IFNalpha/betaR, IFNAR, IFNBR, IFRC, Infar, INFAR1, Interferon Receptor, LOC284829, Type I infr | interferon (alpha, beta and omega) receptor 1 | 204191_at |
| KL | ALPHA KLOTHO, alpha-kl, KLOTHO | klotho | 205978_at |
| NAIP | AV364616, BIRC1, BIRC1A, Birc1b, Birc1e, BIRC1F, D13Lsd1, FLJ18088, FLJ42520, FLJ58811, LGN1, LOC652755, Naip-rs1, Naip-rs3, Naip-rs4, Naip-rs4A, Naip1, Naip2, Naip5, Naip6, NLRB1, psiNAIP, RGD1559914 | NLR family, apoptosis inhibitory protein | 204861_s_at |
| NDST1 | 1200015G06RIK, HSNST, HSST, HSST1, NST1 | N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 | 202608_s_at |
| PPP1R15A (includes EG:23645) | 9630030H21, GADD34, MYD116, Myeloid Differentiation, Peg-3, PP1 REGULATORY SUBUNIT, Ppp1r15a | protein phosphatase 1, regulatory (inhibitor) subunit 15A | 202014_at |
| PTGIS | CYP8, CYP8A1, MGC126858, MGC126860, Pcs, Pgi2, PGIS, PROSTACYCLIN, Prostacyclin Synthase, PTGI | prostaglandin I2 (prostacyclin) synthase | 211892_s_at |
| RAD23B | 0610007D13Rik, AV001138, HHR23B, HR23B, MGC112630, mHR23B, P58 | RAD23 homolog B (S. cerevisiae) | 201223_s_at |
| SP1 | 1110003E12RIK, AA450830, AI845540, Sp1 (trans spliced isoform), SP1-1, Trans-acting transcription factor 1 | Sp1 transcription factor | 214732_at |
| TACSTD2 | C80403, EGP-1, GA733, GA733-1, Ly97, M1S1, MGC141612, MGC141613, MGC72570, Prp1, TROP2 | tumor-associated calcium signal transducer 2 | 202286_s_at |
| TUBB3 | 3200002H15Rik, beta-4, M(beta)3, M(beta)6, MC1R, Nst, Tub beta3, TUBB4, Tubulin beta-3, Tubulin beta-III, Tuj1 | tubulin, beta 3 | 202154_x_at |
| ZNF10 | KOX1 | zinc finger protein 10 | 216350_s_at |
Table 6. The 34 cancer related genes of the 100 features selected by NBC-MSC on original training group of MAQC-II breast cancer data for erpos prediction.
| Symbol | Synonym(s) | Entrez Gene Name | Affymetrix |
| APP | A beta 25–35,A-BETA 40,A-BETA 42,AAA,ABETA,ABPP,AD1,Adap,AL024401,AMYLOID BETA,AMYLOID BETA 40,AMYLOID BETA 40 HUMAN,AMYLOID BETA 42,Amyloid beta A4,AMYLOID BETA PEPTIDE 40,Amyloid precursor,Amyloidogenic glycoprotein,App alpha,APPI,appican,BETAAPP,CTFgamma,CVAP,E030013M08RIK,Nexin II,P3,PN2,PreA4,PROTEASE NEXIN2 | amyloid beta (A4) precursor protein | 214953_s_at |
| CCDC28A | 1700009P13Rik,AI480677,C6ORF80,CCRL1AP,DKFZP586D0623,MGC116395,MGC131913,MGC19351,RGD1310326 | coiled-coil domain containing 28A | 209479_at |
| CEACAM1 | bb-1,BGP,BGP1,BGPA,Bgpd,BGPI,BGPR,C-CAM,C-CAM1,CCAM105,CD66,CD66A,Cea-1,Cea-7,CEACAM1-4L,ECTO ATPASE,HV2,mCEA1,Mhv-1,MHVR,MHVR1,mmCGM1,mmCGM1a,mmCGM2,Pp120 | carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | 211883_x_at |
| CHRNB4 | Acrb-4,NACHR BETA4 | cholinergic receptor, nicotinic, beta 4 | 207516_at |
| CXCL9 | BB139920,CMK,crg-10,Humig,MGC105312,MIG,SCYB9 | chemokine (C-X-C motif) ligand 9 | 203915_at |
| EIF1 | A121,EIF1A,ISO1,MGC101938,MGC6503,SUI1,SUI1-RS1 | eukaryotic translation initiation factor 1 | 212130_x_at |
| EMP1 | CL-20,ENP1MR,EPITHELIAL MEMBRANE PROTEIN 1,MGC93627,TMP | epithelial membrane protein 1 | 201325_s_at |
| EPOR | EP-R,ERYTHROPOIETIN RECEPTOR,MGC108723,MGC138358 | erythropoietin receptor | 215054_at |
| ETV5 | 1110005E01Rik,8430401F14Rik,ERM | ets variant 5 | 203349_s_at |
| FBLN1 | FBLN,FIBULIN 1 | fibulin 1 | 207834_at |
| FBXL7 | AL023057,D230018M15Rik,FBL6,FBL7,FBP7,MGC102204 | F-box and leucine-rich repeat protein 7 | 213249_at |
| GHR | AA986417,GHBP,GHR/BP,GROWTH HORMONE RECEPTOR,MGC124963,MGC156665 | growth hormone receptor | 205498_at |
| GPC3 | DGSX,Glypican 3,GTR2-2,MGC93606,OCI-5,SDYS,SGB,SGBS,SGBS1 | glypican 3 | 209220_at |
| GPS2 | AI505953,AMF-1,MGC104294,MGC119287,MGC119288,MGC119289 | G protein pathway suppressor 2 | 209350_s_at |
| IGF2BP3 | 2610101N11Rik,AA522010,Ab2-255,AL022933,AU045931,DKFZp686F1078,IMP-3,KOC1,Koc13,mimp3,Neilsen,RGD1306512,VICKZ3 | insulin-like growth factor 2 mRNA binding protein 3 | 203820_s_at |
| ING4 | D6Wsu147e,D6Xrf92,MGC12557,MGC156688,my036,p29ING4,p33ING1 ISOLOG | inhibitor of growth family, member 4 | 218234_at |
| ITGB4 | AA407042,C230078O20,CD104,INTEGRIN-BETA 4 | integrin, beta 4 | 204989_s_at |
| KLF5 | 4930520J07Rik,BTEB2,CKLF,IKLF,Kruppel-like factor 5,mBTEB2 | Kruppel-like factor 5 (intestinal) | 209211_at |
| MFAP3L | 4933428A15RIK,5430405D20Rik,AI461995,AW125052,KIAA0626,mKIAA0626,NYD-sp9 | microfibrillar-associated protein 3-like | 210493_s_at |
| NCOR1 | 5730405M06RIK,A230020K14RIK,hCIT529I10,hN-CoR,KIAA1047,MGC104216,MGC116328,mKIAA1047,N-COR,RIP13,Rxrip13,TRAC1 | nuclear receptor co-repressor 1 | 200854_at |
| PA2G4 | 38kDa,AA672939,EBP1,HG4-1,Itaf45,MGC94070,p38-2G4,Plfap,PROLIFERATION ASSOCIATED 2G4,Proliveration-associated protein 1 | proliferation-associated 2G4, 38kDa | 214794_at |
| PCNA | MGC8367,Pcna/cyclin,PCNAR | proliferating cell nuclear antigen | 217400_at |
| PLD1 | AA536939,C85393,Pc-Plc,Phospholipase D1,PLD1A,PLD1B,Plda,Pldb | phospholipase D1, phosphatidylcholine-specific | 215723_s_at |
| RPL13A | 1810026N22Rik,23-KD HIGHLY BASIC,MGC107571,Ribol13a,Ribosomal protein l13a,Tstap198-7,tum-antigen | ribosomal protein L13a | 211942_x_at |
| SCGB1D2 | BU101,LIPB,LIPOPHILIN B,LPHB | secretoglobin, family 1D, member 2 | 206799_at |
| SEL1L | AW493766,IBD2,KIAA4137,mKIAA4137,PRO1063,SEL1,SEL1-LIKE,SEL1H | sel-1 suppressor of lin-12-like (C. elegans) | 202063_s_at |
| SERPINA6 | AI265318,AV104445,CBG,MGC112780 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 | 206325_at |
| SMA4 | b55C20.2,FLJ36702,MGC22265,MGC60382,SMA3 | glucuronidase, beta pseudogene | 214850_at |
| SMC4 | 2500002A22Rik,C79747,CAP-C,DKFZP434F205,HCAP-C,MGC125078,SMC4L1 | structural maintenance of chromosomes 4 | 215623_x_at |
| SOS1 | 9630010N06,AI449023,GF1,GGF1,GINGF,HGF,MSOS1,NS4,Sos | son of sevenless homolog 1 (Drosophila) | 212780_at |
| TNFRSF11A | CD265,FEO,LOH18CR1,Ly109,MGC112793,mRANK,ODAR,ODFR,OFE,OPGL receptor,OPTB7,OSTS,PDB2,RANK,RGD1563614,TRANCE-R | tumor necrosis factor receptor superfamily, member 11a, NFKB activator | 207037_at |
| TNFSF12 | APO3L,DR3L,DR3LG,MGC129581,MGC20669,TWEAK | tumor necrosis factor (ligand) superfamily, member 12 | 205611_at |
| TRPV6 | ABP/ZF,Cac,CAT,CAT-L,CAT1,Crac,ECAC2,HSA277909,LP6728,Otrpc3,ZFAB | transient receptor potential cation channel, subfamily V, member 6 | 206827_s_at |
| USP7 | 2210010O09Rik,AA409944,AA617399,AU019296,AW548146,C80752,HAUSP,TEF1 | ubiquitin specific peptidase 7 (herpes virus-associated) | 222032_s_at |
Table 7. The 33 cancer related genes of the 100 features selected by NMSC-MSC on original training group of MAQC-II breast cancer data for erpos prediction.
| Symbol | Synonym(s) | Entrez Gene Name | Affymetrix |
| BAG1 | BAG1L, Bag1s, RAP46 | BCL2-associated athanogene | 202387_at |
| CCDC28A | 1700009P13Rik, AI480677, C6ORF80, CCRL1AP, DKFZP586D0623, MGC116395, MGC131913, MGC19351, RGD1310326 | coiled-coil domain containing 28A | 209479_at |
| CEACAM1 | bb-1, BGP, BGP1, BGPA, Bgpd, BGPI, BGPR, C-CAM, C-CAM1, CCAM105, CD66, CD66A, Cea-1, Cea-7, CEACAM1-4L, ECTO ATPASE, HV2, mCEA1, Mhv-1, MHVR, MHVR1, mmCGM1, mmCGM1a, mmCGM2, Pp120 | carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | 211883_x_at |
| CHRNB4 | Acrb-4, NACHR BETA4 | cholinergic receptor, nicotinic, beta 4 | 207516_at |
| EIF1 | A121, EIF1A, ISO1, MGC101938, MGC6503, SUI1, SUI1-RS1 | eukaryotic translation initiation factor 1 | 212130_x_at |
| EPOR | EP-R, ERYTHROPOIETIN RECEPTOR, MGC108723, MGC138358 | erythropoietin receptor | 215054_at |
| FBLN1 | FBLN, FIBULIN 1 | fibulin 1 | 207834_at |
| GBP1 (includes EG:2633) | 5830475C06, GBP1, GBPI, IFI67-K, MAG-1, MGC124334, Mpa-1 | guanylate binding protein 1, interferon-inducible, 67kDa | 202270_at |
| GHR | AA986417, GHBP, GHR/BP, GROWTH HORMONE RECEPTOR, MGC124963, MGC156665 | growth hormone receptor | 205498_at |
| GPC3 | DGSX, Glypican 3, GTR2-2, MGC93606, OCI-5, SDYS, SGB, SGBS, SGBS1 | glypican 3 | 209220_at |
| GPS2 | AI505953, AMF-1, MGC104294, MGC119287, MGC119288, MGC119289 | G protein pathway suppressor 2 | 209350_s_at |
| IMPDH2 | ENSMUSG00000071041, Imp dehydrogenase 2, IMPD, IMPD2, Impdh, IMPDH-II, inosine 5′-phosphate dehydrogenase 2, MGC72938, OTTMUSG00000019498 | IMP (inosine monophosphate) dehydrogenase 2 | 201892_s_at |
| ING4 | D6Wsu147e, D6Xrf92, MGC12557, MGC156688, my036, p29ING4, p33ING1 ISOLOG | inhibitor of growth family, member 4 | 218234_at |
| ITGB4 | AA407042, C230078O20, CD104, INTEGRIN-BETA 4 | integrin, beta 4 | 204990_s_at |
| KLF5 | 4930520J07Rik, BTEB2, CKLF, IKLF, Kruppel-like factor 5, mBTEB2 | Kruppel-like factor 5 (intestinal) | 209211_at |
| MAP3K12 | DLK, MUK, PK, ZPK, ZPKP1 | mitogen-activated protein kinase kinase kinase 12 | 205447_s_at |
| MFAP3L | 4933428A15RIK, 5430405D20Rik, AI461995, AW125052, KIAA0626, mKIAA0626, NYD-sp9 | microfibrillar-associated protein 3-like | 210493_s_at |
| NCOR1 | 5730405M06RIK, A230020K14RIK, hCIT529I10, hN-CoR, KIAA1047, MGC104216, MGC116328, mKIAA1047, N-COR, RIP13, Rxrip13, TRAC1 | nuclear receptor co-repressor 1 | 200854_at |
| PA2G4 | 38kDa, AA672939, EBP1, HG4-1, Itaf45, MGC94070, p38-2G4, Plfap, PROLIFERATION ASSOCIATED 2G4, Proliveration-associated protein 1 | proliferation-associated 2G4, 38kDa | 214794_at |
| PCNA | MGC8367, Pcna/cyclin, PCNAR | proliferating cell nuclear antigen | 217400_at |
| PLA2G16 | ADPLA, C78643, H-REV107-1, HRASLS3, HREV107, HREV107-3, MGC118754 | phospholipase A2, group XVI | 209581_at |
| PLD1 | AA536939, C85393, Pc-Plc, Phospholipase D1, PLD1A, PLD1B, Plda, Pldb | phospholipase D1, phosphatidylcholine-specific | 215723_s_at |
| PTPN14 | C130080N23RIK, MGC126803, OTTMUSG00000022087, PEZ, PTP36, PTPD2 | protein tyrosine phosphatase, non-receptor type 14 | 205503_at |
| SCGB1D2 | BU101, LIPB, LIPOPHILIN B, LPHB | secretoglobin, family 1D, member 2 | 206799_at |
| SEL1L | AW493766, IBD2, KIAA4137, mKIAA4137, PRO1063, SEL1, SEL1-LIKE, SEL1H | sel-1 suppressor of lin-12-like (C. elegans) | 202063_s_at |
| SERPINA6 | AI265318, AV104445, CBG, MGC112780 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 | 206325_at |
| SMC4 | 2500002A22Rik, C79747, CAP-C, DKFZP434F205, HCAP-C, MGC125078, SMC4L1 | structural maintenance of chromosomes 4 | 215623_x_at |
| SPAM1 (includes EG:6677) | 4933439A12Rik, HYA1, HYAL1, HYAL3, HYAL5, MGC108951, MGC26532, PH-20, SPAG15, SPAM1, TESTICULAR HYALURONIDASE | sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding) | 210536_s_at |
| TCF3 | A1, AA408400, ALF2, AW209082, bHLHb21, E12, E12/E47, E2-5, E2A, E47, ITF1, ME2, MGC129647, MGC129648, PAN1, Pan2, Tcfe2a, TRANSCRIPTION FACTOR 3, VDIR | transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | 213730_x_at |
| TGFA | RATTGFAA, TFGA, TGF ALPHA, TGFAA, wa-1 | transforming growth factor, alpha | 211258_s_at |
| TNFRSF11A | CD265, FEO, LOH18CR1, Ly109, MGC112793, mRANK, ODAR, ODFR, OFE, OPGL receptor, OPTB7, OSTS, PDB2, RANK, RGD1563614, TRANCE-R | tumor necrosis factor receptor superfamily, member 11a, NFKB activator | 207037_at |
| TNFSF12 | APO3L, DR3L, DR3LG, MGC129581, MGC20669, TWEAK | tumor necrosis factor (ligand) superfamily, member 12 | 205611_at |
| USP7 | 2210010O09Rik, AA409944, AA617399, AU019296, AW548146, C80752, HAUSP, TEF1 | ubiquitin specific peptidase 7 (herpes virus-associated) | 222032_s_at |
Table 8. The 40 cancer related genes of the 100 features selected by GLGS on original training group of MAQC-II breast cancer data for erpos prediction.
| Symbol | Synonym(s) | Entrez Gene Name | Affymetrix |
| ADRA2A | ADRA-2, ADRA2R, ADRAR, ADRENOCEPTOR ALPHA2A, alpha(2A)AR, ALPHA-2A ADRENERGIC RECEPTOR, ALPHA2-C10, alpha2A, Alpha2a Adrenoceptor, ALPHA2A-AR, AW122659, RATRG20, RG20, ZNF32 | adrenergic, alpha-2A-, receptor | 209869_at |
| ANGPTL4 | ANGIOPOIETIN-LIKE 4, ANGPTL2, ARP4, Bk89, Fasting Induced Adipose Factor, FIAF, Harp, HFARP, LOC362850, Ng27, NL2, PGAR, PGARG, pp1158, PPARG | angiopoietin-like 4 | 221009_s_at |
| BAG1 | BAG1L, Bag1s, RAP46 | BCL2-associated athanogene | 202387_at |
| CCNE1 | AW538188, CCNE, CYCLE, CYCLIN E, Cyclin E1 | cyclin E1 | 213523_at |
| CHRNA5 | AChR alpha5, Acra-5, ALPHA5 NACHR, ALPHA5 NICOTINIC RECEPTOR, LNCR2, MGC124059, MGC124168, nAChR alpha5, sialoprotein | cholinergic receptor, nicotinic, alpha 5 | 206533_at |
| CHRNB4 | Acrb-4, NACHR BETA4 | cholinergic receptor, nicotinic, beta 4 | 207516_at |
| CTSL2 | 1190035F06Rik, Cathepsin l, CATHEPSIN V, CATHL, CATL2, Ctsl, Ctsl1, CTSU, CTSV, fs, MEP, MGC125957, nkt | cathepsin L2 | 210074_at |
| EGFR | 9030024J15RIK, AI552599, EGF RECEPTOR, EGF-TK, EGFR1, EGFRec, ER2, ERBB, ERBB1, Errp, HER1, MENA, PIG61, wa-2, Wa5 | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 201983_s_at |
| EPHA2 | AW545284, ECK, ECKR, Myk2, Sek-2 | EPH receptor A2 | 203499_at |
| EPOR | EP-R, ERYTHROPOIETIN RECEPTOR, MGC108723, MGC138358 | erythropoietin receptor | 215054_at |
| FBLN1 | FBLN, FIBULIN 1 | fibulin 1 | 207834_at |
| GPC3 | DGSX, Glypican 3, GTR2-2, MGC93606, OCI-5, SDYS, SGB, SGBS, SGBS1 | glypican 3 | 209220_at |
| GPS2 | AI505953, AMF-1, MGC104294, MGC119287, MGC119288, MGC119289 | G protein pathway suppressor 2 | 209350_s_at |
| HMGA1 | AL023995, HMG-I(Y), HMG-R, Hmg-y/i, HMGA1A, Hmgi, Hmgi/y, HMGIY, HMGY, MGC102580, MGC12816, MGC4242, MGC4854 | high mobility group AT-hook 1 | 206074_s_at |
| HNRPDL | AA407431, AA959857, D5Ertd650e, D5Wsu145e, HNRNP, HNRNP-D LIKE, hnRNP-DL, JKTBP, JKTBP1, JKTBP2, laAUF1, MGC125262 | heterogeneous nuclear ribonucleoprotein D-like | 209068_at |
| HSD17B2 | 17Hsd, AI194836, AI194967, AI255511, EDH17B2, HSD17, SDR9C2 | hydroxysteroid (17-beta) dehydrogenase 2 | 204818_at |
| ICAM1 | BB2, CD54, ICAM, INTERCELLULAR ADHESION MOLECULE 1, Ly-47, M90551, MALA-2, Melanoma Progression Associated Antigen, MGC6195, P3.58 | intercellular adhesion molecule 1 | 202637_s_at |
| IGF2BP3 | 2610101N11Rik, AA522010, Ab2-255, AL022933, AU045931, DKFZp686F1078, IMP-3, KOC1, Koc13, mimp3, Neilsen, RGD1306512, VICKZ3 | insulin-like growth factor 2 mRNA binding protein 3 | 203820_s_at |
| ITGB4 | AA407042, C230078O20, CD104, INTEGRIN-BETA 4 | integrin, beta 4 | 204989_s_at |
| MAP3K12 | DLK, MUK, PK, ZPK, ZPKP1 | mitogen-activated protein kinase kinase kinase 12 | 205447_s_at |
| MFAP3L | 4933428A15RIK, 5430405D20Rik, AI461995, AW125052, KIAA0626, mKIAA0626, NYD-sp9 | microfibrillar-associated protein 3-like | 210493_s_at |
| PCM1 | 2600002H09Rik, 9430077F19Rik, C030044G17Rik, LOC100044052, MGC170660, PTC4 | pericentriolar material 1 | 214118_x_at |
| PCNA | MGC8367, Pcna/cyclin, PCNAR | proliferating cell nuclear antigen | 217400_at |
| PLD1 | AA536939, C85393, Pc-Plc, Phospholipase D1, PLD1A, PLD1B, Plda, Pldb | phospholipase D1, phosphatidylcholine-specific | 215723_s_at |
| PML | 1200009E24Rik, AI661194, MYL, PP8675, Retinoic acid receptor, RGD1562602, RNF71, TRIM19 | promyelocytic leukemia | 211013_x_at |
| PPP3CB | 1110063J16Rik, Calcineurin A Beta, CALCINEURIN A1, CALNA2, CALNB, CnA-gamma, CnAbeta, Protein phosphatase 3 catalytic subunit beta isoform | protein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform | 202432_at |
| PURA | CAGER-1, PUR-ALPHA, PUR1, ssCRE-BP, VACSSBF1 | purine-rich element binding protein A | 204020_at |
| PVR | 3830421F03Rik, CD155, D7ERTD458E, FLJ25946, HVED, mE4, NECL5, NECTIN 2 ALPHA, NECTIN-2, Pe4, PVS, TAA1, TAGE4 | poliovirus receptor | 212662_at |
| RBM5 | D030069N10RIK, FLJ39876, G15, H37, LUCA15, RMB5 | RNA binding motif protein 5 | 201395_at |
| SEL1L | AW493766, IBD2, KIAA4137, mKIAA4137, PRO1063, SEL1, SEL1-LIKE, SEL1H | sel-1 suppressor of lin-12-like (C. elegans) | 202063_s_at |
| SERPINA6 | AI265318, AV104445, CBG, MGC112780 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 | 206325_at |
| SLC5A3 | AA623876, BF642829, SMIT, Smit1, SMIT2 | solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 | 213164_at |
| SLC7A5 | 4f2 light chain, 4F2LC, CD98, CD98 LIGHT CHAIN, CD98LC, D0H16S474E, D16S469E, E16, E16/TA1, hLAT1, LAT1, MPE16, TA1 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 201195_s_at |
| SMC4 | 2500002A22Rik, C79747, CAP-C, DKFZP434F205, HCAP-C, MGC125078, SMC4L1 | structural maintenance of chromosomes 4 | 215623_x_at |
| SOS1 | 9630010N06, AI449023, GF1, GGF1, GINGF, HGF, MSOS1, NS4, Sos | son of sevenless homolog 1 (Drosophila) | 212780_at |
| TGFA | RATTGFAA, TFGA, TGF ALPHA, TGFAA, wa-1 | transforming growth factor, alpha | 205016_at |
| TNFRSF11A | CD265, FEO, LOH18CR1, Ly109, MGC112793, mRANK, ODAR, ODFR, OFE, OPGL receptor, OPTB7, OSTS, PDB2, RANK, RGD1563614, TRANCE-R | tumor necrosis factor receptor superfamily, member 11a, NFKB activator | 207037_at |
| UBE2B | 17-kDa Ubiquitin-Conjugating Enzyme E2, 2610301N02RIK, E2-14K, E2-17kDa, E2b, HHR6B, HR6B, LOC81816, RAD6B, UBC2 | ubiquitin-conjugating enzyme E2B (RAD6 homolog) | 211763_s_at |
| USP7 | 2210010O09Rik, AA409944, AA617399, AU019296, AW548146, C80752, HAUSP, TEF1 | ubiquitin specific peptidase 7 (herpes virus-associated) | 222032_s_at |
| VAMP2 | FLJ11460, mVam2, RATVAMPB, RATVAMPIR, SYB, SYB2, SYNAPTOBREVIN 2, Vamp ii | vesicle-associated membrane protein 2 (synaptobrevin 2) | 201557_at |
Table 9. The 44 cancer related genes of the 100 features selected by LOOCSFS on original training group of MAQC-II breast cancer data for erpos prediction.
| Symbol | Synonym(s) | Entrez Gene Name | Affymetrix |
| ACP1 | 4632432E04Rik, AI427468, HAAP, LMPTP, LMW-PTP, MGC103115, MGC111030, MGC132904, MGC3499 | acid phosphatase 1, soluble | 215227_x_at |
| BAG1 | BAG1L, Bag1s, RAP46 | BCL2-associated athanogene | 202387_at |
| BCL2 | AW986256, Bcl2 alpha, C430015F12Rik, CED9, D630044D05RIK, D830018M01RIK, LOC100046608, ORF16 | B-cell CLL/lymphoma 2 | 203685_at |
| BTG2 | AA959598, Agl, An, an-1, APRO1, B-cell translocation gene 2, antiproliferative, MGC126063, MGC126064, PC3, TIS21 | BTG family, member 2 | 201236_s_at |
| CCDC28A | 1700009P13Rik, AI480677, C6ORF80, CCRL1AP, DKFZP586D0623, MGC116395, MGC131913, MGC19351, RGD1310326 | coiled-coil domain containing 28A | 209479_at |
| CCNA2 | AA408589, CCN1, CCNA, CYCA, CYCLIN A2, CYCLIN-A, MGC156527, p60 | cyclin A2 | 213226_at |
| CD47 | 9130415E20RIK, AA407862, AI848868, AW108519, B430305P08RIK, CD47 ANTIGEN, CDw149, IAP, Itgp, Locuslink 71587, MER6, MGC93490, OA3 | CD47 molecule | 211075_s_at |
| CFD | ADIPSIN, ADN, Complement Factor D, DF, EVE, FACTOR D, PFD | complement factor D (adipsin) | 205382_s_at |
| CHRNA5 | AChR alpha5, Acra-5, ALPHA5 NACHR, ALPHA5 NICOTINIC RECEPTOR, LNCR2, MGC124059, MGC124168, nAChR alpha5, sialoprotein | cholinergic receptor, nicotinic, alpha 5 | 206533_at |
| CTSL2 | 1190035F06Rik, Cathepsin l, CATHEPSIN V, CATHL, CATL2, Ctsl, Ctsl1, CTSU, CTSV, fs, MEP, MGC125957, nkt | cathepsin L2 | 210074_at |
| CXCL10 | C7, CRG-2, GAMMA-IFN INDUCIBLE EARLY RESPONSE, gIP-10, IFI10, IFNG INDUCIBLE PROTEIN-10, INP10, Interferon-inducible protein-10, IP-10, mob-1, SCYB10, SMALL INDUCIBLE CYTOKINE SUBFAMILY B (Cys-X-Cys), MEMBER 10 | chemokine (C-X-C motif) ligand 10 | 204533_at |
| CYB5A | 0610009N12Rik, CB5, CYB5, Cytb5, CYTOCHROME B5, MCB5, MGC108694, MGC128769 | cytochrome b5 type A (microsomal) | 209366_x_at |
| DDX17 | 2610007K22RIK, A430025E01Rik, AI047725, C80929, DEAD-box protein p72, DEAD/H box RNA helicase p72, DKFZp761H2016, Gm926, MGC109323, MGC79147, P72, RH70 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | 208718_at |
| DNMT3B | DNA MTase HsaIIIB, ICF, M.HsaIIIB, MGC124407 | DNA (cytosine-5-)-methyltransferase 3 beta | 220668_s_at |
| EPHA2 | AW545284, ECK, ECKR, Myk2, Sek-2 | EPH receptor A2 | 203499_at |
| ERBB4 | C-erb-b4, Erbb4 Cyt2, ERBB4 JM-A, HER4, MGC138404, p180erbB4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | 214053_at |
| ESR1 | AA420328, Alpha estrogen receptor, AU041214, DKFZp686N23123, ER, ER ALPHA, Er alpha (46 kDa isoform), ER66, ERA, ER[a], ESR, ESRA, ESTR, ESTRA, ESTROGEN RECEPTOR ALPHA, ESTROGEN RECEPTOR1, NR3A1, RNESTROR, TERP-1 | estrogen receptor 1 | 205225_at |
| ETV5 | 1110005E01Rik, 8430401F14Rik, ERM | ets variant 5 | 203349_s_at |
| FAM134B | 1810015C04RIK, AU015349, FLJ20152, FLJ22155, FLJ22179 | family with sequence similarity 134, member B | 218510_x_at |
| FBLN1 | FBLN, FIBULIN 1 | fibulin 1 | 207834_at |
| GREB1 | 5730583K22Rik, 9130004E13, AF180470, AU023194, KIAA0575, mKIAA0575 | GREB1 protein | 205862_at |
| IL6ST | 5133400A03Rik, AA389424, Ac1055, BB405851, CD130, CDw130, D13Ertd699e, GP130, GP130-RAPS, Il6 transd, IL6R-beta | interleukin 6 signal transducer (gp130, oncostatin M receptor) | 211000_s_at |
| IRS1 | ENSMUSG00000022591, G972R, HIRS-1, IRS1IRM | insulin receptor substrate 1 | 204686_at |
| ITPR1 | D6Pas2, I145TR, InsP3R, INSP3R1, IP3 RECEPTOR, Ip3 Receptor Type 1, IP3R, IP3R1, opt, P400, Pcd6, Pcp-1, SCA15, SCA16 | inositol 1,4,5-triphosphate receptor, type 1 | 203710_at |
| LRP8 | 4932703M08Rik, AA921429, AI848122, APOER2, HSZ75190, LR8B, MCI1 | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor | 208433_s_at |
| MAP3K12 | DLK, MUK, PK, ZPK, ZPKP1 | mitogen-activated protein kinase kinase kinase 12 | 205447_s_at |
| MAPT | AI413597, ALZ50, AW045860, DDPAC, FLJ31424, FTDP-17, MAPTL, MGC134287, MGC138549, MGC156663, MSTD, Mtapt, MTBT1, MTBT2, PHF TAU, PPND, pTau, RNPTAU, TAU, Tau 3r, Tau-1, TAU-FACTOR, Tau40, TAU4R | microtubule-associated protein tau | 203929_s_at |
| MCM7 | AI747533, CDABP0042, CDC47, D16Mgi24, mCDC47, MCM2, Mcmd7, MGC93853, P1.1-MCM3, P1CDC47, P85MCM, PNAS-146 | minichromosome maintenance complex component 7 | 208795_s_at |
| MYO10 | AW048724, D15Ertd600e, FLJ10639, FLJ21066, FLJ22268, FLJ43256, KIAA0799, MGC131988, mKIAA0799, Myo10 (predicted), myosin-X | myosin X | 201976_s_at |
| NCAPG (includes EG:64151) | CAP-G, CHCG, FLJ12450, HCAP-G, MGC126525, NCAPG, NY-MEL-3, RGD1562646 | non-SMC condensin I complex, subunit G | 218663_at |
| NDRG1 | CAP43, CMT4D, DRG1, GC4, HMSNL, N-myc Downstream Regulated 1, N-myc downstream regulatory protein 1, NDR1, NDRL, NMSL, PROXY1, RIT42, RTP, TARG1, TDD5 | N-myc downstream regulated 1 | 200632_s_at |
| NOTCH3 | AW229011, CADASIL, CASIL | Notch homolog 3 (Drosophila) | 203238_s_at |
| NPY1R | MGC109393, NPY receptor Y1, NPY Y1 receptor, NPY-1, NPYIR, NPYR, Y1, Y1 RECEPTOR, Y1-R | neuropeptide Y receptor Y1 | 205440_s_at |
| PCM1 | 2600002H09Rik, 9430077F19Rik, C030044G17Rik, LOC100044052, MGC170660, PTC4 | pericentriolar material 1 | 214118_x_at |
| PDZK1 | 1700023D20Rik, 2610507N21Rik, 4921513F16Rik, AI267131, AI314638, AL022680, CAP70, CLAMP, D3Ertd537e, mPDZK1, NHERF3, PDZD1, Sodium sulfate cotransporter | PDZ domain containing 1 | 205380_at |
| PTPN14 | C130080N23RIK, MGC126803, OTTMUSG00000022087, PEZ, PTP36, PTPD2 | protein tyrosine phosphatase, non-receptor type 14 | 205503_at |
| PVR | 3830421F03Rik, CD155, D7ERTD458E, FLJ25946, HVED, mE4, NECL5, NECTIN 2 ALPHA, NECTIN-2, Pe4, PVS, TAA1, TAGE4 | poliovirus receptor | 212662_at |
| RPS15 | EG633683, MGC111130, RIG | ribosomal protein S15 | 200819_s_at |
| RRM2 | AA407299, MGC113712, MGC116120, R2, Ribonucleoside-diphosphate reductase M2 subunit, Ribonucleotide reductase non-heme subunit, RIBONUCLEOTIDE REDUCTASE SMALL SUBUNIT, Rnr-r2, RNRII, RR2, RR2M | ribonucleotide reductase M2 polypeptide | 209773_s_at |
| SEL1L | AW493766, IBD2, KIAA4137, mKIAA4137, PRO1063, SEL1, SEL1-LIKE, SEL1H | sel-1 suppressor of lin-12-like (C. elegans) | 202063_s_at |
| SLC39A6 | Ermelin, LIV-1 | solute carrier family 39 (zinc transporter), member 6 | 202088_at |
| TMBIM4 | 0610007H07Rik, AU022431, CGI-119, GAAP, MGC73002, S1R, ZPRO | transmembrane BAX inhibitor motif containing 4 | 219206_x_at |
| UBE2B | 17-kDa Ubiquitin-Conjugating Enzyme E2, 2610301N02RIK, E2-14K, E2-17kDa, E2b, HHR6B, HR6B, LOC81816, RAD6B, UBC2 | ubiquitin-conjugating enzyme E2B (RAD6 homolog) | 211763_s_at |
| VEGFA | Gd-vegf, MGC70609, VEGF, Vegf-3, VEGF1, VEGF120, VEGF164, Vegf165, Vegf188, Vegfa 188, VPF, VPF/VEGF | vascular endothelial growth factor A | 210513_s_at |
Table 10. The 40 cancer related genes of the 100 features selected by SVMRFE on original training group of MAQC-II breast cancer data for erpos prediction.
| Symbol | Synonym(s) | Entrez Gene Name | Affymetrix |
| ANGPTL4 | ANGIOPOIETIN-LIKE 4, ANGPTL2, ARP4, Bk89, Fasting Induced Adipose Factor, FIAF, Harp, HFARP, LOC362850, Ng27, NL2, PGAR, PGARG, pp1158, PPARG | angiopoietin-like 4 | 221009_s_at |
| APP | A beta 25–35, A-BETA 40, A-BETA 42, AAA, ABETA, ABPP, AD1, Adap, AL024401, AMYLOID BETA, AMYLOID BETA 40, AMYLOID BETA 40 HUMAN, AMYLOID BETA 42, Amyloid beta A4, AMYLOID BETA PEPTIDE 40, Amyloid precursor, Amyloidogenic glycoprotein, App alpha, APPI, appican, BETAAPP, CTFgamma, CVAP, E030013M08RIK, Nexin II, P3, PN2, PreA4, PROTEASE NEXIN2 | amyloid beta (A4) precursor protein | 214953_s_at |
| CHRNB4 | Acrb-4, NACHR BETA4 | cholinergic receptor, nicotinic, beta 4 | 207516_at |
| CREBL2 | AI046348, B230205M03, MGC109304, MGC117311, MGC130380, MGC130381, MGC138362 | cAMP responsive element binding protein-like 2 | 201990_s_at |
| CTSL2 | 1190035F06Rik, Cathepsin l, CATHEPSIN V, CATHL, CATL2, Ctsl, Ctsl1, CTSU, CTSV, fs, MEP, MGC125957, nkt | cathepsin L2 | 210074_at |
| CX3CL1 | AB030188, ABCD-3, AI848747, C3Xkine, CX3C, CX3CL, CXC3, CXC3 CHEMOKINE PRECURSOR, CXC3C, D8Bwg0439e, FK, FKN, FRACTALKINE, NEUROTACTIN, NTN, NTT, SCYD1 | chemokine (C-X3-C motif) ligand 1 | 823_at |
| DACH1 | AI182278, Dac, DACH, E130112M23Rik, FLJ10138 | dachshund homolog 1 (Drosophila) | 205471_s_at |
| E2F1 | E2f, E2f1 predicted, KIAA4009, mKIAA4009, RBAP1, RBBP3, RBP3 | E2F transcription factor 1 | 204947_at |
| EPOR | EP-R, ERYTHROPOIETIN RECEPTOR, MGC108723, MGC138358 | erythropoietin receptor | 215054_at |
| ERBB4 | C-erb-b4, Erbb4 Cyt2, ERBB4 JM-A, HER4, MGC138404, p180erbB4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | 214053_at |
| ESR1 | AA420328, Alpha estrogen receptor, AU041214, DKFZp686N23123, ER, ER ALPHA, Er alpha (46 kDa isoform), ER66, ERA, ER[a], ESR, ESRA, ESTR, ESTRA, ESTROGEN RECEPTOR ALPHA, ESTROGEN RECEPTOR1, NR3A1, RNESTROR, TERP-1 | estrogen receptor 1 | 217190_x_at |
| FAM152A | 5830417C01Rik, C1orf121, CGI-146, DKFZp586C1019, FLJ21998, PNAS-4 | family with sequence similarity 152, member A | 212371_at |
| FBLN1 | FBLN, FIBULIN 1 | fibulin 1 | 207834_at |
| GATA3 | HDR, MGC2346, MGC5199, MGC5445 | GATA binding protein 3 | 209602_s_at |
| GHR | AA986417, GHBP, GHR/BP, GROWTH HORMONE RECEPTOR, MGC124963, MGC156665 | growth hormone receptor | 205498_at |
| GPC3 | DGSX, Glypican 3, GTR2-2, MGC93606, OCI-5, SDYS, SGB, SGBS, SGBS1 | glypican 3 | 209220_at |
| GREB1 | 5730583K22Rik, 9130004E13, AF180470, AU023194, KIAA0575, mKIAA0575 | GREB1 protein | 205862_at |
| IGF2BP3 | 2610101N11Rik, AA522010, Ab2-255, AL022933, AU045931, DKFZp686F1078, IMP-3, KOC1, Koc13, mimp3, Neilsen, RGD1306512, VICKZ3 | insulin-like growth factor 2 mRNA binding protein 3 | 203820_s_at |
| IL6ST | 5133400A03Rik, AA389424, Ac1055, BB405851, CD130, CDw130, D13Ertd699e, GP130, GP130-RAPS, Il6 transd, IL6R-beta | interleukin 6 signal transducer (gp130, oncostatin M receptor) | 212195_at |
| IRS1 | ENSMUSG00000022591, G972R, HIRS-1, IRS1IRM | insulin receptor substrate 1 | 204686_at |
| ITGB4 | AA407042, C230078O20, CD104, INTEGRIN-BETA 4 | integrin, beta 4 | 204990_s_at |
| ITPR1 | D6Pas2, I145TR, InsP3R, INSP3R1, IP3 RECEPTOR, Ip3 Receptor Type 1, IP3R, IP3R1, opt, P400, Pcd6, Pcp-1, SCA15, SCA16 | inositol 1,4,5-triphosphate receptor, type 1 | 211323_s_at |
| LMO4 | A730077C12Rik, Crp3, Etohi4, MGC105593 | LIM domain only 4 | 209205_s_at |
| MFAP3L | 4933428A15RIK, 5430405D20Rik, AI461995, AW125052, KIAA0626, mKIAA0626, NYD-sp9 | microfibrillar-associated protein 3-like | 210493_s_at |
| MGMT | AGAT, AGT, AI267024, ATase, MGC107020, O6-ALKYLGUANINE DNA ALKYLTRANSFERASE | O-6-methylguanine-DNA methyltransferase | 204880_at |
| PCNA | MGC8367, Pcna/cyclin, PCNAR | proliferating cell nuclear antigen | 217400_at |
| PGR | 9930019P03Rik, BB114106, ENSMUSG00000074510, LOC360433, NR3C3, Pgrb, PR, PR BETA, PR-A, PR-B, PROGESTERONE RECEPTOR, Progesterone receptor A | progesterone receptor | 208305_at |
| PLD1 | AA536939, C85393, Pc-Plc, Phospholipase D1, PLD1A, PLD1B, Plda, Pldb | phospholipase D1, phosphatidylcholine-specific | 215723_s_at |
| PML | 1200009E24Rik, AI661194, MYL, PP8675, Retinoic acid receptor, RGD1562602, RNF71, TRIM19 | promyelocytic leukemia | 211013_x_at |
| PVR | 3830421F03Rik, CD155, D7ERTD458E, FLJ25946, HVED, mE4, NECL5, NECTIN 2 ALPHA, NECTIN-2, Pe4, PVS, TAA1, TAGE4 | poliovirus receptor | 212662_at |
| RARRES1 | 5430417P09RIK, AI662122, TIG1, Tig1/retinoic acid receptor responder 1 | retinoic acid receptor responder (tazarotene induced) 1 | 206391_at |
| SCGB1D2 | BU101, LIPB, LIPOPHILIN B, LPHB | secretoglobin, family 1D, member 2 | 206799_at |
| SEL1L | AW493766, IBD2, KIAA4137, mKIAA4137, PRO1063, SEL1, SEL1-LIKE, SEL1H | sel-1 suppressor of lin-12-like (C. elegans) | 202063_s_at |
| SERPINA5 | 4933415L04, MGC93420, PAI-3, PCI, Pi5 alpha1, PLANH3, PROCI | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 | 209443_at |
| SERPINA6 | AI265318, AV104445, CBG, MGC112780 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 | 206325_at |
| SLC7A5 | 4f2 light chain, 4F2LC, CD98, CD98 LIGHT CHAIN, CD98LC, D0H16S474E, D16S469E, E16, E16/TA1, hLAT1, LAT1, MPE16, TA1 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 201195_s_at |
| SPAM1 (includes EG:6677) | 4933439A12Rik, HYA1, HYAL1, HYAL3, HYAL5, MGC108951, MGC26532, PH-20, SPAG15, SPAM1, TESTICULAR HYALURONIDASE | sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding) | 210536_s_at |
| TBC1D9 | 4933431N12RIK, AI847101, AW490653, C76116, KIAA0882, MDR1, RGD1308221 | TBC1 domain family, member 9 (with GRAM domain) | 212956_at |
| TFRC | 2610028K12Rik, AI195355, AI426448, AU015758, CD71, E430033M20Rik, Mtvr-1, p90, TFNR, TFR, TFR1, TRANSFERRIN RECEPTOR, TRFR | transferrin receptor (p90, CD71) | 208691_at |
| TRPV6 | ABP/ZF, Cac, CAT, CAT-L, CAT1, Crac, ECAC2, HSA277909, LP6728, Otrpc3, ZFAB | transient receptor potential cation channel, subfamily V, member 6 | 206827_s_at |
Figure 1 lists the testing accuracy, MCC values, and AUC errors on prediction of erpos by using two RFA methods: NBC-MSC and NMSC-MSC, as well as GLGS, LOOCSFS, and SVMRFE with the four learning classifiers. Regarding the prediction performance evaluated using testing accuracy and MCC values (left column and middle column, Fig. 1), on average the two RFA methods NBC-MSC and NMSC-MSC outperform other compared gene selection methods. The advantage of RFA by using NMSC and UDC is especially noticeable.
Figure 1. Comparison of different gene selection methods for prediction of erpos status of MAQC-II breast cancer dataset with different learning classifiers.
X-axis shows the number of used features and Y-axis shows average values of the testing accuracy (left column), MCC values (middle column), and AUC errors (right column) of twenty-time experiments, respectively.
We notice that the performance evaluation using testing accuracy (left column) and MCC (middle column) is consistent, but the performance evaluation using AUC measurement is not always consistent with the evaluation using testing accuracy and MCC. For instance, in applying UDC to the feature sets, RFA methods have the best prediction performance evaluated using testing accuracy and MCC values; however, with respect to performance evaluated using AUC errors RFA is not the best. Regarding the testing performance measured by using AUC errors, the best results have been obtained by using RFA with NMSC classifier. The AUC errors are as low as 0.08, which is much better than the results by using other methods of gene selection.
The prediction results on pCR endpoint, shown in Figure 2, also demonstrate the advantage of RFA methods over other compared methods. All the best prediction results, evaluated by using AUC, MCC and testing accuracy, are obtained by using RFA methods.
Figure 2. Comparison of different gene selection methods for prediction of pCR status of MAQC-II breast cancer dataset with different learning classifiers.
X-axis shows the number of used features and Y-axis shows average values of the testing accuracy (left column), MCC values (middle column), and AUC errors (right column) of twenty-time experiments, respectively.
Tables 11 and 12 list the average testing under the best training, MS_HR, and the best testing under the best training, HS_HR, for the predictions on erpos and pCR, respectively. Figure 3 and Figure 4 show box-plots of MS_HR values for the predictions on erpos and pCR. The results indicate that the prediction performance depends not only on gene selection but also on learning classifier. In other words, gene selection and classifier are coupled in determining prediction performance. On average, RFA gene selection methods deliver the best performance, followed by GLGS; NMSC classifier outperforms the others with respect to the performance and the consistency across the three measurements. The combination of RFA with NMSC has the best overall prediction performance.
Table 11. Mean values and standard errors of HS_HR and MS_HR on ERPOS prediction in the study of MAQC-II breast cancer dataset. In applying each classifier with each measurement, the best prediction value is in bold; the best prediction value of the results using the four learning classifiers is in bold and italic.
| MEASURE-MENTS | GENE SELECTION METHOD | MEAN(HS_HR)±STD(HS_HR), % | MEAN(MS_HR)±STD(MS_HR), % | ||||||
| NMSC | SVM | NBC | UDC | NMSC | SVM | NBC | UDC | ||
| Testing Accuracy | NBC-MSC | 89.5±2.2 | 79.3±2.2 | 82.9±1.1 | 91.6±1.7 | 88.5±1.4 | 76.5±0.5 | 81.7±0.3 | 91.0±1.6 |
| NMSC-MSC | 90.6±2.8 | 73.3±0 | 85.4±2.8 | 93.0±0.8 | 86.9±1.5 | 68.3±1.6 | 83.7±1.9 | 91.6±1.1 | |
| GLGS | 89.7±2.1 | 83.8±2.1 | 86.8±1.8 | 90.5±1.4 | 89.2±1.7 | 80.3±0.9 | 85.7±1.7 | 90.2±1.3 | |
| LOOCSFS | 86.4±0.8 | 66.7±1.9 | 86.9±1.4 | 86.3±1.6 | 86.1±0.8 | 65.1±0.9 | 86.2±1.0 | 85.8±1.4 | |
| SVMRFE | 77.3±7.2 | 84.4±0 | 77.6±6.6 | 77.5±8.9 | 75.6±6.7 | 78.4±0.1 | 76.7±6.1 | 75.8±7.6 | |
| MCC values | NBC-MSC | 76.7±4.5 | 56.1±4.7 | 63.1±2.3 | 81.9±3.7 | 75.3±3.3 | 50.5±1.0 | 60.4±0.7 | 81.0±3.4 |
| NMSC-MSC | 79.1±5.8 | 44.8±0 | 66.6±5.6 | 84.5±2.4 | 72.2±4.1 | 34.4±3.5 | 64.4±4.2 | 82.7±2.5 | |
| GLGS | 77.4±3.6 | 67.1±4.9 | 71.3±3.9 | 79.5±3.2 | 76.9±3.4 | 59.2±2.3 | 69.7±3.8 | 79.1±2.7 | |
| LOOCSFS | 70.8±1.7 | 27.0±5.6 | 70.9±3.3 | 69.7±3.3 | 70.5±1.8 | 24.4±3.0 | 70.2±2.9 | 69.6±3.3 | |
| SVMRFE | 51.9±14.5 | 66.6±0 | 53.9±11.9 | 53.8±16.6 | 51.0±13.9 | 53.8±0.3 | 53.5±11.5 | 52.3±15.0 | |
| AUC errors | NBC-MSC | 8.5±0.4 | 19.5±2.2 | 22.2±0.5 | 25.7±4.4 | 8.5±0.3 | 22.2±0.7 | 22.2±0.5 | 25.7±4.4 |
| NMSC-MSC | 8.4±0.7 | 23.5±0.4 | 23.1±1.3 | 17.1±3.6 | 8.4±0.7 | 28.2±2.3 | 23.1±1.3 | 17.1±3.6 | |
| GLGS | 8.0±0.4 | 10.3±1.8 | 16.7±1.0 | 15.1±1.5 | 8.0±0.4 | 13.4±1.1 | 16.7±1.0 | 15.1±1.5 | |
| LOOCSFS | 11.1±2.2 | 32.3±2.7 | 15.5±0.5 | 17.5±2.6 | 11.1±2.2 | 32.9±1.8 | 15.5±0.5 | 17.5±2.6 | |
| SVMRFE | 17.3±6.1 | 14.1±0 | 15.3±0.6 | 17.3±2.9 | 17.3±6.1 | 19.1±0.1 | 15.3±0.6 | 17.3±2.9 | |
Table 12. Mean values and standard errors of HS_HR and MS_HR on pCR prediction in the study of MAQC-II breast cancer dataset. In applying each classifier with each measurement, the best prediction value is in bold; the best prediction value is in bold and italic of the results by using four classifiers.
| MEASURE-MENTS | GENE SELECTION METHOD | MEAN(HS_HR)±STD(HS_HR), % | MEAN(MS_HR)±STD(MS_HR), % | ||||||
| NMSC | SVM | NBC | UDC | NMSC | SVM | NBC | UDC | ||
| Testing Accuracy | NBC-MSC | 88.7±2.3 | 88.9±0 | 90.9±1.0 | 88.6±0.8 | 87.5±1.9 | 83.1±0.3 | 86.6±0.6 | 87.7±1.6 |
| NMSC-MSC | 90.4±2.1 | 86.7±0.0 | 85.3±2.2 | 79.6±7.7 | 88.6±2.7 | 79.8±0.7 | 82.9±2.0 | 78.9±7.4 | |
| GLGS | 81.2±4.1 | 81.9±0.8 | 79.4±1.0 | 77.0±5.2 | 80.8±4.3 | 76.2±0.6 | 79.1±1.0 | 76.5±5.2 | |
| LOOCSFS | 77.0±1.1 | 82.1±0.5 | 72.1±1.7 | 74.1±3.1 | 76.6±1.2 | 78.1±2.5 | 71.4±1.8 | 73.2±2.7 | |
| SVMRFE | 81.9±2.9 | 88.9±0 | 79.1±2.6 | 80.7±1.9 | 80.9±2.7 | 85.3±0.2 | 77.9±2.3 | 80.3±1.8 | |
| MCC values | NBC-MSC | 62.5±6.7 | 63.9±0 | 69.7±3.4 | 60.7±2.8 | 61.3±5.9 | 48.8±0.6 | 54.7±2.1 | 59.2±5.0 |
| NMSC-MSC | 68.4±6.5 | 55.2±0 | 47.7±7.4 | 43.9±16.0 | 63.3±7.9 | 37.8±1.9 | 40.9±5.0 | 42.2±14.9 | |
| GLGS | 36.2±9.5 | 43.0±3.5 | 29.3±3.5 | 30.3±10.2 | 35.4±9.8 | 29.2±1.2 | 28.4±2.8 | 30.0±10.3 | |
| LOOCSFS | 23.4±4.9 | 38.5±3.5 | 18.7±4.7 | 21.6±11.0 | 23.0±5.1 | 26.3±5.1 | 18.2±5.0 | 21.1±10.7 | |
| SVMRFE | 40.9±7.9 | 62.5±0.0 | 32.6±6.9 | 41.1±4.7 | 39.4±7.4 | 49.1±0.7 | 30.5±5.5 | 40.7±4.9 | |
| AUC errors | NBC-MSC | 17.2±2.2 | 23.2±1.1 | 40.0±5.6 | 34.8±6.3 | 17.2±2.2 | 27.5±0.3 | 40.0±5.6 | 34.8±6.3 |
| NMSC-MSC | 14.4±3.0 | 19.1±0 | 47.9±3.7 | 46.0±5.1 | 14.4±3.0 | 28.2±1.1 | 47.9±3.7 | 46.0±5.1 | |
| GLGS | 22.6±1.3 | 29.3±0 | 37.4±1.8 | 36.7±1.8 | 22.6±1.3 | 33.5±0.3 | 37.4±1.8 | 36.7±1.8 | |
| LOOCSFS | 27.5±1.3 | 22.8±1.4 | 45.8±3.8 | 41.6±3.1 | 27.5±1.3 | 27.1±2.2 | 45.8±3.8 | 41.6±3.1 | |
| SVMRFE | 20.0±4.4 | 18.8±5.7 | 38.7±3.4 | 38.4±2.0 | 20.0±4.3 | 25.6±0.2 | 38.7±3.4 | 38.4±2.0 | |
Figure 3. Average erpos prediction performance by using MAQC-II breast cancer dataset with the measurements testing accuracy (left column), MCC values (middle column), and AUC errors (right column), respectively.
Classification models are setup based on the best training. In each column, the best combination of gene selection and classifier is highlighted by a red dash circle. If there are multiple best combinations, or the difference of these combinations is not conspicuous, multiple circles are placed.
Figure 4. Average pCR prediction performance by using MAQC-II breast cancer dataset with the measurements testing accuracy (left column), MCC values (middle column), and AUC errors (right column), respectively.
Classification models are setup based on the best training. In each column, the best combination of gene selection and classifier is highlighted by a dash circle.
Results on MAQC-II Multiple Myeloma Dataset
Figures 5 and 6 show the box-plots of average testing values for EFSMO and OSMO, the classification models are based on the best training. We did not apply GLGS because it would take an excessive amount of time for the identification of the feature sets on the multiple myeloma dataset. Experimental results again manifest that gene selection is strictly coupled to learning classifier in performance measurement. On average, RFA methods and LOOCSFS are superior to SVMRFE.
Figure 5. Average EFSMO prediction performance by using MAQC-II multiple myeloma dataset with the measurements testing accuracy (left column), MCC values (middle column), and AUC errors (right column), respectively.
Classification models are setup based on the best training. In each column, the best combination of gene selection and classifier is highlighted by a dash circle.
Figure 6. Average OSMO prediction performance by using MAQC-II multiple myeloma dataset with the measurements testing accuracy (left column), MCC values (middle column), and AUC errors (right column), respectively.
Classification models are setup based on the best training. In each column, the best combination of gene selection and classifier is highlighted by a dash circle. If there are multiple best combinations, or the difference of these combinations is not conspicuous, multiple circles are placed.
Discussion
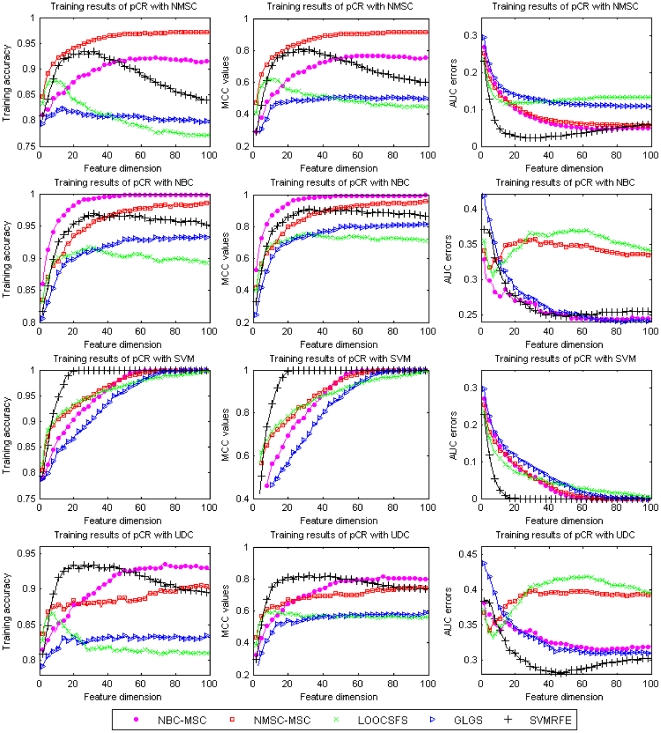
Due to a huge number of variables and small sample size, there are complicated interactions and relations among genes as well as high redundancy information with microarray data. The selection of predictive models that depend on selected features and employed classifiers is extremely important for the classification of microarray data and for the further biological function analysis/validation. Machine learning and data mining techniques provide us with a powerful approach to the study of the relationship among genes. Based on supervised learning and similarity measurements, we propose a Recursive Feature Addition (RFA), recursively employ supervised learning to obtain the highest training accuracy and add a subsequent gene based on the similarity between the chosen features and the candidates to minimize the redundancy within the feature set. We believe this RFA method captures more informative and differently expressed genes than other methods. Experimental comparisons are performed by using two MAQC-II microarray datasets, breast cancer and multiple myeloma. Our studies show that the method of gene selection is strictly paired with learning classifier, which determines the final predictive model by using training data. In other words, the best classification models under different learning classifiers are associated with different methods of gene selection. Using several popular learning classifiers including NMSC, NBC, SVM, and UDC, on average, the best method of gene selection is RFA, followed by GLGS, LOOCSFS, and SVMRFE. Regarding compared learning classifiers, NMSC outperforms the others with respect to testing performance, stabilization, and consistency.
Biological function analysis based on MAQC-II breast cancer dataset finds that our applied feature selection methods including RFA, GLGS, LOOCSFS, and SVMRFE can generate features containing a significant portion of known cancer related genes for both pCR and erpos endpoints (Tables 1– 10). Although the cancer related gene number is not absolutely correlated with the prediction performance of various methods of feature selection, the remarkable cancer related genes in the features indicate that the feature selection methods including RFA, GLGS, LOOCSFS, and SVMRFE could produce biologically meaningful features, which will convince the users to apply them for phenotype prediction.
In all results of the five gene selection methods with the four learning classifiers, on average, the combination of gene selections of NMSC-MSC and NBC-MSC with the learning classifier of NMSC has performed the best, regarding the comprehensive evaluation criteria of testing accuracy, MCC values, and AUC errors. It should be noted that the gene selection methods of NMSC-MSC and NBC-MSC are not always the best over the four learning classifiers, in other words, the best models among different learning classifiers are associated with different gene selection methods. The selection of the best model with the use of a specific learning classifier is normally based on the training and the evaluation criteria. Under an evaluation criterion with the use of some learning classifier, the best training model among the five gene selection methods is selected as the best model under the learning classifier. To select the best model among the four learning classifiers, the best models among the four learning classifiers are compared and the model obtaining the highest evaluation score is generally selected the best among the five gene selection methods and the four learning classifiers. For instance, Figure 7 demonstrates the average training performance of MAQC-II breast cancer dataset over twenty times, with the measurements of training accuracy, MCC values, and AUC errors, for the classification of pCR endpoint status. Regarding the comprehensive evaluation of the three criteria, the best classification models are obtained by using gene selection method NMSC-MSC for learning classifier NMSC, NBC-MSC for NBC classifier, and NBC-MSC and SVMRFE for UDC classifier. With the use of SVM classifier, although the gene selection method of SVMRFE first hits the best training as the feature dimension increases, almost all gene selection methods achieve the best before the feature number increases to 100, in such case, it is hard for us to determine the best model with the use of learning classifier SVM. In the limit of feature number, we can say that SVMRFE is the best for SVM classifier. The comparison between the training (Figure 7) and the testing (Figure 2) shows that such selection of the best model among the various methods of gene selection and various learning classifiers is reasonable.
Figure 7. Comparison of different gene selection methods for the training of pCR endpoint of MAQC-II breast cancer dataset using the four classifiers.
X-axis shows the number of used features and Y-axis shows average values of the training accuracy (left column), MCC values (middle column), and AUC errors (right column) of twenty-time experiments, respectively.
Regarding different evaluation criteria, the gene selection associated with the best model under a learning classifier may be different, as shown in Figure 7; with the use of learning classifier NMSC, the best model is obtained by using gene selection method of NMSC-MSC, evaluated by training accuracy and MCC values, the best model is associated with SVMRFE, evaluated by AUC errors. Overall, NMSC-MSC is the best for learning classifier NMSC. Figure 7 shows that dozens of the best training models are obtained by using different methods of gene selection with the use of SVM classifier. One possible solution for the selection of the best model under SVM classifier is to divide all data points into training set, validation set, and testing set. The training set is used to construct training models, the validation set is used to select the best model by applying the validation data to the best training models and selecting the best training model that produce the best validation result. The testing set is used for prediction or testing. The selection of the best model using SVM classifier is very interesting and challenging, especially in the case of small data points and huge number of features. Although the topic is beyond the scope of this paper, it is worthy to be explored in the future study.
Materials and Methods
MAQC-II Breast Cancer Dataset
The breast cancer dataset used in the MAQC-II project is used to predict the pre-operative treatment response (pCR) and estrogen receptor status (erpos) [28]. The normalization was provided by MAQC-II project using standard procedure (i.e., MAS 5.0 for Affymetrix platform). It was originally grouped into two groups: a training set containing 130 samples (33 positives and 97 negatives for pCR, 80 positives and 50 negatives for erpos), and a validation set containing 100 samples (15 positives and 85 negatives for pCR, 61 positives and 39 negatives for erpos).
MAQC-II Multiple Myeloma Dataset
We take the MAQC-II multiple myeloma dataset to predict the overall survival milestone outcome (OSMO, 730-day cutoff) and to predict event-free survival milestone outcome (EFSMO, 730-day cutoff). For OSMO label information, there are 51 positives and 289 negatives in original training set, 27 positives and 187 negatives in original validation set; as for EFSMO label information, there are 84 positives and 256 negatives in original training set, 34 positives and 180 negatives in original validation set [28]. The normalization was provided by MAQC-II project research group.
Feature Selection
Supervised recursive learning
Our method of recursive feature addition (RFA) employs supervised learning to achieve the best training accuracy and uses statistical similarity measures to choose the next variable with the least dependence on, or correlation to, the already identified variables as follows:
Insignificant genes are removed according to their statistical insignificance. Specifically, a gene with a high p-value is usually not differently expressed and therefore has little contribution to classification of microarray data. To reduce the computational load, those insignificant genes are removed.
Each individual gene is selected by supervised learning. A gene with highest classification accuracy is chosen as the most important feature or the first element of the feature set. If multiple genes achieve the same highest classification accuracy, the one with the lowest p-value measured by test-statistics (e.g., score test), is identified as the first element. At this point the chosen feature set, G 1, contains just one element, g 1, corresponding to the feature dimension one.
The (N+1)st dimensional feature set, G N +1 = {g 1, g 2, …, g N, g N +1} is obtained by adding g N +1 to the Nth dimensional feature set, G N = {g 1, g 2, …, g N}. The choice of g N+1 is processed as follows:
Add each gene gi (gi
 G
N) into G
N and have the classification accuracy of the feature set G
N
G
N) into G
N and have the classification accuracy of the feature set G
N
 {gi}. The gi (gi
{gi}. The gi (gi
 G
N) associated with the group, G
N
G
N) associated with the group, G
N
 {gi} that obtains the highest classification accuracy, is the candidate for g
N+1 (not yet g
N+1). Considering the large number of variables, it is very likely that multiple features correspond to the same highest classification accuracy, these multiple candidates are placed into the set C, but only one candidate in C will be identified as g
N+1. How to make the selection is described next.
{gi} that obtains the highest classification accuracy, is the candidate for g
N+1 (not yet g
N+1). Considering the large number of variables, it is very likely that multiple features correspond to the same highest classification accuracy, these multiple candidates are placed into the set C, but only one candidate in C will be identified as g
N+1. How to make the selection is described next.
Candidate feature addition
To find a most informative (or least redundant) candidate for g N+ 1, we measure the statistical similarity between the chosen features and each candidate. We design a similarity measurement with the use of a widely-used Pearson's correlation coefficient [30].
Suppose gn (gn
 G
N, n = 1, 2, …, N) is a chosen gene, gc (gc
G
N, n = 1, 2, …, N) is a chosen gene, gc (gc
 C) is a candidate gene, and cor stands for the function of Pearson's correlation coefficient. The sum of the square of the correlation SC that is calculated as follows:
C) is a candidate gene, and cor stands for the function of Pearson's correlation coefficient. The sum of the square of the correlation SC that is calculated as follows:
| (1) |
Then selection of g N+1 is based on Minimal value of the Square of the Correlation (MSC), that is,
| (2) |
In the methods mentioned above, a feature is recursively added to the chosen feature set based on supervised learning and the similarity measurement. In our experiments we choose naïve bayes classier (NBC) and nearest means scale classifier (NMSC) [29] for supervised learning, NBC and NMSC-based RFA feature selection methods are denoted as NBC-MSC and NMSC-MSC, respectively.
Model Implementation and Comparison
Cross-validation is a technique for estimating how accurately a predictive model will perform in practice. Generally, the data are partitioned into complementary subsets, one subset (called the training set) is used for constructing the predictive model and the other subset (called the validation set or testing set) is used for validation. To reduce variability, multiple rounds are performed using different partitions, and the validation results are averaged over all rounds. There are three common types of cross-validation:
Repeated random sub-sampling validation. This technique randomly splits the dataset intro training and testing data. The results are then averaged over the splits. The advantage over K-fold cross validation (described below) is that the portion of the training/testing split is not dependent on the number of iterations (folds);
K-fold cross-validation. The original sample is partitioned into K subsamples. A single subsample is retained as the validation data for testing the model and the remaining K-1 subsamples are used as training data. The cross-validation process is repeated K times with each of the K subsamples used exactly once as the validation data;
Leave-one-out cross-validation. It uses a single observation from the original sample as the validation data and the remaining observations as the training data. It is the same as a K-fold cross validation with K being equal to the number of observations in the original sample. Leave-one-out cross-validation is often computationally expensive.
Considering the high computational requirement of leave-one-out cross-validation and the insufficiency of one time K-fold cross-validation, we took the strategy of repeated random sub-sampling validation. In the model implementation, we mixed all the training data points and validation points. In each experiment, we randomly chose 80% of samples for training and the remaining 20% of samples for testing. Twenty experiments were performed (this strategy is approximately equal to performing 5-fold cross validation four times). The average testing performances, evaluated in terms of testing accuracy, MCC values, and AUC errors, were compared. The learning classifiers UDC, NBC, NMSC, and SVM were employed for training and testing.
Acknowledgments
The authors gratefully acknowledge Drs. Matteo Masotti and E Ke Tang for offering their codes of RfeRanking, GLGS, and LOOCSFS; Drs. Weita Tong and Leming Shi at the National Center for Toxicological Research, US Food and Drug Administration, for their invaluable help during this study; and Ms. Kimberly Lawson at the Department of Radiology of the Brigham and Women's Hospital and Harvard Medical School for her excellent assistance in revising the manuscript. The MAQC-II consortium for making the data sets available to this study is gratefully appreciated.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The first two authors gratefully appreciate the support for this study from the Institute for Complex Additive Systems Analysis, a research division of New Mexico Tech. The last author highly appreciate the support under the grant of NIH/NCMHD/RIMI, P20MD002725. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chen Z, McGee M, Liu Q, Scheuermann RH. A distribution free summarization method for affymetrix genechip arrays. Bioinformatics. 2007;23(3):321–327. doi: 10.1093/bioinformatics/btl609. [DOI] [PubMed] [Google Scholar]
- 2.Hand DJ, Heard NA. Finding groups in gene expression data. J Biomed Biotechnol. 2005;2005(2):215–225. doi: 10.1155/JBB.2005.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin Z. Clustering microarray gene expression data using weighted Chinese restaurant process. Bioinformatics. 2006;22(16):1988–1997. doi: 10.1093/bioinformatics/btl284. [DOI] [PubMed] [Google Scholar]
- 4.Quackenbush J. Computational analysis of microarray data. Nature Rev Genetic. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- 5.Segal E, Friedman N, Kaminski N, Regev A, Koller D. From signatures to models: understanding cancer using microarrays. Nature Genetics. 2005;37:S38–45. doi: 10.1038/ng1561. [DOI] [PubMed] [Google Scholar]
- 6.Torrente A, Kapushesky M, Brazma A. A new algorithm for comparing and visualizing relationships between hierarchical and flat gene expression data clusterings. Bioinformatics. 2005;21(21):3993–3999. doi: 10.1093/bioinformatics/bti644. [DOI] [PubMed] [Google Scholar]
- 7.Brazma A, Krestyaninova M, Sarkans U. Standards for system biology. Nat Rev Gent. 2006;7:593–605. doi: 10.1038/nrg1922. [DOI] [PubMed] [Google Scholar]
- 8.Bo T, Jonassen I. New feature subset selection procedures for classification of expression profiles. Genome Biology. 2002;3(4):research0017. doi: 10.1186/gb-2002-3-4-research0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long A, Mangalam H, Chan B, Tolleri L, Hatfield G, et al. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. J Biol Chem. 2001;276:19937–19944. doi: 10.1074/jbc.M010192200. [DOI] [PubMed] [Google Scholar]
- 10.Newton MA, Kendziorski CM, Richmond CS, Blattner FR, Tsui KW. On differential variability of expression ratios: improving statistical inference about gene expression changes from microarray data. Journal of Computational Biology. 2001;8(1):37–52. doi: 10.1089/106652701300099074. [DOI] [PubMed] [Google Scholar]
- 11.Yu J, Chen XW. Bayesian neural network approaches to ovarian cancer identification from high-resolution mass spectrometry data. Bioinformatics. 2005;21(suppl-1):i487–i494. doi: 10.1093/bioinformatics/bti1030. [DOI] [PubMed] [Google Scholar]
- 12.Pavlidis P, Noble WS. Analysis of strain and regional variation in gene expression in mouse brain. Genome Biology. 2001;2(10):research0042.1–0042.15. doi: 10.1186/gb-2001-2-10-research0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inza I, Sierra B, Blanco R, Larranaga P. Gene selection by sequential search wrapper approaches in microarray cancer class prediction. Journal of Intelligent and Fuzzy Systems. 2002;12(1):25–33. [Google Scholar]
- 14.Guyon I, Weston J, Barnhill S, Vapnik VN. Gene selection for cancer classification using support vector machines. Machine Learning. 2002;46(1–3):389–422. [Google Scholar]
- 15.Liu Q, Sung AH, Chen Z, Xu J. Feature mining and pattern classification for steganalysis of LSB matching steganography in grayscale images. Pattern Recognition. 2008;41(1):56–66. [Google Scholar]
- 16.Monari G, Dreyfus G. Withdrawing an example from the training set: an analytic estimation of its effect on a nonlinear parameterized model. Neurocomputing Letters. 2000;35:195–201. [Google Scholar]
- 17.Rivals I, Personnaz L. MLPs (mono-layer polynomials and multi-layer perceptrons) for nonlinear modeling. Journal of Machine Learning Research. 2003;3:1383–1398. [Google Scholar]
- 18.Tibshirani R. Regression shrinkage and selection via the lasso. J Royal Statist Soc B. 1996;58(1):267–288. [Google Scholar]
- 19.Tibshirani R. The lasso method for variable selection in the cox model. Statistics in Medicine. 1997;16:385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 20.Krishnapuram B, Carin L, Figueiredo M, Hartemink A. Sparse multinomial logistic regression: fast algorithms, and generalization bounds. IEEE Trans Pattern Analysis and Machine Intelligence. 2005;27(6):957–968. doi: 10.1109/TPAMI.2005.127. [DOI] [PubMed] [Google Scholar]
- 21.Liang Y, Kelemen A. Temporal gene expression classification with regularised neural network. International Journal of Bioinformatics Research and Applications. 2005;1(4):399–413. doi: 10.1504/IJBRA.2005.008443. [DOI] [PubMed] [Google Scholar]
- 22.Zhou X, Mao KZ. LS bound based gene selection for DNA microarray data. Bioinformatics. 2005;21(8):1559–1564. doi: 10.1093/bioinformatics/bti216. [DOI] [PubMed] [Google Scholar]
- 23.Tang EK, Suganthan PN, Yao X. Gene selection algorithms for microarray data based on least square support vector machine. BMC Bioinformatics. 2006;7:95. doi: 10.1186/1471-2105-7-95. doi: 10.1186/1471-2105-7-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diaz-Uriarte R, de Andres SA. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 26.Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 27.Baldi P, Brunak S, Chauvin Y, Andersen CA, Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- 28.Shi L, et al. The MAQC-II project: a comprehensive study of common practices for the development and validation of microarray-based predictive models. Nature biotechnology. doi: 10.1038/nbt.1665. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heijden F, Duin R, Ridder D, Tax D. Classification, parameter estimation and state estimation. John Wiley; 2004. [Google Scholar]
- 30.Tan P, Steinbach M, Kumar V. Introduction to data mining. Addison-Wesley; 2005. pp. 76–79. [Google Scholar]