Abstract
BACKGROUND
The authors previously showed that increased tumor expression levels of B7-H1, survivin, and Ki-67 are independent predictors of poor outcome for patients with clear cell renal cell carcinoma (ccRCC). In the current study, they described the creation of a scoring system based on this panel of biomarkers that can be used in tandem with existing clinicopathologic features and algorithms to improve ccRCC outcome prediction.
METHODS
The authors used immunohistochemistry to determine tumor expression levels of B7-H1, survivin, and Ki-67 for 634 consecutive ccRCC patients. A multivariate model verified that each biomarker was independently associated with RCC-specific death after adjusting for the remaining 2. A biomarker-based panel, termed BioScore, was generated to predict the likelihood of RCC-specific death. BioScore was tested for its ability to enhance the performance of several clinicopathologic features and algorithms.
RESULTS
Patients with high BioScores were 5 times more likely to die from RCCcompared with patients with low BioScores (hazard ratio, 5.03; 95% confidence interval, 3.82–6.61; P < .001). Multivariate adjustment for individual clinicopathologic features or existing prognostic algorithms failed to attenuate this positive association. Moreover, an examination of concordance indexes revealed that BioScore significantly enhanced the prognostic ability of each of the individual prognostic features or algorithms studied.
CONCLUSIONS
The authors described the creation of BioScore, a biomarker-based scoring system that can be used in tandem with established prognostic algorithms to further enhance ccRCC outcome prediction. The need for external validation notwithstanding, they envision that BioScore can be readily updated as new biomarkers are identified.
Keywords: kidney neoplasms, carcinoma, renal cell, tumor biomarkers, biological, survival
Annually, renal cell carcinoma (RCC) and cancer of the renal pelvis account for more than 50,000 new cases and nearly 13,000 cancer deaths in the United States alone.1 The most common RCC histologic subtype is clear cell RCC (ccRCC), which behaves more aggressively than the papillary and chromophobe subtypes, particularly among patients with localized tumors, and accounts for the majority of RCC-related deaths. Surveillance, Epidemiology, and End Results data further indicate that a recent rise in RCC incidence is likely attributable to increased image-based detection of organ-confined tumors that are seemingly curable by surgical resection.2,3 Despite this, 10% to 30% of localized RCC tumors will still progress, typically within the first 3 years after surgery.4–6 Progression from localized to metastatic RCC results in a precipitous decline in 5-year survival from 60% to <10%.7 As such, considerable emphasis has been placed on the development of predictive tools that can help forecast outcomes for surgically treated RCC patients, guide postoperative surveillance protocols, and more accurately pinpoint high-risk patients who might benefit from off-label or clinical trial-based adjunctive therapy to preempt cancer relapse.
Several clinicopathologic scoring systems (also referred to as nomograms or algorithms) have been reported to predict outcomes for surgically treated RCC patients. Such algorithms include the 2002 American Joint Committee on Cancer (AJCC) TNM stage groupings, 8 the UCLA Integrated Scoring System (UISS),9 nomograms from Memorial Sloan-Kettering Cancer Center, 10,11 and the Mayo Clinic stage, size, grade, and necrosis (SSIGN) score.12,13 In general, the features entered into these prediction algorithms can be extracted from standard-of-care patient history and pathology reports, rendering them easily accessible to clinicians. These scoring systems, however, do not fully account for the varied outcomes associated with RCC, and fail to reveal the molecular basis for RCC aggressiveness or rational targets for therapy. As a result, there is considerable interest in the identification of tumor-associated biomarkers that might enhance RCC prognostication and guide development of new therapies.
Understanding how biomarker data might best be applied to enhance the predictive capabilities of existing clinicopathologic algorithms, however, has evolved slowly and remains largely exploratory in nature. One approach has been to integrate biomarkers and clinicopathologic information into a single multivariate model to predict outcome. For example, Kim et al14 recently developed a hybrid algorithm that integrates biomarker data into a statistically retooled form of the UISS. Unquestionably, this approach ties together biomarker and clinicopathologic information in an effort to improve assessment of RCC prognosis. There are, however, notable drawbacks to this approach. For instance, incorporation of new and nonvalidated biomarker data into a statistically revised version of the UISS algorithm, by definition, necessitates revalidation of the entire algorithm. In addition, proxy usage of biomarker readouts, in lieu of traditional clinicopathologic features of RCC, places such integrated algorithms outside of standard-of-care practice, ultimately adding to expense and complexity, while lending only modest increments in overall prognostic capability. Moreover, integrated algorithms cannot be easily updated or judiciously applied in a stepwise fashion commensurate with patient need or risk. Lastly, and perhaps most importantly, complete integration of biomarker readouts into a clinicopathologic algorithm causes the algorithm to become biomarker-specific and, conversely, biomarkers to become algorithm-specific, thereby restricting widespread use of these tools for RCC prognostication.
Herein, we describe an alternative approach to combining biomarkers with existing clinicopathologic algorithms as a means of improving outcome prediction. Our sequential approach permits biomarker data to be judiciously used only for those patients in greatest need of prognostic refinement. Moreover, our approach allows for use of biomarkers in tandem with a variety of existing clinicopathologic scoring systems, thus broadening their overall applicability while obviating the need to retool and revalidate these systems. Specifically, we combine tumor expression levels of 3 biomarkers (B7-H1, survivin, and Ki-67), each previously reported by our group as an independent predictor of RCC outcome, into a single scoring panel, collectively termed BioScore, that can be used to refine outcome prediction provided by existing clinicopathologic algorithms.
MATERIALS AND METHODS
Patient Selection and Centralized Review of Pathologic Features
After institutional review board approval, we identified 818 patients treated with radical nephrectomy or nephron-sparing surgery for unilateral, sporadic, noncystic ccRCC between 1990 and 1999 from the Mayo Clinic Nephrectomy Registry. Of these, 634 (77.5%) patients had representative paraffin-embedded tissue blocks available for immunohistochemical staining. We noted no statistically significant differences in overall survival (P=.60) or cancer-specific survival (P=.20) between patients with and without tissue available for analysis.
The clinicopathologic features evaluated included age at surgery, sex, symptoms at presentation, Eastern Co-operative Oncology Group (ECOG) performance status, histologic subtype, the 2002 AJCC primary tumor classification, regional lymph node involvement, distant metastases, the 2002 AJCC TNM stage groupings, tumor size, nuclear grade, and presence of coagulative tumor necrosis. To obtain these pathologic features in a standardized fashion, 1 urologic pathologist centrally reviewed all hematoxylin and eosin–stained specimens without knowledge of patient outcome.
Tumor B7-H1, Survivin, and Ki-67 Expression
A detailed description of our methods for immunohistochemical staining of tumor B7-H1, survivin, and Ki-67 expression can be found in Thompson et al,15 Parker et al,16,17 and Tollefson et al,18 respectively. Briefly, we identified a paraffin-embedded block with representative tumor tissue for each patient in our cohort. From each block, we obtained 3 5-μ-thick slides for immunostaining, which was performed using monoclonal antibodies and the respective protocols for each biomarker as previously reported.
The study pathologist (Y.S.) reviewed the stained slides to determine tumor expression levels of the 3 biomarkers. The membranous staining pattern of B7-H1 was quantified as the percentage of positive tumor cells in 5% to 10% increments. The nuclear staining patterns of survivin and Ki-67 were quantified as the number of positive tumor cells in each of 5 representative high-powered fields using a Leica DMR microscope (Leica Microsystems, Wetzlar, Germany). With a 10/25 eyepiece and a × 40 objective, the Leica DMR has an object field diameter of 0.625 mm2, resulting in a high-powered field of 0.307 mm2. As such, survivin and Ki-67 expression were quantified as the number of positive tumor cells per mm2.
Statistical Methods
Cancer-specific survival was estimated using the Kaplan-Meier method and compared among groups using log-rank tests. The associations of biomarker expression and clinicopathologic features with RCC-specific death were estimated using Cox proportional hazards regression models and summarized with hazard ratios (HRs) and 95% confidence intervals (CIs). For these models, B7-H1 expression was dichotomized as positive versus negative, because nearly 85% of the tumors were B7-H1 negative. As described in a previous publication, a tumor is considered B7-H1 positive if there is histologic evidence of cell-surface membrane staining in at least 5% of tumor cells; otherwise, a tumor is considered B7-H1 negative.15 Survivin and Ki-67 expression were analyzed as continuous variables (ie, as the number of survivin-positive and Ki-67–positive tumor cells per mm2) and as categorical variables in an attempt to simplify the interpretation of the association of these biomarkers with patient outcome. We categorized survivin and Ki-67 expression based on the quartiles of their distributions, and dichotomized expression as high versus low. As established in previous publications, 16–18 survivin expression was classified as low or high (<15 vs ≥15 survivin-positive tumor cells per mm2), and Ki-67 expression was classified as low or high (<50 vs ≥50 Ki-67–positive tumor cells per mm2). A Poisson regression approach19 to observe the functional form of the associations of survivin and Ki-67 expression (as continuous variables) with cancer-specific survival further supported our choice of cut points for dichotomizing these 2 biomarkers.
We first estimated the univariate association of each biomarker with time to RCC-specific death. Survivin and Ki-67 expression were modeled as continuous variables, using the quartiles of their distributions, and as high versus low. Once the positive association of each biomarker with RCC-specific death was verified, we evaluated the potential for pairwise interactions. Given that we did not detect evidence of significant interactions (all interaction P values were >.85), we evaluated multivariate models containing all 3 biomarkers to assess the association of each biomarker with RCC-specific death after adjusting for the effects of the other 2. As with our univariate analyses, survivin and Ki-67 expression were modeled as continuous variables, using the quartiles of their distributions, and as high versus low. The concordance (c) index was used to compare the predictive ability of these multivariate models. After confirming the independent predictive ability of each biomarker after adjusting for the other 2, we used regression coefficients from the model containing dichotomous versions of B7-H1, survivin, and Ki-67 to develop a biomarker-based scoring algorithm, termed BioScore, to predict the likelihood of RCC-specific death. Lastly, we constructed a series of models containing either individual clinicopathologic features or established RCC prognostic systems, including the TNM stage groupings, the UISS, and the Mayo Clinic SSIGN score. We then added BioScore to each of these models and summarized the increase in the c index to evaluate the ability of BioScore to enhance the prognostic ability of individual features and commonly used algorithms. Throughout, all reported c indexes were internally validated using a bootstrap methodology proposed by Harrell et al,20 and therefore represent optimism-corrected estimates of prognostic accuracy. Statistical analyses were performed using the SAS (SAS Institute, Cary, NC) and S-Plus (Insightful Corporation, Seattle, Wash) software packages. All tests were 2-sided and P values <.05 were considered statistically significant.
RESULTS
Patient Characteristics and Outcome
Clinicopathologic features for the 634 patients under study are summarized in Table 1. At last follow-up, 359 patients had died, including 211 who died from RCC at a mean of 3.3 years after surgery (median, 2.1 years; range, 0.1–14.0 years). Among the 275 patients who were still alive at last follow-up, the mean duration of follow-up was 10.4 years (median, 10.3 years; range, 0.1–17.2 years); only 9 (3.3%) patients had fewer than 2 years of follow-up. Estimated cancer-specific survival rates (standard error, number still at risk) at 1, 3, 5, 7, and 10 years after surgery were 89.8% (1.2%, 543), 79.0% (1.7%, 456), 73.1% (1.8%, 394), 69.1% (1.9%, 347), and 64.4% (2.1%, 190), respectively.
Table 1.
Clinicopathologic Features for 634 Patients With ccRCC
| Feature | No. (%) |
|---|---|
| Age at Surgery, y | |
| <65 | 322 (50.8) |
| ≥65 | 312 (49.2) |
| Sex | |
| Women | 221 (34.9) |
| Men | 413 (65.1) |
| Symptoms | |
| Absent | 228 (36.0) |
| Present | 406 (64.0) |
| Constitutional symptoms | |
| Absent | 472 (74.5) |
| Present | 162 (25.6) |
| ECOG performance status | |
| 0 | 572 (90.2) |
| ≥1 | 62 (9.8) |
| Primary tumor classification | |
| pT1a | 163 (25.7) |
| pT1b | 182 (28.7) |
| pT2 | 103 (16.3) |
| pT3a | 59 (9.3) |
| pT3b | 109 (17.2) |
| pT3c | 10 (1.6) |
| pT4 | 8 (1.3) |
| Regional lymph node involvement | |
| pNX and pN0 | 607 (95.7) |
| pN1 and pN2 | 27 (4.3) |
| Distant metastases | |
| pM0 | 564 (89.0) |
| pM1 | 70 (11.0) |
| TNM stage groupings | |
| I | 331 (52.2) |
| II | 81 (12.8) |
| III | 142 (22.4) |
| IV | 80 (12.6) |
| Tumor size, cm | |
| <5 | 216 (34.1) |
| ≥5 | 418 (65.9) |
| Nuclear grade | |
| 1 | 43 (6.8) |
| 2 | 293 (46.2) |
| 3 | 240 (37.9) |
| 4 | 58 (9.1) |
| Coagulative tumor necrosis | |
| Absent | 442 (69.7) |
| Present | 192 (30.3) |
| UISS | |
| I | 224 (35.3) |
| II | 322 (50.8) |
| III | 17 (2.7) |
| IV | 68 (10.7) |
| V | 3 (0.5) |
| SSIGN score | |
| Low, 0–2 | 279 (44.0) |
| Intermediate, 3–6 | 200 (31.6) |
| High, 7+ | 155 (24.4) |
ccRCC indicates clear cell renal cell carcinoma; ECOG, Eastern Cooperative Oncology Group; UISS, UCLA Integrated Scoring System; SSIGN, Mayo Clinic stage, size, grade, and necrosis.
Association of B7-H1, Survivin, and Ki-67 With RCC-specific Death
Of the 634 patients evaluated, 97 (15.3%) had B7-H1–positive tumors. The mean level of survivin expression was 15.5 survivin-positive tumor cells per mm2 (median, 7.6 cells; range, 0.0–157.0 cells); 198 (31.2%) patients had survivin-high tumors (≥15 survivin-positive tumor cells per mm2). The mean level of Ki-67 expression was 63.6 Ki-67–positive tumor cells per mm2 (median, 36.7 cells; range, 0.8–501.3 cells); 245 (38.6%) patients had Ki-67-high tumors (≥50 Ki-67–positive tumor cells per mm2).
Estimates of the univariate and multivariate associations of each biomarker with RCC-specific death are summarized in Table 2. For these analyses, survivin and Ki-67 expression were modeled as continuous variables, using the quartiles of their distributions, and as high versus low. In a multivariate setting, each biomarker remained significantly associated with an increased risk of RCC-specific death even after adjusting for the effects of the other 2. The optimism-corrected c indexes from the 3 multivariate models presented were 0.752, 0.745, and 0.733, respectively. Because our goal was to obtain a biomarker-based scoring algorithm that was both easy to use and easy to interpret, the remaining analyses used the dichotomized versions of survivin and Ki-67 expression with minimal loss of information. The distribution of coexpression levels of these 3 biomarkers is shown in Table 3; 50.5% of patients had tumors with negative B7-H1 expression and low survivin and Ki-67 expression.
Table 2.
Univariate and Multivariate Associations of B7-H1, Survivin, and Ki-67 Expression With RCC-specific Death for 634 Patients With ccRCC
| Biomarker | Univariate | Multivariate* | ||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Survivin and Ki-67 as continuous | ||||
| B7-H1 | ||||
| Negative | 1.0 (reference) | 1.0 (reference) | ||
| Positive | 3.98 (2.96–5.33) | <.001 | 1.97 (1.41–2.75) | <.001 |
| Survivin, 5-unit increase | 1.17 (1.15–1.20) | <.001 | 1.11 (1.08–1.14) | <.001 |
| Ki-67, 5-unit increase | 1.04 (1.03–1.05) | <.001 | 1.02 (1.02–1.03) | <.001 |
| Survivin and Ki-67 as quartiles | ||||
| B7-H1 | ||||
| Negative | 1.0 (reference) | 1.0 (reference) | ||
| Positive | 3.98 (2.96–5.33) | <.001 | 2.14 (1.54–2.96) | <.001 |
| Survivin | ||||
| 1st quartile, 0–2.9 | 1.0 (reference) | 1.0 (reference) | ||
| 2nd quartile, 3.0–7.9 | 1.40 (0.84–2.33) | .193 | 1.19 (0.71–1.99) | .517 |
| 3rd quartile, 8.0–19.9 | 1.92 (1.16–3.18) | .011 | 1.28 (0.75–2.20) | .360 |
| 4th quartile, ≥20 | 6.74 (4.28–10.64) | <.001 | 2.85 (1.66–4.87) | <.001 |
| Ki-67 | ||||
| 1st quartile, 0–14.9 | 1.0 (reference) | 1.0 (reference) | ||
| 2nd quartile, 15.0–34.9 | 1.62 (0.94–2.78) | .080 | 1.50 (0.86–2.62) | .152 |
| 3rd quartile, 35.0–74.9 | 2.34 (1.41–3.90) | .001 | 1.81 (1.06–3.09) | .031 |
| 4th quartile, ≥75 | 6.36 (3.98–10.19) | <.001 | 3.17 (1.85–5.43) | <.001 |
| Survivin and Ki-67 as dichotomous | ||||
| B7-H1 | ||||
| Negative | 1.0 (reference) | 1.0 (reference) | ||
| Positive | 3.98 (2.96–5.33) | <.001 | 2.16 (1.57–2.96) | <.001 |
| Survivin | ||||
| Low, <15 | 1.0 (reference) | 1.0 (reference) | ||
| High, ≥15 | 4.56 (3.46–6.00) | <.001 | 2.76 (2.00–3.80) | <.001 |
| Ki-67 | ||||
| Low, <50 | 1.0 (reference) | 1.0 (reference) | ||
| High, ≥50 | 3.42 (2.59–4.52) | <.001 | 1.95 (1.42–2.67) | <.001 |
RCC indicates renal cell carcinoma; ccRCC, clear cell renal cell carcinoma; HR, hazard ratio; CI, confidence interval.
Multivariate HRs for each biomarker were adjusted for the effects of the remaining 2.
Table 3.
Distribution of Combined Tumor Expression of B7-H1, Survivin, and Ki-67 Among 634 Patients With ccRCC
| B7-H1 | Survivin | Ki-67 | No. (%) |
|---|---|---|---|
| Negative | Low | Low | 320 (50.5) |
| Negative | Low | High | 91 (14.4) |
| Negative | High | Low | 42 (6.6) |
| Negative | High | High | 84 (13.3) |
| Positive | Low | Low | 15 (2.4) |
| Positive | Low | High | 10 (1.6) |
| Positive | High | Low | 12 (1.9) |
| Positive | High | High | 60 (9.5) |
ccRCC indicates clear cell renal cell carcinoma.
Development and Evaluation of BioScore
To create BioScore, we used regression coefficients from the model in Table 2 containing the dichotomized versions of survivin and Ki-67 expression. We first divided the regression coefficient for each of the biomarkers by the coefficient for high survivin expression, multiplied that number by 3, and rounded to the nearest integer. As a result, a patient with a B7-H1–negative, survivin-low, and Ki-67-low tumor would define a baseline patient with a BioScore of 0. However, if the patient’s tumor was B7-H1 positive, 2 points would be added to their BioScore. Similarly, patients with survivin-high and Ki-67–high tumors would have 3 and 2 points added to their BioScore, respectively. These biomarker weights, which collectively constitute our BioScore panel, are summarized in Table 4. The maximum BioScore possible is 7; BioScores of 1 and 6 are not possible.
Table 4.
Biomarker Weights Used to Calculate BioScore
| Biomarker | Score |
|---|---|
| B7-H1 | |
| Negative | 0 |
| Positive | 2 |
| Survivin | |
| Low, <15 | 0 |
| High, ≥15 | 3 |
| Ki-67 | |
| Low, <50 | 0 |
| High, ≥50 | 2 |
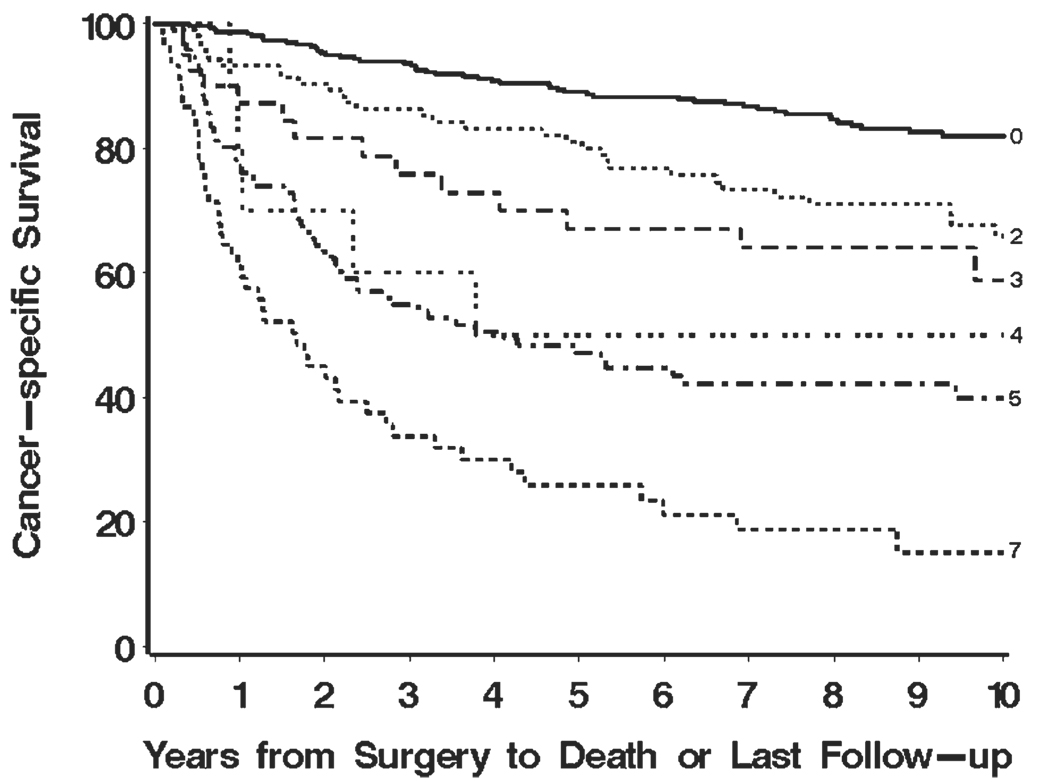
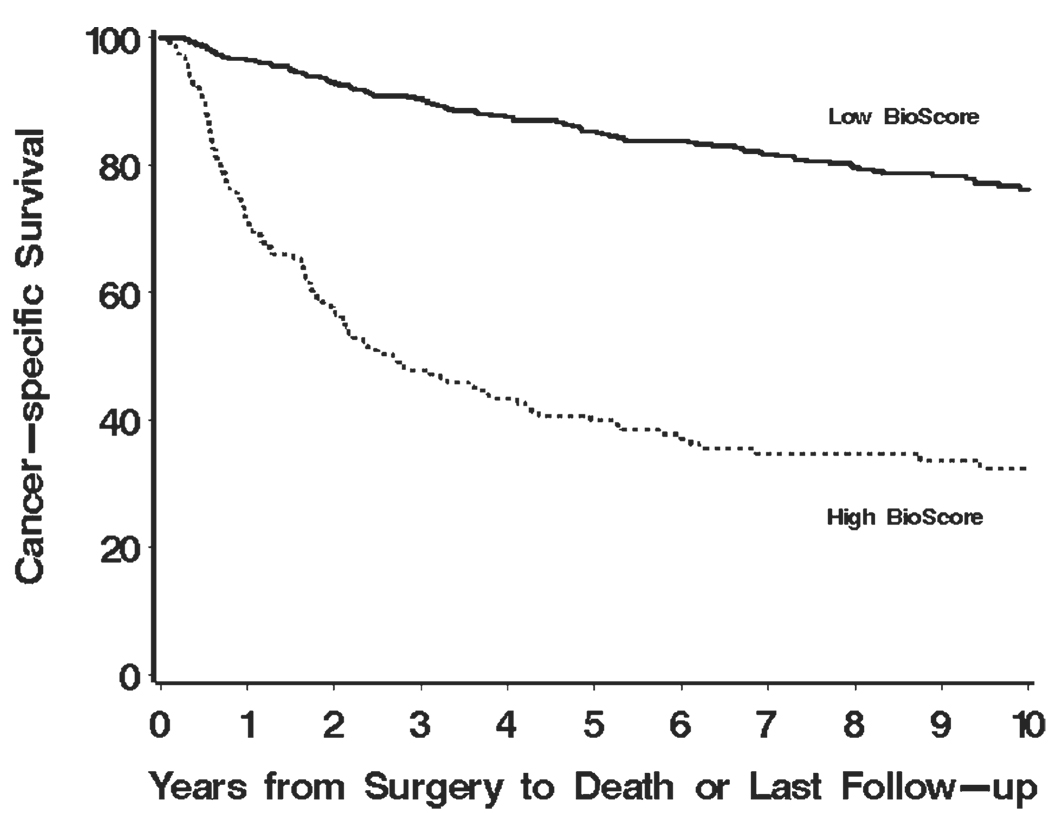
The mean BioScore for our study cohort was 2.0 (median, 0; range, 0–7). Figure 1 illustrates that higher BioScores are associated with poorer cancer-specific survival (P < .001). For easier interpretation, we also dichotomized BioScore into 2 groups: those with scores of 0, 2, or 3 (herein referred to as low BioScore) and those with scores of 4, 5, or 7 (herein referred to as high Bio-Score). Using this designation, there were 468 (73.8%) patients with low BioScores and 166 patients (26.2%) with high BioScores. Figure 2 displays the disparity in cancer-specific survival for patients with low and high BioScores (P < .001). Patients with high BioScores were observed to be 5 times more likely to experience RCC-specific death compared with patients with low BioScores (HR, 5.03; 95% CI, 3.82–6.61).
FIGURE 1.
Estimated cancer-specific survival after surgery by BioScore is shown for 634 patients with clear cell renal cell carcinoma.
FIGURE 2.
Estimated cancer-specific survival after surgery by BioScore is shown for 634 patients with clear cell renal cell carcinoma.
Ability of BioScore to Enhance Clinicopathologic Prognostic Features
Table 5 summarizes the ability of BioScore to enhance ccRCC patient outcome prediction for a variety of individual clinicopathologic features as well as for 3 established multivariate scoring systems, including the TNM stage groupings, the UISS, and the Mayo Clinic SSIGN score. We provide a comparison of the optimism-corrected c index from a model containing each feature or algorithm alone and the optimism-corrected c index from a model containing each feature or algorithm plus the dichotomized BioScore. In each case, the addition of the dichotomized BioScore provided additional prognostic information to the model. For example, when we added BioScore to a model with nuclear grade alone, the c index increased from 0.768 to 0.792. Similarly, when we added BioScore to a model with UISS alone, the c index increased from 0.774 to 0.819. The c indexes from models that contained the SSIGN score alone and the SSIGN score plus BioScore were 0.821 and 0.837, respectively. Even after adjusting for the SSIGN score, patients with high BioScores were twice as likely to die from RCC compared with patients with low BioScores (HR, 2.00; 95% CI, 1.48–2.72; P < .001). When we repeated this analysis using BioScore as a continuous variable instead of the dichotomized version, we noted even greater improvements in c index values. Furthermore, the optimism-corrected estimates of slope shrinkage20 for the 3 models listed above were 0.99, 0.98, and 0.99, respectively, indicating that these models were well calibrated (data not shown).
Table 5.
Univariate and BioScore-adjusted Associations of Clinicopathologic Features and Algorithms With RCC-specific Death for 634 Patients With ccRCC
| Feature | Univariate* | BioScore-adjusted* | ||
|---|---|---|---|---|
| HR (95% CI) | c Index | HR (95% CI) | c Index | |
| Age at surgery, y | ||||
| <65 | 1.0 | 1.0 | ||
| ≥65 | 0.97 (0.74–1.28) | 0.502 | 0.80 (0.61–1.06) | 0.692 |
| Sex | ||||
| Women | 1.0 | 1.0 | ||
| Men | 1.18 (0.88–1.58) | 0.510 | 1.16 (0.87–1.55) | 0.701 |
| Symptoms | ||||
| Absent | 1.0 | 1.0 | ||
| Present | 2.55 (1.83–3.57) | 0.595 | 2.01 (1.43–2.82) | 0.722 |
| Constitutional symptoms | ||||
| Absent | 1.0 | 1.0 | ||
| Present | 2.74 (2.08–3.60) | 0.614 | 2.12 (1.60–2.81) | 0.726 |
| ECOG performance status | ||||
| 0 | 1.0 | 1.0 | ||
| ≥1 | 0.91 (0.56–1.50) | 0.502 | 0.95 (0.58–1.57) | 0.695 |
| Primary tumor classification | ||||
| pT1a | 1.0 | 1.0 | ||
| pT1b | 4.71 (2.20–10.09) | 3.91 (1.82–8.40) | ||
| pT2 | 11.09 (5.22–23.56) | 10.74 (5.05–22.85) | ||
| pT3a | 18.22 (8.47–39.17) | 13.02 (6.01–28.21) | ||
| pT3b, pT3c, and pT4 | 23.83 (11.52–49.28) | 0.759 | 15.57 (7.45–32.52) | 0.800 |
| Regional lymph node involvement | ||||
| pNX and pN0 | 1.0 | 1.0 | ||
| pN1 and pN2 | 4.72 (3.00–7.43) | 0.546 | 3.65 (2.30–5.78) | 0.700 |
| Distant metastases | ||||
| pM0 | 1.0 | 1.0 | ||
| pM1 | 7.99 (5.90–10.81) | 0.639 | 6.04 (4.41–8.27) | 0.750 |
| TNM stage groupings | ||||
| I | 1.0 | 1.0 | ||
| II | 3.06 (1.84–5.10) | 3.43 (2.06–5.73) | ||
| III | 7.34 (4.94–10.90) | 5.58 (3.72–8.35) | ||
| IV | 20.17 (13.41–30.34) | 0.791 | 16.21 (10.65–24.66) | 0.816 |
| Tumor size, cm | ||||
| <5 | 1.0 | 1.0 | ||
| ≥5 | 5.43 (3.52–8.38) | 0.645 | 4.48 (2.89–6.93) | 0.746 |
| Nuclear grade | ||||
| 1 and 2 | 1.0 | 1.0 | ||
| 3 | 7.22 (4.96–10.52) | 5.83 (3.95–8.61) | ||
| 4 | 21.14 (13.55–32.97) | 0.768 | 11.96 (7.27–19.67) | 0.792 |
| Coagulative tumor necrosis | ||||
| Absent | 1.0 | 1.0 | ||
| Present | 7.22 (5.42–9.61) | 0.737 | 5.02 (3.66–6.88) | 0.769 |
| UISS | ||||
| I | 1.0 | 1.0 | ||
| II | 9.52 (5.26–17.24) | 7.94 (4.37–14.44) | ||
| III | 18.21 (8.02–41.33) | 16.91 (7.43–38.45) | ||
| IV and V | 51.96 (27.84–96.96) | 0.774 | 35.76 (18.89–67.69) | 0.819 |
| SSIGN score | ||||
| Low, 0–2 | 1.0 | 1.0 | ||
| Intermediate, 3–6 | 5.59 (3.31–9.44) | 5.34 (3.16–9.03) | ||
| High, 7+ | 30.61 (18.58–50.41) | 0.821 | 22.55 (13.39–37.97) | 0.837 |
RCC indicates renal cell carcinoma; ccRCC, clear cell renal cell carcinoma; HR, hazard ratio; CI, confidence interval; c, concordance; ECOG, Eastern Cooperative Oncology Group; UISS, UCLA Integrated Scoring System; SSIGN, Mayo Clinic stage, size, grade, and necrosis.
All P values <.001 except for age (univariate P =.84; BioScore-adjusted P =.12); sex (univariate P=.26; BioScore-adjusted P=.32); and ECOG performance status (univariate P =.72; BioScore-adjusted P =.85).
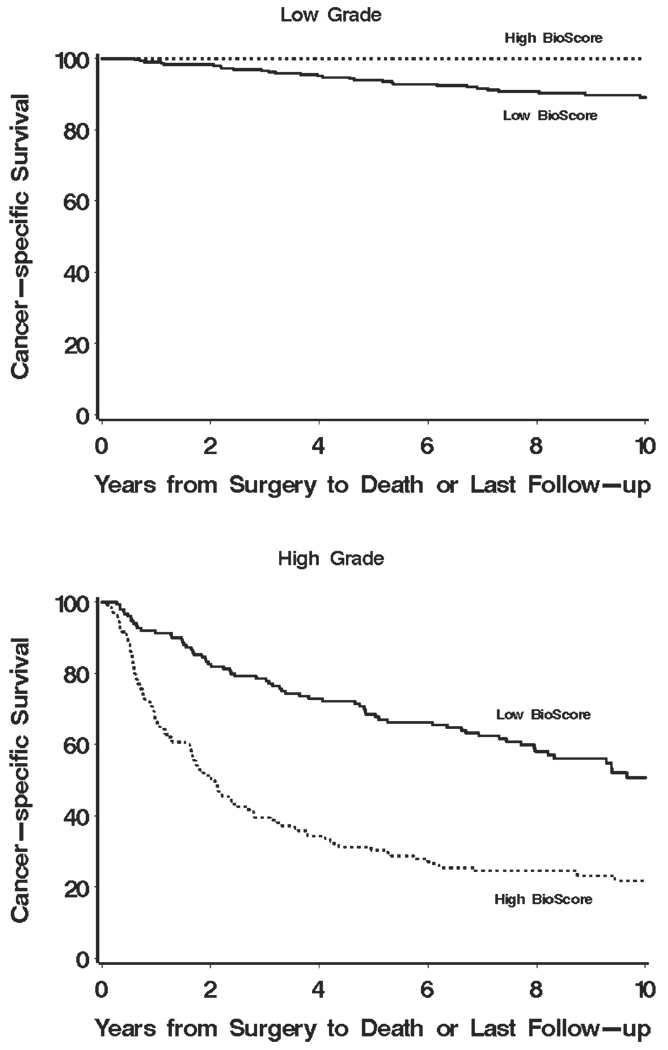
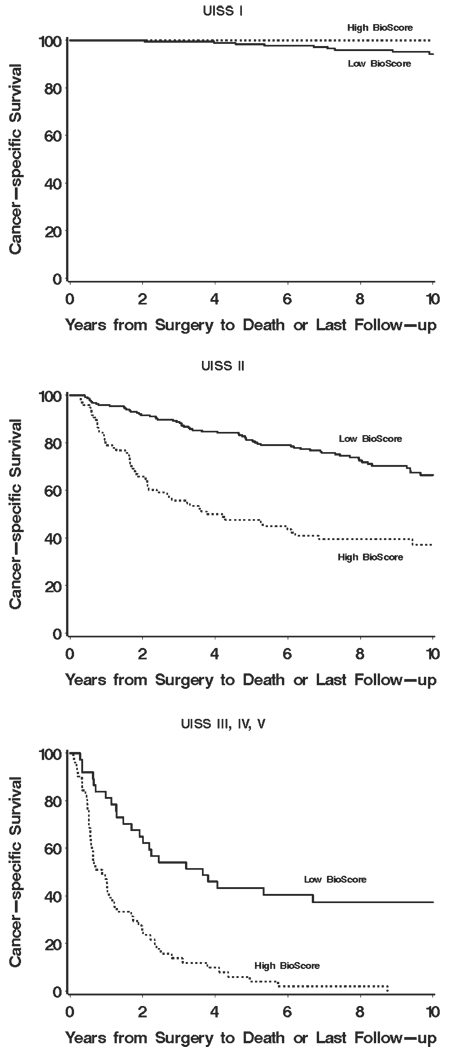
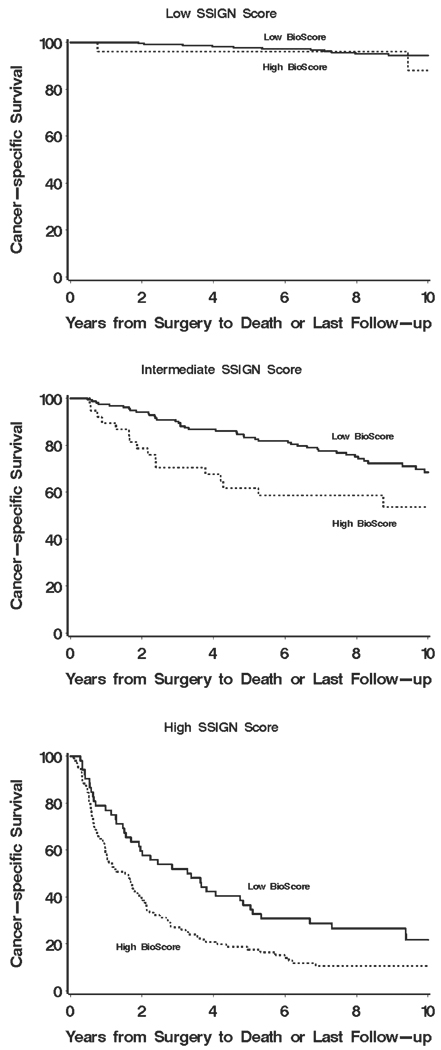
To better illustrate our proposed sequential approach, we have provided a series of Kaplan-Meier curves supporting the overall concept of how BioScore (or any biomarker-based algorithm) can be used in tandem with established prognostic features and algorithms to improve ccRCC outcome prediction. Figure 3 shows how BioScore can be used to further refine risk categories already defined by nuclear grade. Little information was gained by applying BioScore to those patients with low (1 or 2) grade tumors (Fig. 3 Top; P = .75). In contrast, the application of BioScore to those patients with high (3 or 4) grade tumors clearly stratified patients into 2 distinct survival groups (Fig. 3 Bottom; P < .001). Moving from individual features to the setting of more sophisticated prognostic algorithms, Figure 4 demonstrates the potential for BioScore to further refine outcome stratification as initially provided by the UISS. Similarly, it is apparent that BioScore provided little improvement in the ability to further stratify patients already classified to be at low risk of RCC-specific death (Fig. 4 Top; UISS I; P = .93). In contrast, among those patients classified in the more moderate (UISS II) and severe (UISS III, IV, and V) categories, BioScore was able to provide additional information to further stratify patient outcome (Fig. 4 Middle and Bottom; P < .001 for both). We noted similar results regarding the ability of BioScore to further stratify patients after initial classification by the Mayo Clinic SSIGN score. Indeed, Figure 5 (Top) shows that BioScore had limited ability to improve upon stratification among patients already predicted to be at low risk of RCC-specific death based on the SSIGN score (Fig. 5 Top; P = .06). In contrast, among patients initially predicted to be at intermediate risk and high risk by the SSIGN score, BioScore provided a significant degree of further stratification (Fig. 5 Middle and Bottom; P=.009 and P=.003, respectively). Cancer-specific survival rates at 1, 3, 5, 7, and 10 years after surgery for Figure 1–Figure 5 are available from the authors upon request.
FIGURE 3.
Estimated cancer-specific survival after surgery by BioScore is shown among patients classified as low grade (Top) and high grade (Bottom).
FIGURE 4.
Estimated cancer-specific survival after surgery by BioScore is shown among patients classified as UCLA Integrated Scoring System (UISS) I (Top), UISS II (Middle), and UISS III-V (Bottom).
FIGURE 5.
Estimated cancer-specific survival after surgery by BioScore is shown among patients with low (Top), intermediate (Middle), and high (Bottom) Mayo Clinic stage, size, grade, and necrosis scores.
DISCUSSION
RCC patients find little solace in knowing that they are at “variable risk” for developing metastatic disease after surgery, particularly when faced with limited prospects for effective therapy. As such, a key advancement in the management of RCC patients will be to more accurately pinpoint an individual’s postoperative risk for cancer progression, to better formulate patient surveillance schedules and to streamline clinical trial testing of promising agents.21 A second obvious advancement will be elucidation of rational molecular targets for novel RCC therapy development.22 It is principally for these 2 reasons that studies pertaining to identification and validation of RCC biomarkers have recently flourished.
Before discussing the exploitation of biomarkers for the purpose of enhancing RCC outcome prediction, it is important to clarify that many reported RCC biomarkers tend to exhibit only univariate associations with RCC outcome, in essence mirroring disease severity. Such biomarkers may demarcate potentially promising molecular targets or pathways for treatment, but should not be empirically construed as biomarkers that can be used to enhance the prognostic performance of existing RCC algorithms. Conversely, a few biomarkers have been reported to convey independent prognostic information even after multivariate adjustment for established prognostic clinicopathologic features of RCC.15–18,23–25 Examples of such biomarkers include, but are not limited to, B7-H1, survivin, and Ki-67—3 biomarkers that we selected for this study based on their potential to refine clinicopathologic algorithms for RCC outcome prediction.
The optimal approach for use of biomarkers to enhance RCC outcome prediction remains in question. Is it better to integrate biomarker readouts into reformulated clinicopathologic algorithms, as some groups in the field have reported? Or should biomarker information be developed into standalone scoring tools that can be sequentially applied (when necessary) to further refine outcome prediction estimates already provided by existing, validated clinicopathologic algorithms?
An example of the full integration of biomarker information into an existing clinicopathologic algorithm, namely the UISS, is provided by Kim et al.14 Specifically, these authors first screened 8 tumor biomarkers (gelsolin, p53, CAIX, Ki-67, vimentin, CA12, EpCAM, and PTEN) for their potential to function as predictors of RCC outcome. On the basis of their screening, the authors concluded that tumor expression levels of gelsolin, p53, and vimentin, as well as the presence of metastases and the interaction between tumor CAIX expression and metastases, represented key independent predictors of RCC outcome. In a subsequent step, the authors statistically integrated these biomarkers into the UISS algorithm, which is comprised of the TNM stage groupings, ECOG performance status, and nuclear grade. During this integration process, gelsolin expression and nuclear grade were dropped as predictive variables, leaving T stage, presence of metastases at surgery, ECOG performance status, tumor expression of p53 and vimentin, and an interaction between CAIX and metastases in the final algorithm. Of note, the authors reported that this biomarker integration resulted in a new scoring system with improved predictive ability over UISS alone (c indexes of 0.79 and 0.75, respectively). Given that the original UISS includes nuclear grade (which is a potent predictive feature for RCC outcome), whereas their hybrid biomarker-UISS algorithm does not, one might argue that tumor expression of p53, vimentin, and CAIX may be serving as surrogates for pathologic components of the original UISS, as opposed to additional prognostic features that can enhance overall prediction. Nevertheless, this study encompasses an important first step toward the use of biomarkers to improve the capabilities of established clinicopathologic algorithms to predict RCC outcome.
Herein, we evaluate the feasibility and advantages of a sequential approach, rather than the fully integrated approach described above. Further impetus for our approach is provided by the following considerations. Specifically, multiple clinicopathologic algorithms for RCC constructed from standard-of-care patient and pathology data have already been reported, validated, and more or less assimilated by clinicians. As a result, reconfiguration of these algorithms to accommodate biomarker information necessitates their revalidation as prognostic instruments. Related to this, statistical retooling of existing algorithms already regarded as somewhat cumbersome by many clinicians causes these algorithms to grow even more unwieldy. Integration of biomarker data into existing algorithms has 2 additional unfavorable effects. First, integrated algorithms cannot be readily updated upon discovery of new prognostic biomarkers and, once again, must be revalidated with the introduction of each new biomarker. Second, the full integration of biomarker readouts into clinicopathologic algorithms causes algorithms to become biomarker-specific and, conversely, the incorporated biomarkers to become algorithm-specific. This precludes widespread use of such scoring instruments across institutions, because many clinicians tend to have a favorite parent algorithm that they use to assess RCC patient risk. Finally, as we demonstrate in this investigation, not all RCC patients require biomarker analysis to refine prognosis. As such, it can be argued that integrated algorithms that fully incorporate biomarkers and clinicopathologic features into 1 monolithic model will ultimately prove unnecessarily wasteful for certain patients, especially those who are already at very low risk of RCC-specific death.
Given its freestanding design, 1 advantage of Bio-Score is that it can be selectively applied to patients in greatest need of refined risk characterization. Specifically, we demonstrate that BioScore provides little to no advantage for patients at low risk for RCC-specific death as defined by nuclear grade, the UISS, or the Mayo Clinic SSIGN score, a finding that is not particularly surprising given that low-risk patients experience 5-year cancer-specific survival rates exceeding 95%. Conversely, BioScore appears to markedly enhance nuclear grade, UISS, or SSIGN score prediction for patients who fall into intermediate-risk or high-risk categories. Thus, BioScore can be discriminately applied to refine outcome prediction for intermediate-risk to high-risk RCC patients, while sparing low-risk patients from unnecessary laboratory analysis. Lastly, as we demonstrate, BioScore can theoretically be used across a variety of existing clinicopathologic prognostic RCC algorithms without any requirements for statistical remodeling or algorithm revalidation.
Several limitations pertaining to our current study merit further discussion. For instance, although we show that BioScore can be used to enhance risk assessment provided by the TNM and UISS, our analysis of BioScore to modify prognostication rendered by the SSIGN score remains incomplete. Given that the SSIGN score was generated using clinicopathologic features from 1800 patients, we estimate that it will require an analysis of nearly that many to fully test the ability of BioScore to refine every prognostic category defined by the SSIGN score. In addition, the patients in the current study represent the practice of a tertiary referral center and are nearly all Caucasian, which limits the generalizability of our findings. Lastly, the ability of BioScore to enhance prognostication provided by clinicopathologic tools will need to be more stringently established, preferably through the conduct of large-scale, prospective, and independent validation.
Conclusions
Distinct from a prior study in which biomarkers were immutably blended into a particular clinicopathologic algorithm to predict RCC outcome, BioScore encompasses a freestanding biomarker panel that can be readily updated and selectively used, in tandem and across a variety of existing clinicopathologic prognostic algorithms, to enhance RCC outcome prediction. Large-scale, prospective studies will be needed to externally validate BioScore and test its utility for RCC outcome prediction in the clinical trial and clinical practice setting.
Footnotes
Conflict of Interest Disclosures
Some of the authors have filed patent applications pertaining to cancer prognostic markers, including B7-H1, survivin, and Ki-67.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Chow WH, Devesa SS, Warren JL, et al. Rising incidence of renal cell cancer in the United States. JAMA. 1999;281:1628–1631. doi: 10.1001/jama.281.17.1628. [DOI] [PubMed] [Google Scholar]
- 3.Wallen EM, Pruthi RS, Joyce GF, et al. Urologic Diseases in America Project. Kidney cancer. J Urol. 2007;177:2006–2018. doi: 10.1016/j.juro.2007.01.126. [DOI] [PubMed] [Google Scholar]
- 4.Ljungberg B, Alamdari FI, Rasmuson T, et al. Follow-up guidelines for nonmetastatic renal cell carcinoma based on the occurrence of metastases after radical nephrectomy. BJU Int. 1999;84:405–411. doi: 10.1046/j.1464-410x.1999.00202.x. [DOI] [PubMed] [Google Scholar]
- 5.Levy DA, Slaton JW, Swanson DA, et al. Stage specific guidelines for surveillance after radical nephrectomy for local renal cell carcinoma. J Urol. 1998;159:1163–1167. [PubMed] [Google Scholar]
- 6.Lam JS, Leppert JT, Figlin RA, et al. Surveillance following radical or partial nephrectomy for renal cell carcinoma. Curr Urol Rep. 2005;6:7–18. doi: 10.1007/s11934-005-0062-x. [DOI] [PubMed] [Google Scholar]
- 7.Figlin R, Pierce WC, Kaboo R, et al. Treatment of metastatic renal cell carcinoma with nephrectomy, interleukin-2 and cytokine-primed or CD8(+) selected tumor infiltrating lymphocytes from primary tumor. J Urol. 1997;158:740–745. doi: 10.1097/00005392-199709000-00012. [DOI] [PubMed] [Google Scholar]
- 8.Ficarra V, Galfano A, Mancini M, et al. TNM staging system for renal-cell carcinoma: current status and future perspectives. Lancet Oncol. 2007;8:554–558. doi: 10.1016/S1470-2045(07)70173-0. [DOI] [PubMed] [Google Scholar]
- 9.Zisman A, Pantuck AJ, Dorey F, et al. Improved prognostication of renal cell carcinoma using an integrated staging system. J Clin Oncol. 2001;19:1649–1657. doi: 10.1200/JCO.2001.19.6.1649. [DOI] [PubMed] [Google Scholar]
- 10.Kattan MW, Reuter V, Motzer RJ, et al. A postoperative nomogram for renal cell carcinoma. J Urol. 2001;166:63–67. [PubMed] [Google Scholar]
- 11.Sorbellini M, Kattan MW, Snyder ME, et al. A postoperative prognostic nomogram predicting recurrence for patients with conventional clear cell renal cell carcinoma. J Urol. 2005;173:48–51. doi: 10.1097/01.ju.0000148261.19532.2c. [DOI] [PubMed] [Google Scholar]
- 12.Frank I, Blute ML, Cheville JC, et al. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. J Urol. 2002;168:2395–2400. doi: 10.1016/S0022-5347(05)64153-5. [DOI] [PubMed] [Google Scholar]
- 13.Ficarra V, Martignoni G, Lohse C, et al. External validation of the Mayo Clinic stage, size, grade and necrosis (SSIGN) score to predict cancer specific survival using a European series of conventional clear cell renal cell carcinoma. J Urol. 2006;175:1235–1239. doi: 10.1016/S0022-5347(05)00684-1. [DOI] [PubMed] [Google Scholar]
- 14.Kim HL, Seligson D, Liu X, et al. Using tumor markers to predict the survival of patients with metastatic renal cell carcinoma. J Urol. 2005;173:1496–1501. doi: 10.1097/01.ju.0000154351.37249.f0. [DOI] [PubMed] [Google Scholar]
- 15.Thompson RH, Kuntz SM, Leibovich BC, et al. Tumor B7-H1 is associated with poor prognosis in renal cell carcinoma patients with long-term follow-up. Cancer Res. 2006;66:3381–3385. doi: 10.1158/0008-5472.CAN-05-4303. [DOI] [PubMed] [Google Scholar]
- 16.Parker AS, Kosari F, Lohse CM, et al. High expression levels of survivin protein independently predict a poor outcome for patients who undergo surgery for clear cell renal cell carcinoma. Cancer. 2006;107:37–45. doi: 10.1002/cncr.21952. [DOI] [PubMed] [Google Scholar]
- 17.Parker AS, Lohse CM, Leibovich BC, et al. Comparison of digital image analysis versus visual assessment to assess survivin expression as an independent predictor of survival for patients with clear cell renal cell carcinoma. Hum Pathol. 2008;39:1176–1184. doi: 10.1016/j.humpath.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tollefson MK, Thompson RH, Sheinin Y, et al. Ki-67 and coagulative tumor necrosis are independent predictors of poor outcome for patients with clear cell renal cell carcinoma and not surrogates for each other. Cancer. 2007;110:783–790. doi: 10.1002/cncr.22840. [DOI] [PubMed] [Google Scholar]
- 19.Therneau T, Grambsch P. Modeling Survival Data: Extending the Cox Model. 1st ed. Ann Arbor, MI: Springer-Verlag; 2000. pp. 87–92. [Google Scholar]
- 20.Harrell FE, Lee KL, Mark DB. Multivariate prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Atkins MB, Avigan DE, Bukowski RM, et al. Innovations and challenges in renal cancer: consensus statement from the first international conference. Clin Cancer Res. 2004;10(18 pt 2):6277S–6281S. doi: 10.1158/1078-0432.CCR-040720. [DOI] [PubMed] [Google Scholar]
- 22.Lam JS, Shvarts O, Leppert JT, et al. Renal cell carcinoma 2005: new frontiers in staging, prognostication and targeted molecular therapy. J Urol. 2005;173:1853–1862. doi: 10.1097/01.ju.0000165693.68449.c3. [DOI] [PubMed] [Google Scholar]
- 23.Tunuguntla HS, Jorda M. Diagnostic and prognostic molecular markers in renal cell carcinoma. J Urol. 2008;179:2096–2102. doi: 10.1016/j.juro.2008.01.083. [DOI] [PubMed] [Google Scholar]
- 24.George S, Bukowski RM. Biomarkers in clear cell renal cell carcinoma. Exp Rev Anticancer Ther. 2007;7:1737–1747. doi: 10.1586/14737140.7.12.1737. [DOI] [PubMed] [Google Scholar]
- 25.Ljungberg B. Prognostic markers in renal cell carcinoma. Curr Opin Urol. 2007;17:303–308. doi: 10.1097/MOU.0b013e328277f180. [DOI] [PubMed] [Google Scholar]