Abstract
Circulating tumor cells (CTC) are emerging as a powerful prognostic and predictive biomarker in several types of cancer, including breast, colon, and prostate. Studies of CTC in metastasis and further development of CTC as a biomarker in cancer have been limited by the inability to repetitively monitor CTC in mouse models of cancer. We have validated a method to enumerate CTC in blood samples obtained from living mice using a modified version of an in vitro diagnostic system for quantifying CTC in patients. Different routes of blood collection were tested to identify a method to reproducibly recover CTC from tumor-bearing mice without interference from contaminating normal murine epithelial cells. CTC are present in blood samples from mice bearing orthotopic xenografts of several different breast cancer cell lines and primary breast cancer cells from patient biopsies. We also show that this technology can be used for serial monitoring of CTC in mouse xenograft models of human breast cancer. These results establish a new method for studying CTC in mouse models of epithelial cancer, providing the foundation for studies of molecular regulation of CTC in cancer and CTC as biomarker for therapeutic efficacy.
Introduction
Epithelial tumor cells were first identified in the blood of a breast cancer patient over 150 years ago (1). Since then, circulating tumor cells (CTC) have been shown to be a critical link between primary cancer, a disease stage at which cure is possible, and metastatic disease, which continues to be the leading cause of death for most malignancies. Studies by our group have shown that CTC, as measured by an automated immunomagnetic and fluorescent system (CellTracks), are a powerful prognostic and predictive biomarker in metastatic breast cancer (2–5), and similar findings have been reported in prostate (6, 7) and colorectal cancer (8). These data show that CTC are representative of the underlying biology driving metastatic cancer and suggest that further cellular and molecular analyses of these cells will reveal new insights into molecular regulation of metastasis and response to therapy.
Research on CTC in metastasis and further development of CTC as a biomarker for pharmacodynamics of therapy have been limited by the inability to repetitively monitor CTC in preclinical mouse models of breast and other cancers. Previous studies of CTC in animal models have used samples obtained when mice are euthanized, precluding serial analysis of temporal changes in CTC during tumor progression and response to therapy in living mice (9, 10). Recently, a two-photon system for in vivo flow cytometry was used for repetitive measurements of CTC in living mice (11). This technology currently is limited to detection of cancers that overexpress the folate receptor, and there is no direct translational link to clinical oncology. Adapting existing technology used to serially assay CTC in humans for studies in mice would permit integration of CTC assessments into preclinical models and allow for direct translation to clinical investigations. Building upon technology approved for human studies also permits early development of “companion” diagnostic assays for targeted therapies in mouse models of cancer, accelerating translation of new assay protocols into clinical trials in patients and ultimately into clinical practice.
Materials and Methods
Human breast cancer cell lines
MDA-MB-231, MCF-7, and SKBR-3 cells were obtained from American Type Culture Collection. SUM-159 cells were from Stephen Ethier (Karmanos Cancer Center), and MCF-7 cells stably transfected with fibroblast growth factor (MCF-7–FGF) were obtained from Francis Kern (Georgetown University). We cultured MDA-MB-231, MCF-7, MCF-7–FGF, and SKBR-3 cells in DMEM with 10% fetal bovine serum (FBS), 1% l-glutamine, and 0.1% penicillin/streptomycin. SUM-159 cells were cultured in Ham’s F12 medium (Invitrogen) supplemented with 5% FBS, 5 µg/mL insulin, 1 µg/mL hydrocortisone, and 0.1% penicillin/streptomycin. Cells were maintained at 37° in a 5% CO2 incubator. For selected experiments, we transduced MDA-MB-231 cells with the lentiviral vector pSico (12) to establish cells that stably express enhanced green fluorescent protein (GFP). Efficiency of transduction was 100%, as determined by phase-contrast and fluorescence microscopy.
Modification of CellTracks system for small volumes of murine blood
The CellTracks System (Immunicon) has previously been used to isolate and enumerate circulating epithelial tumor cells from human blood samples (13). The system is composed of an automated sample preparation system (CellTracks II AutoPrep) that enriches for epithelial cells using antibodies to epithelial cell adhesion molecule (EpCAM) coupled to magnetic beads. Isolated cells then are stained with the fluorescent nucleic acid dye 4,2-diamidino-2-phenylindole dihydrochloride (DAPI) to identify nucleated cells. Recovered cells subsequently are stained with fluorescently labeled monoclonal antibodies to murine CD45 (APC channel) and cytokeratin 8 (CK8), CK18, CK19 (PE channel) to distinguish epithelial cells from leukocytes. EpCAM+, CK+, CD45−, DAPI+ cells that fulfill morphological criteria are counted as circulating tumor cells by a technician using the semiautomated CellTracks Analyzer II. There is an additional fluorescence channel for FITC that is not part of the standard CellTracks assay and may be used for further characterization of tumor cells.
To verify that human breast cancer cells can be detected reproducibly in the small volumes of blood that can be obtained from living mice, we diluted 500 MDA-MB-231 or 231-GFP human breast cancer cells into ≈100 µL mouse blood. Samples then were diluted into 300 µL CellSave solution (Immunicon) and shipped overnight at room temperature to Immunicon for processing on the CellTracks system. Human breast cancer epithelial cells were isolated from these samples, using GFP as additional confirmation that recovered epithelial cells were the added 231-GFP cells.
Processing blood samples for CTC
Blood samples from mice were diluted into 300 µL CellSave solution and processed for numbers of CTC as described above for validation of the CellTracks system.
Primary human breast cancer specimens
Biopsy specimens from patients with primary breast cancer were obtained after informed consent according to a protocol approved by the University of Michigan Institutional Review Board. Specimens were digested with collagenase and hyaluronidase solution (Stem Cell Technologies) diluted 1:10 in medium 199 (Invitrogen). Digestion was stopped with 5% FBS. Samples were centrifuged to separate single cells from organoids and then washed twice with HBSS with 5% FBS to obtain an enriched suspension of single malignant cells for direct implantation into mice.
Routes of blood collection
To identify a method of blood collection that could be used repetitively for serial studies of circulating tumor cells, we obtained blood from the lateral tail vein, retroorbital venous plexus, jugular vein, or left ventricle of the heart. For lateral tail vein samples, mice were restrained in a plastic holder and a vein was punctured with a 26-G needle. Blood (≈25 µL) was collected in a heparinized capillary tube. Blood samples from the retroorbital venous plexus, jugular vein, and left ventricle were obtained from mice anesthetized with 1% to 1.5% isoflurane. The retroorbital venous plexus was punctured with a heparinized capillary tube to collect ≈50 to 100 µL of blood. Jugular vein samples were obtained using a 22-G needle (14). Cardiac blood samples (≈75–100 µL) were collected using a 26-G needle inserted into the chest over the point of maximal impulse from the heart. This procedure was performed without assistance from a needle holder or other external guidance system. Cardiac puncture was used for serial studies of CTC, and blood samples (≈0.5–1 mL) were collected by this same route when mice were euthanized at the end of some experiments. Mice without tumor xenografts were used as controls to test for contaminating epithelial cells after collecting blood from various anatomic sites.
Tumor xenografts
Female Ncr nude (MDA-MB-231; Taconic) or SCID (SUM-159, SKBR-3; Jackson) mice (5–6 wk old) were used for all experiments. To generate tumor xenografts from cell lines, 1 × 106 cells were injected orthotopically into bilateral inguinal mammary fat pads (15). For tumor xenografts with clinical isolates of human breast cancer cells, single-cell suspensions of tumor cells were implanted into the fourth inguinal mammary fat pad of mice (1–5 × 105 cells per implant). Mice implanted with clinical breast cancer isolates also received a subcutaneous pellet of 60-d sustained release17-β-estradiol (0.72 mg/pellet; Innovative Research of America). Tumors were measured in two dimensions with calipers, and tumor volumes were calculated as width (mm) × width (mm) × length (mm) × 0.52. For serial studies of CTC, blood samples were collected from the left ventricle at approximately weekly intervals, as shown in the figure legend.
Data analysis
Mean values for CTC and t test values were calculated using GraphPad Prism software. Significant differences were regarded as P < 0.05.
Results and Discussion
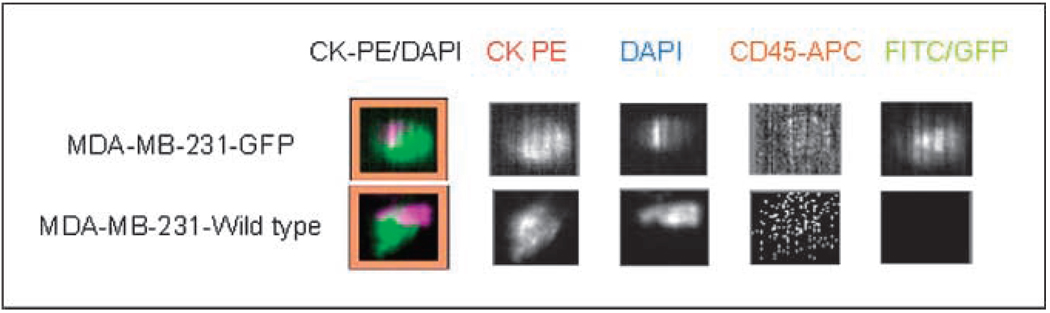
To serially monitor CTC in mouse models of human breast cancer, we modified a Food and Drug Administration (FDA)–cleared assay (CellTracks, Immunicon) for immunomagnetic isolation of epithelial cells from blood and immunofluorescent staining to further differentiate epithelial cancer cells from leukocytes (13). Because CellTracks originally was developed to process 7.5 to 30 mL samples of human blood, we first had to establish that human epithelial breast cancer cells could be reliably recovered from small volumes of mouse blood using this assay. To accomplish this, 500 MDA-MB-231 breast cancer cells were spiked into 100 µL blood samples collected from mice without tumors. Because the clinical version of the assay requires blood to be drawn into a vacuum tube (CellSave) containing both an anticoagulant and a preservative, a proportionately reduced amount of CellSave solution was added to specimens. Spiked specimens then were prepared (CellTracks II AutoPrep) and quantified with an automated system (CellTracks Analyzer II). As a positive control, additional samples using MDA-MB-231 cells stably transduced with GFP were prepared. Fluorescence from GFP was detected in an open channel (FITC) of the system to confirm that all cells quantified as epithelial cells corresponded with 231-GFP cells added to mouse blood (Fig. 1A). As a negative control, mouse blood samples without cancer cells were collected, processed in an identical manner, and analyzed. Of the 500 cells added to mouse blood (n = 4 samples), 482 to 526 cells per specimen were recovered, which is within the range of the dilution error for spike-in experiments at this concentration. For samples using 231-GFP cells, all cells identified as epithelial cells also expressed GFP, verifying that these were human breast cancer cells and not contaminating murine epithelial cells. No epithelial cells were recovered in blood samples from normal mice, confirming specificity of the assay.
Figure 1.
Quantification of human breast cancer cells in mouse blood samples. A, MDA-MB-231 human breast cancer cells with or without stable transduction of GFP were added to 100 µL blood samples from mice without tumor xenografts. Samples were fixed, and epithelial cells were enriched by immunomagnetic bead isolation using an antibody-to-epithelial cell adhesion molecule. Recovered cells then were stained with an antibody to cytokeratin (CK8, CK18, and CK19) to identify epithelial cells (CK-PE) and distinguish them from leukocytes stained with CD45 (CD45-APC). Nucleated cells were identified by staining with the fluorescent nucleic acid dye DAPI. GFP in cancer cells was detected in the FITC channel. Representative images of recovered MDA-MB-231 and 231-GFP breast cancer cells. These cells stain positively for CK-PE and DAPI and negatively for CD45-APC. GFP fluorescence in 231-GFP cells is detected in the FITC channel. The merged image of all fluorescent channels is shown as CK-PE/DAPI.
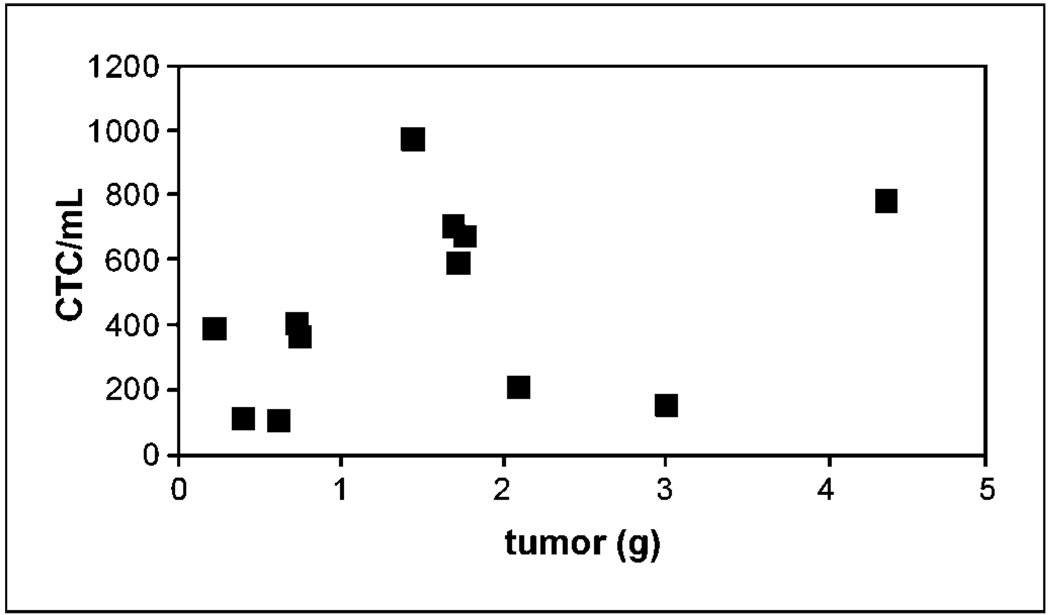
Having validated the assay for samples added to breast cancer cells in vitro, we then used the system to identify CTC that spontaneously intravasated into the circulation from orthotopic tumor xenografts of MDA-MB-231 cells. Small volume (0.7–1 mL) blood samples were collected from each mouse by puncture of the left ventricle when animals were euthanized for tumor burden at 10 weeks. Total numbers of CTC ranged from ~ 100 to 1,000 cells/mL blood (Fig. 2). No CTC were recovered from blood samples collected from mice without tumor xenografts (data not shown). The number of CTC did not correlate with size of the primary tumor. These data suggest that numbers of CTC reflect the underlying biology of various primary tumors, which is consistent with previous studies showing that MDA-MB-231 cells contain subpopulations with differing metastatic potential (16). Using the same method, CTC also were detectable in mice with tumor xenografts of MCF-7, MCF-7 cells stably transfected with FGF, SUM-159, and SKBR-3 human breast cancer cell lines (data not shown).
Figure 2.
CTC recovered from mice with MDA-MB-231 tumor xenografts. Blood samples from mice bearing MDA-MB-231 human breast cancer cells were obtained by cardiac puncture at the time animals were euthanized. CTC were quantified and plotted versus tumor weight in grams.
Whereas the system was successful in detecting CTC using cardiac puncture to collect blood, this procedure is relatively invasive compared with other sites of blood sampling in mice. Because the goal of the project was to repetitively draw blood samples for analysis of CTC, we compared recovery of CTC from blood samples obtained from the lateral tail vein, retroorbital venous plexus, or jugular vein in mice with or without orthotopic MDA-MB-231 tumor xenografts. No epithelial cells were detected in any of the lateral tail vein samples, independent of the presence of a tumor xenograft (Fig. 3A). One possible explanation for the failure to detect CTC in tumor-bearing mice was the small volume of blood (≤25 µL) that could be collected from the lateral tail vein. Although larger volumes of blood (50–75 µL) could be obtained from the retroorbital venous plexus, three of three blood samples from this site contained epithelial cells (5–500 cells) in mice without tumors. These contaminating cells were normal murine epithelial cells dislodged by the microcapillary tube during blood collection, which would make it impossible to reliably identify CTC in tumor-bearing mice (Fig. 3B). Larger blood samples (75–100 µL) also could be collected from the jugular vein, but these samples occasionally were contaminated by normal epithelial cells in mice without tumors and had low sensitivity for detecting CTC in tumor-bearing mice. By comparison, there were no CTC in blood samples obtained by cardiac puncture in mice without tumor xenografts, but CTC could be detected in blood obtained via left ventricle cardiac puncture in mice with MDA-MB-231 xenografts.
Figure 3.
Comparison of different routes of blood collection. A, blood samples were collected from normal or tumor-bearing mice using tail vein, retroorbital, jugular vein, or cardiac routes of sampling. Numbers of mice with recovered epithelial cells are shown out of the total number of mice analyzed. B, representative images of contaminating normal murine epithelial cells in blood samples from retroorbital venous plexus. These cells stain positively for cytokeratin (CK-PE) and DAPI and negatively for CD45 (CD45-APC), as shown in the merged image. The FITC channel is unused in this assay and shows only background signal. Such murine epithelial cells are indistinguishable from CTC produced by human breast cancer xenografts.
After validating the assay and route of blood collection, we then determined the feasibility of detecting temporal changes in CTC in mice implanted with orthotopic tumor xenografts of SUM-159 (n = 3) or SKBR-3 (n = 4) cells. We collected 75 to 100 µL blood samples by cardiac puncture approximately once per week for 1 month until mice were euthanized because of tumor burden. This experiment entailed 37 separate blood draws on these seven mice, only one of which in a mouse bearing a SKBR-3 xenograft resulted in death of the animal (<3% mortality rate from the procedure). Low levels of CTC (0–7 cells) were detected in earlier samples (days 8–23; Fig. 4A). Numbers of CTC increased significantly on day 30 in six of seven mice (26–55 cells; P < 0.05), corresponding with an increase in tumor volume. Microscopic metastases were evident in lung and liver of each mouse (data not shown). These studies establish that the assay can be used successfully for serial studies of CTC in mouse models of metastatic breast cancer.
Figure 4.
Serial analysis of CTC in mice. Mice were implanted with orthotopic tumor xenografts of SUM-159 or SKBR-3 human breast cancer cells. Approximately, 100 µL blood samples were obtained via cardiac puncture from mice anesthetized with 1.5% isoflurane. Samples were obtained at approximately weekly intervals until mice were euthanized because of tumor burden. CTC data (open symbols) were normalized to 100 µL volume and plotted against tumor volume (filled symbols) for individual animals. Representative data are shown from one mouse each with SUM-159 or SKBR-3 tumors, respectively.
To further validate the mouse-adapted CTC assay for use in preclinical studies of human breast cancer, CTC were measured in mice implanted with xenografts of primary breast cancer cells obtained from patient biopsy specimens. Blood samples (200– 800 µL) were collected via cardiac puncture at the time animals were euthanized because of tumor burden. Breast cancer cells from seven different patients formed tumors in mice, and two of these tumors produced CTC (72 and 75 CTC/µL blood, respectively). Presence of detectable CTC was not affected by the period of time tumors were in place or volumes of primary tumors (data not shown). Notably, none of these animals without or with CTC had overt or histologically detectable metastases (data not shown), suggesting that CTC produced by primary clinical specimens may not be capable of forming metastases in mice or humans. These data show that xenografts of clinical breast cancer isolates can produce CTC in mice and may provide a model system for investigating properties and subpopulations of human breast cancer cells involved in metastasis.
In conclusion, we have established a method to quantify serial changes in CTC in mouse xenograft tumor models of human breast cancer, using both established cell lines and primary clinical isolates of breast cancer cells. This technology is the basis for ongoing and future studies examining therapeutic response and investigating biological questions involving CTC-associated molecules and signaling pathways that regulate metastasis. The information from these investigations should facilitate discovery of new therapeutic targets and expedite development of existing drugs and their companion diagnostics. We expect this preclinical model system to accelerate development of protocols for biological profiling of CTC, which will greatly expand the diagnostic capabilities of this assay in clinical oncology. Whereas we have focused on breast cancer, this method for quantifying CTC should be applicable to other mouse models of human epithelial cancers. Because the protocol for recovering and analyzing CTC in mice is based on an FDA-cleared system for patients, innovations in detection and molecular profiling of CTC validated in preclinical models can be translated directly to clinical trials in patients with breast and other cancers. To facilitate general implementation of the technology, a simplified system for manual sample preparation has been developed and is currently being evaluated in our laboratory.
Acknowledgments
Grant support: Sidney Kimmel Foundation, Kathy Bruk Pearce Fund from University of Michigan Comprehensive Cancer Center, Fashion Footwear Association of New York/QVC’s “FFANY Shoes on Sale,” and Immunicon Corporation.
Footnotes
Disclosure of Potential Conflicts of Interest
M. Repollet: employment, Immunicon Corporation. M. Wicha: commercial research grant, Merck; ownership interest, OncoMed. G.V. Doyle: ownership interest, Immunicon Corporation. The other authors disclosed no potential conflicts of interest.
References
- 1.Ashworth A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aus Med J. 1869;14:149. [Google Scholar]
- 2.Cristofanilli M, Budd G, Ellis M, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 3.Hayes D, Cristofanilli M, Budd G, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12:4218–4224. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- 4.Budd G, Cristofanilli M, Ellis M, et al. Circulating tumor cells versus imaging-predicting overall survival in metastatic breast cancer. Clin Cancer Res. 2006;12:6403–6409. doi: 10.1158/1078-0432.CCR-05-1769. [DOI] [PubMed] [Google Scholar]
- 5.Cristofanilli M, Hayes D, Budd G, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol. 2005;23:1420–1430. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 6.Moreno J, Miller M, Gross S, Allard W, Gomella L, Terstappen L. Circulating tumor cells predict survival in patients with metastatic prostate cancer. Urology. 2005;65:713–718. doi: 10.1016/j.urology.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Shaffer D, Leversha M, Danila D, et al. Circulating tumor cell analysis in patients with progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:2023–2029. doi: 10.1158/1078-0432.CCR-06-2701. [DOI] [PubMed] [Google Scholar]
- 8.Cohen S, Alpaugh R, Gross S, et al. Isolation and characterization of circulating tumor cells in patients with metastatic colorectal cancer. Clin Colorectal Cancer. 2006;6:125–132. doi: 10.3816/CCC.2006.n.029. [DOI] [PubMed] [Google Scholar]
- 9.Aslakson C, Miller F. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 10.Wyckoff J, Jones J, Condeelis J, Segall J. A critical step in metastasis: in vivo analysis of intravasation at the primary tumor. Cancer Res. 2000;60:2504–2511. [PubMed] [Google Scholar]
- 11.He W, Wang H, Hartmann L, Cheng J, Low P. In vivo quantitation of rare circulating tumor cells by multiphoton intravital flow cytometry. Proc Natl Acad Sci U S A. 2007;104:11760–11765. doi: 10.1073/pnas.0703875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ventura A, Meissner A, Dillon C, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allard W, Matera J, Miller M, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 14.Hoff J. Methods of blood collection in the mouse. Lab Animal. 2000;29:47–53. [Google Scholar]
- 15.Smith M, Luker K, Garbow J, et al. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64:8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- 16.Minn A, Kang Y, Serganova I, et al. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest. 2005;115:44–55. doi: 10.1172/JCI22320. [DOI] [PMC free article] [PubMed] [Google Scholar]