Abstract
Background
In recent years, the phenotypes of leukodystrophies linked to mutations in the eukaryotic initiation factor 2B genes have been extended, classically called CACH/VWM (Childhood ataxia with cntral hypomyélination/vanishing white matter disorder). The large clinical spectrum observed from the more severe antenatal forms responsible for fetal death to milder adult forms with an onset after 16 years old and restricted to slow cognitive impairment have lead to the concept of eIF2B-related disorders. The typical MRI pattern with a diffuse CSF-like aspect of the cerebral white matter can lack particularly in the adult forms whereas an increasing number of patients with clinical and MRI criteria for CACH/VWM disease but without eIF2B mutations are found. Then we propose the use of biochemical markers to help in this difficult diagnosis. The biochemical diagnosis of eIF2B-related disorder is difficult as no marker, except the recently described asialotransferrin/transferrin ratio measured in cerebrospinal fluid, has been proposed and validated until now. Decreased eIF2B GEF activity has been previously reported in lymphoblastoid cell lines from 30 eIF2B-mutated patients. Our objective was to evaluate further the utility of this marker and to validate eIF2B GEF activity in a larger cohort as a specific diagnostic test for eIF2B-related disorders.
Methodology/Principal Findings
We performed eIF2B GEF activity assays in cells from 63 patients presenting with different clinical forms and eIF2B mutations in comparison to controls but also to patients with defined leukodystrophies or CACH/VWM-like diseases without eIF2B mutations. We found a significant decrease of GEF activity in cells from eIF2B-mutated patients with 100% specificity and 89% sensitivity when the activity threshold was set at ≤77.5%.
Conclusion
These results validate the measurement of eIF2B GEF activity in patients' transformed-lymphocytes as an important tool for the diagnosis of eIF2B-related disorders.
Introduction
Mutations in the EIF2B1-5 genes (OMIM 606686, 606454, 606273, 606687, 603945) encoding the subunits of the ubiquitously expressed eukaryotic initiation factor 2B (eIF2B) have been reported in a group of clinically heterogeneous leukodystrophies termed eIF2B-related disorders [1], [2], [3], [4]. Disease severity is correlated with age at disease onset, with stress onset trigger or aggravating factors [5], [6]. A large clinical spectrum is observed and several distinct forms have been proposed: i) the classical childhood ataxia with central hypomyelination/vanishing white matter disease (CACH/VWM, OMIM 603896), with progressive neurological deterioration between age 2-5 years [1], [3], ii) the infantile severe forms with disease onset <2 years and rapid fatal evolution [7], iii) the most severe antenatal forms responsible for fetal death [8], and iv) the milder forms with disease onset >5 years and restricted to slow cognitive impairment [6], [9].
The typical MRI pattern shows a diffuse CSF-like aspect of the cerebral white matter of the cerebral hemispheres and this pattern recognition permits the selection of patients eligible for the EIF2B1-5 genes sequencing [10]. But MRI can lack particularly in the adult forms whereas an increasing number of patients with clinical and MRI criteria for CACH/VWM disease but without EIF2B1-5 genes mutations (CACH/VWM-like) are found, underlining the necessity to have biochemical markers to help in the diagnosis process and in the selection of patients eligible for EIF2B1-5 direct sequencing.
Two biochemical markers have been recently proposed as potential tools for the screening of eIF2B-related disorders: the decrease of asialotransferrin/total transferrin in CSF [11], [12], and the decrease of the eIF2B GEF activity measured in Epstein-Barr Virus (EBV)-transformed lymphocytes or lymphoblasts (LLB) [13]. In fact, eIF2B is a key regulator of the protein synthesis particularly under cellular stresses through its nucleotide guanine exchange (GEF) activity: it converts the initiation factor 2 (eIF2) from an inactive GDP-bound form to an active eIF2-GTP complex [14]. Measurement of eIF2B GEF activity in patients' LLB has been proposed as a potential diagnostic tool relating to a previous work showing decrease of this GEF activity in LLB from 30 affected eIF2B-mutated patients in comparison to controls. In order to further evaluate the specificity and sensitivity of eIF2B GEF activity in patients' LLB regarding the diagnosis of eIF2B-related disorders, we extended this initial cohort of 30 to 63 mutated patients and compared the results not only to healthy non mutated subjects but also to patients with defined leukodystrophies or CACH/VWM-like diseases.
Methods
Objectives
Our hypothesis is that eIF2B GEF activity measurement in LLB is a specific and sensitive marker that would be powerful to select patients eligible for the EIF2B1-5 genes sequencing and then to help in the molecular diagnosis of eIF2B-related disorders. Our objective is then to extend our initial cohort of 30 [13] to 63 eIF2B-mutated patients and to compare the results to healthy subjects but also to CACH/VWM-like patients, in order to evaluate the specificity and sensitivity of eIF2B GEF activity in patients' LLB regarding the diagnosis of eIF2B-related disorders.
Participants
The GEF activity was measured in LLB from 63 eIF2B-mutated patients (including 30 patients already reported [13] and 13 patients never reported todate, (Table 1), 18 clinically healthy subjects (controls) and 38 patients with leukodystrophies of other causes, termed eIF2B-unrelated leukodystrophic group (Table 2). This last group included:
Table 1. Clinical data and eIF2B GEF activity measured in lymphoblasts from the 63 eIF2B-mutated patients.
| Patienta | Age at disease onset (y) | Disease evolution (y) | Score of disabilityb | Mutated gene | Amino-acid changec | eIF2B GEF activity (%)d | % ATe |
| 1187-1 | 57 | 11 | 3 | EIF2B3 | p.Ser14Phe/p.Ala87Val | 111.6±3.6 | NA |
| 1074-1 | 8 | NA | NA | EIF2B5 | p.Arg113His/p.Arg113His | 108±11.2 | NA |
| 971-1 | 5 | 11 | 1 | EIF2B3 | p.Glu136Pro/? | 105.5±6.8 | NA |
| 1135-1 | 5 | 1.1 | 2 | EIF2B5 | p.Arg113His/p.Arg315Cys | 99±7.4 | NA |
| *432-1 | 3 | 8.2 | 5 | EIF2B5 | p.Arg113His/p.Trp628X | 90.4±1.8 | 2.11 |
| 356-1 | 3 | 16 | 5 | EIF2B4 | p.Pro243Leu/p.Pro243Leu | 80.4±2.8 | NA |
| 736-1 | 8 | 4 | 1 | EIF2B4 | p.Arg374Cys/p.Arg374Cys | 80±0 | NA |
| 630-1 | 4.5 | 10.5 | 4 | EIF2B5 | p.Arg113His/p.Arg113His | 77.5±2.5 | 5.50 |
| *370-2 | 4.5 | 5 | 3 | EIF2B5 | p.Arg113His/p.Arg195His | 77±2.5 | NA |
| 1304-1 | 18 | 32 | 2 | EIF2B5 | p.Arg113His/p.Arg113His | 76.1±4.3 | NA |
| 1008-1 | 18 | 7 | 5 | EIF2B5 | p.Arg113His/p.Arg113His | 76.1±2.6 | NA |
| 954-1 | 3.5 | 4 | 4 | EIF2B5 | p.Arg113His/p.Arg269Leu | 75.9±1.6 | 7.77 |
| *76-1 | 17 | 8 | 1 | EIF2B2 | p.Glu213Gly/p.Lys273Arg | 75.5±10 | NA |
| 299-1 | 11 | 17 | 4 | EIF2B5 | p.Arg113His/p.Arg113His | 75.2±5.5 | NA |
| 338-1 | 10 | 16 | 5 | EIF2B5 | p.Arg113His/p.Arg113His | 75.2±1.5 | NA |
| 1407-1 | 24 | 2 | 1 | EIF2B5 | p.Arg113His/p.Arg222Trp | 74.9±0.8 | NA |
| *370-1 | 3.5 | 7 | 3 | EIF2B5 | p.Arg113His/p.Arg195His | 71.5±9 | NA |
| 73-1 | 4 | 11 | 3 | EIF2B5 | p.Arg113His/p.Arg113His | 70.8±7 | NA |
| 904-1 | 2 | 1.5 | 1 | EIF2B5 | p.Arg113His/p.Gly481fs493X | 70±1.2 | 4.52 |
| 1014-1 | 16 | 0.5 | 0 | EIF2B5 | p.Arg113His/p.Arg195Cys | 68±4 | NA |
| *470-2 | 2 | 4 | 1 | EIF2B5 | p.Glu81Lys/p.Arg113His | 67.9±1.8 | NA |
| 1240-1 | 17 | 4 | 0 | EIF2B3 | p.Ala202Thr/p.Arg438X | 67.7±2 | NA |
| 1627-1 | 7 | 24 | 3 | EIF2B5 | p.Pro87Leu/p.Arg113His | 67.1±1.8 | NA |
| *76-2 | 7 | 11 | 3 | EIF2B2 | p.Glu213Gly/p.Kys273Arg | 67±5.1 | NA |
| 807-1 | 6 | 4.5 | 3 | EIF2B5 | p.Arg113His/p.Arg113His | 67±4.3 | NA |
| 435-1 | 3 | 5.5 | 3 | EIF2B5 | p.Arg113His/p.Arg422X | 67±3 | NA |
| 1467-1 | 25 | 19 | 1 | EIF2B5 | p.Tyr483Cys/p.Arg195His | 66.3±0.6 | NA |
| 1232-1 | 28 | 3 | 1 | EIF2B5 | g.IVS8+59A/G/? | 64.3±7.8 | NA |
| *648-2 | 7 | 3 | 1 | EIF2B2 | p.Glu213Gly/p.Glu213Gly | 64±4 | 4.11 |
| 1108-1 | 3.5 | 1.5 | 4 | EIF2B5 | p.Arg113His/c.[+2081delG] | 63.6±2.3 | NA |
| 1115-1 | 2 | 6 | 2 | EIF2B5 | p.Pro427Leu/p.Pro427Leu | 63.4±8.2 | NA |
| 1441-1 | 2.8 | 3.2 | 4 | EIF2B5 | p.Leu106Phe/p.Arg113His | 63.2±2 | NA |
| 997-1 | 7 | 4 | 1 | EIF2B5 | p.Arg113His/p.Arg113His | 61.3±0.3 | NA |
| *823-1 | 4 | 2 | 4 | EIF2B3 | p.His341Gln/p.His341Gln | 61±6 | NA |
| *576-2 | 7 | 5 | 0 | EIF2B5 | p.Tyr343Cys/p.Ile385Val | 61±0.2 | NA |
| 1012-1 | 14 | 10 | 5 | EIF2B2 | p.Glu213Gly/p.Glu213Gly | 60±2 | NA |
| 663-1 | 3.5 | 1.5 | 1 | EIF2B5 | p.Arg113His/p.Arg113His | 59.9±3.8 | NA |
| 1004-1 | 2 | 1 | 1 | EIF2B2 | p.Glu213Gly/p.Glu213Gly | 59.9±0.8 | NA |
| *375-2 | 2.5 | 3 | 5 | EIF2B5 | p.Arg113His/p.Glu650Leu | 59.4±0.7 | NA |
| *648-1 | 7 | 14 | 4 | EIF2B2 | p.Glu213Gly/p.Glu213Gly | 59±1 | NA |
| 442-1 | 2.5 | NA | NA | EIF2B5 | p.Glu81Lys/p.Arg113His | 58.8±0.3 | NA |
| 308-1 | 5 | 8 | 4 | EIF2B5 | p.Arg113His/p.Arg113His | 57.1±6.1 | NA |
| *576-1 | 8 | 11 | 1 | EIF2B5 | p.Val73Gly/p.Arg113His | 56±4 | NA |
| 359-1 | 6 | 1.5 | 5 | EIF2B4 | p.Pro243Leu/p.Pro243Leu | 54±6 | NA |
| 522-1 | 3.5 | 3.5 | 3 | EIF2B5 | p.Ala16Asp/p.Arg113His | 54±6 | 4.80 |
| *432-2 | 2 | 8.2 | 5 | EIF2B5 | p.Arg113His/W628stop | 53.9±0.9 | NA |
| *570-2 | 1.5 | 6.5 | 4 | EIF2B4 | p.Arg209Gln/p.Arg209Gln | 52±3 | NA |
| *470-1 | 3 | 4 | 1 | EIF2B5 | p.Glu81Lys/p.Arg113His | 51.5±4.5 | NA |
| 984-1 | 5 | 4 | 2 | EIF2B2 | p.Glu213Gly/p.Glu213Gly | 51.5±0.5 | NA |
| *571-2 | 1.2 | 9 | 5 | EIF2B5 | p.Phe56Val/p.Arg315His | 50±5 | NA |
| 928-1 | 3 | 5 | 1 | EIF2B5 | p.Arg113His/p.Arg113His | 49±3 | NA |
| 995-1 | 1 | 2 | 4 | EIF2B5 | p.Arg113His/p.Leu425Arg | 48±2 | NA |
| *590-2 | 1 | 4 | 5 | EIF2B5 | p.Tyr343Cys/p.Ile385Val | 45.8±2.2 | 4.36 |
| 949-1 | 4 | 9 | 4 | EIF2B4 | p.Pro243Leu/p.Pro243Leu | 45.6±1.8 | NA |
| *590-1 | 3 | 4 | 3 | EIF2B5 | p.Tyr343Cys/p.Ile385Val | 44.9±4.3 | NA |
| 357-1 | 2 | 2 | 5 | EIF2B5 | p.Arg136Cys/p.Arg339Trp | 44.5±4.5 | NA |
| 291-1 | 1.5 | 6.5 | 4 | EIF2B5 | p.Arg113His/p.Arg113His | 41.5±6 | NA |
| *571-1 | 0.8 | 9 | 5 | EIF2B5 | p.Phe56Val/p.Arg315His | 40±3 | NA |
| 569-1 | 3.5 | 19.5 | 4 | EIF2B5 | p.Arg113His/p.256_281del | 40±2 | NA |
| 1388-1 | 22 | 17 | 1 | EIF2B5 | p.His214Arg/p.Arg269X | 39±7 | NA |
| 942-2 | 6 | 7 | 1 | EIF2B4 | p.Cys465Arg/p.Tyr489TThr | 34.5±1 | NA |
| 137-1 | 5 | 7.5 | 1 | EIF2B2 | p.Glu213Gly/p.Glu213Gly | 33.9±3.2 | NA |
| 1036-1 | 0.8 | 0.5 | 5 | EIF2B5 | p.Pro323Ser/p.Pro427Leu | 30±7.5 | NA |
* = Familial form: two affected children in the same family; underlined: new patients whose genotype has not been reported in our previous studies [5], [13].
score of disability as previously described [5]: 0: no neurologic signs, 1: stiff gait, 2: walk with help, 3: wheelchair-bound, 4: help for daily living, 5: death.
Amino-acid numbers refer to the eIF2B peptide corresponding sequence.? = mutation not yet identified.
Expressed as % control value ± standard deviation (assays performed in triplicate); underlined: new eIF2B GEF activity measured and not reported in [13].
Table 2. eIF2B GEF activity measured in lymphoblasts from the 38 leukodystrophic patients (19 OL and 19 CACH/VWM-like) and the 18 controls.
| Patient | Diseasea | Mutated gene | Mutationb | eIF2B GEF activity (%)c |
| 81 | PMD | PLP1 | duplication | 108.6±10.4 |
| 1437 | AD | GFAP | p.Asp114Glu | 103.8±6.6 |
| 940 | MLC | MLC1 | p.Leufs | 109.6±0.4 |
| 002 | PMD | PLP1 | duplication | 98.4±1.3 |
| 672 | AD | GFAP | p.Asn77Tyr | 133.6±9 |
| 958 | AD | GFAP | p.Arg407Met | 113.2±5.1 |
| 150 | PMD | PLP1 | duplication | 103.4±4.5 |
| 42 | PMD | PLP1 | duplication | 86.2±4.4 |
| 726 | AD | GFAP | p.Arg239Cys | 119.8±1.6 |
| 989 | AD | GFAP | p.Met74Lys | 101.5±8.4 |
| 256 | MLC | MLC1 | NA | 102.6±4.3 |
| 737 | AD | GFAP | p.Arg88Cys | 101±14.8 |
| 1047 | MLC | MLC1 | p.Ser289Tyr/? | 98.7±13 |
| 750-1 | AD | GFAP | p.Arg79His | 105.6±1.8 |
| 750-2 | AD | GFAP | p.Arg79His | 79.9±3.9 |
| 613 | MLC | MLC1 | NA | 101.8±2.5 |
| 968 | PMD | PLP1 | duplication | 105.9±6.3 |
| 1207 | MLC | MLC1 | p.Cys46fs | 107.7±5.3 |
| 758 | AD | GFAP | p.Arg88Cys | 96.4±4.4 |
| 1011-1 | CACH-L | - | - | 104.43±6.5 |
| 1242-1 | CACH-L | - | - | 105.29±8.8 |
| 1469-1 | CACH-L | - | - | 103.30±1.7 |
| 1479-1 | CACH-L | - | - | 99.75±2.2 |
| 1082-1 | CACH-L | - | - | 89.5±8.8 |
| 1196-1 | CACH-L | - | - | 110.4±3.3 |
| 1290-2 | CACH-L | - | - | 105.9±2.7 |
| 1211-1 | CACH-L | - | - | 87.4±5.1 |
| 1200-1 | CACH-L | - | - | 108.5±2 |
| 798-1 | CACH-L | - | - | 119.0±2.3 |
| 1253-1 | CACH-L | - | - | 107.6±6.6 |
| 1206-1 | CACH-L | - | - | 94.67±8.2 |
| K80 | CACH-L | - | - | 89.27±2.6 |
| K128 | CACH-L | - | - | 84.76±1.2 |
| K112 | CACH-L | - | - | 100.41±2.5 |
| 1313-1 | CACH-L | - | - | 82.02±1.2 |
| 1283-1 | CACH-L | - | - | 101.40±6.8 |
| 1454-1 | CACH-L | - | - | 111±16.6 |
| 331-1 | CACH-L | - | - | 88.3±9.5 |
| N1 | C | - | - | 100 |
| N2 | C | - | - | 99 |
| N3 | C | - | - | 101 |
| N4 | C | - | - | 100 |
| N5 | C | - | - | 102 |
| N6 | C | - | - | 100 |
| N7 | C | - | - | 98 |
| N8 | C | - | - | 100 |
| N9 | C | - | - | 94.3±7 |
| N10 | C | - | - | 100±0.3 |
| N11 | C | - | - | 95.3±7.5 |
| N12 | C | - | - | 88.5±1.5 |
| N13 | C | - | - | 97.5±2.5 |
| N14 | C | - | - | 99.5±7 |
| N15 | C | - | - | 99.3±6.1 |
| N16 | C | - | - | 93.5±0.5 |
| N17 | C | - | - | 96.5±0.5 |
| N18 | C | - | - | 102±2 |
OL: PMD: Pelizaeus-Merzbacher disease, AD: Alexander disease, MLC: Megalencephalic Leukoencephalopathy with Cysts; and CACH/VWM-like: CACH-L.
?: second mutation not yet identified; NA: not available.
Expressed as % control value ± standard deviation (assays performed in triplicate); underlined: new eIF2B GEF activity measured and not reported in [13].
19 leukodystrophic patients with an identified genetic defect and termed OL (other leukodystrophy)-patients: 9 GFAP-mutated patients (with Alexander disease, OMIM 203450), 5 MLC1-mutated patients (with megalencephalic leukoencephalopathy with cysts, OMIM 604004) and 5 PLP1-mutated patients (with Pelizaeus-Merzbacher disease, PMD, OMIM 312080);
19 patients with the presence of clinical and/or MRI features observed in eIF2B-related disorders but screened negative for mutations in the coding regions of the five EIF2B1-5 genes genes, and termed CACH-VWM-like patients.
Measurement of eIF2B GEF Activity in Transformed Lymphocytes
The direct GEF activity of the eIF2B complex was measured in triplicate in protein extracts from patients'EBV-transformed lymphocytes (LLB) as already described [13], [15].
Ethics
An Institutional Review Board of the participating centers (Comité de Protection des Personnes Sud-Est VI, 2009-A00188-49) approved the use of human subjects for this study. A written informed consent was obtained from all patients.
Statistical Methods
The eIF2B GEF activity was considered as a continuous variable and results are displayed as mean ± SEM (standard error of the mean). Since the normality of eIF2B GEF (assessed by a Kolmogorov-Smirnov test) was not rejected, a one-way analysis of variance (ANOVA 1) was performed to assess the links between GEF activity and the patients'groups. On condition of a significant F-test for the ANOVA 1, a post-hoc multiple comparisons procedure was performed controlling for a 5% family-wise type I error using the Tukey honestly significant difference (THSD) test. A Spearman correlation coefficient (r) was calculated between GEF activity and age at disease onset (correleted to disease severity [5]). The Receiver Operating Characteristic, or ROC curve analysis (graphical plot of the sensitivity versus (1-specificity) for a binary classifier system as its discrimination threshold is varied), was performed on MedCalc® (v10.4, Mariakerke, Belgium) to determine the optimal threshold of GEF activity which best discriminates between eIF2B-mutated (n = 63) and eIF2B-unrelated leukodystrophic patients (n = 38), aiming a 100% specificity and the best associated sensitivity. The area under the ROC curve (AUC) was also estimated (with its 95% confidence limits) and tested towards 0.5. All remaining statistical analyses were performed on SAS® (v9.1, Cary, USA) with a type I error set at 5%.
Results
eIF2B GEF Activity in 63 eIF2B-Mutated Patients in Comparison to the Other Groups
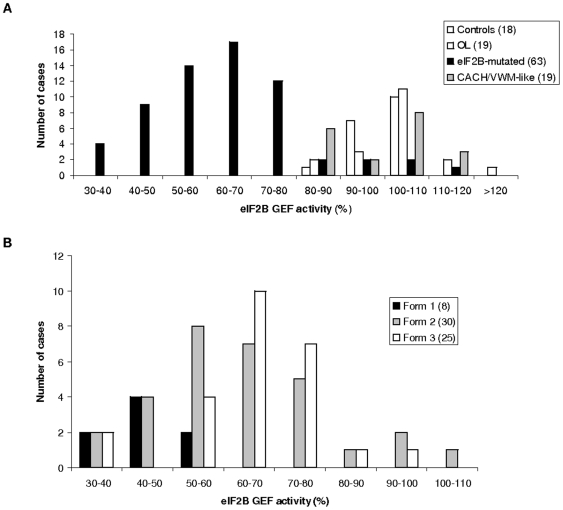
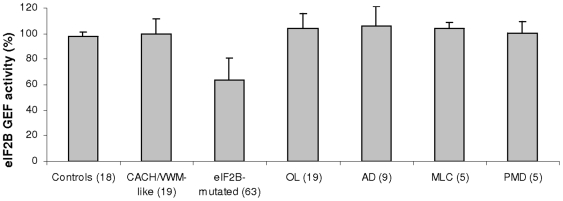
The eIF2B GEF activity was measured in LLB of 63 affected patients exhibiting various eIF2B mutations, age of onset and disease severity (Table 1). Despite the wide range of GEF activity in eIF2B-mutated cells (63.2±17.1%, range: 30–111.6%) (Figure 1A), multiple comparisons demonstrated a significant difference between the group of eIF2B-mutated patients (n = 63) and the three other groups of patients [OL-patients (104.1±11.4%, range: 80–133.6%, n = 19), CACH/VWM-like (99.6±10.3%, range: 82–119%, n = 19) and controls (98±3.3%, range: 88.5–102%, n = 18)] (F(3,115) = 71.1, p<0.001) (Tables 1 and 2, Figures 1A and 2). The OL, CACH/VWM-like and controls groups had similar GEF activity (THSD test). The THSD test further detailed that only the eIF2B-mutated group had significantly lower GEF activity compared with each of the three other groups (OL, CACH/VWM-like and controls).
Figure 1. Distribution of patients per classes of eIF2B GEF activity.
A. Distribution of the patients'groups per classes of eIF2B GEF activity in %. The patients' groups are healthy controls, eIF2B-mutated, others leukodystrophies (OL) and CACH-VMW-like. B. Distribution of the 63 eIF2B-mutated patients per classes of eIF2B GEF activity. The mutated patients have been classified into three clinical groups depending of their clinical severity, according to previous studies [5]. Form 1: disease onse before 2 years, form 2: disease onset berween 2 and 5 years, form 3: disease onset >5 years.
Figure 2. Decreased eIF2B GEF activity in eIF2B-mutated patients' lymphoblasts (LLB).
The eIF2B GEF activity was measured in LLB from 63 eIF2B-mutated patients in comparison to 8 healthy controls, 19 patients carrying other leukodystrophies (OL: 9 patients with Alexander disease, AD, 5 patients with Pelizaeus-Merzbacher disease, PMD, and 5 patients with Megalencephalic leukoencephalopathy with cysts, MLC), and 19 CACH/VWM-like patients without eIF2B mutations. Experiments were carried out in triplicate. Data are presented as percentage of exchange activity of control LLB. The statistical multiple comparisons analysis between all the seven groups showed significant differences (*) only for the eIF2B-mutated group (p<0.001).
Pooling CACH/VWM-like patients and OL-patients (group of eIF2B-unrelated leukodystrophic patients, n = 38), the aforementioned comparison of GEF activity led still to a significant result (F(2,116) = 106.1, p<0.0001) and the THSD test showed again the same significantly lower GEF activity in the eIF2B-mutated group as compared with each of the two other groups (eIF2B-unrelated leukodystrophic and controls). Then difference in GEF activity remained statistically significant between eIF2B-mutated patients and the other groups tested.
Correlation between Age at Disease Onset and GEF Activity
A weak correlation was found between GEF activity measured in the eIF2B-mutated LLB and age at disease onset (r = 0.4309, p = 0.0004). Two patients 356–1 and 432–1 (Table 1) exhibited high GEF activities (respectively 80.4±2.8% and 90.4±1.8%) despite a classical clinical form (disease onset at 3 years followed by severe disability within 3 years and death occurring after 16 years in one patient). On the other hand, patient 1388–1 affected with an adult onset, slowly progressive form (only stiff gait after 17 years of disease progression), exhibited a low GEF activity (39±7%) (Table 1). However, the eight patients with onset ≤2 years (disease severity form 1) always had eIF2B GEF activity <55% (45.1±7.8%, mean age at disease onset: 1.2 year) (Table 1B). The correlation coefficient is higher in this group of eight patients (r = 0.68, p = 0.06) compared to the 30 patients carrying the clinical severity form 2 with onset between 2 and 5 years (r = 0.11, p = 0.52), and to the 25 patients carrying the severity form 3 with onset >5 years (r = 0.45, p = 0.0218) (Table 1B).
ROC Curve Analysis of eIF2B GEF Activity in the Leukodystrophic Groups
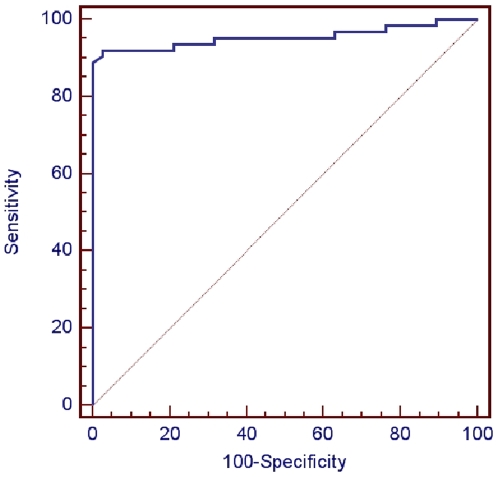
Three out of the 63 eIF2B-mutated patients (4.8%) had GEF activity >100%. They expressed a mild juvenile/adult form of the disease with onset ≥5 years of age (Table 1). The ROC curve analysis of eIF2B GEF activity performed towards differential diagnosis between eIF2B-related (n = 63) and eIF2B-unrelated (n = 38) leukodystrophy patients lead to a pathognomonic threshold “≤77.5% of GEF activity”, achieving 100% specificity (95% CL = 90.7–100%) and 88.9% sensitivity (95% CL = 78.4–95.4%), with an almost perfect discrimination (AUC = 0.96±0.024, p<0.0001) (Figure 3). Only 7/63 (11.1%) eIF2B-mutated patients had GEF activities >77.5%. These patients presented with classic to milder clinical forms with onset ranging from 3 to 57 years, slow disease progression, and carrying mutations in different EIF2B1-5 genes (Table 1). This group illustrates that the high level of GEF activity found is not linked to the type of mutated gene nor to a specific degree of clinical severity. However, none of these patients had a disease onset <3 years.
Figure 3. ROC (Receiver Operating Characteristic) curve of eIF2B GEF activity regarding the diagnosis of eIF2B-related disorders.
The ROC curve analysis was performed to determine the optimal threshold of GEF activity which best discriminates between eIF2B-mutated (n = 63) and eIF2B-unrelated leukodystrophic patients (n = 38). The ≤77.5% threshold achieves 100% specificity (95% CL = 90.7–100%) and 88.9% sensitivity (95% CL = 78.4–95.4%). The area under the ROC curve (AUC) = 0.955; standard error = 0.0244; 95% confidence interval: 0.894 to 0.986; test for the: AUC = 0.5, p = 0.0001.
Discussion
Analysis of this extended cohort showed that eIF2B GEF activity measured in patients' LLB distinguishes eIF2B-mutated patients from those with eIF2B-unrelated leukodystrophies with 100% positive predictive value (PPV) and 89% negative predictive value at ≤77.5% threshold. At this threshold, the assay systematically excludes patients without eIF2B mutations. Therefore, it represents an interesting screening tool to select patients for a direct sequencing of the EIF2B1-5 genes. For leukodystrophic patients with >77.5% GEF activity, the probability to find eIF2B mutations is 15% (7/45) and increases to 26.9% (7/26) if patients have clinical and/or MRI features typical to eIF2B-related disorder. Therefore, EIF2B1-5 sequencing is still indicated for patients who are clinically suspected of eIF2B-related disorder with GEF activity >77.5%, particularly in milder forms.
The wider range of disease onset reported in this cohort in comparison to previous work [13] may explain the weaker correlation found between age at disease onset and GEF activity. Such correlation is better in severe forms with GEF activity <55% for patients with onset <2 years (Figure 1B). Discrepancies in the GEF activity values were found among patients of the same group of disease onset or with the same mutations such as siblings 432–1 and 432–2 (GEF activities at respectively 90% and 53.9%), confirming that GEF activity in LLB is modulated by factors other than eIF2B mutations.
Determination of the asialotransferrin/total transferrin ratio in CSF is also a reliable marker to distinct eIF2B-mutated from non-mutated patients with 100% sensitivity and 94% specificity associated to 8%-ratio threshold [12]. This marker has been determined in parallel for five of our 63 eIF2B-mutated patients and a decreased ratio has been found in CSF (Table 1) in all five, including patient 432–1 with a surprisingly normal eIF2B GEF activity (90.4±1.8%) compared to his sibling. This suggests that these two biomarkers, if available for the same patient, may be complementary in order to assess with 100% sensitivity, specificity and PPV.
Limitations
Limitations of this study are:
the relatively small number of patients overall, as eIF2B-related disorders are rare disorders;
the age of disease onset, that may be imprecise for some patients, since it is determined retrospectively.
Acknowledgments
We gratefully acknowledge the participation of the patients' families. We acknowledge Scot Kimball to provide us the eIF2 substrate.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the ELA Foundation (2006–08). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schiffmann R, Moller JR, Trapp BD, Shih HH, Farrer RG, et al. Childhood ataxia with diffuse central nervous system hypomyelination. Ann Neurol. 1994;35:331–40. doi: 10.1002/ana.410350314. [DOI] [PubMed] [Google Scholar]
- 2.van der Knaap MS, Barth PG, Gabreels FJ, Franzoni E, Begeer JH, et al. A new leukoencephalopathy with vanishing white matter. Neurology. 1997;48:845–55. doi: 10.1212/wnl.48.4.845. [DOI] [PubMed] [Google Scholar]
- 3.Leegwater PA, Vermeulen G, Konst AA, Naidu S, Mulders J, et al. Subunits of the translation initiation factor eIF2B are mutant in leukoencephalopathy with vanishing white matter. Nat Genet. 2001;29:383–8. doi: 10.1038/ng764. [DOI] [PubMed] [Google Scholar]
- 4.van der Knaap MS, Leegwater PA, Könst AA, Visser A, Naidu A, et al. Mutations in each of the five subunits of translation initiation factor eIF2B can cause leukoencephalopathy with vanishing white matter. Ann Neurol. 2002;51:264–70. doi: 10.1002/ana.10112. [DOI] [PubMed] [Google Scholar]
- 5.Fogli A, Schiffmann R, Bertini E, Ughetto S, Combes P, et al. The effect of genotype on the natural history of eIF2B-related leukodystrophies. Neurology. 2004;62:1509–17. doi: 10.1212/01.wnl.0000123259.67815.db. [DOI] [PubMed] [Google Scholar]
- 6.Labauge P, Horzinski L, Ayrignac X, Blanc P, Vukusic S, et al. Natural history of adult-onset eIF2B-related disorders: a multi-centric survey of 16 cases. Brain. 2009;132:2161–9. doi: 10.1093/brain/awp171. [DOI] [PubMed] [Google Scholar]
- 7.Fogli A, Wong K, Eymard-Pierre E, Wenger J, Bouffard JP, et al. Cree leukoencephalopathy and CACH/VWM disease are allelic at the EIF2B5 locus. Ann Neurol. 2002;52:506–10. doi: 10.1002/ana.10339. [DOI] [PubMed] [Google Scholar]
- 8.van der Knaap MS, van Berkel CG, Herms J, van Coster R, Baethmann M, et al. eIF2B-related disorders: antenatal onset and involvement of multiple organs. Am J Hum Genet. 2003;73:1199–207. doi: 10.1086/379524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fogli A, Rodriguez D, Eymard-Pierre E, Bouhour F, Labauge P, et al. Ovarian failure related to eukaryotic initiation factor 2B mutations. Am J Hum Genet. 2003;72:1544–50. doi: 10.1086/375404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiffmann R, van der Knaap Invited article: an MRI-based approach to the diagnosis of white matter disorders. Neurology. 2009;72:750–9. doi: 10.1212/01.wnl.0000343049.00540.c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanderver A, Schiffmann R, Timmons M, Kellersberger KA, Fabris D, et al. Decreased asialotransferrin in cerebrospinal fluid of patients with Childhood-onset ataxia and central nervous system hypomyelination/Vanishing white matter disease. Clin Chem. 2005;51:2031–42. doi: 10.1373/clinchem.2005.055053. [DOI] [PubMed] [Google Scholar]
- 12.Vanderver A, Hathout Y, Maletkovic J, Gordon ES, Mintz M, et al. Sensitivity and specificity of decreased CSF asialotransferrin for eIF2B-related disorder. Neurology. 2008;70:2226–32. doi: 10.1212/01.wnl.0000313857.54398.0e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fogli A, Schiffmann R, Hugendubler L, Combes P, Bertini E, et al. Decreased guanine nucleotide exchange factor activity in eIF2B-mutated patients. Eur J Hum Genet. 2004;12:561–6. doi: 10.1038/sj.ejhg.5201189. [DOI] [PubMed] [Google Scholar]
- 14.Gomez E, Pavitt GD. Identification of domains and residues within the epsilon subunit of eukaryotic translation initiation factor 2B (eIF2Bepsilon) required for guanine nucleotide exchange reveals a novel activation function promoted by eIF2B complex formation. Mol Cell Biol. 2000;20:3965–76. doi: 10.1128/mcb.20.11.3965-3976.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neitzel H. A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum Genet. 1986;73:320–36. doi: 10.1007/BF00279094. [DOI] [PubMed] [Google Scholar]