Abstract
Currently, the Oxidative Stress (or Free Radical) Theory of Aging is the most popular explanation of how aging occurs at the molecular level. While data from studies in invertebrates (e.g., C. elegans and Drosophila) and rodents show a correlation between increased lifespan and resistance to oxidative stress (and in some cases reduced oxidative damage to macromolecules), direct evidence showing that alterations in oxidative damage/stress play a role in aging are limited to a few studies with transgenic Drosophila that overexpress antioxidant enzymes. Over the past eight years, our laboratory has conducted an exhaustive study on the effect of under- or overexpressing a large number and wide variety of genes coding for antioxidant enzymes. In this review, we present the survival data from these studies together. Because only one (the deletion of the Sod1 gene) of the 18 genetic manipulations we studied had an effect on lifespan, our data calls into serious question the hypothesis that alterations in oxidative damage/stress play a role in the longevity of mice.
Keywords: Antioxidant defense, oxidative stress, oxidative damage, knockout mice, transgenic mice, longevity
1. Introduction
The Free Radical Theory of Aging proposed in the 1950s by Denham Harman [1], postulates that oxygen free radicals formed endogenously from normal metabolic processes play a role in the aging process because of an increase in oxidative damage to macromolecules. The Free Radical Theory of Aging has sense been modified to the Oxidative Stress Theory of Aging because oxygen species such as peroxides and aldehydes, which are not technically free radicals, also play a role in oxidative damage to cells. The imbalance between prooxidants and antioxidants leads to an accumulation of oxidative damage in a variety of macromolecules with age resulting in a progressive loss in functional cellular processes, leading to the aging phenotype [2].
Several lines of evidence support the oxidative stress theory of aging. First, the levels of oxidative damage to lipid, DNA, and protein have been reported to increase with age in a wide variety of tissues and animal models [3]. Second, studies with animal models showing increased longevity are consistent with the Oxidative Stress Theory of Aging; the longer-lived animals show reduced oxidative damage and/or increased resistance to oxidative stress. For example, early studies on caloric restriction, which is the first and most studied experimental manipulation shown to increase lifespan and retard aging, showed that oxidative damage to lipid, DNA, and protein was reduced in caloric restricted rodents compared to rodents fed ad libitum (for review, see [3]). Subsequently, caloric restricted mice also were shown to be more resistant to oxidative stress [2, 4, 5]. In the 1990s, investigators showed that mutations in the insulin/IGF-1 signaling pathways (age-1, daf-2, and daf-16 mutants) increased the lifespan of C. elegans that was correlated with increased resistance to oxidative stress [6-8] and reduced oxidative damage [9, 10]. More recently, several genetic mouse models of longevity have been reported, e.g., Ames and Snell dwarf mice, p66sch-/-3mice, and Igf1r+/- female mice (for a review, see [11]), and the increased lifespan of these models has been correlated to increased resistance of either the cells from these mice or the mice to oxidative stress in dwarf mice [12, 13]; p66sch-/- mice [14], and Igf1r+/- female mice [15]. Thus, the observation that the experimental manipulations that increase lifespan in invertebrates and rodents correlate to increased resistance to oxidative stress or reduced oxidative damage provides strong evidence in support of the Oxidative Stress Theory of Aging. However, all of the experimental manipulations that increase lifespan also alter processes other than oxidative stress/damage; therefore, the increase in longevity in these animal models could arise through another mechanism.
A direct experimental test of the Oxidative Stress Theory of Aging is to alter the level of oxidative stress/damage and determine how these alterations affect lifespan. Using DNA recombinant technology, investigators over the past fifteen years have studied the effect of altering the expression of various components of the antioxidant defense system on lifespan; these studies are described in the Discussion. Below, we bring together all of the lifespan data that our group has conducted on transgenic/knockout mice with alterations in a wide variety of genes involved in the antioxidant defense system. These data demonstrate that almost all alterations in the antioxidant system of mice have no effect on lifespan.
2. Methods
2.1 Animals
All mice were maintained under pathogen-free, barrier conditions using microisolator cages in a temperature controlled environment as previously described [16, 17]. Mice were housed four per cage following weaning and fed ad libitum with commercial mouse chow (Teklad Diet LM485). The mice were genotyped at 4 to 5 weeks of age by PCR analysis of DNA obtained from tail clips. Mice were assigned to survival groups at 2 months of age and allowed to live out their entire lifespan, i.e., there was no censoring of the mice when measuring survival. All procedures followed the guidelines approved by the Institutional Animal Care and Use Committee at the University of Texas Health Science Center at San Antonio and South Texas Veterans Health Care System, Audie L. Murphy Division. A list and description of the genetic manipulations in the antioxidant defense system that were used in the studies described in this review are given in Table 1.
Table 1.
List of Knockout/Transgenic Mice Studied
| Gene | Description of Genetic Manipulation | Abbreviation | Reference |
|---|---|---|---|
| Knockout Mice | |||
| Mn-Superoxide Dismutase | Deletion of exon 3 of Sod2 gene | Sod2+/- | [21] |
| Cu/Zn-Superoxide Dismutase | Deletion of exon 3 and 4 of Sod1 gene | Sod1-/- | [32] |
| Glutathione Peroxidase 1 | Deletion of exon 2 of Gpx1 gene | Gpx1-/- | [35] |
| Glutathione Peroxidase 4 | Deletion of exon 3, 4, 5, 6, and 7 of Gpx4 gene | Gpx4+/- | [16] |
| Methionine Sulfoxide Reductase A | Deletion of exon 2 on MsrA gene | MsrA+/- | [45] |
| Thioredoxin 2 | Mutational insertion in exon 1 of Trx2 gene | Trx2+/- | [48] |
| Transgenic Mice | |||
| Cu/Zn-Superoxide Dismutase | 64 Kb genomic fragment of the human SOD1 gene containing 27 Kb of 5' and 3'-flanking sequences. | SOD1 Tg | [53] |
| Mn-Superoxide Dismutase | 13Kb of the genomic fragment of Sod2 mouse gene. | Sod2 Tg | [55] |
| Catalase | 80Kb genomic fragment of human catalase gene containing 41 Kb of 5' and 6Kb of the 3'flanking sequences. | CAT Tg | [53] |
| Glutathione Peroxidase 4 | 53Kb genomic fragment of human Gpx4 gene containing 30Kb of 5' and 20Kb of 3' flanking sequences. | GPX4 Tg | [58] |
The descriptions of the genetic manipulation for each animal model described in Table 1 were taken from the references given.
2.2 Analysis of Lifespan
Mice in the survival groups were allowed to live out their life, and the lifespan for individual mice was determined by recording the age of spontaneous death. The survival curves were compared statistically using the log-rank test [18], and the median, mean, 90th percentile (when 90% of the mice died), and maximum survivals were calculated for each group. Mean survivals (± SEM) for each experimental group were compared to the respective wild type (WT) group by performing a Student's t-test upon log-transformed survival times. The median and 90th percentile survivals for each group were compared to the WT group using a score test adapted from Wang et al. [19]. All comparisons were made individually between each experimental group and the WT group, and in the case where multiple experimental groups were compared to one WT group, Holm's method [20] was used to correct for multiple comparisons.
3. Genetic Manipulated Mice Models
3.1 Knockout mice
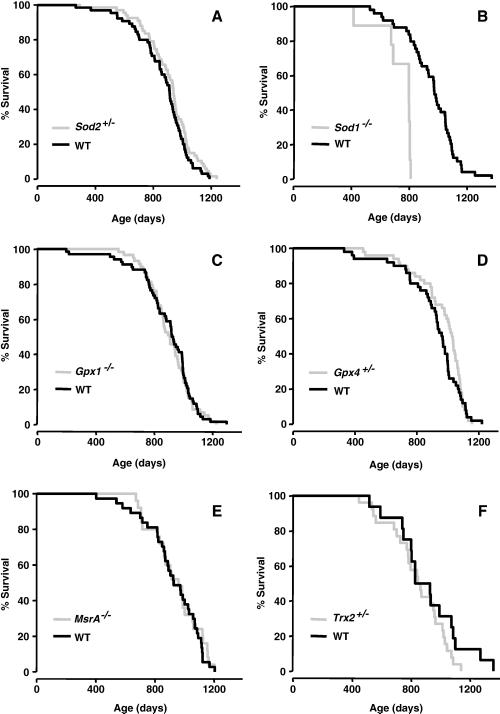
The first mouse model we studied was mice deficient in MnSOD (Sod2), which plays a major role in the detoxification of superoxide anions generated in the mitochondria. Mice lacking MnSOD die within days or weeks after birth from cardiomyopathy or neurodegeneration, depending on the genetic background [21-23]. Sod2+/- mice show reduced (~50%) MnSOD in all tissues studied [17] and both embryonic fibroblasts from these mice and the whole animals [17] are more sensitive to oxidative stress. Furthermore, tissues from these mice show a significant increase in oxidative damage to DNA[17]. As the data in Figure 1A and Table 2 show, the Sod2+/- mice show no difference in lifespan relative to WT mice [17].
Figure 1. Lifespans of knockout mice with a deficiency in various antioxidant enzymes.
The survival curves of Sod2+/- (Graph A; [17]), Sod1-/- (Graph D; unpublished), Gpx1-/- (Graph C; [43], Gpx4+/- (Graph B; [16]), MsrA-/- (Graph E; [42], and Trx2+/- (Graph F; unpublished) mice are shown compared to their WT cohorts. The genetic background, number, sex and survival data for these curves are given in Table 2.
Table 2.
Survival Data for Mice Deficient in Antioxidant Enzymes
| Genotype | Strain | Sex | N | Survival Curve (p=) | Mean | Median | 90% | Maximum | Reference |
|---|---|---|---|---|---|---|---|---|---|
| WT | C57BL/6 | Female | 66 | 894 ± 19 | 913 (855-943) | 1034 (1002-1009) | 1189 | [17] | |
| Sod2+/- | C57BL/6 | Female | 68 | 0.12 | 918 ± 19 | 940 (900-959) | 1088 (1044-1196) | 1239 | [17] |
| WT | C57BL/6 | Female | 50 | 915 ± 27 | 963 (923-996) | 1145 (1071-1220) | 1220 | [16] | |
| Gpx4+/- | C57BL/6 | Female | 50 | 0.32 | 964 ± 23 | 1029 (979-1050)* | 1026 (1085-1157) | 1157 | [16] |
| WT | C57BL/6 | Mixed | 68 | 891 ± 24 | 926 (874-994) | 1091 (1040-1188) | 1298 | [38] | |
| Gpx1-/- | C57BL/6 | Mixed | 59 | 0.70 | 903 ± 19 | 908 (853-963) | 1063 (1031-1183) | 1226 | [38] |
| Sod2+/- X Gpx1+/- | C57BL/6 | Mixed | 25 | 0.61 | 880 ± 33 | 877 (791-943) | 1057 (1027-1298) | 1283 | [38] |
| Sod2+/- X Gpx1-/- | C57BL/6 | Mixed | 33 | 0.76 | 905 ± 25 | 911 (833-983) | 1121 (1069-1248) | 1248 | [38] |
| Controla | Mixedb | Male | 37 | 925 ± 32 | 926 (868-1030) | 1120 (1092-1204) | 1204 | [46] | |
| MsrA-/- | Mixedb | Male | 25 | 0.87 | 942 ± 34 | 959 (858-1056) | 1156 (1122-1203) | 1203 | [46] |
| WT | C57BL/6 | Mixed | 50 | 902 ± 23 | 921 (874-993) | 1076 (1035-1298) | 1298 | UP | |
| Sod1-/- | C57BL/6 | Mixed | 10 | < 0.001 | 693 ± 41* | 755 (638-761) | 762 (761-767) | 767 | UP |
| Gpx1-/-XSod1-/- | C57BL/6 | Male | 11 | 0.004 | 725 ± 48 | 773*(674-823) | 828 (799-868) | 868 | UP |
| Gpx4+/-XSod1-/- | C57BL/6 | Male | 16 | 0.004 | 667 ± 42* | 672*(563-817) | 866 (817-883) | 883 | UP |
| Sod1-/-XSod2+/- | C57BL/6 | Mixed | 11 | < 0.001 | 730 ± 53 | 778 (674-823) | 886 (817-883) | 908 | UP |
| Gpx4+/-XGpx1-/- | C57BL/6 | Mixed | 40 | 0.85 | 933 ± 25 | 917 (866-1013) | 1124 (1086-1248) | 1248 | UP |
| Gpx4+/-XSod2+/- | C57BL/6 | Mixed | 11 | 0.85 | 906 ± 33 | 918 (824-1017) | 1025 (938-1099) | 1099 | UP |
| WT | Mixedb | Female | 16 | 913 ± 56 | 879 (802-1074) | 1186 (1086-1359) | 1359 | UP | |
| Trx2+/- | Mixedb | Female | 26 | 0.23 | 846 ± 36 | 855 (781-959) | 1059 (1020-1139) | 1139 | UP |
The survival data were taken from references given (UP = unpublished) and are presented together in the groups of mice in which the survival experiments were conducted. The survival data are expressed in days as mean ± SEM, median and 90% (when 90% of the mice have died) with 95% confidence interval in parenthesis, and maximum (age when the oldest mouse in the cohort died). The survival curves for the WT and knockout mice were statistically analyzed by the log-rank test, and the p values are given under survival curve column.
Control mice contained both MsrA+/+ and MsrA+/- mice.
The genetic background is a mixture of C57BL/6 and 129
Values significantly different from the WT mice at the p < 0.05 level.
CuZnSOD (Sod1) is the major superoxide dismutase isozyme found in cells and is localized in the cytosol and the intermembrane space of the mitochondria [24, 25]. Mice null for CuZnSOD are viable and appear normal at birth [26]. Studies by other laboratories have reported a number of moderate to more severe pathologies in the Sod1-/- mice [27-29]. For example, Sod1-/- females are almost totally infertile due to ovarian-dysfunction [27, 30], and Sod1-/- mice show very high levels of oxidative stress in several tissues and plasma and an accelerated loss of hind limb muscle mass with age that is associated with a phenotype consistent with distal axonopathy [31]. In 2005, Huang's laboratory reported that Sod1-/- mice show a decrease in lifespan, approximately 30%, which is associated with a high incidence of hepatocellular carcinoma [32]. We observed a ~30% decrease in mean, median, and maximum lifespan of the Sod1-/- mice (Figure 1B and Table 2). Neither Huang's laboratory [33] nor our laboratory found any decrease in the lifespan of the Sod1+/- mice compared to WT mice (unpublished data). Currently there is no information about the levels of oxidative stress/damage in the Sod1+/- animals.
Glutathione peroxidase 1 (Gpx1) is also viewed as one of the major cellular scavengers of hydrogen peroxide (H2O2) and alkyl hydroperoxides in the cell [34]. Mice null for Gpx1 are viable and have normal development [35]. However, they are highly sensitive to both paraquat and diquat [36, 37] and show increased oxidative damage to DNA [38]. Mice null for Gpx1 develop a high incidence of cataracts at a young age [39], suggesting an accelerated aging phenotype. The data in Figure 1C and Table 2 show that Gpx1-/- mice show no difference in lifespan compared to WT mice.
We studied the effect of glutathione peroxidase 4 (Gpx4) deficiency on lifespan because this enzyme plays a unique role in the detoxification of lipid peroxides in membranes. Gpx4 is widely expressed in tissues at low levels compared to Gpx1; it is found in the cytosol, mitochondrial and nuclear fractions [40]. Among all glutathione peroxidases, Gpx4 is the only peroxidase that can catalyze the reduction of complex lipid hydroperoxides, e.g., phospholipid hydroperoxides as well as hydroperoxides of cholesterol esters [41, 42]. Null mice for Gpx4 are embryonic lethal (abnormal embryo development at stage E7.5; [43]). Gpx4+/- mice showed reduced Gpx4 protein levels and activity in the cytosolic and mitochondrial fractions from all tissues studied, and whole animals as well as embryonic fibroblasts from these mice were more sensitive to oxidative stress [43]. In addition, oxidative damage was increased in embryonic fibroblasts from the Gpx4+/- mice, as indicated by increased levels of F2-isoprostanes and 8-oxo-2-deoxyguanosine in these cells [44]. Based on the oxidative stress theory, we predicted that the Gpx4+/- mice would show reduced lifespan because of the reduced ability to repair oxidative damage to membranes. However, we actually observed a slight (~7%), but significant, extension in median lifespan (Table 2), which appears to be due to a delay in the incidence of cancer in the Gpx4+/- mice [16]; neither the survival curve (Figure 1D), mean, nor 90% survival was significantly altered in the Gx4+/- mice.
Methionine sulfoxide reductase-A (MsrA) repairs oxidized methionine residues within proteins and also may function as a general antioxidant. In 2001, Moskovitz et al. reported that MsrA-/- mice have increased sensitivity to hyperoxia and show a major decrease (~40%) in lifespan [45]. We also observed that embryonic fibroblasts from MsrA-/- mice as well as whole animals showed increased sensitivity to oxidative stress [46]. However, as shown in Figure 1E and Table 2, we did not observe any decrease in the lifespan of MsrA-/- mice. These contradictory data show the importance of replicating lifespan studies, and the possible reason for these contradictory data are presented in the Discussion.
Thioredoxin 2 (Trx2), which is the mitochondrial form of thioredoxin, is the electron donor for several antioxidant enzymes (perroxiredoxins, MsrA, etc.), but also plays a major role in repairing the oxidation of cysteine residues in proteins [47]. Previous studies demonstrated that Trx2 null mice are embryonic lethal at stage E 8.5, and the histological assessment of these embryos indicate a high incidence of apoptosis [48]. Trx2+/- mice were viable and showed reduced levels of Trx2 in all tissues studied. In addition, the Trx2+/- mice display diminished mitochondrial functions (decreased ATP synthesis and increased ROS production) in several tissues studied. Additionally, we found increased levels of oxidative damage to DNA, lipids and protein [49]. The Trx2+/- mice showed a slight decrease (7%) in mean lifespan and a 16% decrease in maximum lifespan (Figure 1F and Table 2). However, the lifespan curves were not significantly different, and the decrease in mean, median, and 90% survival were not significantly different. This difference in lifespan might be significant with a greater sample size.
In addition to studying the effect of a reduction or complete deletion of a single antioxidant enzyme gene on lifespan, we also studied the effect of deletions in more than one antioxidant gene on lifespan, and these data are presented in Table 2. We hypothesized that reducing the expression of more than one antioxidant enzyme at a time, i.e., reducing two pathways in the antioxidant defense system, would affect longevity when reducing one pathway might not. In an attempt to test this hypothesis, we measured the lifespan of mice that have decreased levels of the following pairs of antioxidant enzymes: CuZnSOD and MnSOD (Sod1-/- / Sod2+/-); CuZnSOD and Gpx1 (Sod1-/- / Gpx1-/-); CuZnSOD and Gpx4 (Sod1-/- / Gpx4+/-); MnSOD and Gpx1 (Sod2+/- and Gpx1-/-); MnSOD and Gpx4 (Sod2+/- / Gpx4+/-) and Gpx1 and Gpx4 (Gpx1-/- / Gpx4+/-). Only the double mutants that that are null for Sod1 show a significant differences in the survival curves compared to WT mice (Table 2), and these mice show an ~20% decrease in mean lifespan and a more than 30% decrease in maximum lifespan. These data show that a lack of CuZnSOD consistently reduces lifespan no matter what changes occur in other antioxidant enzymes.
3.2 Transgenic mice
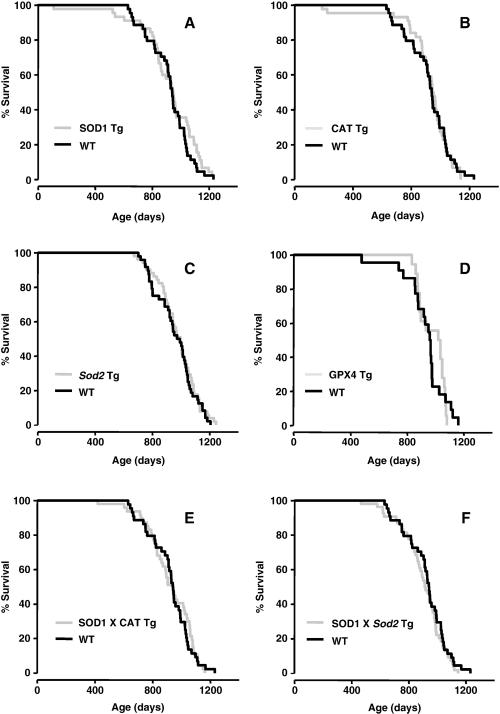
Overexpressing CuZnSOD has been shown to increase the lifespan of Drosophila [50-52]. CuZnSOD transgenic mice (SOD1 Tg) were generated using a large fragment of human genomic DNA containing the SOD1 gene (Table 1), and we studied the effect of overexpressing CuZnSOD on the lifespan of mice (Figure 2A). We showed that the activity of CuZnSOD was two- to five-fold higher in tissues of the SOD1 Tg compared to WT mice [53]. Embryonic fibroblasts from the SOD1 Tg mice were more resistance to paraquat toxicity ([54] and Figure 3). Moreover, data from whole animal indicate that SOD1Tg mice are more resistant to oxidative stress induced by paraquat, and levels of lipid peroxidation (measured as 8-isoprostane) induced by diquat treatment are lower in SOD1Tg mice compared to WT mice, as would be predicted from an increase in CuZnSOD expression (unpublished data). The data in Figure 2A and Table 3 show that the lifespan of SOD1 Tg mice was not significantly different from the lifespan of WT mice. These data are in agreement with the previous study from Epstein's laboratory using a different transgenic mouse model in which Huang et al. [33] showed that the lifespan of transgenic mice overexpressing CuZnSOD (two- to five-fold increase) was similar to WT mice.
Figure 2. Lifespans of transgenic mice overexpressing different antioxidant enzymes.
The survival curves of SOD1 Tg (Graph A; [103], CAT Tg (Graph B; [103]), Sod2 Tg (Graph C; [53]), GPX4 Tg (Graph D; unpublished), SOD1/CAT Tg (Graph E; [103]), and SOD1/Sod2 Tg (Graph F; [103]) mice are shown compared to their WT cohorts. The genetic background, number, sex and survival data for these curves are given in Table 3.
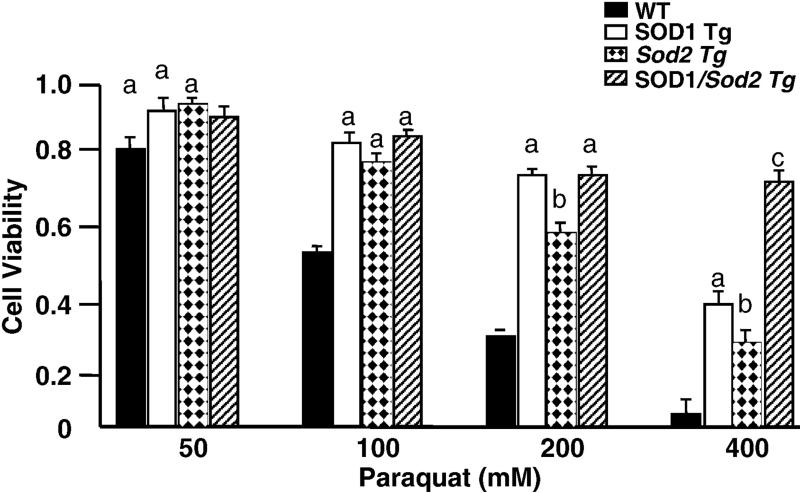
Figure 3. Sensitivity to transgenic mice to oxidative stress.
The sensitivity of embryonic fibroblasts isolated from WT, SOD1 Tg, Sod2 Tg, and SOD1/Sod2 Tg mice to paraquat (48 hrs) was determined as previously described [59]. The data are the mean ± SEM of experiments repeated with fibroblasts derived from three animals and were analyzed by two-way ANOVA. Values that are significant different (p< 0.05) from each other are shown with different subscripts.
Table 3.
Survival Data for Mice Overexpressing Antioxidant Enzymes
| Genotype | Strain | Sex | N | Survival Curve (p=) | Mean | Median | 90% | Maximum | Reference |
|---|---|---|---|---|---|---|---|---|---|
| WT | C57BL/6 | Male | 47 | 982±20 | 960 (920-1028) | 1128 (1080-1206) | 1206 | [57] | |
| Sod2 Tg | C57BL/6 | Male | 50 | 0.48 | 997±12 | 977 (943-1035) | 1165 (1092-1245) | 1245 | [57] |
| WT | C57BL/6 | Male | 22 | 933±31 | 963 (879-975) | 1106 (1026-1161) | 1161 | UP | |
| Gpx4 Tg | C57BL/6 | Male | 18 | 0.84 | 977±21 | 1028 (888-1062) | 1072 (1062-1080) | 1080 | UP |
| WT | C57BL/6 | Male | 44 | 922±22 | 941 (911-991) | 1090 (1038-1235) | 1231 | [103] | |
| SOD1 Tg | C57BL/6 | Male | 45 | 0.27 | 915±40 | 944 (859-1038) | 1131 (1111-2120) | 1229 | [103] |
| CAT Tg | C57BL/6 | Male | 44 | 0.87 | 916±28 | 949 (904-978) | 1099 (1036-1139) | 1139 | [103] |
| SOD1 Tg X CAT Tg | C57BL/6 | Male | 47 | 0.75 | 922±23 | 945 (863-1025) | 1098 (1078-1163) | 1163 | [103] |
| SOD1 Tg X Sod2 Tg | C57BL/6 | Male | 54 | 0.41 | 899±20 | 914 (868-967) | 1075 (1038-1117) | 1144 | [103] |
The survival data were taken from references (UP = unpublished) given and are presented together in the groups of mice in which the survival experiments were conducted. The survival data are expressed in days as mean ± SEM, median and 40% (when 90% of the mice have died) with 95% confidence interval in parenthesis, and maximum (age when the oldest mouse in the cohort died). The survival curves for the WT and transgenic mice were statistically analyzed by the log-rank test, and the p values are given under survival curve column.
Catalase is an antioxidant enzyme found in all aerobic cells that catalyzes the decomposition of hydrogen peroxide to oxygen and water. Catalase transgenic (CAT Tg) mice were generated in our laboratory using a large genomic fragment of human DNA containing the catalase gene (Table 1). Catalase activity is two- to four-fold higher in the tissues of the CAT Tg mice and is expressed in the peroxisomes [53]. Embryonic fibroblasts from CAT Tg mice are more resistant to hydrogen peroxide toxicity [54]. Data obtained from whole animal indicate that CAT Tg mice have less DNA oxidation (measured by 8oxodG) in all the tissues studied compared to WT mice (unpublished data). As shown in Figure 2B and Table 3, we observed no difference in lifespan of CAT Tg mice compared to WT mice.
In 2001, Epstein's laboratory generated transgenic mice overexpressing MnSOD (Sod2 Tg) using a genomic fragment of the mouse Sod2 gene (Table 1). These mice show a ~two-fold overexpression of MnSOD in all tissues examined [55]. Hu et al., reported that overexpression of MnSOD increased the maximum lifespan of transgenic mice using a transgene with the Sod2 cDNA fused to β-actin promoter (two- to four- fold increase, except liver) [56]. We showed that embryonic fibroblasts from the Sod2 Tg mice are more resistant to paraquat (Figure 3C) and that oxidative damage (protein carbonyls and F2-isoprostanes) is attenuated in old Sod2 Tg mice [57]. However, as shown in Figure 2C and Table 3, no difference was observed in the lifespans of the Sod2-Tg and WT mice.
In 2004, we generated Gpx4 transgenic mice (GPX4 Tg) using a large fragment of human genomic DNA containing the GPX4 gene (Table 1). Expression of the Gpx4 protein is two- to three fold higher than in the GPX4 Tg mice compared to WT mice. Embryonic fibroblasts from the GPX4 Tg mice were more resistant to t-butylhydroperoxide and diquat, and diquat-induced liver damage and lipid peroxidation were significantly reduced in vivo in the GPX4 Tg mice [58]. In addition, diquat-induced caspase 3 activation and cytochrome c release from the mitochondria were significantly reduced in GPX4 Tg mice showing that overexpressing Gpx4 protected cells from oxidative stress induced apoptosis [58]. The data in Figure 2D and Table 3 show that the lifespan of GPX4 Tg mice was not significantly different from WT mice.
We also have determined the effect of overexpressing multiple antioxidant genes, hypothesizing that antioxidant enzymes might work synergistically. First, we studied mice overexpressing both CuZnSOD and catalse (SOD1/CAT Tg), predicting that the increased hydrogen peroxide generated by overexpressing CuZnSOD would be converted to water by overexpressing catalase. Embryonic fibroblasts from SOD1/CAT Tg mice were resistant to both paraquat and hydrogen peroxide while embryonic fibroblasts from either SOD1 Tg or CAT Tg mice were resistant to only paraquat or hydrogen peroxide, respectively [54, 59]. The data in Figure 2E and Table 3 show that the lifespan of the SOD1/CAT Tg was essentially the same as that of WT, SOD1 Tg, or CAT Tg mice. In other words, there was no benefit of simultaneously overexpressing both CuZnSOD and catalase on lifespan. Second, we studied the effect of simultaneously overexpressing CuZnSOD and MnSOD (SOD1/Sod2 Tg mice) on lifespan because SOD1/Sod2 Tg mice would be predicted to have enhanced detoxification of superoxide anions in both the cytosol and mitochondria. The data in Figure 3 show that embryonic fibroblasts from SOD1/Sod2 Tg mice were more resistance to paraquat citotoxicity than embryonic fibroblasts from either SOD1 Tg or Sod1 Tg mice. As shown in Figure 2F and Table 3, the lifespan of SOD1/Sod2 Tg mice transgenic was not significantly different than the lifespan of WT, SOD1 Tg, or Sod2 Tg mice. Thus, simultaneous overexpression of superoxide dismutase in both the cytosol and mitochondria does not have a beneficial effect on longevity.
4. Discussion
The Oxidative Stress Theory of Aging has become the predominant theory to explain aging at the molecular level. Although there is a large amount of research over the past five decades supporting this theory, almost all the research has been correlative, e.g., a correlation between increased oxidative damage and age and a correlation between manipulations that increase lifespan and a reduction in oxidative damage and/or increase in resistance to oxidative stress. However, direct evidence showing that oxidative damage/stress alters aging was limited until the advent of genetic technology allowing investigators to alter the expression of antioxidant enzymes, which, because of their role in detoxification of free radicals and reactive oxygen species, can alter the sensitivity of the organism to oxidative stress and the levels of oxidative damage in cells and tissues.
Longevity or lifespan is the most acceptable parameter that has been used for several years to study aging, for example, crucial data obtained in several animals model with genetic mutations e.g. C. elegans (age-1, daf-2, daf-16 mutants), yeast (sir2), Drosophila (methusela and its ligand, stunted), and mice (Ames dwarf mice), etc; have used lifespan as determinant of aging. Ideally, would be better to determine other parameters involved in changes in the basic mechanisms of aging or healthspan. However, nowadays there is not consensus about how to define healthspan and how to measure this parameter in all of these model systems. Nevertheless, we do know that it is possible to retard aging in multiples animal models and simultaneously obtain lengthen in lifespan, for example, when mice and rats are fed restricted amount of food, aging mechanism appear to be delayed and the animal live longer. It is possible that the genetic alterations being studied may have slowed the rate of aging in certain tissues even though the mice were not living longer, however the pathology obtained from some of these lines, showed no major differences between mice genetically manipulated and WT mice, with the exception of MnSOD heterozygous mice, where this deficiency in MnSOD in mice resulted in a significant increase of incidence of tumor, however we did not observed differences in lifespan [17].
In this review article, we present data we have generated over the past eight years on the lifespan of mice with alterations in various enzymes in the antioxidant defense system. When using lifespan to determine whether an experimental manipulation alters aging, it is critical that lifespan be determined under optimal husbandry conditions to eliminate/minimize deaths from non-aging causes, e.g., infectious disease, inflammation, stress, etc. This is particularly important for studies using genetic manipulations in the antioxidant defense system because these genetic manipulations have the potential of altering survival when animals are maintained under sub-optimal husbandry conditions where the animals are exposed to increased stress/inflammation. The husbandry conditions used in our studies were optimal as shown by the long lifespans of the WT mice used in these studies, e.g., the mean and maximum survivals of more than 30 and 40 months, respectively, which is as long if not longer than the lifespans of similar inbred mice reported by other laboratories, including the aging colony maintained by the NIA [11, 16]. For example, data published by Jackson's Laboratory show that C57B6 mice have a mean and maximal lifespans of 27 and 40 months, respectively (mean lifespan approximately 3 months shorter compared to our data) [60, 61]. However, the comparisons of others parameters such as food intake, body weight, fecundity and incidence of tumors, resulted to be similar to the data coming from our animal facility [60-62]. Therefore, altogether indicate that our husbandry has optimal healthy conditions that allow us to observe an extension in lifespan of mice without changes in other parameters, such as body weight and food consumption.
Thus, the lifespan data we have generated in our studies allow us to determine with a high degree of accuracy whether the genetic manipulations in various components of the antioxidant defense system alter mouse longevity.
In the first series of experiments, we studied the effect of targeted genetic deletions in antioxidant enzymes on lifespan with the prediction that mice deficient or lacking one or more antioxidant enzymes would show an increase in the sensitivity of cells/tissues to oxidative stress resulting in increased oxidative damage and reduced lifespan. Previous studies with invertebrates have given mixed results with respect to the effect of reducing antioxidant gene expression on lifespan. In yeast, both clonal and chronological lifespan was shortened by deleting either CuZnSOD [63, 64], MnSOD [64, 65], MsrA, or MsrB [66]. However, under anaerobic conditions, deletion or overexpression of MsrA or MsrB had no effect on clonal lifespan [66]. In C. elegans, Doonan et al. [67], Yang et al. [68], Yen et al. [69], and Van Raamsdonk et al [70] demonstrated that the absence of SOD genes (both the cytosolic and mitochondrial isoforms) had no effect on lifespan, either in WT or in long-lived mutants despite the fact that these genetic manipulations increased both the sensitivity of the nematodes to oxidative stress (paraquat) and levels of oxidative damage to proteins. Drosophila lacking either CuZnSOD (SOD1) or MnSOD (SOD2) show a severe phenotype, e.g., deletion in CuZnSOD resulted in 80% reduction in lifespan [71, 72]; deletion in Sod2 induced postnatal lethality [73, 74]; Drosophila heterozygous for the SOD1 gene show no significant differences, and SOD2 gene show a slight reduction in lifespan [71, 74, 75].
In mice, two previous studies have measured the lifespan of mice lacking an antioxidant enzyme. In 2001, Moskovitz et al. [45] reported an ~40% decrease in mean and maximum lifespans in MsrA-/- mice, and in 2005, Elchuri et al. [32] reported that Sod1-/- mice have a 30 % reduction in mean lifespan and an ~40% in maximum lifespan. We have replicated the study by Elchuri et al. [32] showing that the mean, median, 90%, and maximum lifespans of Sod1-/- mice are reduced 20% to 40% compared to WT mice. However, we were unable to replicate the study by Moskovitz et al. [45]; we found no difference in the lifespans of MsrA-/- and WT/MsrA+/- mice. The difference in lifespan observed in the two studies is not due to the genetic background of the mice because the mice used in both studies were a mixture of C57BL/6 and 129. Rather, the small sample size and sub-optimum conditions used in the study by Moskovitz et al. [45] are a more likely cause of this discrepancy. Our larger sample size lessens the influence that each animal has on the overall survival, i.e., the survival data are less likely to be distorted by outliers that arise from maternal- or paternal-specific effects on lifespan [11, 76]. Moskovitz et al. used the following number of mice in their study: 17 MsrA-/-, 8 MsrA+/-, and 14 WT mice. However, the most likely reason for the discrepancy between our studies is that the mice in used in the study by Moskovitz et al. [45] appear to have been maintained under sub-optimum conditions, i.e., the mice were relatively short-lived. The mean lifespan of WT mice in the study by Moskovitz et al. was 680 days compared to the mean lifespan of 925 days for WT mice in our study. In other words, the husbandry conditions used in our study resulted in a ~35% longer lifespan. Thus, the reduction in lifespan of MsrA-/- mice observed by Moskovitz et al. could be due to the environment under which the mice were studied rather than due to accelerated aging because no difference in lifespan is observed when mice are maintained under conditions where they are able to live out their entire lifespan.
We also studied the effect of reduced expression of other antioxidant enzymes on lifespan, e.g., Sod2+/-, Gpx1-/-, Gpx4+/-, and Trx2+/- mice, and found no significant differences in the lifespans of these mice compared with their WT littermates. In fact, we found that reduced expression of Gpx4 resulted in a slight but significant increase in the median lifespan [16]. Furthermore, we studied the effect of reduced expression of various combinations of antioxidant enzymes on lifespan and found that only those mice lacking CuZnSOD showed a decreased in lifespan, which was similar to the lifespan for Sod1-/- mice. Therefore, our studies with knockout mice demonstrate that only mice that lack CuZnSOD show a reduction in lifespan. Although these mice exhibit some phenotypes of accelerating aging, i.e., increased age-related hearing loss [77-79], macular degeneration [80], early incidence of cataracts [81], vascular hypertrophy [82, 83], and increased age-related muscle atrophy [31], they also had high incidence of hepatocellular carcinoma [32], which is never observed in C57BL/6 mice [62, 84]. Therefore, it is uncertain whether the reduced lifespan observed in the Sod1-/- mice is due to accelerated aging or is from novel pathology arising from the genetic mutation.
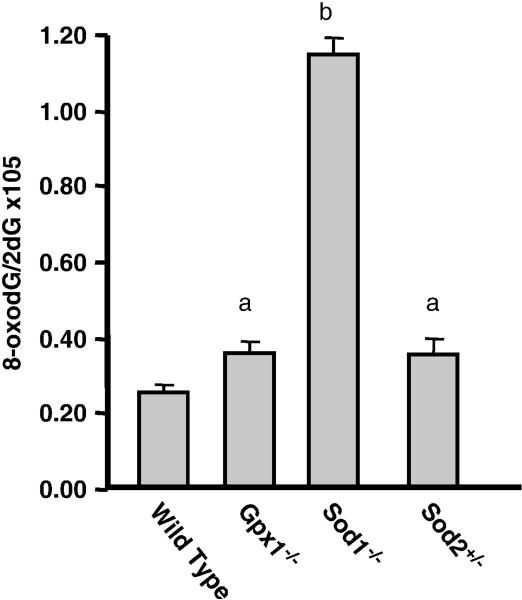
Why do Sod1-/- mice show reduced lifespan while all the other mice we studied that were deficient in other antioxidant enzymes show no effect on lifespan? All the knockout mouse models we studied show increased sensitivity to oxidative stress, i.e., they exhibit the phenotype expected from the reduced expression of the antioxidant enzyme(s). However, the Sod1-/- mice appear to have greater endogenous stress as observed by oxidative damage and changes in gene expression. For example, we showed that the genetic expression profile in the livers of Sod1-/- mice was differed from the profiles observed in WT or Gpx1-/- mice and showed that many of the changes in gene expression were similar to those found in the livers of WT or Gpx1-/- mice after induction of oxidative stress by diquat [37]. As shown in Figure 4, DNA oxidation is four- to five-fold higher in the livers of Sod1-/- mice than WT mice; DNA oxidation is ~40% higher in the Gpx1-/- or Sod2+/- mice compared to WT mice. In other words, the level of DNA oxidation in the Sod1-/- mice is two-fold higher than that observed in the Gpx1-/- or Sod2+/- mice. It should be noted that, the levels of oxidative damage observed in the Sod1-/- mice are much greater than the normally observed in tissues of old mice. For example, we have observed that oxidative damage to DNA increases with age ~40% in the livers of mice [17, 85], which is one-third that observed in the Sod1-/- mice but is similar to the increase in DNA oxidation observed in the Gpx1-/- or Sod2+/- mice. Thus, while Sod1-/- mice show an increase in oxidative damage and a decrease in lifespan, as would be predicted by the oxidative stress theory of aging, the levels of oxidative damage in the Sod1-/- mice are much higher than that observed even in old mice. In contrast, the levels of oxidative damage in Sod2+/- [17] and Gpx1-/- [43] mice are similar to those observed in old WT mice; however, and the lifespan of the Gpx1-/- and Sod2+/- mice are essentially identical to WT mice.
Figure 4. Oxidative damage to macromolecules in knockouts mice.
The levels of DNA oxidation measured as 8-oxodG/2dG in livers from young WT, Gpx1-/-, Sod1-/-, and Sod2+/- mice was determined as described previously [85]. The data are expressed as the mean ± SEM from 3 to 5 mice and were analyzed by the non-parametric test of ANOVA. Values that are significant different (p< 0.05) from each other are shown with different subscripts.
One of the problems in determining whether oxidative stress plays a role in aging using knockout mice to accelerate aging is that many manipulations can shorten lifespan that would not have any effect on aging, for example, peroxiredoxin-1 (Prdx1) knockout mice (B6), showed to have a significantly shortened lifespan, however this effect is due mainly to a higher incidence in cancer burden (osteosarcoma, fibrosarcoma) and hemolytic anemia, which are not pathologies found in this strain of mice [86]. Therefore, most gerontologists agree that a manipulation that increases lifespan gives the greatest insight in to the mechanism of aging. In other words, determining whether an increase in the antioxidant defense system would increase lifespan would be more powerful evidence for oxidative stress/free radicals playing a role in aging than showing that a reduction in the antioxidant system decreases lifespan.
The effect of overexpressing antioxidant enzymes on lifespan in invertebrates has been mixed. The lifespan (clonal and chronological) of yeast has been reported to be increased by the overexpression of MsrA [66] and MsrB [66], and MnSOD [65, 87]. Initial studies in Drosophila using P-element mediated transformation reported that overexpressing either CuZnSOD [72, 88, 89] or catalase [90] had no effect on lifespan. Later, Orr and Sohal [89] reported that overexpression of both CuZnSOD and catalase significantly increased the lifespan of Drosophila. However, the site of the P-element insertion can alter lifespan independently [91], and in a subsequent study with a large number of transgenic lines of Drosophila, Orr et al. [92] found that neither the overexpression of CuZnSOD and catalase, MnSOD, nor thioredoxin reductase did not significantly alter the lifespans of long-lived Drosophila strains. Using inducible systems to overexpress antioxidant genes to avoid the problems associated with the site of transgene insertion, Parkes et al. [51] and Sun and Tower [93] reported that overexpressing CuZnSOD increased the lifespan of Drosophila. Sun and Tower also reported that overexpression of catalase had no effect on lifespan and that there was no added benefit of overexpressing both catalase and CuZnSOD [93]. Subsequently, Sun et al. [94] reported that overexpression of MnSOD also increased the lifespan of Drosophila and that the simultaneous overexpression of MnSOD and CuZnSOD had an additional increase in lifespan effect [95]. Using 10 different genetic backgrounds of Drosophila of both sexes, Promislow's laboratory [96] reported that overexpression of CuZnSOD in motorneurones increased lifespan of long-lived flies; however, effect of CuZnSOD overexpression varied considerably with different genetic backgrounds. The effect of overexpressing CuZnSOD on lifespan was sex dependent, e.g., an increase in lifespan was observed in six of the strains of female drosophila, but only one strain of male drosophila showed an increase in lifespan [96]. The overexpression of MsrA has also been reported to increase the lifespan of Drosophila [97-99].
Our data show that transgenic mice overexpressing CuZnSOD, catalase, MnSOD, or Gpx4 have lifespans similar to WT mice even though cells/tissues from these mice show increased resistance to oxidative stress. Our data with SOD1 Tg mice confirm the previous study by Huang et al. [33], which used a different transgenic mouse model overexpressing CuZnSOD in a different strain of mice. Schriner et al. [100] previously reported that transgenic mice overexpressing catalase showed an ~21% increase in lifespan; however, in this study catalase overexpression was targeted to the mitochondria while in our study, catalase overexpression occurred in the peroxisomes [53], where catalase is normally expressed [101]. Moreover, data from the same group also showed that the cytosolic expression of catalase had no effect on lifespan [100]. Hu et al. [56] reported that transgenic mice overexpressing MnSOD showed an 18% increase in maximum lifespan (1095 days vs 1290 days); however, the mean survival of the transgenic mice overexpressing MnSOD was only 4% longer than the WT mice. However, Hu et al. [56] presented no statistical analysis of the survival data. We observed no statistical difference in the survival curves or in the mean or median survival. The maximum survival of the Sod2-Tg mice in our study was 3% longer than WT mice; however, to statistically assess whether changes in maximum lifespan are significant, it is necessary to compare survival ratios at some quantile where an adequate number of animals are still alive. The 90th percentile (when 90% of the mice have died and only 10% remain) is used for this purpose. We found no significant difference in the 90% survival between the Sod2-Tg and WT mice in our study. We also found that overexpressing both CuZnSOD and catalase or CuZnSOD and MnSOD had no effect on lifespan even though cells from the SOD1/CAT-Tg and SOD1/Sod2-Tg mice showed greater resistance to oxidative stress than increased expression of one of the antioxidant enzymes.
In summary, our research with 18 different genetic manipulations in the antioxidant defense system show that only mouse model null for Sod1 had an effect on lifespan that would be predicted from the Oxidative Stress Theory of Aging. One could argue that we failed to observe an effect on lifespan because the cells/tissues of the knockout/transgenic mice up- or down-regulate other components of the antioxidant defense system that counter the reduced or increased expression of the specific antioxidant enzyme(s). Except for the Sod1-/- mice [32], we have no evidence that any of the other manipulations showed an alteration in any of the other major antioxidant enzymes [16, 17, 38, 49, 54, 57-59, 102]. However, one can always argue that some minor component of the antioxidant enzyme system is altered in response to the changes in the genetic manipulation. To our knowledge, the only detailed study of gene expression in mice with an alteration in the antioxidant defense system was conducted by our group for Sod1-/- and Gpx1-/- mice [37]. We showed that neither knockout mouse model resulted in an up-regulation of any classical antioxidant genes in liver; however, the Sod1-/- mice showed an up-regulation of thiol antioxidants (e.g., metallothione, glutathione, thioredoxin, sulfiredoxin, etc.). Interestingly, the Sod1 null mice showed the reduction in lifespan even though these genes were up-regulated. The strongest evidence that the knockout/transgenic mouse models we studied exhibit the phenotype predicted from the genetic manipulation is that cells from these mouse models show alterations in sensitivity to oxidative stress. Therefore, we believe the fact that the lifespan was not altered in the majority, if not most, of the knockout/transgenic mice is strong evidence against oxidative stress/damage playing a major role in the molecular mechanism of aging in mice.
Acknowledgements
Financial support for the lifespan data generated over the past eight years was provided by National Institutes of Health (grants R01-AG-015908, R01-AG-023843, P01-AG19316, P01AG020591, and R37-AG026557) and the Department of Veterans Affairs (Merit Grants and a Research Enhancement Award Program). Especial acknowledgement is given to the Animal Core of the San Antonio Nathan Shock Center for Excellence in the Basic Biology of Aging (P30-AG-13319) directed by Dr. James Nelson, Dr. Randy Strong, and Vivian Diaz.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- [2].Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bokov A, Chaudhuri A, Richardson A. The role of oxidative damage and stress in aging. Mech Ageing Dev. 2004;125:811–26. doi: 10.1016/j.mad.2004.07.009. [DOI] [PubMed] [Google Scholar]
- [4].Yu BP. Aging and oxidative stress: modulation by dietary restriction. Free Radic Biol Med. 1996;21:651–68. doi: 10.1016/0891-5849(96)00162-1. [DOI] [PubMed] [Google Scholar]
- [5].Barja G. Endogenous oxidative stress: relationship to aging, longevity and caloric restriction. Ageing Res Rev. 2002;1:397–411. doi: 10.1016/s1568-1637(02)00008-9. [DOI] [PubMed] [Google Scholar]
- [6].Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A. 1995;92:7540–4. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Johnson TE, Lithgow GJ, Murakami S. Hypothesis: interventions that increase the response to stress offer the potential for effective life prolongation and increased health. J Gerontol A Biol Sci Med Sci. 1996;51:B392–5. doi: 10.1093/gerona/51a.6.b392. [DOI] [PubMed] [Google Scholar]
- [8].Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. Faseb J. 1999;13:1385–93. [PubMed] [Google Scholar]
- [9].Ishii N, Goto S, Hartman PS. Protein oxidation during aging of the nematode Caenorhabditis elegans. Free Radic Biol Med. 2002;33:1021–5. doi: 10.1016/s0891-5849(02)00857-2. [DOI] [PubMed] [Google Scholar]
- [10].Yasuda K, Adachi H, Fujiwara Y, Ishii N. Protein carbonyl accumulation in aging dauer formation-defective (daf) mutants of Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci. 1999;54:B47–51. doi: 10.1093/gerona/54.2.b47. discussion B52-3. [DOI] [PubMed] [Google Scholar]
- [11].Liang H, Masoro EJ, Nelson JF, Strong R, McMahan CA, Richardson A. Genetic mouse models of extended lifespan. Exp Gerontol. 2003;38:1353–64. doi: 10.1016/j.exger.2003.10.019. [DOI] [PubMed] [Google Scholar]
- [12].Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the ageing process. Nature. 1996;384:33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- [13].Murakami S, Salmon A, Miller RA. Multiplex stress resistance in cells from long-lived dwarf mice. Faseb J. 2003;17:1565–6. doi: 10.1096/fj.02-1092fje. [DOI] [PubMed] [Google Scholar]
- [14].Migliaccio E, Giorgio M, Mele S, et al. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–13. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- [15].Holzenberger M, Dupont J, Ducos B, et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–7. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- [16].Ran Q, Liang H, Ikeno Y, et al. Reduction in glutathione peroxidase 4 increases life span through increased sensitivity to apoptosis. J Gerontol A Biol Sci Med Sci. 2007;62:932–42. doi: 10.1093/gerona/62.9.932. [DOI] [PubMed] [Google Scholar]
- [17].Van Remmen H, Ikeno Y, Hamilton M, et al. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- [18].Andersen PH, Richelsen B, Bak J, et al. Influence of short-term dexfenfluramine therapy on glucose and lipid metabolism in obese non-diabetic patients. Acta Endocrinol (Copenh) 1993;128:251–8. doi: 10.1530/acta.0.1280251. [DOI] [PubMed] [Google Scholar]
- [19].Wang C, Li Q, Redden DT, Weindruch R, Allison DB. Statistical methods for testing effects on “maximum lifespan”. Mech Ageing Dev. 2004;125:629–32. doi: 10.1016/j.mad.2004.07.003. [DOI] [PubMed] [Google Scholar]
- [20].Holm S. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics. 1979;6:65–70. [Google Scholar]
- [21].Li Y, Huang TT, Carlson EJ, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11:376–81. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- [22].Huang TT, Carlson EJ, Kozy HM, et al. Genetic modification of prenatal lethality and dilated cardiomyopathy in Mn superoxide dismutase mutant mice. Free Radic Biol Med. 2001;31:1101–10. doi: 10.1016/s0891-5849(01)00694-3. [DOI] [PubMed] [Google Scholar]
- [23].Lebovitz RM, Zhang H, Vogel H, et al. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc Natl Acad Sci U S A. 1996;93:9782–7. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Okado-Matsumoto A, Fridovich I. Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu, Zn-SOD in mitochondria. J Biol Chem. 2001;276:38388–93. doi: 10.1074/jbc.M105395200. [DOI] [PubMed] [Google Scholar]
- [25].Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. A fraction of yeast Cu, Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J Biol Chem. 2001;276:38084–9. doi: 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- [26].Reaume AG, Elliott JL, Hoffman EK, et al. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet. 1996;13:43–7. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- [27].Ho YS, Gargano M, Cao J, Bronson RT, Heimler I, Hutz RJ. Reduced fertility in female mice lacking copper-zinc superoxide dismutase. J Biol Chem. 1998;273:7765–9. doi: 10.1074/jbc.273.13.7765. [DOI] [PubMed] [Google Scholar]
- [28].Shefner JM, Reaume AG, Flood DG, et al. Mice lacking cytosolic copper/zinc superoxide dismutase display a distinctive motor axonopathy. Neurology. 1999;53:1239–46. doi: 10.1212/wnl.53.6.1239. [DOI] [PubMed] [Google Scholar]
- [29].Flood DG, Reaume AG, Gruner JA, et al. Hindlimb motor neurons require Cu/Zn superoxide dismutase for maintenance of neuromuscular junctions. Am J Pathol. 1999;155:663–72. doi: 10.1016/S0002-9440(10)65162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Matzuk MM, Dionne L, Guo Q, Kumar TR, Lebovitz RM. Ovarian function in superoxide dismutase 1 and 2 knockout mice. Endocrinology. 1998;139:4008–11. doi: 10.1210/endo.139.9.6289. [DOI] [PubMed] [Google Scholar]
- [31].Muller FL, Song W, Liu Y, et al. Absence of CuZn superoxide dismutase leads to elevated oxidative stress and acceleration of age-dependent skeletal muscle atrophy. Free Radic Biol Med. 2006;40:1993–2004. doi: 10.1016/j.freeradbiomed.2006.01.036. [DOI] [PubMed] [Google Scholar]
- [32].Elchuri S, Oberley TD, Qi W, et al. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24:367–80. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- [33].Huang TT, Carlson EJ, Gillespie AM, Shi Y, Epstein CJ. Ubiquitous overexpression of CuZn superoxide dismutase does not extend life span in mice. J Gerontol A Biol Sci Med Sci. 2000;55:B5–9. doi: 10.1093/gerona/55.1.b5. [DOI] [PubMed] [Google Scholar]
- [34].Halliwell B, Gutteridge JM. Lipid peroxidation in brain homogenates: the role of iron and hydroxyl radicals. J Neurochem. 1997;69:1330–1. doi: 10.1046/j.1471-4159.1997.69031330.x. [DOI] [PubMed] [Google Scholar]
- [35].Ho YS, Magnenat JL, Bronson RT, et al. Mice deficient in cellular glutathione peroxidase develop normally and show no increased sensitivity to hyperoxia. J Biol Chem. 1997;272:16644–51. doi: 10.1074/jbc.272.26.16644. [DOI] [PubMed] [Google Scholar]
- [36].Fu Y, Cheng WH, Porres JM, Ross DA, Lei XG. Knockout of cellular glutathione peroxidase gene renders mice susceptible to diquat-induced oxidative stress. Free Radic Biol Med. 1999;27:605–11. doi: 10.1016/s0891-5849(99)00104-5. [DOI] [PubMed] [Google Scholar]
- [37].Han ES, Muller FL, Perez VI, et al. The in vivo gene expression signature of oxidative stress. Physiol Genomics. 2008;34:112–26. doi: 10.1152/physiolgenomics.00239.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang Y, Ikeno Y, Qi W, Chaudhuri A, Li Y, Bokov A, Epstein C, Richardson A, Van Remmen H. Mice deficient in both manganese superoxide dismutase and glutathione peroxidase-1 have increased oxidative damage and a greater incidence of pathology that do not lead to a reduction in longevity. J. Gerontol Biol Sci Med Sci. doi: 10.1093/gerona/glp132. Submitted to. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wolf N, Penn P, Pendergrass W, et al. Age-related cataract progression in five mouse models for anti-oxidant protection or hormonal influence. Exp Eye Res. 2005;81:276–85. doi: 10.1016/j.exer.2005.01.024. [DOI] [PubMed] [Google Scholar]
- [40].Savaskan NE, Ufer C, Kuhn H, Borchert A. Molecular biology of glutathione peroxidase 4: from genomic structure to developmental expression and neural function. Biol Chem. 2007;388:1007–17. doi: 10.1515/BC.2007.126. [DOI] [PubMed] [Google Scholar]
- [41].Ursini F, Bindoli A. The role of selenium peroxidases in the protection against oxidative damage of membranes. Chem Phys Lipids. 1987;44:255–76. doi: 10.1016/0009-3084(87)90053-3. [DOI] [PubMed] [Google Scholar]
- [42].Maiorino M, Chu FF, Ursini F, Davies KJ, Doroshow JH, Esworthy RS. Phospholipid hydroperoxide glutathione peroxidase is the 18-kDa selenoprotein expressed in human tumor cell lines. J Biol Chem. 1991;266:7728–32. [PubMed] [Google Scholar]
- [43].Yant LJ, Ran Q, Rao L, et al. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic Biol Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- [44].Ran Q, Van Remmen H, Gu M, et al. Embryonic fibroblasts from Gpx4+/- mice: a novel model for studying the role of membrane peroxidation in biological processes. Free Radic Biol Med. 2003;35:1101–9. doi: 10.1016/s0891-5849(03)00466-0. [DOI] [PubMed] [Google Scholar]
- [45].Moskovitz J, Bar-Noy S, Williams WM, Requena J, Berlett BS, Stadtman ER. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc Natl Acad Sci U S A. 2001;98:12920–5. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Salmon AB, Pérez VI, Bokov A, et al. Lack of Methionine Sulfoxide Reductase A in Mice Increases Sensitivity to Oxidative Stress but Does not Diminish Lifespan. FASEB J. doi: 10.1096/fj.08-127415. Submitted to. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lillig CH, Holmgren A. Thioredoxin and related molecules--from biology to health and disease. Antioxid Redox Signal. 2007;9:25–47. doi: 10.1089/ars.2007.9.25. [DOI] [PubMed] [Google Scholar]
- [48].Nonn L, Williams RR, Erickson RP, Powis G. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol Cell Biol. 2003;23:916–22. doi: 10.1128/MCB.23.3.916-922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Perez VI, Lew CM, Cortez LA, et al. Thioredoxin 2 haploinsufficiency in mice results in impaired mitochondrial function and increased oxidative stress. Free Radic Biol Med. 2008;44:882–92. doi: 10.1016/j.freeradbiomed.2007.11.018. [DOI] [PubMed] [Google Scholar]
- [50].Sohal RS, Agarwal A, Agarwal S, Orr WC. Simultaneous overexpression of copper- and zinc-containing superoxide dismutase and catalase retards age-related oxidative damage and increases metabolic potential in Drosophila melanogaster. J Biol Chem. 1995;270:15671–4. doi: 10.1074/jbc.270.26.15671. [DOI] [PubMed] [Google Scholar]
- [51].Parkes TL, Elia AJ, Dickinson D, Hilliker AJ, Phillips JP, Boulianne GL. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat Genet. 1998;19:171–4. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- [52].Phillips JP, Parkes TL, Hilliker AJ. Targeted neuronal gene expression and longevity in Drosophila. Exp Gerontol. 2000;35:1157–64. doi: 10.1016/s0531-5565(00)00117-0. [DOI] [PubMed] [Google Scholar]
- [53].Chen X, Mele J, Giese H, et al. A strategy for the ubiquitous overexpression of human catalase and CuZn superoxide dismutase genes in transgenic mice. Mech Ageing Dev. 2003;124:219–27. doi: 10.1016/s0047-6374(02)00161-6. [DOI] [PubMed] [Google Scholar]
- [54].Mele J, Van Remmen H, Vijg J, Richardson A. Characterization of transgenic mice that overexpress both copper zinc superoxide dismutase and catalase. Antioxid Redox Signal. 2006;8:628–38. doi: 10.1089/ars.2006.8.628. [DOI] [PubMed] [Google Scholar]
- [55].Raineri I, Carlson EJ, Gacayan R, et al. Strain-dependent high-level expression of a transgene for manganese superoxide dismutase is associated with growth retardation and decreased fertility. Free Radic Biol Med. 2001;31:1018–30. doi: 10.1016/s0891-5849(01)00686-4. [DOI] [PubMed] [Google Scholar]
- [56].Hu D, Cao P, Thiels E, et al. Hippocampal long-term potentiation, memory, and longevity in mice that overexpress mitochondrial superoxide dismutase. Neurobiol Learn Mem. 2007;87:372–84. doi: 10.1016/j.nlm.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Jang YC, Pérez VI, Song W, Lustgarten MS, Salmon AB, Mele J, Qi W, Liu Y, Liang H, Chaudhuri A, Ikeno Y, Epstein CJ, Van Remmen H, Richardson A. Overexpression of Mn Superoxide Dismutase Protects against Oxidative Stress but Does Not Increase Lifespan in Mice. J. Gerontol Biol Sci Med Sci. doi: 10.1093/gerona/glp100. Accepted in. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ran Q, Liang H, Gu M, et al. Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. J Biol Chem. 2004;279:55137–46. doi: 10.1074/jbc.M410387200. [DOI] [PubMed] [Google Scholar]
- [59].Chen X, Liang H, Van Remmen H, Vijg J, Richardson A. Catalase transgenic mice: characterization and sensitivity to oxidative stress. Arch Biochem Biophys. 2004;422:197–210. doi: 10.1016/j.abb.2003.12.023. [DOI] [PubMed] [Google Scholar]
- [60].Rowlatt C, Chesterman FC, Sheriff MU. Lifespan, age changes and tumour incidence in an ageing C57BL mouse colony. Lab Anim. 1976;10:419–42. doi: 10.1258/002367776780956917. [DOI] [PubMed] [Google Scholar]
- [61].Mewissen DJ, Rust JH, Ugarte A, Haren DJ. Time sequence of cancer occurrence. Implications in low level radiation risk assessment. C R Acad Sci III. 1999;322:183–96. doi: 10.1016/s0764-4469(99)80043-2. [DOI] [PubMed] [Google Scholar]
- [62].Ikeno Y, Hubbard GB, Lee S, et al. Housing density does not influence the longevity effect of calorie restriction. J Gerontol A Biol Sci Med Sci. 2005;60:1510–7. doi: 10.1093/gerona/60.12.1510. [DOI] [PubMed] [Google Scholar]
- [63].Wawryn J, Krzepilko A, Myszka A, Bilinski T. Deficiency in superoxide dismutases shortens life span of yeast cells. Acta Biochim Pol. 1999;46:249–53. [PubMed] [Google Scholar]
- [64].Longo VD, Gralla EB, Valentine JS. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J Biol Chem. 1996;271:12275–80. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- [65].Fabrizio P, Pletcher SD, Minois N, Vaupel JW, Longo VD. Chronological aging-independent replicative life span regulation by Msn2/Msn4 and Sod2 in Saccharomyces cerevisiae. FEBS Lett. 2004;557:136–42. doi: 10.1016/s0014-5793(03)01462-5. [DOI] [PubMed] [Google Scholar]
- [66].Koc A, Gasch AP, Rutherford JC, Kim HY, Gladyshev VN. Methionine sulfoxide reductase regulation of yeast lifespan reveals reactive oxygen species-dependent and -independent components of aging. Proc Natl Acad Sci U S A. 2004;101:7999–8004. doi: 10.1073/pnas.0307929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Doonan R, McElwee JJ, Matthijssens F, et al. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev. 2008;22:3236–41. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Yang W, Li J, Hekimi S. A Measurable increase in oxidative damage due to reduction in superoxide detoxification fails to shorten the life span of long-lived mitochondrial mutants of Caenorhabditis elegans. Genetics. 2007;177:2063–74. doi: 10.1534/genetics.107.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Yen K, Patel HB, Lublin AL, Mobbs CV. SOD isoforms play no role in lifespan in ad lib or dietary restricted conditions, but mutational inactivation of SOD-1 reduces life extension by cold. Mech Ageing Dev. 2009;130:173–8. doi: 10.1016/j.mad.2008.11.003. [DOI] [PubMed] [Google Scholar]
- [70].Van Raamsdonk JM, Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000361. doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Phillips JP, Campbell SD, Michaud D, Charbonneau M, Hilliker AJ. Null mutation of copper/zinc superoxide dismutase in Drosophila confers hypersensitivity to paraquat and reduced longevity. Proc Natl Acad Sci U S A. 1989;86:2761–5. doi: 10.1073/pnas.86.8.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Reveillaud I, Phillips J, Duyf B, Hilliker A, Kongpachith A, Fleming JE. Phenotypic rescue by a bovine transgene in a Cu/Zn superoxide dismutase-null mutant of Drosophila melanogaster. Mol Cell Biol. 1994;14:1302–7. doi: 10.1128/mcb.14.2.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Kirby K, Hu J, Hilliker AJ, Phillips JP. RNA interference-mediated silencing of Sod2 in Drosophila leads to early adult-onset mortality and elevated endogenous oxidative stress. Proc Natl Acad Sci U S A. 2002;99:16162–7. doi: 10.1073/pnas.252342899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Duttaroy A, Paul A, Kundu M, Belton A. A Sod2 null mutation confers severely reduced adult life span in Drosophila. Genetics. 2003;165:2295–9. doi: 10.1093/genetics/165.4.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Muller FL, Lustgarten MS, Jang Y, Richardson A, Van Remmen H. Trends in oxidative aging theories. Free Radic Biol Med. 2007;43:477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- [76].Priest NK, Mackowiak B, Promislow DE. The role of parental age effects on the evolution of aging. Evolution. 2002;56:927–35. doi: 10.1111/j.0014-3820.2002.tb01405.x. [DOI] [PubMed] [Google Scholar]
- [77].McFadden SL, Ding D, Reaume AG, Flood DG, Salvi RJ. Age-related cochlear hair cell loss is enhanced in mice lacking copper/zinc superoxide dismutase. Neurobiol Aging. 1999;20:1–8. doi: 10.1016/s0197-4580(99)00018-4. [DOI] [PubMed] [Google Scholar]
- [78].McFadden SL, Ding D, Burkard RF, et al. Cu/Zn SOD deficiency potentiates hearing loss and cochlear pathology in aged 129, CD-1 mice. J Comp Neurol. 1999;413:101–12. [PubMed] [Google Scholar]
- [79].Ohlemiller KK, Dugan LL. Elevation of reactive oxygen species following ischemia-reperfusion in mouse cochlea observed in vivo. Audiol Neurootol. 1999;4:219–28. doi: 10.1159/000013845. [DOI] [PubMed] [Google Scholar]
- [80].Imamura Y, Noda S, Hashizume K, et al. Drusen, choroidal neovascularization, and retinal pigment epithelium dysfunction in SOD1-deficient mice: a model of age-related macular degeneration. Proc Natl Acad Sci U S A. 2006;103:11282–7. doi: 10.1073/pnas.0602131103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Reddy VN, Kasahara E, Hiraoka M, Lin LR, Ho YS. Effects of variation in superoxide dismutases (SOD) on oxidative stress and apoptosis in lens epithelium. Exp Eye Res. 2004;79:859–68. doi: 10.1016/j.exer.2004.04.005. [DOI] [PubMed] [Google Scholar]
- [82].Baumbach GL, Didion SP, Faraci FM. Hypertrophy of cerebral arterioles in mice deficient in expression of the gene for CuZn superoxide dismutase. Stroke. 2006;37:1850–5. doi: 10.1161/01.STR.0000227236.84546.5a. [DOI] [PubMed] [Google Scholar]
- [83].Didion SP, Kinzenbaw DA, Schrader LI, Faraci FM. Heterozygous CuZn superoxide dismutase deficiency produces a vascular phenotype with aging. Hypertension. 2006;48:1072–9. doi: 10.1161/01.HYP.0000247302.20559.3a. [DOI] [PubMed] [Google Scholar]
- [84].Frith CH, Wiley LD. Morphologic classification and correlation of incidence of hyperplastic and neoplastic hematopoietic lesions in mice with age. J Gerontol. 1981;36:534–45. doi: 10.1093/geronj/36.5.534. [DOI] [PubMed] [Google Scholar]
- [85].Hamilton ML, Guo Z, Fuller CD, et al. A reliable assessment of 8-oxo-2-deoxyguanosine levels in nuclear and mitochondrial DNA using the sodium iodide method to isolate DNA. Nucleic Acids Res. 2001;29:2117–26. doi: 10.1093/nar/29.10.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Neumann CA, Krause DS, Carman CV, et al. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature. 2003;424:561–5. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- [87].Harris N, Costa V, MacLean M, Mollapour M, Moradas-Ferreira P, Piper PW. Mnsod overexpression extends the yeast chronological (G(0)) life span but acts independently of Sir2p histone deacetylase to shorten the replicative life span of dividing cells. Free Radic Biol Med. 2003;34:1599–606. doi: 10.1016/s0891-5849(03)00210-7. [DOI] [PubMed] [Google Scholar]
- [88].Seto NO, Hayashi S, Tener GM. Overexpression of Cu-Zn superoxide dismutase in Drosophila does not affect life-span. Proc Natl Acad Sci U S A. 1990;87:4270–4. doi: 10.1073/pnas.87.11.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Orr WC, Sohal RS. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 1994;263:1128–30. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]
- [90].Orr WC, Sohal RS. The effects of catalase gene overexpression on life span and resistance to oxidative stress in transgenic Drosophila melanogaster. Arch Biochem Biophys. 1992;297:35–41. doi: 10.1016/0003-9861(92)90637-c. [DOI] [PubMed] [Google Scholar]
- [91].Kaiser M, Gasser M, Ackermann R, Stearns SC. P-element inserts in transgenic flies: a cautionary tale. Heredity. 1997;78(Pt 1):1–11. doi: 10.1038/hdy.1997.1. [DOI] [PubMed] [Google Scholar]
- [92].Orr WC, Mockett RJ, Benes JJ, Sohal RS. Effects of overexpression of copper-zinc and manganese superoxide dismutases, catalase, and thioredoxin reductase genes on longevity in Drosophila melanogaster. J Biol Chem. 2003;278:26418–22. doi: 10.1074/jbc.M303095200. [DOI] [PubMed] [Google Scholar]
- [93].Sun J, Tower J. FLP recombinase-mediated induction of Cu/Zn-superoxide dismutase transgene expression can extend the life span of adult Drosophila melanogaster flies. Mol Cell Biol. 1999;19:216–28. doi: 10.1128/mcb.19.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Sun J, Folk D, Bradley TJ, Tower J. Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster. Genetics. 2002;161:661–72. doi: 10.1093/genetics/161.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Sun J, Molitor J, Tower J. Effects of simultaneous over-expression of Cu/ZnSOD and MnSOD on Drosophila melanogaster life span. Mech Ageing Dev. 2004;125:341–9. doi: 10.1016/j.mad.2004.01.009. [DOI] [PubMed] [Google Scholar]
- [96].Spencer CC, Howell CE, Wright AR, Promislow DE. Testing an `aging gene' in long-lived drosophila strains: increased longevity depends on sex and genetic background. Aging Cell. 2003;2:123–30. doi: 10.1046/j.1474-9728.2003.00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Chavous DA, Hake LE, Lynch RJ, O'Connor CM. Translation of a unique transcript for protein isoaspartyl methyltransferase in haploid spermatids: implications for protein storage and repair. Mol Reprod Dev. 2000;56:139–44. doi: 10.1002/(SICI)1098-2795(200006)56:2<139::AID-MRD3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- [98].Chavous DA, Jackson FR, O'Connor CM. Extension of the Drosophila lifespan by overexpression of a protein repair methyltransferase. Proc Natl Acad Sci U S A. 2001;98:14814–8. doi: 10.1073/pnas.251446498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Ruan H, Tang XD, Chen ML, et al. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc Natl Acad Sci U S A. 2002;99:2748–53. doi: 10.1073/pnas.032671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Schriner SE, Linford NJ, Martin GM, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–11. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- [101].Zamocky M, Koller F. Understanding the structure and function of catalases: clues from molecular evolution and in vitro mutagenesis. Prog Biophys Mol Biol. 1999;72:19–66. doi: 10.1016/s0079-6107(98)00058-3. [DOI] [PubMed] [Google Scholar]
- [102].Van Remmen H, Qi W, Sabia M, et al. Multiple deficiencies in antioxidant enzymes in mice result in a compound increase in sensitivity to oxidative stress. Free Radic Biol Med. 2004;36:1625–34. doi: 10.1016/j.freeradbiomed.2004.03.016. [DOI] [PubMed] [Google Scholar]
- [103].Perez VI, Van Remmen H, Bokov A, Epstein CJ, Vijg J, Richardson A. The Overexpression of Major Antioxidant Enzymes Does Not Extend the Lifespan of Mice. Aging Cell. 2008 doi: 10.1111/j.1474-9726.2008.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]