Abstract
The T box transcription antitermination riboswitch is one of the main regulatory mechanisms utilized by Gram-positive bacteria to regulate genes that are involved in amino acid metabolism. The details of the antitermination event, including the role that Mg2+ plays, in this riboswitch have not been completely elucidated. In these studies, details of the antitermination event were investigated utilizing 2-aminopurine to monitor structural changes of a model antiterminator RNA when it was bound to model tRNA. Based on the results of these fluorescence studies, the model tRNA binds the model antiterminator RNA via an induced fit. This binding is enhanced by the presence of Mg2+, facilitating the complete base pairing of the model tRNA acceptor end with the complementary bases in the model antiterminator bulge.
Keywords: RNA, Riboswitch, T box, Transcription, 2-Aminopurine, Fluorescence
The T box riboswitch is a transcriptional control mechanism found primarily in Gram-positive bacteria [1–3], and its prevalence indicates that it is one of the main regulatory mechanisms for genes involved in amino acid metabolism [4]. The 5′ untranslated leader region (5′-UTR) of the T box genes is composed of a complex set of conserved primary sequence and secondary structural elements that allow the nascent mRNA to sense the charging ratio of the gene’s cognate tRNA [5, 6]. Control of T box genes hinges on the formation of one of two mutually-exclusive, competing structural elements – the terminator and the antiterminator [1–3]. The uncharged cognate tRNA is capable of factor-independent binding with the mRNA via a two-point interaction: the tRNA anticodon with the mRNA specifier sequence and the tRNA acceptor end with the complementary bases in the antiterminator (Fig. 1A) [1, 5, 7–9]. The stabilization of the antiterminator through this interaction allows transcription of the gene, while the formation of the competing terminator (in the absence of uncharged, cognate tRNA) results in transcription termination. The specificity of the interaction between the tRNA and mRNA is dictated by the co-variation of the tRNA anticodon and the mRNA specifier sequence [1] and by the co-variation of the tRNA discriminator base and the mRNA variable base, which is located in the bulge of the antiterminator element [5].
Fig. 1.
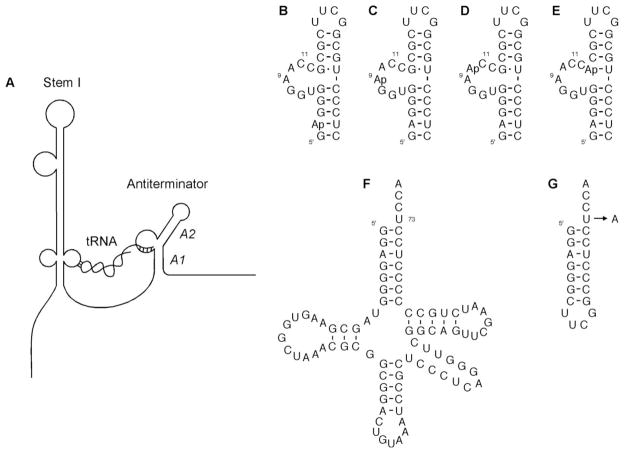
(A) Schematic of T box gene mRNA leader region with cognate uncharged tRNA. The tRNA binds to the leader region via two points of contact: 1) tRNA anticodon with the specifier sequence in Stem 1 of the leader region [1], and 2) tRNA acceptor end with the complementary bases in the 5′ end of the antiterminator bulge [5]. (A1 and A2 refer to the helical regions in the antiterminator). Antiterminator and tRNA model RNAs. (B) 2-AP-AM1A; (C) 9-AP-AM1A [30]; (D) 10-AP-AM1A; (E) 13-AP-AM1A; (F) tRNA-UCCA [28]; (G) mh-UCCA [28] and mh-ACCA [28].
Interesting facets of this unique interaction between the 5′-UTR of the mRNA and the tRNA have been elucidated via in vitro biochemical studies. In one study involving the B. subtilis glyQS 5′-UTR, Mg2+ was implicated as an important requirement for efficient in vitro antitermination [9, 10]. Near maximal tRNA binding occurs with 5 mM Mg2+, but maximal antitermination is not achieved until 15 mM Mg2+. In another in vitro study utilizing this same model system, it was found that the tRNA is able to be competitively displaced prior to the formation of the antiterminator. However, once the tRNA is bound to the mRNA at both points of contact, the tRNA is resistant to competitive displacement [10].
The 2-aminopurine purine base analog (AP) provides local nucleic acid structural information via steady-state [11–15] and time-resolved [16–19] fluorescence experiments. The fluorophore senses local stacking and polarity changes, yielding changes in emission intensity and/or emission wavelength [20–22]. In addition, AP is capable of base pairing with uracil in a manner that is similar to that of adenine [23–25]. These characteristics of AP have allowed the substitution of the fluorophore for a variety of nucleotides, especially guanine and adenine, to monitor nucleic acid dynamics and interactions with minimal structural perturbation to double-stranded motifs and a variety of single-stranded RNA motifs [11, 13–18, 21, 24–27].
Past studies from this lab have raised some interesting hypotheses regarding the interaction between the tRNA acceptor end and the antiterminator. The NMR solution structure of an antiterminator model RNA, AM1A [28], revealed structural flexibility in the antiterminator that was hypothesized to allow for tRNA binding via an induced-fit and/or tertiary structure capture. A more recent study identified a preferred binding site for Mg2+ along helix A1 in AM1A [29]. From this work, it was hypothesized that the Mg2+ binding at this site might facilitate tRNA binding by reducing electrostatic repulsion. In the current work, AP was utilized to probe structural changes in T box antiterminator model RNA upon its binding with model tRNA. The results of these studies will provide evidence in support of the two hypotheses presented above. In addition, while much work has been done to increase the general understanding of the nature of AP in DNA dynamics, these studies will contribute to the understanding of the nature of AP in RNA dynamics.
Materials and methods
Model RNAs
Local environmental changes within the structure of the antiterminator model RNA, AM1A, were monitored using the 2-aminopurine fluorescent purine base analog (AP). The AM1A model RNA motif has been demonstrated, both in vitro and in vivo, to be structurally and functionally relevant [28]. Four different AP-labeled AM1As (Figs. 1B-E) were designed to monitor changes in the A1 helix, the bulge, and the hinge region. In the A1 helix, AP was substituted for the adenine at position 2 (2-AP-AM1A). For probing the bulge, AP was substituted for the adenines at positions 9 and 10 (9-AP-AM1A [26, 30] and 10-AP-AM1A, respectively). Finally, AP was substituted for the guanine at position 13 in the hinge region (13-AP-AM1A). Purified, fluorescently-labeled RNAs were obtained from Dharmacon Research, Inc.
An unlabeled model tRNA [28] (tRNA-UCCA, Fig. 1F) and two unlabeled tRNA acceptor stem/end microhelix model RNAs [28] (mh-UCCA and mh-ACCA, Fig. 1G) were bound with the AP-AM1A RNAs in titration and fluorescence decay experiments. The mh-UCCA and tRNA-UCCA have an acceptor end sequence that is fully complementary to the four bases at the 5′ end of the AM1A bulge. The mh-ACCA model is the single base mismatch analog, in which the discriminator base is not complementary to the AM1A variable base. All of these model tRNAs, whose functional relevance were previously demonstrated [26, 28], were synthesized via in vitro transcription with T7 RNA polymerase [31–33], followed by purification with denaturing gel electrophoresis and electroelution.
All RNAs were dialyzed into 10 mM sodium phosphate pH 6.5, 0.01 mM EDTA and were renatured prior to use. In addition, all RNA stock solutions were stored in standard microcentrifuge tubes, as opposed to low-binding microcentrifuge tubes. During this investigation, it was discovered that previously reported preliminary results [26] were contaminated by trace impurities that resulted from the use of low-binding microcentrifuge tubes for RNA stock solution storage (data not shown). Due to the large amount of the unlabeled microhelix RNAs that was necessary for those previously reported fluorescence titration studies, an emission band at 394 nm was observed upon excitation of these trace impurities at 310 nm. The absence of the trace impurities in the RNA stock solutions for the current studies was verified by obtaining the fluorescence emission profiles of all unlabeled RNAs (data not shown). In light of the results from the current work, the previous trace impurities did not affect the binding of the model antiterminator and model tRNAs in the previous work.
Steady-State Fluorescence
The steady-state fluorescence spectra were obtained by titrating 400 μL of 100 nM AP-RNA (50 mM sodium phosphate pH 6.5, 0.01 mM EDTA, 50 mM NaCl, 15 mM or 5 mM MgCl2) with 2 μL aliquots (up to 20 μL total) of the model tRNA or either of the microhelix model tRNAs. The experiments were performed with a JY Horiba Spex Fluoromax-3, which was equipped with a temperature-controlled sample holder that was set to 20 °C. The labeled RNAs were excited at 310 nm (5 nm slit width), and the fluorescence emission was monitored over the range of 330 to 600 nm (5 nm slit width). Spectra were obtained for the unbound, labeled RNAs and after each successive addition of the model tRNAs. The buffer background was subtracted from each spectrum to remove the water Raman signal at 347 nm.
Time-Resolved Fluorescence
Fluorescence decay spectra were obtained with a PTI LaserStrobe spectrofluorometer that was equipped with a PTI GL-3300 nitrogen laser as its light source. The 2-aminopurine fluorophore was excited at 337 nm, utilizing a frequency of 10 Hz and 5 laser shots per time point. Fluorescence emission decays were obtained at 370 nm for 1 μM free 2-aminopurine deoxyribonucleoside (APdNS) and at both 370 nm and 420 nm for 1 μM 9-AP-AM1A by itself and a 99% bound complex of 9-AP-AM1A and mh-UCCA. The concentration of mh-UCCA that was needed to achieve the 99% (1521 μM) bound state was calculated based on the previously reported Kd of 16 μM for 5′-Fl-AM1A and mh-UCCA [26]. All RNA solutions had a volume of 400 μL and contained 50 mM sodium phosphate pH 6.5, 0.01 mM EDTA, 50 mM NaCl, 15 mM MgCl2. The decay spectra were collected with start and end delays of 50 and 100 ns, respectively. The data points were obtained with an integration time of 1000 μs and were an average of 7 reads over 200 channels. The excitation and emission slit widths were adjusted to maximize the intensity of the 9-AP-AM1A fluorescence intensity at 370 nm. The instrument response function was collected at 337 nm, utilizing an empty cuvette and a neutral density filter in the emission path.
Decays were deconvoluted with the TimeMaster software (PTI, Inc.), and the goodness of fit was based on the reduced χ2 and the randomness of the autocorrelation function of the weighted residuals. The time-resolved fluorescence spectra were fit to the following decay function:
where I(t) is the fluorescence intensity as a function of time, αi is the amplitude for each component, t is the time, τi is the fluorescence lifetime of each component, and N is the number of components. In order to determine the minimum number of components that were required to fit each of the decay spectra, each spectrum was fit utilizing N = 1, 2, 3, and 4.
Results
Steady-state fluorescence
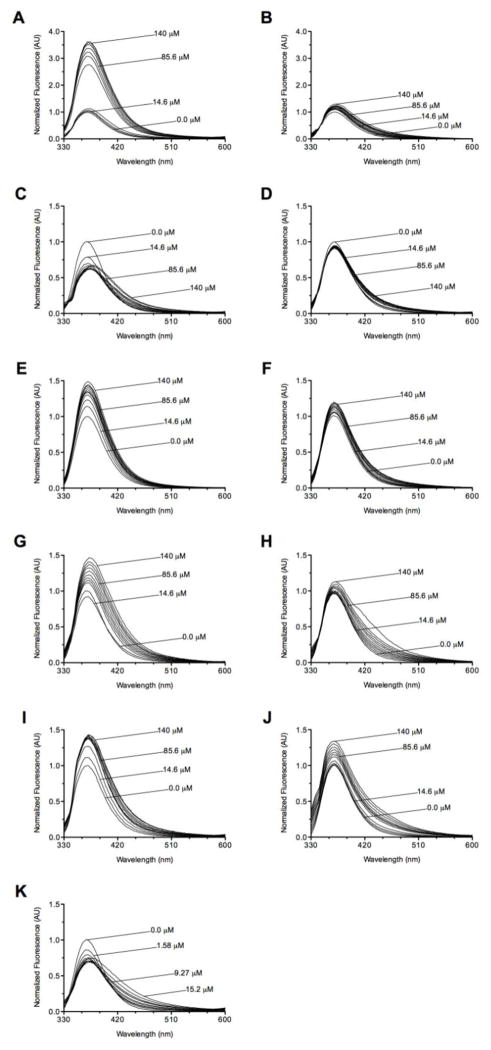
Addition of mh-UCCA in the presence of 15 mM MgCl2 led to strong fluorescence enhancements for the antiterminator model RNAs 2-AP-AM1A, 10-AP-AM1A, and 13-AP-AM1A (Figs. 2A, 2E, 2G). An enhancement of the fluorescence intensity likely indicates a destabilization of the stacking around the fluorophore, since stacking quenches fluorescence [34–36]. When each of these AP-AM1As was titrated with mh-ACCA, similar results were observed; however, the intensity enhancements were less pronounced (Figs. 2B, 2F, 2H). The titrations of 2-AP-AM1A and 10-AP-AM1A with either of the microhelix RNAs yielded no significant emission wavelength shift, but a 6 nm bathochromic shift was observed when 13-AP-AM1A was titrated with either mh-UCCA or mh-ACCA.
Fig. 2.
Fluorescence spectra for titrations of AP-AM1A with tRNA model RNAs in 50 mM sodium phosphate pH 6.5, 0.01 mM EDTA, 50 mM NaCl, 15 mM MgCl2 (except where noted). (A) 2-AP-AM1A with mh-UCCA, (B) 2-AP-AM1A with mh-ACCA, and (C) 9-AP-AM1A with mh-UCCA; (D) 9-AP-AM1A with mh-ACCA; (E) 10-AP-AM1A with mh-UCCA; (F) 10-AP-AM1A with mh-ACCA; (G) 13-AP-AM1A with mh-UCCA; (H) 13-AP-AM1A with mh-ACCA; (I) 9-AP-AM1A with mh-UCCA in 5 mM MgCl2; (J) 9-AP-AM1A with mh-ACCA in 5 mM MgCl2; and (K) 9-AP-AM1A with tRNA-UCCA. With the exception of 13-AP-AM1A with mh-ACCA, all observed enhancements were more significant than the respective buffer control enhancements.
The titration experiments involving 9-AP-AM1A were intriguingly different from the other AP-labeled antiterminator model RNAs. When 9-AP-AM1A was titrated with mh-UCCA at 15 mM MgCl2, no net intensity enhancement was observed (Fig. 2C). Instead, the increasing concentrations of mh-UCCA induced a 12 nm bathochromic shift of the emission wavelength and a broadening of the emission peak. Comparable results were observed when 9-AP-AM1A was titrated with tRNA-UCCA in the presence of 15 mM MgCl2. Namely, no net intensity enhancement, a broadening of the emission peak, and a 6 nm bathochromic shift of the emission wavelength were observed (Fig. 2K). As with the other three AP-AM1A titration experiments, titration of 9-AP-AM1A with the mismatched mh-ACCA at 15 mM MgCl2 (Fig. 2D) yielded effects that were similar to those of mh-UCCA, but were less pronounced.
The microhelix titrations of 9-AP-AM1A were also performed at 5 mM MgCl2. In contrast to all the spectra with 15 mM MgCl2, the spectra from mh-UCCA and mh-ACCA titrations of 9-AP-AM1A at 5 mM MgCl2 exhibited similar intensity enhancements for mh-UCCA compared to enhancements for mh-ACCA, while there was no shift of the emission wavelength induced by either microhelix (Figs. 2I–J).
Time-resolved fluorescence
Time-resolved fluorescence measurements were employed to better understand the nature of the peak broadening and the 12 nm bathochromic shift of the 9-AP-AM1A emission at 15 mM MgCl2 with mh-UCCA. The time-resolved fluorescence data are summarized in Table 2 and the decay spectra can be found in Supplementary data. The decay spectra for APdNS were best fit with a monoexponential function, yielding a decay lifetime of 8.4 ns. This is similar to previously-reported decay lifetimes for various free AP species [17, 37–39]. The spectrum for 9-AP-AM1A was best fit with a biexponential function at 370 nm. For 9-AP-AM1A, the shorter of the two decay lifetimes was in the range of 2 ns, while the longer of the two lifetimes was in the range of 7.6–9.5 ns. At 420 nm, the decay profile of the 99% complex was best fit with a monoexponential function, and that single lifetime (2.7 ns) was comparable to the shorter lifetime of the 9-AP-AM1A at 370 nm. The decay lifetimes that corresponded to 9-AP-AM1A at 420 nm, as well as for the 99% complex at 370 nm, were disregarded due to the potentially inaccurate nature of the data that resulted from their very low fluorescence intensities.
Discussion
Induced-fit binding
For each AP-labeled antiterminator model RNA, greater fluorescence enhancements or λmax shifts were observed when titrated with mh-UCCA, compared to titration with the mismatched mh-ACCA. These results indicate that base pairing of the tRNA discriminator and antiterminator variable bases induces a structural change throughout the antiterminator. The most dramatic differences were with 2-AP-AM1A and 9-AP-AM1A. In the case of 9-AP-AM1A, structural studies of uncomplexed AM1A indicated that A9 is stacked in a hydrophobic pocket with the Watson-Crick face not readily accessible for base pairing [40]. The more significant λmax shift in 9-AP-AM1A that was induced by the matched discriminator base (mh-UCCA), as compared with the shift induced by the mismatched discriminator base (mh-ACCA), is consistent with a structural rearrangement involving A9 (the variable base). This rearrangement can be correlated to the identity of the discriminator base and may be necessary to accommodate the base pairing of the variable base with the model tRNA discriminator base. In the case of 2-AP-AM1A, previous studies have shown that the phylogenetically-conserved 5′-side of helix A1 is a diffuse Mg2+ binding site that may facilitate tRNA binding [29]. The more significant enhancement of 2-AP-AM1A fluorescence in the presence of the matched discriminator base, as opposed to the mismatched discriminator base, and the data indicating that the discriminator base-variable base pair formation in 9-AP-AM1A is facilitated by higher Mg2+ concentrations are both consistent with a structural change occurring in the region of the second nucleotide in the diffuse Mg2+ binding site of helix A1 to facilitate tRNA binding.
Mg2+ facilitation of binding
The data indicate that Mg2+ likely plays a role in facilitating complete base pairing of model tRNA with AM1A (including the discriminator base-variable base pairing). The titration of 9-AP-AM1A with mh-UCCA at 5 mM MgCl2 led to a simple fluorescence intensity enhancement. This is in marked contrast to the bathochromic shift of the emission band observed for the same titration at 15 mM MgCl2. A proposed model of these different binding modes is shown in Fig. 3.
Fig. 3.
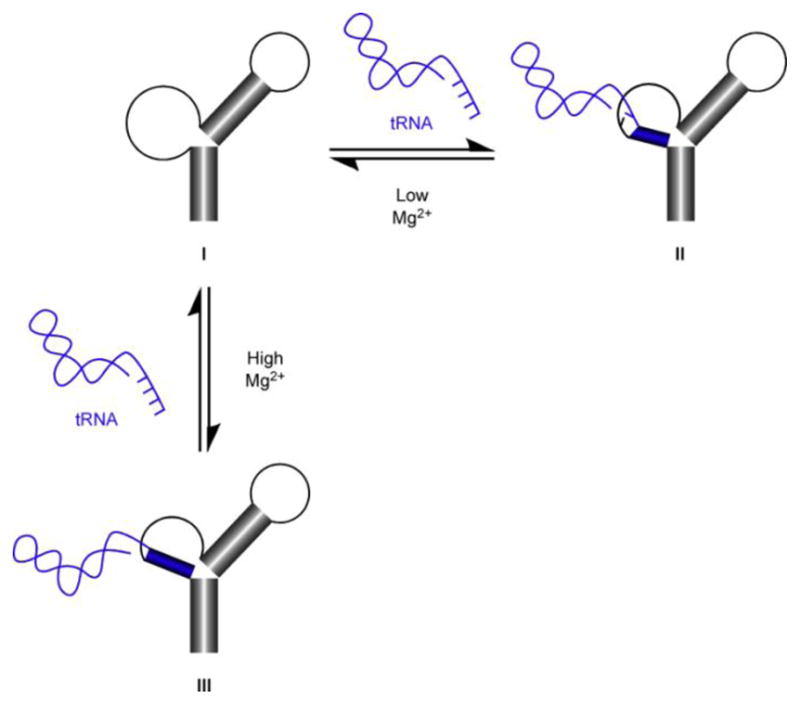
Proposed binding model for formation of tRNA-antiterminator RNA complex. In the presence of low concentrations of Mg2+, unbound antiterminator RNA (I) binds the 5′-CCA-3′ acceptor end nucleotides of tRNA to form II. In the presence of high concentrations of Mg2+, unbound antiterminator RNA (I) binds the entire 5′-UCCA-3′ acceptor end nucleotides of tRNA to form III.
The results of the steady-state fluorescence titration studies at 5 mM MgCl2 may arise from incomplete binding of the mh-UCCA with 9-AP-AM1A (II in Fig. 3). This is supported by the steady-state fluorescence data comparing the mh-UCCA-induced emission effects with the mh-ACCA-induced emission effects. The fact that the mh-ACCA-induced effects were not dissimilar from the mh-UCCA-induced effects likely indicates incomplete binding, as opposed to a lack of binding.
The steady-state fluorescence data and the time-resolved fluorescence data at 15 mM MgCl2 indicate that mh-UCCA is possibly binding AM1A differently than it does at 5 mM MgCl2. The structural change that is likely induced throughout the antiterminator, as was intimated from the steady-state fluorescence data involving the four different AP-AM1A models, may be indicative of a more complete binding event than is observed at 5 mM MgCl2 (III in Fig. 3). The apparent peak broadening and bathochromic shift of the emission wavelength that was observed for the titration of 9-AP-AM1A with mh-UCCA at 15 mM MgCl2 more closely resembled the appearance of a long wavelength emission band (~420 nm), coupled with the disappearance of the typical AP emission band at 370 nm. The fluorescence decay profile differences for the emission bands at 370 nm (9-AP-AM1A) and 420 nm (99% complex) support the hypothesis that the emission bands are likely arising from two different emitting conformational species.
Peak broadening and long-wavelength shifts have been observed previously in DNA [34, 41–43]. From recent studies with AP-labeled single-stranded and duplex DNA, the emission at the longer wavelength was proposed to result from the formation of a highly-stacked nucleodimer between AP and its neighboring base [20]. A similar conclusion was reached in base flipping studies involving the EcoKI and the M. TaqI methyltransferases [20, 43, 44]. In these studies, the longer wavelength fluorescence of AP was correlated with the stacking of the extrahelical fluorophore with an aromatic sidechain within the enzyme. In light of these other studies, the long wavelength emission band that arises from the titration of 9-AP-AM1A with mh-UCCA at 15 mM MgCl2 could indicate that AP is assuming a highly-stacked arrangement in the complex.
Conclusions
These studies provide insight into the mechanistic details of how the T box riboswitch antiterminator RNA structurally responds to base pairing with the tRNA acceptor end and of the discernment of the tRNA discriminator base by the antiterminator element. The magnesium-dependent results are consistent with the B. subtilis glyQS 5′-UTR in vitro studies in which 5 mM MgCl2 was sufficient to promote the proper folding and binding of the tRNA with the leader sequence, but a minimum of 15 mM MgCl2 was necessary for maximum efficient transcription antitermination [9]. The results of this investigation also support the hypothesis that binding of the mh-UCCA with AM1A involves an induced-fit structural rearrangement of the antiterminator model RNA. This structural change accompanies the Mg2+-facilitated base pairing of the antiterminator variable base with the tRNA discriminator base. The induced-fit may play a role in the tRNA-induced stabilization of the antiterminator and the previously-observed resistance of bound tRNA to displacement.
Supplementary Material
Table 1.
Fluorescence decay lifetimes and amplitudes for free 2-aminopurine deoxyribonucleoside (APdNS; at 370 nm), 9-AP-AM1A (at 370 and 420 nm) and 99% 9-AP-AM1A/mh-UCCA complex (at 370 and 420 nm).
| Sample | α1 | τ1 (ns) | α2 | τ2 (ns) | χ2 |
|---|---|---|---|---|---|
| APdNS (370 nm) | - | - | 0.028±0.0003 | 8.4±0.1 | 1.04 |
| 9-AP-AM1A (370 nm) | 0.014±0.001 | 2.1±0.3 | 0.003±0.001 | 9.5±1.7 | 1.29 |
| 9-AP-AM1A (420 nm) | n.a.a | n.a.a | n.a.a | n.a.a | n.a.a |
| 99% Complex (370 nm) | n.a.a | n.a.a | n.a.a | n.a.a | n.a.a |
| 99% Complex (420 nm) | 0.007±0.001 | 2.7±0.3 | - | - | 0.94 |
Fluorescence decays were measured at 370 nm and 420 nm for each sample and were analyzed individually. Due to very low fluorescence intensities, the data for 9-AP-AM1A at 420 nm and the data for 99% Complex at 370 nm were not utilized, see text.
Acknowledgments
This work was supported by NIH grant R01-GM61048 and the Office of the Vice President for Research, Ohio University.
Appendix A. Supplementary data
Supplementary data associated with this article can be found online.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 2.Grundy FJ, Henkin TM. Conservation of a transcription antitermination mechanism in aminoacyl-tRNA synthetase and amino acid biosynthesis genes in Gram-positive bacteria. J Mol Biol. 1994;235:798–804. doi: 10.1006/jmbi.1994.1038. [DOI] [PubMed] [Google Scholar]
- 3.Henkin TM, Glass BL, Grundy FJ. Analysis of the Bacillus subtilis tyrS gene: conservation of a regulatory sequence in multiple transfer-RNA synthetase genes. J Bacteriol. 1992;174:1299–1306. doi: 10.1128/jb.174.4.1299-1306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vitreschak AG, Mironov AA, Lyubetsky VA, Gelfand MS. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA. 2008;14:717–735. doi: 10.1261/rna.819308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grundy FJ, Rollins SM, Henkin TM. Interaction between the acceptor end of tRNA and the T-box stimulates antitermination in the Bacillus subtilis tyrS gene: a new role for the discriminator base. J Bacteriol. 1994;176:4518–4526. doi: 10.1128/jb.176.15.4518-4526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rollins SM, Grundy FJ, Henkin TM. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol Microbiol. 1997;25:411–421. doi: 10.1046/j.1365-2958.1997.4851839.x. [DOI] [PubMed] [Google Scholar]
- 7.Grundy FJ, Hodil SE, Rollins SM, Henkin TM. Specificity of tRNA-mRNA interactions in Bacillus subtilis tyrS antitermination. J Bacteriol. 1997;179:2587–2594. doi: 10.1128/jb.179.8.2587-2594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundy FJ, Winkler WC, Henkin TM. tRNA-mediated transcription antitermination in vitro: codon-anticodon pairing independent of the ribosome. Proc Natl Acad Sci USA. 2002;99:11121–11126. doi: 10.1073/pnas.162366799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yousef MR, Grundy FJ, Henkin TM. Structural transitions induced by the interaction between tRNAGly and the Bacillus subtilis glyQS T box leader RNA. J Mol Biol. 2005;349:273–287. doi: 10.1016/j.jmb.2005.03.061. [DOI] [PubMed] [Google Scholar]
- 10.Grundy FJ, Yousef MR, Henkin TM. Monitoring uncharged tRNA during transcription of the Bacillus subtilis glyQS gene. J Mol Biol. 2005;346:73–81. doi: 10.1016/j.jmb.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 11.Bradrick TD, Marino JP. Ligand-induced changes in 2-aminopurine fluorescence as a probe for small molecule binding to HIV-1 TAR RNA. RNA. 2004;10:1459–1468. doi: 10.1261/rna.7620304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gondert ME, Tinsley RA, Rueda D, Walter NG. Catalytic core structure of the transacting HDV ribozyme is subtly influenced by sequence variation outside the core. Biochemistry. 2006;45:7563–7573. doi: 10.1021/bi052116j. [DOI] [PubMed] [Google Scholar]
- 13.Harris DA, Rueda D, Walter NG. Local conformational changes in the catalytic core of the trans-acting hepatitis delta virus ribozyme accompany catalysis. Biochemistry. 2002;41:12051–12061. doi: 10.1021/bi026101m. [DOI] [PubMed] [Google Scholar]
- 14.Lafontaine DA, Wilson TJ, Zhao ZY, Lilley DMJ. Functional group requirements in the probable active site of the VS ribozyme. J Mol Biol. 2002;323:23–34. doi: 10.1016/s0022-2836(02)00910-5. [DOI] [PubMed] [Google Scholar]
- 15.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clerte C, Hall KB. Global and local dynamics of the U1A polyadenylation inhibition element (PIE) RNA and PIE RNA-U1A complexes. Biochemistry. 2004;43:13404–13415. doi: 10.1021/bi049117g. [DOI] [PubMed] [Google Scholar]
- 17.Kaul M, Barbieri CM, Pilch DS. Fluorescence-based approach for detecting and characterizing antibiotic-induced conformational changes in ribosomal RNA: comparing aminoglycoside binding to prokaryotic and eukaryotic ribosomal RNA sequences. J Am Chem Soc. 2004;126:3447–3453. doi: 10.1021/ja030568i. [DOI] [PubMed] [Google Scholar]
- 18.Neely RK, Daujotyte D, Grazulis S, Magennis SW, Dryden DTF, Klimasauskas S, Jones AC. Time-resolved fluorescence of 2-aminopurine as a probe of base flipping in M. HhaI-DNA complexes. Nucleic Acids Res. 2005;33:6953–6960. doi: 10.1093/nar/gki995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tleugabulova D, Reha-Krantz LJ. Probing DNA polymerase-DNA interactions: examining the template strand in exonuclease complexes using 2-aminopurine fluorescence and acrylamide quenching. Biochemistry. 2007;46:6559–6569. doi: 10.1021/bi700380a. [DOI] [PubMed] [Google Scholar]
- 20.Bonnist EYM, Jones AC. Long-wavelength fluorescence from 2-aminopurine-nucleobase dimers in DNA. Chem Phys Chem. 2008;9:1121–1129. doi: 10.1002/cphc.200700813. [DOI] [PubMed] [Google Scholar]
- 21.Rachofsky EL, Osman R, Ross JBA. Probing structure and dynamics of DNA with 2-aminopurine: effects of local environment on fluorescence. Biochemistry. 2001;40:946–956. doi: 10.1021/bi001664o. [DOI] [PubMed] [Google Scholar]
- 22.Zagórowska I, Adamiak RW. 2-Aminopurine labeled RNA bulge loops. Synthesis and thermodynamics. Biochimie. 1996;78:123–130. doi: 10.1016/0300-9084(96)82644-7. [DOI] [PubMed] [Google Scholar]
- 23.Kirk SR, Luedtke NW, Tor Y. 2-Aminopurine as a real-time probe of enzymatic cleavage and inhibition of hammerhead ribozymes. Bioorg Med Chem. 2001;9:2295–2301. doi: 10.1016/s0968-0896(01)00123-7. [DOI] [PubMed] [Google Scholar]
- 24.O’Neill MA, Barton JK. 2-aminopurine: a probe of structural dynamics and charge transfer in DNA and DNA:RNA hybrids. J Am Chem Soc. 2002;124:13053–13066. doi: 10.1021/ja0208198. [DOI] [PubMed] [Google Scholar]
- 25.Rist MJ, Marino JP. Association of an RNA kissing complex analyzed using 2-aminopurine fluorescence. Nucleic Acids Res. 2001;29:2401–2408. doi: 10.1093/nar/29.11.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Means JA, Wolf S, Agyeman A, Burton JS, Simson CM, Hines JV. T box riboswitch antiterminator affinity modulated by tRNA structural elements. Chem Biol Drug Des. 2007;69:139–145. doi: 10.1111/j.1747-0285.2007.00476.x. [DOI] [PubMed] [Google Scholar]
- 27.Nelson AR, Henkin TM, Agris PF. tRNA regulation of gene expression: Interactions of an mRNA 5′-UTR with a regulatory tRNA. RNA. 2006;12:1254–1261. doi: 10.1261/rna.29906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerdeman MS, Henkin TM, Hines JV. In vitro structure-function studies of the Bacillus subtilis tyrS mRNA antiterminator: evidence for factor-independent tRNA acceptor stem binding specificity. Nucleic Acids Res. 2002;30:1065–1072. doi: 10.1093/nar/30.4.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jack KD, Means JA, Hines JV. Characterizing riboswitch function: Identification of Mg2+ binding site in T box antiterminator RNA. Biochem Biophys Res Commun. 2008;370:306–310. doi: 10.1016/j.bbrc.2008.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Means JA, Katz S, Nayek A, Anupam R, Hines JV, Bergmeier SC. Structure-activity studies of oxazolidinone analogs as RNA-binding agents. Bioorg Med Chem Lett. 2006;16:3600–3604. doi: 10.1016/j.bmcl.2006.03.068. [DOI] [PubMed] [Google Scholar]
- 31.Kao C, Zheng M, Rudisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3′ terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999;5:1268–1272. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA-polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA-polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 34.Guest CR, Hochstrasser RA, Sowers LC, Millar DP. Dynamics of mismatched base-pairs in DNA. Biochemistry. 1991;30:3271–3279. doi: 10.1021/bi00227a015. [DOI] [PubMed] [Google Scholar]
- 35.Scheit KH, Rackwitz HR. Synthesis and physicochemical properties of two analogs of poly(dA): poly(2-aminopurine-9-β-D-deoxyribonucleotide) and poly 2-amino-deoxyadenylic acid. Nucleic Acids Res. 1982;10:4059–4069. doi: 10.1093/nar/10.13.4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward DC, Reich E, Stryer L. Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2, 6-diaminopurine riboside, and their derivatives. J Biol Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
- 37.Neely RK, Jones AC. Influence of base dynamics on the conformational properties of DNA: observation of static conformational states in rigid duplexes at 77 K. J Am Chem Soc. 2006;128:15952–15953. doi: 10.1021/ja064390m. [DOI] [PubMed] [Google Scholar]
- 38.Somsen OJG, Keukens LB, de Keijzer MN, van Hoek A, van Amerongen H. Structural heterogeneity in DNA: temperature dependence of 2-aminopurine fluorescence in dinucleotides. Chem Phys Chem. 2005;6:1622–1627. doi: 10.1002/cphc.200400648. [DOI] [PubMed] [Google Scholar]
- 39.Tsujikawa L, Strainic MG, Watrob H, Barkley MD, deHaseth PL. RNA polymerase alters the mobility of an A-residue crucial to polymerase-induced melting of promoter DNA. Biochemistry. 2002;41:15334–15341. doi: 10.1021/bi026539m. [DOI] [PubMed] [Google Scholar]
- 40.Gerdeman MS, Henkin TM, Hines JV. Solution structure of the Bacillus subtilis T-box antiterminator RNA: seven nucleotide bulge characterized by stacking and flexibility. J Mol Biol. 2003;326:189–201. doi: 10.1016/s0022-2836(02)01339-6. [DOI] [PubMed] [Google Scholar]
- 41.Neely RK, Magennis SW, Parsons S, Jones AC. Photophysics and X-ray structure of crystalline 2-aminopurine. Chem Phys Chem. 2007;8:1095–1102. doi: 10.1002/cphc.200600593. [DOI] [PubMed] [Google Scholar]
- 42.Rist M, Wagenknecht HA, Fiebig T. Exciton and excimer formation in DNA at room temperature. Chem Phys Chem. 2002;3:704–707. doi: 10.1002/1439-7641(20020816)3:8<704::AID-CPHC704>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 43.Su TJ, Connolly BA, Darlington C, Mallin R, Dryden DTF. Unusual 2-aminopurine fluorescence from a complex of DNA and the EcoKI methyltransferase. Nucleic Acids Res. 2004;32:2223–2230. doi: 10.1093/nar/gkh531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lenz T, Bonnist EYM, Pljevaljčić G, Neely RK, Dryden DTF, Scheidig AJ, Jones AC, Weinhold E. 2-Aminopurine flipped into the active site of the adenine-specific DNA methyltransferase M. Taql: crystal structures and time-resolved fluorescence. J Am Chem Soc. 2007;129:6240–6248. doi: 10.1021/ja069366n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.