It is now well-established and widely, if not universally, accepted that virtually all cervical cancer and its immediate precancerous lesions arise from persisting cervical infections by approximately 15 cancer-associated (high-risk or hr) human papillomavirus (hrHPV) genotypes (1,2). The most important of these HPV genotypes are HPV16 and HPV18, which account for approximately 70% of all invasive cervical cancers with minor variations in this percentage between continents (3). A new paradigm of cervical carcinogenesis replaces an older model of stepwise progression from low-grade to high-grade morphological changes and can now be summarized as four reliably measured stages: 1) HPV acquisition, 2) HPV persistence (vs. clearance), 3) progression of a persisting infection to cervical precancer (with incidental co-occurrence of both conditions), and 4) invasion (4,5).
Based on this nearly absolute etiologic link between carcinogenic HPV and cervical cancer, testing for hrHPV is now being considered as an alternative for cytology-based cervical cancer screening. However, before cost-effective implementation of population-based hrHPV testing in cervical cancer screening and prevention can be envisaged, any candidate HPV testing technologies must offer an optimal balance between clinical sensitivity and specificity for detection of cervical intraepithelial neoplasia grade 2 or 3 and treatable cancer (≥CIN 2) to minimize redundant or excessive follow-up procedures. Reliable clinical performance needs to be established before any candidate screening test is widely disseminated and adopted into clinical practice or in organized screening programmes. Data from various studies can be used to guide the translation of hrHPV testing into clinical practice by setting standards of test performance and characteristics. Based on these data, guidelines for hrHPV DNA test requirements and use in primary cervical cancer screening can be developed, as outlined below.
The key issue for hrHPV DNA testing in cervical screening is to detect hrHPV infections that are associated with or develop into ≥CIN 2 and to differentiate them from transient hrHPV infections. This implies that there should be a balance between clinical sensitivity and specificity for detection of ≥CIN 2. Currently, two tests, i.e. the U.S. Food and Drug Administration-approved Hybrid Capture 2 (hc2; Qiagen, Gaithersburg, MD, USA) and GP5+/6+-PCR enzyme immunoassay (GP5+/6+-PCR EIA) have repeatedly demonstrated clinical sensitivity of about 90–95% for the detection of ≥CIN 2 in large prospective cohorts or randomized controlled trials (6–11, 26).
We caution against misguided attempts to increase the clinical sensitivity for HPV assays, as the adverse effect of a small gain in sensitivity will be a dramatic increase in the number of false positives (i.e., hrHPV positives without ≥CIN 2) (14). Given the low prevalence ≥CIN 2 in the screened populations even small reductions in clinical specificity will have dramatic effects on the number of unneeded follow-up procedures and associated costs. Changes in analytic sensitivity to improve clinical sensitivity require formal evaluation and validation, using receiver-operator curve (ROC) or other analytic approaches that permit thoughtful consideration of the balance between true and false positives (15).
For example, in a case-control format the hrHPV GP5+/6+-PCR EIA was compared with an ultra-sensitive commercially available PCR-based broad spectrum HPV assay in women with normal cytology over 29 years of age (16). The application of the latter did not lead to an increase in clinical sensitivity for ≥CIN 2, but instead resulted in a substantial decrease in clinical specificity compared to that of the GP5+/6+-PCR. The extra positivity scored by the ultra-sensitive assay mainly involved infections characterized by a very low viral load that were not associated with ≥CIN 2. Conversely, insufficient analytic sensitivity will translate to unacceptably low clinical sensitivity. One example is a commercially available DNA in situ hybridization (ISH) assay, which showed a substantially lower sensitivity for prevalent ≥CIN 2 than hc2 (17). Since in situ hybridization positivity was only found in samples displaying relatively high viral loads, as deduced from the corresponding hc2 RLU/CO values that are semi-quantitative measures of HPV viral load (18), it can be concluded that the ISH assay used suffered from a low sensitivity to detect HPV, resulting in a clinical sensitivity for ≥CIN2 of less than 77%.
Both the hrHPV hc2 and GP5+/6+-PCR assays have shown to yield considerably better results than cytology in large screening studies (6,8–9, 11–14,20).
In particular, evidence has been collected that women testing negative for hrHPV hc2 or GP5+/6+-PCR have a 3 to 5 year risk of ≥CIN 2 that is 40–50% lower than of women testing cytologically negative (7,13,14). Consequently, hrHPV hc2 and GP5+/6+-PCR method can be considered as clinically validated for use in cervical screening. Both hc2 and GP5+/6+-PCR target 13 hrHPV types (ie. HPV16, −18, −31, −33, −35, −39, −45, −51, −52, −56, −58, −59, and −68). GP5+/6+-PCR additionally targets HPV66, which hc2 detects as the result of coincidental cross-reactivity with types genetically related to targeted HPV genotypes (21). Comparison of the automated version of the hc2 assay and GP5+/6+ PCR-EIA in a cross-sectional study of women participating in a population-based cervical screening trial, revealed that the assays had nearly similar sensitivities for high-grade CIN or cervical cancer (22). There is no consensus on the minimal number of hrHPV types that a HPV detection assay should be able to detect as there is still debate about the carcinogenicity of certain HPV types that have been rarely detected in carcinomas (1,,23,24), although it is widely accepted that the detectable types should include the 14 listed above. However, as long as the HPV detection assay complies with the criteria listed below and performs well as a screening test, it is of minor importance to what extent uncommon hrHPV types are actually targeted.
Since both median and clinically relevant viral load levels in women of a screening population differ markedly across the various hrHPV types (26) clinical test requirements cannot easily be translated into analytical test requirements in terms of setting universal assay cut-off points that can be used by candidate test manufacturers. Therefore, clinical criteria should form the basis for guidelines for hrHPV test requirements for primary cervical screening. Here, a proposal for such guidelines in the European setting is presented as well as a clinical validation strategy in order to determine whether new tests fulfill these guidelines. Since in large prospective screening trials both hc2 and GP5+/6+-PCR have been shown to be superior to cytology the hrHPV test requirements are deduced from data obtained with these assays (9,11,13,14).
The HPV test requirements are formulated relative to the performance of hc2 because, in contrast to GP5+/6+ PCR, hc2 is approved by the US Food and Drug Administration (FDA) and commercially available. In line with FDA, we only consider HPV testing in women of 30 years and older. In younger women, the specificity of the HPV test is relatively low (9,10) and the use of HPV testing is not undisputed.
In addition, indications for quality assurance (26) of hrHPV testing by the laboratories over time are proposed. We believe that these guidelines would also be useful for non-European countries seeking to adopt HPV testing as part of their screening programmes.
Requirements of HPV tests in primary cervical screening
In a primary cervical screening setting a HPV detection assay should fulfill the following requirements:
The candidate test should have a clinical sensitivity for ≥CIN2 not less than 90% of the clinical sensitivity of the hc2 in women of at least 30 years. This recommendation is based on recent meta-analyses that reported a pooled sensitivity for hc2 of 97.9% (95%CI: 95.9%–99.9%) in primary screening in Europe and North-America (6) and a pooled sensitivity of hc2 and GP5+/6+ PCR in European studies of 96.1% (95%CI: 94.2%–97.4%)(12). In addition, in recent prospective screening trials, hc2 sensitivities were 97.3% (95%CI 90.7%–99.7%) (9) and 94.6% (95%CI 84.2–100) (11) and the GP5+/6+-PCR-EIA sensitivity was 94.1% (95%CI: 91.7%–95.9%) (14). This high sensitivity translates into a very high negative predictive value (reassurance) of the HPV detection assay, allowing for extending screening intervals for test negative women, who are typically the majority of participants in a screening programme.
Acceptable standards for clinical specificity are more difficult to define because prevalences of the targeted HPV genotypes vary across populations. Notwithstanding that, we suggest a clinical specificity for ≥CIN2 of the candidate test not less than 98% of the clinical specificity of the hc2 in women of at least 30 years of age The rationale for the high lower bound on the clinical specificity is that a high clinical specificity will limit the number of test positives that would possibly trigger increased surveillance and unnecessarily stigmatized women as HPV positive. In North America and Europe, the pooled specificity of hc2 was 91.3% (95% CI: 89.5–93.1%; range: 85–95%) In European trials, the hc2 and GP5+/6+ PCR pooled clinical specificity was 93.3% (95%CI 92.9% – 93.6%) for women 35–49 years of age and 90.7% (95%CI 90.4%–91.1%) for all women (12). In recent trials, the hc2 specificities were 93.2% (95%CI 92.8–93.6) for women 35–60 years of age (19) and 94.1% (95%CI 93.4–94.8%) for women 30–69 years of age (11) and the specificity of GP5+/6+-PCR was 97.3% (95%CI 97.1%–97.5%) for women 30–60 years of age (8).
When the hc2 cut-off is increased to 2–3 RLU/CO the clinical specificity of hc2 increases and approaches that of GP5+/6+-PCR (9,20,21,27), partly due to the reduction in cross-reactivity of hc2 with low-risk HPV genotypes (21). .
To ensure a robust and highly reliable performance of the test in clinical practice the candidate test should display intra-laboratory reproducibility (ie. agreement in test result when the same specimens are tested more than once) and inter-laboratory agreement with a lower confidence bound not less than 87%. The hc2 and GP5+/6+-PCR revealed high inter-laboratory agreements of at least 92% (28–30).
Validation guidelines for candidate HPV assays
It is obvious from the requirements outlined above that validation of a candidate assay for clinical application in cervical screening requires a comparative analysis with a clinically validated reference HPV test on samples that originate from a population-based screening cohort.
The following validation strategy is advised:
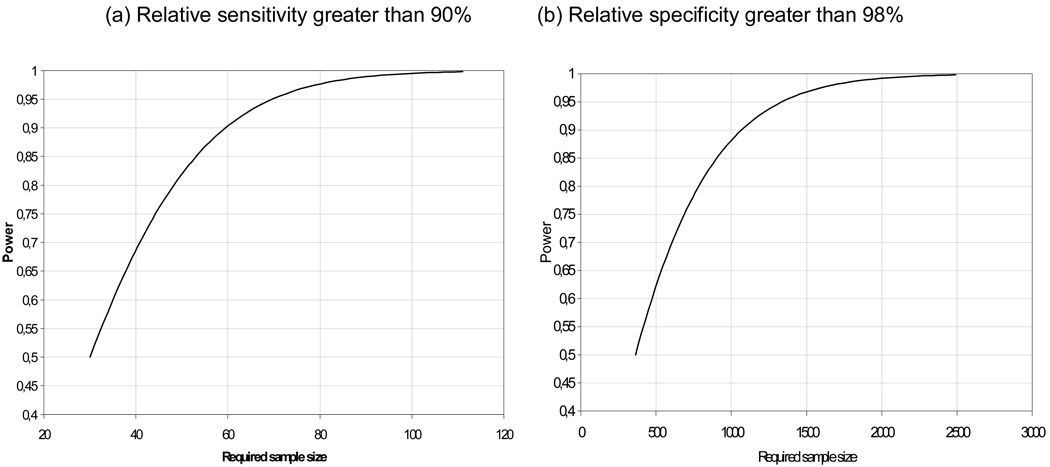
The sensitivity of the candidate test for ≥CIN2 should be at least 90% of the sensitivity of the hc2 (i.e. relative sensitivity of at least 90%) as assessed by a non-inferiority score test (31). A description of the non-inferiority test is given in the Appendix. Samples should be derived from a representative set of women in a population-based screening cohort, tested by hc2, either or not combined with cytology, and the candidate test that had a histologically confirmed ≥CIN2 detected through either of these tests. The non-inferiority test has been shown to perform well if the number of samples is 50 or more. The power of the non-inferiority test, obtained under the assumption that the candidate test and the reference test have equal sensitivity and that the agreement between the tests is moderate/good (kappa value of 0.7), is presented in Figure 1. To achieve a power of 80%, 60 samples should be tested with both the new test and hc2. The power increases with the sample size and is greater than 99% when 100 samples are tested.
The specificity of the candidate test for ≥CIN 2 should be at least 98% of the specificity of hc2. This should be determined by applying the non-inferiority test to a random sample of women of at least 30 years of age from a population-based screening cohort, tested by hc2, either or not combined with cytology, and the candidate test and that did not have histologically confirmed ≥CIN 2 To achieve a power of 80% under the assumption that the new test and the reference test have equal specificity and that the agreement between the tests is moderate/good (kappa value of 0.7), a sample size of 800 samples is required. When 2500 samples are tested, the power is greater than 99% (Figure 1).
The intra-laboratory reproducibility in time and inter-laboratory agreement should be determined by evaluation of at least 500 samples, 30% of which tested positive in a reference laboratory using a clinically validated assay. This should result in a percentage of agreement with a lower confidence bound not less than 87% (kappa value of at least 0.5 in this series of samples including 30% positives). The same intra-laboratory reproducibility should be reached after testing the same set of samples several weeks later.
Figure 1.
Power of the non-inferiority score test for assessing a relative sensitivity greater than 0.9 (a) or a relative specificity greater than 0.98. It is assumed that the new test and the reference test have equal sensitivities (specificities) and moderate/good agreement (kappa=0.7).
All the above mentioned samples can be obtained either through new studies where women are tested by hc2, either or not combined with cytology, or by exploiting well preserved archived material from previously conducted studies with the described features as long as this material is qualitatively adequate for applying the candidate test (26).
Example: validation strategy applied to GP5+/6+-PCR EIA assay indicating non-inferiority of sensitivity and specificity
As indicated above, the GP5+/6+-PCR EIA method can be considered clinically validated on the basis of data collected in large prospective screening trials As an example of applying the validation strategy we here illustrate the non-inferiority of the GP5+/6+-PCR EIA as compared to the hc2 reference test. To assess non-inferiority of the sensitivity (ie. relative sensitivity not lower than 90%), 75 cervical scrapes of women with ≥CIN2 were tested. The cervical samples were obtained from a population-based screening study where women were referred for colposcopy-guided biopsy on the basis of positive hc2 and/or cytology result (VUSA-SCREEN study, the Netherlands). The data are presented in Table 1a. The null hypothesis of inferiority is rejected (T = 2.68, p-value 0.0037) and hence the sensitivity of the GP5+/6+-PCR-EIA is not inferior to the sensitivity of the hc2. To assess non-inferiority of the specificity of the GP5+/6+-PCR EIA (i.e., relative specificity not lower than 98%), 8040 samples of women without ≥CIN2 were tested (Table 1b). The samples were obtained from a population-based screening study where all women aged >35 years were tested for hc2 and GP5+/6+-PCR EIA (POBASCAM II study). The null hypothesis is rejected (T=16.57, p-value <0.00001) and hence the specificity of the GP5+/6+-PCR-EIA (i.e. 96.0%) is judged not inferior to the specificity of the hc2 (i.e. 94.1%; Table 1b).
Table 1.
Comparison hc2 and GP5+/6+-PCR Test results
| (a): VUSA-SCREEN trial: women with ≥CIN2 | |||
|---|---|---|---|
| hc2 + | hc2 − | Total | |
| GP5+/6+-PCR EIA + | 73 | 1 | 74 |
| GP5+/6+-PCR EIA − | 1 | 0 | 1 |
| Total | 74 | 1 | 75 |
| (b): POBASCAM II trial: women without ≥CIN2 | |||
|---|---|---|---|
| hc2 + | hc2 − | Total | |
| GP5+/6+-PCR EIA + | 284 | 34 | 318 |
| GP5+/6+-PCR EIA − | 193 | 7529 | 7722 |
| Total | 477 | 7563 | 8040 |
Laboratory guidelines for HPV testing
Laboratories performing HPV testing for clinical and screening purposes should comply with quality assurance (QA), including internal quality control (IQC), external quality assessment (EQA) and quality improvement (QI). To realize QA, at least the following items should be fulfilled:
The laboratory should have a specific infrastructure in case nucleic acid amplification technology is used. This includes separate laboratories for preparation of test reagents, sample identification/preparation and DNA extraction, and DNA amplification and detection.
The laboratory should have accreditation for clinical molecular testing and should comply with standard operation procedures (SOP) and good laboratory practice (GLP) guidelines.
The HPV test performance and sample processing of the laboratory should be monitored by proficiency testing including regular intra-laboratory evaluation (32,33). Inter-laboratory performance might be evaluated by sending proficiency panels to the laboratories.
In practise large volume labs have been associated with higher proficiency in molecular testing than small volume labs. However, the laboratory requirements set forward here are formulated independent of the size of the laboratory and can in principle also be fulfilled by laboratories that process a small number of smears
In conclusion, within a cervical cancer screening setting hrHPV tests should exhibit specific requirements to assure high clinical sensitivity for detection of cervical precancer and cancer and at the same time high clinical specificity to limit unnecessary procedures and follow-up of HPV test positive women. Most of the candidate hrHPV assays mainly differ in clinical specificity. These differences in clinical specificity can be largely attributed to differences in the detection rate of transient HPV infections characterized by low viral loads. Such infections do not cause malignancies, are therefore clinically irrelevant, and potentially harmful in a screening setting because they trigger unnecessary follow-up of HPV positive women and redundant anxiety and costs coupled thereto. At present, hc2 and GP+/6+-PCR-EIA fulfill the listed requirements; it is our hope that other HPV tests with proven, reliable clinical performance are forthcoming.
While we focused on ≥CIN2 as an endpoint, we note that there is increasing recognition that histologic CIN2 is an equivocal diagnosis of cervical precancer, representing a mixture of CIN3 and low-grade lesions resulting from productive human papillomavirus (HPV) infections by both low-risk and high-risk HPV genotypes (4,5) Consequently, European guidelines have consistently promoted the separate reporting of CIN2 and CIN3 (26, 34) and also the WHO classifies these two histologic entities separately, supporting this distinction. There is universal agreement that CIN3 is the best surrogate marker for risk of progression to invasive cancer (4).
It must be acknowledged that in the future, as molecular screening tests become more accurate for true precancerous lesions, some CIN2 will test negative because in fact some are not truly precancerous lesions. This will create quandary of whether some test results for CIN2 are false or true negative. In evaluating the performance of the next generation of tests, it will be important to have an a priori plan to adjudicate cases of test-negative CIN2 by pathology review, adjunctive molecular markers, and/or tissue-based testing for HPV genotype presence and E6/E7 oncoprotein expression.
It can be envisioned that future implementation of hrHPV testing in primary cervical cancer screening is accelerated once this proposal for guidelines for HPV tests has received international consensus. These guidelines should prove useful for national and supra-national regulatory bodies in the approval of new HPV tests for public health and clinical use in cervical cancer screening. This paper has been written with the aim to achieve such international consensus by defining criteria that should be fulfilled by a new test before it can be used in primary cervical screening.
Appendix
Non-inferiority test
Suppose n samples have been tested with the new test and the hc2 reference test. The results are presented in Table 2.
Table 2.
Test results
| hc2 + | hc2 − | Total | |
|---|---|---|---|
| New test + | a | b | a+b |
| New test − | c | d | c+d |
| Total | a+c | b+d | n |
Under the null hypothesis, the relative sensitivity (when comparing the new test to hc2) is δ0 and under the alternative hypothesis, the relative sensitivity is greater than δ0. According to the present guidelines ∂0 should be set to 0.90 for sensitivity and to 0.98 for specificity. The test statistic is defined as
where
with A = n (1 + δ0), B = (a + c)δ02 −(a + b + 2c), and C = c (1−δ0)(a + b + c)/n. The null hypothesis is rejected at nominal significance level α if T is equal to or greater than the 100 × (1-α) percentile point of the standard normal distribution (T is interpreted as a z statistic).
Reference List
- 1.Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ International Agency for Research on Cancer Multicenter Cervical Cancer Study Groupl. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl.J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 2.IARC. Human Papillomaviruses. vol. 90. Lyon: International Agency for Research on Cancer; IARC Monographs on the evaluation of carcinogenic risks to humans. In press. [Google Scholar]
- 3.Smith JS, Lindsay L, Hoots B, Keys J, Franceschi S, Winer R, Clifford GM. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: a meta-analysis update. Int J Cancer. 2007;121:621–632. doi: 10.1002/ijc.22527. [DOI] [PubMed] [Google Scholar]
- 4.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 5.Snijders PJ, Steenbergen RD, Heideman DA, Meijer CJ. HPV-mediated cervical carcinogenesis: concepts and clinical implications. J Pathol. 2006;208:152–164. doi: 10.1002/path.1866. [DOI] [PubMed] [Google Scholar]
- 6.Arbyn M, Sasieni P, Meijer CJ, Clavel C, Koliopoulos G, Dillner J. Chapter 9: Clinical applications of HPV testing: A summary of meta-analyses. Vaccine. 2006;24 Suppl 3:S78–S89. doi: 10.1016/j.vaccine.2006.05.117. [DOI] [PubMed] [Google Scholar]
- 7.Sherman ME, Lorincz AT, Scott DR, Wacholder S, Castle PE, Glass AG, Mielzynska- Lohnas I, Rush BB, Schiffman M. Baseline cytology, human papillomavirus testing, and risk for cervical neoplasia: a 10-year cohort analysis. J Natl Cancer Inst. 2003;95:46–52. doi: 10.1093/jnci/95.1.46. [DOI] [PubMed] [Google Scholar]
- 8.Bulk S, Bulkmans NW, Berkhof J, Rozendaal L, AJ PB, Verheijen RH, Snijders PJ, Meijer CJ. Risk of high-grade cervical intra-epithelial neoplasia based on cytology and high-risk HPV testing at baseline and at 6-months. Int.J.Cancer. 2007;121:361–367. doi: 10.1002/ijc.22677. [DOI] [PubMed] [Google Scholar]
- 9.Ronco G, Segnan N, Giorgi-Rossi P, Zappa M, Casadei GP, Carozzi F, Dalla Palma P, Del Mistro A, Folicaldi S, Gillio-Tos A, Nardo G, Naldoni C, et al. New Technologies for Cervical Cancer Working Group. Human Papillomavirus testing and liquid-based cytology: results at recruitment from the New Technologies for Cervical Cancer randomized controlled trial. J Natl. Cancer Inst. 2006;98:765–774. doi: 10.1093/jnci/djj209. [DOI] [PubMed] [Google Scholar]
- 10.Ronco G, Giorgi-Rossi P, Carozzi F, Dalla Palma P, Del Mistro A, De Marco L, de Lillo M, Naldoni C, Pierotti P, Rizzolo R, Segnan N, Schincaglia, et al. New Technologies for Cervical Cancer Working Group. Human papillomavirus testing and liquid-based cytology in primary screening of women younger than 35 years: results at recruitment for a randomised controlled trial. Lancet Oncol. 2006;7:547–555. doi: 10.1016/S1470-2045(06)70731-8. [DOI] [PubMed] [Google Scholar]
- 11.Mayrand MH, Duarte-Franco E, Rodrigues I, Walter SD, Hanley J, Ferenczy A, Ratnam S, Coutlée F, Franco EL Canadian Cervical cancer Screening Trial Study Group. Human papillomavirus DNA versus Papanicolaou screening tests for cervical cancer. N Engl J Med. 2007;357:1579–1588. doi: 10.1056/NEJMoa071430. [DOI] [PubMed] [Google Scholar]
- 12.Cuzick J, Clavel C, Petry KU, Meijer CJ, Hoyer H, Ratnam S, Szarewski A, Birembaut P, Kulasingam S, Sasieni P, Iftner T. Overview of the European and North American studies on HPV testing in primary cervical cancer screening. Int J Cancer. 2006;119:1095–1101. doi: 10.1002/ijc.21955. [DOI] [PubMed] [Google Scholar]
- 13.Naucler P, Ryd W, Tornberg S, Strand A, Wadell G, Elfgren K, Radberg T, Strander B, Forslund O, Hanson BG, Rylander E, Dillner J. Human papillomavirus and Papanicolaou tests to screen for cervical cancer. N Engl J Med. 2007;357:1589–1597. doi: 10.1056/NEJMoa073204. [DOI] [PubMed] [Google Scholar]
- 14.Bulkmans N, Berkhof J, Rozendaal L, van Kemenade F, Boeke A, Bulk S, Voorhorst FJ, Verheijen RH, van Groningen K, Boon ME, Ruitinga W, van Ballegooijen M, et al. Human papillomavirus DNA testing for the detection of cervical intraepithelial neoplasia grade 3 and cancer: 5-year follow-up of a randomised controlled implementation trial. Lancet. 2007;370:1764–1772. doi: 10.1016/S0140-6736(07)61450-0. [DOI] [PubMed] [Google Scholar]
- 15.Schiffman M, Khan MJ, Solomon D, Herrero R, Wacholder S, Hildesheim A, Rodriguez AC, Bratti MC, Wheeler CM, Burk RD PEG Group; ALTS Group. A study of the impact of adding HPV types to cervical cancer screening and triage tests. J Natl Cancer Inst. 2005;97:147–150. doi: 10.1093/jnci/dji014. [DOI] [PubMed] [Google Scholar]
- 16.Stoler MH, Castle PE, Solomon D, Schiffman M. American Society for Colposcopy and Cervical Pathology. The expanded use of HPV testing in gynecologic practice per ASCCP-guided management requires the use of well-validated assays. Am J Clin Pathol. 2007;127:335–337. doi: 10.1309/RNF3C01JKADQCLKP. [DOI] [PubMed] [Google Scholar]
- 17.Hesselink AT, van Ham MAPC, Heideman DAM, Groothuismink ZMA, Rozendaal L, Berkhof J, Massuger LAFG, Melchers WJG, Meijer CJLM, Snijders PJF. Comparison of the clinical performance of GP5+/6+-PCR-EIA and SPF10-LiPa to detect high-grade cervical lesions in women with normal cytology. 2008 doi: 10.1128/JCM.00476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hesselink AT, van den Brule AJC, Brink AATP, Berkhof J, van Kemenade FJ, Verheijen RHM, Snijders PJF. Comparison of hybrid capture 2 with in situ hybridization for the detection of high-risk human papillomavirus in liquid-based cervical samples. Cancer Cytopathology. 2004;102:11–18. doi: 10.1002/cncr.11904. [DOI] [PubMed] [Google Scholar]
- 19.Gravitt PE, Burk RD, Lorincz A, Herrero R, Hildesheim A, Sherman ME, Bratti MC, Rodriguez AC, Helzlsouer KJ, Schiffman M. A comparison between real-time polymerase chain reaction and hybrid capture 2 for human papillomavirus DNA quantitation. Cancer Epidemiol Biomarkers Prev. 2003;12:477–484. [PubMed] [Google Scholar]
- 20.Ronco G, Giorgi-Rossi P, Carozzi F, Confortini M, Dalla Palma P, Del Mistro A, Gillio-Tos A, Minucci D, Naldoni C, Rizzolo R, Schincaglia P, Volante R, Zappa M, Zorzi M, Cuzick J, Segnan N New Technologies for Cervical Cancer Screening Working Group. Results at recruitment from a randomized controlled trial comparing human papillomavirus testing alone with conventional cytology as the primary cervical cancer screening test. J Natl Cancer Inst. 2008;100:492–501. doi: 10.1093/jnci/djn065. [DOI] [PubMed] [Google Scholar]
- 21.Castle PE, Schiffman M, Burk RD, Wacholder S, Hildesheim A, Herrero R, Bratti MC, Sherman ME, Lorincz A. Restricted cross-reactivity of hybrid capture 2 with nononcogenic human papillomavirus types. Cancer Epidemiol Biomarkers Prev. 2002;11:1394–1399. [PubMed] [Google Scholar]
- 22.Hesselink AT, Bulkmans NW, Berkhof J, Lorincz AT, Meijer CJ, Snijders PJ. Cross-sectional comparison of an automated hybrid capture 2 assay and the consensus GP5+/6+PCR method in a population-based cervical screening program. J Clin Microbiol. 2006;44:3680–3685. doi: 10.1128/JCM.02078-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cogliano V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F WHO International Agency for Research on Cancer Monograpgh Working Group. Carcinogenicity of human papillomaviruses. Lancet Oncol. 2005;6:204. doi: 10.1016/s1470-2045(05)70086-3. [DOI] [PubMed] [Google Scholar]
- 24.Muñoz N, Bosch FX, Castellsagué X, Díaz M, de Sanjose S, Hammouda D, Shah KV, Meijer CJ. Against which human papillomavirus types shall we vaccinate and screen? The international perspective. Int J Cancer. 2004;111:278–285. doi: 10.1002/ijc.20244. [DOI] [PubMed] [Google Scholar]
- 25.Snijders PJF, Hogewoning CJA, Hesselink AT, Berkhof J, Voorhorst FJ, Bleeker MCG, Meijer CJLM. Determination of viral load thresholds in cervical scrapings to rule out CIN 3 in HPV 16, 18, 31 and 33-positive women with normal cytology. Int J Cancer. 2006;119:1102–1107. doi: 10.1002/ijc.21956. [DOI] [PubMed] [Google Scholar]
- 26.Arbyn M, Anttila A, Jordan J, Ronco G, Schenck U, Segnan N, Wiener H, Daniel J, von Karsa L, editors. European Guidelines for Quality Assurance in Cervical Cancer Screening. 2nd ed. Luxembourg: Office for Official Publications of the European Communities; 2008. pp. 1–291. [Google Scholar]
- 27.Cuzick J, Szarewski A, Cubie H, Hulman G, Kitchener H, Luesley D, McGoogan E, Menon U, Terry G, Edwards R, Brooks C, Desai M, et al. Management of women who test positive for high-risk types of human papillomavirus: the HART study. Lancet. 2003;362:1871–1876. doi: 10.1016/S0140-6736(03)14955-0. [DOI] [PubMed] [Google Scholar]
- 28.Castle PE, Wheeler CM, Solomon D, Schiffman M, Peyton CL ALTS Group. Interlaboratory reliability of Hybrid Capture 2. Am J Clin Pathol. 2004;122:238–245. doi: 10.1309/BA43-HMCA-J26V-WQH3. [DOI] [PubMed] [Google Scholar]
- 29.Carozzi FM, Del Mistro A, Confortini M, Sani C, Puliti D, Trevisan R, De Marco L, Tos AG, Girlando S, Palma PD, Pellegrini A, Schiboni ML, et al. Reproducibility of HPV DNA Testing by Hybrid Capture 2 in a Screening Setting. Am J Clin Pathol. 2005;124:716–721. doi: 10.1309/84E5-WHJQ-HK83-BGQD. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs MV, Snijders PJ, Voorhorst FJ, Dillner J, Forslund O, Johansson B, von Knebel Doberitz M, Meijer CJ, Meyer T, Nindl I, Pfister H, Stockfleth E, et al. Reliable high risk HPV DNA testing by polymerase chain reaction: an intermethod and intramethod comparison. J Clin Pathol. 1999;52:498–503. doi: 10.1136/jcp.52.7.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang NS, Tang ML, Chan IS. On tests of equivalence via non-unity relative risk for matched-pair design. Stat Med. 2003;22:1217–1233. doi: 10.1002/sim.1213. [DOI] [PubMed] [Google Scholar]
- 32.Quint WG, Pagliusi SR, Lelie N, De Villiers E-M, Wheeler CM WHO Human Papillomavirus DNA International Collaborative Study Group. Results of the first world health organization international collaborative study of detection of human papillomavirus DNA. J Clin Microbiol. 2006;44:571–579. doi: 10.1128/JCM.44.2.571-579.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dillner L, Dillner J. International quality assurance of human papillomavirus testing. Cent Eur J Public Health. 2008;16:S18–S20. [Google Scholar]
- 34.Herbert A, Arbyn M, Bergeron C. Why CIN3 and CIN2 should be distinguished on histological reports. Cytopathology. 2008;19:63–64. doi: 10.1111/j.1365-2303.2006.00539.x. [DOI] [PubMed] [Google Scholar]