Abstract
Evolution of facial morphology arises from variation in the activity of developmental regulatory networks that guide the formation of specific craniofacial elements. Importantly, the acquisition of novel morphology must be integrated with a phylogenetically inherited developmental program. We have identified a unique region of the secondary palate associated with the periodic formation of rugae during the rostral outgrowth of the face. Rugae function as SHH signaling centers to pattern the elongating palatal shelves. We have found that a network of signaling genes and transcription factors is spatially organized relative to palatal rugae. Additionally, the first formed ruga is strategically positioned at the presumptive junction of the future hard and soft palate that defines anterior-posterior differences in regional growth, mesenchymal gene expression and cell fate. We propose a molecular circuit integrating FGF and BMP signaling to control proliferation and differentiation during the sequential formation of rugae and inter-rugae domains in the palatal epithelium. The loss of p63 and Sostdc1 expression and failed rugae differentiation highlight that coordinated epithelial mesenchymal signaling is lost in the Fgf10 mutant palate. Our results establish a genetic program that reiteratively organizes signaling domains to coordinate the growth of the secondary palate with the elongating midfacial complex.
Keywords: craniofacial evolution, palate development, periodic patterning, developmental module, signaling network, FGF, BMP, SHH, p63
Introduction
Craniofacial development requires the outgrowth and precisely choreographed movements of multiple facial primordia. Bilateral maxillary prominences fuse along the midline with the frontonasal process to frame the upper jaw and face. Outgrowth and patterning of the facial prominences depends on the immigration of cranial neural crest (CNC) cells that delaminate from the neural folds at the time of neural tube closure. Fate mapping and heterospecific transplantation studies show that distinct populations of CNC cells, defined by rostro-caudal level of origin and path of migration, are prepatterned as to the skeletal elements into which they will ultimately differentiate (Lee et al., 2004; Noden, 1983; Santagati and Rijli, 2003; Schneider and Helms, 2003). Conversely, evidence from genetic studies indicate a critical role for epithelial signals and support that refinement of facial form is derived from local tissue interactions between CNC and surface epithelia (Haworth et al., 2004; Haworth et al., 2007; Shigetani et al., 2000; Tyler and Koch, 1977; Yamagishi et al., 2006). These local interactions regulate cellular behaviors such as proliferation, migration, apoptosis, and differentiation within individual facial primordia and are mediated by developmental signaling pathways including the Bone morphogenic protein (BMP), Sonic hedgehog (SHH), Fibroblast growth factor (FGF), Retinoic acid, and WNT pathways (Ahlgren and Bronner-Fraser, 1999; He et al., 2008; Hu and Helms, 1999; Jeong et al., 2004; Liu et al., 2005; Schneider et al., 2001; Szabo-Rogers et al., 2008).
While critical roles have been demonstrated for individual transcription factors and signaling molecules, developmental control of craniofacial morphogenesis is achieved through the integration of molecular activity within transiently organized signaling centers. Mutations in genes which result in altered craniofacial development are often associated with syndromes that affect the formation of other anatomical structures such as the limb, pointing to a conservation of underlying regulatory interactions and morphogenetic processes that define the activity of signaling centers (Barrow et al., 2002; Ibrahimi et al., 2001; Schneider et al., 1999; Stanier and Moore, 2004). Studies support that species-specific craniofacial morphology is generated by spatiotemporal differences in the activity of discrete signaling centers within individual facial primordia (Abzhanov et al., 2004; Wu et al., 2006b). For example, reciprocal FGF/SHH/BMP interactions establish the frontonasal ectodermal zone (FEZ), a signaling center positioned at the distal tip of the frontonasal mass that guides outgrowth of the midfacial complex (Abzhanov et al., 2007; Hu et al., 2003; Marcucio et al., 2005). The FEZ is organized relative to the adjacent expression domains of Shh and Fgf8 and the mutual antagonism between these two pathways (Abzhanov et al., 2007; Hu and Marcucio, 2009a). Signals from the FEZ also regulate Bmp4 expression within the adjacent mesenchyme, thereby directing CNC proliferation that sculpts the final size and shape of the upper jaw (Hu and Marcucio, 2009a; Hu and Marcucio, 2009b; Hu et al., 2003). Importantly, differences in the spatial organization of the FEZ and its associated signaling correlate with species-specific patterns of growth and final shape of the midfacial complex (Abzhanov et al., 2004; Hu and Marcucio, 2009b; Wu et al., 2006a; Wu et al., 2006b).
Many vertebrates, notably mammals, generate a derivative of the maxillary prominences, the secondary palate that separates the oral and nasal cavities. Palate development involves a series of rotation and elevation movements that accompany the outgrowth of the bilateral palatal shelves that meet and fuse along the midline to form the roof of the oral cavity. The molecular and genetic basis of medial outgrowth and fusion of the palatal shelves has been studied extensively (Gritli-Linde, 2007; Lan et al., 2004; Stanier and Moore, 2004). However, less is known about growth control and patterning during the anterior extension of the palate. The anterior and posterior palate have been shown to exhibit differential competence to respond to signaling input, the anterior palate is BMP responsive whereas the posterior palate is considered to be permissive to FGF signaling (Shigetani et al., 2000; Zhang et al., 2002). Gene expression patterns also highlight molecular differences between the anterior and posterior palate (He et al., 2008; Hilliard et al., 2005; Li and Ding, 2007). In the mesenchyme, signaling and transcription factors including Bmp4, Fgf10, Msx1 and Shox2 are expressed in anterior domains while Barx1, Tbx22, and Mn1 expression is restricted to posterior domains (Liu et al., 2008; Welsh et al., 2007; Yu et al., 2005). These regional differences in signaling and gene expression are ultimately translated into differences in cell fate, whereby the anterior mesenchyme forms the bony hard palate while posterior mesenchyme contributes to the muscular soft palate.
Reciprocal signaling between mesenchyme and the overlying epithelium also plays a critical role in growth of the secondary palate (Gritli-Linde, 2007; Rice et al., 2004; Tyler and Koch, 1977). Both Fgf10 and Bmp4 are required for the epithelial expression of Shh, which is in turn involved in a cascade directing proliferation of the underlying mesenchyme (Rice et al., 2004; Zhang et al., 2002). Shh expression is restricted to the palatal rugae, epithelial thickenings that form the transverse ridges on the roof of the oral cavity (Bitgood and McMahon, 1995). Finally, highlighting the role of epithelial-mesenchymal feedback in integrated signaling and tissue patterning, recent work by Lan and Jiang demonstrates that epithelial SHH signaling acts to restrict Bmp4 expression but is required to maintain Fgf10 expression in the mesenchyme (Lan and Jiang, 2009).
Importantly, the elongation of the secondary palate must coincide with the rostral extension of the midfacial complex. However, a defined domain of molecular interactions that pattern the outgrowth of the secondary palate, comparable to the FEZ, has not been appreciated. Our previous finding that the anterior growth of the palate involves periodic addition of Shh expressing rugae suggests that outgrowth is guided by a dynamic sequence of interactions between a regionalized mesenchyme and a highly patterned surface epithelium (Welsh et al., 2007). In this study we define a region of critical morphogenetic activity positioned at the junction of the future anterior-posterior (A-P) palate that serves to integrate FGF/SHH/BMP signaling to direct the differentiation of rugae signaling centers during the outgrowth of the anterior palate. The periodic formation of rugae provides a novel reference frame for studies of the spatial and temporal organization of a network of signaling genes and transcription factors that direct patterning and growth of the secondary palate.
Materials and methods
Mice
Fgf10 +/− mice, originally described by Sekine (Sekine et al., 1999), were provided by Dr. J. Greer (University of Alberta, Canada) and maintained on a C57Bl/6J background. Genomic DNA from tail biopsies was isolated for genotyping mice with the following primers: wildtype fwd., (5’-CTTCCAGTATGTTCCTTCTGATGAGAC-3’); wildtype rev., (5’-GTACGGACAGTCTTCTTCTTGGTCCC-3’); mutant fwd., (5’-ACGACGGGCGTTCCTTGCGCAGCTGTG-3’); mutant rev., (5’-TCAGAAGAACCGTCAAGAAGGCGATA-3’). Occasional ectopic fusions of the palate to the tongue or mandible in Fgf10 mutants were noted at the time of dissection and only samples without fusions were selected for expression analysis. The Sostdc1shk/J (Sharkey) mouse is a spontaneous mutation resulting from a single base pair deletion in exon 2 of Sostdc1 which introduces a premature stop codon. Sharkey mice were acquired from the craniofacial mutant resource at the Jackson Laboratory (Bar Harbor, Maine) and maintained as a homozygous population.
RNA isolation and Quantitative RT-PCR
Individual palatal shelves were dissected free of surrounding tissue including the molar tooth bud, snap frozen in RNAlater (Invitrogen), and stored at −80°C until processing. RNA from each palatal shelf was purified using the RNeasy Micro kit (Qiagen) and RNA yield and quality were assessed via Nanodrop UV spectrophotometry. Individual cDNAs were generated from each sample via reverse transcription of 1.0 µg of total RNA using the ABI high capacity archive kit. Quantitative real-time PCR on an ABI 7500 platform was carried out on triplicate reactions of each sample using 5.0 ng of cDNA template. β-Actin expression was used as an endogenous control and cDNA derived from a pool of 1.0 µg each of E8.5-E18.5 whole embryo RNA was used to calibrate relative expression levels in both wildtype (n=3) and mutant (n=4). The following ABI TaqMan probes were used in this analysis: Bmp4, Mm_00432087_m1; Etv5, Mm_00465816_m1; Fgf9, 00442759_m1; Jag2, Mm_00439935_m1; ΔNp63, Mm_01169470_m1; Tap63, Mm_01150797_m1; Shh, Mm_00436527_m1; Sostdc1, Mm_00840254_m1.
In situ hybridization
Whole mount and section in situ hybridization was performed with digoxygenin (DIG) or dinitrophenol (DNP) labeled antisense riboprobes. Processing of whole mount samples was carried out on an Intavis InsituPro VS robotic platform. Samples for section in situ were fresh frozen in OCT compound and sectioned at 20µm thickness. For experiments involving mutants, wildtype and mutants were processed together and detected for an identical period. Gene expression was analyzed in 3 wildtype and 4 mutant samples. Detailed protocols and a complete list of sequences cloned to generate in situ probes are available upon request.
BrdU labeling
Embryos were labeled with BrdU (Roche) by injecting pregnant females with 10mM BrdU (20µl/gram of body weight) 1 hour prior to sacrifice. Heads were embedded in paraffin and serial 6µm sagittal sections collected. DNA was denatured with 2N HCl for one hour prior to incubation with anti-BrdU (Roche) followed by Cy3 (Molecular Probes) and counterstaining with DAPI. 3 sections through the region of the RGZ of E13.5 embryos (n=4) were used to count the number of BrdU positive cells and the total number of cells to determine the mitotic index for each domain. The RGZ was divided into two equal domains defined by drawing a vertical line midway between the anterior extent of R1 and the posterior boundary of the first inter-rugae domain.
Results
Periodic rugae formation during outgrowth of the anterior palate
A significant number of genes are known to be required for palate development. However, the lack of orienting landmarks has made interpreting the integrated expression patterns of these genes difficult. In this study we have used rugae formation as a reference to examine the spatiotemporal dynamics of gene expression in the secondary palate. Rugae, which are formed in a defined sequence during the anterior extension of the palate, act as SHH signaling centers involved in epithelial-mesenchymal interactions required to coordinate palate outgrowth and patterning (Lan and Jiang, 2009; Pantalacci et al., 2008; Welsh et al., 2007). A first stripe of Shh rugae expression (R1) is evident at E12.0 and by E15.5, as the palatal shelves contact and fuse, the full complement of 8 rugae have formed (Fig. 1). In order to better appreciate the temporal dynamics of rugae formation in relation to palate development, we used in situ hybridization to generate a reconstructed time series. The number of rugae present and the extent of nascent Shh expression provide a proxy for ordering the developmental progression of a collection of palatal shelves. From 134 samples, dissected between E12.0 and E15.5 of development and hybridized for Shh expression, we ordered a series of 45 palates that capture the sequential formation of R1–R8. This reconstructed time series represents the full sequence of rugae formation during the anterior extension, medially directed outgrowth, and midline fusion of the palatal shelves (Fig. 1B and C).
Figure 1.
Periodic rugae formation and rostral growth of the palate. (A) Shh expression marking rugae at E15.5 (oral view of palates, anterior is towards top). Rugae (R1–R8) are labeled with respect to the order of their formation (shown in B). Shh expression posterior to the first formed rugae gives rise to a lateral line of taste buds termed the geschmacksstreifen and punctate Shh domains in developing sensory papilla overlaying the posterior soft palate. (B) A reconstructed time series showing the formation of R1–R8 highlights regional differences in the growth of the anterior and posterior palate. Around E12.0, the R1 ruga arises as a distinct band of Shh expression immediately posterior to the forming molar tooth bud signaling center. During rostral extension of the palate, the spatial relationship between R1 and the developing molar tooth bud remains unchanged (asterisks in left most panel of each row). Following the formation of R1, the remaining rugae (R2–R8) are generated anterior to R1. R3 and R4 form in quick succession anterior and posterior to R2 respectively. The domain of R4–R8 formation can be referenced to three distinct landmarks: anterior to R1; posterior to the most recently formed ruga; and medial to the molar tooth bud. (C) Variation in the morphology of R7 and R8 appears to be associated with the extent to which R6 extends posteriorly. gs, geschmacksstreifen; mtb, molar tooth bud; pd, posterior domain of Shh expression; sp, sensory papilla.
This data set demonstrates that at approximately E12.0, the first ruga (R1) arises as a distinct band of Shh expression that separates posteriorly from the forming molar tooth bud (mtb) signaling center. This molecular data supports earlier observations of Peterkova based on detailed histological analysis that suggest the rugae and teeth share a common developmental origin in the odontogenic epithelium (Peterkova, 1985). Following the formation of R1, the remaining rugae (R2–R8) are generated anterior to R1. Furthermore, with the exception of R3, new rugae formation occurs via an interposition process (Fig. 1B). R3 actually forms anterior to R2 but is closely followed by the formation of R4 between R1 and R2. All subsequent rugae form between R1 and the most recently formed ruga (R1+n). Previously, rugae have been numbered relative to their position along the A-P axis of the palate, with the anterior most ruga labeled as R1 (Peterkova, 1985). However, the labeling of rugae, numbered in the order in which they are formed would result in the sequence: anterior<R3, R2, R4, R5, R6, R7, R8, R1>posterior.
The domain of periodic rugae formation can be referenced to three distinct landmarks: anterior to R1; medial to the position of the mtb; and posterior to R1+n (Fig. 1B). Following formation at this site, each new ruga is displaced with the anterior elongation of the palate. Thus, periodic rugae formation is localized to a specific domain and is intimately associated with the rostral outgrowth of the anterior palate. We refer to this distinct region of the palate as the rugae growth zone (RGZ).
Shh expression in R1 exhibits a characteristic morphology and dynamic that is distinct from the remaining rugae. R1 initiates as a posteriorly angled band of expression (during R2–R3 formation) that then becomes broader and chevron shaped (during R4–R5 formation) prior to flattening along its A-P axis (during R6–R8 formation). This flattening of R1is accompanied by the posterior regression of the site of nascent rugae formation with respect to the mtb such that R4 forms at the anterior limit of the mtb, while R7 and R8 form at the mid-posterior level of the mtb. The increasingly posterior formation of R7 and R8 is accompanied by the elongation of the Shh expression domain in the mtb as well as the flattening of R1. Variability in the morphology of R7 and R8 has been noted previously. It is interesting to speculate that the position of nascent rugae induction and the variability in R7/R8 morphology may be related to the dynamics and interplay between three signaling domains, R1, the mtb, and R1+n. This hypothesis is supported by the observation that the formation of a posteriorly angled R6 is associated with a laterally shortened R7 ruga (Fig. 1C).
A network of genes is organized about rugae signaling centers
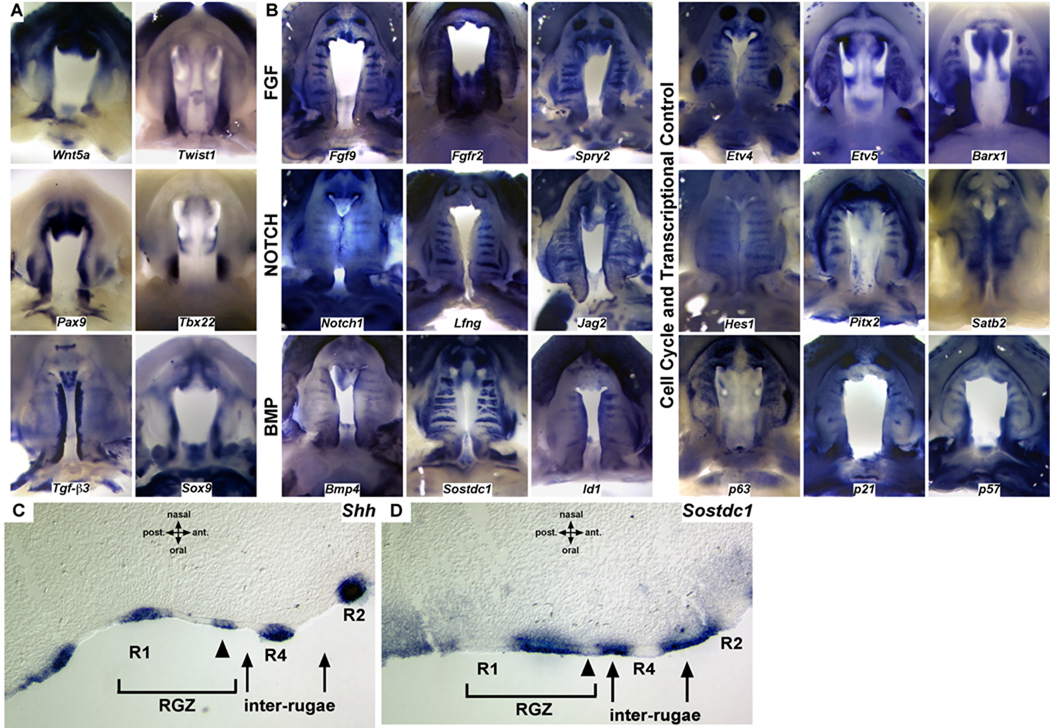
The dynamics of Shh expression demonstrate that periodic patterning generates a series of signaling domains in the epithelium of the elongating palatal shelves. To further investigate patterning in relation to rugae formation and the anterior extension of the palatal shelves, we surveyed the expression of a number of key signaling molecules and transcription factors. Regionally restricted expression of genes such as Wnt5a, Twist1, Pax9, Tbx22, Tgf-β3, and Sox9 along the anterior-posterior and medial-lateral axis, highlight the spatial heterogeneity of gene expression in the developing palate (Fig. 2A). Significantly, we also found that the expression of a number of transcription factors and members of several developmental signaling pathways (FGF, BMP, SHH, and NOTCH) is segmentally organized with respect to rugae and inter-rugae domains in the developing palate (Fig. 2B–D). Restricted expression within rugae has been reported for components of the SHH pathway as well as Barx1, Etv5, and Bmp4 (Rice et al., 2006; Welsh et al., 2007; Lee et al., 2007). However, the segmental expression of genes critical for orofacial development such as Jag2, Fgfr2, and Pitx2 has not been reported (Casey et al., 2006; Liu et al., 2003; Rice et al., 2004). Our survey demonstrates that several components of the FGF, BMP, and NOTCH signaling pathways exhibit expression that is restricted to either rugae (Fgf9, Spry2, Etv4, Notch1, Lfng, Hes1) or inter-rugae domains (Fgfr2, Etv5, Sostdc1, Id1) in the palatal epithelium or mesenchyme (Fig. 2B). The complementary organization of gene expression domains for components of these signaling pathways and transcription factors provide information of the directionality of cellular signaling acting during palate development. For example, the NOTCH receptor Notch1, its signaling modulator Lfng, and downstream transcriptional mediator Hes1 are all expressed within rugae, whereas the ligand Jag2 is expressed in the adjacent inter-rugae domains.
Figure 2.
A survey of spatial gene expression during the outgrowth and fusion of the palate. (A) Regional differences in expression domains, particularly with respect to the anterior-posterior and medial-lateral axes, highlight domains of expression in the developing palate. (B) Gene expression domains in the developing palate are organized relative to the developing rugae. In addition to SHH, multiple components of a network of signaling genes and transcription factors that are critical for palate development, including the FGF, NOTCH, and BMP pathways, exhibit restricted expression to either rugae or inter-rugae expression domains. The distribution of rugae signaling centers, generated during anterior growth, provide a mechanism to integrate A-P differences in gene expression with localized sources of patterning information. (C) Sagittal section in situ showing Shh expression restricted to the thickened epithelium of established rugae as well as the anterior edge of the RGZ epithelium prior to epithelial thickening. (D) Epithelial expression of the dual BMP/WNT antagonist Sostdc1 is restricted to the RGZ epithelium and inter-rugae domains but is downregulated at the site of rugae formation (arrowheads in C and D).
We also note that the segmental expression of certain genes appears to be stage specific. For example at E12.5 and E13.5, transcripts for Notch1, Bmp4, and the transcription factor Satb2 appear to be broadly distributed in the palate (data not shown) however, beginning at ∼E14.5 expression becomes rugae associated (Fig. 2B). Moreover, changes in the epithelial expression of Etv5 during rugae formation and maturation point to dynamic epithelial-mesenchymal signaling along the A-P axis of the palate (Fig. S1). Our expression survey uncovers a previously unappreciated organization of gene expression and signaling domains in the developing palate. Further studies to localize and refine gene expression domains with respect to the RGZ, rugae, and inter-rugae domains will provide additional insights into the spatial organization of genetic networks in the palate.
The R1 ruga coincides with gene expression domains defining anterior and posterior palate
Our expression studies reveal that molecular signals in the palate are organized relative to the developing rugae. We next investigated the spatial relationship between the region of rugae formation and A-P domains in the developing palate. Prior to the formation of the secondary palate, mesenchyme of the maxillary prominence is patterned into anterior Msx1 and posterior Barx1 expression domains (Barlow et al., 1999; Zhang et al., 2002). We found that A-P differences in the expression of Msx1 and Barx1 are maintained from the earliest stages of the secondary palatal shelf development as they form along the medial aspect of the maxillary prominence (Fig. 3). At E11.5, restricted Msx1 expression in the anterior most aspect of the palatal shelf is initiated coincident with the formation of the primary choanae on the roof of the stomodeum (Fig. 3A, D). Choanae are bilateral involutions that will form the nasal cavity and provide landmarks for the junction between the primary and secondary palate (Tamarin, 1982). At E11.5, the mesenchymal expression of Barx1 extends nearly along the entire length of the palatal shelf and abuts the posterior limit of the primary choanae (Fig. 3B, E). Significantly, the site of R1 formation is positioned at the junction of the mesenchymal expression domains of Msx1 and Barx1 at the posterior limit of the choanae (Fig. 3C&F). As previously reported, epithelial expression of Barx1 is restricted to the inter-rugae domains of the anterior palate (Welsh et al., 2007). As palate development progresses, growth of the palate along the A-P axis results in the relative expansion of the mesenchymal Msx1 and inter-rugae Barx1 expression domains anterior to R1 compared to the mesenchymal domain of Barx1 posterior to R1 (Fig. 3G–L). Up to E12.5, mesenchymal Msx1 and Barx1 share a posterior and anterior boundary respectively with R1 (Fig. 3G, H, I). However, by E13.5 continued expansion of the mesenchyme anterior to R1 begins to shift the posterior boundary of Msx1 expression away from R1 (Fig. 3J, K, L). Therefore, the RGZ and the sequential formation of rugae signaling centers provide a reference frame to directly visualize the formation of the anterior palate as it extends away from R1 at the boundary of the presumptive soft palate.
Figure 3.
The first formed ruga (R1) highlights regional growth of the anterior palate and is coincident with anterior-posterior differences in mesenchymal gene expression. (A–C) Oral view of E11.5 wildtype embryos hybridized for Msx1 (A), Barx1 (B), and Shh (C). White dashed line marks the lamboidal junction between the maxillary process and the frontonasal mass. White and black filled arrowheads mark the anterior and posterior limit of the palatal process respectively. Mesenchyme of the maxillary prominence is patterned into anterior Msx1 (A, D, G, J, between white and open arrowheads) and posterior Barx1 (B, E, H, K, between open and black arrowheads) expression domains. Shh expression (C, F, I, L) in R1 is coincident with the mesenchymal boundary between Msx1 and Barx1 (open arrowheads) and provides a landmark to visualize the anterior outgrowth of the Msx1 positive palate (region between white and open arrowheads). Restricted inter-rugae expression of Barx1 in the epithelium expands with the elongating anterior palate (H and K) whereas the posterior mesenchymal domain remains a constant size (region between open and black arrowheads)
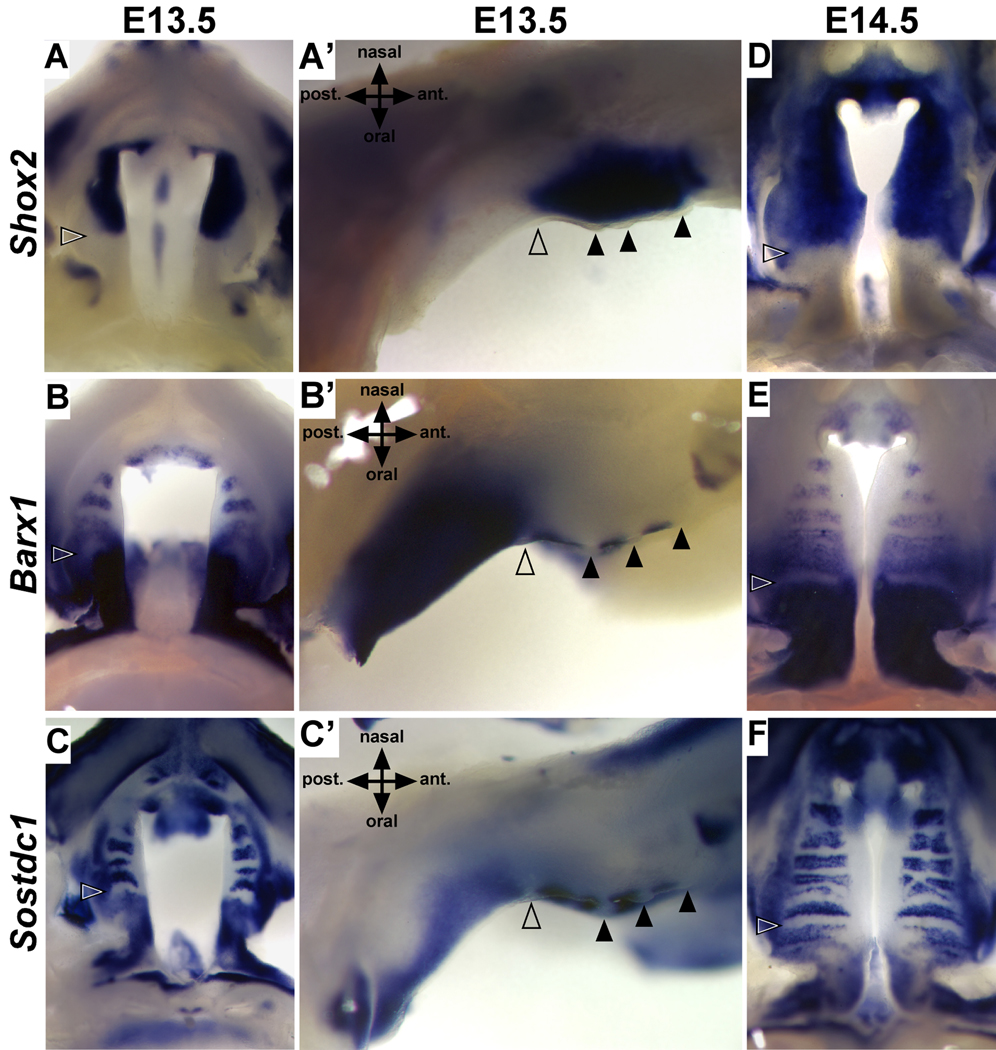
The directed growth of the anterior palate away from R1 correlates with known differences in signaling responsiveness and cell fate (Hilliard et al., 2005; Yu et al., 2005; Zhang et al., 2002). To further investigate the relationship between A-P patterning in relation to the position of R1, we compared the expression of Barx1 with that of Shox2, a marker of the anterior palate, between E12.5 and E15.5 (Li and Ding, 2007; Yu et al., 2005). Similar to Msx1, the expression of Shox2 is initiated in the anterior-most mesenchyme upon rostral extension of the anterior palate (Yu et al., 2005). In contrast to the posterior expansion of Shox2 expression reported by Li and Ding, we found that the posterior boundary of Shox2 remains coincident with R1 and the anterior boundary of mesenchymal Barx1 expression throughout palate development (Fig. 4). Interestingly, expression of Shox2 in the anterior mesenchyme is dependent on BMP signaling, while Barx1 expression is inhibited by BMP signaling (Yu et al., 2005). Consistent with the differential regulation of Shox2 and Barx1 by BMP signaling, we found the expression of the BMP signaling antagonist Sostdc1, is also spatially organized relative to R1. Similar to Barx1, Sostdc1 is expressed in the mesenchyme posterior to R1, while anterior expression is restricted to domains of inter-rugae epithelium (Fig. 4C,C’, and F). These data support that R1 and the RGZ are key features defining differences in patterning, signaling competence, and growth along the A-P axis of the palatal shelves.
Figure 4.
R1 marks the anterior-posterior boundary of mesenchymal cell fate. During the outgrowth (E13.5) and fusion (E14.5) of the palatal shelves, the mesenchymal expression of Shox2 (A,A’, and D) and Barx1 (B,B’, and E) share a common boundary defined by the location of R1 (open arrowheads) in the overlying epithelium. Expression of Shox2 and Barx1 is positively or negatively regulated by BMP signaling respectively. The expression of the BMP signaling antagonist Sostdc1 is consistent with the establishment of BMP permissive or restricted signaling domains in both the anterior and posterior palate (C,C’, and F). Black arrowheads mark position of anterior rugae.
Mesenchymal Fgf10 is expressed in a posterior-anterior gradient adjacent to the RGZ
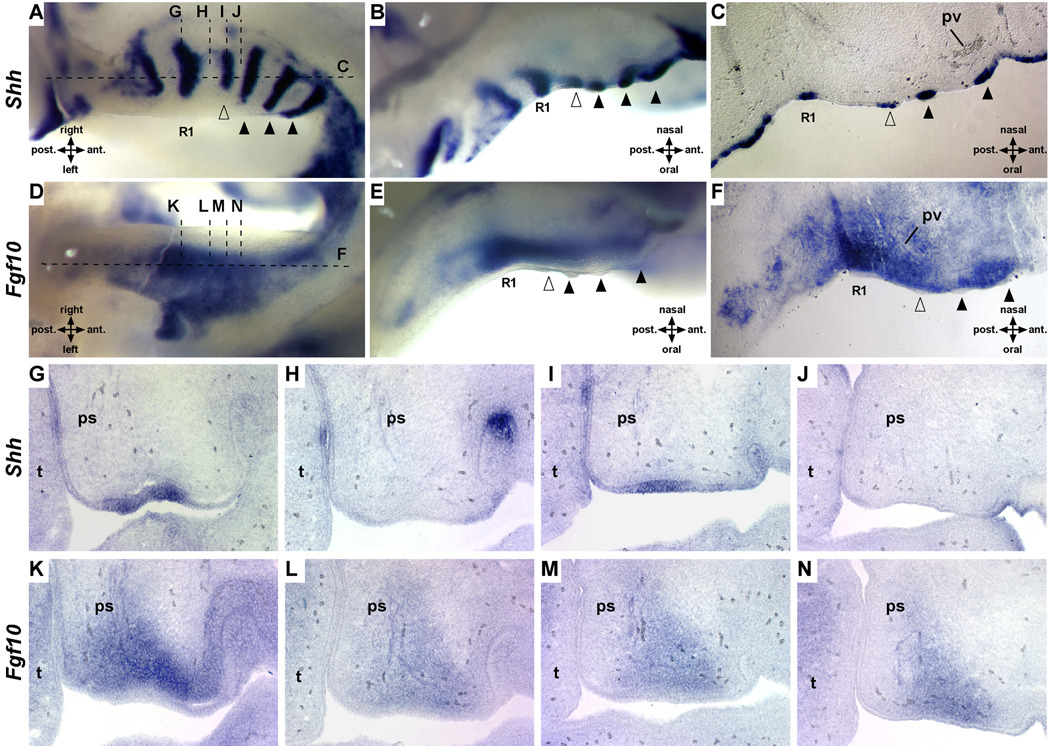
The sequential generation and relative spacing of rugae are consistent with an activation-inhibition mechanism that regulates the formation and patterning of ectodermal appendages (Pispa and Thesleff, 2003). Mesenchyme often provides the first instructive signal for the formation of ectodermal organs that develop through reciprocal epithelial and mesenchymal interactions. Previously, we demonstrated that Shh expression, rugae morphology and palate closure are disrupted in mice lacking the FGF signaling antagonist Spry2 (Welsh et al., 2007). Therefore, we examined Fgf10 expression in the mesenchyme adjacent to the RGZ. We performed whole mount in situ hybridization of Shh and Fgf10 on paired sets of right and left palatal shelves from individual embryos as well as on adjacent serial frontal and sagittal sections. These data show that contrary to the findings of Pantalacci et el. nascent Shh expression at the anterior edge of the RGZ actually precedes the overt epithelial thickening that defines rugae (Fig. 5A–C) (Pantalacci et al., 2008). Surprisingly, we found that Fgf10 expression forms a gradient within the mesenchyme of the RGZ. Fgf10 is most highly expressed in the condensed mesenchyme directly adjacent to R1 but expression diminishes anteriorly towards the site of nascent rugae formation (Fig. 5). Consistent with the recently demonstrated positive feedback of Fgf10 expression by epithelial SHH signaling (Lan and Jiang, 2009), Fgf10 expression is again upregulated anterior to the R1+n rugae, although not to the level seen adjacent to R1 (Fig. 5F).
Figure 5.
Fgf10 is expressed in a gradient defined by R1 and the site of nascent Shh expression at the anterior of the RGZ. Whole mount in situ hybridization on the palatal shelves of a single embryo detecting expression of Shh (right shelf, A and B) or Fgf10 (left shelf, D and E). Dashed horizontal and vertical lines indicate approximate plane of section shown in C and F or G–N respectively (open arrowheads mark nascent ruga, black arrowheads mark definitive rugae). Section in situ hybridization for Shh and Fgf10 on adjacent sagittal (C and F) or serial frontal sections (G–N) of E13.5 wildtype palates show robust posterior expression of Fgf10 in the mesenchyme adjacent to Shh in R1 compared to the mid-RGZ (H and L), anterior RGZ (I and M), and recently formed inter-rugae domain (J and N). ps, palatal shelf; pv, palantine vein; t, tongue.
Analysis of H&E stained sections through the RGZ show that compared to the thickened and protruding epithelium of mature rugae and the markedly thinner abutting inter-rugae domain, the oral epithelium throughout the RGZ adjacent to the Fgf10 gradient is of an intermediate thickness forming a placode that extends anteriorly from R1 (Fig. 6A). We also note that mesenchymal cells corresponding to the domain of highest levels of Fgf10 expression exhibit a distinct polarity orthogonal to adjacent epithelium of R1 (Fig. 6B). Therefore, the RGZ is not only defined by distinct gene expression domains but also unique differences in the cellular organization of both the mesenchyme and epithelium. Thus, a gradient of FGF10 signaling potentially provides an important inductive cue from the mesenchyme that together with inhibitory signals from the epithelium control the timing and spatial positioning of epithelial differentiation to establish rugae and inter-rugae domains. A candidate source for an inhibitory signal is the R1+n ruga that moves away from the RGZ during palate elongation (Pantalacci et al., 2008).
Figure 6.
Domains of cell proliferation and gene expression associated with rugae formation in the RGZ. (A) H&E stained sagittal section of an E13.5 palate shows differences in the organization of both the mesenchyme and epithelium in the region of the RGZ. The epithelium within the RGZ is of an intermediate thickness to that in rugae and inter-rugae domains (arrow) that extends anteriorly from R1 (arrowhead marks anterior RGZ and site of rugae formation). The condensed mesenchyme adjacent to R1 shows a polarity orthogonal relative to the epithelium (dashed box enlarged in B). (C) Cell proliferation detected by BrdU incorporation (BrdU: red, nuclei counterstained with DAPI (blue), dashed white line separates mesenchyme and epithelium) is uniform in the palatal mesenchyme. Proliferation is high in inter-rugae domains (arrows) and in the posterior RGZ and reduced in the thickened epithelium of rugae and nascent rugae (white arrowhead). (D) Summary of regional differences in epithelial proliferation in the E13.5 palate (error bars represent SEM, n=4). Sagittal sections of E13.5 palates hybridized for ΔNp63 (E and F) p21 (G) and Sostdc1 (H) expression supports that rugae morphogenesis involves cell cycle exit. Within the RGZ, ΔNp63 is strongly expressed immediately anterior to R1 (E) but is periodically downregulated at the site of nascent rugae formation (arrowhead, F). Expression of ΔNp63 (E and F) and the cell cycle inhibitor p21 (G) becomes progressively upregulated in maturing rugae. (H) Sostdc1 expression overlaps with ΔNp63 within the RGZ and is also periodically downregulated at the site of rugae formation (arrowhead) but is restricted to inter-rugae domains in the anterior palate. Abbreviations: md, mandible; pv, palatine vessel.
Molecular signals maintaining proliferation versus cell cycle exit during rugae differentiation
We considered that rugae differentiation involves an FGF10-dependent program that triggers Shh expression and cell cycle exit in a localized population of cells within the RGZ. In a series of BrdU labeling experiments, we confirmed that epithelial cell proliferation is diminished in established rugae, including R1 and the most recently formed rugae (R1+n). In contrast, interrugae epithelium, including the domain between the anterior boundary of the RGZ and the R1+n rugae display high levels of proliferation. Notably, we detected non-uniform proliferation within the RGZ (Fig. 6C, D). Epithelial cells within the posterior half of the RGZ adjacent to R1 exhibit an elevated mitotic index relative to that of cells in the anterior half of the RGZ. Significantly, this region of diminished proliferation within the RGZ corresponds to cells that initiate Shh expression prior to the overt epithelial differentiation into rugae (see Fig. 5).
The proliferation pattern within the RGZ suggests that rugae and inter-rugae domains are established through the spatial organization of molecular signals controlling cell cycle exit and maintenance, respectively. The p53 related factor p63 is thought to integrate the activity of multiple signaling pathways and act as a switch to regulate molecular cascades that promote the maintenance of epithelial progintors versus cell cycle exit and differentiation (Yang et al., 1999). p63 has been shown to maintain epithelial cell "stemness" through the positive regulation of Fgfr2b and Jag2 (Candi et al., 2007). We established that Fgfr2b and Jag2 expression is localized to inter-rugae domains (see Fig. 2). Complex regulation of the p63 locus results in the expression of at least six protein variants exhibiting distinct and often opposing regulation of target genes (Vigano et al., 2006; Wu et al., 2003; Yang et al., 1998). Alternative promoter usage generates two N-terminal isoforms, a longer transactivating (TA) domain containing TAp63 and a truncated ΔNp63 lacking the TA domain, while alternative splicing of the C-terminus generates α, β, and γ isoforms. Notably, the ΔNp63 isoform has been shown to be the predominant isoform expressed during craniofacial development and to be required for the differentiation and maintenance of signaling centers in the oral epithelium (Laurikkala et al., 2006; Mikkola, 2007; Mills et al., 1999). Furthermore, FGF10 has been shown to be a potent inducer of p63 during ectodermal organogenesis and the periodic patterning of skin appendages (Tao et al., 2002). Therefore, we examined the spatial organization of ΔNp63 expression in the developing palate epithelium.
We found that in the developing palate, ΔNp63 expression is notably dynamic with respect to the sequence of rugae induction and maturation. ΔNp63 is not expressed in R1 or the R1+n rugae but its expression becomes rugae specific in more mature anterior rugae. Significantly ΔNp63 is strongly expressed within the RGZ epithelium immediately anterior to R1, however as the R1+n rugae is displaced by the expansion of the newly formed inter-rugae domain, expression is downregulated at the site of nascent rugae formation (Fig. 6E and F). ΔNp63 has been shown to inhibit the expression of the cell cycle regulator p21, a factor induced by BMP4 that promotes growth arrest during signaling center formation (Jernvall et al., 1998; Nguyen et al., 2006; Okuyama et al., 2007). Our expression analysis demonstrates that p21 is strongly expressed in R1 but is excluded from the RGZ epithelium, a pattern complementary to that of ΔNp63. Interestingly, similar to ΔNp63 in R1+n and more mature anterior rugae, p21 expression becomes progressively upregulated (Fig. 6G). In the developing tooth bud, BMP4 induces its own antagonist Sostdc1, and expression of Shh and p21 (Laurikkala et al., 2003). Sostdc1 acts to inhibit BMP4 signaling, while SHH signaling acts to locally inhibit the expression of Sostdc1. In this way, reciprocal epithelial-mesenchymal signaling serves to restrict the field of cells competent to respond to a BMP threshold dependent induction of signaling center differentiation. Similar to ΔNp63 and consistent with its role in tooth development, Sostdc1 is also periodically down regulated at the site of rugae induction (Fig. 6H). Therefore, within the RGZ ΔNp63 is coexpressed with Sostdc1, while in established rugae expression overlaps with p21. Thus, while p63 expression within the RGZ likely inhibits p21 expression to sustain the cell cycle, periodic down regulation of ΔNp63 and Sostdc1 expression at the anterior RGZ may facilitate growth arrest and signaling center differentiation. These data suggest that during the rostral extension of the anterior palate, a dynamic molecular circuit coordinates the sequential formation of both rugae and inter-rugae epithelium from a common precursor population in the RGZ.
Loss of Fgf10 results in failure to maintain the RGZ and loss of coordinating epithelial-mesenchymal signaling
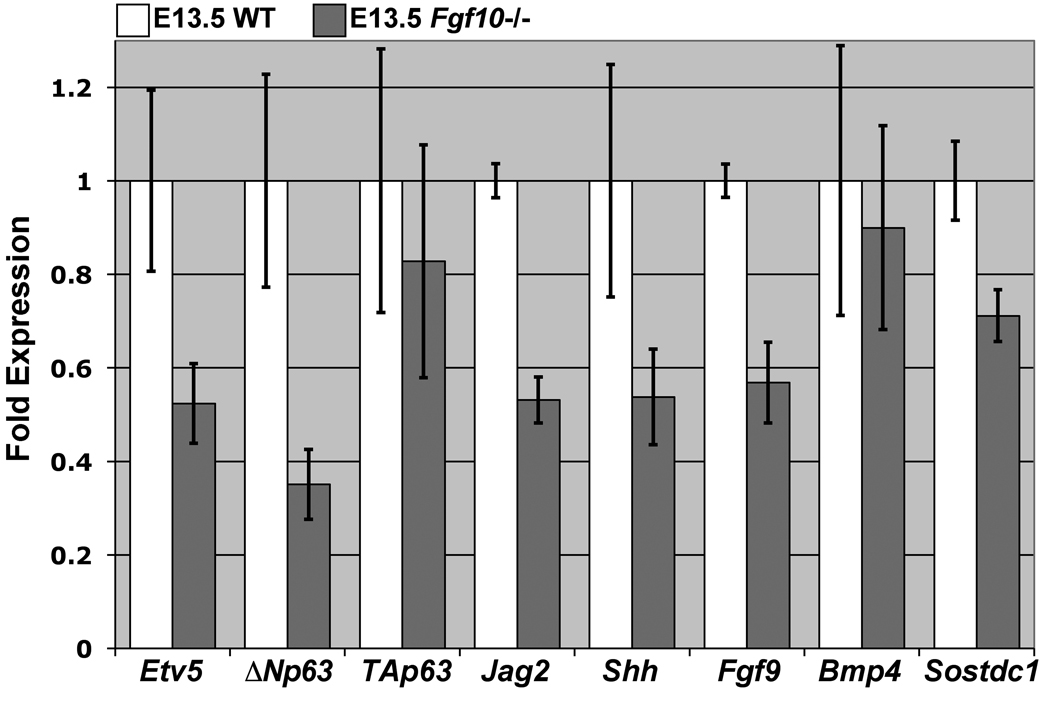
Our data suggest that p63 expression and rugae morphogenesis are dependent on FGF10 signaling from the mesenchyme. The targeted disruption of Fgf10 in mice results in cleft palate and loss of Shh expression (Alappat et al., 2005; Rice et al., 2004). We sought to further examine palate defects in Fgf10 mutants with respect to signaling in the RGZ. Using QRT-PCR analysis, we compared the palatal expression of several genes proposed to mediate rugae morphogenesis between E13.5 wildtype (n=3) and Fgf10 mutants (n=4) (Fig. 7). Consistent with its role as a mediator of FGF signaling during craniofacial development and dynamic epithelial expression in the palate (Firnberg and Neubuser, 2002) we found that Etv5 expression is reduced to approximately 50 percent wildtype levels in Fgf10 mutant palates. Laurikkala et al previously showed that ΔNp63 is the predominant isoform of p63 expressed in the oral epithelium during development (Laurikkala et al., 2006). We detected no significant differences in the expression levels of the TAp63 isoform, however, ΔNp63 expression is significantly reduced in the palate of Fgf10 mutants. Furthermore, expression levels of Jag2, a p63 target expressed in the inter-rugae epithelium, as well as Fgf9 that is coexpressed with Shh, are also reduced. Interestingly, although we detected normal levels of Bmp4 expression in the palates of E13.5 Fgf10 mutants, we detected a consistent reduction in the expression levels of the BMP antagonist Sostdc1.
Figure 7.
QRT-PCR analysis detects altered expression of both rugae and inter-rugae specific genes in Fgf10 mutant palates. Relative to wildtype (n=3), E13.5 Fgf10 mutants (n=4) exhibit reduced expression of Etv5, ΔNp63, Jag2, Shh and Fgf9, and Sostdc1, but not the TA isoform of p63 or Bmp4 (error bars represent SEM).
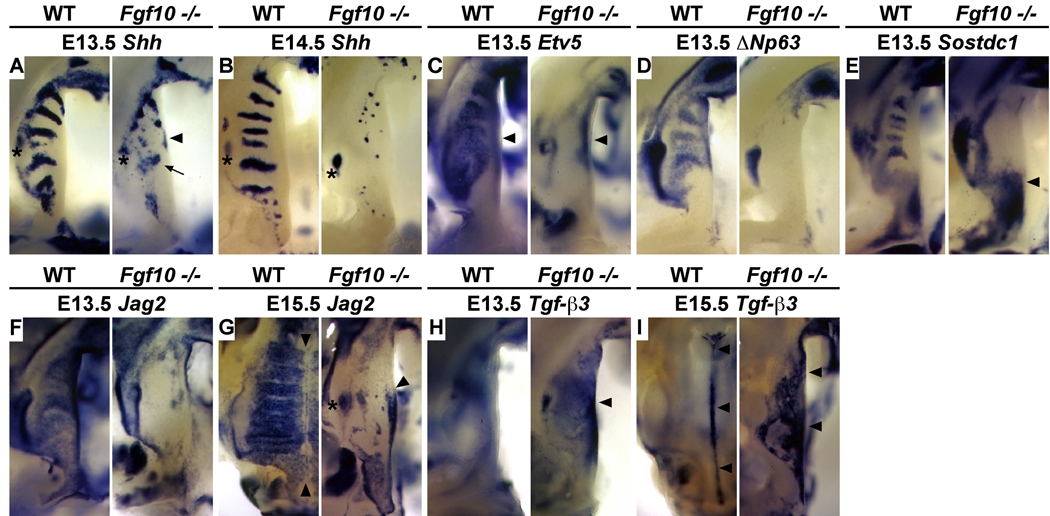
We next sought to investigate the impact of these quantitative changes in gene expression on the spatial organization of the RGZ and rugae development. Consistent with the QRT-PCR data, in situ hybridization confirmed that rugae formation is severely disrupted in the Fgf10 mutant palate. The organized Shh expression domains that highlight established and forming rugae progressively deteriorate between E13.5 - E14.5 (Fig. 8A, B). Furthermore, the expression of both Etv5 and ΔNp63 is specifically lost in the epithelium of the palatal shelves of Fgf10 mutants, while Etv5 expression in the mesenchyme of the medial palatal shelf appears unaffected (Fig. 8C, D). Although QRT-PCR indicated a moderate reduction in expression levels, we found that Sostdc1 expression in the anterior epithelium to be completely lost (Fig. 8E). Surprisingly, we also detected significant upregulation of Sostdc1 in the mesenchyme posterior to R1 and in the molar tooth bud, suggesting a loss of coordinated epithelial-mesenchymal signaling via the BMP and possibly WNT pathways in Fgf10 mutant palates (Fig. 8E).
Figure 8.
Altered patterning of the palatal epithelium associated with loss of the RGZ in Fgf10 mutants. (A) The RGZ is reduced in size (distance between R1 and the molar tooth bud) and Shh expression in R1 (arrow) is diminished in E13.5 Fgf10 mutant palates. Shh is highly disorganized in the anterior palate and ectopically expressed in the medial edge epithelium (arrowheads). (B) In the E14.5 Fgf10 mutant, Shh expression is reduced to small puncta of expression in the anterior palate and all evidence of R1 and the RGZ has been lost. At E13.5 Fgf10 mutants exhibit specific loss of gene expression in the RGZ and inter-rugae epithelium of the anterior palate including that of: (C) Etv5, a transcriptional mediator of the FGF/MAPK pathway (arrowheads mark expression in the mesenchyme), (D) ΔNp63, and (E) Sostdc1, a negative feedback inhibitor of BMP signaling. Mesenchymal expression of Sostdc1 (E) in the presumptive soft palate and adjacent tooth anlage is elevated (arrowhead). (F–I) Patterning of the medial edge epithelium (MEE) is also altered in Fgf10 mutants. (F) In E13.5 wildtype, Jag2 is expressed in inter-rugae domains of the anterior palate and the medial edge epithelium (MEE) but excluded from rugae. Jag2 expression is absent within the RGZ and anterior oral epithelium of E13.5 Fgf10 mutants but weak expression is seen medially. (G) Jag2 expression is downregulated within the MEE of E15.5 wildtype palates and the medial boundary of inter-rugae Jag2 expression abuts the MEE (arrowheads). In E15.5 Fgf10 mutants, Jag2 is ectopically expressed in the MEE (arrowhead) with an anterior boundary that is aligned with the normal anterior extent of the RGZ (asterisk marks the molar tooth bud). (H) Tgf-β3 expression is significantly upregulated in Fgf10 mutant palates at E13.5, particularly in the MEE adjacent to the region of the RGZ (arrowhead). (I) In the fusing palates of E15.5 wildtype embryos, Tgf-β3 expression is restricted to the medial epithelial seem (MES) (arrowheads). Tgf-β3 expression in E15.5 Fgf10 mutants is ectopically expressed across the oral surface of the palatal shelf but reduced in the region showing highest levels of precocious expression at E13.5 and ectopic Jag2 at E15.5 (arrowheads).
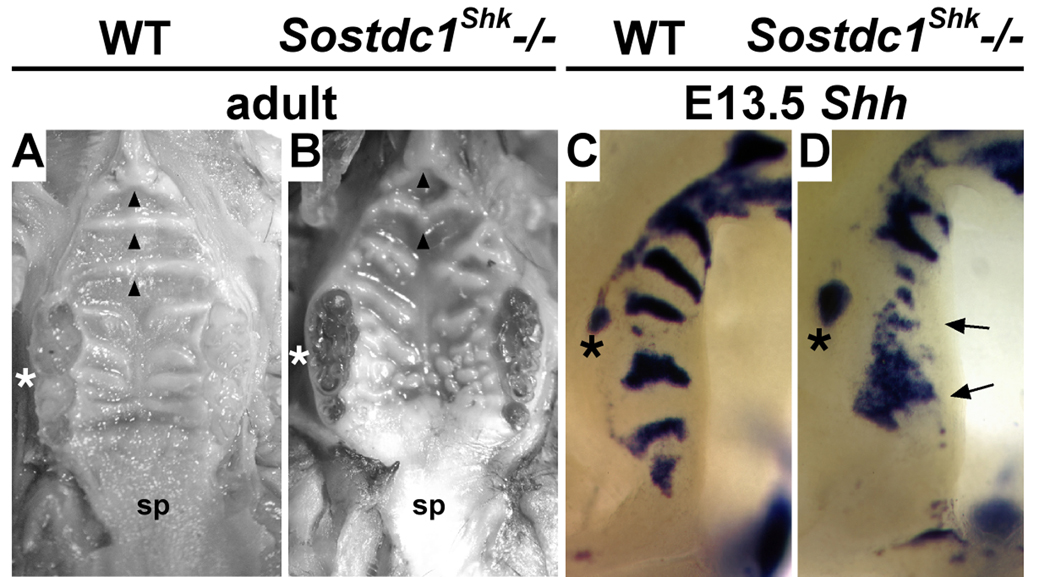
Significantly, loss of p63 function has recently been shown to result in failed outgrowth of the anterior palate and is associated with elevated levels of Bmp4 and loss of Shh expression in the maxillary process (Thomason et al., 2008). During tooth development, Sostdc1 integrates the BMP, SHH, and FGF pathways and is required to regulate epithelial responsiveness to Bmp4 induction of Shh and p21 expressing signaling centers (Kassai et al., 2005; Laurikkala et al., 2003). Furthermore, Sostdc1 (also called Wise, Ectodin, and USAG-1) has recently been shown to coordinate the BMP and WNT pathways via its interaction with the WNT co-receptor Lrp4 (Ohazama et al., 2008). In order to determine whether Sostdc1 plays a similar role during rugae formation we analyzed Shh expression in the palates of Sostdc1Shk (Sharkey) mutants. Sharkey was recovered as a spontaneous mutation resulting in supernumerary teeth. We have observed that rugae are disorganized in the Sharkey mutant palate (Fig. 9A, B). Characterization by The Jackson Laboratory Craniofacial Mutant Resource group confirmed Sharkey as a new null mutation in Sosdtc1 resulting from a single base pair deletion in exon 2 (Craniofacial Resource, The Jackson Laboratory, www.jax.org/cranio/index.html). Significantly, we found that Shh is ectopically expressed throughout the RGZ of Sostdc1Shk mutant palates at E13.5 (Fig. 9C, D). This data supports that similar to its role in tooth cusp patterning, Sostdc1 in the palate acts to regulate induction of signaling centers.
Figure 9.
Altered rugae formation and gene expression in the Sostdc1Shk mutant palate. (A, B) Midline fusion of anterior rugae (R2–4, arrowheads) and normal rugae morphology in the wildtype palate compared with failed fusion of anterior ruga (R4) and disorganized rugae morphology in the Sharkey (Sostdc1Shk) mutant palate. (C, D) Loss of Sostdc1 function in the Sharkey mutant results in expanded expression of Shh in the molar tooth bud, ectopic expression throughout the RGZ (arrows), and an increased distance between these two expression domains. fused molars (asterisk A, B); molar tooth bud (asterisk C, D); soft palate (sp).
The epithelial patterning defects in Fgf10 mutants are not limited to the organization of gene expression domains along the A-P axis of the palate. We note that from E14.5 to E15.5 as the palatal shelves make contact and fuse, the medial edge of palatal rugae share a common boundary with the lateral extent of medial edge epithelia (MEE). Only after shelf fusion at E16.5 do the anterior-most rugae (R3, R2, and R4) fuse across the midline. Down-regulation of Jag2 and the restricted expression of Tgf-β3 within the medial edge MEE is part of an intrinsic program to pattern palatal epithelium and localize the tissue remodeling that promotes shelf fusion in the region of midline contact (Jin et al., 2008). Interestingly, we found that the inter-rugae expression of Jag2 is lost in Fgf10 mutants but is expressed ectopically in the MEE (Fig. 8F, G) while Tgf-β3 is precociously and ectopically expressed across the oral epithelium of Fgf10 mutant palates (Fig. 8H, I). Therefore, failed rugae morphogenesis and altered epithelial patterning resulting from loss of Fgf10 impacts both the anterior-posterior and medial-lateral axis of the palatal shelves.
Collectively our data suggests that a highly integrated molecular circuit coordinates epithelial-mesenchymal signaling within the RGZ in order to direct the periodic differentiation of SHH expressing signaling centers during palate outgrowth. Mesenchymal Fgf10 acting upstream of both p63 and Sostdc1 thus provides a mechanism to couple both FGF10 and BMP4 regulation of Shh expression by restricting induction of Shh expression to the anterior edge of the Fgf10 gradient in the RGZ.
Discussion
The RGZ and spatial organization of a network of genes that directs palate development
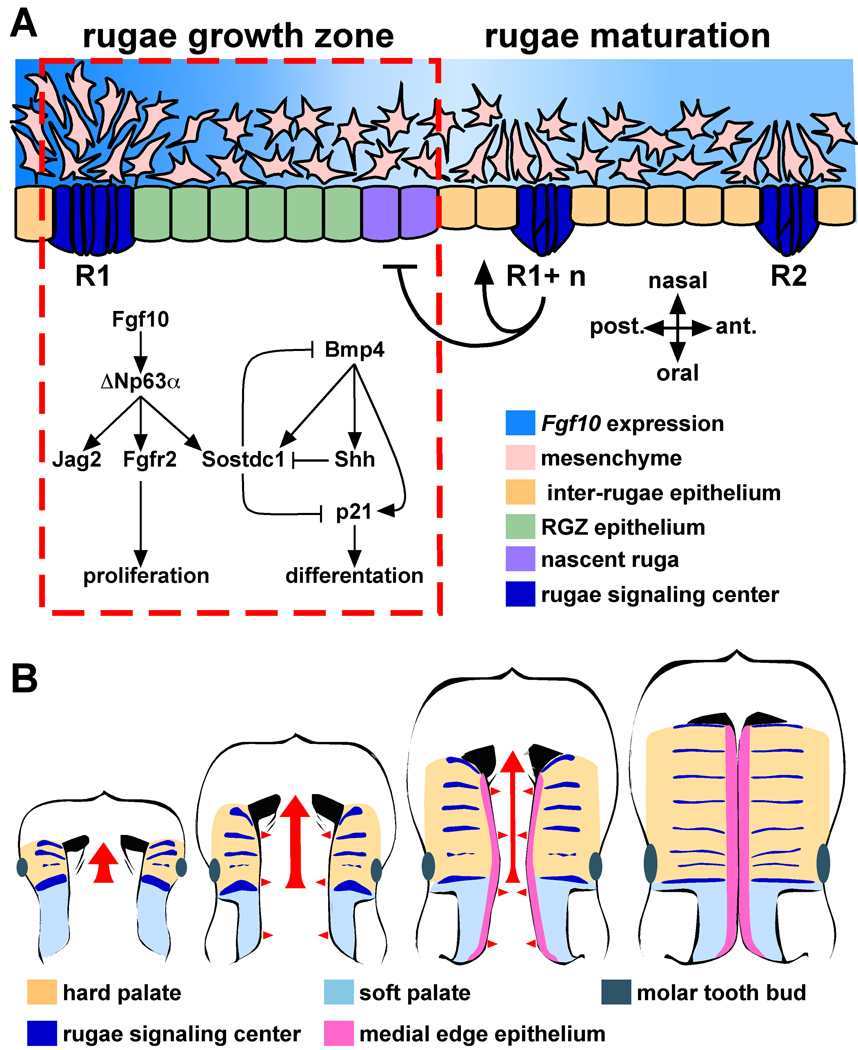
In this study we have identified a novel region of morphogenetic activity, the RGZ, that functions to integrate multiple signaling pathways critical for guiding palate morphogenesis. Signaling via the FGF, BMP, and SHH pathways is essential for multiple aspects of craniofacial development (Abzhanov and Tabin, 2004; Hu and Helms, 1999; Kim et al., 1998; Trumpp et al., 1999). The spatial organization of molecular interactions amongst these pathways has been shown to establish the frontonasal ectodermal zone (FEZ) that directs mid-facial outgrowth (Abzhanov et al., 2007; Abzhanov et al., 2004; Hu et al., 2003). In the secondary palate, both Fgf10 and Bmp4 are required for the expression of Shh (Rice et al., 2004; Zhang et al., 2002). However, a mechanism explaining their combined regulation of Shh has been lacking. We propose a molecular model involving p63 and Sostdc1 that integrates FGF10 and BMP4 signaling that balances epithelial proliferation and differentiation to control the sequential induction of Shh expression in the palate (Fig. 10A). Epithelial SHH signaling to the mesenchyme completes a positive feedback loop that maintains Fgf10 expression and is required for directing patterning and outgrowth of the palate (Lan and Jiang, 2009). In this way, the periodic formation of rugae signaling centers supplies a register of signaling cues to the underlying mesenchyme as the palate extends anteriorly. The periodicity of rugae formation will facilitate a more detailed dissection of the timing of molecular and cellular events that establish and maintain this critical epithelial-mesenchymal feedback loop.
Figure 10.
Models of the molecular and morphogenetic activity associated with the RGZ. (A) We suggest a model of molecular interactions integrating FGF10 and BMP4 signaling during rugae formation within the RGZ. Both Fgf10 and Bmp4 are required for epithelial expression of Shh. The RGZ acts as a source of both rugae and inter-rugae epithelium. Within the RGZ, Fgf10 is expressed in a gradient extending from R1 to the site of nascent rugae formation. We propose that FGF10 signaling, mediated through ΔNp63α and its targets Jag2 and Fgfr2b, maintains proliferation of epithelial progenitors at the posterior end of the RGZ. Epithelial expression of the Bmp4 antagonist Sostdc1 in the anterior palate also requires Fgf10. We found that similar to its role in tooth cusp patterning, Sostdc1 acts to restrict induction of Shh in the RGZ. We propose that the relative balance between FGF10 and BMP4 signaling is one component defining the A-P position of rugae formation. Induction of Shh and p21 expression results in cell cycle exit and epithelial differentiation while continued proliferation of inter-rugae epithelium moves the R1+n rugae away from the RGZ. Signals from the R1+n rugae (bar and arrow) are also proposed to influence the fate of RGZ epithelium. (B) The anterior growth of the anterior palate (tan) proceeds from the first formed rugae (red arrow) and is coincident with the establishment of segmental signaling domains (rugae). Fusion of the bilateral shelves requires medially directed growth (red arrowheads) and patterning of the medial edge epithelium (MEE, pink). The lateral boundary of the MEE coincides with the medial edge of the rugae, suggesting that signals from the rugae also participate in the intrinsic program that patterns the MEE. Thus, rugae and the RGZ provide a reference frame for visualizing the organization of signaling domains with respect to the anterior–posterior and medial-lateral patterning and growth of the palatal shelves.
The early disorganized and ectopic expression of Shh in Fgf10 mutant palates, followed by progressive loss of Shh expression, demonstrates that Shh is not directly dependent on Fgf10. Instead, our data indicates that mesenchymal Fgf10 is required to organize and maintain the RGZ and rugae morphogenesis, possibly through p63 mediated maintenance of an epithelial progenitor population in the posterior RGZ (Harada et al., 2002). In addition to Shh, loss of RGZ activity in Fgf10 mutants results in the absence or altered expression of Etv5, p63, and Fgf9. Similar to p63, epithelial expression of the BMP signaling antagonist Sostdc1 is completely lost in the anterior palate while expression posterior to the RGZ and in the molar tooth bud is actually elevated. Loss of p63 results in elevated BMP4 signaling, loss of Shh expression and failed outgrowth of the anterior palate (Thomason et al., 2008). Altered expression in Fgf10 mutants suggest that Sostdc1 is a downstream target through which p63 regulates Bmp4.
We have identified complementary rugae and inter-rugae specific expression of ligands, receptors, and transcriptional mediators in both the epithelium and mesenchyme providing new insight into the directionality, organization, and spatial integration of signaling interactions that guide palate morphogenesis. For example, both loss and gain of function mutations of Fgf9 have been reported to result in cleft palate, however the etiology underlying the defect has not been analyzed (Colvin et al., 2001; Harada et al., 2009; Murakami et al., 2002). Rugae specific expression of Fgf9 in the epithelium along with Etv4 (Pea3) in the underlying mesenchyme suggests the presence of a reciprocal epithelial-mesenchymal FGF feedback loop commonly observed during the development of other structures (del Moral et al., 2006; Sun et al., 2000; Zhang et al., 2006). Our data support a model where rugae signaling centers, established through the dynamic activity of the RGZ, provide a framework to organize a network of genes that pattern the three dimensional growth and patterning of the palatal shelves (Fig. 10B).
The RGZ and regional differences in A-P growth of the palate
The RGZ, positioned at the A-P junction of the future soft and elongating hard palate, provides a landmark for understanding regional differences in palate outgrowth and patterning. Elongation of the maxillary prominence is accompanied by growth of the anterior palate. A recent report from Li and Ding compared the dynamic expression of anteriorly restricted Shox2 with Meox2, a marker of posterior palatal fate, during A-P elongation of the palatal shelves (Jin and Ding, 2006; Li and Ding, 2007). Focusing on the changes in the relative size of the Shox2 and Meox2 expression domains during palate development, the authors argue that the growth of the anterior palate involves a posteriorly directed expansion of Shox2 expression and conversion of once posterior Meox2 expressing cells into an anterior fate. However, in the present study we demonstrate that the position of R1 remains constant relative to Shox2 and Barx1 expression, markers of anterior and posterior mesenchymal fate respectively. This observation is inconsistent with a posteriorly directed expansion of the Shox2 expression domain. Furthermore, we show that rostral extension of the anterior epithelium is achieved by interposition of additional rugae and inter-rugae domains between R1 and the R1+n rugae, implying the presence of an epithelial progenitor population in the posterior RGZ that is maintained by high levels of FGF10 (Harada et al., 2002). Differences in mesenchymal proliferation rates along the A-P axis of the palate have not been documented (Li and Ding, 2007; Liu et al., 2008). This suggests that an alternative mechanism provides a source of mesenchyme to accompany the growth of the anterior palate immediately anterior to R1.
Recent analysis of cleft palate in Wnt5a mutants identified the presence of directed cell migration in the developing palate (He et al., 2008; Li and Ding, 2007; Liu et al., 2008). Isotopic grafting of EGFP expressing anterior or posterior palatal mesenchyme into wildtype or Wnt5a mutant palates demonstrated that posterior palatal mesenchyme preferentially migrates anteriorly whereas anterior mesenchyme tends to migrate towards the lateral palate. Interestingly, the authors showed that directed migration is dependent on a Wnt5a expression gradient in the anterior palate that is complementary to the Fgf10 gradient that we report here. Furthermore, in addition to Wnt5a He et al. found that FGF10 also acts as a potent chemoattractant for palatal mesenchyme. Therefore, the strong expression domain of Fgf10 at the junction of the anterior and posterior palate and the complementary Wnt5a expression gradient in the anterior palate (see Fig. 2A) suggest that a complex and combinatorial control of cell movements may be involved in the rostral outgrowth of the palatal shelves. Further investigation of the differences in migratory behavior between anterior and posterior palatal mesenchyme in reference to the landmarks provided by the RGZ will likely prove informative.
Modularity and integration of signaling domains during craniofacial outgrowth
Craniofacial development involves the precisely coordinated outgrowth and midline fusion of multiple bud-like prominences and requires positional and patterning information provided by spatially separated signaling centers. In avians and mammals, formation of the beak or nasal capsule and primary palate is coordinated by FEZ signals that direct the growth of the frontonasal mass (Hu and Marcucio, 2009b; Wu et al., 2006b). The mammalian secondary palate represents an elaboration of the skeletodontal elements of the upper jaw within the vertebrate craniofacial body plan. We have identified the RGZ, strategically positioned relative to the A-P axis of the palate, as the location of periodic generation of signaling centers during the rostral outgrowth of the palate. We observe that at E11.5 Shh expression in the FEZ is contiguous with the adjacent region of the maxillary prominence that will give rise to R1 (see Fig. 3C and F). During the initial stages of secondary palate development, the site of R1 formation is closely associated with the formation of the primary choanae that presages the site of fusion between the primary palate with the anterior secondary palate (Tamarin, 1982). Therefore, growth of the anterior hard palate occurs via the expansion of the domain intervening the FEZ and R1. How these two morphogenetic domains may be integrated to coordinate craniofacial development is an important question with the potential to significantly advance our understanding of the etiology of an important class of birth defects, clefting of the primary and secondary palate (cleft lip and cleft palate).
To address the question of coordination between morphogenic domains during craniofacial development, Depew and Simpson have proposed a “hinge and caps” model to explain how spatially distributed sources of positional information may be integrated to achieve a “global” solution to the outgrowth of the numerous facial primordia that form the oral cavity in gnathostomes (Depew and Simpson, 2006). This model is based upon the interplay between prepatterned populations of CNC with the activity of proximo-distal positioned epithelial signaling centers. In the model, proximal “hinge” and distal “caps” derived signals coordinate the outgrowth and patterning of intervening tissue to achieve midline fusion of paired facial primordia as well as maintain proper registration between elements of the upper and lower jaws (Depew and Compagnucci, 2008; Depew and Simpson, 2006). “Hinge” defining signals, including Fgf8, Ptx2, and the nested expression of the Dlx gene family members, emanate from the junction of the maxillary and mandibular components of the first branchial arch. “Caps” are sources of positional information located most distally from the “hinge” region, such as the distal mandibular arch of the lower jaw and the lamboidal junction between the distal maxillary arch and frontonasal mass of the upper jaw (i.e. the FEZ). The juxtaposition of proximal and distal sources of positional information imposes an inherent polarity upon the developing facial primordia. A prediction that follows from this model is the potential to establish coordinating domains or “developmental modules” within the facial prominences that are defined in response to “hinge” and “caps” signaling. Such "developmental modules" would provide a mechanism for coordinating the growth of autonomous components of the upper and lower jaw.
The FEZ-directed rostral growth of the midfacial complex is accompanied by the extension of the anterior palate. We propose that the modularity of rugae signaling domains provides a distributed system of common instructional cues that maintain growth of the secondary palate in proper registration with the surrounding elements of the upper jaw. Furthermore, integration of the unique periodic activity of the RGZ within the hierarchy of the “hinge and caps” model would also provide a mechanism to maintain evolutionary plasticity while meeting the morphogenic requirements specific to palate closure. If RGZ dynamics are coupled with adjacent “hinge and caps” signaling domains, evolutionary variation in facial form resulting from species-specific activity of the FEZ would conceivably be accompanied by corresponding output from the RGZ. As evidence in support of this hypothesis, we note that species-specific variations in rugae number from 3–4 in human, 8 in mouse, and 18 in the horse correlate with striking differences in the rostral extension of the face and the underlying skeletal elements. Therefore, signaling dynamics within the RGZ potentially provide a readily modulated mechanism that satisfies the need to coordinate a phylogenetically inherited body plan for skull development with the palate morphogenetic program while also accommodating evolutionary adaptation of facial form.
Supplementary Material
Acknowledgements
We thank Drs. Mark Roberson, Kevin Peterson and Randal Babiuk for critical reading of the manuscript along with Jim Hagarman for helpful discussions and Yingying Zhao for technical assistance. We thank Dr. Leah-Rae Donahue and Julie Hurd at the Craniofacial Mutant Resource at The Jackson Laboratory for assistance with Sharkey mice. This work was supported by National Institutes of Health (NIH) HD41066 (T.P.O.) and a Presidential Life Sciences fellowship from Cornell University (I.C.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Whole mount in situ hybridization of a palate from an E15.5 mouse embryo showing Shh expression on the oral surface of the fusing palatal shelves. In the anterior palate, Shh expression is restricted to lateral stripes of thickened epithelium called rugae. A distinct region of the secondary palate, the rugae growth zone (RGZ), is associated with the periodic formation of rugae during palate outgrowth. Rugae function as SHH signaling centers to pattern the palatal shelves and coordinate the growth of the secondary palate with the elongating midfacial complex.
References
- Abzhanov A, Cordero DR, Sen J, Tabin CJ, Helms JA. Cross-regulatory interactions between Fgf8 and Shh in the avian frontonasal prominence. Congenit Anom (Kyoto) 2007;47:136–148. doi: 10.1111/j.1741-4520.2007.00162.x. [DOI] [PubMed] [Google Scholar]
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Abzhanov A, Tabin CJ. Shh and Fgf8 act synergistically to drive cartilage outgrowth during cranial development. Dev Biol. 2004;273:134–148. doi: 10.1016/j.ydbio.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Ahlgren SC, Bronner-Fraser M. Inhibition of sonic hedgehog signaling in vivo results in craniofacial neural crest cell death. Curr Biol. 1999;9:1304–1314. doi: 10.1016/s0960-9822(00)80052-4. [DOI] [PubMed] [Google Scholar]
- Alappat SR, Zhang Z, Suzuki K, Zhang X, Liu H, Jiang R, Yamada G, Chen Y. The cellular and molecular etiology of the cleft secondary palate in Fgf10 mutant mice. Dev Biol. 2005;277:102–113. doi: 10.1016/j.ydbio.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Barlow AJ, Bogardi JP, Ladher R, Francis-West PH. Expression of chick Barx-1 and its differential regulation by FGF-8 and BMP signaling in the maxillary primordia. Dev Dyn. 1999;214:291–302. doi: 10.1002/(SICI)1097-0177(199904)214:4<291::AID-AJA2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Barrow LL, van Bokhoven H, Daack-Hirsch S, Andersen T, van Beersum SE, Gorlin R, Murray JC. Analysis of the p63 gene in classical EEC syndrome, related syndromes, and non-syndromic orofacial clefts. J Med Genet. 2002;39:559–566. doi: 10.1136/jmg.39.8.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitgood MJ, McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- Candi E, Rufini A, Terrinoni A, Giamboi-Miraglia A, Lena AM, Mantovani R, Knight R, Melino G. DeltaNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc Natl Acad Sci U S A. 2007;104:11999–12004. doi: 10.1073/pnas.0703458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey LM, Lan Y, Cho ES, Maltby KM, Gridley T, Jiang R. Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development. Dev Dyn. 2006;235:1830–1844. doi: 10.1002/dvdy.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin JS, White AC, Pratt SJ, Ornitz DM. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- del Moral PM, De Langhe SP, Sala FG, Veltmaat JM, Tefft D, Wang K, Warburton D, Bellusci S. Differential role of FGF9 on epithelium and mesenchyme in mouse embryonic lung. Dev Biol. 2006;293:77–89. doi: 10.1016/j.ydbio.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Compagnucci C. Tweaking the hinge and caps: testing a model of the organization of jaws. J Exp Zoolog B Mol Dev Evol. 2008;310:315–335. doi: 10.1002/jez.b.21205. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Simpson CA. 21st century neontology and the comparative development of the vertebrate skull. Dev Dyn. 2006;235:1256–1291. doi: 10.1002/dvdy.20796. [DOI] [PubMed] [Google Scholar]
- Firnberg N, Neubuser A. FGF signaling regulates expression of Tbx2, Erm, Pea3, and Pax3 in the early nasal region. Dev Biol. 2002;247:237–250. doi: 10.1006/dbio.2002.0696. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A. Molecular control of secondary palate development. Dev Biol. 2007;301:309–326. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- Harada H, Toyono T, Toyoshima K, Yamasaki M, Itoh N, Kato S, Sekine K, Ohuchi H. FGF10 maintains stem cell compartment in developing mouse incisors. Development. 2002;129:1533–1541. doi: 10.1242/dev.129.6.1533. [DOI] [PubMed] [Google Scholar]
- Harada M, Murakami H, Okawa A, Okimoto N, Hiraoka S, Nakahara T, Akasaka R, Shiraishi Y, Futatsugi N, Mizutani-Koseki Y, Kuroiwa A, Shirouzu M, Yokoyama S, Taiji M, Iseki S, Ornitz DM, Koseki H. FGF9 monomer-dimer equilibrium regulates extracellular matrix affinity and tissue diffusion. Nat Genet. 2009;41:289–298. doi: 10.1038/ng.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haworth KE, Healy C, Morgan P, Sharpe PT. Regionalisation of early head ectoderm is regulated by endoderm and prepatterns the orofacial epithelium. Development. 2004;131:4797–4806. doi: 10.1242/dev.01337. [DOI] [PubMed] [Google Scholar]
- Haworth KE, Wilson JM, Grevellec A, Cobourne MT, Healy C, Helms JA, Sharpe PT, Tucker AS. Sonic hedgehog in the pharyngeal endoderm controls arch pattern via regulation of Fgf8 in head ectoderm. Dev Biol. 2007;303:244–258. doi: 10.1016/j.ydbio.2006.11.009. [DOI] [PubMed] [Google Scholar]
- He F, Xiong W, Yu X, Espinoza-Lewis R, Liu C, Gu S, Nishita M, Suzuki K, Yamada G, Minami Y, Chen Y. Wnt5a regulates directional cell migration and cell proliferation via Ror2-mediated noncanonical pathway in mammalian palate development. Development. 2008;135:3871–3879. doi: 10.1242/dev.025767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard SA, Yu L, Gu S, Zhang Z, Chen YP. Regional regulation of palatal growth and patterning along the anterior-posterior axis in mice. J Anat. 2005;207:655–667. doi: 10.1111/j.1469-7580.2005.00474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Helms JA. The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis. Development. 1999;126:4873–4884. doi: 10.1242/dev.126.21.4873. [DOI] [PubMed] [Google Scholar]
- Hu D, Marcucio RS. A SHH-responsive signaling center in the forebrain regulates craniofacial morphogenesis via the facial ectoderm. Development. 2009a;136:107–116. doi: 10.1242/dev.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Marcucio RS. Unique organization of the frontonasal ectodermal zone in birds and mammals. Dev Biol. 2009b;325:200–210. doi: 10.1016/j.ydbio.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Marcucio RS, Helms JA. A zone of frontonasal ectoderm regulates patterning and growth in the face. Development. 2003;130:1749–1758. doi: 10.1242/dev.00397. [DOI] [PubMed] [Google Scholar]
- Ibrahimi OA, Eliseenkova AV, Plotnikov AN, Yu K, Ornitz DM, Mohammadi M. Structural basis for fibroblast growth factor receptor 2 activation in Apert syndrome. Proc Natl Acad Sci U S A. 2001;98:7182–7187. doi: 10.1073/pnas.121183798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 2004;18:937–951. doi: 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernvall J, Aberg T, Kettunen P, Keranen S, Thesleff I. The life history of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development. 1998;125:161–169. doi: 10.1242/dev.125.2.161. [DOI] [PubMed] [Google Scholar]
- Jin JZ, Ding J. Analysis of Meox-2 mutant mice reveals a novel postfusion-based cleft palate. Dev Dyn. 2006;235:539–546. doi: 10.1002/dvdy.20641. [DOI] [PubMed] [Google Scholar]
- Jin JZ, Li Q, Higashi Y, Darling DS, Ding J. Analysis of Zfhx1a mutant mice reveals palatal shelf contact-independent medial edge epithelial differentiation during palate fusion. Cell Tissue Res. 2008;333:29–38. doi: 10.1007/s00441-008-0612-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassai Y, Munne P, Hotta Y, Penttila E, Kavanagh K, Ohbayashi N, Takada S, Thesleff I, Jernvall J, Itoh N. Regulation of mammalian tooth cusp patterning by ectodin. Science. 2005;309:2067–2070. doi: 10.1126/science.1116848. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Rice DP, Kettunen PJ, Thesleff I. FGF-, BMP- and Shh-mediated signalling pathways in the regulation of cranial suture morphogenesis and calvarial bone development. Development. 1998;125:1241–1251. doi: 10.1242/dev.125.7.1241. [DOI] [PubMed] [Google Scholar]
- Lan Y, Jiang R. Sonic hedgehog signaling regulates reciprocal epithelial-mesenchymal interactions controlling palatal outgrowth. Development. 2009;136:1387–1396. doi: 10.1242/dev.028167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Ovitt CE, Cho ES, Maltby KM, Wang Q, Jiang R. Odd-skipped related 2 (Osr2) encodes a key intrinsic regulator of secondary palate growth and morphogenesis. Development. 2004;131:3207–3216. doi: 10.1242/dev.01175. [DOI] [PubMed] [Google Scholar]
- Laurikkala J, Kassai Y, Pakkasjarvi L, Thesleff I, Itoh N. Identification of a secreted BMP antagonist, ectodin, integrating BMP, FGF, and SHH signals from the tooth enamel knot. Dev Biol. 2003;264:91–105. doi: 10.1016/j.ydbio.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Laurikkala J, Mikkola ML, James M, Tummers M, Mills AA, Thesleff I. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development. 2006;133:1553–1563. doi: 10.1242/dev.02325. [DOI] [PubMed] [Google Scholar]
- Lee JM, Kim JY, Cho KW, Lee MJ, Cho SW, Zhang Y, Byun SK, Yi CK, Jung HS. Modulation of cell proliferation during palatogenesis by the interplay between Tbx3 and Bmp4. Cell Tissue Res. 2007;327:285–292. doi: 10.1007/s00441-006-0271-8. [DOI] [PubMed] [Google Scholar]
- Lee SH, Bedard O, Buchtova M, Fu K, Richman JM. A new origin for the maxillary jaw. Dev Biol. 2004;276:207–224. doi: 10.1016/j.ydbio.2004.08.045. [DOI] [PubMed] [Google Scholar]
- Li Q, Ding J. Gene expression analysis reveals that formation of the mouse anterior secondary palate involves recruitment of cells from the posterior side. Int J Dev Biol. 2007;51:167–172. doi: 10.1387/ijdb.062212ql. [DOI] [PubMed] [Google Scholar]
- Liu W, Lan Y, Pauws E, Meester-Smoor MA, Stanier P, Zwarthoff EC, Jiang R. The Mn1 transcription factor acts upstream of Tbx22 and preferentially regulates posterior palate growth in mice. Development. 2008;135:3959–3968. doi: 10.1242/dev.025304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Selever J, Lu MF, Martin JF. Genetic dissection of Pitx2 in craniofacial development uncovers new functions in branchial arch morphogenesis, late aspects of tooth morphogenesis and cell migration. Development. 2003;130:6375–6385. doi: 10.1242/dev.00849. [DOI] [PubMed] [Google Scholar]
- Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, Martin JF. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–1461. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- Marcucio RS, Cordero DR, Hu D, Helms JA. Molecular interactions coordinating the development of the forebrain and face. Dev Biol. 2005;284:48–61. doi: 10.1016/j.ydbio.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Mikkola ML. p63 in skin appendage development. Cell Cycle. 2007;6:285–290. doi: 10.4161/cc.6.3.3798. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Murakami H, Okawa A, Yoshida H, Nishikawa S, Moriya H, Koseki H. Elbow knee synostosis (Eks): a new mutation on mouse Chromosome 14. Mamm Genome. 2002;13:341–344. doi: 10.1007/s00335-001-2143-6. [DOI] [PubMed] [Google Scholar]
- Nguyen BC, Lefort K, Mandinova A, Antonini D, Devgan V, Della Gatta G, Koster MI, Zhang Z, Wang J, Tommasi di Vignano A, Kitajewski J, Chiorino G, Roop DR, Missero C, Dotto GP. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006;20:1028–1042. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noden DM. The role of the neural crest in patterning of avian cranial skeletal, connective, and muscle tissues. Dev Biol. 1983;96:144–165. doi: 10.1016/0012-1606(83)90318-4. [DOI] [PubMed] [Google Scholar]
- Ohazama A, Johnson EB, Ota MS, Choi HY, Porntaveetus T, Oommen S, Itoh N, Eto K, Gritli-Linde A, Herz J, Sharpe PT. Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS One. 2008;3:e4092. doi: 10.1371/journal.pone.0004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama R, Ogawa E, Nagoshi H, Yabuki M, Kurihara A, Terui T, Aiba S, Obinata M, Tagami H, Ikawa S. p53 homologue, p51/p63, maintains the immaturity of keratinocyte stem cells by inhibiting Notch1 activity. Oncogene. 2007;26:4478–4488. doi: 10.1038/sj.onc.1210235. [DOI] [PubMed] [Google Scholar]
- Pantalacci S, Prochazka J, Martin A, Rothova M, Lambert A, Bernard L, Charles C, Viriot L, Peterkova R, Laudet V. Patterning of palatal rugae through sequential addition reveals an anterior/posterior boundary in palatal development. BMC Dev Biol. 2008;8:116. doi: 10.1186/1471-213X-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterkova R. The common developmental origin and phylogenetic aspects of teeth, rugae palatinae, and fornix vestibuli oris in the mouse. J Craniofac Genet Dev Biol. 1985;5:89–104. [PubMed] [Google Scholar]
- Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Dev Biol. 2003;262:195–205. doi: 10.1016/s0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- Rice R, Connor E, Rice DP. Expression patterns of Hedgehog signalling pathway members during mouse palate development. Gene Expr Patterns. 2006;6:206–212. doi: 10.1016/j.modgep.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, Thesleff I, Rice DP. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J Clin Invest. 2004;113:1692–1700. doi: 10.1172/JCI20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagati F, Rijli FM. Cranial neural crest and the building of the vertebrate head. Nat Rev Neurosci. 2003;4:806–818. doi: 10.1038/nrn1221. [DOI] [PubMed] [Google Scholar]
- Schneider RA, Helms JA. The cellular and molecular origins of beak morphology. Science. 2003;299:565–568. doi: 10.1126/science.1077827. [DOI] [PubMed] [Google Scholar]
- Schneider RA, Hu D, Helms JA. From head to toe: conservation of molecular signals regulating limb and craniofacial morphogenesis. Cell Tissue Res. 1999;296:103–109. doi: 10.1007/s004410051271. [DOI] [PubMed] [Google Scholar]
- Schneider RA, Hu D, Rubenstein JL, Maden M, Helms JA. Local retinoid signaling coordinates forebrain and facial morphogenesis by maintaining FGF8 and SHH. Development. 2001;128:2755–2767. doi: 10.1242/dev.128.14.2755. [DOI] [PubMed] [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S. Fgf10 is essential for limb and lung formation. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- Shigetani Y, Nobusada Y, Kuratani S. Ectodermally derived FGF8 defines the maxillomandibular region in the early chick embryo: epithelial-mesenchymal interactions in the specification of the craniofacial ectomesenchyme. Dev Biol. 2000;228:73–85. doi: 10.1006/dbio.2000.9932. [DOI] [PubMed] [Google Scholar]
- Stanier P, Moore GE. Genetics of cleft lip and palate: syndromic genes contribute to the incidence of non-syndromic clefts. Hum Mol Genet. 2004;13(Spec No 1):R73–R81. doi: 10.1093/hmg/ddh052. [DOI] [PubMed] [Google Scholar]
- Sun X, Lewandoski M, Meyers EN, Liu YH, Maxson RE, Jr, Martin GR. Conditional inactivation of Fgf4 reveals complexity of signalling during limb bud development. Nat Genet. 2000;25:83–86. doi: 10.1038/75644. [DOI] [PubMed] [Google Scholar]
- Szabo-Rogers HL, Geetha-Loganathan P, Nimmagadda S, Fu KK, Richman JM. FGF signals from the nasal pit are necessary for normal facial morphogenesis. Dev Biol. 2008;318:289–302. doi: 10.1016/j.ydbio.2008.03.027. [DOI] [PubMed] [Google Scholar]
- Tamarin A. The formation of the primitive choanae and the junction of the primary and secondary palates in the mouse. Am J Anat. 1982;165:319–337. doi: 10.1002/aja.1001650308. [DOI] [PubMed] [Google Scholar]
- Tao H, Yoshimoto Y, Yoshioka H, Nohno T, Noji S, Ohuchi H. FGF10 is a mesenchymally derived stimulator for epidermal development in the chick embryonic skin. Mech Dev. 2002;116:39–49. doi: 10.1016/s0925-4773(02)00131-4. [DOI] [PubMed] [Google Scholar]
- Thomason HA, Dixon MJ, Dixon J. Facial clefting in Tp63 deficient mice results from altered Bmp4, Fgf8 and Shh signaling. Dev Biol. 2008;321:273–282. doi: 10.1016/j.ydbio.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler MS, Koch WE. In vitro development of palatal tissues from embryonic mice. III. Interactions between palatal epithelium and heterotypic oral mesenchyme. J Embryol Exp Morphol. 1977;38:37–48. [PubMed] [Google Scholar]
- Vigano MA, Lamartine J, Testoni B, Merico D, Alotto D, Castagnoli C, Robert A, Candi E, Melino G, Gidrol X, Mantovani R. New p63 targets in keratinocytes identified by a genome-wide approach. EMBO J. 2006;25:5105–5116. doi: 10.1038/sj.emboj.7601375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh IC, Hagge-Greenberg A, O’Brien TP. A dosage-dependent role for Spry2 in growth and patterning during palate development. Mech Dev. 2007;124:746–761. doi: 10.1016/j.mod.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Nomoto S, Hoque MO, Dracheva T, Osada M, Lee CC, Dong SM, Guo Z, Benoit N, Cohen Y, Rechthand P, Califano J, Moon CS, Ratovitski E, Jen J, Sidransky D, Trink B. DeltaNp63alpha and TAp63alpha regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 2003;63:2351–2357. [PubMed] [Google Scholar]
- Wu P, Jiang TX, Shen JY, Widelitz RB, Chuong CM. Morphoregulation of avian beaks: Comparative mapping of growth zone activities and morphological evolution. Developmental Dynamics. 2006a;235:1400–1412. doi: 10.1002/dvdy.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Jiang TX, Shen JY, Widelitz RB, Chuong CM. Morphoregulation of avian beaks: comparative mapping of growth zone activities and morphological evolution. Dev Dyn. 2006b;235:1400–1412. doi: 10.1002/dvdy.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi C, Yamagishi H, Maeda J, Tsuchihashi T, Ivey K, Hu T, Srivastava D. Sonic hedgehog is essential for first pharyngeal arch development. Pediatr Res. 2006;59:349–354. doi: 10.1203/01.pdr.0000199911.17287.3e. [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yu L, Gu S, Alappat S, Song Y, Yan M, Zhang X, Zhang G, Jiang Y, Zhang Z, Zhang Y, Chen Y. Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development. 2005;132:4397–4406. doi: 10.1242/dev.02013. [DOI] [PubMed] [Google Scholar]
- Zhang X, Stappenbeck TS, White AC, Lavine KJ, Gordon JI, Ornitz DM. Reciprocal epithelial-mesenchymal FGF signaling is required for cecal development. Development. 2006;133:173–180. doi: 10.1242/dev.02175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen Y. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development. 2002;129:4135–4146. doi: 10.1242/dev.129.17.4135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.