Abstract
Phagocytosis is an important process for the removal of apoptotic cells or cellular debris. Eat-me signals control the initiation of phagocytosis and hold the key for in-depth understanding of its molecular mechanisms. However, because of difficulties to identify unknown eat-me signals, only a limited number of them have been identified and characterized. Using a newly-developed functional cloning strategy of open-reading-frame (ORF) phage display, we identified 9 putative eat-me signals, including tubby-like protein 1 (Tulp1). This further led to the elucidation of tubby as the second eat-me signal in the same protein family. Both proteins stimulated phagocytosis of retinal pigment epithelium (RPE) cells and macrophages. Tubby-conjugated fluorescent microbeads facilitated RPE phagocytosis. Tubby and Tulp1, but not other family members, enhanced the uptake of membrane vesicles by RPE cells in synergy. Retinal membrane vesicles of Tubby mice and Tulp1−/− mice showed reduced activities for RPE phagocytosis, which were compensated by purified tubby and Tulp1, respectively. These data reveal a novel activity of tubby and Tulp1, and demonstrate that unbiased identification of eatme signals by the broadly applicable strategy of ORF phage display can provide detailed insights into phagocyte biology.
Keywords: Eat-me signal, phagocytosis, tubby, Tulp1, phage display, retinal pigment epithelium
Introduction
Phagocytosis of apoptotic cells and cellular debris, also called engulfment, is critical for many biological processes, including cell homeostasis, morphogenesis, tissue injury repair, immune tolerance and resolution of inflammation [1, 2]. However, the molecular mechanisms of phagocytosis have only been limitedly defined. “Eat-me” signals or “phagocytosis stimulating molecules” control the initiation of phagocytosis and hold a key to phagocyte biology. In contrast to phosphatidylserine as a well-characterized eat-me signal, much less is known about protein ligands [3]. Although several protein ligands, such as growth arrest-specific gene 6 (Gas6), protein S, milk fat globule epidermal growth factor 8 (MFG-E8), have been identified [4], it is unknown how many more eat-me signals are yet to be identified.
Eat-me signals are traditionally identified on a case-by-case basis with daunting challenges. A live screen of accumulated cell corpses in Caenorhabditis elegans by optics microscopy was described ~26 years ago to identify chemically-induced random genetic mutations critical for phagocytosis [5]. However, this strategy was more designed to identify intracellular signaling molecules, rather than eat-me signals [1]. Furthermore, it is not applicable to organisms that are not transparent for live screening by optics microscopy.
Previous studies showed that phage display with antibody (Ab) libraries can identify Abs with internalization capacity in cancer cells [6, 7]. The question is whether this strategy can be used to identify unknown eat-me signals. We recently demonstrated that T7 phage displaying MEG-E8 or Gas6, two well-characterized eat-me signals, was preferentially phagocytosed and enriched by retinal pigment epithelium (RPE) cells or macrophages, but not by non-phagocytes [8], suggesting that the strategy of phagocytosis-based phage selection is feasible for functional cloning of unknown eat-me signals. However, unlike Ab libraries, cDNA repertoires with unpredictable reading frames and stop codons may not be expressed in correct frames as phage capsid fusion proteins [9]. Consequently, more than 90% of bait-binding clones identified from conventional phage display cDNA libraries encoded out-of-frame unnatural short peptides with minimal implications in protein networks [10, 11]. To overcome this challenge, we have recently engineered open-reading-frame (ORF) cDNA phage display libraries with most identified clones encoding real proteins [12, 13].
In this study, we describe the first case of unbiased identification of eat-me signals by phage display. Among 9 putative eat-me signals identified was tubby-like protein 1 (Tulp1), which in turn led to the identification of tubby as the second eat-me signal in the same protein family.
Materials and methods
Cell lines and culture conditions
Human ARPE19 RPE cells were cultured as previously described [8]. Rat RPE-J and human D407 RPE cell lines, gifts from Dr. G. Hoppe (Cleveland Clinic, Ohio), were cultured in Dulbecco's modified essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 2 mM L-glutamine. Mouse primary RPE cells were isolated from postnatal day 10 mice and cultured as previously described [14]. Culture conditions for Neuro-2a, J774 and HeLa cells were described previously [8].
Phage selection
An ORF phage display cDNA library was generated from adult mouse eyes as previously described [12, 13]. Phage selection for eat-me signals was carried out with ARPE19 cells as previously described [8]. After 4 rounds of selection, enriched phages were selected twice with immobilized streptavidin for ORF phage clones. Total phagocytosed phages at each round were quantified by plaque assay. Phage lysates at each round were analyzed by PCR using T7-F (5′-CCAAGCGGACCAGATTATCG-3′) and T7-R (5′-GAGCGCATATAGTTC CTCCT-3′) primers to monitor phage enrichment. Individual phage clones were randomly picked from phage plates, amplified, and re-analyzed for their uptake by ARPE19 cells in 12-well plates. The cDNA inserts of positive clones were amplified by PCR with T7-F and T7-R primers, and identified by sequencing. All procedures for T7 phage display, including ORF phage library construction, phage packaging, phage plaque assay, phage liquid amplification and plating with BLT5615 E. coli, were previously described [8, 12, 13].
Clonal phage construction
Full-length coding sequence for tubby was generated by reverse transcription-PCR (RTPCR) from mouse eye with primers 5′-TAGCGGCCGCTAATGACTTCCAAGCCGCATTC-3′ and 5′-AGCTCGAGCTCGCAGGCCAGCTTGCTGTC-3′ (underlined sequences for NotI and XhoI sites), digested with NotI and XhoI, ligated into T7Bio3C vector [12] at the same sites, and packaged into T7 phage to generate tubby-phage, as previously described [8]. Tulp1-phage was generated in a similar manner. Control phage had no cDNA insert [8]. All phages were verified by sequencing.
Plasmid construction
Full-length coding sequences for tubby, Tulp1, 2 and 3 (Tulps) were generated from mouse eye or testis by RT-PCR. The cDNA for mouse tubby IVS11+1G→T splice site mutation with extra 22 amino acids (aa) to replace its C-terminal 44 aa [15, 16] was generated by PCR using overlapping primers. The sequence of Intron 14 for IVS14+1G→A mutation in human Tulp1 (hTulp1) [16, 17] was generated by PCR. These DNA fragments were fused to truncated tubby at (Asp461 or hTulp1 at Asp498 by PCR with overlapping primers. Other missense mutations in human Tulp1 were generated by PCR using overlapping primers with mutations embedded in primers appropriately. All full-length Tulps and their mutants with deletion or missense mutation were cloned into pcDNA3 (Invitrogen, Carlsbad, CA) with an N-terminal FLAG tag. All plasmids were verified by DNA sequencing. All plasmids for FLAG-Tulps were further verified by Western blot using anti-FLAG M2 monoclonal antibody (mAb) (Sigma, St. Louis, MO).
A membrane-targeting epitope derived from protein GAP-43 was generated by PCR with overlapping primers [18], digested with BglII and BamHI, and fused to the N-terminus of green fluorescent protein (GFP) in pEGFP-N1 plasmid (Clontech, Mountain View, CA) at BamHI site to yield membrane-targeted GFP (mGFP) plasmid. GFP-FLAG was generated by fusing FLAG tag to the C-terminus of GFP in pEGFP-N1 plasmid using a PCR strategy with overlapping primers.
Microbead phagocytosis assay
Glutathione S-transferase-tubby (GST-tubby) and GST-Tulp3 fusion proteins were expressed in BL21(DE3) bacteria, and purified as previously described [19]. After dialysis, purified proteins were conjugated to carboxylate-modified yellow-green fluorescent microbeads (Invitrogen) according to the manufacturer's protocol. Ligand-conjugated microbeads were incubated with ARPE19 cells in 2-chamber slides (Nunc, Rochester, NY) (~300 μg beads with ~15 μg conjugated protein per chamber) at 37°C for 3 h, washed, and incubated at 37°C for additional 15 h in 293 SFM II medium (Invitrogen) for phagocytosis. After washing, phagocytosed fluorescent microbeads were analyzed by Leica TCS SP5 confocal microscopy with diode laser for excitation at 405 nm and emission at 480 nm for 4′,6-diamidino-2-phenylindole (DAPI) and argon laser for excitation at 488 nm and emission at 510-570 nm for FITC. All data for fluorescence-labeled phagocytosis were derived from the middle sections of ARPE19 cells by confocal microscopy with DAPI staining. Relative fluorescence intensity per cell was measured using Leica Application Suite software by manually tracing the outline of individual phagocytes under bright field channel with the corresponding fluorescence quantified in the FITC channel. More than 100 cells per group were analyzed.
Membrane vesicle phagocytosis assay
Neuro-2a cells at 80% confluence were co-transfected with two plasmids, mGFP plasmid and Tulp-expressing plasmid, by calcium phosphate method. The plasma membrane was prepared at 48 h post-transfection by sucrose gradient centrifugation, as previously described [20]. Briefly, co-transfected cells were scraped off from culture plates, homogenized and purified by sucrose gradient centrifugation at 105,000 × g for 1 h. Plasma membrane vesicles at the interface of 20% and 27% sucrose were collected, washed twice with phosphate buffered saline (PBS) and resuspended in PBS. Purified plasma membrane vesicles were incubated with ARPE19 cells in the chamber slides (50 μg membrane protein/ml/chamber) and analyzed for phagocytosis as described above.
Apoptotic cell phagocytosis
Jurkat cells were labeled with 0.5 pM of 5(6)-carboxyfluorescein diacetate N-succinimidyl ester (CFSE) (Sigma) for 10 min at 37°C, stopped with 2% FBS in PBS, washed, induced for apoptosis with etoposide and verified by annexin V/propidium iodide staining, as previously described [21]. Small amount of GST-Tulp1 was purified from soluble E. coli lysate as described above. Labeled apoptotic cells were incubated with J774 macrophage cells in the presence of GST-tubby, GST-Tulp1 and GST control (50 nM each) for 3 h. Phagocytosis was analyzed by confocal microscopy. Moreover, macrophage cells with phagocytosed apoptotic cells were labeled with APC/Cy7-conjugated anti-mouse CD11b mAb (Biolegend, San Diego, CA) and quantified by flow cytometry. All the cells were gated with the forward scatter and side scatter to eliminate possible surface-bound apoptotic cells.
Tubby and Tulp1 knockout mice
Animal procedures were approved by University of Miami Institutional Animal Care and Use Committee. C57BL/6 mice and the breeder pair of tubby knockout mice on the same genetic background were purchased from the Jackson Laboratory (Bar Harbor, ME). Tulp1−/− mice were gifts from Dr. Patsy Nishina (The Jackson Laboratory). Genotyping was carried out by PCR using gene-specific primers [22]. All neonatal mice were raised in the dark to slowdown photoreceptor degeneration in homozygotes. Eyes were enucleated from euthanized homozygous mice at postnatal day 19. Retinas were collected and homogenized for the preparation of membrane vesicles as described above. Purified membrane vesicles were labeled with CFSE as described above. After washing, the labeled membrane vesicles were analyzed for phagocytosis in ARPE19 cells as described above in the presence or absence of indicated FLAG-tagged recombinant proteins (10 μg/ml). Recombinant adenoviruses expressing FLAG-tagged recombinant tubby, Tulp1 and GFP were constructed as previously described [23]. The recombinant proteins were purified with anti-FLAG mAb affinity columns (Sigma), eluted with FLAG peptide and dialyzed against PBS according to the manufacturer's protocol.
Tubby association with photoreceptor outer segments (POS) vesicles
Fresh retinas were dissected from euthanized pigs, vortexed gently in PBS with 20% sucrose to detach POS. Porcine POS were purified by sucrose gradient centrifugation as described above, washed and analyzed by Western blot using anti-tubby (T-19) Ab (Santa Cruz Biotechnology, Santa Cruz, CA) or anti-β-actin mAb.
Data analysis
All experiments were repeated independently at least 3 times and analyzed by Student's t-test.
Results
Functional cloning of eat-me signals
Putative eat-me signals were functionally identified by ORF phage display, as depicted in Fig. 1A. The ORF library (~1 × 1011 pfu) was incubated with human ARPE19 cells at 4°C without phagocytosis. After washing, the cells were incubated at 37°C for 30 min to phagocytose the surface-bound phages. Unphagocytosed phages were removed from cell surface by a low pH isotonic stripping buffer. The internalized phages were released by cell lysis, amplified in bacteria and used as input for the next round of phage selection. After 4 rounds of selection, enriched phages showed more than 20-fold increase in internalization activity (Fig. 1B). Phage enrichment was further monitored by PCR analysis of cDNA inserts with several dominant PCR bands emerged at Round 3, indicating the enrichment of specific phage clones (Fig. 1C). A total of 192 individual phage clones were randomly picked from phage plates, amplified, analyzed for their internalization capacity in ARPE19 cells (Fig. 1D), and identified by DNA sequencing. A total of 9 putative eat-me signals were identified, including Tulp1 (Table 1).
Fig. 1.
Functional cloning of eat-me signals by ORF phage display. (A) Phage selection scheme. ARPE19 cells were incubated with the library at 4°C without phagocytosis, washed and incubated at 37°C for 30 min for phagocytosis. After stripping off unphagocytosed surface-bound phages, internalized phages were released by cell lysis, amplified and used as input for the next round of selection. A total of 5 rounds of selection were performed. (B) Total phagocytosed phages were quantified by plaque assay. Control phage had no cDNA insert. (C) Phage enrichment was analyzed by PCR with T7-F and T7-R primers. As a negative control, the unselected library with cDNA inserts in various sizes appeared as diffused PCR products. (D) Screening of individual phage clones. A total of 192 phage clones were randomly picked from phage plates at Round 4, amplified and analyzed for their internalization activity as in (B). Only the first 22 phage clones are shown.
Table 1.
Identified putative eat-me signals for RPE phagocytosis. Phagocytosis index = (total phagocytosed clonal phage)/(total phagocytosed control phage).
| Phage clone |
Protein name |
Accession number |
Matched aa residues |
Phagocytosis index |
|---|---|---|---|---|
| PH013 | ATP-binding cassette, sub-family F (GCN20), member 1 (Abcf1) | NM_013854 | 29K-176G | ~46X |
| PH019 | Ly1 antibody reactive clone (LyAR) | NM_025281 | 100Q-220R | ~25X |
| PH054 | Tubby-like protein 1 (Tulp1) | NM_021478 | 79A-199T | ~22X |
| PH140 | Max protein (Max) | NM_008558 | 35R-154K | ~31X |
| PH137 | Predicted gene, EG666160 (EG666160) | XR 033995 | 575L-701P | ~25X |
| PH138 | UPF3 regulator of nonsense transcripts homolog A (Upf3a) | NM_025924 | 173P-257E | ~22X |
| PH213 | Growth arrest specific 6 (Gas6) | NM_039455 | 43Q-143D | ~14X |
| PH227 | Coiled-coil domain containing 55 (Ccdc55) | NM_001012309 | 223E-365E | ~11X |
| PH228 | SPRY domain containing 3 (Spryd3) | NM_001033277 | 31I-215S | ~15X |
Tubby as the second eat-me signals in the family
Tulp1 belongs to a well-characterized tubby protein family with 4 members (tubby, Tulp1, 2 and 3), which share a highly conserved C-terminal “tubby domain” of ~260 aa [24, 25]. Humans with Tulp1 mutations and Tulp1−/− mice develop autosomal recessive retinal degeneration [16, 26], and Tubby mice with a spontaneous autosomal recessive mutation in the tubby gene manifest adult-onset obesity, progressive retinal and cochlear degeneration [15]. Pathological mechanisms for both proteins are undefined.
We chose to further characterize Tulp1 because of its role in retinal degeneration. The question is whether tubby in the same family with a similar phenotype in retinal degeneration is also capable of stimulating RPE phagocytosis. We constructed clonal tubby-phage and Tulp1-phage expressing the full-length proteins, and analyzed the phage uptake by RPE cells. Similar to Tulp1, tubby also facilitated phage internalization in ARPE19 cells (Fig. 2A). To independently verify the finding, we expressed GST fusion proteins of all 4 Tulps in E. coli. While GST-Tulp1 and GST-Tulp2 were minimally expressed in cell lysates possibly due to formation of inclusion body, GST-tubby and GST-Tulp3 were successfully expressed, purified and covalently conjugated to fluorescent microbeads (~2 μm in diameter). RPE phagocytosis analysis showed that the conjugated GST-tubby, but not GST-Tulp3 or GST control, facilitated microbead uptake by RPE cells (Fig. 2B-C). Most phagocytosed microbeads were ~1-3 μm in diameter, suggesting they were internalized individually, rather than as aggregated forms. Superimposition of intracellular confocal image and cognate bright field image revealed that most signals for GST-tubby-conjugated microbeads were intracellular, rather than surface-bound, suggesting that the microbeads were phagocytosed by RPE cells.
Fig. 2.
Tubby stimulates RPE phagocytosis. (A) Uptake of tubby-phage and Tulp1-phage by RPE cells. Tulp179-199, full-length tubby and Tulp1 displaying on the surface of clonal phages facilitated phage uptake by ARPE19 cells. The phagocytosis index is the ratio of phagocytosed clonal phage (pfu) vs. phagocytosed control phage (± SEM, n=3, * p<0.001, vs. control phage). (B) Phagocytosis of tubby-conjugated fluorescent microbeads by RPE cells. Purified GST-tubby, GST-Tulp3 and GST control were covalently conjugated to fluorescent microbeads (~2 μm in diameter) and analyzed for their phagocytosis in ARPE19 cells. Phagocytosed microbeads (green signals) were analyzed by confocal microscopy. Nuclei were stained with DAPI (blue signals). Bottom 3 panels are intracellular Z-stack confocal images superimposed with the cognate bright field image to illustrate phagocytosed microbeads. The superimposed image of Tulp3 is not shown because of few phagocytosed signals. Bar = 10 μm. (C) Relative fluorescence intensity per cells in (B) was quantified with more than 100 cells per group (± SEM, n>100, *p<0.001, vs. GST).
Independent verification of tubby and Tulp1 as eat-me signals
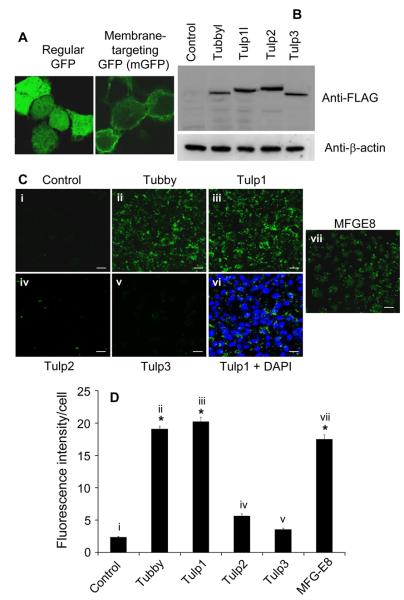
To further verify tubby and Tulp1 as eat-me signals, we analyzed the phagocytosis of membrane vesicles by RPE cells in the presence or absence of tubby or Tulp1. Instead of preparing membrane vesicles from the retina, we prepared membrane vesicles from Neuro-2a cells. mGFP (Fig. 3A) was co-expressed in Neuro-2a cells with one of the Tulps. All Tulps were expressed at a similar level (Fig. 3B). mGFP-labeled plasma membrane vesicles were prepared from the cells and analyzed for phagocytosis in ARPE19 cells. The results showed that Tulp1-expressing membrane vesicles (Tulp1-vesicles) and tubby-vesicles triggered robust phagocytosis in the RPE cells (Fig. 3C-D). However, Tulp2-vesicles, Tulp3-vesicles and control vesicles elicited minimal phagocytosis. Most internalized membrane vesicles were ~1-4 μm in size. Similar results with membrane vesicles prepared from HEK293 cells expressing FLAG-Tulps were obtained (not shown).
Fig. 3.
Tubby and Tulp1 facilitate membrane vesicle phagocytosis by RPE cells. (A) Comparison of regular GFP and mGFP. GFP and mGFP were expressed in Neuro-2a cells and analyzed for their subcellular distribution by confocal microscopy. (B) Expression level of FLAG-tagged Tulps. FLAG-tagged Tulps were expressed in Neuro2a cells and analyzed by Western blot using anti-FLAG mAb (~9 × 104 cells/lane). Western blot using anti-β-actin Ab is included as loading controls. (C) Plasma membrane vesicles were prepared from Neuro-2a cells co-expressing mGFP and the indicated protein, and incubated with ARPE19 cells. Phagocytosed fluorescent signals were analyzed by confocal microscopy. vi is identical to iii but with DAPI nuclear staining. For better visual perception, DAPI signals are not shown in all other figures. MFG-E8 (50 nM, R&D Systems) was added to control vesicles as a positive control. Bar = 10 μm. (D) Relative fluorescence intensity per cell in (C) was quantified with more than 100 cells per group (± SEM, n>100, * p<0.001, vs. control).
Tubby mice develop autosomal recessive retinal degeneration due to IVS11+1G→T mutation, leading to mRNA alternative splicing and replacement of the C-terminal 44-aa chain with a 22-aa polypeptide derived from Intron 11 (Tubby-ΔC44) [16]. The important role of tubby in RPE phagocytosis was supported by the significant reduction in the phagocytosis of fluorescence-labeled membrane vesicles prepared from the retina of Tubby mice (Fig. 4A-B). Retinal membrane vesicles prepared from Tulp1−/− mice, which also manifest autosomal recessive retinal degeneration, had similar reduction in RPE phagocytosis (Fig. 4A-B). Purified recombinant tubby and Tulp1 were able to rescue the phagocytosis, suggesting that both proteins facilitate RPE phagocytosis. Association of tubby with POS vesicles was demonstrated with purified pig POS (Fig. 4C).
Fig. 4.
Tubby and Tulp1 facilitate POS vesicle phagocytosis. (A) Reduced RPE phagocytosis for the retinal membrane vesicles of Tubby mice or Tulp1−/− mice. Membrane vesicles were prepared from the retinas of wild-type, Tubby mice or Tulp1−/− mice at postnatal day 19, labeled with CFSE, analyzed and quantified for RPE phagocytosis as in Fig. 3C-D in the presence or absence of purified FLAG-tagged proteins. Recombinant GFP-FLAG was included as a negative control. Bar = 10 μm. (B) Relative fluorescence intensity per cell in (A) was quantified with more than 100 cells per group (± SEM, n>100). (C) Tubby association with purified POS vesicles. POS vesicles (~30 μg protein/lane) were purified and analyzed by Western blot using anti-tubby Ab. The homogenate of total pig retina was included as a control. The lysates of Neuro-2a cells expressing FLAG-tubby and FLAG-Tulp1 were included as positive and negative controls. Western blot using anti-β-actin Ab was included as loading controls.
Defective RPE phagocytosis and mutations in tubby and Tulp1
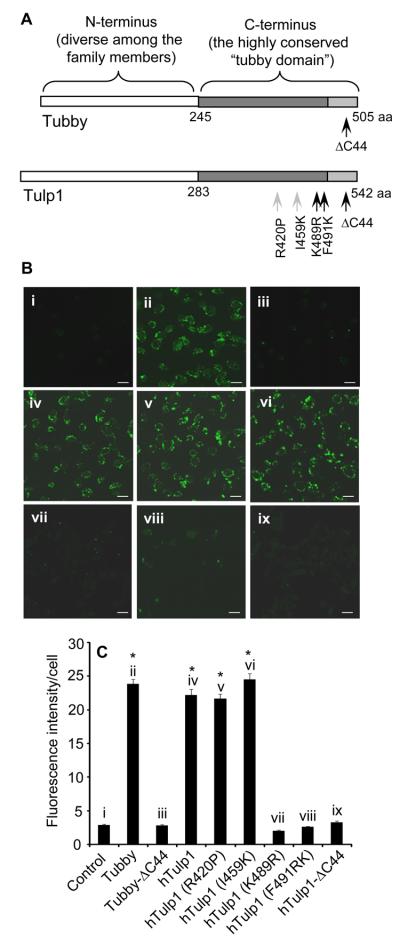
Similar to the mutation in Tubby mice, IVS14+1G→A mutation in hTulp1 is associated with retinal degeneration [16, 17], presumably affecting mRNA splicing by deleting the C-terminal 44 aa in a similar fashion (Tulp1-ΔC44 with 499P-542E deletion). We constructed plasmids expressing Tubby-ΔC44 and Tulp1-ΔC44. Both mutants failed to stimulate membrane vesicle phagocytosis by RPE cells (Fig. 5). Moreover, several missense mutations in hTulp1, including R420P, I459K, K489R or F491L, were reported to associate with retinal degeneration [16, 27]. While R420P or I459K mutation had no effect on RPE phagocytosis, K489R or F491L mutation near the C-terminal end of hTulp1 abolished its stimulation of RPE phagocytosis (Fig. 5).
Fig. 5.
The effect of tubby or Tulp1 mutations on RPE phagocytosis. (A) Illustration of tubby and Tulp1 mutations analyzed for RPE phagocytosis. Mutations indicated by black arrows reduced RPE phagocytosis, whereas mutations with grey arrows had no effect. (B) FLAG-tagged tubby and Tulp1 mutants were expressed in Neuro-2a cells. mGFP-labeled plasma membrane vesicles were prepared and analyzed for RPE phagocytosis as in Fig. 3C. Bar = 10 μm. (C) Relative fluorescence intensity per cell in (A) was quantified with more than 100 cells per group (± SEM, n>100, * p<0.001, vs. control).
Tubby and Tulp1 stimulate RPE phagocytosis in synergy
To elucidate intracellular and extracellular functions of Tulps, we recently identified 16 new tubby-binding proteins by ORF phage display [12]. A serendipitous finding was Tulp1 as a tubby-binding protein. Tubby-Tulp1 interaction was further verified by yeast two-hybrid assay [12] and protein pull-down assay (Fig. 6A), suggesting that Tulp1 and tubby form a heterodimer or heterooligomer. Their interaction was functionally revealed by their synergistic stimulation of RPE phagocytosis by mixing two membrane vesicles expressing individual proteins (Fig. 6B-C). However, Tubby-ΔC44 failed to synergistically facilitate Tulp1-mediated phagocytosis, and vice versa for Tulp1-ΔC44.
Fig. 6.
Tubby and Tulp1 stimulate RPE phagocytosis in synergy. (A) Tubby specifically interacts with Tulp1. Purified GST-tubby was incubated with the cell lysates of FLAG-tagged Tulps or GFP-FLAG, pull-downed with glutathione resin, analyzed by Western blot using anti-FLAG mAb. (C) and (D) Tubby and Tulp1 synergistically stimulate RPE phagocytosis. Mixture of indicated membrane vesicles expressing tubby, Tulp1 or their mutants were analyzed for RPE phagocytosis and quantified as in Fig. 3C-D. Total amount of vesicles in each group was the same (50 μg membrane protein/ml/chamber) (±SEM, n>100, *p<0.001, iv vs. ii, all others vs. i).
Tubby and Tulp1 stimulate phagocytosis in other phagocytes
Since the molecular pathways of phagocytosis are highly conserved among different species and phagocytes, the question is whether tubby and Tulp1 are capable of stimulating other phagocytes like macrophages. The analysis of retinal membrane vesicles prepared from Tubby mice or Tulp1−/− mice showed that both proteins were necessary to facilitate J774 macrophage phagocytosis, but failed to stimulate the vesicle uptake by HeLa cells (Fig. 7A). However, because Tulp1 is specifically expressed in the photoreceptors, its importance for other phagocytes in extraocular tissues is yet to be defined. Similarly, Tulp1-phage was preferentially internalized by all RPE cells tested, including mouse primary RPE cells, ARPE19, D407 and RPE-J cell lines, but failed to be internalized by other epithelium cell lines, such as HeLa and MCF7 (Fig. 7B).
Fig. 7.
Tubby and Tulp1 specifically stimulate phagocytes, but not non-phagocytes. (A) Tubby and Tulp1 stimulate macrophage phagocytosis. Membrane vesicles were prepared from the retinas of wild-type, Tubby mice or Tulp1−/−, and analyzed for phagocytosis in J774 macrophage cell line, as in Fig. 4A. HeLa cells were included as a negative control. Bar = 10 μm. (B) Specific uptake of Tulp1-phage by all RPE cells tested, but not by other epithelial cells. Tulp1-phage was internalized by mouse RPE primary cells, ARPE19, RPE-J and D407 cell lines, but not by HeLa and MCF7 cells. HeLa and MCF7 are human cervix epithelial and mammary gland epithelial cell lines, respectively (± SEM, n=3, *p<0.05, **p<0.001, vs. control phage). (C) Tubby and Tulp1 facilitate macrophage phagocytosis of apoptotic cells. CFSE-labeled apoptotic Jurkat cells were incubated with J774 cells in the presence of GST-tubby, GST-Tulp1 or GST control (50 nM each) and analyzed for phagocytosis by confocal microscopy. The images of the intracellular CFSE and DAPI signals (Z-stack) were superimposed with the cognate bright fields to show the phagocytosed apoptotic cells. Bar = 10 μm (D) Flow cytometry analysis of macrophage phagocytosis. Macrophages with phagocytosed apoptotic Jurkat cells were labeled with anti-CD11b mAb and quantified by flow cytometry. The numbers on the upright corners indicate the percentage of macrophages with phagocytosed apoptotic cells.
Moreover, the capacity of tubby and Tulp1 to facilitate macrophage phagocytosis of apoptotic cells was analyzed by confocal microscopy and flow cytometry. Tubby and Tulp1 substantially stimulated the phagocytosis of CFSE-labeled apoptotic Jurkat cells by J774 cells in confocal analysis (Fig. 7C). Phagocytosed Jurkat cells were clearly visible inside the macrophages. The percentage of macrophages with phagocytosed Jurkat cells was quantified by flow cytometry with 84.0% for tubby, 89.5% for Tulp1 and 35.5% for control (Fig. 7D). These data suggest that both proteins facilitate phagocytosis in different phagocytes, but not in non-phagocytes.
Discussion
Identification of unknown eat-me signals has proven to be challenging. All the known eat-me signals, including Gas6, protein S and MFG-E8, were identified in macrophages on a case-by-case basis [3]. Subsequently, they were verified in RPE phagocytosis [28, 29]. We have recently characterized the feasibility of identifying eat-me signals by phage display with two well-characterized eat-me signals, Gas6 and MFG-E8, in RPE cells and macrophages [8]. Although phagocytosis and pinocytosis are often arbitrarily defined by the sizes of internalized particles [30], we believe that our mechanism-based studies with the well-characterized eat-me signals should be more reliable and relevant to demonstrate the feasibility of the strategy.
This study is to extend our recent finding by functionally cloning endogenous eat-me signals for RPE phagocytosis from a cDNA library by a recently-developed technology of ORF phage display with minimal reading frame issue [12]. ORF phage display is a versatile technology, applicable not only to protein-protein interactions but also to protein interactions with non-protein molecule like phosphatidylserine [12, 13]. Here we used RPE cells as multimolecular bait to functionally clone putative eat-me signals in the absence of knowledge of their phagocytic receptors or specific blocking reagents by ORF phage display. Unlike previous studies with phage display Ab libraries [6, 7], phage clones identified from the ORF cDNA library in this study encode real endogenous proteins with biological implications in protein networks. For example, identification of Tulp1 further led to the elucidation of tubby as the second eat-me signal in the same family.
Some well-characterized eat-me signals, such as MFG-E8, as well as tubby were failed to be identified by the functional cloning, possibly because of the following two reasons. First, MFG-E8 is mainly expressed by RPE cells and may not be abundant in the library generated from whole eye. Tubby expression in the eye seems lower than Tulp1 based on previous data of immunohistochemistry [31]. Moreover, we have identified five putative receptor-binding consensus motifs in Tulp1, whereas tubby has only one such motif (unpublished data, Caberoy et al.). Second, all eat-me signals identified in this study are derived from a limited-scale screening of only 192 phage clones. It is possible that other known eat-me signals will be identified with a larger-scale screening.
An important criterion of eat-me signals is that they should have physiological or pathological access to phagocytic targets and cognate receptors on phagocyte surface. Tubby and Tulp1 with no classical signal peptide were apparently detected in photoreceptors by immunohistochemistry [31]. Tulp1 is predominantly expressed in the inner segments [32]. How can an intracellular protein in the inner segments facilitate POS phagocytosis? Photoreceptors are susceptible to degeneration induced by a number of factors, including various genetic mutations or constant light exposure [33]. Tubby and Tulp1 may be released from damaged photoreceptors to facilitate phagocytic removal of apoptotic cells or cellular debris either by RPE cells or recruited macrophages [34]. Moreover, our recent data revealed that tubby and Tulp1 are secreted through unconventional pathways [35]. Soluble molecules, such as Gas6, protein S and MFG-E8, have been demonstrated to function as bridging molecules with two functional domains, one binding to their cognate receptor on phagocytes and the other interacting with phosphatidylserine abnormally displayed on apoptotic cell surface [36]. We speculate that secreted tubby and Tulp1 may function in a similar fashion. This speculation is partially verified by the identification of MerTK, an essential phagocytic receptor for RPE phagocytosis [36, 37], as their common receptor and the demonstration of their binding to membrane vesicles through a phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2]-independent mechanism (unpublished data, Caberoy et al.). It is possible that some unknown phosphatidylserine-like molecules or proteins are abnormally displayed on the surface of the shed POS vesicles or membrane vesicles prepared from healthy cells. These molecules mediate tubby and Tulp1 interaction with the vesicles. However, since tubby lacks phosphatidylserine-binding activity [38], the exact mechanisms of tubby and Tulp1 association with membrane vesicles are yet to be elucidated.
Like the well-characterized unconventional secretion of galectin-3 and fibroblast growth factor-2 [39], the secretion of tubby and Tulp1 is independent of classical endoplasmic reticulum (ER)-Golgi pathway [35]. An essential secretory signal has been mapped to tubby N-terminal region between Asn51 and Arg100 with its C-terminal PI(4,5)P2-binding domain partially contributing to the unconventional secretion. Tubby and Tulp1 have been characterized with a number of intracellular functions [24, 25, 40, 41]. The question is whether they can have an additional extracellular function. In fact, it is not uncommon that a protein plays multiple functional roles both intracellularly and extracellularly. One of the examples is galectin-3 with a number of well-characterized intracellular and extracellular functions [42].
Mutations in either tubby or Tulp1 associate with autosomal recessive retinal degeneration with unknown mechanisms. Coincidently, both tubby and Tulp1, but not their two other family members, are eat-me signals for RPE phagocytosis. Because RPE phagocytosis has been previously demonstrated to be critical to prevent retinal degeneration in Royal College of Surgeons (RCS) rats with the mutation in a well-characterized phagocytic receptor MerTK [36, 37], it is appealing to assume that tubby and Tulp1 are essential eat-me signals to prevent retinal degeneration. An intriguing question is why these two functionally redundant proteins fail to compensate each other in their autosomal recessive mutations. One of the possible explanations is that tubby and Tulp1 synergistically stimulate RPE phagocytosis. The failure of mutant tubby or Tulp1 to stimulate phagocytosis in synergy (Fig. 6) may be responsible for the lack of their functional compensation (Fig. 4A), leading to autosomal recessive retinal degeneration with mutation in either gene. However, the caveat is that the data in this study are sufficient only to support tubby and Tulp1 as eat-me signals, but not enough to support a claim that they are essential eat-me signals to prevent retinal degeneration. For example, pathogenic mutants of tubby and Tulp1 are only partially correlated to the loss of their stimulatory activity on RPE phagocytosis (Fig. 5). More importantly, possible accumulation of unphagocytosed vesicles in POS is not observed in Tubby mice and Tulp1−/− mice during retinal degeneration [22, 26, 43]. Unlike MerTK mutation in RCS rats [44], ablation of its ligand Gas6 results in minimal defect in phagocytosis and retinal homeostasis [28, 45], possibly due to functional compensation by other MerTK ligand like protein S [3, 36]. Our unpublished data (Caberoy et al.) revealed that both tubby and Tulp1 bind to and activate MerTK. Hence, the lack of accumulation of unphagocytosed POS debris in Tubby mice or Tulp1−/− mice could be due to functional compensation by other MerTK ligands. A body of evidence, including previously-described intracellular functions of tubby and Tulp1, 16 new tubby-binding proteins identified by our ORF phage display [12] and 4 distinct clinical manifestations of retinal degeneration associated with 23 different hTulp1 mutations [16], suggests that tubby and Tulp1 are proteins with diverse functions. Stimulation of RPE phagocytosis is only one of their important functions. Their critical role as eat-me signals in retinal homeostasis is yet to be delineated, as their other intracellular functions [24]
In summary, this study identified and characterized tubby and Tulp1 as eat-me signals for RPE phagocytosis by ORF phage display. This versatile technology should be broadly applicable to professional and non-professional phagocytes, such as macrophages, microglia, RPE cells and Sertoli cells, and will stimulate the field of phagocyte biology with newly-identified eat-me signals.
Acknowledgements
This project was supported by NIH R01EY016211, P30-EY014801 and an institutional grant from Research to Prevent Blindness. N.B.C is a recipient of a Fight for Sight postdoctoral fellowship. We thank Dr. George Inana for discussion, Dr. A Hackam for manuscript review, and G. Gaidosh at the confocal core facility of Bascom Palmer Eye Institute.
Abbreviations
- Ab
antibody
- Gas6
growth arrest-specific gene 6
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- hTulp1
human tubby-like protein 1
- MFG-E8
milk fat globule epidermal growth factor 8
- mGFP
plasma membrane-targeted green fluorescent protein
- ORF
open reading frame
- pfu
plaque forming unit
- POS
photoreceptor outer segments
- RPE
retinal pigment epithelium
- Tulp1
tubby-like protein 1
- Tulps
tubby, Tulp1, 2 and 3.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reddien PW, Horvitz HR. The engulfment process of programmed cell death in caenorhabditis elegans. Annu Rev Cell Dev Biol. 2004;20:193–221. doi: 10.1146/annurev.cellbio.20.022003.114619. [DOI] [PubMed] [Google Scholar]
- 2.Erwig LP, Henson PM. Immunological consequences of apoptotic cell phagocytosis. Am J Pathol. 2007;171:2–8. doi: 10.2353/ajpath.2007.070135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ravichandran KS, Lorenz U. Engulfment of apoptotic cells: signals for a good meal. Nat Rev Immunol. 2007;7:964–974. doi: 10.1038/nri2214. [DOI] [PubMed] [Google Scholar]
- 4.Lauber K, Blumenthal SG, Waibel M, Wesselborg S. Clearance of apoptotic cells: getting rid of the corpses. Mol Cell. 2004;14:277–287. doi: 10.1016/s1097-2765(04)00237-0. [DOI] [PubMed] [Google Scholar]
- 5.Hedgecock EM, Sulston JE, Thomson JN. Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science. 1983;220:1277–1279. doi: 10.1126/science.6857247. [DOI] [PubMed] [Google Scholar]
- 6.Becerril B, Poul MA, Marks JD. Toward selection of internalizing antibodies from phage libraries. Biochem Biophys Res Commun. 1999;255:386–393. doi: 10.1006/bbrc.1999.0177. [DOI] [PubMed] [Google Scholar]
- 7.Goenaga AL, Zhou Y, Legay C, Bougherara H, Huang L, Liu B, Drummond DC, Kirpotin DB, Auclair C, Marks JD, Poul MA. Identification and characterization of tumor antigens by using antibody phage display and intrabody strategies. Mol Immunol. 2007;44:3777–3788. doi: 10.1016/j.molimm.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caberoy NB, Zhou Y, Li W. Can phage display be used as a tool to functionally identify endogenous eat-me signals in phagocytosis? J Biomol Screen. 2009;14:653–661. doi: 10.1177/1087057109335679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paschke M. Phage display systems and their applications. Appl Microbiol Biotechnol. 2006;70:2–11. doi: 10.1007/s00253-005-0270-9. [DOI] [PubMed] [Google Scholar]
- 10.Kalnina Z, Silina K, Meistere I, Zayakin P, Rivosh A, Abols A, Leja M, Minenkova O, Schadendorf D, Line A. Evaluation of T7 and lambda phage display systems for survey of autoantibody profiles in cancer patients. J Immunol Methods. 2008;334:37–50. doi: 10.1016/j.jim.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 11.Lin HS, Talwar HS, Tarca AL, Ionan A, Chatterjee M, Ye B, Wojciechowski J, Mohapatra S, Basson MD, Yoo GH, Peshek B, Lonardo F, Pan CJ, Folbe AJ, Draghici S, Abrams J, Tainsky MA. Autoantibody approach for serum-based detection of head and neck cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2396–2405. doi: 10.1158/1055-9965.EPI-07-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caberoy NB, Zhou Y, Jiang X, Li W. Efficient identification of tubby-binding proteins by an improved system of T7 phage display. J Mol Recognit. 2009 doi: 10.1002/jmr.983. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caberoy NB, Zhou Y, Alvarado G, Fan X, Li W. Efficient identification of phosphatidylserine-binding proteins by ORF phage display. Biochem Biophys Res Commun. 2009;386:197–201. doi: 10.1016/j.bbrc.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbs D, Williams DS. Isolation and culture of primary mouse retinal pigmented epithelial cells. Adv Exp Med Biol. 2003;533:347–352. doi: 10.1007/978-1-4615-0067-4_44. [DOI] [PubMed] [Google Scholar]
- 15.Noben-Trauth K, Naggert JK, North MA, Nishina PM. A candidate gene for the mouse mutation tubby. Nature. 1996;380:534–538. doi: 10.1038/380534a0. [DOI] [PubMed] [Google Scholar]
- 16.Abbasi AH, Garzozi HJ, Ben-Yosef T. A novel splice-site mutation of TULP1 underlies severe early-onset retinitis pigmentosa in a consanguineous Israeli Muslim Arab family. Mol Vis. 2008;14:675–682. [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee P, Kleyn PW, Knowles JA, Lewis CA, Ross BM, Parano E, Kovats SG, Lee JJ, Penchaszadeh GK, Ott J, Jacobson SG, Gilliam TC. TULP1 mutation in two extended Dominican kindreds with autosomal recessive retinitis pigmentosa. Nat Genet. 1998;18:177–179. doi: 10.1038/ng0298-177. [DOI] [PubMed] [Google Scholar]
- 18.Zuber MX, Strittmatter SM, Fishman MC. A membrane-targeting signal in the amino terminus of the neuronal protein GAP-43. Nature. 1989;341:345–348. doi: 10.1038/341345a0. [DOI] [PubMed] [Google Scholar]
- 19.Li W, Handschumacher RE. Identification of two calcineurin B-binding proteins: tubulin and heat shock protein 60. Biochim Biophys Acta. 2002;1599:72–81. doi: 10.1016/s1570-9639(02)00402-8. [DOI] [PubMed] [Google Scholar]
- 20.Molday RS, Hicks D, Molday L. Peripherin. A rim-specific membrane protein of rod outer segment discs. Invest Ophthalmol Vis Sci. 1987;28:50–61. [PubMed] [Google Scholar]
- 21.Lillis AP, Greenlee MC, Mikhailenko I, Pizzo SV, Tenner AJ, Strickland DK, Bohlson SS. Murine low-density lipoprotein receptor-related protein 1 (LRP) is required for phagocytosis of targets bearing LRP ligands but is not required for C1q-triggered enhancement of phagocytosis. J Immunol. 2008;181:364–373. doi: 10.4049/jimmunol.181.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda S, Shiva N, Ikeda A, Smith RS, Nusinowitz S, Yan G, Lin TR, Chu S, Heckenlively JR, North MA, Naggert JK, Nishina PM, Duyao MP. Retinal degeneration but not obesity is observed in null mutants of the tubby-like protein 1 gene. Hum Mol Genet. 2000;9:155–163. doi: 10.1093/hmg/9.2.155. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Huber BT, Grand RJ, Li W. Recombinant adenovirus coexpressing covalent peptide/MHC class II complex and B7-1: in vitro and in vivo activation of myelin basic protein-specific T cells. J Immunol. 2001;167:1297–1305. doi: 10.4049/jimmunol.167.3.1297. [DOI] [PubMed] [Google Scholar]
- 24.Carroll K, Gomez C, Shapiro L. Tubby proteins: the plot thickens. Nat Rev Mol Cell Biol. 2004;5:55–63. doi: 10.1038/nrm1278. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda A, Nishina PM, Naggert JK. The tubby-like proteins, a family with roles in neuronal development and function. J Cell Sci. 2002;115:9–14. doi: 10.1242/jcs.115.1.9. [DOI] [PubMed] [Google Scholar]
- 26.Hagstrom SA, Duyao M, North MA, Li T. Retinal degeneration in tulp1−/− mice: vesicular accumulation in the interphotoreceptor matrix. Invest Ophthalmol Vis Sci. 1999;40:2795–2802. [PubMed] [Google Scholar]
- 27.Hagstrom SA, North MA, Nishina PL, Berson EL, Dryja TP. Recessive mutations in the gene encoding the tubby-like protein TULP1 in patients with retinitis pigmentosa. Nat Genet. 1998;18:174–176. doi: 10.1038/ng0298-174. [DOI] [PubMed] [Google Scholar]
- 28.Hall MO, Obin MS, Heeb MJ, Burgess BL, Abrams TA. Both protein S and Gas6 stimulate outer segment phagocytosis by cultured rat retinal pigment epithelial cells. Exp Eye Res. 2005;81:581–591. doi: 10.1016/j.exer.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 29.Nandrot EF, Anand M, Almeida D, Atabai K, Sheppard D, Finnemann SC. Essential role for MFG-E8 as ligand for alphavbeta5 integrin in diurnal retinal phagocytosis. Proc Natl Acad Sci U S A. 2007;104:12005–12010. doi: 10.1073/pnas.0704756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Shapiro JI. Endocytosis and signal transduction: basic science update. Biol Res Nurs. 2003;5:117–128. doi: 10.1177/1099800403256860. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda S, He W, Ikeda A, Naggert JK, North MA, Nishina PM. Cell-specific expression of tubby gene family members (tub, Tulp1,2, and 3) in the retina. Invest Ophthalmol Vis Sci. 1999;40:2706–2712. [PubMed] [Google Scholar]
- 32.Milam AH, Hendrickson AE, Xiao M, Smith JE, Possin DE, John SK, Nishina PM. Localization of tubby-like protein 1 in developing and adult human retinas. Invest Ophthalmol Vis Sci. 2000;41:2352–2356. [PubMed] [Google Scholar]
- 33.Paskowitz DM, LaVail MM, Duncan JL. Light and inherited retinal degeneration. Br J Ophthalmol. 2006;90:1060–1066. doi: 10.1136/bjo.2006.097436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wenzel A, Grimm C, Samardzija M, Reme CE. Molecular mechanisms of light-induced photoreceptor apoptosis and neuroprotection for retinal degeneration. Prog Retin Eye Res. 2005;24:275–306. doi: 10.1016/j.preteyeres.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 35.Caberoy NB, Zhou Y, Li W. Unconventional secretion of tubby and tubby-like protein 1. FEBS Lett. 2009 doi: 10.1016/j.febslet.2009.08.015. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemke G, Rothlin CV. Immunobiology of the TAM receptors. Nat Rev Immunol. 2008;8:327–336. doi: 10.1038/nri2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D'Cruz PM, Yasumura D, Weir J, Matthes MT, Abderrahim H, LaVail MM, Vollrath D. Mutation of the receptor tyrosine kinase gene Mertk in the retinal dystrophic RCS rat. Hum Mol Genet. 2000;9:645–651. doi: 10.1093/hmg/9.4.645. [DOI] [PubMed] [Google Scholar]
- 38.Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L. G-protein signaling through tubby proteins. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- 39.Nickel W, Seedorf M. Unconventional Mechanisms of Protein Transport to the Cell Surface of Eukaryotic Cells. Annu Rev Cell Dev Biol. 2008 doi: 10.1146/annurev.cellbio.24.110707.175320. [DOI] [PubMed] [Google Scholar]
- 40.Xi Q, Pauer GJ, Marmorstein AD, Crabb JW, Hagstrom SA. Tubby-like protein 1 (TULP1) interacts with F-actin in photoreceptor cells. Invest Ophthalmol Vis Sci. 2005;46:4754–4761. doi: 10.1167/iovs.05-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xi Q, Pauer GJ, Ball SL, Rayborn M, Hollyfield JG, Peachey NS, Crabb JW, Hagstrom SA. Interaction between the photoreceptor-specific tubby-like protein 1 and the neuronal-specific GTPase dynamin-1. Invest Ophthalmol Vis Sci. 2007;48:2837–2844. doi: 10.1167/iovs.06-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dumic J, Dabelic S, Flogel M. Galectin-3: an open-ended story. Biochim Biophys Acta. 2006;1760:616–635. doi: 10.1016/j.bbagen.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 43.Ohlemiller KK, Hughes RM, Lett JM, Ogilvie JM, Speck JD, Wright JS, Faddis BT. Progression of cochlear and retinal degeneration in the tubby (rd5) mouse. Audiol Neurootol. 1997;2:175–185. doi: 10.1159/000259242. [DOI] [PubMed] [Google Scholar]
- 44.Bok D, Hall MO. The role of the pigment epithelium in the etiology of inherited retinal dystrophy in the rat. J Cell Biol. 1971;49:664–682. doi: 10.1083/jcb.49.3.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Angelillo-Scherrer A, de Frutos P, Aparicio C, Melis E, Savi P, Lupu F, Arnout J, Dewerchin M, Hoylaerts M, Herbert J, Collen D, Dahlback B, Carmeliet P. Deficiency or inhibition of Gas6 causes platelet dysfunction and protects mice against thrombosis. Nat Med. 2001;7:215–221. doi: 10.1038/84667. [DOI] [PubMed] [Google Scholar]