Abstract
Neuronal morphogenesis, the growth and arborization of neuronal processes, is an essential component of brain development. Two important but seemingly disparate components regulating neuronal morphology have previously been described. In the hippocampus, neurotrophins, particularly brain-derived neurotrophic factor (BDNF) and neurotrophin-3 (NT3), act to enhance cell growth and branching, while activity-induced branching was shown to be dependent upon intracellular β-catenin. We now describe a molecular link between NT3 stimulation and β-catenin increase in developing neurons and demonstrate that this process is required for the NT3-mediated increase in process branching. Here, we show that β-catenin is rapidly increased specifically in growth cones following NT3 stimulation. This increase in β-catenin is protein synthesis dependent and requires the activity of cytoplasmic polyadenylation element-binding protein-1 (CPEB1), an mRNA-binding protein that regulates mRNA translation. We find that CPEB1 protein binds β-catenin mRNA in a CPE-dependent manner and that both localize to growth cones of developing hippocampal neurons. Both the NT3-mediated rapid increase in β-catenin and process branching are abolished when CPEB1 function is inhibited. In addition, the NT3-mediated increase in β-catenin in growth cones is dependent upon internal calcium and the activity of CaMKII (calcium/calmodulin-dependent kinase II). Together, these results suggest that CPEB1 regulates β-catenin synthesis in neurons and may contribute to neuronal morphogenesis.
Introduction
During development, neuronal process growth, or morphogenesis, is regulated through signaling in the growth cone. Undoubtedly, neuronal morphogenesis is governed by the coordination of gene expression, trophic cues, synaptic inputs, cell adhesion, and cytoskeletal organization. For example, the neurotrophins brain-derived neurotrophic factor (BDNF) and neurotrophin-3 (NT3) can both induce increased branching in neurons in culture (McAllister et al., 1995; Labelle and Leclerc, 2000). In addition, there is clearly a role for the cadherin–catenin complex in dendritic branching, as changing the levels of these proteins also results in changes in morphology (Yu and Malenka, 2003). One likely target of morphogenic signaling is protein metabolism within the growth cone itself.
Localization of proteins to specific cellular compartments is critical to cell function. Mounting evidence now suggests that regulation of the transport and translation of mRNA in neurons plays an important role in axonal growth and guidance (Campbell and Holt, 2001; Zhang et al., 2001; Ming et al., 2002; Wu et al., 2005; Lin and Holt, 2007; Hengst et al., 2009). Since β-catenin plays such an important role in neuronal morphogenesis, we sought to determine how β-catenin mRNA is regulated in developing hippocampal neurons.
mRNA translation targeted to a specific cellular compartment is thought to have at least three regulable steps: the transport of selective mRNA, the translational silencing of this mRNA while en route and the ability to activate translation in response to a signal (Bramham and Wells, 2007). The transport and translational regulation of targeted mRNAs is thought to be mediated by mRNA-binding proteins (Wells, 2006). For example, the zipcode-binding protein-1 (ZBP1) and heterologous nuclear ribonucleoprotein R (hnRNPR), along with its binding partner survival motor neuron (SMN), are thought to regulate β-actin synthesis in growth cones (Zhang et al., 2001; Rossoll et al., 2003). The characterization of the functional cohort of mRNAs in growth cones and how they are regulated will be essential to our understanding of normal development and regeneration.
Here, we show that β-catenin mRNA contains binding sites for the mRNA-binding protein cytoplasmic polyadenylation element-binding protein-1 (CPEB1). CPEB1 was first characterized in Xenopus oocytes, where it both silences translation of bound mRNAs and activates translation in a mechanism that involves cytoplasmic mRNA polyadenylation (Mendez and Richter, 2001). In mature neurons, CPEB1 regulates glutamate-dependent mRNA polyadenylation and translation (Wu et al., 1998; Wells et al., 2001; Shin et al., 2004; McEvoy et al., 2007) and may play a role in mRNA localization (Huang et al., 2003).
We show that β-catenin mRNA and CPEB1 localize to growth cones of hippocampal neurons. Further, following NT3 stimulation, β-catenin protein accumulates rapidly and exclusively in the growth cones, and this increase is dependent on new protein synthesis. This increase in β-catenin is correlated with the phosphorylation of CPEB1 at T171, which activates the translation of bound mRNA (Mendez et al., 2000b), and can be inhibited by blocking mRNA polyadenylation. Finally, disrupting CPEB1 function blocks the increase in β-catenin and alters the growth and branching of hippocampal processes caused by prolonged exposure to NT3. This work demonstrates for the first time a role for CPEB1-mediated protein synthesis in the morphogenesis of hippocampal neurons.
Materials and Methods
Cell cultures.
Rat hippocampal neuron cultures were made as previously described with slight modifications (Wells et al., 2001). Briefly, the hippocampus was removed from rat embryonic day 18 embryos (E18), trypsinized (0.25%), triturated, and plated on poly-l-lysine-coated coverslips (1 mg/ml) at 50,000 cells/ml for 3 h. The coverslips were then transferred to dishes containing a monolayer of glial cells in growth media containing 10% horse serum. At 1 d in vitro (DIV), the media was replaced with serum-free N2.1 media.
Stimulation protocol.
Individual coverslips with hippocampal neurons (1 DIV) were transferred to 12-well plates in fresh N2.1 media and equilibrated at 37°C for 4 h. NT3 (Cell Sciences) was added to the media at a final concentration of 50 ng/ml for 15 min before processing for immunocytochemistry. This concentration of NT3 was derived from data indicating an increase in dendritic branching when cells were grown in NT for 48 h (Labelle and Leclerc, 2000). With the exception of BAPTA-AM, which was added 5 min before stimulation, all drugs were added to the media 15 min before NT3 stimulation and remained there for the duration of the stimulation experiment. For experiments involving Western blot analysis, the same stimulation protocol was used and the cells were harvested in sample buffer at the end of the stimulation. Western blots were probed with an anti-pCPEB1 antibody (1:2000) (supplemental Fig. 1, available at www.jneurosci.org as supplemental material) (Atkins et al., 2004). The same blots were then stripped and reprobed with an anti-CPEB1 antibody (1:1000) (supplemental Fig. 1, available at www.jneurosci.org as supplemental material) (Shin et al., 2004). Blots were analyzed using Epi Chemi II Darkroom (UVP), and densitometry analysis was performed using Labworks software to obtain a pCPEB1/CPEB1 ratio.
Immunocytochemistry.
Immunocytochemistry (ICC) on hippocampal neurons (1 DIV) was performed as described previously (Wu et al., 1998; Wells et al., 2001). CPEB1 was detected using a rabbit anti-CPEB1 antibody (1:500) (Shin et al., 2004). β-Catenin was detected using a mouse anti-β-catenin antibody (1:500; Millipore Bioscience Research Reagents). Fluorescent and phase-contrast images were captured using a Nikon Eclipse E800 microscope equipped with a CCD camera and processed using Adobe Photoshop 7.0 (Adobe Systems).
Quantification of fluorescence.
In stimulation experiments, growth cones were selected under phase-contrast optics to obtain growth cones of approximately similar morphologies (e.g., see Fig. 3A). Fluorescent images were taken of immunoreactivity in the control group. Experimental group images were then acquired using the same magnification and exposure time. To quantify immunofluorescence in growth cones, the area of each growth cone was defined in the phase image. This area was then outlined on the unprocessed fluorescent image, and the mean pixel intensity in the growth cone was calculated using the histogram function in Adobe Photoshop 7.0. Background fluorescence varied with different treatments and antibodies used, so to correct for background fluorescence, the pixel intensity of the same size area was measured immediately adjacent to the growth cone, and this value was subtracted. To verify the accuracy of quantifying the mean fluorescent intensity, some images were processed using ImageJ (NIH, Bethesda, MD) with identical results. In the same cells in which growth cone fluorescence was determined, cell body and process fluorescence was determined using the same technique. Processes >20 μm long were quantified by selecting under phase optics a 20 μm region midway between the cell body and growth cone and measuring the mean pixel intensity. An area of equal size immediately adjacent to the process was subtracted to correct for background fluorescence.
Figure 3.

NT3-mediated increase in β-catenin protein in growth cones is protein synthesis dependent. A, Cultured hippocampal neurons (1 DIV) were stimulated with 50 ng/ml NT3 for 15 min and processed for ICC with antibodies directed against β-catenin and CPEB1. NT3 induces a 42% increase in β-catenin within growth cones, while CPEB1 levels remain unchanged (n = 69 growth cones, 4 experiments). The increase in β-catenin is completely blocked by the protein synthesis inhibitor cycloheximide (CHX, 100 μm, n = 51 growth cones, 3 experiments) but not by actinomycin D, which still showed a 35% increase in β-catenin (ActD, 5 μm, n = 66 growth cones, 3 experiments). Bright-field, phase-contrast (Phase) images of the growth cones are also shown and were used to quantify immunofluorescence in the growth cone (see Materials and Methods). Scale bar: 5 μm. B, NT3 stimulation as in A in the presence of a polyadenylation inhibitor cordycepin (CDY, 20 μm) also abolishes the increase in β-catenin in growth cones (n = 44 growth cones, 3 experiments). Scale bar: 5 μm. C, Quantification of results in A and B along with the adenosine (Ade, 20 μm) control data (left graph), with β-catenin (solid bars) and CPEB1 (clear bars); ***p < 0.001. In a subset of neurons, fluorescence was quantified in the cell body, all processes, and growth cones of axons and dendrites. Data represent abundance of β-catenin following NT3 compared with the fluorescence intensity in unstimulated cells. In individual cells, an increase in β-catenin is only detected within the growth cones; n = 32 cells from 3 experiments for cell body and processes, 24 axon growth cones, and 59 dendritic growth cones; **p < 0.01, ***p < 0.001.
In situ probe preparation.
We used DNAStar software to design antisense probes directed against two regions of the β-catenin 3′-UTR. Target regions were selected based upon their selectivity for β-catenin, high G/C content, and low self-complementarity. Two oligonucleotide probes (50 nt each) were synthesized for each target mRNA. Oligonucleotide probes were end-labeled with Digoxin (DIG) according to manufacturer specifications (DIG oligonucleotide tailing kit second generation, Roche Applied Science). Two similarly selected oligonucleotide probes complementary to eGFP mRNA were also prepared as controls. The probe sequences were as follows: β-catenin1: 5′-ctccctaccaagtctttctggagttctgcaggcagagtaaagtattcacc-3′; β-catenin2: 5′-gcgcaggtgaccacatttatatcatcagaacccagaagctgcactagagt-3′; eGFP1: 5′-atggcggacttgaagaagtcgtgctgcttcatgtggtcggggtag-3′; eGFP2: 5′-cgagatctgagtccggacttgtacagctcgtccatgccgagagtga-3′.
In situ hybridization.
Fluorescence in situ hybridization (FISH) for β-catenin mRNA was performed according to Shin et al. (2004) with minor modifications. Neurons (1 DIV) were fixed for 15 min at room temperature with 4% paraformaldehyde in 1× PBS and 5 mm MgCl2. Cells were washed in 1×PBS with 5 mm MgCl2 and permeabilized for 5 min at room temperature with 0.1% Triton X-100 in 1×PBS with 5 mm MgCl2, before equilibration for 20 min at room temperature in 1× SSC, 50% formamide, and 10 mm sodium phosphate, pH 7.0. Coverslips were hybridized face down for 5 h at 37°C on a 40 μl drop of probe/hybridization buffer on Parafilm (40 ng of β-catenin probes in 50% formamide, 2× SSC, 10% dextran sulfate, 40 μg of tRNA, 0.2% BSA, 10 μg of salmon sperm DNA, 20 μm VRC, and 10 mm sodium phosphate). After hybridization, cells were washed for 20 min at 37°C in 50% formamide with 1× SSC. Cells were then washed 3 × 10 min at room temperature in 1× SSC on a rocker. Neurons were blocked for 1 h at room temperature in 0.1 m Tris, pH 7.5, 0.15 m NaCl, and 1% BSA, and incubated overnight at 4°C with a 1:300 dilution of mouse anti-DIG-cy3 in block solution. Fluorescent signal was quantified by selecting random growth cones under phase microscopy and determining the fluorescence intensity as described for ICC above.
Immunoprecipitation.
CPEB1 immunoprecipitation (IP) was performed from embryonic rat brain homogenate as described previously (Shin et al., 2004) with minor modifications (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). Two E18 rat brains were homogenized in 2 ml of ice-cold buffer (200 mm NaCl, 10 mm HEPES, pH 7.4, 30 mm EDTA) with 200 U/ml RNase inhibitor (Roche). Brain homogenate was centrifuged for 5 min at 2000 × g, and supernatants were spun at 16,000 × g for 30 min. Five hundred microliters of the resulting supernatant were mixed with rabbit anti-CPEB1 (1:100) for 30 min before the addition of Protein A beads (50 μl) for 30 min. Beads were pelleted and washed three times, and RNA was isolated by phenol:chloroform extraction and ethanol precipitation. Pelleted RNA was resuspended in water and treated with DNase I (50 U/ml; Ambion) for 15 min at 37°C followed by reverse transcription (RT) and PCR analysis to detect β-catenin and Histone H1 (HisH1) mRNAs. Primer sequences were as follows: β-catenin: (forward) 5′-cttatccgatgcagcgactaa-3′ and (reverse) 5′-gtccataatgaaggcgaacgg-3′; HisH1: (forward) 5′-ggtggctttcaagaagaccaa-3′ and (reverse) 5′-tgaggtctgtttgctgtcctt-3′.
CPEB1-RNA pull-down experiments.
COS cells were grown to ∼80% confluence in 100 ml culture in DMEM supplemented with 10% fetal calf serum (FCS). Cells were cotransfected (using Lipofectamine 2000) with 2.5 μg each of a construct encoding a His-tagged CPEB1-RNA-binding domain (His-RBD) (Jones et al., 2008) and GFP (Clontech) linked to the distal 3′-UTR of β-catenin. The 3′-UTR of β-catenin contained either the WT version or a sequence where the CPE sequences had been mutated.
The distal portion of the β-catenin 3′-UTR (∼250 nt) was amplified from mouse cDNA using the following primers: 3′-cttgtccgatgcagcgactaa-5′ and 3′-gtccataatgaaggcgaacgg-5′. This fragment was ligated into the PCR TOPO2.1 vector (Invitrogen), digested with XhoI and BamHI, and ligated into the pEGFP-C1 vector (Clontech) precut with the same enzymes. Mutagenesis of the three CPE sequences was performed using the Stratagene QuikChange Kit according to manufacturer specifications. The following pairs of primers were used to mutate the CPE sequences: CPE1: gcggttatagaaatggttcagaattaaacttcccattcattc and gaatgaatgggaagtttaattctgaaccatttctataaccgc; CPE2: cagtataccagtgccttccctcccaaagttgttg and caacaactttgggagggaaggcactggtatactg; CPE3: ggtccgattagtttcctttcccatatgcttaaaataagc and gcttattttaagcatatgggaaaggaaactaatcggacc. Constructs were sequenced to confirm insert and mutagenesis.
Twenty four hours after transfection, cells were harvested using a scraper, and two dishes were pooled for each pull-down experiment. Pull-down and RNA isolation were performed as described previously (Jones et al., 2008). mRNA was reverse transcribed using an oligodT probe and PCR amplified using the following primers specific for GFP: 5′-cgagggcgagggcgatgccaccta-3′ and 5′-agaacacccccatcggcgacg-3′.
Transfections.
Two separate protocols were used to transfect constructs into cells depending upon the age of the neurons. Amaxa nucleofection was used for experiments addressing β-catenin synthesis in growth cones, and Lipofectamine 2000 (Invitrogen) was used on older neurons where branching was assessed.
To obtain expression of either GFP or GFP-CPEB1-RBD (GFP-RBD) using Amaxa nucleofection, 4 × 106 neurons in suspension were nucleofected with the mouse neuron solution on program 0-003 using either 1.5 μg of GFP-CPEB1-RBD or pmaxGFP supplied by the manufacturer. The cloning of GFP-RBD has been described by Jones et al. (2008). Neurons were then plated on 18 mm coverslips at ∼3 × 105 cells/ml for 3 h in MEM containing 10% horse serum. After 3 h, coverslips were flipped into 60 mm dishes with a monolayer of glia feeder cells. Cells were further allowed to recover from the transfection for 48 h before stimulated with NT3 for 15 min as described above. Cells were then processed for β-catenin immunoreactivity and quantified as described above.
To assess branching, hippocampal neurons (1 DIV) were transfected on coverslips with Lipofectamine 2000 for 3 h in 12-well plates. Either GFP alone or GFP-RBD was expressed using 500 ng of each construct. At the end of the transfection, coverslips were placed back into their dishes either with fresh N2.1 media or with fresh media containing 50 ng/ml NT3 for the duration of the experiment (48 h). Neurons were fixed for morphological analysis in 4% paraformaldehyde at 3 DIV (48 h after transfection). Images of GFP- and GFP-RBD-transfected cells were taken with a Nikon Eclipse 800 microscope. Although the majority of the fluorescent signal in the GFP-RBD-expressing cells was contained within the cell body, some fluorescence was detected throughout the entire cell, consistent with other reports (Huang et al., 2003). Therefore, for these cells exposure times were adjusted to visualize all processes. The presence of fluorescence in all processes was confirmed in every cell by phase optics, however, all measurements were performed using GFP images. The length of processes were traced and measured using Scion Image (NIH). To quantify process branching, a branch was defined as any process >5 μm in length. Branching was analyzed by visually marking and tallying the number of branch points per cell.
Results
β-catenin mRNA localizes to growth cones and interacts with CPEB1
To determine whether β-catenin mRNA might be translationally regulated, we analyzed the 3′-UTR for potential binding sites for regulatory proteins. We identified three potential CPE sequences that were conserved in most vertebrate species examined (Fig. 1A). Although many mRNAs contain putative CPE elements, we chose to focus on β-catenin based on the role it plays in growth and branching of hippocampal dendrites (Yu and Malenka, 2003, 2004), and its role in neurotrophin-induced axon growth in DRG neurons (Zhou et al., 2004).
Figure 1.

CPEB1 protein interacts with the β-catenin mRNA. A, β-Catenin mRNA contains conserved 3′-UTR CPE sequences in rat (GenBank GeneID 84353), mouse [GenBank accession (Acc.) #. NM_007614], and human (GenBank Acc. #: NM_001904). The diagram highlights three conserved CPE sequences in the mouse and human mRNA, two in the rat, along with the hexanucleotide AUUAAA (HEX) sequence. The distal region of the mouse 3′-UTR was fused to eGFP-C1 and used for pull-down analysis along with a mutated CPE construct (bottom). B, Western blot analysis of E18 rat brain homogenates immunoprecipitated with either an affinity-purified CPEB1 antibody (CPEB1), or control (Con) shows CPEB1 present only when the specific antibody is used (left panel; see also supplemental Fig. 1, available at www.jneurosci.org as supplemental material). mRNA isolated from the immunoprecipitated material was subjected to RT-PCR analysis with primers specific to β-catenin and a non-CPE-containing mRNA, Histone H1 (HisH1; right panel) (n = 4). C, COS cells were cotransfected with His-CPEB1-RBD and either a construct encoding eGFP linked to the β-catenin 3′-UTR or a mutated β-catenin 3′-UTR (mutCPE) construct. Pull-down of the His-CPEB1-RBD and subsequent RT-PCR analysis with probes specific to GFP (left), β-catenin (middle), or GAPDH (right) was performed. The mRNA encoding GFP was only pulled down when it contained CPE sequences, and the non-CPE-containing GAPDH was never associated with CPEB1. As a positive control that the pull-down was effective in mutCPE-expressing cells, endogenous β-catenin was still pulled down (n = 3). D, Hippocampal neurons were processed for FISH using DIG-labeled oligonucleotide β-catenin probes alone (β-catenin mRNA) or together with 100× excess of unlabeled probes (control). Fluorescent images were overlaid onto phase-contrast images. FISH revealed β-catenin mRNA localization through processes and accumulating in growth cones (arrow). Boxed regions (rotated in the case of β-catenin) are enlarged to the right (n = 4). Scale bars: 10 μm.
To assess the ability of CPEB1 protein to interact with β-catenin mRNA, we immunoprecipitated CPEB1 from E18 brain homogenate (Fig. 1B) and isolated the mRNAs that coimmunoprecipitated. Using gene-specific primers for RT-PCR analysis, we demonstrated that β-catenin mRNA is present in the CPEB1 IP. This association is specific in that β-catenin does not immunoprecipitate with a control antibody and a non-CPE-containing mRNA, Histone H1 (HisH1), is not present in the CPEB1 IP (Fig. 1B). To confirm the specificity of this interaction and to determine the role of CPE sequences, we coexpressed a His-tagged CPEB1 RNA-binding domain (RBD) with an eGFP construct fused to the distal portion of the mouse β-catenin 3′-UTR harboring either the wild-type CPE sequences or with all three CPE sequences mutated (Fig. 1A). Pull-down of the His-RBD protein and subsequent RT-PCR analysis of bound mRNAs revealed that the CPEB1 RBD interacts selectively with the eGFP encoding mRNA only if the wild-type β-catenin 3′-UTR is present (Fig. 1C). Thus, CPEB1 binding to β-catenin mRNA is dependent upon the short CPE sequences.
To determine the localization of β-catenin mRNA in developing neurons, we used FISH with DIG-labeled probes specific to β-catenin on hippocampal neurons in culture. We observed β-catenin mRNA in the cell body and in puncta throughout the processes and into growth cones (Fig. 1D). This pattern is reminiscent of the localization of β-actin mRNA, whose presence in the growth cone is well established (supplemental Fig. 2, available at www.jneurosci.org as supplemental material) (Zhang et al., 2001). The fluorescent signal detected using probes for β-catenin was 3.61 ± 0.61-fold higher than that detected in controls (excess unlabeled probe; n = 18 growth cones/group; p < 0.0005). Localization of CPEB1 protein was examined using an antibody specific for CPEB1 (supplemental Fig. 1, available at www.jneurosci.org as supplemental material) and showed a distribution that was similar to that of β-catenin mRNA (Fig. 2). In hippocampal neurons 3–6 DIV, the process at least twice as long as the other processes will become the axon (Shi et al., 2003; Yoshimura et al., 2005). Using this criterion, CPEB1 protein and β-catenin mRNA localized to growth cones of both axons and dendrites.
Figure 2.

CPEB1 localizes to hippocampal growth cones. A, Rat hippocampal neurons (1 DIV) were processed for immunocytochemistry (ICC) with an anti-CPEB1 antibody. CPEB1 immunoreactivity is detected in the cell body and in neuronal processes in puncta (arrows) that extend into growth cones (arrowheads). This image represents the CPEB1 fluorescence overlaid on the phase-contrast image. B, The boxed growth cone in A is expanded to show CPEB1 enriched in the central region of the growth cone. Scale bars: 10 μm.
NT3 stimulates β-catenin synthesis in neurons
Several growth and guidance factors are known to stimulate mRNA transport and translation in neurons and represent possible stimuli to elicit CPEB1-mediated protein synthesis in hippocampal neurons. Since NT3 induces β-actin mRNA transport and translation in cultured cortical neurons (Zhang et al., 1999, 2001), and prolonged NT3 treatment leads to both growth and branching of hippocampal axons and dendrites (Labelle and Leclerc, 2000; Yoshimura et al., 2005), we examined the effect of NT3 stimulation on β-catenin levels in hippocampal neurons. We observed a significant 42% increase in β-catenin immunoreactivity in growth cones 15 min following NT3 application (Fig. 3). To control for nonspecific or volume effects, we examined the levels of CPEB1 immunoreactivity in the same growth cones and observed no significant change (Fig. 3). In addition, growth cones from unstimulated and stimulated neurons did not show a difference in growth cone area (control 79.6 ± 5.77 μm2; NT3 stimulated 82.89 ± 5.71 μm2). Nor was this increase likely due to a sparing of β-catenin protein from degradation, since inhibiting the proteasome with MG132 (10 μm) had no effect on β-catenin protein levels (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). Furthermore, the NT3-mediated increase in β-catenin is sensitive to the protein synthesis inhibitor cycloheximide (CHX, Sigma-Aldrich) but not to the transcription inhibitor actinomycin D (ActD) (Fig. 3A), suggesting that the increase in β-catenin immunoreactivity is due to an increase in the translation of mRNA present at the time of stimulation. One characteristic of CPEB1-induced translation is mRNA polyadenylation, and β-catenin mRNA can undergo stimulation-dependent polyadenylation in non-neural cells (Oh et al., 2000; Jones et al., 2008). Therefore, we treated the cultures with the polyadenylation inhibitor cordycepin (Cdy, Sigma-Aldrich) 15 min before NT3 stimulation and revealed that the rapid β-catenin increase in growth cones was completely eliminated (Fig. 3B). Since Cdy is an adenosine analog, we stimulated with NT3 in the presence of adenosine but saw no change in the NT3 effect (Fig. 3B).
To address the possibility that β-catenin is synthesized in the cell body and transported to growth cones, we quantified the β-catenin immunoreactivity in the cell bodies, axons, and dendrites in a subset of the same neurons shown in Figure 3. Again setting the control (unstimulated) pixel intensity to 1 (Fig. 3C), we could detect no increase of β-catenin in either the cell body or the processes 15 min after NT3 stimulation (cell body pixel intensity 1.04 ± 0.05, p = 0.62; processes 1.07 ± 0.08, p = 0.51), but detected a similar and significant increase of β-catenin in the growth cones of axons (1.41 ± 0.10, p < 0.01) and dendrites (1.32 ± 0.05, p < 0.001). Thus, we detect an NT3-stimulated increase in β-catenin mRNA translation exclusively in the growth cones of both axons and dendrites.
β-Catenin synthesis is dependent on CPEB function
To assess the role of CPEB1 in mediating NT3-dependent β-catenin translation, we inhibited CPEB1 function by expressing a truncated CPEB1 mutant encoding only the RBD fused with eGFP (Jones et al., 2008). This mutant protein binds CPE-containing mRNAs and acts as a dominant negative in oocytes, astrocytes, and neurons (Mendez et al., 2000a; Huang et al., 2003; Bestman and Cline, 2008; Jones et al., 2008). Hippocampal neurons were electroporated (Amaxa) before plating and grown in culture for 2 d before 15 min NT3 stimulation. In these experiments, NT3 stimulated a statistically significant increase in β-catenin in the growth cones relative to unstimulated, untransfected growth cones (Fig. 4). In cells expressing the unmodified GFP vector, NT3-stimulated growth cones also showed a significant increase in β-catenin in growth cones relative to untransfected, unstimulated growth cones. In contrast, GFP-CPEB1-RBD-expressing cells show a slight but statistically insignificant reduction in β-catenin levels relative to untransfected growth cones on the same coverslips (95% of control). However, CPEB1-RBD expression completely blocks the NT3-dependent increase in β-catenin that is detected in untransfected cells within the same dish (Fig. 4B,C). One potentially confounding factor is that CPEB1-RBD may inhibit other mRNA-binding proteins. However, these data in combination with the Cdy data (Fig. 3), and the inhibition of CPEB1 phosphorylation shown below, together implicate CPEB1 in the regulation of NT3-mediated synthesis in β-catenin.
Figure 4.

CPEB1 mediates NT3-induced β-catenin synthesis. A, Hippocampal neurons expressing GFP stimulated with NT3 for 15 min show growth cone β-catenin levels equaling those of untransfected cells (GFP: n = 20 growth cones, 3 experiments; untransfected: n = 72 growth cones, 7 experiments). Arrows denote growth cones shown at higher magnification overlaid with the phase image in the right panel. Scale bars: left, 20 μm; right, 5 μm. B, Neurons expressing GFP-RBD show 30% less β-catenin in growth cones compared with untransfected cells in the same dish following NT3 stimulation (n = 54 growth cones, 4 experiments). Arrows denote growth cones shown at higher magnification overlaid with the phase image in right panel. Scale bars: left, 20 μm; right, 5 μm. C, Quantification of results with untransfected cells (black bars), GFP-expressing cells (gray bars), and GFP-RBD-expressing cells (tan bars). Data represent β-catenin immunoreactivity in growth cones normalized to levels in untransfected, unstimulated growth cones. **p < 0.01.
Signal transduction pathway leading to β-catenin synthesis
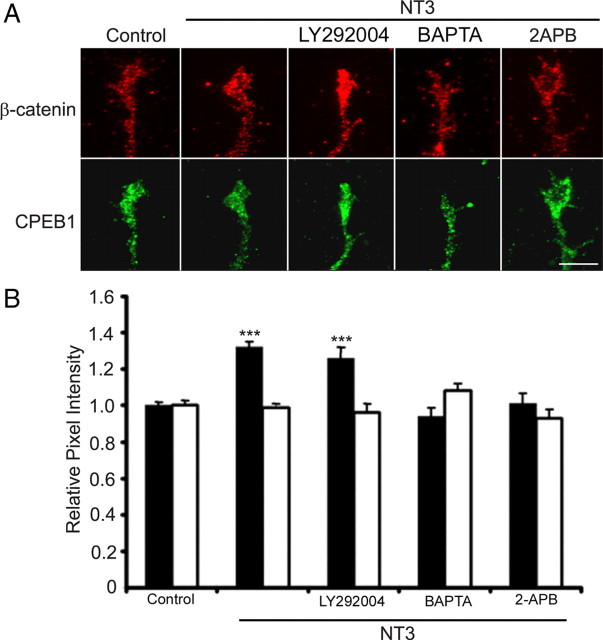
To begin to address the intracellular signal transduction pathway from NT3 stimulation to CPEB1 activation, we used a pharmacological approach. We first treated neurons with the phosphoinositide 3-kinase (PI3-kinase) inhibitor LY294002 (Sigma-Aldrich) before NT3 stimulation, since NT3 is known to activate PI3-kinase in cultured hippocampal neurons (Yoshimura et al., 2005). However, LY294002 did not block the NT3-mediated β-catenin increase in growth cones, suggesting that PI3-kinase is not involved directly in this pathway (Fig. 5). To determine whether the NT3-mediated β-catenin synthesis was Ca2+ dependent, we stimulated neurons with NT3 for 15 min in the presence of the membrane-permeable Ca2+ chelator BAPTA-AM (50 μm). BAPTA-AM completely blocked the NT3-mediated increase in β-catenin (Fig. 5). Similarly, application of 2-aminoethoxydiphenyl borate (2-APB; Calbiochem; 100 μm), an inositol 1,4,5-trisphosphate (IP3) receptor inhibitor (Maruyama et al., 1997), also blocked the NT3-dependent β-catenin increase in growth cones. Together, this indicates that the increase in β-catenin is likely activated by release of calcium from internal stores.
Figure 5.
NT3-mediated β-catenin increase is calcium dependent. A, Hippocampal neurons stimulated with 50 ng/ml NT3 resulted in a 32% increase in β-catenin (n = 224 growth cones, 14 experiments). In the presence of the PI3-kinase inhibitor LY294002 (20 μm), NT3 still yields a 26% increase in β-catenin (n = 64 growth cones, 6 experiments). However, in the presence of BAPTA-AM (BAPTA; 50 μm), NT3 had no effect on β-catenin levels (n = 72 growth cones, 5 experiments). Similarly, treatment with the IP3 receptor inhibitor, 2-APB (100 μm) resulted in no change in β-catenin levels (n = 84 growth cones, 4 experiments). Scale bar: 5 μm. B, Quantification of results with β-catenin (solid bars) and CPEB1 (clear bars) immunoreactivity intensity compared with unstimulated cells (control). ***p < 0.001.
As noted earlier, CPEB1 can both inhibit and stimulate protein synthesis. The activation of translation is mediated by phosphorylation of CPEB1 at T171 (Mendez and Richter, 2001). Two kinases have been implicated in this phosphorylation: Aurora A kinase (Mendez et al., 2000b; Huang et al., 2002; McEvoy et al., 2007) and calcium/calmodulin-dependent kinase II (CaMKII) (Atkins et al., 2004; Atkins et al., 2005). To test the possibility that one or both of these kinases plays a role in CPEB1-mediated translation in hippocampal growth cones, we stimulated neurons with NT3 in the presence of the Aurora kinase inhibitor ZM447439 (200–500 nm; Tocris Bioscience) or the CaMKII inhibitor KN93 (5 μg/ml; Calbiochem), and analyzed β-catenin and CPEB1 immunofluorescence (Fig. 6). ZM447439 inhibits Aurora kinase with an IC50 of 110 nm (Ditchfield et al., 2003); however, even high concentration (500 nm) did not alter the NT3-dependent increase in β-catenin. In contrast, KN93 abolished the β-catenin increase (Fig. 5). To confirm the specificity of the KN93 response, we treated cells with KN92 (5 μg/ml; Calbiochem), a compound structurally related to KN93 but unable to inhibit CaMKII, and saw no effect on the β-catenin increase (Fig. 6). To verify CaMKII involvement, we used tatCN21, a cell-permeable version of the highly specific CaMKIIN peptide inhibitor of CaMKII (Chang et al., 1998; Vest et al., 2007). NT3 was unable to induce β-catenin synthesis in the presence of tatCN21 peptides, while the reverse-sequence peptide (CNRev) had no effect (Fig. 7). Finally, we confirmed that NT3 induced CPEB1 phosphorylation in a CaMKII-dependent process by using a phosphospecific CPEB1 antibody (Atkins et al., 2004). NT3 induced a rapid phosphorylation of CPEB1 in hippocampal neurons that was inhibited by KN93 but not ZM447439 (Fig. 8). We observed a rapid 35% increase in pCPEB1 levels 5 min following NT3 application that remained high (56% higher than unstimulated) at 15 min after stimulation (Fig. 8). This time course is consistent with the rapid increase of β-catenin protein in growth cones.
Figure 6.

KN-93 inhibits the NT3-mediated β-catenin synthesis in growth cones. A, Hippocampal neurons (1 DIV) stimulated with NT3 showed a 33% increase in β-catenin (n = 103 growth cones, 6 experiments). In the presence of the Aurora kinase inhibitor, ZM447439 (500 nm), NT3 still elicits a 32% increase β-catenin in growth cones (n = 93 growth cones, 5 experiments). In contrast, the NT3-dependent increase in growth cone β-catenin immunoreactivity was blocked by the CaMKII inhibitor KN93 (5 μg/ml; 51 growth cones, 4 experiments). While the inactive analog KN92 (5 μg/ml) had no effect (44% increase; n = 55 growth cones, 4 experiments). Scale bar: 5 μm. B, Quantification of results with β-catenin (solid bars) and CPEB1 (clear bars) immunoreactivity intensity compared with unstimulated cells (control). ***p < 0.001.
Figure 7.

NT3-mediated increase in β-catenin mediated by CaMKII. A, Hippocampal neurons (1 DIV) stimulated with NT3 show an increase in β-catenin in growth cones (n = 85 growth cones, 5 experiments). Treatment with a membrane-permeable CaMKII peptide inhibitor, tatCN21 (CN21, 5 μm), drastically reduced this β-catenin increase (n = 95 growth cones, 5 experiments). However, in the presence of a control peptide with the reverse sequence (CNRev, 5 μm), NT3 still yielded a 32% increase in β-catenin in growth cones (n = 90 growth cones, 5 experiments). Scale bar: 5 μm. B, Quantification of results with β-catenin (solid bars) and CPEB1 (clear bars) immunoreactivity intensity compared with unstimulated cells (control). ***p < 0.001.
Figure 8.
NT3 stimulation leads to CPEB1 phosphorylation. Hippocampal neurons (1 DIV) were stimulated with NT3 for 5 (n = 7 experiments) or 15 min (n = 18 experiments), and analyzed by Western blot using an anti-pCPEB1 (Atkins et al., 2004). Blots were then stripped and reprobed with an anti-CPEB1 antibody. Treatment with KN93 inhibited the NT3-mediated pCPEB1 increase at both 5 and 15 min (n = 6 experiments), while treatment with ZM447439 (ZM, n = 5 experiments) had no effect on CPEB1 phosphorylation. Graph illustrates the quantification of pCPEB1 levels relative to total CPEB1 revealing a statistically significant increase in pCPEB1 of 35% at 5 min and 56% at 15 min (**p < 0.01, ***p < 0.001).
NT3-induced branching is dependent on CPEB function
Prolonged NT3 treatment of cultured hippocampal neurons results in increased growth and branching of processes (Labelle and Leclerc, 2000; Yoshimura et al., 2005). To determine whether CPEB1-mediated protein synthesis plays a role in mediating NT3-induced growth and branching in hippocampal neurons, we again expressed the CPEB1 mutant GFP-CPEB1-RBD. Hippocampal neurons (1 DIV) were transfected with either GFP alone or GFP-CPEB1-RBD, and grown in the presence or absence of NT3 for 48 h before analyzing their morphology (Fig. 9). In GFP-expressing cells, fluorescence was evenly distributed throughout the cell. In GFP-RBD-expressing cells fluorescence was primarily localized to the cell body, however, weaker fluorescence could be detected in the processes. Therefore, image exposure times were increased for morphological analysis of these cells (see Materials and Methods). Consistent with previous reports, GFP-expressing neurons grown in the presence of NT3 had 40% longer processes and a 95% increase in branch number compared with neurons grown in the absence of NT3 (Fig. 9). However, in neurons expressing the GFP-CPEB1-RBD, NT3's effect on growth and branching was muted. In fact, there was a statistically significant decrease in branching of GFP-RBD-expressing neurons compared with GFP alone in the presence of NT3 (Fig. 9).
Figure 9.

Expression of CPEB1-RBD inhibits NT3-mediated process growth and branching. A, Hippocampal neurons (1 DIV) were transfected with either eGFP alone or with eGFP-tagged CPEB1-RBD. After transfection, coverslips were placed back in their dishes in the presence or absence of 50 ng/ml NT3. Scale bar: 10 μm. B, Quantification of results in A with process branching (clear bars) and process length (solid bars) of GFP and GFP-CPEB1-RBD (GFP-RBD)-expressing cells. GFP-transfected neurons grown in NT3 (n = 32 cells) exhibited a statistically significant 40% increase in process length (*p < 0.05) and 95% increase in branching (**p < 0.01) compared with control neurons (n = 30 cells). In the absence of NT3, expression of GFP-RBD (n = 22 cells) resulted in a modest reduction in both branching and length compared with cells expressing GFP alone. Additionally, GFP-RBD-expressing cells grown in the presence of NT3 (n = 30 cells) trend toward an increase in branching (34%) and length (14%) compared with unstimulated GFP-RBD-expressing cells, but these increases were not statistically significant (branching p = 0.24; length p = 0.42).
Discussion
The ability to synthesize new proteins within the axon and growth cone was first suggested by the identification of ribonucleic acid in developing axons by ultraviolet microscopy (Hydén, 1943) and the subsequent visualization of ribosomes within the growth cone (Tennyson, 1970). Substantial evidence now shows that local regulation of protein synthesis is an important mechanism by which neurons regulate how processes grow in response to different stimuli. Guidance factors including netrin-1, slit2, and Sema3A and growth factors BDNF and NT3 are thought to stimulate the synthesis of proteins such as β-actin and RhoA in growth cones (Zhang et al., 2001; Wu et al., 2005; Leung et al., 2006; Piper et al., 2006; Yao et al., 2006). In this study, we describe a mechanism for neurons to regulate cell morphology. Here, an mRNA-binding protein CPEB1 is localized to growth cones and regulates the morphological changes induced by the neurotrophin NT3. In addition, we show that one of the mRNAs regulated by CPEB1 is that encoding β-catenin, a protein already implicated in dendritic and axonal branching (Elul et al., 2003; Yu and Malenka, 2003, 2004).
The molecular regulation of mRNA transport and translation in growth cones has been subject to intense investigation. Numerous mRNA-binding proteins including ZBP1, hnRNPR, and HuD have been shown to regulate mRNA transport to axonal growth cones, and many likely play a role in regulating translation (Zhang et al., 2001; Aronov et al., 2002; Rossoll et al., 2003). ZBP1 binds β-actin mRNA and is thought to maintain it in a translationally dormant state until ZBP1 is phosphorylated by Src kinase, allowing for β-actin protein synthesis (Hüttelmaier et al., 2005). Similarly, a recent study showed that Grb7 binds and translationally silences the κ-opioid receptor (KOR) mRNA in DRG neurons (Tsai et al., 2007). Upon netrin-1 stimulation, focal adhesion kinase (FAK) phosphorylates Grb7, causing it to dissociate from KOR mRNA, thus activating translation (Tsai et al., 2007). HuD, an mRNA-binding protein that binds to AU-rich element (ARE), facilitates the translation of target mRNAs such as GAP43 and Tau by stabilizing the mRNAs (Deschênes-Furry et al., 2006). In addition, several studies have shown the involvement of factors that globally affect translation such as eukaryotic initiation factor 4E (eIF4E) and eIF4E-binding protein (4E-BP) in the regulation of protein synthesis in growth cones (Campbell and Holt, 2001, 2003; Piper et al., 2006).
Our data support a model in which CPEB1 is bound to β-catenin mRNA in growth cones and regulates the translation of β-catenin locally following NT3 stimulation. Several findings support our conclusion that the increase in β-catenin is likely the result of local synthesis. First, the increase is protein synthesis dependent but not a result of new transcription; thus, a product of translation from mRNA already present at the time of stimulation. Second, there is a clear abundance of β-catenin mRNA in the growth cones. Third, NT3 receptors are highly enriched at growth cones (Menn et al., 2000). Fourth, CPEB1 binds β-catenin mRNA and is expressed in growth cones, and the increase is dependent upon CPEB1 function. To determine CPEB1 dependence, we inhibited its function by three independent approaches: expression of a mutant protein (CPEB1-RBD), inhibition of polyadenylation (cordycepin), and inhibition of CPEB1 phosphorylation (KN93). Although each approach may inhibit multiple mechanisms, the only demonstrable common mechanism we are aware of is CPEB1. In each case, the NT3-mediated increase in β-catenin was inhibited. Finally, the rapid time course (15 min) and increase in β-catenin protein restricted to growth cones (not in either the cell body or along the process) suggest that protein transport does not play a significant role. While together these data suggest a role for local synthesis of β-catenin protein, other factors could contribute to the NT3-induced increase in β-catenin mRNA transport into growth cones.
The translation of β-catenin mRNA following NT3 stimulation is mediated by the mRNA-binding protein CPEB1. CPEB1 binding to mRNA inhibits translation, and this may allow for transport of the complex to distal regions of the cell in a translationally dormant state (Huang et al., 2003; Bramham and Wells, 2007). However, when CPEB1 is phosphorylated at T171, the bound mRNA is polyadenylated and translation is stimulated (Mendez and Richter, 2001). We show that CPEB1 binds to β-catenin mRNA specifically through CPE sequences in the 3′-UTR and that both are localized to growth cones (Figs. 1, 2). Technical limitations of the polyadenylation assay made direct measurement of changes of the polyadenylation state of β-catenin in primary hippocampal neurons in culture unobtainable. We believe this is due to the limited amount of mRNA we can isolate from these cultures. However, we recently demonstrated that β-catenin mRNA can be polyadenylated in non-neural cells (Jones et al., 2008), and we show here that the increase in β-catenin synthesis is blocked by cordycepin, a polyadenylation inhibitor (Fig. 3). In addition, we show that NT3 induces a phosphorylation of CPEB1 and that the increase in β-catenin in growth cones and the increase in branching are both inhibited by blocking CPEB1 function (Figs. 4, 9). This is consistent with a recent report showing that CPEB1 function is required for dendritic arbor development and circuit formation in the Xenopus retinotectal system (Bestman and Cline, 2008). By expressing either the CPEB1-RBD or a phospho-mutant of CPEB1, Bestman and Cline (2008) showed that activity-induced dendritic arbor growth in vivo was severely impaired. As noted above, it is possible that in addition to inhibiting CPEB1-mediated translation, CPEB1-RBD could be blocking mRNA transport into growth cones. In fact, NT3 induces an increase in β-actin mRNA transport in chick forebrain neurons that is thought to be mediated by ZBP1 (Zhang et al., 2001). Indeed, since the CPEB1-RBD is primarily localized to the cell body (Fig. 9) (Huang et al., 2003), the resulting inhibition could be mediated by blocking mRNA transport, translation, or both. In either case, β-catenin mRNA translation is being regulated by CPEB1 in developing hippocampal neurons.
Surprisingly, we find that the NT3-mediated increase of β-catenin in growth cones is independent of a recognized PI3-kinase activation downstream of NT3 stimulation (Fig. 5). Previously, PI3-kinase activation and subsequent Gsk-3β inactivation had been implicated in NT3-dependent growth and branching in hippocampal neurons (Yoshimura et al., 2005). However, in our studies the dependence upon internal calcium and the activation of CaMKII appear to be consistent with other findings showing NT3 activation of CaMKII. At the Xenopus neuromuscular junction, vesicle release is potentiated by NT3 via a PI3-kinase-dependent pathway and an independent Ca2+- and CaMKII-dependent pathway (He et al., 2000; Yang et al., 2001). Further, CaMKII has been shown to be important in the regulation of process growth and guidance. In cultured cortical neurons, bath application of netrin-1 stimulates axonal growth and branching in a mechanism that requires CaMKII and the release of calcium from internal stores (Tang and Kalil, 2005). Consistent with our results, blocking CaMKII function reduces dendrite growth and branching in hippocampal neurons (Fink et al., 2003). In growth cone turning assays, where intracellular Ca2+ is known to be an important mediator of attractive or repulsive responses, the balance between CaMKII and calcineurin activity plays an essential role in determining the direction of a turn (Wen et al., 2004). Intriguingly, the balance of CaMKII and calcineurin activity regulates the phosphorylation state and activity of CPEB1 in neurons (Atkins et al., 2005). Based on the results presented in this study, regulation of CPEB1-mediated protein synthesis may provide an important mechanism by which CaMKII mediates growth, guidance, and branching effects in developing neurons.
The results presented here show that inhibition of CPEB1 function disrupts the growth and branching of processes in the presence of NT3 (Fig. 9). Similarly, inhibition of β-catenin binding to cadherins and α-catenin also disrupts growth and branching of hippocampal dendrites and axons of Xenopus retinal ganglion axons (Elul et al., 2003; Yu and Malenka, 2003). These findings suggest that inhibiting synthesis of β-catenin may be part of the defect observed with CPEB1-RBD expression. However, it is important to note that numerous mRNAs contain putative CPE elements in their 3′-UTRs, and the full functional consequence of inhibiting CPEB1 with the RBD surely represents the misregulation of a population of different mRNAs. Elucidation of the complement of mRNAs bound and regulated by CPEB1 in neurons will provide valuable insight into its function in different physiological processes.
Footnotes
This work was supported by the F. W. and Elsie Heyl Foundation (M.K.), National Institute of Mental Health (R01 MH66274, to D.G.W.), and The Ellison Medical Foundation (D.G.W.). We thank Dr. K. Ulrich Bayer (University of Colorado Health Sciences Center) for the tatCN21 forward and reverse peptides and Drs. Thomas Soderling (Vollum Institute, Portland, OR) and Naohito Nozaki (Kanagawa Dental College, Yokosuka, Kanagawa, Japan) for the pCPEB1 antibody.
References
- Aronov S, Aranda G, Behar L, Ginzburg I. Visualization of translated tau protein in the axons of neuronal P19 cells and characterization of tau RNP granules. J Cell Sci. 2002;115:3817–3827. doi: 10.1242/jcs.00058. [DOI] [PubMed] [Google Scholar]
- Atkins CM, Nozaki N, Shigeri Y, Soderling TR. Cytoplasmic polyadenylation element binding protein-dependent protein synthesis is regulated by calcium/calmodulin-dependent protein kinase II. J Neurosci. 2004;24:5193–5201. doi: 10.1523/JNEUROSCI.0854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins CM, Davare MA, Oh MC, Derkach V, Soderling TR. Bidirectional regulation of cytoplasmic polyadenylation element-binding protein phosphorylation by Ca2+/calmodulin-dependent protein kinase II and protein phosphatase 1 during hippocampal long-term potentiation. J Neurosci. 2005;25:5604–5610. doi: 10.1523/JNEUROSCI.5051-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestman JE, Cline HT. The RNA binding protein CPEB regulates dendrite morphogenesis and neuronal circuit assembly in vivo. Proc Natl Acad Sci U S A. 2008;105:20494–20499. doi: 10.1073/pnas.0806296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32:1013–1026. doi: 10.1016/s0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- Campbell DS, Holt CE. Apoptotic pathway and MAPKs differentially regulate chemotropic responses of retinal growth cones. Neuron. 2003;37:939–952. doi: 10.1016/s0896-6273(03)00158-2. [DOI] [PubMed] [Google Scholar]
- Chang BH, Mukherji S, Soderling TR. Characterization of a calmodulin kinase II inhibitor protein in brain. Proc Natl Acad Sci U S A. 1998;95:10890–10895. doi: 10.1073/pnas.95.18.10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschênes-Furry J, Perrone-Bizzozero N, Jasmin BJ. The RNA-binding protein HuD: a regulator of neuronal differentiation, maintenance and plasticity. Bioessays. 2006;28:822–833. doi: 10.1002/bies.20449. [DOI] [PubMed] [Google Scholar]
- Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elul TM, Kimes NE, Kohwi M, Reichardt LF. N- and C-terminal domains of β-catenin, respectively, are required to initiate and shape axon arbors of retinal ganglion cells in vivo. J Neurosci. 2003;23:6567–6575. doi: 10.1523/JNEUROSCI.23-16-06567.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink CC, Bayer KU, Myers JW, Ferrell JE, Jr, Schulman H, Meyer T. Selective regulation of neurite extension and synapse formation by the beta but not the alpha isoform of CaMKII. Neuron. 2003;39:283–297. doi: 10.1016/s0896-6273(03)00428-8. [DOI] [PubMed] [Google Scholar]
- He X, Yang F, Xie Z, Lu B. Intracellular Ca(2+) and Ca(2+)/calmodulin-dependent kinase II mediate acute potentiation of neurotransmitter release by neurotrophin-3. J Cell Biol. 2000;149:783–792. doi: 10.1083/jcb.149.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengst U, Deglincerti A, Kim HJ, Jeon NL, Jaffrey SR. Axonal elongation triggered by stimulus-induced local translation of a polarity complex protein. Nat Cell Biol. 2009;11:1024–1030. doi: 10.1038/ncb1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Jung MY, Sarkissian M, Richter JD. N-methyl-d-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. EMBO J. 2002;21:2139–2148. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Carson JH, Barbarese E, Richter JD. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003;17:638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Hydén H. Protein metabolism in the nerve cell during growth and function. Acta Physiol Scand. 1943;6(Suppl 17):1–136. [Google Scholar]
- Jones KJ, Korb E, Kundel MA, Kochanek AR, Kabraji S, McEvoy M, Shin CY, Wells DG. CPEB1 regulates beta-catenin mRNA translation and cell migration in astrocytes. Glia. 2008;56:1401–1413. doi: 10.1002/glia.20707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labelle C, Leclerc N. Exogenous BDNF, NT-3 and NT-4 differentially regulate neurite outgrowth in cultured hippocampal neurons. Brain Res Dev Brain Res. 2000;123:1–11. doi: 10.1016/s0165-3806(00)00069-9. [DOI] [PubMed] [Google Scholar]
- Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AC, Holt CE. Local translation and directional steering in axons. EMBO J. 2007;26:3729–3736. doi: 10.1038/sj.emboj.7601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Kanaji T, Nakade S, Kanno T, Mikoshiba K. 2APB, 2-aminoethoxydiphenyl borate, a membrane-penetrable modulator of Ins(1,4,5)P3-induced Ca2+ release. J Biochem. 1997;122:498–505. doi: 10.1093/oxfordjournals.jbchem.a021780. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15:791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- McEvoy M, Cao G, Llopis PM, Kundel M, Jones K, Hofler C, Shin C, Wells DG. Cytoplasmic polyadenylation element binding protein 1-mediated mRNA translation in Purkinje neurons is required for cerebellar long-term depression and motor coordination. J Neurosci. 2007;27:6400–6411. doi: 10.1523/JNEUROSCI.5211-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Mendez R, Murthy KG, Ryan K, Manley JL, Richter JD. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell. 2000a;6:1253–1259. doi: 10.1016/s1097-2765(00)00121-0. [DOI] [PubMed] [Google Scholar]
- Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000b;404:302–307. doi: 10.1038/35005126. [DOI] [PubMed] [Google Scholar]
- Menn B, Timsit S, Represa A, Mateos S, Calothy G, Lamballe F. Spatiotemporal expression of noncatalytic TrkC NC2 isoform during early and late CNS neurogenesis: a comparative study with TrkC catalytic and p75NTR receptors. Eur J Neurosci. 2000;12:3211–3223. doi: 10.1046/j.1460-9568.2000.00215.x. [DOI] [PubMed] [Google Scholar]
- Ming GL, Wong ST, Henley J, Yuan XB, Song HJ, Spitzer NC, Poo MM. Adaptation in the chemotactic guidance of nerve growth cones. Nature. 2002;417:411–418. doi: 10.1038/nature745. [DOI] [PubMed] [Google Scholar]
- Oh B, Hwang S, McLaughlin J, Solter D, Knowles BB. Timely translation during the mouse oocyte-to-embryo transition. Development. 2000;127:3795–3803. doi: 10.1242/dev.127.17.3795. [DOI] [PubMed] [Google Scholar]
- Piper M, Anderson R, Dwivedy A, Weinl C, van Horck F, Leung KM, Cogill E, Holt C. Signaling mechanisms underlying Slit2-induced collapse of Xenopus retinal growth cones. Neuron. 2006;49:215–228. doi: 10.1016/j.neuron.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverte CG, Ahearn MD, Hake LE. CPEB degradation during Xenopus oocyte maturation requires a PEST domain and the 26S proteasome. Dev Biol. 2001;231:447–458. doi: 10.1006/dbio.2001.0153. [DOI] [PubMed] [Google Scholar]
- Rossoll W, Jablonka S, Andreassi C, Kröning AK, Karle K, Monani UR, Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112:63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- Shin CY, Kundel M, Wells DG. Rapid, activity-induced increase in tissue plasminogen activator is mediated by metabotropic glutamate receptor-dependent mRNA translation. J Neurosci. 2004;24:9425–9433. doi: 10.1523/JNEUROSCI.2457-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Kalil K. Netrin-1 induces axon branching in developing cortical neurons by frequency-dependent calcium signaling pathways. J Neurosci. 2005;25:6702–6715. doi: 10.1523/JNEUROSCI.0871-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson VM. The fine structure of the axon and growth cone of the dorsal root neuroblast of the rabbit embryo. J Cell Biol. 1970;44:62–79. doi: 10.1083/jcb.44.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai NP, Bi J, Wei LN. The adaptor Grb7 links netrin-1 signaling to regulation of mRNA translation. EMBO J. 2007;26:1522–1531. doi: 10.1038/sj.emboj.7601598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vest RS, Davies KD, O'Leary H, Port JD, Bayer KU. Dual mechanism of a natural CaMKII inhibitor. Mol Biol Cell. 2007;18:5024–5033. doi: 10.1091/mbc.E07-02-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DG. RNA-binding proteins: a lesson in repression. J Neurosci. 2006;26:7135–7138. doi: 10.1523/JNEUROSCI.1795-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DG, Dong X, Quinlan EM, Huang YS, Bear MF, Richter JD, Fallon JR. A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J Neurosci. 2001;21:9541–9548. doi: 10.1523/JNEUROSCI.21-24-09541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Guirland C, Ming GL, Zheng JQ. A CaMKII/calcineurin switch controls the direction of Ca(2+)-dependent growth cone guidance. Neuron. 2004;43:835–846. doi: 10.1016/j.neuron.2004.08.037. [DOI] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- Yang F, He X, Feng L, Mizuno K, Liu XW, Russell J, Xiong WC, Lu B. PI-3 kinase and IP3 are both necessary and sufficient to mediate NT3-induced synaptic potentiation. Nat Neurosci. 2001;4:19–28. doi: 10.1038/82858. [DOI] [PubMed] [Google Scholar]
- Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci. 2006;9:1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- Yoshimura T, Kawano Y, Arimura N, Kawabata S, Kikuchi A, Kaibuchi K. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell. 2005;120:137–149. doi: 10.1016/j.cell.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Yu X, Malenka RC. Beta-catenin is critical for dendritic morphogenesis. Nat Neurosci. 2003;6:1169–1177. doi: 10.1038/nn1132. [DOI] [PubMed] [Google Scholar]
- Yu X, Malenka RC. Multiple functions for the cadherin/catenin complex during neuronal development. Neuropharmacology. 2004;47:779–786. doi: 10.1016/j.neuropharm.2004.07.031. [DOI] [PubMed] [Google Scholar]
- Zhang HL, Singer RH, Bassell GJ. Neurotrophin regulation of beta-actin mRNA and protein localization within growth cones. J Cell Biol. 1999;147:59–70. doi: 10.1083/jcb.147.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- Zhou FQ, Zhou J, Dedhar S, Wu YH, Snider WD. NGF-induced axon growth is mediated by localized inactivation of GSK-3beta and functions of the microtubule plus end binding protein APC. Neuron. 2004;42:897–912. doi: 10.1016/j.neuron.2004.05.011. [DOI] [PubMed] [Google Scholar]