Abstract
Microbial diversity of sediments from the northern slope of the South China Sea was studied by constructing bacterial and archaeal 16S rRNA gene clone libraries. Fourteen bacterial phylogenetic groups were detected, including Gammaproteobacteria, Deltaproteobacteria, Planctomycetes, Alphaproteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, Chloroflexi, Acidobacteria, Betaproteobacteria, Nitrospirae, candidate divisions OP8 and OP11, and an unknown group. Gammaproteobacteria was the predominant group in bacterial libraries with the percentage ranging from 31.8% to 63.2%. However, archaeal libraries had relatively lower diversity, with most clones belonging to marine archaeal group I uncultured Crenarchaeota. In addition, two novel euryarchaeal clones were detected not to match any culture-dependent or -independent isolates. Compared with other gas hydrate-rich ecosystems and different areas of the South China Sea, a distinct microbial community was revealed in this study.
Keywords: 16S rRNA, Library, Diversity, South China Sea
INTRODUCTION
The South China Sea close to the West Pacific “warm pool” is the biggest and deepest sea in China, as well as one of the largest marginal seas in the world (Lai et al., 2006; Li et al., 2008b). Gas hydrates in deep marine environments are solid compounds, which contain mainly methane and water, formed due to high pressure, low temperature, abundant gas, and other unknown factors under deep-sea conditions (Mills et al., 2005). The South China Sea exhibits a great potential for gas hydrates presence. The northern slope covers an area of 21×104 km2, which takes up approximately 6% of total area of the South China Sea (Yu et al., 2004). Like many other continental slope margins, it is an important component of gas hydrate-bearing area in the South China Sea (Lin et al., 2005; Wu et al., 2008).
Microorganisms play a significant role in the formation of gas hydrates (Kvenvolden, 1995). Microbial communities can be apparently influenced by the presence of gas hydrates (Inagaki et al., 2006). However, only a few studies have been carried out to survey the microbial diversity of the South China Sea, including Qiongdongnan Basin (Jiang et al., 2007), Xisha Trough (Li et al., 2008b), northern slope (17°57.70′ N, 114°57.33′ E) (Wang and Li, 2008), and southern slope (Li et al., 2008a). The diverse microbial communities living in vast areas of the South China Sea are still poorly known. In this study, we investigated the diversity of bacteria and archaea in marine sediments from three sites on the northern slope of the South China Sea.
MATERIALS AND METHODS
Sampling site information
Characteristics of three sampling sites were described in Table 1. Sediment samples from the upper 35 cm of the seabed were collected by multicorer on the northern slope of the South China Sea in the summer of 2006, and stored at −80 °C until transported to laboratory for storage at −20 °C. Muddy sediment cores with depths of 0~5 cm (approximately 50 g wet weight) from three sampling sites were used for diversity analysis. All processes were aseptic to avoid contamination.
Table 1.
Information of sampling sites
| Sites | Longitude | Latitude | Water depth (m) | Seafloor |
| S0610 | 118°53′ | 22°08′ | 546 | Mud |
| S0604 | 118°40′ | 21°57′ | 1211 | Mud |
| S0615 | 119°06′ | 22°05′ | 1285 | Mud |
DNA extraction and polymerase chain reaction amplification
Total genomic DNA in a 500-mg sediment from each sample was extracted directly using FastDNA® Spin Kit for soil (Q-BIOgene, Carlsbad, CA, USA), as described previously (Polymenakou et al., 2005). The DNA extracts were diluted 10-fold prior to polymerase chain reaction (PCR) amplification to reduce inhibition by contaminants. Bacterial 16S rRNA gene was amplified by PCR with universal primer 1492r (5′-GGTTACCTTGTTACGACTT-3′) (Eden et al., 1991; Dojka et al., 1998) and bacterial specific primer 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) (Dojka et al., 1998; Tanner et al., 1998). Primers A571F (5′-GCCTAAAGCGTCCGTAGC-3′) and UA1204R (5′-TTCGGGGCATACTGACCT-3′) were used to amplify archaeal partial 16S rRNA gene (around 650 bp) (Baker et al., 2003). PCR reaction mixtures contained 1 to 4 ng diluted DNA extracts, 1× PCR buffer (1.5 mmol/L MgCl2, 10 mmol/L Tris-HCl pH 8.3, 50 mmol/L KCl), 0.4 μmol/L of each primer, 200 μmol/L dNTPs, and 2 U Taq DNA polymerase (TaKaRa, Japan), and Mill-Q water added to a final volume of 50 μl. Thermal cycling for both bacteria and archaea modified to reduce PCR bias was 94 °C for 5 min, followed by 29 cycles of 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 1.5 min, and a final extension at 72 °C for 10 min. Each site was amplified in two replicate PCR reactions of 50 μl. Negative (no-template) control was used to exclude contamination. PCR products were electrophoresed in 1.5% (w/v) low-melting agarose gels and extracted with the AxyPrep™ DNA Gel Extraction Kit (Axygen, USA).
Cloning and sequencing
Bacterial and archaeal libraries were constructed for each site, including bS0610, bS0615 and bS0604 for bacterial libraries, and aS0610, aS0615 and aS0604 for archaeal libraries. The equivalent amount of PCR products of each sample was cloned into pMD19-T vectors (TaKaRa, Japan), and transformed into Escherichia coli DH5α competent cells. Transformants were screened out by the blue-white screening system and picked out randomly for plasmid extraction. Recombinant plasmids were identified by agarose gel electrophoresis after digestion with two restriction enzymes BamHI and HindIII (TaKaRa, Japan), and sequenced using primer 27f for bacterial clones and A571F for archaeal clones on an ABI 3730 sequencer at Chinese National Human Genome Center at Shanghai. Sequences over 600 bp with sound quality were used for further analysis. Sequences were checked for chimeras with CHECK_CHIMERA software of Ribosomal Database Project II (Maidak et al., 2001), and cross-checked with Pintail program (Ashelford et al., 2005).
Statistical analysis of diversity and differences between libraries
Chimera-free sequences in each library were aligned separately by CLUSTALW online, and the Phylip output format files were used to calculate distance matrices by DNADIST program contained in Phylip 3.67 program package. Distance-based OTU and richness program (DOTUR) (Schloss and Handelsman, 2005) was used to assign OTUs and calculate diversity indices including abundance-based coverage estimator (ACE) (Chao, 1987), Chao1 (Kemp and Aller, 2004), Simpson and Shannon (Zhang et al., 2008).
To determine the significance of differences between bacterial libraries, web LIBSHUFF version 0.96 (http://libshuff.mib.uga.edu/) was used following Singleton et al.(2001)’s method. A LIBSHUFF comparison of libraries yielded the following formula using the Bonferroni correction:
| 0.05=1−(1−a)k(k−1), |
where a is the critical P-value and k is the number of libraries. The critical P-value is 0.0085 when three libraries are compared. If any comparison of two libraries has a lower P-value below or at 0.0085, then there is a 95% confidence to believe that those two libraries are significantly different in community composition.
Phylogenetic analysis
One representative clone was chosen for each OTU, and then submitted to BLAST program and Ribosomal Database Project II program online to obtain the closest published relatives. Phylogenetic trees were constructed by MEGA software version 4.0 using Neighbor-Joining method (Saitou and Nei, 1987) with Kimura 2-parameter model.
GenBank accession numbers
All partial 16S rRNA gene sequences determined in the present study were deposited in GenBank under the accession numbers EU886378 to EU886464 for bacterial clones, and GQ180871 to GQ180903 for archaeal clones.
RESULTS
Diversity analysis and differences between bacterial libraries
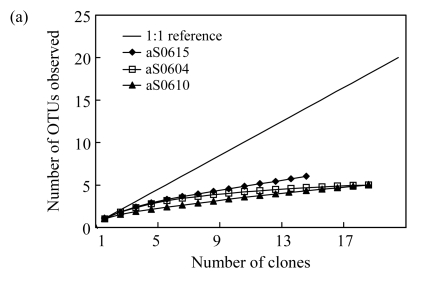
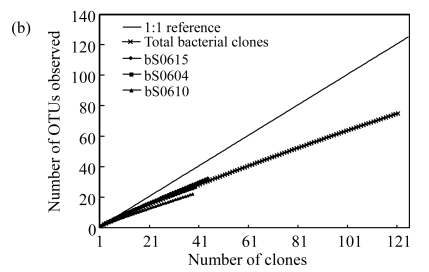
OTUs and diversity estimators were all determined at 3% 16S rDNA sequence difference level by DOTUR program (Table 2). In all, 22, 26, and 32 OTUs were obtained in 3 bacterial libraries bS0610, bS0604, and bS0615, respectively. However, archaeal libraries had much fewer OTUs, with only 5, 5, and 6 OTUs assigned in libraries aS0610, aS0604, and aS0615, respectively. As shown in Fig.1a, rarefaction curves for archaeal libraries almost reach asymptote, indicating that the archaeal community was well sampled with low diversity. In contrast, rarefaction curves for bacterial libraries fail to approach a plateau (Fig.1b), indicating a high bacterial diversity. Previous studies suggested that the rarefaction curves were not saturated even if hundreds of bacterial 16S rRNA gene clones were retrieved. But they did reveal important information about the relative diversity. The curves for bacterial libraries (Fig.1b) reveal the same tendency with the curves for archaeal libraries (Fig.1a) in which diversity appears to increase with depth, supported by the diversity estimators (Table 2).
Table 2.
Diversity indices (calculated at 0.03 difference level) of bacterial and archaeal libraries
| Library | No. of Clones | No. of OTUs | ACE | Chao1 | Shannon | Simpson |
| bS0610 | 38 | 22 | 232.000 | 117 | 2.434 | 0.172 |
| bS0604 | 39 | 26 | 156.114 | 79 | 2.965 | 0.057 |
| bS0615 | 44 | 32 | 422.395 | 235 | 3.154 | 0.052 |
| aS0610 | 18 | 5 | 11.000 | 7 | 0.961 | 0.517 |
| aS0604 | 18 | 5 | 5.628 | 5 | 1.382 | 0.249 |
| aS0615 | 14 | 6 | 13.656 | 12 | 1.475 | 0.231 |
Fig.1.
Rarefaction analysis comparing the relative richness of (a) archaea and (b) bacteria among three libraries
The observed numbers of OTUs identified by DOTUR program at 3% difference level are plotted against number of clones in library. The curve of 1:1 reference means that each sequenced clone belongs to a unique OTU. Forks curve represents the rarefaction curve for total bacterial clones from three libraries in (b)
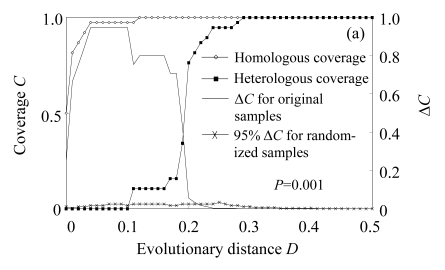
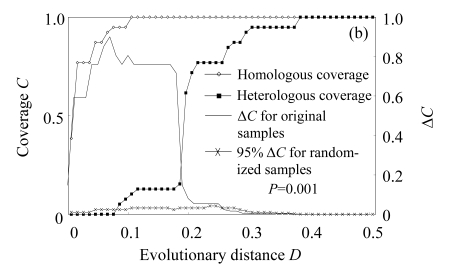
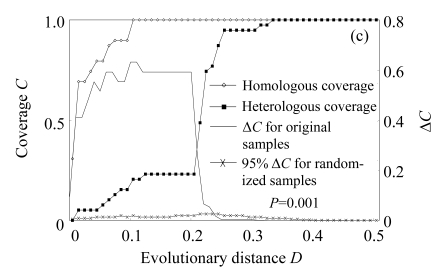
The web LIBSHUFF program was used to determine the significance of differences between libraries. P-value of pairwise comparisons was 0.001 (below the critical P-value 0.0085) in our comparisons, indicating libraries were significantly different in community composition, with a 95% confidence. Besides, the difference between homologous and heterologous coverage curves was determined by the distribution of delta-C (ΔC) as a function of evolutionary distance (D) (Fig.2). If two libraries are identical, the value of delta-C (ΔC) will be very small. Our results of all the comparisons showed significant differences between libraries, with considerable ΔC values at D below 0.2. All these results supported a conclusion that bacterial libraries had significantly different community composition.
Fig.2.
Results of selected LIBSHUFF comparisons of (a) libraries bS0610 (X) to bS0615 (Y), (b) libraries bS0604 (X) to bS0610 (Y) and (c) libraries bS0604 (X) to bS0615 (Y)
Analysis of bacterial and archaeal libraries
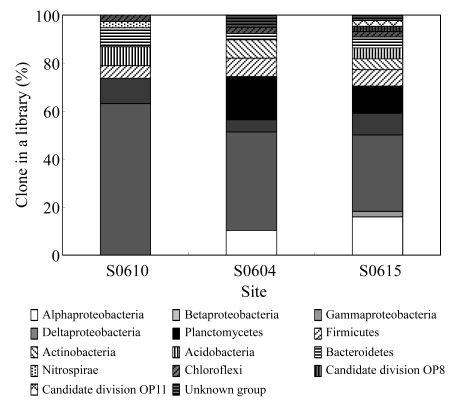
A total of 121 sequences were generated from 3 bacterial libraries, with 14 different phylogenetic groups being identified (Table 3). Proteobacteria (with 76 clones) dominated in the bacterial community, Gammaproteobacteria in particular, accounting for 63.2%, 41.0%, and 31.8% in libraries bS0610, bS0604, and bS0615, respectively (Fig.3). Gammaproteobacteria, Deltaproteobacteria, Firmicutes, Bacteroidetes, and Chloroflexi were commonly detected in three libraries, while Betaproteobacteria, Nitrospirae, and candidate divisions OP8 and OP11 were seldom detected, with only one clone in each group. Taking three libraries as a whole represent of the South China Sea, bacteria affiliated with Alphaproteobacteria, Gammaproteobacteria, Deltaproteobacteria, and Planctomycetes took up 9.1%, 44.6%, 8.3%, and 9.9% of the total 121 clones, respectively. The percentages of remaining phylogenetic groups ranged from 0.8% to 6.6%.
Table 3.
16S rDNA phylotype distribution in three bacterial libraries
| Phylogenetic groups | Bacterial librariesa |
||
| bS0610 | bS0604 | bS0615 | |
| Alphaproteobacteria | 0 | 4 | 7 |
| Betaproteobacteria | 0 | 0 | 1 |
| Gammaproteobacteria | 24 | 16 | 14 |
| Deltaproteobacteria | 4 | 2 | 4 |
| Planctomycetes | 0 | 7 | 5 |
| Firmicutes | 2 | 3 | 3 |
| Actinobacteria | 0 | 3 | 2 |
| Acidobacteria | 3 | 0 | 2 |
| Bacteroidetes | 3 | 1 | 2 |
| Nitrospirae | 1 | 0 | 0 |
| Chloroflexi | 1 | 1 | 1 |
| Candidate division OP8 | 0 | 0 | 1 |
| Candidate division OP11 | 0 | 0 | 1 |
| Unknown group | 0 | 2 | 1 |
Number of clones affiliated with each phylogenetic group
Fig.3.
16S rDNA phylotype comparison of three bacterial libraries
Archaeal libraries were much simpler with a lower diversity. Marine archaeal group I was the dominant group, which took up 100%, 83%, and 78.6% of aS0604, aS0610, and aS0615, respectively. Furthermore, Candidatus Nitrososphaera, deep-sea archaeal group (DSAG), and an unknown group were found in Crenarchaea. Crenarchaea took up 96% of the total archaeal clones. Merely two clones from library aS0610 (aS0610-10 and aS0610-16) belonged to Euryarchaea.
Phylogenetic analysis of bacterial libraries
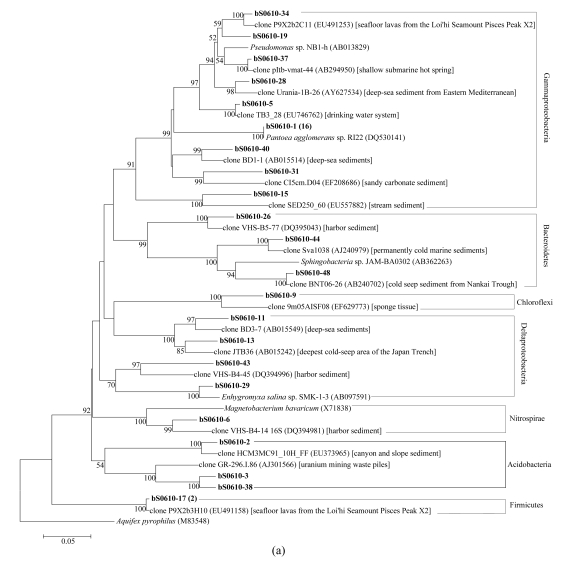
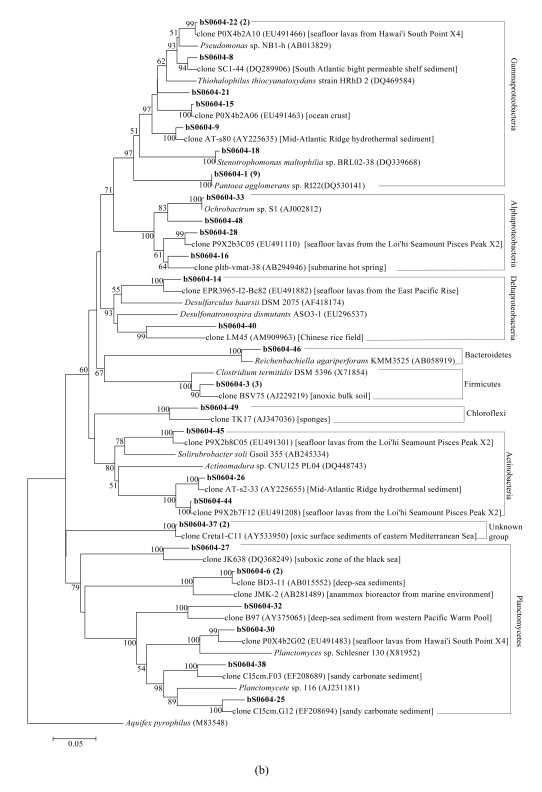
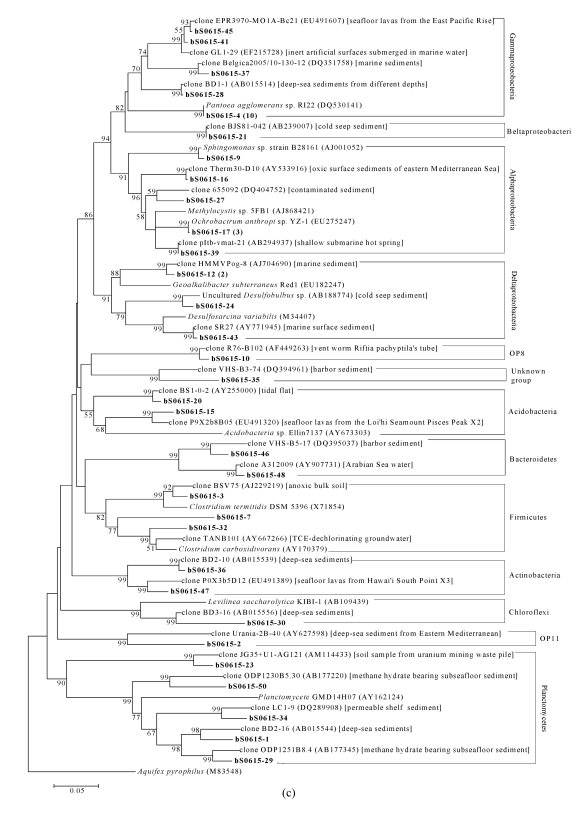
In total, 7, 9, and 13 phylogenetic groups were identified in bacterial libraries bS0610, bS0604, and bS0615, respectively (Fig.4). The most abundant OTU was affiliated to Gammaproteobacteria with the closest relative Pantoea agglomerans sp. RI22 (DQ530141). A few OTUs were affiliated with established groups that contained isolated representatives, including Pantoea, Pseudomonas, and Stenotrophomonas in Gammaproteobacteria, Ochrobactrum and Sphingomonas in Alphaproteobacteria, Desulfobulbus and Enhygromyxa in Deltaproteobacteria, and Clostridium in Firmicutes. However, the majority of bacterial clones were closely related to uncultured clones from marine environments, except for a few ones from non-marine environments such as Chinese rice field and uranium mining waste piles. Two novel OTUs (bS0604-37 and bS0615-35), assigned to unknown group, were found in libraries bS0604 and bS0615. OTU bS0604-37 (two related clones) were the closest to clone Creta1-C11 (AY533950) obtained from oxic surface sediments, and formed an independent branch that was far away from the remaining groups (Fig.4b). OTU bS0615-35, distantly related to clone VHS-B3-74 (DQ394961) from harbor sediment with 86% identity, formed a sister branch with candidate division OP8 (Fig.4c).
Fig.4.
Phylogenetic trees showing the relationship of bacterial 16S rDNA sequences in libraries (a) bS0610, (b) bS0604, and (c) bS0615 to relatives in GenBank
The trees were constructed by bootstrap neighbor-joining method in MEGA 4.0. Clones in bold were obtained in the present study, and numbers in parentheses showed the number of related clones. The environments where relative clones were obtained from were given in square brackets. Bootstrap values under 50% were not shown. Aquifex pyrophilus was used as the outgroup. Bar, 0.05 substitutions per nucleotide position
Phylogenetic analysis of archaeal libraries
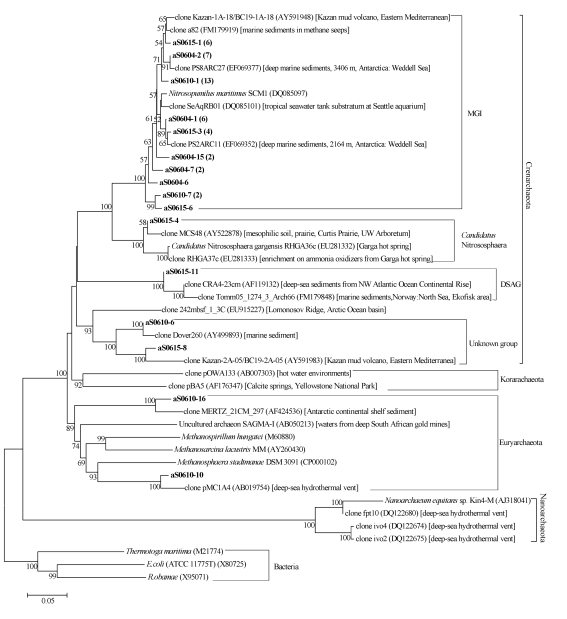
A distinct diversity of archaeal community was revealed (Fig.5). In total, 100%, 83%, and 78.6% of archaeal clones in libraries aS0604, aS0610, and aS0615 belonged to marine archaeal group I, respectively. Relatives of this group included uncultured clones retrieved from surface sediments at 2164 m and 3406 m depth in the Weddell Sea, Antarctica (Gillan and Danis, 2007) and the isolate Nitrosopumilus maritimus SCM1 (DQ085097). All clones of marine archaeal group I shared high similarity (>96%) with each other, and formed a sister branch with Candidatus Nitrososphaera containing clone aS0615-4. In addition, DSAG and an unknown group were detected in Crenarchaeota. Only two clones, aS0610-16 and aS0610-10, were grouped into Euryarchaeota, and shared low similarity with other isolates or established groups in Euryarchaeota.
Fig.5.
Phylogenetic trees showing the relationship of archaeal 16S rDNA sequences in libraries aS0610, aS0604, and aS0615 to relatives in GenBank
The trees were constructed by bootstrap neighbor-joining method in MEGA 4.0. Clones in bold were obtained in the present study, and numbers in parentheses showed the number of related clones. The environments which relative clones were obtained from were given in square brackets. Bootstrap values under 50% were not shown. Bar, 0.05 substitutions per nucleotide position
DISCUSSION
Proteobacteria are the dominant bacteria in all three libraries, which is consistent with previous diversity investigation of marine sediments from the South China Sea (Xu et al., 2004; Lai et al., 2006; Li et al., 2008a; 2008b). Because they are the most metabolically diverse bacteria by far, Proteobacteria appear in various environments and play crucial roles in cycling of chemical elements. Even in deep-sea environments, they can still dominate in the bacterial community, which is also suggested by our results.
The northern slope of the South China Sea is considered to contain large amount of oil/gas/gas-hydrate resources (Jiang et al., 2007). The microbial community may be special in such an environment. In the present study, Deltaproteobacteria were found in all three libraries. A Desulfobulbus related sequence (bS0615-24), which shares 95% identity with the relative uncultured Desulfobulbus sp., was identified in library bS0615. Some species isolated from oilfields or water-oil separation system belong to Desulfobulbus (Lien et al., 1998). Furthermore, it is an interesting discovery that sulfate-reducing bacteria (SRB) are syntrophically associated with uncultured anaerobic methane-oxidizing archaea (ANME) to form a complex consortia in methane-rich deep marine sediments, and exhaust a large portion of methane from marine ecosystem (Pernthaler et al., 2008). Desulfobulbus is a commonly found syntrophic partner in the consortia (Niemann et al., 2006). Thus, the detection of Desulfobulbus may give some clues to understand the particular role of bacterial community inhabiting our sampling environment. These related clones may participate in sulfur cycling through sulfate reduction, and cooperate with ANME group to consume methane. However, no ANME group is detected in our archaeal libraries, except two Euryarchaeota clones that are distantly related to methanogenic bacteria such as Methanosphaera. Previously, ANME group from methane hydrate sites was not detected, although sulfate-reducing bacteria were observed (Inagaki et al., 2006). A possible explanation is that bacterial biomass is much greater than archaeal biomass in these ecosystems, which leads to the miss of ANME group in culture-independent analysis.
Bacterial JS1 candidate group is a major methane-associated group recovered from Pacific Ocean margins including Peru margin and Cascadia margin, where methane hydrates are in great concentrations, and acts as an indicator of methane presence (Inagaki et al., 2006). Planctomycetes were also found abundant in these methane hydrate-rich sediments. However, no bacterial clone is affiliated with JS1 group in our libraries. Referring to previous studies of the South China Sea (Xu et al., 2004; Jiang et al., 2007; Lai et al., 2006; Li et al., 2008a; 2008b; Wang and Li, 2008), we conclude that no JS1 group has ever been discovered in the South China Sea margins. We, thus, hypothesize that JS1 group is location-specific and cannot be used as a universal indicator for methane presence in different marine ecosystems. In our study, Planctomycetes was detected as the most abundant group in addition to Proteobacteria in two libraries (bS0604 and bS0615) constructed from deeper sediments (>1 200 m water depth), but was absent in library bS0610 constructed from shallow sediment (546 m water depth). This result suggests that Planctomycetes prefer deeper depth in our sampling environment.
Chloroflexi (or green non-sulfur bacteria) can be detected in all three libraries with only one clone in each library. A previous study of late Pleistocene organic-rich sediments (sapropels) from the eastern Mediterranean Sea has shown that as high as 70% of total bacteria belonged to uncultured green non-sulfur bacteria, and this high percentage of green non-sulfur bacteria was associated with organic-rich sediments (Coolen et al., 2002). Chloroflexi was also found to be abundant in organic-rich, hydrate-free sites of the Pacific Ocean margins, but in a very small portion in hydrate-rich sediment (Inagaki et al., 2006). The detection of few Chloroflexi clones in the present study is consistent with the character of methane hydrate-bearing ecosystem.
Candidate divisions OP8 and OP11 are unique in library bS0615 constructed from the deepest sediment samples. The candidate division OP series were firstly discovered in a hot spring Obsidian Pool in Yellowstone National Park of America (Hugenholtz et al., 1998). These bacteria are distantly related to known isolates and clones, and have not been cultured till now. Our discovery of the candidate divisions OP8 and OP11 is consistent with previous studies in which these OP series were also identified in hydrocarbon-containing soil samples under methanogenic conditions (Dojka et al., 1998; Hugenholtz et al., 1998) and sediments from the South China Sea (Li et al., 2008a). This consistency may indicate a non-negligible role of these candidate divisions in hydrocarbon or methane-bearing environments.
Besides, some OTUs are affiliated with clones obtained from carbonate sediments (bS0610-31, bS0604-25, and bS0604-38), and methane hydrate-bearing sediments (bS0615-50 and bS0615-29). These environments are characterized by a high content of carbon, indicating a possible role in carbon cycling of these related clones.
Archaeal community inhabiting the northern slope of the South China Sea has a low diversity. Only two clones (aS0610-10 and aS0610-16) in library aS0610 belong to Euryarchaeota, while the remaining clones are all grouped in Crenarchaeota. Our results are consistent with previous studies that have discovered that Euryarchaeota are more abundant in upper marine water column, while Crenarchaeota are predominant in deeper sediments (Massana et al., 1997; Karner et al., 2001; Church et al., 2003). Marine archaeal group I is the most dominant part in Crenarchaeota, which accounts for 83%, 78.6%, and 100% in libraries aS0610, aS0615, and aS0604, respectively. Marine archaeal group I, also named as archaebacterium group 1, crenarchaeal group I, and marine group 1 Crenarchaeota, was first found in oxygenated coastal surface waters of North America and abundant in marine environments (Delong, 1992). Previous studies have revealed that marine archaeal group I Crenarchaeota, including the first isolated ammonia-oxidizing archaeon Nitrosopumilus maritimus (Könneke et al., 2005), play important roles in nitrogen cycling (Leininger et al., 2006; Gillan and Danis, 2007). Due to the ubiquity of the marine archaeal group I in various environments, the roles of these Crenarchaeota are supposed to be more versatile and key. Marine archaeal group I is possibly the most abundant archaeal group on Earth (Wang et al., 2005). Unexpectedly, marine archaeal group I was also found in a large proportion in methane hydrate-rich sediments from Peru margin and Cascadia margin. Thus, it is believed that marine archaeal group I may be an unusual group that plays an unknown role in methane metabolism.
Besides marine archaeal group I, Candidatus Nitrososphaera and DSAG (also named as marine benthic group B) were also detected in Crenarchaeota. Candidatus Nitrososphaera is the first described thermophilic ammonia-oxidizing Crenarchaea obtained from Garga hot spring enrichments (Hatzenpichler et al., 2008), and plays an important role in nitrogen cycling. DSAG is a dominant group detected in methane hydrates sites of Peru margin and Cascadia margin (Inagaki et al., 2006). DSAG also presents in the current study, although there is only one related clone.
No methanogens were found in our archaeal libraries. Previous studies showed that only a small proportion of methanogens in hydrate-bearing sediments could be detected using methanogen-specific primers (Inagaki et al., 2006). Thus, methanogens might be missed by universal archaeal primers.
Therefore, we propose that microbial community inhabiting sampling sediments from northern slope of the South China Sea may participate in nitrogen, carbon and sulfur cycling, with the dominance of nitrogen cycling in archaeal community. Further study is needed to capture more organisms. Since our clones are mostly close to uncultured relatives from environments, culture-dependent experiments should be taken to obtain isolates. Novel bacteria and archaea detected in the present study are also worth analyzing further. This study provides a primary knowledge of microbial diversity in sediments from the northern slope of the South China Sea, and indicates a distinct microbial community that contains potential novel species.
CONCLUSION
To conclude, both bacterial diversity and archaeal diversity were studied by 16S rRNA gene clone libraries constructed from sediments on the northern slope of the South China Sea at different depths. Fourteen phylogenetic groups including an unknown group were detected among three bacterial libraries. Proteobacteria dominated in the bacterial community, followed by Planctomycetes and Firmicutes. Most clones obtained in the present study were affiliated with uncultured bacteria from marine ecosystems, including methane hydrate-bearing environment, deep-sea sediments, hydrothermal vents, and so on. Archaeal community was much simpler with a lower diversity, and marine archaeal group I dominated significantly in archaeal libraries. Besides, two novel clones were found in Euryarchaeota. Microbial communities in the sampling sediments from northern slope of the South China Sea played an important role in nitrogen, carbon, and sulfur cyclings. The present study disclosed a distinct microbial community, and provided a primary analysis of microbial diversity of this special marine environment.
Acknowledgments
The authors thank Prof. M. Y. LONG (University of Chicago, USA) and Dr. Y. CAO (Zhejiang University, China) for English editing on the manuscript.
Footnotes
Project supported by the Open Fund of the Key Laboratory of Marine Geology and Environment, China Academy of Sciences (No. MGE2008KG05), the National Basic Research Program (973) of China (No. 2004CB719604-3), and the High-Tech Research and Development (863) of China (No. 2007AA021305)
References
- 1.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71(12):7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Meth. 2003;55(3):541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Chao A. Estimating the population-size for capture recapture data with unequal catchability. Biometrics. 1987;43(4):783–791. doi: 10.2307/2531532. [DOI] [PubMed] [Google Scholar]
- 4.Church MJ, Delong EF, Ducklow HW, Karner MB, Preston CM, Karl DM. Abundance and distribution of planktonic Archaea and Bacteria in the waters west of the Antarctic Peninsula. Limnol Oceanogr. 2003;48(5):1893–1902. [Google Scholar]
- 5.Coolen MJL, Cypionka H, Sass AM, Sass H, Overmann J. Ongoing modification of Mediterranean Pleistocene sapropels mediated by prokaryotes. Science. 2002;296(5577):2407–2410. doi: 10.1126/science.1071893. [DOI] [PubMed] [Google Scholar]
- 6.Delong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89(12):5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dojka MA, Hugenholtz P, Haack SK, Pace NR. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64(10):3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eden PA, Schmidt TM, Blakemore RP, Pace NR. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int J Syst Bacteriol. 1991;41(2):324–325. doi: 10.1099/00207713-41-2-324. [DOI] [PubMed] [Google Scholar]
- 9.Gillan DC, Danis B. The archaebacterial communities in Antarctic bathypelagic sediments. Deep-Sea Res Part II-Top Stud Oceanogr. 2007;54(16-17):1682–1690. doi: 10.1016/j.dsr2.2007.07.002. [DOI] [Google Scholar]
- 10.Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105(6):2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180(2):366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A, Suzuki M, Takai K, Delwiche M, Colwell FS, et al. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc Natl Acad Sci USA. 2006;103(8):2815–2820. doi: 10.1073/pnas.0511033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang HC, Dong HL, Ji SS, Ye Y, Wu NY. Microbial diversity in the deep marine sediments from the Qiongdongnan Basin in South China Sea. Geomicrobiol J. 2007;24:505–517. [Google Scholar]
- 14.Karner MB, Delong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409(6819):507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 15.Kemp PF, Aller JY. Estimating prokaryotic diversity: when are 16S rDNA libraries large enough? Limnol Oceanogr-Meth. 2004;2:114–125. [Google Scholar]
- 16.Könneke M, Bernhard AE, De La Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437(7058):543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 17.Kvenvolden KA. A review of the geochemistry of methane in natural gas hydrate. Org Geochem. 1995;23(11-12):997–1008. doi: 10.1016/0146-6380(96)00002-2. [DOI] [Google Scholar]
- 18.Lai XT, Zeng XF, Fang S, Huang YL, Cao LX, Zhou SN. Denaturing gradient gel electrophoresis (DGGE) analysis of bacterial community composition in deep-sea sediments of the South China Sea. World J Microbiol Biotechnol. 2006;22(12):1337–1345. doi: 10.1007/s11274-006-9181-x. [DOI] [Google Scholar]
- 19.Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442(7104):806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 20.Li T, Wang P, Wang PX. Bacterial and archaeal diversity in surface sediment from the south slope of the South China Sea. Wei Sheng Wu Xue Bao. 2008;48(3):323–329. (in Chinese) [PubMed] [Google Scholar]
- 21.Li T, Wang P, Wang PX. Microbial diversity in surface sediments of the Xisha Trough, the South China Sea. Acta Ecol Sin. 2008;28(3):1166–1173. doi: 10.1016/S1872-2032(08)60036-0. [DOI] [Google Scholar]
- 22.Lien T, Madsen M, Steen IH, Gjerdevik K. Desulfobulbus rhabdoformis sp. nov., a sulfate reducer from a water-oil separation system. Int J Syst Bacteriol. 1998;48(2):469–474. doi: 10.1099/00207713-48-2-469. [DOI] [PubMed] [Google Scholar]
- 23.Lin WD, Shen P, Xu YC, Zhou YZ. Geochemical characteristics and source of hydrocarbon gases of recent sediment from South China Sea. Acta Sedimentol Sin. 2005;23(1):170–174. (in Chinese) [Google Scholar]
- 24.Maidak BL, Cole JR, Lilburn TG, Parker CT, Saxman PR, Farris RJ, Garrity GM, Olsen GJ, Schmidt TM, Tiedje JM. The RDP-II (Ribosomal Database Project) Nucleic Acids Res. 2001;29(1):173–174. doi: 10.1093/nar/29.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massana R, Murray AE, Preston CM, Delong EF. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63(1):50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills HJ, Martinez RJ, Story S, Sobecky PA. Characterization of microbial community structure in gulf of Mexico gas hydrates: comparative analysis of DNA- and RNA-derived clone libraries. Appl Environ Microbiol. 2005;71(6):3235–3247. doi: 10.1128/AEM.71.6.3235-3247.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niemann H, Lösekann T, de Beer D, Elvert M, Nadalig T, Knittel K, Amann R, Sauter EJ, Schluter M, Klages M, et al. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature. 2006;443(7113):854–858. doi: 10.1038/nature05227. [DOI] [PubMed] [Google Scholar]
- 28.Pernthaler A, Dekas AE, Brown CT, Goffredi SK, Embaye T, Orphan VJ. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci USA. 2008;105(19):7052–7057. doi: 10.1073/pnas.0711303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polymenakou PN, Bertilsson S, Tselepides A, Stephanou EG. Links between geographic location, environmental factors, and microbial community composition in sediments of the Eastern Mediterranean Sea. Microb Ecol. 2005;49(3):367–378. doi: 10.1007/s00248-004-0274-5. [DOI] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71(3):1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singleton DR, Furlong MA, Rathbun SL, Whitman WB. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl Environ Microbiol. 2001;67(9):4374–4376. doi: 10.1128/AEM.67.9.4374-4376.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanner MA, Goebel BM, Dojka MA, Pace NR. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol. 1998;64(8):3110–3113. doi: 10.1128/aem.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang P, Li T. Phylogenetic analysis of bacterial community in deep-sea sediment from northern slope of the South China Sea. Marine Sciences. 2008;32(4):36–39. (in Chinese) [Google Scholar]
- 35.Wang P, Xiao X, Wang F. Phylogenetic analysis of Archaea in the deep-sea sediments of west Pacific Warm Pool. Extremophiles. 2005;9(3):209–217. doi: 10.1007/s00792-005-0436-5. [DOI] [PubMed] [Google Scholar]
- 36.Wu NY, Yang SX, Zhang HQ, et al. Preliminary Discussion on Gas Hydrate Reservoir System of Shenhu Area, North Slope of South China Sea. Proceedings of the 6th International Conference on Gas Hydrates (ICGH 2008); July 6~10, 2008; Vancouver, British Columbia, Canada. 2008. [Google Scholar]
- 37.Xu F, Dai X, Chen YQ, Zhou H, Cai JH, Qu HH. Phylogenetic diversity of bacteria and archaea in the Nansha marine sediments, as determined by 16S rDNA analysis. Oceanologia et Limnologia Sinica. 2004;35(1):89–94. (in Chinese) [Google Scholar]
- 38.Yu XH, Zhang ZJ, Su X, Chen F, Li Y. Primary discussion on accumulation conditions for sedimentation of gas hydrate and its distribution in South China Sea. Earth Science Frontiers (China University of Geosciences, Beijing) 2004;11(1):311–315. (in Chinese) [Google Scholar]
- 39.Zhang W, Ki JS, Qian PY. Microbial diversity in polluted harbor sediments I: bacterial community assessment based on four clone libraries of 16S rDNA. Estuar Coast Shelf Sci. 2008;76(3):668–681. doi: 10.1016/j.ecss.2007.07.040. [DOI] [Google Scholar]