Abstract
The fungal pathogen Phoma clematidina is used as a biological agent to control the invasive plant species Clematis vitalba in New Zealand. Research conducted on P. clematidina as a potential biocontrol agent against C. vitalba, led to the discovery of two perithecial-forming strains. To assess the diversity of P. clematidina and to clarify the teleomorph-anamorph relationship, phylogenetic analyses of 18 P. clematidina strains, reference strains representing the Phoma sections in the Didymellaceae and strains of related species associated with Clematis were conducted. Partial sequences of the ITS1, ITS2 and 5.8S rRNA gene, the ß-tubulin gene and 28S rRNA gene were used to clarify intra- and inter-species relationships. These analyses revealed that P. clematidina resolves into three well-supported clades which appear to be linked to differences in host specificity. Based on these findings, Didymella clematidis is newly described and the descriptions of P. clematidina and D. vitalbina are amended.
Keywords: Ascochyta vitalbae, ß-tubulin, Clematis, Didymella clematidis, Didymella vitalbina, DNA phylogeny, ITS, LSU, taxonomy
INTRODUCTION
The genus Clematis (Ranunculaceae) accommodates (semi-) woody, climbing plants and shrubs. Species of Clematis occur throughout the temperate regions of the northern and southern hemispheres and can also be found in the tropics and mountainous regions. Clematis contains more than 400 species, and more than 600 varieties are grown commercially. In the 19th century the cultivation of Clematis became popular but soon after the start of its large scale cultivation, a widespread destructive disease which caused high yield losses emerged in Europe and America (van de Graaf et al. 2001). This disease was referred to as Clematis wilt, exhibiting symptoms of stem rot and wilting of above-ground plant parts (Gloyer 1915). Ascochyta clematidina and Coniothyrium clematidis-rectae were identified as the causal organisms of Clematis wilt (Gloyer 1915, Blok 1965).
On the basis of new circumscriptions of Phoma and Ascochyta (Boerema & Bollen 1975), A. clematidina was transferred to Phoma as P. clematidina (Boerema & Dorenbosch 1979). Phoma clematidina is presently regarded as a widespread pathogen of Clematis spp. Incidentally, P. clematidina has also been isolated from plants other than Clematis, including a cultivated Selaginella sp. (Boerema & Dorenbosch 1979). Gloyer (1915) inoculated a series of plant species such as bean, pea, muskmelon, pumpkin, eggplant and elm with P. clematidina to assess its host range and found no development of disease symptoms. However, in the necrotic tissue at the point of inoculation developing pycnidia could be observed, indicating that P. clematidina may survive as a saprobe on different plant hosts.
Clematis vitalba (old man’s beard) is a vine that is native to Europe but has become widespread primarily due to its introduction as an ornamental. As an invasive plant species, C. vitalba is a threat to native trees and shrubs, as it reduces light levels and smothers crowns of trees with its prolific foliage (Gourlay et al. 2000). In New Zealand, C. vitalba is regarded as a serious pest, and much research has been undertaken in order to save the native forest remnants from disappearing due to smothering caused by C. vitalba (Hume et al. 1995, Ogle et al. 2000, Hill et al. 2001, 2004, Paynter et al. 2006). After extensive laboratory tests, a virulent strain of P. clematidina, which was originally isolated from an American C. ligusticifolia, was introduced to New Zealand in 1996 as a biological control agent of C. vitalba (Gourlay et al. 2000). Remarkably, a teleomorph was observed to develop on agar slants in vitro after storage for approximately 2 yr. A similar finding was observed in a strain isolated from C. vitalba from Switzerland.
In the present study the sexual strains of P. clematidina are phylogenetically and morphologically compared to reference strains housed in the culture collections of the Centraalbureau voor Schimmelcultures (CBS) and the Dutch Plant Protection Service (PD). The aims of this study were to assess the variation within this species, and to clarify the morphology of its potential sexual state.
MATERIALS AND METHODS
Fungal isolation and DNA extraction
Small fragments (< 1.0 mm2) of necrotic leaf tissue were removed with a dissecting needle and plated onto filtered V8-juice agar (V8) (Gams et al. 2007), and incubated at 20 °C under a 12 h near-ultraviolet / 12 h dark photo period. After 7 d, the colonies were subcultured onto fresh media. Strain CBS 123707 was isolated from leaves of C. vitalba plants at Gampelsteg, Swiss Valley, Switzerland (Table 1). Strain CBS 123705 was isolated from leaves of C. ligusticifolia at Toppenish, Washington State, USA (Table 1). Isolates were stored on V8 agar slants at 3 °C.
Table 1.
Isolates included in the phylogenetic analyses.
| Species | Accession no. 1 | Host | Origin | GenBank no. |
||
|---|---|---|---|---|---|---|
| ITS | TUB | LSU | ||||
| Coniothyrium clematidis‐rectae | CBS 507.63, PD 07/03486747 | Clematis sp. | Netherlands | FJ515606 | FJ515624 | FJ515647 |
| PD 95/1958 | Clematis sp. | Netherlands | FJ515607 | FJ515625 | FJ515648 | |
| Didymella vitalbina | CBS 454.64 | Clematis vitalba | France | FJ515605 | FJ515623 | FJ515646 |
| Phoma clematidina | CBS 201.49 | Clematis sp. | Netherlands | FJ426991 | FJ427102 | FJ515628 |
| CBS 195.64 | Clematis jackmannii | Netherlands | FJ426990 | FJ427101 | FJ515629 | |
| CBS 102.66 | Clematis sp. | England | FJ426988 | FJ427099 | FJ515630 | |
| CBS 520.66, PD 64/657 | Selaginella sp. | Netherlands | FJ426992 | FJ427103 | FJ515631 | |
| CBS 108.79, PD 78/522 | Clematis sp. | Netherlands | FJ426989 | FJ427100 | FJ515632 | |
| CBS 911.87 | Clematis vitalba | Germany | FJ515592 | FJ515610 | FJ515633 | |
| CBS 123705, PD 97/13460.1, ICMP 13664 | Clematis ligusticifolia | USA | FJ515593 | FJ515611 | FJ515634 | |
| CBS 123706, PD 08/04373904.5 | Clematis vitalba | Netherlands | FJ515594 | FJ515612 | FJ515635 | |
| CBS 123707, PD 97/13460.2, ICMP 13663 | Clematis vitalba | Switzerland | FJ515595 | FJ515613 | FJ515636 | |
| PD 75/294 | Clematis sp. | Unknown | FJ515596 | FJ515614 | FJ515637 | |
| PD 80/683 | Clematis sp. | Netherlands | FJ515597 | FJ515615 | FJ515638 | |
| PD 91/1865 | Clematis sp. | Netherlands | FJ515598 | FJ515616 | FJ515639 | |
| PD 95/895 | Clematis sp. | Netherlands | FJ515599 | FJ515617 | FJ515640 | |
| PD 97/12061 | Clematis cv. Purple spider | Netherlands | FJ515600 | FJ515618 | FJ515641 | |
| PD 97/12062 | Clematis cv. New Dawn | Netherlands | FJ515601 | FJ515619 | FJ515642 | |
| PD 99/2069 | Clematis sp. | England | FJ515602 | FJ515620 | FJ515643 | |
| PD 08/04373904.2B | Clematis vitalba | Netherlands | FJ515603 | FJ515621 | FJ515644 | |
| PD 08/04417700.3 | Clematis vitalba | Netherlands | FJ515604 | FJ515622 | FJ515645 | |
| Phoma complanata | CBS 268.92, PD 75/3 | Angelica sylvestris | Netherlands | FJ515608 | FJ515626 | EU754180 |
| Phoma exigua var. exigua | CBS 431.74, PD 74/2447 | Solanum tuberosum | Netherlands | FJ427001 | FJ427112 | EU754183 |
| Phoma glaucii | CBS 114.96, PD 94/888 | Chelidonium majus | Netherlands | FJ515609 | FJ515627 | FJ515649 |
| Phoma glomerata | CBS 528.66, PD 63/590 | Chrysanthemum sp. | Netherlands | FJ427013 | FJ427124 | EU754184 |
| Phoma herbarum | CBS 615.75, PD 73/665, ATCC 2499, IMI 199779 | Rosa multiflora | Netherlands | FJ427022 | FJ427133 | EU754186 |
| Phoma zeae‐maydis | CBS 588.69 | Zea mays | USA | FJ427086 | FJ427190 | EU754192 |
1ATCC: American Type Culture Collection, Virginia, USA; CBS: CBS Fungal Biodiversity Centre, Utrecht, The Netherlands; ICMP: International Collection of Micro-organisms from Plants, Auckland, New Zealand; IMI: International Mycological Institute, CABI-Bioscience, Egham, United Kingdom; PD: Dutch Plant Protection Service, Wageningen, The Netherlands.
For the phylogenetic study of the two Phoma strains isolated (CBS 123705, CBS 123707), all P. clematidina strains which were available from the CBS and PD collection, one Didymella vitalbina strain and six Phoma reference strains were included (Table 1). The Phoma reference strains represent the type species of the five Phoma sections recently classified in the Didymellaceae (de Gruyter et al. 2009), including the type species of the genus Phoma, P. herbarum (CBS 615.75). Phoma clematidina has been placed in Phoma sect. Heterospora (Boerema et al. 1997), however, the type species of this section, Phoma heteromorphospora proved not to be related to the Didymellaceae (de Gruyter et al. 2009). Therefore Phoma glaucii (CBS 114.96) was used as the reference strain for this section. Two C. clematidis-rectae strains were also included, as they are closely related to Phoma and have also been found associated with wilting symptoms of Clematis (Table 1).
DNA extraction from all isolates was performed using the Ultraclean Microbial DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA, USA), according to the manufacturer’s instructions. All DNA extracts were diluted 10× in milliQ water and stored at 4 °C before their use as PCR templates.
DNA amplification and phylogenetic analyses
For phylogenetic analyses, parts of the ITS1, ITS2 and 5.8S rRNA gene (ITS), the ß-tubulin gene (TUB) and 28S rRNA gene (LSU) were analysed. The primers V9G (de Hoog & Gerrits van den Ende 1998) and ITS4 (White et al. 1990) were used for the amplification of the ITS region, primers Btub2Fd (5’-GTB CAC CTY CAR ACC GGY CAR TG-3’) and Btub4Rd (5’-CCR GAY TGR CCR AAR ACR AAG TTG TC-3’) for the TUB region (J.Z. Groenewald, CBS) and primers LR0R (Rehner & Samuels 1994) and LR7 (Vilgalys & Hester 1990) for the LSU region. The LSU PCR was performed as described by de Gruyter et al. 2009. The ITS and TUB PCR mixtures both contained 0.5 units of Taq polymerase E (Genaxxon Bioscience, Biberach, Germany), 0.2 μM of each primer and 1× PCR buffer E incomplete (Genaxxon Bioscience). The remaining PCR mixture consisted of 0.5 μL diluted genomic DNA, 0.04 mM dNTPs and 1 mM MgCl2 for the ITS region and 1.0 μL diluted genomic DNA, 0.02 mM dNTPs and 2 mM MgCl2 for the TUB region. The amplification reactions were performed in a 2720 Thermal Cycler (Applied Biosystems, Foster City, California, USA) and had a total volume of 12.5 μL. Conditions for PCR amplification were comparable for both regions and consisted of an initial denaturation step of 5 min at 94 °C, followed by 35 cycles of denaturation, annealing and elongation and a final elongation step of 7 min at 72 °C. For the ITS region the 35 cycles consisted of 30 s at 94 °C, 30 s at 48 °C and 60 s at 72 °C, for the TUB region 30 s at 94 °C, 30 s at 52 °C and 30 s at 72 °C.
PCR amplicons were visualised by electrophoresis and sequenced as described by de Gruyter et al. (2009). DNA sequences obtained from forward and reverse primers were used to obtain consensus sequences using Bionumerics v. 4.60 (Applied Maths, St-Marthens-Lathem, Belgium) and phylogenetic analyses of the sequence data were conducted in PAUP v. 4.0b10 (Swofford 2003). To test whether the three different loci could be used in combined analyses, a partition homogeneity test was executed (Farris et al. 1995). Phylogenetic analyses consisted of Neighbour-Joining analysis with the uncorrected “p”, Jukes-Cantor and Kimura 2-parameter substitution models, and a parsimony analysis using the heuristic search option with 100 random taxa additions. Alignment gaps were set as fifth state, and tree bisection and reconstruction (TBR) was used as the branch-swapping algorithm. The robustness of the most parsimonious tree was evaluated by 1 000 bootstrap replicates (Hillis & Bull 1993). The resulting trees were printed with TreeView v. 1.6.6 (Page 1996) and are deposited in TreeBASE (www.treebase.org).
Morphology
Cultural characteristics of the strains (Table 1) were studied on oatmeal agar (OA) and malt extract agar (MEA) (Gams et al. 2007) as described by Boerema et al. 2004. The growth rates on both plates were examined after 7 and 14 d of incubation. Colony colours were determined after 2 wk using the colour charts of Rayner (1970).
Sexual structures were studied on V8 whereas other morphological features were described from OA as soon as sporulation occurred. Fruiting bodies were mounted in water and examined with the aid of a Nikon 80i light microscope. Pycnidial wall structure and the shape of the conidiogenous cells were studied using microtome sections of 9 μm thickness that were prepared with a Leica CM3050 freezing microtome and mounted in lactic acid. For electron microscopy, small segments (5 × 5 mm) of agar with pycnidia/perithecia were fixed in 3 % glutaraldehyde and 2 % formaldehyde in 0.1 M phosphate buffer (Karnovsky 1965) and prepared for scanning and transmission electron microscopy as previously described (Spiers & Hopcroft 1992).
RESULTS
Phylogenetic analyses of ITS, TUB and LSU
The partition homogeneity test indicated that the DNA sequence data from the three loci were combinable (P = 0.737). Concatenated sequences were thus used in all phylogenetic analyses. The combined alignment consisted of 2 150 bp (ITS 490 bp, TUB 333 bp, LSU 1327 bp), of which 1 978 characters were constant, 60 were parsimony uninformative and 112 were parsimony informative. The Neighbour-Joining trees obtained with the three different substitution models, and the single most parsimonious tree exhibited identical topology. The most parsimonious tree is presented in Fig. 1 (TL = 314 steps, CI = 0.697, RI = 0.888, RC = 0.619). This phylogenetic tree supports division of the P. clematidina strains into three distinct and well-supported groups (Fig. 1). A first group (clade A) contains the representative culture of P. clematidina CBS 108.79 (Boerema & Dorenbosch 1979) and strains isolated from symptomatic Clematis species and hybrids. A second group (clade B) comprises strains isolated from C. vitalba, including the freshly isolated strain CBS 123707, producing perithecia in pure culture, and the D. vitalbina strain CBS 454.64. Strain CBS 123705 is closely related to strains in clade B but forms a distinct clade (C) on its own. Strain PD 99/2069 clusters with the two Coniothyrium clematidis-rectae strains (100 % bootstrap support) among the other clades. The morphological characters of this strain proved to be similar to those of both C. clematidis-rectae strains and therefore strain 99/2069 requires renaming.
Fig. 1.
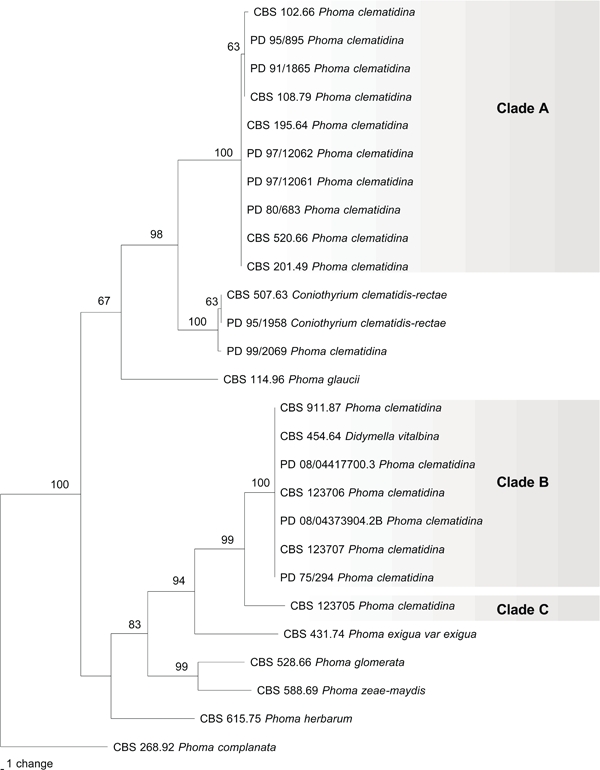
Parsimony tree obtained from a heuristic search with 100 random taxon additions of the combined ITS, BT and LSU sequences alignment. Scale bar indicates 1 change and bootstrap support values from 1 000 replicates are shown in percentages at the nodes.
Taxonomy
Clade A
Phoma clematidina (Thüm.) Boerema. Versl. Meded. Plziektenk. Dienst Wageningen 153 (Jaarb. 1978): 17. 1979
Basionym. Ascochyta clematidina Thüm., Bull Soc. Imp. Naturalistes Moscou 55: 98. 1880.
= Phyllosticta clematis Brunaud, Ann. Soc. Sci. Nat. Charente-Infér. 26: 9. 1889.
= Phyllosticta clematis Ellis & Dearn., Canad. Rec. Sci 5: 268. 1893; not Phyllosticta clematis Brunaud, see above.
= Ascochyta indusiata Bres., Hedwigia 35: 199. 1896.
= Ascochyta davidiana Kabát & Bubák, Oesterr. Bot. Z. 54: 25. 1904.
Description in vitro (amended from Boerema 1993). Pycnidia subglobose, mostly solitary on the agar surface, 110–120 μm diam, or larger, up to 350 μm diam, glabrous or with some hyphal outgrows around the ostioli. Ostioli 1(–3), papillate, relatively wide, up to 50 μm diam. Pycnidial wall 1–4 cells thick, pseudoparenchymatous, composed of isodiametric, somewhat elongated cells, dark pigmented around the ostioli. Conidiogenous cells phialidic, hyaline, simple, smooth, flask-shaped, 6–7.5 × 5.5–7 μm. Conidia ellipsoidal, occasionally slightly allantoid, thin-walled, smooth, hyaline, mostly aseptate, (3.5–)4–8.5(–9) × 2–3(–3.5) μm, occasionally larger and 1-septate, 9–13 × 3–4 μm, usually guttulate. Conidial matrix honey to salmon. Chlamydospores usually scanty, uni- or multicellular, where unicellular usually intercalary in short strains, guttulate, thick-walled, green-brown, 8–10 μm diam, where multicellular irregular dictyo/phragmosporous, often somewhat botryoid and in combination with unicellular chlamydospores, tan to dark brown, 3–50 × 12–25 μm.
Cultural characteristics — Colonies on OA: growth rate 50–65 mm diam after 7 d, with entire margin. Aerial mycelium present in irregular zones, felty or scarcely floccose, white to olivaceous-grey. Colonies olivaceous to iron-grey. Reverse similar. A rosy-buff discoloration of the agar medium often occurs due to the presence of anthraquinone needle-shaped crystals which persist after application of NaOH. Colonies on MEA: growth rate variable, 30–55 mm diam after 7 d, with entire margin. Aerial mycelium felty, white to pale olivaceous-grey, or absent near centre. Colonies rosy-buff to rosy-vinaceous. Reverse similar.
Specimens examined. Russia, Minussinsk, on leaves of Clematis glaucae, N. Martianoff, isotype LE 40082. – The Netherlands, Spaubeek, on the stem of Clematis sp., July 1978, G.H. Boerema, epitype designated here CBS H-16193, culture ex-epitype CBS 108.79 = PD 78/522.
Notes — The holotype has apparently been lost, and is not in LE or LEP. The isotype is selected here, with similar host, location and collector. The specimen and associated strain designated here as epitype represent the modified taxonomy of this species.
Clade B
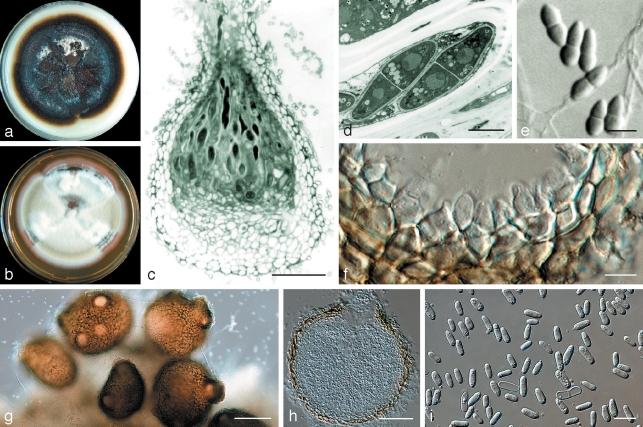
Didymella vitalbina Petr., Ann. Mycol. 38: 348. 1940 — Fig. 2
Fig. 2.
Didymella vitalbina (CBS 123707) a. Colony on OA after 14 d; b. colony on MEA after 14 d; c. longitudinal section through a perithecium; d. ascus; e. ascospores; f. pycnidial wall with conidiogenous cells; g. pycnidia; h. longitudinal section through a pycnidium; i. conidia. — Scale bars: c, h = 50 μm; d, f = 5 μm; e, i = 10 μm; g = 100 μm.
Anamorph. ‘Ascochyta’ vitalbae Briard & Har. apud Briard, Rev. Mycol. (Toulouse) 13: 17. 1891.
≡ Diplodina vitalbae (Briard & Har.) Allesch., Rabenh. Krypt.-Fl., ed. 2. Pilze 6 (Lief. 69): 683. 1900 (vol. dated 1901).
= Diplodina clematidina Fautrey & Roum. apud Roum., Rev. Mycol. (Toulouse) 14: 105. 1892.
Description in vitro. Perithecia superficial, solitary or clustered, globose/subglobose to pyriform, (75–)200–300 μm, with prominent, ostiolate, elongated neck, 30–60 μm. Perithecial wall black, textura globulosa, 6.5–10 μm, ectal excipulum 3–4 layers of elongated cells (c. 8 × 3 μm), medullary excipulum 8–10 layers of globular cells (5.5 × 5 μm), integrated with 6–8 basal layers of smaller globular cells (5 × 3 μm). Ascospore mass white. Asci bitunicate, 8-spored uniseriate, cylindrical with club-shaped base, 50–80 × 6.5–9.5 μm, paraphyses septate, but not obvious. Ascospores hyaline, septate, ovate to obpyriform, smooth, 9–15 × 3–5.5 μm (av. 11.2 × 4.1 μm). Pycnidia solitary or confluent, highly variable in shape and size, (sub)globose, to elongated or flask-shaped, glabrous, dark brown, superficial on the agar (105–)135–290(–330) × 95–210(–250) μm. Ostioli (1–)2–3(–5), prominent, 11–22 μm diam on an elongated neck. Pycnidial wall pseudoparenchymatous, thin, 5.5–9.5 μm, consisting of up to only 2 cell layers, outer cells isodiametric to oblong. Conidiogenous cells phialidic, hyaline, simple, smooth, variable in shape and size, 6.5–8.5 × 7–11 μm. Conidia ellipsoidal, hyaline, smooth, mainly aseptate, (5.5–)6.5–10(–11) × 2–4 μm, or 1-septate up to 18 × 4 μm, usually guttulate. Conidial matrix honey to rosy- buff/salmon. Chlamydospores absent.
Cultural characteristics — Colonies on OA: growth rate 50–60 mm diam after 7 d, with entire, smooth, sharp margins. Aerial mycelium absent or with some floccose tufts, white to (pale) olivaceous-grey. Colonies olivaceous to iron-grey. Reverse similar. Colonies on MEA: growth rate 45–55 mm diam after 7 d, with entire margin or undulate, smooth. Aerial mycelium felty, white to rosy-buff, near colony margin iron-grey. Colonies iron-grey to olivaceous. Reverse similar.
Specimens examined. Austria, Vienna, Gaisberg, on stem of Clematis vitalba, April 1939, F. Petrak, holotype 2644. – France, Var, Jouques, on leaves of Clematis vitalba, Dec. 1964, E. Müller, CBS H-11972, culture ETH2672 = CBS 454.64. – Switzerland, Gampel-Steg, on leaves of Clematis vitalba, 10 Oct. 1991, A.G. Spiers, epitype designated here PDD69378, culture ex-epitype ICMP 13663 isolate 9 = PD 97/13460-2 = CBS 123707.
Notes — The first observations of the teleomorph in vitro were made on V8 subcultures obtained from V8 slants stored at 3 °C for 2 yr. It is not likely that the teleomorph will be observed in vitro after routine cultivation. The anamorph of Didymella vitalbina would be more appropriately accommodated in Phoma than in Ascochyta. However, the priority of the teleomorph name makes a new combination in the anamorph superfluous.
Clade C
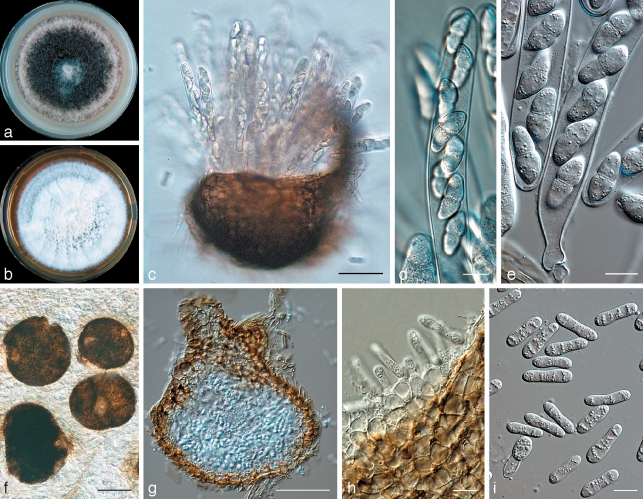
Didymella clematidis Woudenberg, Spiers & Gruyter, sp. nov. — MycoBank MB513003; Fig. 3
Fig. 3.
Didymella clematidis (CBS 123705). a. Colony on OA after 14 d; b. colony on MEA after 14 d; c. perithecium with asci; d, e. asci with ascospores; f. pycnidia; g. longitudinal section through a pycnidium; h. pycnidial wall with conidiogenous cells; i. conidia. — Scale bars: c, f = 100 μm; d, e, h, i = 10 μm; g = 50 μm.
Anamorph. Ascochyta sp.
Asci bitunicati, octospori, cylindracei, 65–125 × 10–20 μm. Paraphyses septatae, inconspicuae, 2 μm latae. Ascosporae hyalinae, septatae, ovatae usque ad obpyriformes, laeves, 15–22 × 4.5–8 μm (av. 19 × 5.7 μm).
Etymology. Named after its host, Clematis.
Description in vitro. Perithecia superficial, solitary or clustered, globose/subglobose to pyriform, (130–)250–370 μm, with prominent elongated neck, up to 75 μm, with central ostiole. Perithecial wall black, textura globulosa 6–10 μm, ectal excipulum up to several layers of elongated cells (8 × 5 μm), medullary excipulum 4–5 layers of globular cells (5.5 × 8 μm), integrated with 2–3 layers of smaller globular cells (5 × 5.5 μm). Ascospore mass white. Asci bitunicate, 8-spored uniseriate/biseriate, cylindrical with club-shaped base, 65–125 × 10–20 μm, paraphyses septate, inconspicuous, 2 μm wide. Ascospores hyaline, septate, ovate to obpyriform, smooth, 15–22 × 4.5–8 μm (av. 19 × 5.7 μm). Pycnidia mostly solitary but also confluent, globose to subglobose or irregular, glabrous, sienna to brown, superficial on the agar but also immersed or in aerial mycelium, (100–)130–360(–560) × 110–340(–475) μm. Ostioli 1(–2) or up to 5, 20–50 μm diam, initially non-papillate but forming an elongated neck in a later stage. Pycnidial wall pseudoparenchymatous, thin, 8–12 μm, consisting of up to 5 cell layers, outer cells isodiametric to oblong. Conidiogenous cells phialidic, hyaline, simple, smooth, globose or flask-shaped, c. 7–9.5 × 5.5–8.5 μm. Conidia elongate, sometimes slightly allantoid, constricted in the middle, hyaline, smooth, mostly uniseptate, (14.5–)16–23(–30) × 4–7(–7.5) μm, with numerous guttules. Only incidentally smaller, aseptate conidia occur, c. 6–8 × 2–3 μm. Conidial matrix saffron to salmon. Chlamydospores absent.
Cultural characteristics — Colonies on OA: growth rate 55–65 mm diam after 7 d, with entire, smooth, sharp margins. Aerial mycelium woolly to floccose, pale olivaceous-grey. Colonies olivaceous-grey. Reverse iron-grey to (pale) olivaceous-grey. Colonies on MEA: growth rate 55–65 mm diam after 7 d, with entire, smooth, sharp margins. Aerial mycelium woolly to floccose, white. Colonies olivaceous-grey. Reverse iron-grey, with greyish sepia tinges or striated olivaceous-grey zones.
Specimens examined. USA, Washington State, Toppenish, on leaves of Clematis ligusticifolia, 7 Sept. 1991, A.G. Spiers, holotype PDD69379, culture ex-holotype ICMP 13664 isolate 30/32 = PD 97/13460-1 = CBS 123705.
Notes — Didymella clematidis produces large 2-celled conidia both in vitro and in vivo. Strain CBS 123705 produced both the teleomorph and anamorph state in pure culture. The species is highly virulent on C. vitalba.
DISCUSSION
The present study is the first to assess the diversity of P. clematidina by means of DNA sequence comparisons. The reconstructed phylogeny (Fig. 1) indicates that multiple taxa are present within the morphological variation understood to represent P. clematidina. Three distinct and well-supported groups were identified which are elevated to species level. A first clade (clade A) includes the representative culture of P. clematidina CBS 108.79 (Boerema & Dorenbosch 1979). This species is characterised by chlamydospore production and a wide ostiolar opening. Thus far, no teleomorph connection has been established with this species. In contrast, Phoma strains from clade B, which were identified as anamorphs of D. vitalbina, were lacking chlamydospore production but regularly formed an elongated neck with ostiole. A third novel taxon (clade C), of which we currently have only one single isolate, is described here as D. clematidis. Due to the presence of only two-celled conidia both in vitro and in vivo, the anamorph stage of D. clematidis is classified in the genus Ascochyta. This species has been applied in New Zealand as a biological control agent of the environmental weed old man’s beard (Gourlay et al. 2000). Its phytopathological value as a highly aggressive strain may be not only due to it being a genetically different entity, but also to its distinct geographical origin. Clematis vitalba originates from Europe and therefore may not have been adapted to this D. clematidis, which is found in the USA. Further research on a larger set of strains originating from various geographical origins is required to obtain more detailed information about the genetic diversity of this species. This may also provide a better assessment of the variation within clade A. The isolates in this clade have been obtained from various Clematis species and hybrids, however, almost exclusively originated from the Netherlands.
The observed difference in susceptibility to Clematis wilt caused by P. clematidina between cultivated and wild Clematis spp. (van de Graaf et al. 2001) may be explained by the existence of three genetically distinct fungal species. Strains that now belong to the newly defined P. clematidina have been isolated from Clematis hybrids, whereas D. vitalbina is recorded exclusively from C. vitalba. Another feature that suggested a high level of variability within P. clematidina is their resistance against benzimidazole fungicides. Van Kuik & Brachter (1997) and van de Graaf et al. (2003) have reported on two groups of P. clematidina within their collections that clearly responded differently to these fungicides. Van de Graaf et al. 2003 could link these groups to slight differences in morphological appearance in culture and to differences in pathogenicity. Although resistance studies could not be conducted within this study, it is worthwhile to conduct further research on the phytopathological features of all taxonomic groups observed.
The previous misidentification of C. clematidis-rectae PD 99/2069 as P. clematidina and the introduction of the Ascochyta anamorph of D. clematidis, previously reported as the P. clematidina biocontrol agent, illustrates the difficulties within the Phoma generic complex. Coniothyrium is characterised by holoblastic, annellidic conidiogenous cells and brown conidia (Crous et al. 2007, Damm et al. 2008). In contrast, Phoma spp. produce enteroblastic, phialidic conidiogenous cells with hyaline conidia (Sutton 1980). Differences in conidiogenesis between these genera are best observed by means of electron microscopy. The difference in conidial pigmentation is also sometimes hard to observe as the conidia of several Phoma spp. have been reported to darken with time (Boerema et al. 2004), whereas (young) conidia of some Coniothyrium species may appear almost hyaline (Taylor & Crous 2001, Verkley et al. 2004). Moreover, both C. clematidis-rectae and P. clematidina can be simultaneously isolated from infected material. The main difference between Ascochyta and Phoma are the annellidic conidiogenesis and distoseptation of Ascochyta (Boerema & Bollen 1975). When septa occur in Phoma, they are secondary. In Ascochyta spp. the septation is an essential part of the conidial maturation, which explains why mature conidia are nearly always septate, both in vivo and in vitro.
As reported by de Gruyter et al. 2009, the distinction among the different coelomycete genera based on morphological features is not always supported by molecular studies. Some species of the anamorph genera such as Coniothyrium, Ascochyta, Ampelomyces and Microsphaeropsis cluster with Phoma species in the Didymellaceae. This is also seen in our study where strains of C. clematidis-rectae and the Ascochyta anamorph of D. clematidis cluster amidst Phoma isolates within the Didymellaceae. It is therefore recommended to improve the current classification of the anamorphic Pleosporales by further evaluating the Phoma, Ascochyta and Coniothyrium complexes and strive to establish monophyletic groups.
Acknowledgments
This research was supported by the Dutch Ministry of Agriculture, Nature and Food Quality through an endowment of the FES programme ‘Versterking Infrastructuur Plantgezondheid’. The authors would like to thank the following persons: Mrs K. Rosendahl (PD) for providing strains, Dr J.Z. Groenewald (CBS) for designing the TUB primers which we used for our research, Mrs M. Vermaas (CBS) for helping with the photo plates, Dr D.H. Hopcroft (Electron Microscope Unit, Massey University, Palmerston North) for the scanning and transmission electron microscopy pictures, and Dr V. Mel’nik (V.L. Komarov Botanical Institute of the Russian Academy of Sciences, St. Petersburg, Russia), for assisting us in obtaining specimens.
REFERENCES
- Blok I. 1965. Verwelkingsziekte in Clematis. Jaarboek Proefstation voor de boomkwekerij te Boskoop 1965: 66 [Google Scholar]
- Boerema GH. 1993. Contributions towards a monograph of Phoma (Coelomycetes) – II. Section Peyronellaea. Persoonia 15: 197 – 221 [Google Scholar]
- Boerema GH, Bollen GJ. 1975. Conidiogenesis and conidial septation as differentiating criteria between Phoma and Ascochyta. Persoonia 8: 111 – 144 [Google Scholar]
- Boerema GH, Dorenbosch MMJ. 1979. Mycologisch-taxonomisch onderzoek in Jaarboek 1978. Verslagen en Mededelingen Plantenziektenkundige Dienst 153: 17 – 21 [Google Scholar]
- Boerema GH, Gruyter J de, Noordeloos ME. 1997. Contributions towards a monograph of Phoma (Coelomycetes) – IV. Section Heterospora: Taxa with large sized conidial dimorphs, in vivo sometimes as Stagonosporopsis synanamorphs. Persoonia 16: 335 – 371 [Google Scholar]
- Boerema GH, Gruyter J de, Noordeloos ME, Hamers MEC. 2004. Phoma identification manual. Differentiation of specific and infra-specific taxa in culture CABI Publishing, Wallingford, United Kingdom: [Google Scholar]
- Crous PW, Braun U, Groenewald JZ. 2007. Mycosphaerella is polyphyletic. Studies in Mycology 58: 1 – 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Verkley GJM, Crous PW, Fourie PH, Haegi A, Riccioni L. 2008. Novel Paraconiothyrium species on stone fruit trees and other woody hosts. Persoonia 20: 9 – 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris JS, Källersjo M, Kluge AG, Bult C. 1995. Testing significance of incongruence. Cladistics 10: 315 – 319 [Google Scholar]
- Gams W, Verkley GJM, Crous PW. 2007. CBS Course of Mycology, 5th ed.Centraalbureau voor Schimmelcultures, Utrecht, Netherlands: [Google Scholar]
- Gloyer WO. 1915. Ascochyta clematidina, the cause of stem-rot and leaf-spot of clematis. Journal of Agricultural Research 4: 331 – 342 [Google Scholar]
- Gourlay AH, Wittenberg R, Hill RL, Spiers AG, Fowler SV. 2000. The biological control programme against Clematis vitalba in New Zealand . In: Spencer NR. (ed), Proceeding of the 10th International Symposium on Biological Control of Weeds: 709 – 718 Montana State University, Bozeman, MT, USA: [Google Scholar]
- van de Graaf P, O’Neill TM, Chartier-Hollis JM, Joseph ME. 2001. Susceptibility of clematis varieties and species to stem infection by Phoma clematidina as an indicator for resistance to wilt. European Journal of Plant Pathology 107: 607 – 614 [Google Scholar]
- van de Graaf P, O’Neill TM, Chartier-Hollis JM, Joseph ME. 2003. Aspects of the biology and control of benzimidazole resistant isolates of Phoma clematidina, cause of leaf spot and wilt in Clematis. Journal of Phytopathology 151: 442 – 450 [Google Scholar]
- Gruyter J de, Aveskamp MM, Woudenberg JHC, Verkley GJM, Groenewald JZ, Crous PW. 2009. Molecular phylogeny of Phoma and allied anamorph genera: towards a reclassification of the Phoma complex. Mycological Research doi:10.1016/j.mycres.2009.01.002. [DOI] [PubMed]
- Hill RL, Fowler SV, Wittenberg R, Barton J, Casonato S, Gourlay AH, Winks C. 2004. Phytomyza vitalbae, Phoma clematidina, and insect-plant pathogen interactions in the biological control of weeds . In: Cullen JM, Briese DT, Kriticos DJ, Lonsdale WM, Morin L, Scott JK. (eds), Proceedings of the 11th International Symposium of Biological Control of Weeds: 48 – 56 CSIRO Entomology, Canberra, Australia: [Google Scholar]
- Hill RL, Wittenberg R, Gourlay AH. 2001. Biology and host range of Phytomyza vitalbae and its establishment for the biological control of Clematis vitalba in New Zealand. Biocontrol Science and Technology 11: 459 – 473 [Google Scholar]
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182 – 192 [Google Scholar]
- Hoog GS de, Gerrits van den Ende AH. 1998. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses 41: 183 – 189 [DOI] [PubMed] [Google Scholar]
- Hume LJ, West CJ, Watts HM. 1995. Nutritional requirements of Clematis vitalba L. (old man’s beard). New Zealand Journal of Botany 33: 301 – 313 [Google Scholar]
- Karnovsky JEM. 1965. A formaldehyde-glutaraldehyde fixative of high osmolarity for use in electron microscopy. Journal of Cellular Biology 27: 137 – 138 [Google Scholar]
- van Kuik A, Brachter E. 1997. Schimmelziekte bij Clematis steekt opnieuw de kop op. De boomkwekerij 9: 18 – 19 [Google Scholar]
- Ogle CC, la Cock GD, Arnold G, Mickleson N. 2000. Impact of an exotic vine Clematis vitalba (F. Ranunculaceae) and of control measures on plant biodiversity in indigenous forest, Taihape, New Zealand. Austral Ecology 25: 539 – 551 [Google Scholar]
- Page RDM. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12: 357 – 358 [DOI] [PubMed] [Google Scholar]
- Paynter Q, Waipara N, Peterson P, Hona S, Fowler S, Gianotti A, Wilkie P. 2006. The impact of two introduced biocontrol agents, Phytomyza vitalbae and Phoma clematidina, on Clematis vitalba in New Zealand. Biological Control 36: 350 – 357 [Google Scholar]
- Rayner RW. 1970. A mycological colour chart Commonwealth Mycological Institute and British Mycological Society, Kew, United Kingdom: [Google Scholar]
- Rehner SA, Samuels GJ. 1994. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycological Research 98: 625 – 634 [Google Scholar]
- Spiers AG, Hopcroft DH. 1992. Some electron microscope observations of conidium ontogeny of Sphaceloma murrayae on Salix. New Zealand Journal of Botany 30: 353 – 358 [Google Scholar]
- Sutton BC. 1980. The coelomycetes. Fungi imperfecti with pycnidia, acervuli and stromata Commonwealth Mycological Institute, Kew, United Kingdom: [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sunderland, Massachusetts, USA: Sinauer Associates; 2003. [Google Scholar]
- Taylor JE, Crous PW. 2001. Morphological variation and cultural characteristics of Coniothyrium leucospermi associated with leaf spots of Proteaceae. Mycoscience 42: 265 – 271 [Google Scholar]
- Verkley GJM, da Silva M, Wicklow DT, Crous PW. 2004. Paraconiothyrium, a new genus to accommodate the mycoparasite Coniothyrium minitans, anamorphs of Paraphaeosphaeria, and four new species. Studies in Mycology 50: 323 – 335 [Google Scholar]
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238 – 4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics . In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR protocols: a guide to methods and applications: 315 – 322Academic Press, San Diego, California, USA: [Google Scholar]