Abstract
The domestic dog exhibits greater diversity in body size than any other terrestrial vertebrate. We used a strategy that exploits the breed structure of dogs to investigate the genetic basis of size. First, through a genome-wide scan, we identified a major quantitative trait locus (QTL) on chromosome 15 influencing size variation within a single breed. Second, we examined genetic variation in the 15-megabase interval surrounding the QTL in small and giant breeds and found marked evidence for a selective sweep spanning a single gene (IGF1), encoding insulin-like growth factor 1. A single IGF1 single-nucleotide polymorphism haplotype is common to all small breeds and nearly absent from giant breeds, suggesting that the same causal sequence variant is a major contributor to body size in all small dogs.
Size variation in the domestic dog is extreme and surpasses that of all other living and extinct species in the dog family, Canidae (1, 2). However, the genetic origin of this diversity is obscure. Explanations include increased recombination or mutation rates (3, 4), a unique role of short repeat loci near genes (3), expansion of specific short interspersed nuclear elements (5), regulatory gene variation (6, 7), or a readily altered developmental program (1, 6). The domestic dog descended from the gray wolf at least 15,000 years ago (8–10), but the vast majority of dog breeds originated over the past few hundred years (11). Understanding the genetic basis for the rapid generation of extreme size variability in the dog would provide critical tests of alternative genetic mechanisms and insight into how evolutionary diversification in size could occur rapidly during adaptive radiations (12).
To investigate the genetic basis for size variation in dogs and understand how change in size might occur rapidly in dogs and other canids, we first initiated sequence-based marker discovery across a 15–megabase (Mb) interval on chromosome 15 in the Portuguese water dog (PWD), a breed that is allowed large variation in skeletal size by the American Kennel Club (13). Previously, based on 92 radiographic skeletal measurements for size and shape, we found that two QTL (FH2017 at 37.9 Mb and FH2295 at 43.5 Mb) within this region were strongly associated with body size in 463 PWDs from a well-characterized extended pedigree (13, 14). We discovered 302 single-nucleotide polymorphisms (SNPs) and 34 insertion/deletion polymorphisms by sequencing 338 polymerase chain reaction (PCR) amplicons in four large and four small PWDs and in nine dogs from small and giant breeds (<9 and >30 kg average breed mass, respectively). We then measured the association between 116 SNPs and skeletal size in a sample of 463 PWDs and identified a single peak within 300 kb of the insulin-like growth factor 1 gene (IGF1) (Fig. 1A), confirming the FH2295 QTL. IGF1 is an excellent candidate gene known to influence body size in both mice and humans (15–17).
Fig. 1.
Relationships of skeletal size, SNP markers, IGF1 haplotype, and serum levels of the IGF1 protein in PWDs. (A) A mixed-model test for association between size and genotype. The association of three genotype categories (A1A1, A1A2, and A2A2) with skeletal size measurements was calculated with the use of all pairwise coefficients of consanguinity for 376 dogs. Each point represents a single SNP position on canine chromosome 15 and negative log P value for the association statistic. (B) PWD IGF1 haplotypes and mean skeletal size. Haplotypes were inferred for 20 markers spanning the IGF1 gene (chromosome 15: 44,212,792 to 44,278,140, CanFam1). Out of the 720 chromosomes with successful inference, 96% carry one of just two haplotypes, B and I, identical to haplotypes inferred for small and giant dogs, respectively (Fig. 3). Data are graphed as a histogram for each genotype: B/B (closed triangle, black line), B/I (open square, dashed line), and I/I (closed circle, gray line). (C) Serum levels of IGF1 protein (ng/ml) as a function of haplotype. Serum levels of IGF1 protein were assayed in 31 PWDs carrying haplotypes B and I. Box plots show the median (center line in box), first and third quartile (box ends), and maximum and minimum values (whiskers) obtained for each category: homozygous B/B (n = 15), heterozygous B/I (n = 7), and homozygous I/I (n = 9).
Haplotype analysis of 20 SNPs spanning IGF1 further supported a role for the locus in determining body size. We observed that 889 of the 926 (96%) PWD chromosomes carry one of just two haplotypes, termed B and I. Dogs homozygous for haplotype B have a smaller median skeletal size [Fig. 1B; P < 3.27 × 10−7, analysis of variance (ANOVA)] and mass (fig. S1) than dogs homozygous for I and a lower level of IGF1 protein in blood serum (Fig. 1C; P < 9.34 × 10−4, ANOVA). In PWDs, 15% of the variance in skeletal size is explained by the IGF1 haplotype. Linkage disequilibrium around IGF1 in PWDs is too extensive to allow fine mapping, presumably because of the breed’s recent origin and small population size (18, 19). However, if a mutation at IGF1 in general underlies genetic differences in size among dog breeds, comparison of breeds of different sizes that have distinct genealogical histories may allow fine mapping of the mutation. Moreover, because size has been the target of strong selection by dog breeders, we would expect to find a signature of selection surrounding the QTL in breeds of extreme small or giant size.
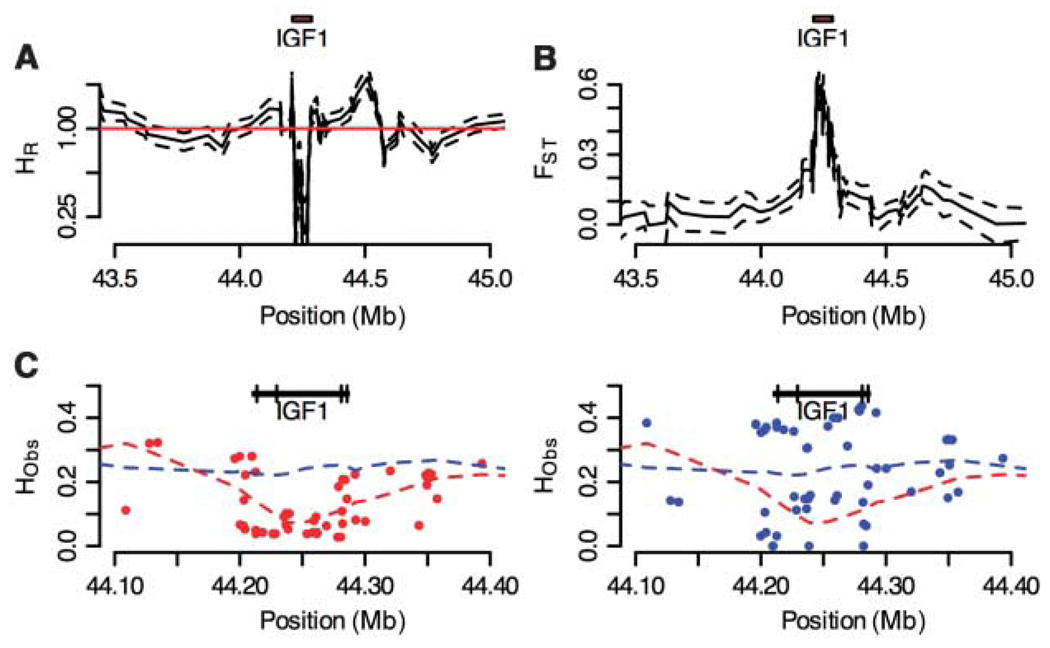
To test these predictions, we surveyed genetic variation for the same 116 SNPs in 526 dogs from 23 small (<9 kg) and 20 giant (>30 kg) breeds. To obtain an empirical distribution of our association mapping test statistics, we also surveyed variation in 83 SNPs with no known association to body size on canine chromosomes 1, 2, 3, 34, and 37. These data were analyzed first to determine if intense artificial selection on body size has resulted in a “selective sweep” (20), reducing variability and increasing allele frequency divergence near IGF1. We found a marked reduction in marker heterozygosity and increased genetic differentiation between small and giant dogs centered on IGF1 (Fig. 2). Specifically, near IGF1, average heterozygosity in small dogs is only 25% of that in large dogs, genetic differentiation (FST, where ST represents subpopulation) peaks significantly at 0.6, and overall heterozygosity is sharply reduced (Fig. 2B) (figs. S2 to S5). Together, these results suggest that a narrow and precisely defined genomic region holds the variant (or variants) responsible for small size in a disparate set of small dog breeds.
Fig. 2.
Signatures of recent selection on the IGF1 locus across 22 small and giant dog breeds. (A) Heterozygosity ratio (HR) for small versus giant dogs. (B) Genetic differentiation (FST) for small versus giant dogs. For both (A) and (B), a sliding 10-SNP window across IGF1 was used. Dashed lines delimit the 95% confidence intervals based on nonparametric bootstrap resampling. The IGF1 gene interval is indicated above the graphs as a red box drawn to scale. (C) Observed heterozygosity (HObs) of SNPs near IGF1 typed in small breeds (<9 kg) and giant breeds (>30 kg). Small breeds have a reduction in observed heterozygosity compared with that of giant breeds. Red and blue points are average observed heterozygosity in small and giant breeds, respectively. Dashed lines are locally weighted scatterplot smoothing (LOWESS) best fit to the data. The IGF1 gene is shown as a black bar with exons indicated by vertical lines.
We next tested for association between each SNP and average breed size (Fig. 3A). The null hypothesis of no association between body size and marker frequency across breeds is rejected (Bonferroni-correct P value < 0.05) for 25 contiguous SNPs defining an 84-kb interval spanning the same region that shows evidence of a selective sweep (chromosome 15 base pairs 44,199,850 to 44,284,186) (Fig. 2 and Fig. 3A). The Mann-Whitney U statistic provides a uniform distribution of P values for 83 genomic control markers (fig. S6). Similarly, P values from Fisher’s exact test of association across individuals were smaller than 10−100 in the 84-kb interval; although these P values are clearly biased by confounding population structure (fig. S6), as evidenced by the 83 genomic control markers [for which the minimum P value was 10−20 (fig. S7)], the result is significant.
Fig. 3.
Evidence of association and IGF1 haplotypes for 14 small and 9 giant breeds. (A) Mann-Whitney U (MWU) P values for tests of association between individual SNPs and body size (small versus giant) for 116 SNPs on chromosome 15 and 83 SNPs on five control chromosomes. The dashed line indicates Bonferroni correction for multiple tests. Only breeds with data for at least 10 chromosomes were included (14 small and 9 giant breeds). (B) Haplotypes for the 20 markers spanning the small breed sweep interval near IGF1. The haplotypes were inferred independently in each breed. For each individual, fractional chromosome counts were summed for all haplotypes with at least 5% probability according to the haplotype inference software program PHASE. Chromosome sums for each breed were rounded to integer values; several breeds have odd numbers of chromosomes due to rounding error. Only inferred haplotypes carried by at least three dog chromosomes total (i.e., >0.5% frequency overall) are shown. Sequence reads collected from golden jackal (Canis aureus) were used to determine the ancestral allele for each SNP. The haplotypes are rows labeled A to L, and marker alleles are colored yellow for ancestral state (matching the nucleotide observed in the golden jackal) and blue for derived state. SNP positions within IGF1 are shown at the top with IGF1 introns (horizontal line) and exons (vertical bars) indicated. (C) Breed name and the average size of adult males in kilograms are provided. Small breeds less than 9 kg and giant breeds greater than 30 kg are grouped for totals shown at the far right.
Analysis of specific breed haplotypes shows that a 20-SNP haplotype spanning IGF1 is shared by all 14 sampled small dog breeds (Fig. 3, B and C) and is identical to haplotype B in small PWDs. This haplotype was observed in only three of the nine giant breeds because most giant dogs carry one or both of two distinct haplotypes: F and I. SNP 5, located at base pair position 44,228,468 (Fig. 3B), is the best candidate for being proximate to the causative mutation for the following reasons: (i) It distinguishes haplotypes A, B, and C, associated with small body size, from haplotypes D to L, which are common in large breeds; (ii) an ancestral recombination graph suggests an absence of recombination between SNPs 4 and 5 (fig. S8); and (iii) marker analysis in the golden jackal and gray wolf indicates that the SNP 5 A allele of small breeds is the derived condition (fig. S9) (table S1). To further assess the association between body size and the SNP 5 A allele, we genotyped six tagging SNPs that distinguish all major IGF1 haplotypes in a set of 3241 dogs from 143 breeds (Fig. 4) (table S2). The frequency of the SNP 5 A allele is strongly negatively correlated with breed average mass across this large sample of breeds (Fig. 4, Spearman’s rank correlation coefficient ρ = −0.773; P < 2.2 × 10−16; likelihood ratio test = 2882.3, χ2df=1 < 2 × 10−16, logistic regression of allele frequency on body size). A strong negative correlation remains when the 22 breeds used to discover SNP 5 are removed from the analysis (ρ = −0.729; P < 2.2 × 10−16, Spearman’s rank correlation). Exceptions, such as the large Rottweiler or small whippet breeds, may carry compensatory mutations at other size QTL or recombinants that could aid fine mapping at IGF1. Our results show that a single IGF1 haplotype is common to a large sample of small dogs and strongly imply that the same causal variant (or variants) is a major influence on the phenotype of diminished body size.
Fig. 4.
Association of body size and frequency of the SNP 5 A allele. Binomial regression of allele frequency on square root of mean breed mass. Dashed lines indicate the 95% confidence interval on the predicted equation line as estimated from nonparametric bootstrap resampling. Between 5 and 109 (median = 22) dogs were genotyped for each of 143 breeds. The PWD is highlighted in red along with three giant breeds that have larger breed average masses than is predicted by their SNP 5 allele frequency.
The IGF1 gene is a strong genetic determinant of body size across mammals; mice genetically deficient in IGF1 are just 60% normal birth weight (15), and a human with a homozygous partial deletion of the gene was born 3.9 SD below normal length (16, 17). IGF1 binds the type 1 IGF receptor, a tyrosine kinase signal transducer. This interaction promotes cell growth and organismal longevity (21) and induces cellular differentiation (22). Serum levels of IGF1 protein (23) have been found to correlate with body size in toy, miniature, and standard poodles (24). These studies did not compare IGF1 genetic variation with differences in serum IGF1 protein concentrations; we observed that PWDs carrying the B haplotype of the IGF1 gene have significantly lower serum levels of IGF1 (Fig. 1C).
Finally, to identify possible causative variants, we sequenced the exons of IGF1 in a panel of nine small and giant dogs and found only one variation in coding sequence, a synonymous SNP in exon 3 [chromosome 15 base pair position 44,226,324, Canis familiaris genome assembly 1 (CanFam1)]. Extensive resequencing within introns and flanking genomic sequence was also undertaken (table S3). Several additional SNPs (table S4) and an antisense oriented retrotransposon (table S5) unique to small breeds were identified. Alleles of a dinucleotide CAn microsatellite in the IGF1 promoter were also significantly associated with body size in the PWDs (P < 1.4 × 10−6, ANOVA) and the small and giant breeds (P < 2.2 × 10−14, chi–square test; table S6). All of these variations were in strong linkage disequilibrium and therefore a causative variant could not be definitively identified by this approach. Given the difficulty of developing inbred dog lines segregating small size, future studies will focus on using knock-in mice to explore the effect of these variants on phenotypes.
Our findings suggest that a single IGF1 haplotype substantially contributes to size variation in the domestic dog. Because our sample includes small breeds that are distantly related (25) and reproductively isolated, and because the extent of haplotype sharing at IGF1 is relatively small, the sequence variant or variants probably predate the common origin of the breeds and likely evolved early in the history of dogs. The early appearance of this allele may have facilitated the rapid genesis of size diversity in the domestic dog. The first archaeological record of dogs, beginning about 12,000 to 15,000 years ago (9, 26), shows that size diversity was present early in the history of domestication. For example, dog remains from eastern Russia dated to 14,000 to 15,000 years ago are similar in size and conformation to great Danes, whereas slightly younger dog remains from the Middle East and Europe (10,000 to 12,000 years ago) are similar in size to small terriers (9, 26, 27). The early and widespread appearance of small size suggests that an ancesral small dog IGF1 haplotype was readily spread over a large geographic area by trade and human migration and was maintained in local gene pools by selection. Such early selection on dogs may have been manifest as intentional artificial selection exercised by early humans or as an adaptive trait for coexistence with humans in the more crowded confines of developing villages and cities (28).
The ubiquitous occurrence of the IGF1 B haplotype in a diverse panel of small breeds clearly does not support unorthodox explanations of phenotypic diversity in the dog such as elevated mutation or recombination rates. Rather, we show that a single IGF1 allele is a major determinant of small size in dogs and that intense artificial selection has left a signature in the proximity of IGF1 that can readily be found by genomic scans of breeds sharing a common phenotype. The ability to identify a gene contributing to morphology without doing a genetic cross, but instead by using centuries of dog breeding, highlights the contribution that the study of canine genetics can make to an understanding of mammalian morphogenesis. These results provide a precedent for future studies aimed at identifying the genetic basis for complex traits such as behavior and skeletal morphology in dogs and other species with small populations that have experienced strong artificial or natural selection.
Supplementary Material
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/316/5821/112/DC1
Materials and Methods
Figs. S1 to S9
Tables S1 to S6
References
References and Notes
- 1.Wayne RK. Evolution. 1986;40:243. doi: 10.1111/j.1558-5646.1986.tb00467.x. [DOI] [PubMed] [Google Scholar]
- 2.Wayne RK. J. Morphol. 1986;187:301. doi: 10.1002/jmor.1051870304. [DOI] [PubMed] [Google Scholar]
- 3.Fondon JW, 3rd, Garner HR. Proc. Natl. Acad. Sci. U.S.A. 2004;101:18058. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webber C, Ponting CP. Genome Res. 2005;15:1787. doi: 10.1101/gr.3896805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang W, Kirkness EF. Genome Res. 2005;15:1798. doi: 10.1101/gr.3765505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wayne RK. J. Zool. 1986;210:381. [Google Scholar]
- 7.Saetre P, et al. Brain Res. Mol. Brain Res. 2004;126:198. doi: 10.1016/j.molbrainres.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Savolainen P, Zhang YP, Luo J, Lundeberg J, Leitner T. Science. 2002;298:1610. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- 9.Olsen SJ. Origins of the Domestic Dog. Tucson, AZ: Univ. of Arizona Press; 1985. [Google Scholar]
- 10.Vila C, et al. Science. 1997;276:1687. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- 11.Sampson J, Binns MM. In: The Dog and Its Genome. Ostrander EA, Lindblad-Toh K, Giger U, editors. vol. 44. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 19–30. [Google Scholar]
- 12.Van Valkenburgh B, Wang X, Damuth J. Science. 2004;306:101. doi: 10.1126/science.1102417. [DOI] [PubMed] [Google Scholar]
- 13.Chase K, et al. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9930. doi: 10.1073/pnas.152333099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chase K, Carrier DR, Adler FR, Ostrander EA, Lark KG. Genome Res. 2005;15:1820. doi: 10.1101/gr.3712705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker J, Liu JP, Robertson EJ, Efstratiadis A. Cell. 1993;75:73. [PubMed] [Google Scholar]
- 16.Woods KA, Camacho-Hubner C, Barter D, Clark AJ, Savage MO. Acta Paediatr. Suppl. 1997;423:39. doi: 10.1111/j.1651-2227.1997.tb18367.x. [DOI] [PubMed] [Google Scholar]
- 17.Woods KA, Camacho-Hubner C, Savage MO, Clark AJ. N. Engl. J. Med. 1996;335:1363. doi: 10.1056/NEJM199610313351805. [DOI] [PubMed] [Google Scholar]
- 18.Sutter NB, et al. Genome Res. 2004;14:2388. doi: 10.1101/gr.3147604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindblad-Toh K, et al. Nature. 2005;438:803. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 20.Pollinger JP, et al. Genome Res. 2005;15:1809. doi: 10.1101/gr.4374505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kooijman R. Cytokine Growth Factor Rev. 2006;17:305. doi: 10.1016/j.cytogfr.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Cohen P. Horm. Res. 2006;65:3. [Google Scholar]
- 23.Favier RP, Mol JA, Kooistra HS, Rijnberk A. J. Endocrinol. 2001;170:479. doi: 10.1677/joe.0.1700479. [DOI] [PubMed] [Google Scholar]
- 24.Eigenmann JE, Patterson DF, Froesch ER. Acta Endocrinol. (Copenh.) 1984;106:448. doi: 10.1530/acta.0.1060448. [DOI] [PubMed] [Google Scholar]
- 25.Parker HG, et al. Science. 2004;304:1160. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- 26.Epstein H. The Origin of the Domestic Animals of Africa. New York: Africana Publishing; 1971. [Google Scholar]
- 27.Sablin MV, Khlopachev GA. Curr. Anthropol. 2002;43:795. [Google Scholar]
- 28.Tchernov E, Horwitz LK. J. Anthropol. Archaeol. 1991;10:54. [Google Scholar]
- 29.We thank the hundreds of dog owners who contributed samples; the AKC Canine Health Foundation; S. Hoogstraten-Miller and I. Ginty for assistance at dog shows; P. Cruz for assistance with automated PCR primer designs; S. Kim for analytical assistance, and R. Pelker for assistance with blood serum assays of IGF1. Funded by the National Human Genome Research Institute (E.A.O., N.B.S., E.K., S.D., P.Q., H.G.P., and D.S.M.), the NSF (R.K.W.), NIH grant no. 5 T32 HG002536 (M.M.G.), NSF grant 0516310 (C.D.B. and L.Z.), NSF grant DBI 0606461 (B.P.), NIH grant P50 HG002790 (K.Z. and M.N.), and the National Institute of General Medical Sciences 063056, the Judith Chiara Charitable Trust, and the Nestle Purina Company (K.G.L.).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.