Abstract
The role of nucleocapsid (NC) in the early steps of retroviral replication appears largely that of a facilitator for reverse transcription and integration. Using a wide variety of cell-free assay systems, the properties of mature NC proteins (e.g., HIV-1 p7NC or MLV p10NC) as nucleic acid chaperones have been extensively investigated. The effect of NC on tRNA annealing, reverse transcription initiation, minus-strand transfer, processivity of reverse transcription, plus-strand transfer, strand-displacement synthesis, 3′ processing of viral DNA by integrase, and integrase-mediated strand-transfer has been determined by a large number of laboratories. Interestingly, these reactions can all be accomplished to varying degrees in the absence of NC; some are facilitated by both viral and non-viral proteins and peptides that may or may not be involved in vivo. What is one to conclude from the observation that NC is not strictly required for these necessary reactions to occur? NC likely enhances the efficiency of each of these steps, thereby vastly improving the productivity of infection. In other words, one of the major roles of NC is to enhance the effectiveness of early infection, thereby increasing the probability of productive replication and ultimately of retrovirus survival.
Keywords: Nucleocapsid, Reverse transcription, Integration, Nucleic acid chaperone, Core uncoating, Cell culture
1. Introduction
The nucleocapsid (NC) proteins (Leis et al., 1988) of orthoretroviruses (van Regenmortel et al., 2000) are small (<100 amino acids), highly basic nucleic acid binding proteins (Fig. 1). In addition, these proteins contain either one (gammaretroviruses) (Henderson et al., 1981) or two strictly conserved zinc fingers of the sequence C-X2-C-X4-H-X4-C (CCHC) (Berg, 1986; Covey, 1986; South and Summers, 1990). Spumaviruses, although a genus within the Retroviridae family, maintain NC as a domain of unprocessed Gag and lack the characteristic zinc-finger motifs found in the orthoretroviruses (Linial, 1999). The Ty3 and copia retrotransposons also contain a CCHC zinc finger located at the COOH-terminal region of Gag, while Ty1 and gypsy retrotransposons lack the characteristic zinc-finger motifs (Darlix et al., 1995). These species have been demonstrated to possess in vitro chaperone activities similar to retroviral NC proteins (Cristofari et al., 1999, 2000, 2002; Gabus et al., 2006).
Fig. 1. Comparison of retroviral NC proteins.
The identity, apparent molecular weight, genus, and relative size of several mature NC proteins are shown. A black box indicates the location of the N-terminal zinc finger, and a grey box indicates the location of the C-terminal zinc finger. Slashed boxes indicate the location of the three Gly-Arg boxes in PFV. Although the PFV NC domain of Gag is shown for comparison, it is not cleaved from the Gag precursor during maturation, and is consequently never a discrete protein (Linial, 1999).
In all of the retroviruses containing zinc fingers, the spacing of the zinc-binding residues is absolutely conserved (Table 1). In addition, the surrounding sequences within a particular retroviral species are also highly conserved, as demonstrated, for example, by the various human immunodeficiency virus type-1 (HIV-1) sequences aligned in Table 2 (Leitner et al., 2006/2007). The zinc fingers form stable structures with the NH2- and COOH-terminal regions, being largely unstructured when NC is not bound to nucleic acids (see ribbon diagram of HIV-1 NC, Fig. 2, upper right) (Amarasinghe et al., 2000; Darlix et al., 1995; Lee et al., 1998; Morellet et al., 1992, 1994).
Table 1.
Properties of retroviral NC proteins.
| Virus | (protein) | Size | N-terminal zinc fingera |
C-terminal zinc finger |
Basic Residuesb | Acid Residuesc |
|---|---|---|---|---|---|---|
| RSV | (p12) | 89 a.a. | CYTCGSPGHYNANCd | CNLCNGMGHNAKNCd | 16 (18%) | 3 (3.4%) |
| MLV | (p10) | 60 a.a. | CAYCKEKGHWAKDCe | N.A.f | 14 (23%) | 8 (13%) |
| HTLV-1 | 1 (p15) | 85 a.a. | CFRCGKAGHWSRDCg | CPLCQDPTHWKRDCg | 11 (13%) | 11 (13%) |
| HIV-1 | (p7) | 55 a.a. | CFNCGKEGHIAKNCh | CWKCGKEGHQMKDCh | 15 (27%) | 4 (7.3%) |
| SIV | (p8) | 52 a.a. | CWNCGKEGHSARWCi | CWKCGQMGHVMAKCi | 12 (23%) | 1 (1.9%) |
| FIV | (p10) | 66 a.a. | CFNCKKPGHLARQCj | CNKCGKPGHVAAKCj | 14 (21%) | 1 (1.5%) |
| PFV | (p75) | 142 a.a.k | N.A. | N.A. | 27 (19%) | 8 (5.6%) |
| Copia | 43 a.a.l | CHHCGREGHIKKDCm | N.A. | 14 (32%) | 2 (4.6%) | |
| Ty1 | 103 a.a.n | N.A. | N.A. | 19 (18%) | 8 (7.8%) | |
| Gypsy | 116 a.a.o | N.A. | N.A. | 24 (21%) | 10 (8.6%) | |
| Ty3 | 58 a.a. | CFYCKKEGHRLNECp | N.A. | 18 (31%) | 4 (6.9%) |
Zinc-binding Cys and His residues are highlighted in grey.
The total number of Lys and Arg residues, and in parenthesis, the percentage of basic to total amino acid residues.
The total number of Asp and Glu residues, and in parenthesis, the percentage of acidic to total amino acid residues.
RSV sequence data from GenBank accession number J02342 (Méric and Spahr, 1986).
MLV sequence data from GenBank accession number J02255 (Rein et al., 1994).
N.A., not applicable. MLV, like all gammaretroviruses, has a single zinc finger.
Human T-cell leukemia virus 1 (HTLV-1) sequence data from GenBank accession number D13784 (Derse et al., 2007).
HIV-1 sequence data from GenBank accession number AF324493 (Ottmann et al., 1995).
SIV sequence data from GenBank accession number AY817672 (Gorelick et al., 1999a).
Feline immunodeficiency virus (FIV) sequence data from GenBank accession number M25381 (Manrique et al., 2004).
The number of amino acids in the NC domain of PFV Gag, as NC is not cleaved as with orthoretroviruses (Linial, 1999). GenBank accession number Y07725).
Copia sequence data from GenBank accession number X04456 (Mount and Rubin, 1985). There is no experimental evidence for proteolytic processing to produce a mature NC (Darlix et al., 1995).
Sequence alignment deduced from amino acid sequence (Covey, 1986).
The number of amino acids in the NC domain of Ty1, GenBank accession number M18706 (Boeke et al., 1988). Gag is not processed by the Ty1 PR to produce a mature NC (Merkulov et al., 1996).
The number of amino acids in the NC domain of Gypsy, GenBank accession number M12927 (Marlor et al., 1986). Gag is not processed by PR so NC is always part of Gag (Gabus et al., 2006).
Ty3 sequence data from GenBank accession number M34549 (Hansen and Sandmeyer, 1990). A NC cleavage product has been identified (Kirchner and Sandmeyer, 1993).
Table 2.
Alignment of Group M HIV-1 NC protein sequences.
| N-Terminus | N-terminal zinc finger |
Central | C-terminal zinc finger |
C-Terminus | |
|---|---|---|---|---|---|
| M-Group Consensus | MQRGNFKGQKRIIK | CFNCGKEGHIARNC | RAPRKKG | CWKCGKEGHQMKDC | TERQAN |
| Consensus-A1 | ------R----.-- | ---------L---- | ------- | -------------- | ------ |
| Consensus-A2 | ------R----.-- | ---------L---- | ------- | -------------- | ------ |
| Consensus-B | ------RN-RKTV- | -----------K-- | ------- | -------------- | ------ |
| Consensus-C | ---S----P---V- | -------------- | ------- | -------------- | ------ |
| Consensus-D | --------PRK--- | -----------K-- | ------- | -------------- | ------ |
| Consensus-F1 | --KS-----R--V- | -----------K-- | ------- | -----R-------- | ------ |
| Consensus-G | --KS----PR-T-- | ---------L---- | ------- | -------------- | ------ |
| Consensus-H | --K-----PRK-V- | -------------- | ------- | -----R-------- | ------ |
| Consensus-K | ---------RK--- | -------------- | ------- | -------------- | ------ |
| Consensus-01-AE | -----------.-- | ---------L---- | ------- | -------------- | ------ |
| Consensus-02-AG | ------R--RT.-- | ---------L---- | K------ | -------------- | ------ |
| Consensus-03-AB | --KS--R-P--.-- | ------D--L---- | ------- | -------------- | ------ |
| Consensus-04-CPX | --KS-----R---- | ---------L---- | ------- | -------------- | ------ |
| Consensus-06-CPX | --KS----P--S-- | ---------L---- | ------- | -------------- | ------ |
| Consensus-07-BC | ---S----S---V- | -------------- | ------- | -------------- | ------ |
| Consensus-08-BC | ---S----S---V- | -----------K-- | ------- | -------------- | ------ |
| Consensus-10-CD | --------P-K--- | -----------K-- | ------- | -----R-------- | ------ |
| Consensus-11-CPX | ---S-------.-- | ---------L---- | ------- | -------------- | ------ |
| Consensus-12-BF | --KS-----R--V- | -----------K-- | ------- | -----R-------- | ------ |
| Consensus-14-BG | --KS----PR-N-- | ---------L---- | ------- | -------------- | --SK-- |
| NL4-3 | I-----RN--KTV- | -----------K-- | ------- | -------------- | ------ |
The consensus sequence alignments shown above are from the Los Alamos HIV Sequence Database (http://www.hiv.lanl.gov)(Leitner et al., 2006/2007). The subtype is indicated by a letter, those that are preceded by a number are circulating recombinant forms (CRFs). Dashes indicate amino acid consensus identity, specific amino acids indicate variations, and the presence of a “.” represents a 1 amino acid gap. The generation of the Consensus M-Group sequence does not include CRF; the total number of sequences used to derive the Consensus M-Group are ~1,100 (120 A, ~280 B, ~590 C, ~70 D, ~10 F1, ~20 G, 3 H).
Fig. 2. Events that occur during HIV-1 replication are shown to illustrate the numerous places where NC functions.
A structure of HIV-1 NC, rendered from NMR studies of NC in solution by Summers and coworkers (Lee et al., 1998), is shown in the upper right of the figure. The molecule was drawn using Cn3D version 4.1 (http://www.ncbi.nlm.nih.gov). The backbone of the molecule is shown as a tube structure, with the locations of basic and acidic amino acid residues colored blue and red, respectively. The side chains of aromatic residues are drawn as stick and ball diagrams. The side chains of Cys are shown as space filling pictures. The zinc ions are colored green.
Many of the key processes of viral replication require NC. All events are shown in a single cell for convenience, with infection commencing with receptor binding and ending with budding and maturation. A legend for the viral proteins is presented in the upper left of the figure. Early events start with the virus binding to receptors, subsequent fusion of the virus and cell membranes then the core enters the cytoplasm. After cytoplasmic entry, the core begins to uncoat, a RTC forms with reverse transcription ensuing and it is transported through the cytoplasm to the nuclear membrane. The RTC is transformed via the reverse transcription process into a PIC, where it can be actively transported into the nucleus (in nondividing cells), then is targeted to species-specific regions in the chromosomes where integration occurs. The RTC and PIC are shown in the figure without NC or other factors for clarity. Late events commence with transcription of viral messages from the provirus then translation of viral proteins occurs. Subsequently, packaging of gRNA into assembling virus particles results relying on the ability of the NC domain of the Gag precursor to bind specific regions of the gRNA, forming a nucleation site for the multimerization of Gag upon the RNA scaffold. In this illustration, the assembly depicted to occur at the cell membrane has been described by Booth et al. (2006), Deneka et al. (2007), Finzi et al. (2007), Jolly et al. (2007), Jolly and Sattentau (2007), Jouvenet et al. (2006), Rudner et al. (2005), and Welsch et al. (2007). However, some reports indicate that assembly can occur in intracellular compartments (Jouve et al., 2007; Nydegger et al., 2003; Perlman and Resh, 2006; Sherer et al., 2003). The maturation of virions after budding entails several events that are likely interrelated, from the gRNA dimer maturation, which result in extensive intra- and intermolecular interactions, as well as the condensation of the nucleoprotein core. (Artwork courtesy of Louis E. Henderson, AIDS Vaccine Program, SAIC-Frederick, Inc., NCI-Frederick).
NC is absolutely required for viral replication, and genetic analyses have demonstrated that many, even minor alterations can affect the virus assembly step by disrupting genomic RNA (gRNA) packaging or the formation of infectious virus particles. The NC domain is a key component of assembly processes because it is i) responsible for binding to the RNA scaffold (discussed below) (Campbell and Rein, 1999; Campbell and Vogt, 1995, 1997; Ganser et al., 1999; Muriaux et al., 2001; Zhang et al., 1998), which facilitates the interactions of other regions of Gag with one another and with cellular membranes (Accola et al., 2000; Ivanov et al., 2005; Johnson et al., 2002; Turner and Summers, 1999); and ii) is required for the recognition and packaging of the RNA genome (Barat et al., 1989; Berkowitz et al., 1996; D’Souza and Summers, 2005; Prats et al., 1988).
In early infection processes, NC’s key function is as a nucleic acid chaperone: the processed protein enables nucleic acids to reach the most thermodynamically stable arrangement, i.e., the maximum number of base pairs. Of all the NC proteins studied to date, HIV-1 NC possesses the most effective nucleic acid chaperone activity (Levin et al., 2005) The two molecular properties of NC responsible for this function are the ability to destabilize nucleic acid helices and the ability to elicit nucleic acid aggregation. The basic residues are most responsible for the destabilization activity, while the zinc fingers are largely responsible for the aggregation activity (de Rocquigny et al., 1992; Prats et al., 1991). It should be noted that this chaperone activity has also been observed when NC is still a domain of the Gag precursor, since melting and annealing events occur as the virus is assembling (i.e., tRNA placement) (Feng et al., 1999). For additional details, the reader is referred to a number of excellent reviews discussing NC’s nucleic acid chaperone function (Bampi et al., 2004a, 2004b; Levin et al., 2005; Rein et al., 1998).
For the purposes of this review, it should be pointed out that NC’s function as a nucleic acid chaperone has been demonstrated in several cell-free assay systems. NC facilitates the rapid conversion of single-stranded DNA (ssDNA) to the most stable double-stranded DNA (dsDNA) form (Tsuchihashi and Brown, 1994). Another set of experiments using viral long terminal repeat (LTR) sequences showed that the presence of NC increased the rate of annealing of minus-strand complementary DNA (cDNA) and RNA repeat ® sequences roughly 3 000 fold (You and McHenry, 1994), which is the step required for minus-strand transfer during reverse transcription (Basu et al., 2008; Telesnitsky and Goff, 1997) . Indeed, it has been shown from in vitro reactions that NC increases the processivity of reverse transcription (Drummond et al., 1997; Ji et al., 1996; Klasens et al., 1999; Wu et al., 1996; Zhang et al., 2002).
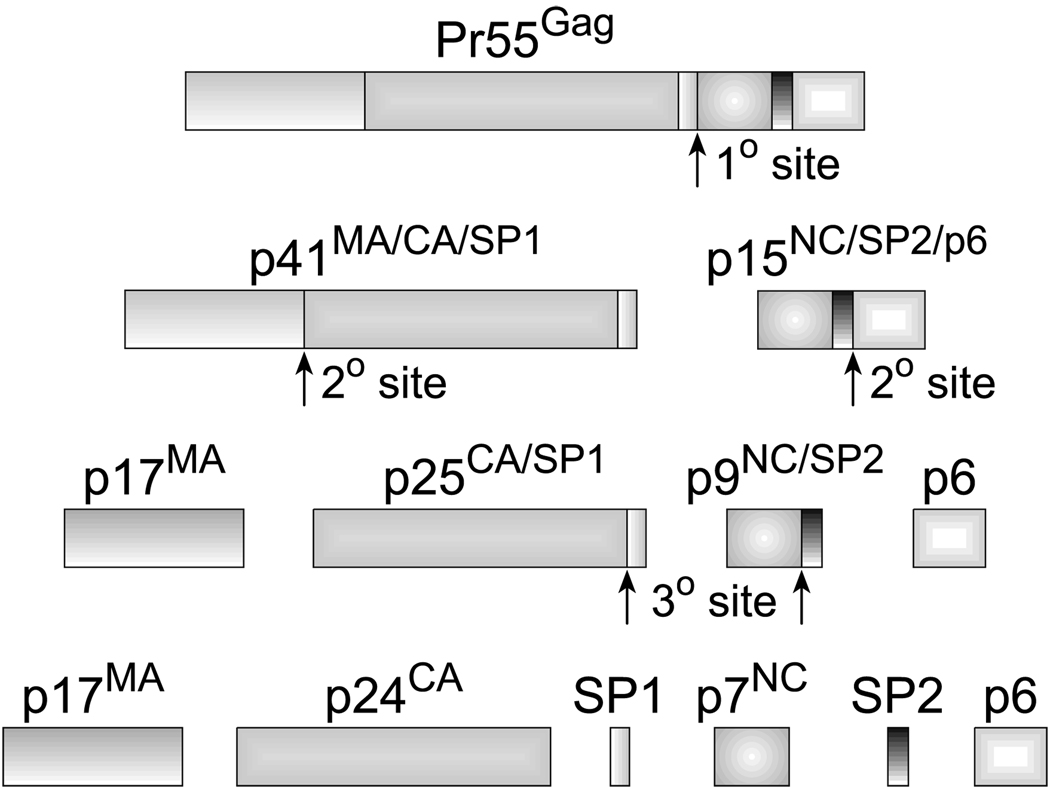
NC is synthesized as a functional domain of the Gag precursor in retroviruses; in the case of HIV-1, this is Pr55Gag (Fig. 3). Mature NC is liberated by a series of discrete retroviral protease (PR)-mediated cleavages that begin later in assembly before virus budding takes place (Kaplan et al., 1994b; Ott et al., 2003). For orthoretroviruses, three main proteins are liberated: matrix (MA), capsid (CA), and NC, as well as additional peptides that are cleaved, depending upon the specific retrovirus (Pettit et al., 1991). The initial PR cleavage of HIV-1 Pr55Gag results in the formation of p15NC, a peptide comprised of the p7NC, p1 (SP2), and p6 domains (Fig. 3) (Shehu-Xhilaga et al., 2001b). The p15NC protein must be bound to RNA for PR to cleave it (Mirambeau et al., 2007; Sheng et al., 1997). The subsequent PR cleavage generates p9NC (p7NC with the SP2 peptide still attached). The ultimate PR cleavage produces the 55 amino acid form of p7NC, the predominant form detected in mature HIV-1 particles (Coren et al., 2007; Henderson et al., 1992; Tanchou et al., 1998). It is important to note that in some studies, p7NC is designated as being either 55 or 72 amino acids (de Rocquigny et al., 1991; Morellet et al., 1992, 1994); these are defined in the present review as p7NC and p9NC, respectively. The actual p9NC intermediate form, comprised of p7NC + SP2 (Fig. 3), is 71 amino acids long (Henderson et al., 1992). PR processing likely results in the presence of small amounts of p9NC or p15NC, as PR processing is unlikely to be 100% efficient: these forms are occasionally observed in Western blots of HIV-1 (Gorelick et al., 1999c; Tanchou et al., 1998). All of the forms of NC, from Pr55Gag to p7NC, exhibit chaperone activity to various degrees and the nature of this activity changes as the protein reaches its more extensively processed forms. For instance, the binding of p7NC to nucleic acids is noncooperative, while p9NC and p15NC binding is cooperative (Cruceanu et al., 2006; Khan and Giedroc, 1994). The HIV-1 Gag precursor is highly cooperative in its binding to single-stranded nucleic acids, a property commensurate with its function in RNA packaging (see below) (Cruceanu et al., 2006).
Fig. 3. Proteolytic processing of HIV-1 Gag by PR.
The pattern and order of PR cleavage sites in Gag are shown. For proteins other than NC, the different species that exist during processing are indicated. The initial cleavage occurs between SP1 and the NC domain, secondary cleavages occur at approximately 10-fold lower rates than the initial cut. The tertiary cleavages occur approximately 400-fold slower than the primary cleavage (Shehu-Xhilaga et al., 2001b). Complete processing of Gag into its mature proteins is efficient and rapid, so that almost immediately after budding, the intermediates are not typically detected by Western blotting (Kaplan et al., 1994b). During proteolysis, NC exists in two intermediate forms, p15NC (partial cleavage product containing NC/SP2/p6), p9NC (partial cleavage product containing NC/SP2), and the fully processed form, p7NC. All three of these proteins exhibit nucleic acid chaperone activities (Cruceanu et al., 2006).
Examination of the steps of viral replication has revealed that mutations in NC can cause defects in the assembly of virus particles, gRNA packaging, RNA dimer maturation, reverse transcription, and integration (Fig. 2). In addition, it is possible that mutations to NC may affect core stability, although this has not been demonstrated to date. Attempts to investigate the roles performed by NC during early infection are difficult because, as a domain of Gag, NC is crucial for the formation of infectious virus particles (Grigorov et al., 2007). Thus, any study of NC function in early infection processes requires the identification of NC mutants that separate defects in the assembly/budding steps (gRNA packaging, RNA dimer maturation, etc.) from defects in early infection events (reverse transcription/integration). For example, if a mutation abolishes gRNA packaging, it is difficult to determine its effects on reverse transcription, as the genome is obviously a key element for reverse transcription.
The main focus of this review is on the role of NC in early infection events as determined in cell-culture systems. Because of this focus, we have limited discussion of in vitro studies of NC. As a consequence, numerous in vitro studies are not cited or discussed; however, a number of recent reviews have covered this information in depth, so we refer the reader to these for more detail (Bampi et al., 2004a, 2004b; Darlix et al., 2000, 2002, 2007; Levin et al., 2005; Rein et al., 1998). In addition, the vast majority of work has been performed with the HIV-1 system, so this review will focus primarily on these studies; however, we will refer to work in other retroviral systems where appropriate.
2. The role of NC in assembly (setting the stage for reverse transcription)
Assembly of infectious virus particles is a necessary prerequisite for the early infection events, reverse transcription and integration. As more is learned about how a virion forms and infects a target cell, the interrelationship between these events becomes clearer. The uncoating of the virus particle after entry of the core into the cytoplasm of an infected cell appears to be an ordered process, so that correct virion assembly is probably required for proper core disassembly (Forshey et al., 2002; Swanstrom and Wills, 1997). Thus, many of the ways in which NC is involved in virus assembly may directly influence how the events of early infection proceed.
For assembly to begin, cellular transcriptional machinery must generate full-length RNA transcripts from proviruses, and these mRNAs are then translated into viral structural proteins. Transcription is controlled by promoters/enhancers in the U3 region of the provirus (Graves et al., 1985; Laimins et al., 1984). It has been proposed that during acute HIV-1 infection, NC from the incoming virus enhances the basal level of transcription, before the production of Tat, which is involved in up-regulation of transcription (Zhang and Crumpacker, 2002; Zhang et al., 2000b).
A number of NC mutants have been generated that severely affect either protein processing, production of virus particles, or gRNA incorporation. Although these mutations have been useful for ascertaining NC’s role in assembly, they unfortunately preclude investigation of early infection events. This section will discuss the roles of NC in assembly and production of virus, an understanding of which is useful for interpretation of early infection data.
2.1. RNA packaging
A key role of NC during particle production is the specific packaging of gRNA into virus particles (Fig. 2) (Berkowitz et al., 1996; D’Souza and Summers, 2005). Genetic analyses have demonstrated that the NC domain in the Gag precursor is critical for specific recognition and packaging of gRNA, since mutations to the zinc fingers (Aldovini and Young, 1990; De Guzman et al., 1998b; Dorfman et al., 1993; Gorelick et al., 1988, 1990, 1993, 1999a, 1999c; Guo et al., 2000; Jentoft et al., 1988; Méric et al., 1988; Mizuno et al., 1996; Poon et al., 1996; Tanchou et al., 1998; Wang and Barklis, 1993; Zhang and Barklis, 1995) or multiple basic residues in HIV-1 (Cimarelli et al., 2000a; Poon et al., 1996) can significantly reduce genome packaging. NMR experiments have shown that the NC zinc fingers are responsible for specific interactions with Psi-site sequences on the gRNA, while the basic residues contribute more to non-specific nucleic acid binding (De Guzman et al., 1998a, 1998b). Elimination of the NC domain from Gag also disrupts specific packaging of HIV-1 gRNA, although packaging of cellular RNAs is supported by basic residues in MA (Ott et al., 2003, 2005; Rulli et al., 2007). The gRNA that is packaged into retroviruses can be the same as that used for translation of Gag (Butsch and Boris-Lawrie, 2002), so that the genome can be co-translationally selected by NC (packaged in cis) as Gag is synthesized (Anderson and Lever, 2006; Poon et al., 2002). In contrast, murine leukemia virus (MLV) genomes appear to be packaged mainly in trans (Dorman and Lever, 2000). In the case of HIV-1 and HIV-1–based vectors, the gRNA can also be packaged in trans (Nikolaitchik et al., 2006).
2.2. Genome dimerization
A necessary step for virus particle maturation is the formation of a stable dimer complex between the two molecules of gRNA present in the retrovirus particle (Fig. 2). Indeed, it has been demonstrated that NC is critical for the formation of this complex (Bieth et al., 1990; Fu et al., 1994; Fu and Rein, 1993; Méric et al., 1988). The mechanism for this stabilization is thought to begin with limited base pairing between the genomes, termed a “kissing loop structure,” which, with the assistance of NC expands to form a more complex, extended structure (Girard et al., 1995; Muriaux et al., 1996; Polge et al., 2000). This process, termed “dimer maturation,” results in the formation of a more thermostable RNA dimer, and occurs during the maturation of the virus particle after budding (Feng et al., 1996; Fu et al., 1994; Fu and Rein, 1993; Laughrea et al., 2001; Song et al., 2007; Takahashi et al., 2001). NC facilitates RNA maturation via its nucleic acid chaperone activity, which assists the RNA to find the most thermodynamically stable annealed structure. The formation of the RNA dimer probably facilitates the complex events of reverse transcription, such as the obligatory strand-transfer steps and the high degree of recombination observed between genomes (Hu and Temin, 1990a, 1990b).
2.3. tRNA placement
Another event that occurs during particle assembly is the incorporation of tRNA molecules into virions: these serve as the primer for the initiation of reverse transcription (Marquet et al., 1995). The tRNA molecules are brought into virions by various means, including coassembly of cognate aminoacyl tRNA synthetases in the cases of HIV-1 (tRNALys,3) and Rous sarcoma virus (RSV) (tRNATrp) (Cen et al., 2002). For MLV, there has been no demonstration of the involvement of a trans-acting factor for the packaging of tRNAPro (Cen et al., 2002; Fu et al., 1997): tRNAPro in wild-type MLV particles is only 2- to 3-fold greater than cellular tRNAPro levels, in contrast to an ∼20-fold enrichment of tRNATrp with avian leukosis virus (Waters and Mullin, 1977) as expected, due to active tRNA incorporation. A portion of these tRNA molecules is complementary to the primer binding site (PBS) on the viral genome. It has been demonstrated from cell-free assays that HIV-1 NC, both in the precursor (Pr55Gag) and mature (p7NC) forms, enables the stable annealing of its cognate tRNA to gRNA sequences, which entails the unfolding of 15 nucleotides of the tRNA (Cen et al., 1999; Feng et al., 1999; Hargittai et al., 2001, 2004; Huang et al., 1998; Rong et al., 2001). Again, this function is associated with the NC’s nucleic acid chaperone activity, which increases the rate of RNA annealing. Interestingly, disruption of the zinc fingers does not eliminate this annealing activity (Hargittai et al., 2004). Enhancement of tRNA annealing to gRNA has also been observed with the NC-containing peptides of Ty3, Ty1, and Gypsy (Cristofari et al., 1999, 2000; Gabus et al., 2006).
2.4. RNA scaffold binding
NC is important for virus assembly: deletion of the NC domain of Gag results in a severe reduction in the production of virus particles (Bowzard et al., 1998; Dawson and Yu, 1998; Ott et al., 2003; Sandefur et al., 1998, 2000). In the case of RSV and MLV, NC is strictly required for particle production (Lee and Linial, 2006; Muriaux et al., 2004). Interestingly, in some viruses, the NC domain could be functionally substituted with a leucine zipper motif (Johnson et al., 2002; Li et al., 2007; Zhang et al., 1998), implying that one of the functions of NC in assembly is to stabilize interactions between Gag molecules. As a result, it was proposed that NC possessed an I (interaction) domain necessary for Gag multimerization, and results obtained from NC deletion mutants indicated that multiple basic residues are required for this activity (Cimarelli et al., 2000a; Sandefur et al., 2000). However, for HIV-1, efficient production of virus particles from an NC deletion mutant was rescued by a mutation that eliminated PR activity, suggesting that the actual defect caused by some NC deletion mutations was accelerated PR processing of Gag (Ott et al., 2003). A similar rescue in particle production by inactivating PR was reported for a NC mutant lacking basic residues (Wang et al., 2004). The rescue of particle production in NC deletion mutants by inactivating PR has not been observed in other systems using different cell lines, suggesting a cell-type specific effect (Ono and Freed, personal communication). Thus, it is likely that the I domain is not simply a protein multimerization domain, as suggested by the ability to replace NC with a leucine zipper motif, but that the binding of NC to gRNA results in the congregation of Gag molecules, which drives multimerization via CA. This idea is termed RNA scaffolding.
The RNA scaffold hypothesis follows from the evidence discussed above, as well as the observation that virus-like particles are produced from cells that only express retroviral Gag proteins (Swanstrom and Wills, 1997). A key observation is that these virus-like particles incorporate cellular RNA in the absence of a Psi-containing RNA (Muriaux et al., 2002; Rulli et al., 2007). In fact, Gag can assemble into virus-like particles in cell-free systems as long as RNA is present, even if it is not viral (Campbell and Vogt, 1997). Thus, an RNA scaffold is required for particle assembly (Fig. 2) (Muriaux et al., 2001). Two regions of HIV-1 Gag contribute to RNA binding and particle assembly: a patch of basic residues in the MA domain and basic residues in the NC domain (Ott et al., 2005; Poon et al., 1998). For other retroviruses, NC is the sole determinant for binding to nucleic acids during assembly, mainly via nonspecific ionic interactions between the basic amino acids and nucleic acid phosphate backbone (Kräusslich, 1991; Lee and Linial, 2004).
Another requisite step in the production of infectious HIV-1 particles is PR processing of Pr55Gag (Fig. 3) and Pr160GagPol. It is evident that the rate and order of the PR-mediated cleavage at the PR recognition sites in Pr55Gag and Pr160GagPol are critical for the production of infectious virus particles (Kräusslich et al., 1995; Pettit et al., 1998; Shehu-Xhilaga et al., 2001b; Wiegers et al., 1998), and even small decreases in PR activity can affect virus infectivity significantly (Kaplan et al., 1993). PR processing and particle maturation are conceptually linked because particles produced by PR-defective virions result in an immature phenotype (Göttlinger et al., 1989), but although PR is necessary, it is not sufficient to produce infectious particles. Interestingly, PR processing appears to be largely complete as soon as virus particles are isolated after budding (Kaplan et al., 1994b), and even blocking the last cleavage step, which is the conversion of p25 to p24 (i.e., the removal of p2 (SP1) from p25) (Li et al., 2003; Pettit et al., 1994; Wiegers et al., 1998), is enough to produce an immature phenotype. Thus, the bulk of the maturation process in HIV-1 particles is the formation of a core by CA, gRNA condensation, and establishment of a proper nucleoprotein structure that is competent for reverse transcription.
3. Reverse transcription processes
The conversion of gRNA to full-length dsDNA is an essential early step in retroviral replication (Fig. 2), but it is a complex process requiring at least two strand-transfers (Basu et al., 2008), and the RNA- and DNA-dependent polymerase transcription of RNA and cDNA sequences, respectively (Fig 4). For a detailed description of general reverse transcription, see Telesnitsky and Goff (1997). The viral nucleic acids used as templates for polymerization possess extensive and complex secondary structures that need to be dealt with as well. Many laboratories have developed cell-free systems to investigate the properties and to determine the mechanism of each of the key steps in this process: it is clear that reverse transcriptase (RT) alone can accomplish many of these steps, albeit inefficiently, with RT pausing and template self-priming being just a couple of the more deleterious consequences (see later).
Fig. 4. Key steps of reverse transcription where NC has been shown to be involved (HIV-1 example presented).
The process of reverse transcription entails the conversion of a plus-strand RNA genome (A) to a double-strand DNA (H), with an LTR duplicated at each end (638 bp in the case of HIV-1) (Telesnitsky and Goff, 1997). The first requirement for reverse transcription is the annealing of a primer tRNA molecule onto the PBS of the gRNA (B). Once annealed, reverse transcriptase synthesizes vDNA to the end of the genome; this product is termed minus-strand strong-stop DNA (C). As RT continues synthesizing the vDNA, the RNase H activity of RT cleaves the gRNA involved in heteroduplex formation into small fragments (Schultz and Champoux, 2008) (C). RNase H digestion allows the transfer of the cDNA repeat ® region, facilitated by NC, to the RNA R region at the 3′ end of the genome, an event termed minus-strand transfer (D). Minus-strand transfer can occur intra- or intermolecularly, so that either genome can contribute genetic material to the final product. Upon transfer, minus-strand synthesis proceeds until it reaches the PBS at the 5′ end of the genome. As RT continues to synthesize the vDNA, RNase H functions to cleave the genome into small fragments. In HIV-1, two regions, the PPT and cPPT (E), are resistant to RNase H cleavage. They both serve as primers for plus-strand DNA synthesis, using the minus-strand DNA as template. As the plus-strand synthesis reaches the 5′ end of the minus-strand DNA, it copies the PBS from the tRNA that remains covalently bound to the 5′ end of the minus strand (E). The two complementary PBS regions on the minus- and plus-DNA strands anneal, forming a circular intermediate (F) (in the case of the intramolecular transfer that is presented in this example), enabling the synthesis of the full plus-strand DNA sequence. Two additional steps are important; completion of the 5′ LTR requires the strand-displacement synthesis activity of RT (G). In an additional region, strand-displacement synthesis occurs in HIV-1 resulting in the central flap, a 99 nt region of ssDNA generated by limited synthesis at the cPPT (not shown).
Of course, in the context of the virus, RT is not the only protein present, and it must perform its functions in the context of gRNA being completely coated with NC (Darlix et al., 2002). It is likely that other cellular and viral proteins are also involved in this process. Interestingly, when NC is added to these various cell-free systems, it typically enhances the efficiency of the reaction by several orders of magnitude, which is thought to be due to its nucleic acid chaperone properties. As mentioned above, a number of excellent reviews have been published, detailing how NC functions as a chaperone in cell-free systems, and highlighting the molecular properties of NC (Bampi et al., 2004a, 2004b; Darlix et al., 2000, 2002, 2007; Levin et al., 2005; Rein et al., 1998). Characterization of NC function during specific events of reverse transcription using highly defined in vitro systems is different from what occurs in the context of virus. Nevertheless, these systems provide critical insights that assist in the interpretation of the processes that take place within an infected cell. They also provide clues regarding possible antiviral therapies by identifying all the possible steps that may be disrupted when NC is targeted.
3.1. NC chaperone activities in vitro
The properties of NC have been examined in many different cell-free assays, developed to investigate its role in reverse transcription. The majority of studies have used either the 55 amino acid form of HIV-1 p7NC or a version termed p7NC (1–72) (de Rocquigny et al., 1991), which is a chemically synthesized form that is actually one amino acid longer than authentic p9NC (comprised of p7NC and SP2) (Henderson et al., 1992). Based on these results, it has been observed that HIV-1 NC can greatly enhance the rate of tRNA primer annealing to the PBS of the gRNA (Fig. 4B) (Barat et al., 1993; Hargittai et al., 2001, 2004; Huang et al., 1998; Khan and Giedroc, 1992; Tisné, 2005). This annealing step involves the partial disruption of tRNA structure so that its 3’ 18 nucleotides can form base pairs with the PBS region on the gRNA. Similar annealing can also be performed using heat to destabilize the intramolecular base pairing of these sequences (Beerens and Berkhout, 2000), indicating the utility of NC in performing this at physiological temperatures. Annealing of the tRNA to gRNA is one requirement for initiation of reverse transcription, so that there is the potential for mutations in NC to prevent even the earliest product of reverse transcription. Reverse transcription from this primer to the 5’ end of the RNA genome forms the DNA product called minus-strand strong-stop DNA (Fig. 4C). Importantly, this gRNA region includes R and U5, which in the case of HIV-1, contain extensive secondary structures. The R region includes TAR, which is a very stable RNA structure that is involved in interactions with the Tat protein and is responsible for transactivation of the HIV-1 LTR promoter (Roebuck and Saifuddin, 1999).
As the minus-strand strong-stop DNA is being synthesized, the RNase H domain of RT digests the RNA template of the newly synthesized heteroduplex into small fragments (Schultz and Champoux, 2008) (Fig. 4C). RNA digestion by RNase H exposes the DNA as a single-stranded region that is complementary to the 3’ end of the RNA genome (Peliska and Benkovic, 1992). This sets the stage for the minus-strand transfer event that is required for continued synthesis of the minus strand viral DNA (vDNA) (Fig. 4D). This strand-transfer is greatly enhanced by NC, both from an acceleration of the annealing of 5′-R cDNA to the 3′-R region of the genome (Allain et al., 1994; Guo et al., 2000; Hu and Temin, 1990b; Peliska et al., 1994; You and McHenry, 1994) and by stimulation of the RNase H activity (Peliska et al., 1994; Roda et al., 2003). Because NC facilitates the formation of nucleic acid structures having the maximum number of base pairs, the RNase H activity will result in the cDNA R region being bound to fragments of gRNA, thus forming fewer base pairs than that present in the 3′-RNA R region (Tsuchihashi and Brown, 1994).
A case that illustrates how the nucleic acid chaperone activity of NC functions is from the work of Jeeninga and collegues. In assays that mimic the minus-strand transfer event, the presence of excess RNA, complementary to the entire minus-strand strong-stop DNA sequence blocked strand transfer, even in the presence of NC, because this RNA formed a greater number of base pairs than would be formed by the transfer event where just the R region is complementary (Jeeninga et al., 1998). However, in vivo, cleavage of the gRNA, as it is copied by RT, prevents such an event since the transfer product would form a more extensively annealed structure than the Rnase H-cleaved vRNA oligonucleotides. In addition, increasing the rate of strand transfer should also reduce the possibility of self-priming, caused by the cDNA R region folding back upon itself, resulting in RT-mediated DNA synthesis of the strong-stop DNA just generated. In vitro reactions show that NC prevents this self-destructive reaction (Driscoll and Hughes, 2000; Guo et al., 1997; Lapadat-Tapolsky et al., 1997; Rascle et al., 1998).
The ability of NC to destabilize nucleic acid secondary structure is advantageous in that it enhances the processivity of reverse transcription (Fig. 4E), resulting in a greater proportion of full-length vDNA products. The enhancement of processivity is coupled with a decrease in the total number of partial products synthesized, since RT continues synthesizing a strand rather than falling off at a pausing site and reinitiating a nascent DNA strand (Drummond et al., 1997; Ji et al., 1996; Tanchou et al., 1995; Wu et al., 1996). During the synthesis of the full-length minus-strand DNA, RNase H continues to digest the RNA template portion of the heteroduplex, but there are polypurine tract (PPT) regions that are resistant to cleavage (Fig. 4E). In most retroviruses, the 3’ PPT defines the 5’ U3 end of the 5’-LTR (Rausch and Le Grice, 2004). In lentiviruses an additional region denoted the central-PPT (cPPT) is important for completion of DNA synthesis in a timely fashion. In addition, DNA synthesis from this region results in the formation of the central flap, a 99 nucleotide ssDNA section that has been implicated in the nuclear import of preintegration complexes (PICs) in non-dividing cells (Arhel et al., 2007; Charneau et al., 1992; Zennou et al., 2000).
In the last obligatory strand-transfer event, plus-strand transfer (Basu et al., 2008; Telesnitsky and Goff, 1997), the minus-strand PBS anneals to the plus-strand PBS, forming a circular intermediate in the case of an intra-strand transfer product (Fig. 4F); an end-to-end linear product would result from an inter-strand transfer event. NC also enhances plus-strand transfer, although the regions of secondary structure are less complex than what was encountered during annealing of the R regions (containing the TAR and cTAR stem-loop regions) when minus-strand transfer took place (Guo et al., 2000; Johnson et al., 2000; Wu et al., 1999, 2007). In addition to enhancing the obligatory strand-transfer events, NC also facilitates internal strand-transfers, thus promoting the high rate of recombination observed between the two RNA genomes (Derebail et al., 2003; Negroni and Buc, 1999; Raja and DeStefano, 1999; Ramirez et al., 2008; Roda et al., 2002).
Completion of reverse transcription requires strand-displacement synthesis, which is strand synthesis through a region of dsDNA (Fig. 4G). For most retroviruses, a single event results in completion of the LTR ends (Fuentes et al., 1996b; Whiting and Champoux, 1998). RT by itself is capable of such synthesis, but again, the presence of NC greatly enhances this process (Fisher et al., 2003; Hameau et al., 2001; Kelleher and Champoux, 1998; Paulson et al., 2007; Urbaneja et al., 2002). The failure to complete strand-displacement synthesis would result in incomplete LTR ends. This, in turn, would create vDNA that cannot integrate. Strand-displacement synthesis is also essential in creating the central flap region that is located in the vicinity of the cPPT (Fuentes et al., 1996a; Hameau et al., 2001) and, as mentioned above, seems necessary for efficient lentiviral replication.
An additional finding showed that interactions of NC in in vitro reverse transcription complexes (RTCs) were able to facilitate nucleotide excision repair by reverse transcriptase. Interestingly, p9NC, but not p7NC, could function in this capacity (Bampi et al., 2006). The mechanism of this is thought to be stabilization of RT on the DNA template (Lener et al., 1998). This observation is correlated with other experiments demonstrating that p9NC i) binds and releases more slowly than p7NC and ii) has a cooperative component in its binding to nucleic acids (Cruceanu et al., 2006).
Properties of NC important for facilitating the many steps of reverse transcription are its ability to destabilize annealed nucleic acid structures and its ability to aggregate nucleic acids. A single HIV-1 NC molecule has multiple nucleic acid binding sites that allow intra- and intermolecular interactions (Fisher et al., 2006), thus providing an excellent explanation for these aggregative properties. Aggregation appears to be beneficial for bringing nucleic acid strands into close proximity, a feature that would certainly aid in the strand-transfer reactions and recombination events. In addition, NC-nucleic acid aggregates have been shown to be dynamic structures. A recent study has illustrated the nature of model NC-nucleic acid aggregates. In this report, p7NC co-aggregated with ssDNA in a Mg2+-dependent manner, which could be visualized by transmission electron microscopy (TEM). It was observed that these aggregates could be dispersed by adding either G-quartet oligonucleotides, which are DNA structures that NC binds with high affinity, or an excess of T4 gene 32 protein, a prototypical single-stranded binding (SSB) protein. Critically, the competition experiments demonstrated that these aggregates are in equilibrium with the components in the solution (Mirambeau et al., 2006). In a second study it was demonstrated that when RT is added to the NC-nucleic acid aggregates in the presence of Mg2+ and deoxynucleotide triphosphates (dNTPs), it was able to convert the primed, ssDNA to dsDNA. Interestingly, as the reverse transcription reaction proceeded, the amount of p7NC bound to the complex progressively decreased, but was not completely eliminated. The decrease in p7NC present is probably because of a relatively low affinity for dsDNA, but the observation that p7NC was still bound reflected that the DNA synthesis reaction also resulted in the formation of an ssDNA flap, with p7NC remaining tightly bound to this region. Using TEM, it was observed that this flap region was extended, in a manner similar to when SSB proteins bind DNA (Mirambeau et al., 2007).
Another aspect of the cell-free reverse transcription reaction on ssDNA-NC aggregates is the effect of PR-digestion of p15NC. When p15NC was added to ssDNA, the protein bound to and extended the nucleic acid strand like a canonical SSB protein. PR subsequently cleaved p15NC, removing p6, then cleaved the p9NC protein, finally forming p7NC (Fig. 3). As PR processing proceeded, a progressive aggregation of single-stranded nucleic acid was observed. As mentioned above, during RT conversion of the primed ssDNA to dsDNA in the presence of Mg2+ and dNTPs, the aggregates dispersed. Dispersal of aggregates was almost complete in the presence of p7NC, but not as extensive when p9NC was present. This study displays how the organization and properties of the nucleoprotein complex can change as a result of the activities of PR and RT, reflecting parallel processes that take place during virion assembly/maturation and early infection events (Mirambeau et al., 2007).
3.2. NC chaperone activities in cell culture
A particular challenge in the interpretation of results obtained from infection experiments is that many concurrent processes are taking place. These may be related or unrelated, competitive or noncompetitive, and the result of one reaction will likely alter the outcome of another. Thus, it is helpful to apply the knowledge obtained from in vitro reactions to what is observed in cells. Cell culture-based studies have been performed to ascertain the progression of reverse transcription, either by Southern blotting, or more recently, by semi-quantitative or quantitative PCR amplification of defined reverse transcription products. A quantitative assay for the detection of strand-specific target sequences has even been recently developed (Thomas et al., 2007a). For PCR analysis, one can amplify different target sequences that correspond to the completion of specific reverse transcription steps, such as detection of minus-strand strong-stop vDNA, by using primers that bind the R-U5 region (Fig. 4C); the presence of this target sequence is a good indicator of whether reverse transcription initiated. In a similar manner, one can examine production of U3-U5 vDNA, which is formed after minus-strand transfer (Fig. 4D); gag vDNA, which is formed during late minus-strand synthesis (Fig. 4E); and R-5’UTR, which is present only after the plus-strand transfer event has taken place (Fig. 4F) (Buckman et al., 2003; Butler et al., 2001; Nagy et al., 1994; Tanchou et al., 1998; Thomas et al., 2006a, 2007a; Zack et al., 1990). Using these techniques, mutations to NC have given rise to a number of different phenotypes: those that block reverse transcription from initiating, those that decrease the progression of reverse transcription, and those that show little apparent effect on reverse transcription.
Infection studies using MLV have identified several NC mutations that appear to cause severe defects in vDNA synthesis. These defects were manifested either by reduced amounts of early vDNA products (Gorelick et al., 1996; Méric and Goff, 1989; Yu and Darlix, 1996), or reduced stability of the newly synthesized vDNA (Gonsky et al., 2001; Yu and Darlix, 1996). In addition, some studies examined the amounts of 2-LTR circles, whose existence indicates near completion of reverse transcription and exposure to nuclear ligases, and observed that some mutations resulted in decreased amounts compared to wild-type virus, while other mutations exhibited evidence of heterogeneous 2-LTR circle populations (Gorelick et al., 1999b). This latter result was verified by PCR amplifying, cloning, and sequencing of the 2-LTR junctions. As expected, these studies showed that PCR detection was more sensitive than Southern blotting, but they also showed that mutations to the zinc-binding residues caused a more severe defect in vDNA synthesis than mutations to other amino acid residues, such as the conserved aromatic amino acid residues in NC (Gorelick et al., 1999b). MLV is a gammaretrovirus, and as such has only a single zinc finger domain; therefore, it is helpful to compare these results with those from HIV-1 and simian immunodeficiency virus (SIV), which, being lentiviruses, have two zinc fingers. In the case of SIVmac239, it was reported that mutations that eliminated zinc binding by either the NH2- or COOH-terminal zinc fingers only slightly impaired synthesis of cDNA, but no plus-strand transfer products were detected. This indicated that these mutations induce a more severe defect on the plusstrand transfer step than on minus-strand transfer or minus-strand synthesis (Akahata et al., 2003).
Results from infections using HIV-1 NC-mutant viruses have shown phenotypes similar to those observed with other retroviruses. There are also some interesting cell-type and viral strain-specific effects. It was reported that several mutations to certain basic residues in the NH2-terminal basic region (Table 3) could greatly reduce vDNA synthesis (Berthoux et al., 1997; Cimarelli et al., 2000b), although, since 2-LTR circles could be readily detected, the R10A/K11A mutants showed less of a defect. In addition, it was observed that the defect associated with the R10A/K11A mutation in NC depended on the virus strain: in the context of HXB2, the mutant was very replication defective, but in the context of NL4-3, the mutant replicated in virtually the same way as wild-type did (Cimarelli and Luban, 2001). Another HIV-1 NC mutant, where amino acids Arg29/Ala30 were replaced with the residues Ser-His-Ala-Trp (Table 3), blocked synthesis of even early vDNA product. However, there was evidence of a cell type-specific defect: in H9 cells this mutant was replication defective, but it did replicate in A3.01 and M8166 cells (Furuta et al., 1997; Kawamura et al., 1998; Koh et al., 2000). The reason for the cell-specific responses is not clear and was not discussed.
Table 3.
Sequences of HIV-1 NC and specific mutants.
| Virus | N-terminus | N-terminal Zinc Finger | Central | C-terminal Zinc Finger | C-terminus |
|---|---|---|---|---|---|
| NL4-3a | IQRGNFRNQKKTVK | CFNCGKEGHIAKNC | RAPRKKG | CWKCGKEGHQMKDC | TERQAN |
| H23Cb | -------------- | --------C----- | ------- | -------------- | ------ |
| H44Cb | -------------- | -------------- | ------- | --------C----- | ------ |
| N17Gc | -------------- | --G----------- | ------- | -------------- | ------ |
| N17Fc | -------------- | --F----------- | ------- | -------------- | ------ |
| N1bd | -------------- | -------------- | SHRW----- | -------------- | ------ |
| NL4-3 R10A,K11Ae | ---------AA--- | -------------- | ------- | -------------- | ------ |
| HXB2 R10A,K11Ae | M--------AAI-- | ---------T-R-- | ------- | -------------- | ------ |
The entire 55 amino acid sequence is displayed, but separated into the 5 standard domains indicated at the top of each column. Identical amino acids are indicated by a dash.
Mutants described in Buckman et al. (2003), Gorelick et al. (1999c), Thomas et al. (2006a).
Mutants described in Thomas et al. (2006b).
Mutant described in Furuta et al. (1997), Kawamura et al. (1998), Koh et al. (2000).
In the study of R10A,K11A in the context of NL4-3 and HXB2, it was determined that the Thr in the N-terminal zinc-finger was responsible for the different phenotypes: NL4-3 R10A,K11A,I24T is replication defective, like HXB2 R10A,K11A (Cimarelli and Luban, 2001). This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Mutations that altered the zinc finger structure while preserving zinc binding and gRNA packaging (NCH23C or NCH44C; Table 3), did not appreciably reduce the initial amount of vDNA synthesized when early and late reverse transcription products were examined (Tanchou et al., 1998; Thomas et al., 2006a). Only a minor defect was detected in plusstrand transfer at early time points after infection (Thomas et al., 2006a). These mutations reduced the apparent intracellular stability of the nascent vDNA so that quantities at 24 hours post-infection were significantly lower than wild-type (Buckman et al., 2003; Tanchou et al., 1998; Thomas et al., 2006a). Some PCR studies detected the presence of 2-LTR circles after infection with these mutants (Gorelick et al., 1999c; Thomas et al., 2006a), although another PCR study using different primers did not detect them (Tanchou et al., 1998). As mentioned earlier, the presence of 2-LTR circles indicates near completion of reverse transcription and exposure to nuclear ligases, and thus provides a snapshot of the LTR ends just before ligation. Interestingly, it was observed that the 2-LTR circles formed after infection exhibited defects at the joined LTR ends. PCR cloning and sequencing showed that both the NCH23C and NCH44C mutations cause insertions and deletions at a higher rate than wild-type (Buckman et al., 2003). The presence of vDNA species with improper ends would not be suitable substrates for the subsequent integration reaction and could account for some of the replication defects observed with these NC mutants.
Recent studies investigating the importance of PR processing of NC (Fig. 3) (p15NC to p9NC + p6, then to p7NC + SP2) (Shehu-Xhilaga et al., 2001a) revealed that mutations that prevent the conversion of p9NC to p7NC (i.e., the removal of the p7NC-SP2 cleavage site) did not have a significant impact on infectivity, replication, or vDNA synthesis. However, mutations that prevented the formation of p9NC by altering the PR cleavage site between SP2-p6, creating only p7NC + SP2-p6 products, caused huge defects in replication but only minor defects in synthesis of reverse transcription products (Coren et al., 2007). Likewise, in another study where the conserved Asn residue at position 17 in the NH2-terminal zinc finger of NC was mutated to Phe or Gly (Table 3), a defect of several orders of magnitude was detected in replication assays, but the processivity of reverse transcription appeared similar to wild-type (Thomas et al., 2006b).
A common phenotype that was observed with NCH23C, NCH44C, NCN17F, and p9NC-deficient mutants discussed above was a decrease in H9-replicative titer compared to wild-type virus, in some cases more than 6 logs. However, when one examines the magnitude of the reverse transcription defects that results from these NC mutations, the effect on vDNA synthesis is much smaller. What has been observed thus far is that these mutations do not block the synthesis of vDNA nearly enough to account for the replication defects. In general, if mutations to NC allowed reverse transcription to begin, the process appeared to go to completion at a reasonably high efficiency (Coren et al., 2007; Thomas et al., 2006a, 2006b). The lack of an effect on reverse transcription processes is in contrast to specific and substantial defects observed in cell-free assays that employ these same mutant NC proteins.
The reason for the discrepancy between the in vitro and cell culture-based results is probably because of several factors. First, the process of reverse transcription, as it occurs in a RTC, is difficult to model in vitro due to technical problems of protein and nucleic acid concentrations. Thus, even if a mutant NC protein is not able to facilitate a particular step in reverse transcription under the in vitro conditions used, the concentrations of NC would be greater in the virion, shifting the equilibrium of the specific reaction. Additionally, the physical state of the nucleoprotein complex within the virion is essentially that of a precipitate or aggregate, and many of the in vitro assays have been developed so that protein and nucleic acid components remain soluble. Perhaps these events could be modeled in the aggregation systems developed by Mirambeau and colleagues (Mirambeau et al., 2006, 2007). Second, in cell culture-based analyses of virus replication processes, it is possible that a number of cellular and viral proteins can facilitate several of these processes in a manner similar to NC. This possibility is based on in vitro evidence of chaperone function and a few examples of viral proteins include Vif (Henriet et al., 2007), Tat (Guo et al., 2003), and even integrase (IN) (Dobard et al., 2007). The cellular prion protein PrP has also been shown to possess NC-like chaperone activities and could facilitate many of the same reactions as NC in vitro (Derrington et al., 2002; Gabus et al., 2001a, 2001b). Other as yet unidentified components may be able to provide the necessary NC-like functions in reverse transcription when the NC protein is altered. The extent to which any or all of these factors in cell culture-based assays supplement NC function is unknown. It is possible that retroviruses may rely on the functional redundancy of these other factors to carry out effective replication.
Interesting findings regarding the role of NC in replication have also been uncovered through experiments using compounds that either compete with NC for nucleic acid binding or eject zinc from the zinc fingers. These types of studies complement observations obtained by genetic analyses, since they allow one to drastically alter NC function after proper assembly of virion particles. Either virions or cells to be infected were treated with these NC-targeting compounds, and then vDNA synthesis was measured. In these types of experiments, it was observed that reverse transcription was either greatly diminished or completely blocked, depending upon the extent to which NC was modified by the compounds (Berthoux et al., 1999; Morcock et al., 2005; Rossio et al., 1998; Srivastava et al., 2004; Turpin et al., 1999) or the concentration of the NC competitors added to the systems (Druillennec et al., 1999). The results of these experiments independently support the hypothesis that NC is critical for reverse transcription.
3.3. NC mutations causing premature reverse transcription
Several HIV-1 NC mutant virions, NCH23C and NCH44C (Table 3), have been extensively studied by this and other laboratories. Kinetic studies indicate that reverse transcription in cultured cells occurred at a reasonable rate, but that the apparent stability of the vDNAs produced after infection with these mutants was lower than that observed by wild-type virus (Thomas et al., 2006a). Subsequently, we observed that much higher levels of vDNA were contained within these mutant virions [unpublished data]. Currently, it is not known whether premature reverse transcription is a phenotype that would be observed in other orthoretroviruses. However, this phenotype is not unheard of since in prototype foamy viruses (PFVs), the particles contain mostly reverse-transcribed vDNA, rather than gRNA (Linial 1999). The fact that these HIV-1 NC mutants are severely replication defective suggests that increasing the efficiency of reverse transcription initiation is detrimental to viral replication. In agreement with this idea, primer activation signal (PAS) mutants that prevent binding of the anti-PAS motif in the TΨC arm of the tRNALys,3 result in increased reverse transcription activity and also cause replication defects (Abbink and Berkhout, 2007; Beerens and Berkhout, 2002; Beerens et al., 2001).
Intriguingly, it is not possible for reverse transcription to occur in virions after budding because the levels of dNTPs present are too low to permit synthesis of the observed amounts of vDNA. This in turn suggests that these NC mutants allow reverse transcription to initiate in the cytoplasm prior to budding. The mechanism behind this observation is not understood at this time, but this scenario is not improbable, since unprocessed Gag-Pol has been demonstrated to carry out efficient endogenous reverse transcription (Kaplan et al., 1994a). Interestingly, these NC mutants show no activity in endogenous reverse transcription reactions [unpublished data], probably because the genome template is consumed during the formation of the vDNA. The RT from these mutant particles is, however, competent in exogenous template reverse transcription assays (Gorelick et al., 1999c).
4. Uncoating and nuclear transport
Receptor binding and virus fusion with cells result in the virion core entering the cytoplasm. Upon entry, there is concomitant core uncoating and reverse transcription, although the details of this process are still poorly understood (Nisole and Saïb, 2004). Reverse transcription occurs with remodeling of the nucleoprotein complex to form the PIC that enters the nucleus, where the integration process occurs (Fig. 2). The various steps and NC’s involvement are described below.
4.1. Entry of the virion core into the cytoplasm
The reverse transcription process is proposed to begin immediately after entry of the virion core into the cytoplasm. The initiation of reverse transcription is dependent on the presence of dNTPs: this is graphically illustrated by observations that reverse transcription proceeds poorly in quiescent cells where intracellular dNTP concentrations are low (Zack et al., 1990, 1992), and simply increasing the concentration of dNTPs permits accumulation of full-length vDNA (O’Brien et al., 1994). In addition, it is possible that the size and structure of cell-free virions preclude significant DNA synthesis before cytoplasmic entry, so that core disassembly is necessary for reverse transcription to occur. There are reports that reverse transcripts are present in extracellular virions, but as no more than 1 in 1 000 particles have minus-strand strong-stop DNA (Lori et al., 1992; Trono, 1992; Zhang et al., 1993; Thomas and Gorelick, unpublished observations), the significance of this in infection processes is tenuous.
For HIV-1, the virion core that enters the cytoplasm and begins this process is thought to consist of a capsid shell comprised of p24CA, which surrounds the nucleocapsid core (dimerized RNA genome, tRNALys,3 primers, and ∼2 000 molecules of p7NC). In addition, purified cores have been shown to contain RT, IN, PR, Vpr, and small amounts of Pr55Gag and p41MA-CA (Miller et al., 1997; Welker et al., 2000). This nucleoprotein assembly is termed the RTC. As the RTC uncoats, reverse transcription proceeds and the complex is transported through the cytoplasm to the nucleus where it is converted into a PIC, and in the case of non-dividing cells with lentiviral infections, is actively transported into the nucleus (Fig. 2) (Dvorin and Malim, 2003; Nisole and Saïb, 2004; Warrilow and Harrich, 2007). For retroviral infections in actively dividing cells, it has previously been shown that PICs gain access to the nucleus by passive diffusion during cell division when the nuclear membrane breaks down during mitosis (Roe et al., 1993).
4.2. RTC vs PIC
One of the challenges to studying early infection events is how to isolate authentic RTCs or PICs so that the exact components of these nucleoprotein complexes can be ascertained. Conceptually, a RTC is a complex where reverse transcription initiates and remains intact until the process is complete. A PIC consists of a complex containing full-length vDNA, the substrate for integration. In practice, it is probably more accurate to view this as a continuum starting with a RTC and ending with a PIC. Experimentally, biochemical isolation of these complexes is complicated by their labile and unsynchronized nature, which makes it difficult to separate the two complexes. For instance, many of the published studies examine RTCs/PICs at 8 hours postinfection, by which point vDNA can be detected in the nucleus, so these complexes are either entirely PICs or very mature RTCs.
Genetic analyses in HIV-1 have provided many suggestive results in that certain mutations to p24CA or p17MA can block either reverse transcription or nuclear entry. During Pr55Gag synthesis, the majority of MA is myristylated at the amino-terminus. This myristylation is necessary for membrane binding of Pr55Gag. However, it has been reported that a subset of Pr55Gag is not myristylated, but instead is phosphorylated on Ser or Tyr residues in the MA domain (Bukrinskaya et al., 1996; Gallay et al., 1995b; Kaushik and Ratner, 2004). In the case of phosphorylation of MA Tyr residues, nuclear import effects appeared to be dependent on multiplicity of infection (Freed et al., 1997; Trono and Gallay, 1997). Phosphorylated MA is associated with the virus core and localizes to the nucleus after infection (Gallay et al., 1995a). Interestingly, mutations that eliminate the Ser or Tyr residues can reduce infectivity by blocking either nuclear entry (Bukrinskaya et al., 1996; Gallay et al., 1995a) or reverse transcription (Kaushik and Ratner, 2004).
In addition, a number of mutations to CA can reduce infectivity (Wacharapornin et al., 2006) also by blocking either nuclear entry (Dismuke and Aiken, 2006; Fitzon et al., 2000; Yamashita and Emerman, 2004) or reverse transcription (Fitzon et al., 2000; Forshey et al., 2002; Tang et al., 2001). The mechanism behind these defects may be due to improper core disassembly, since many of these mutations appear to stabilize the core (Dismuke and Aiken, 2006; Forshey et al., 2002; Kiernan et al., 1998; Wacharapornin et al., 2006). In conceptual agreement with the idea that HIV-1 core uncoating is an important step in infection, the TRIM5α restriction factor from Old World monkeys apparently functions by accelerating core uncoating, which impairs normal reverse transcription (Forshey et al., 2005; Stremlau et al., 2004, 2006). Also in line with the idea that the rate of core uncoating is important, we have observed that certain mutations to NC (NCH23C and NCH44C; Table 3) cause reverse transcription to commence and proceed much faster than for wild-type virus, but the vDNA generated integrates very poorly for some unknown reason (Thomas et al., 2006a). It is possible that accelerated reverse transcription would cause premature disassembly of the core structures, leading to aberrant PIC structures: electron micrographic observations of virions that have undergone natural endogenous reverse transcription because of exposure to dNTPs, exhibit poorly defined cores compared to untreated virions (Zhang et al., 2000a).
4.3. Protection of vDNA by NC
As the RTC is transported to the nucleus, the vDNA is a likely target of attack by cellular nucleases. It has been observed from in vitro assays that nucleic acids coated with NC are protected against nucleases (Krishnamoorthy et al., 2003; Lapadat-Tapolsky et al., 1993; Tanchou et al., 1995). Infection experiments have supported the idea that NC stabilizes nascent vDNA in the cytoplasm (Buckman et al., 2003; Gorelick et al., 1999b; Tanchou et al., 1998; Thomas et al., 2006a). This evidence comes from several observations. It has been observed that these NC mutants often permit reverse transcription to proceed close to wild-type efficiency, but that the ends are unstable. This may mean that the ends were not completed during strand-displacement synthesis (Fig. 4G), or that they were degraded by exonucleases (Buckman et al., 2003; Gorelick et al., 1999b; Tanchou et al., 1998). A kinetic analysis of vDNA synthesis and loss after infection demonstrated that vDNA produced after infection with virus containing either NCH23C or NCH44C mutations was far less stable than wild-type. This is partially due to the integration defect that will be discussed below; the most stable vDNA form is that which is integrated and is replicated as the cell divides.
Circular forms of vDNA (1- and 2-LTR circles) are reasonably stable but are diluted upon successive rounds of cell division because they are not duplicated, and linear forms can be rapidly degraded if not protected effectively. The instability of vDNA was highlighted by two observations: i) there was no apparent increase in accumulation of reverse transcripts at early time points after infection as was observed in wild-type HIV-1, and ii) the reverse transcripts that were synthesized decreased at a faster rate than in wild-type infections (Thomas et al., 2006a). The levels of p7NC associated with the RTC/PIC most likely decrease with time during the conversion of the RNA genome to the double-stranded vDNA, as has been demonstrated in vitro (Mirambeau et al., 2006, 2007). The fact that the mutant NC proteins do not bind to nucleic acids as well as wildtype NC (Cruceanu et al., 2006; Urbaneja et al., 1999) may also lead to a more rapid reduction in vDNA and loss of NC from complexes. Although overall NC levels may decrease, the vDNA may contain specific areas where NC binding is still required. Thus, the decreased binding affinity would result in less NC bound to these specific regions of vDNA as well.
4.4. Changes in NC concentrations as RTCs mature to PICs
The presence of NC in PICs is expected to some extent because of its presence in RTCs and experiments have shown nuclear localization of NC after infection (Gallay et al., 1995b; Risco et al., 1995; Zhang and Crumpacker, 2002). However, there has been a conspicuous inability to detect it in isolated HIV-1 PICs (Nermut and Fassati, 2003; Thomas and Gorelick, unpublished observations). The failure to detect NC may be due either to weak NC-nucleic acid interactions in PICs, or to levels of NC being lower than the limit of detection by current methods such as Western blotting.
Consequently, two models have been suggested to explain how p7NC may affect subsequent steps. The first model suggests that p7NC binds dsDNA non-specifically, forces IN to bind to areas of higher affinity, and/or stabilizes IN binding to LTR ends (Carteau et al., 1997; Poljak et al., 2003; Tanchou et al., 1998). Once IN is bound at the correct position, it may not be necessary for NC to remain present. The second model proposes that NC is bound at low concentrations to specific parts of the full-length vDNA. For example, it has been shown in vitro that HIV-1 p7NC binds ssDNA better than dsDNA, and because NC greatly stimulates strand-displacement synthesis, it would be in the proper location to bind the ssDNA of the central DNA flap. Indeed, TEM visualization of dsDNA containing a central flap shows p7NC bound to this region (Mirambeau et al., 2007). In addition, it is important to keep in mind that PR digestion of p15NC is unlikely to be 100%, so that a small amount of p9NC is expected, and p9NC binds equally well to ssDNA and dsDNA (Mirambeau et al., 2006). Interestingly, it has been demonstrated that p9NC binding has a cooperative component (Cruceanu et al., 2006; Khan and Giedroc, 1994), suggesting that perhaps a small amount of p9NC could assist IN in forming complexes at the LTR ends. To this end, it was observed that p9NC stimulated coordinated integration to a greater degree than p7NC in in vitro integration assays (Gao et al., 2003).
4.5. Central flap and nuclear entry
A distinguishing characteristic of lentiviruses is that they can infect nondividing cells, unlike most other orthoretroviruses, such as MLV (Roe et al., 1993). The large PIC nucleoprotein complex is actively transported into the nucleus of non-dividing cells. There is a great deal of interest in determining the mechanism of nuclear entry, both from the standpoint of basic retrovirology and for the design of retroviral vectors for gene therapy. However, a certain degree of controversy exists within the field (Yamashita and Emerman, 2005). Karyophilic properties have been identified in MA (Bukrinsky et al., 1993; von Schwedler et al., 1994), Vpr (Fouchier et al., 1998; Zhao et al., 1994), IN (Ao et al., 2007; Gallay et al., 1997), and the central flap region (De Rijck and Debyser, 2006; Van Maele et al., 2003; Zennou et al., 2000, 2001). Interestingly, it has also been observed that many, and sometimes all, of these signals can be blocked or eliminated, and this still does not completely eliminate nuclear entry and productive infection (Dvorin et al., 2002; Fouchier et al., 1997; Freed et al., 1997; Gallay et al., 1997; Limón et al., 2002a, 2002b; Marsden and Zack, 2007; Petit et al., 2000; Reil et al., 1998; Yamashita and Emerman, 2005, 2006). One possible explanation for the differences in results is that different studies have used widely differing virus titers to infect cells, implying that a large number of infectious events can overcome the engineered blocks. Another possibility is that there may be other nuclear localization signals not yet identified. Finally, the various karyophilic signals could be additive or synergistic, which may provide a certain level of redundancy to ensure survival of the virus.
Interestingly, in vitro experiments have identified the central flap as a site of strong p7NC-nucleic acid binding (Mirambeau et al., 2007), specifically on the G-quartet regions that can form from sequences on the flap (Kankia et al., 2005; Lyonnais et al., 2002, 2003). This indicates a site where it is possible that low levels of p7NC are tightly bound, and this could be important for nuclear entry. In vitro studies by Lyonnais et al. (2003) examined mutant and wild type recombinant proteins in binding studies with nucleic acids that were modeled after the flap region. In this study it was found that the NCH23C mutant protein (Table 3) formed a limited subset of complexes compared to wild-type NC, indicating a different, possibly defective mode of binding to model flap oligonucleotides. At this time, a role for NC in nuclear entry is only speculative, as nuclear entry has not been specifically examined by any published studies using NC mutants. For instance, although the NCH23C and NCH44C mutants show significant defects in integration, the relative quantities of 2-LTR circles (typically used as surrogates indicating nuclear entry) were comparable to wild-type amounts (Thomas et al., 2006a). However, since these experiments were performed in actively dividing cells, one cannot make any conclusion regarding active nuclear transport. NC binding to the central flap does, however, provide a mechanism for the presence of NC in the nucleus after infection (Gallay et al., 1995b, 1997; Zhang and Crumpacker, 2002).
As mentioned above, a commonly used marker to determine whether vDNA has been exposed to nuclear enzymes as indicative of nuclear entry, is the presence of 1-LTR or 2-LTR circles (Dismuke and Aiken, 2006; Neil et al., 2001; Tanchou et al., 1998). The formation of LTR circles is catalyzed by nuclear enzyme-mediated homologous recombination (1-LTR) or non-homologous end joining (2-LTR) (Jeanson and Mouscadet, 2002; Jeanson et al., 2002; Kilzer et al., 2003; Li et al., 2001). These circular vDNA species are dead-end products and cannot be used as substrates for integration (Brown, 1997; Ellis and Bernstein, 1989; Lobel et al., 1989). However, analyses of their formation and the properties of the 2-LTR junctions are useful in elucidating the progression of infection and for examining the state of the ends of the linear vDNA just before ligation. For example, in wild-type infections the vDNA that is converted into 2-LTR circles typically indicates a defect resulting in its inability to be integrated; this could be a defect at either the DNA level or at the level of the PIC.
5. Integration processes
Integration of full-length vDNA (Fig. 4H and Fig. 5A) into the chromosomal DNA of the infected cell forming the provirus (Fig. 5D) is the final step of early infection (Fig. 2). The events that must occur are understood, but the exact molecular mechanisms of the process are still being elucidated (Fig. 5) (Brown, 1997). In vitro, only three components are strictly required for a strand transfer result; IN, viral LTR ends containing the requisite attachment (att) sites, and a DNA substrate in which to integrate the vDNA (Bushman and Craigie, 1991; Goodarzi et al., 1999; Katz et al., 1990; Masuda et al., 1995; Sinha and Grandgenett, 2005; Vink et al., 1991). The actual reaction involves three specific events, only two of which are performed by IN: the 3’ processing of the LTR ends that entails the removal of a dinucleotide to expose a CA 5’ overhang (Fig. 5B), and the strand-transfer reaction, where the 3’ recessed ends are joined to 5’-PO4 ends created by sequence-nonspecific staggered nicks in the chromosomal DNA (Fig. 5C). The last step in the process is a gap repair event, where the 5’ two nucleotides of the LTR end, which are exposed, are removed and the single-stranded region is repaired, resulting in a direct repeat at each LTR-chromosomal DNA junction. This process is thought to be mediated by cellular repair enzymes (Brown, 1997; Yoder and Bushman, 2000). Although recombinant IN can perform 3’ processing and strand-transfer in vitro, it is part of a large nucleoprotein complex in vivo (Chen and Engelman, 1998; Vora and Grandgenett, 2001; Wei et al., 1997). The identification of viral and cellular cofactors, as well as a more complete understanding of the molecular events of integration is the current direction of much research in this area.
Fig. 5. Integration.
Integration of vDNA into the host chromosome is the final step of early infection (Brown, 1997). The substrate for this reaction is full-length double-strand vDNA (A). Each LTR contains an att site, necessary for the first enzymatic step performed by IN. In this step, 3′ processing of the vDNA ends occur, which generates a free 3′-CA end (B). The host chromosome is cut to generate staggered nicks, 4 nucleotides apart in the case of HIV-1. The 3′ hydroxyl of the processed vDNA joins to the 5′ PO4 in the second enzymatic step of integration, strand-transfer (C). This step results in a 2-nucleotide flap with the 5′ vDNA overhang. These 2 nucleotides are then removed, and the 6 nucleotide single-stranded region is repaired by nuclear enzymes to create direct repeats at each end (Brown, 1997; Yoder and Bushman, 2000). The resulting integrated vDNA is called the provirus (D).
It is currently understood that IN forms a homodimer at the end of each LTR, with IN forming a tetrameric complex (dimer of dimers) (Faure et al., 2005), resulting in the two LTR ends being in close spatial proximity (Esposito and Craigie, 1999; Gao et al., 2001; Jones et al., 1992; Li et al., 2006). An interaction between the IN-LTR end complexes was inferred from observations that one of the key differences between reconstituted integration reactions with recombinant IN and integration reactions performed using purified PICs is the frequency of concerted, or coordinated integration. Concerted integration is a process where both ends integrate at the same time, at the sites of the staggered nicks. Based on the observation that the LTR ends of these PICs are resistant to nuclease digestion, it was concluded that a large nucleoprotein complex, termed the intasome, was present on these ends (Chen and Engelman, 1998; Vora and Grandgenett, 2001; Wei et al., 1997).
The components of these complexes are presumed to contain proteins that stabilize this complex, proteins that aid in localization of the PICs to the requisite chromosomal regions, and perhaps the cellular DNA repair enzymes. Based upon large-scale studies of integration sites into chromosomal DNA by different retroviruses, it has been determined that MLV tends to integrate into promoter regions (Mitchell et al., 2004; Wu et al., 2003), HIV-1 and SIV tend to integrate into actively transcribed genes (Barr et al., 2006; Bushman, 2002; Crise et al., 2005; Mitchell et al., 2004; Schröder et al., 2002), and avian retroviral integration appears to be truly random (Mitchell et al., 2004; Narezkina et al., 2004). Thus, the intasomes of MLV, HIV-1, and SIV are presumed to contain cellular proteins that would help direct each of these PICs to their characteristic sites of integration. Some of the cellular proteins proposed to be involved are LEDGF/p75, HMG I(Y), BAF, and Ini1 (Bushman, 1999; Chen and Engelman, 1998; Farnet and Bushman, 1997; Hindmarsh and Leis, 1999; Hombrouck et al., 2007; Turelli et al., 2001).
Curiously, p7NC is not normally observed in isolated PICs of HIV-1 or MLV when probed by Western blotting (Bowerman et al., 1989; Farnet and Bushman, 1997; Farnet and Haseltine, 1991; Fassati and Goff, 2001; Miller et al., 1997). In agreement with the failure to detect NC by blotting, it was observed that immunodepletion of NC did not diminish the integration potential of isolated PICs (Farnet and Bushman, 1997). Although isolated PICs are deemed to be authentic if they maintain the ability to perform concerted integration, the extent to which they represent the situation within cells cannot be easily confirmed. Critically, it has been reported that coupled integration can be performed in cell-free systems using only purified DNA and recombinant IN (Goodarzi et al., 1999; Sinha and Grandgenett, 2005), which certainly indicates that the failure to detect NC in functional PICs does not justify the assertion that NC is not present. The failure to detect NC could be due to several reasons, none of which is mutually exclusive. The binding of NC to these complexes may be weak enough to be lost with the salt concentrations used for PIC isolation or the amount of NC bound may be below the limit of detection by Western blotting. Conversly, it is possible that NC’s role may solely be at the level of the RTC and not at all in the PIC.
5.1. In vitro integration with NC
As with reverse transcription, a number of groups have developed cell-free reaction systems to study each of the specific steps of integration. Once these systems were developed, different presumed cofactors were added to these integration assays to determine if they enhanced the actual reaction steps of integration. Of course, just because a protein fails to enhance integration in vitro does not prove that it is not involved in the cell, since many aspects to integration may be unrelated to the actual enzymatic reaction steps. However, it has been observed that HIV-1 p7NC is very effective at enhancing in vitro integration reactions. The stimulation of 3’ processing (Fig. 5B) is small, but that of strand-transfer (Fig. 5C) is significantly greater. In addition, concerted integration is greatly stimulated by wild-type and several mutant forms of NC (Carteau et al., 1997, 1999; Gao et al., 2003; Poljak et al., 2003). Some of these assays have indicated that NC’s effect on integration does not seem to be merely the result of a competition with IN for DNA low affinity binding sites, which would thereby increase the specific activity of IN (Carteau et al., 1997).
Additional verification for an NC-mediated enhancement of integration was the observation that NCH23C and NCH44C mutants were not as effective at enhancing integration as wild-type p7NC (Carteau et al., 1999). Another laboratory reported that the binding of p7NC and IN to DNA showed cooperativity, and in a competition experiment, it was observed that the NC-IN-DNA complex was stable, since the addition of a large excess of free DNA did not decrease the number of complexes formed (Poljak et al., 2003). The conclusion from this study is that NC coats the DNA, thus masking nonspecific IN-DNA binding sites. Importantly, it has also been observed that p9NC is more effective at stimulating coordinated integration than p7NC (Gao et al., 2003). The fact that p9NC also exhibits cooperative binding at low concentrations (Cruceanu et al., 2006; Khan and Giedroc, 1994), presumably due to the presence of the SP2 (Fig. 3), suggests that this domain may also interact with IN to stabilize the complexes. This issue is discussed in more detail below.
5.2. Cell culture-based observations of NC and integration
Observations from infection experiments performed with certain NC mutant viruses have provided indirect evidence for the involvement of NC in integration. One mutant in particular, NCH23C, has been key to this proposal. The fact that this mutant virus is profoundly replication defective (≥ 6 log reduction in H9-replicative titer) (Gorelick et al., 1999c), but exhibits only minor defects in reverse transcription (Thomas et al., 2006a), and exhibits little reduction in the conversion of full-length vDNA to 2-LTR circles compared to wild-type (Buckman et al., 2003; Thomas et al., 2006a), implies that the integration step itself is defective. A similar observation was made with the NCH44C mutant, although it was not as replication defective in H9 cells as the NCH23C mutant. This idea was strongly supported by the fact that until recently, the phenotypes of NCH23C or NCH44C could not be readily distinguished from IND116N (Gorelick et al., 1999c), which is an active site mutant of IN that cannot catalyze the 3’ processing or strand-transfer reactions (Engelman et al., 1995).
Of key interest was the observation that the number of unprocessed LTR ends (based on cloning and sequencing of 2-LTR circle junctions) did not change in infections when the NCH23C mutant was coupled to the IND116N mutant, in contrast to what was observed with virus containing wild-type NC. However, the increase in the proportion of unprocessed ends was only 2- to 3-fold more than wild-type, which was small and does not account for the magnitude of the observed replication defects in the mutants (Buckman et al., 2003). It should be noted that the combination NC and IN mutant viruses showed basically the same phenotypes as that of the individual NC mutants: they are all replication defective, show similar quantities of 2-LTR circles, and produce reverse transcripts that do not persist nearly as long as seen in wild-type infections (Thomas et al., 2006a). In contrast, there was a tremendous increase in 2-LTR circles compared to wild-type infections with the IND116N mutant (Thomas et al., 2006a), and the mutant junctions were comprised of a much higher percentage of full-length consensus 2-LTR junction sequences compared to wild type (Buckman et al., 2003).
The most direct way to measure integration at early time points after infection is to use Alu-LTR PCR (Butler et al., 2001). Using this technique, we observed that the NCH23C and NCH44C mutants showed profound defects in integration, >2 logs (Thomas et al., 2006a). In fact, integration was barely detectable in these infections, even with VSV-G pseudotyped virus, which significantly boosts viral titer (Thomas et al., 2007b). Because the conversion of vDNA to 2-LTR circles in infections with these mutants was similar to that observed with wild-type NC (indicating that there is no problem with gaining access to the nucleus), these data strongly support the idea that somehow these mutant NC species cause a specific block to integration.
In contrast to this, we have also observed another phenotype in HIV-1 NC-mutant virus infections, as reported for the NCN17F and NCN17G viruses (Table 3). These mutants were also very replication defective (>5- and >3-log reductions in titer, respectively), but the observed decreases in reverse transcription and integration were very minor. It was determined that there were downstream assembly defects, specific to certain cell types, such as H9, rather than defects in early infection (Thomas et al., 2006b).
Finally, we have observed that when the formation of p9NC was prevented, either by removing the PR-cleavage site between SP2-p6 (Fig. 3), or by preventing the processing of p15NC, a severe reduction in integration efficiency resulted, which was correlated with the reduction in replicative titer (Coren et al., 2007). In contrast, mutations that prevented the processing of p9NC to form p7NC and SP2 showed little or no effect on integration or replicative titer. These observations suggest that p9NC may have a genuine role in viral replication beyond a mere cleavage intermediate of NC (Coren et al., 2007). As mentioned above, p9NC protein was shown to facilitate in vitro integration reactions better than p7NC (Gao et al., 2003). It is likely that there will be some p9NC present, based on the improbability that Gag cleavage by PR proceeds to completion: partially processed Gag has been observed in Western blots of virus particles (Gorelick et al., 1999c; Tanchou et al., 1998). This, coupled with the demonstration that DNA binding by p9NC is cooperative (Cruceanu et al., 2006), and that p9NC binds double-strand DNA as well as single-strand DNA, certainly support this idea (Mirambeau et al., 2007).
5.3. Inferences
Compelling evidence of a role for NC in integration exists, but the mechanism remains elusive, based upon the experimental evidence accumulated thus far. It is certainly known that NC functions during reverse transcription and that if the vDNA is not correctly or completely synthesized, it cannot function as a proper substrate for IN. If NC is directly involved in integration, a number of different possibilities could explain how it can influence this event. First, NC may be directly involved in enhancing the enzymatic steps of integration, as it has been observed to do in cell-free systems. Presumably, this would be due to the nucleic acid chaperone properties of NC so that the strand-transfer reactions occur more efficiently. It may be that at very low concentrations of NC, any small change in its ability to bind nucleic acids and destabilize secondary structures is enough to block this activity. Second, NC may be involved more in the formation of a functional intasome, possibly by assisting IN to bind the LTR ends or by stabilizing the IN nucleoprotein complex at the LTR ends. This possibility is certainly supported by the defect observed in 3' processing after infections with NCH23C or NCH44C. Third, NC may be important for proper PIC structure, in that if the central flap has a role in functional PIC formation (Arhel et al., 2007), perhaps it is important for NC to be bound to the ssDNA for proper PIC structure. It is also important to keep in mind that NC may be functioning in integration as p9NC rather than p7NC, so that the cooperative binding resulting from the presence of the SP2 region may be critical for the required interactions with nucleic acids or proteins.
6. Conclusions
The involvement of NC during early infection is mainly reflected by its activity as a nucleic acid chaperone: NC facilitates the enzymatic steps so that the total efficiency of the early infection process is high enough to permit successful replication. Indeed, investigation of early infection events demonstrates that they are relatively efficient in cells, at least for the reverse transcription step (Thomas et al., 2006a, 2007b). So although it may be asserted that NC is not strictly required for reverse transcription and integration, this small protein does appear to make these processes work well enough for effective replication. Excitingly, the state of knowledge and technology present in the field is such that even the existence of minor nuances can be detected, resulting in a more complete understanding of these events. One of the consequences of NC being important for these processes is that compounds that inactivate NC cause severe impairment to early infection events (Berthoux et al., 1999; De Clercq, 2002; Druillennec et al., 1999; Goel et al., 2002; Morcock et al., 2005; Srivastava et al., 2004; Turpin et al., 1999). Thus, anti-NC chemotherapeutic substances are being actively investigated, since they can provide multiple additional steps in viral replication that can be targeted in the treatment of retroviral infections (Darlix et al., 2007; de Rocquigny et al., 2008; Miller Jenkins et al., 2007; Raja et al., 2006).
Acknowledgments
We thank David E. Ott (AIDS Vaccine Program, SAIC-Frederick, Inc., NCI-Frederick) and Eric O. Freed (HIV-Drug Resistance Program, NCI-Frederick) for their helpful suggestions. We thank Louis E. Henderson (also of the AIDS Vaccine Program) for the artwork presented in Fig. 2. This publication has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Abbreviations
- att
attachment
- CA
capsid
- cDNA
complementary DNA
- dNTP
deoxyribonucleotide triphosphates
- dsDNA
double-stranded DNA
- FIV
feline immunodeficiency virus
- gRNA
genomic RNA
- HIV
human immunodeficiency virus
- HTLV
human T-cell leukemia virus
- IN
integrase
- LTR
long terminal repeat
- MA
matrix
- MLV
murine leukemia virus
- NC
nucleocapsid
- PAS
primer activation signal
- PBS
primer binding site
- PFV
prototype foamy virus
- PIC
preintegration complex
- PPT
polypurine tract
- PR
protease
- R
repeat sequence in LTR
- RSV
Rous sarcoma virus
- RT
reverse transcriptase
- RTC
reverse transcription complex
- SIV
simian immunodeficiency virus
- SP1 and SP2
Gag spacer peptides p2 and p1, respectively
- SSB
single-stranded binding protein
- ssDNA
single-stranded DNA
- vDNA
viral DNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbink TE, Berkhout B. HIV-1 reverse transcription: close encounters between the viral genome and a cellular tRNA. Adv Pharmacol. 2007;55:99–135. doi: 10.1016/S1054-3589(07)55003-9. [DOI] [PubMed] [Google Scholar]
- Accola MA, Strack B, Göttlinger HG. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 2000;74:5395–5402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akahata W, Ido E, Hayami M. Mutational analysis of two zinc-finger motifs in the nucleocapsid protein of simian immunodeficiency virus mac239. J. Gen. Virol. 2003;84:1641–1648. doi: 10.1099/vir.0.18865-0. [DOI] [PubMed] [Google Scholar]
- Aldovini A, Young RA. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain B, Lapadat-Tapolsky M, Berlioz C, Darlix JL. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- Anderson EC, Lever AM. Human immunodeficiency virus type 1 Gag polyprotein modulates its own translation. J. Virol. 2006;80:10478–10486. doi: 10.1128/JVI.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao Z, Huang G, Yao H, Xu Z, Labine M, Cochrane AW, Yao X. Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J. Biol. Chem. 2007;282:13456–13467. doi: 10.1074/jbc.M610546200. [DOI] [PubMed] [Google Scholar]
- Arhel NJ, Souquere-Besse S, Munier S, Souque P, Guadagnini S, Rutherford S, Prévost MC, Allen TD, Charneau P. HIV-1 DNA flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J. 2007;26:3025–3037. doi: 10.1038/sj.emboj.7601740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bampi C, Bibillo A, Wendler M, Divita G, Gorelick RJ, Le Grice SFJ, Darlix J-L. Nucleotide excision-repair and template-independent addition by HIV-1 reverse transcriptase in the presence of nucleocapsid protein. J. Biol. Chem. 2006;281:11736–11743. doi: 10.1074/jbc.M600290200. [DOI] [PubMed] [Google Scholar]
- Bampi C, Jacquenet S, Lener D, Décimo D, Darlix JL. The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance. Curr. HIV Res. 2004a;2:79–92. doi: 10.2174/1570162043485022. [DOI] [PubMed] [Google Scholar]
- Bampi C, Jacquenet S, Lener D, Décimo D, Darlix JL. The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance. Int. J. Biochem. Cell Biol. 2004b;36:1668–1686. doi: 10.1016/j.biocel.2004.02.024. [DOI] [PubMed] [Google Scholar]
- Barat C, Lullien V, Schatz O, Keith G, Nugeyre MT, Grüninger-Leitch F, Barré-Sinoussi F, LeGrice SF, Darlix JL. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989;8:3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barat C, Schatz O, Le Grice S, Darlix JL. Analysis of the interactions of HIV1 replication primer tRNA(Lys,3) with nucleocapsid protein and reverse transcriptase. J. Mol. Biol. 1993;231:185–190. doi: 10.1006/jmbi.1993.1273. [DOI] [PubMed] [Google Scholar]
- Barr SD, Ciuffi A, Leipzig J, Shinn P, Ecker JR, Bushman FD. HIV integration site selection: Targeting in macrophages and the effects of different routes of viral entry. Mol. Ther. 2006;14:218–225. doi: 10.1016/j.ymthe.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA. Strand transfer events during HIV-1 reverse transcription. Virus Res. 2008 doi: 10.1016/j.virusres.2007.12.017. in press. [DOI] [PubMed] [Google Scholar]
- Beerens N, Berkhout B. In vitro studies on tRNA annealing and reverse transcription with mutant HIV-1 RNA templates. J. Biol. Chem. 2000;275:15474–15481. doi: 10.1074/jbc.275.20.15474. [DOI] [PubMed] [Google Scholar]
- Beerens N, Berkhout B. The tRNA primer activation signal in the human immunodeficiency virus type 1 genome is important for initiation and processive elongation of reverse transcription. J. Virol. 2002;76:2329–2339. doi: 10.1128/jvi.76.5.2329-2339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens N, Groot F, Berkhout B. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J. Biol. Chem. 2001;276:31247–31256. doi: 10.1074/jbc.M102441200. [DOI] [PubMed] [Google Scholar]
- Berg JM. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986;232:485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr. Top. Microbiol. Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- Berthoux L, Péchoux C, Darlix JL. Multiple effects of an anti-human immunodeficiency virus nucleocapsid inhibitor on virus morphology and replication. J. Virol. 1999;73:10000–10009. doi: 10.1128/jvi.73.12.10000-10009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthoux L, Péchoux C, Ottmann M, Morel G, Darlix JL. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J. Virol. 1997;71:6973–6981. doi: 10.1128/jvi.71.9.6973-6981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieth E, Gabus C, Darlix JL. A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res. 1990;18:119–127. doi: 10.1093/nar/18.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Eichinger D, Castrillon D, Fink GR. The Saccharomyces cerevisiae genome contains functional and nonfunctional copies of transposon Ty1. Mol. Cell. Biol. 1988;8:1432–1442. doi: 10.1128/mcb.8.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JE, Gould SJ. Exosomes and HIV Gag bud from endosome-like domains of the T-cell plasma membrane. J. Cell Biol. 2006;172:923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B, Brown PO, Bishop JM, Varmus HE. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- Bowzard JB, Bennett RP, Krishna NK, Ernst SM, Rein A, Wills JW. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 1998;72:9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PO. Integration. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Plainview, N. Y.: Cold Spring Harbor Press; 1997. pp. 161–203. [Google Scholar]
- Buckman JS, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid Zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 2003;77:1469–1480. doi: 10.1128/JVI.77.2.1469-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinskaya AG, Ghorpade A, Heinzinger NK, Smithgall TE, Lewis RE, Stevenson M. Phosphorylation-dependent human immunodeficiency virus type 1 infection and nuclear targeting of viral DNA. Proc. Natl. Acad. Sci. U. S. A. 1996;93:367–371. doi: 10.1073/pnas.93.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky MI, Haggerty S, Dempsey MP, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD. Host proteins in retroviral cDNA integration. Adv. Virus Res. 1999;52:301–317. doi: 10.1016/s0065-3527(08)60303-6. [DOI] [PubMed] [Google Scholar]
- Bushman FD. Integration site selection by lentiviruses: biology and possible control. Curr. Top. Microbiol. Immunol. 2002;261:165–177. doi: 10.1007/978-3-642-56114-6_8. [DOI] [PubMed] [Google Scholar]
- Bushman FD, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc. Natl. Acad. Sci. U. S. A. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- Butsch M, Boris-Lawrie K. Destiny of unspliced retroviral RNA: ribosome and/or virion? J. Virol. 2002;76:3089–3094. doi: 10.1128/JVI.76.7.3089-3094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 gag protein lacking the p6 domain. J. Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Vogt VM. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Vogt VM. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J. Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carteau S, Batson SC, Poljak L, Mouscadet JF, de Rocquigny H, Darlix JL, Roques BP, Käs E, Auclair C. Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J. Virol. 1997;71:6225–6229. doi: 10.1128/jvi.71.8.6225-6229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carteau S, Gorelick RJ, Bushman FD. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen S, Huang Y, Khorchid A, Darlix JL, Wainberg MA, Kleiman L. The role of Pr55(gag) in the annealing of tRNA3Lys to human immunodeficiency virus type 1 genomic RNA. J. Virol. 1999;73:4485–4488. doi: 10.1128/jvi.73.5.4485-4488.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen S, Javanbakht H, Kim S, Shiba K, Craven R, Rein A, Ewalt K, Schimmel P, Musier-Forsyth K, Kleiman L. Retrovirus-specific packaging of aminoacyl-tRNA synthetases with cognate primer tRNAs. J. Virol. 2002;76:13111–13115. doi: 10.1128/JVI.76.24.13111-13115.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charneau P, Alizon M, Clavel F. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol. 1992;66:2814–2820. doi: 10.1128/jvi.66.5.2814-2820.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Engelman A. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc. Natl. Acad. Sci. U. S. A. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimarelli A, Luban J. Context-dependent phenotype of a human immunodeficiency virus type 1 nucleocapsid mutation. J. Virol. 2001;75(15):7193–7197. doi: 10.1128/JVI.75.15.7193-7197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimarelli A, Sandin S, Höglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 2000a;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimarelli A, Sandin S, Höglund S, Luban J. Rescue of multiple viral functions by a second-site suppressor of a human immunodeficiency virus type 1 nucleocapsid mutation. J. Virol. 2000b;74:4273–4283. doi: 10.1128/jvi.74.9.4273-4283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coren LV, Thomas JA, Chertova E, Sowder RC, II, Gagliardi TD, Gorelick RJ, Ott DE. Mutational Analysis of the C-Terminal Gag Cleavage Sites in Human Immunodeficiency Virus Type 1. J. Virol. 2007;81:10047–10054. doi: 10.1128/JVI.02496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey SN. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 1986;14:623–633. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crise B, Li Y, Yuan C, Morcock DR, Whitby D, Munroe DJ, Arthur LO, Wu X. Simian immunodeficiency virus integration preference is similar to that of human immunodeficiency virus type 1. J. Virol. 2005;79:12199–12204. doi: 10.1128/JVI.79.19.12199-12204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Bampi C, Wilhelm M, Wilhelm FX, Darlix JL. A 5’-3’ longrange interaction in Ty1 RNA controls its reverse transcription and retrotransposition. EMBO J. 2002;21:4368–4379. doi: 10.1093/emboj/cdf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Ficheux D, Darlix JL. The Gag-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J. Biol. Chem. 2000;275:19210–19217. doi: 10.1074/jbc.M001371200. [DOI] [PubMed] [Google Scholar]
- Cristofari G, Gabus C, Ficheux D, Bona M, Le Grice SF, Darlix JL. Characterization of active reverse transcriptase and nucleoprotein complexes of the yeast retrotransposon Ty3 in vitro. J. Biol. Chem. 1999;274:36643–36648. doi: 10.1074/jbc.274.51.36643. [DOI] [PubMed] [Google Scholar]
- Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, Casas-Finet JR, Rein A, Rouzina I, Williams MC. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza V, Summers MF. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- Darlix JL, Cristofari G, Rau M, Péchoux C, Berthoux L, Roques B. Nucleocapsid protein of human immunodeficiency virus as a model protein with chaperoning functions and as a target for antiviral drugs. Adv. Pharmacol. 2000;48:345–372. doi: 10.1016/s1054-3589(00)48011-7. [DOI] [PubMed] [Google Scholar]
- Darlix JL, Garrido JL, Morellet N, Mély Y, de Rocquigny H. Properties, functions, and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv. Pharmacol. 2007;55:299–346. doi: 10.1016/S1054-3589(07)55009-X. [DOI] [PubMed] [Google Scholar]
- Darlix JL, Lapadat-Tapolsky M, de Rocquigny H, Roques B. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- Darlix JL, Lastra ML, Mély Y, Roques B. Nucleocapsid protein chaperoning of nucleic acids at the heart of HIV structure, assembly and cDNA synthesis. In: Kuiken C, Foley B, Freed E, Hahn B, Korber B, Marx PA, McCutchan F, Mellors JW, Wolinksy S, editors. HIV Sequence Compendium 2002. Los Alamos, NM: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 2002. pp. 71–82. LA-UR 03-3564. [Google Scholar]
- Dawson L, Yu XF. The role of nucleocapsid of HIV-1 in virus assembly. Virology. 1998;251:141–157. doi: 10.1006/viro.1998.9374. [DOI] [PubMed] [Google Scholar]
- De Clercq E. New developments in anti-HIV chemotherapy. Biochim. Biophys. Acta. 2002;1587:258–275. doi: 10.1016/s0925-4439(02)00089-3. [DOI] [PubMed] [Google Scholar]
- De Guzman RN, Turner RB, Summers MF. Protein-RNA recognition. Biopolymers. 1998a;48:181–195. doi: 10.1002/(SICI)1097-0282(1998)48:2<181::AID-BIP7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998b;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- Deneka M, Pelchen-Matthews A, Byland R, Ruiz-Mateos E, Marsh M. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J. Cell Biol. 2007;177:329–341. doi: 10.1083/jcb.200609050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijck J, Debyser Z. The central DNA flap of the human immunodeficiency virus type 1 is important for viral replication. Biochem. Biophys. Res. Commun. 2006;349:1100–1110. doi: 10.1016/j.bbrc.2006.08.141. [DOI] [PubMed] [Google Scholar]
- de Rocquigny H, Ficheux D, Gabus C, Fournié-Zaluski MC, Darlix JL, Roques BP. First large scale chemical synthesis of the 72 amino acid HIV-1 nucleocapsid protein NCp7 in an active form. Biochem. Biophys. Res. Commun. 1991;31:1010–1018. doi: 10.1016/s0006-291x(05)81166-0. [DOI] [PubMed] [Google Scholar]
- de Rocquigny H, Gabus C, Vincent A, Fournié-Zaluski MC, Roques B, Darlix JL. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl. Acad. Sci. U. S. A. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rocquigny H, Shvadchak V, Avilov S, Dong CZ, Dietrich U, Darlix JL, Mély Y. Targeting the viral nucleocapsid protein in anti-HIV-1 therapy. MiniRev. Med. Chem. 2008 doi: 10.2174/138955708783331603. In Press. [DOI] [PubMed] [Google Scholar]
- Derebail SS, Heath MJ, DeStefano JJ. Evidence for the differential effects of nucleocapsid protein on strand transfer in various regions of the HIV genome. J. Biol. Chem. 2003;278:15702–15712. doi: 10.1074/jbc.M211701200. [DOI] [PubMed] [Google Scholar]
- Derrington E, Gabus C, Leblanc P, Chnaidermann J, Grave L, Dormont D, Swietnicki W, Morillas M, Marck D, Nandi P, Darlix JL. PrPC has nucleic acid chaperoning properties similar to the nucleocapsid protein of HIV-1. C. R. Biol. 2002;325:17–23. doi: 10.1016/s1631-0691(02)01388-4. [DOI] [PubMed] [Google Scholar]
- Derse D, Hill SA, Princler G, Lloyd P, Heidecker G. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in NC. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2915–2920. doi: 10.1073/pnas.0609444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dismuke DJ, Aiken C. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J. Virol. 2006;80:3712–3720. doi: 10.1128/JVI.80.8.3712-3720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobard CW, Briones MS, Chow SA. Molecular mechanisms by which human immunodeficiency virus type 1 integrase stimulates the early steps of reverse transcription. J. Virol. 2007;81:10037–10046. doi: 10.1128/JVI.00519-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman T, Luban J, Goff SP, Haseltine WA, Göttlinger HG. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman N, Lever A. Comparison of viral genomic RNA sorting mechanisms in human immunodeficiency virus type 1 (HIV-1), HIV-2, and Moloney murine leukemia virus. J. Virol. 2000;74:11413–11417. doi: 10.1128/jvi.74.23.11413-11417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll MD, Hughes SH. Human immunodeficiency virus type 1 nucleocapsid protein can prevent self-priming of minus-strand strong stop DNA by promoting the annealing of short oligonucleotides to hairpin sequences. J. Virol. 2000;74:8785–8792. doi: 10.1128/jvi.74.19.8785-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druillennec S, Dong CZ, Escaich S, Gresh N, Bousseau A, Roques BP, Fournié-Zaluski MC. A mimic of HIV-1 nucleocapsid protein impairs reverse transcription and displays antiviral activity. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4886–4891. doi: 10.1073/pnas.96.9.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond JE, Mounts P, Gorelick RJ, Casas-Finet JR, Bosche WJ, Henderson LE, Waters DJ, Arthur LO. Wild-type and mutant HIV type 1 nucleocapsid proteins increase the proportion of long cDNA transcripts by viral reverse transcriptase. AIDS Res. Hum. Retroviruses. 1997;13:533–543. doi: 10.1089/aid.1997.13.533. [DOI] [PubMed] [Google Scholar]
- Dvorin JD, Bell P, Maul GG, Yamashita M, Emerman M, Malim MH. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 2002;76:12087–12096. doi: 10.1128/JVI.76.23.12087-12096.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorin JD, Malim MH. Intracellular trafficking of HIV-1 cores: journey to the center of the cell. Curr. Top. Microbiol. Immunol. 2003;281:179–208. doi: 10.1007/978-3-642-19012-4_5. [DOI] [PubMed] [Google Scholar]
- Ellis J, Bernstein A. Retrovirus vectors containing an internal attachment site: evidence that circles are not intermediates to murine retrovirus integration. J. Virol. 1989;63:2844–2846. doi: 10.1128/jvi.63.6.2844-2846.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito D, Craigie R. HIV integrase structure and function. Adv. Virus Res. 1999;52:319–333. doi: 10.1016/s0065-3527(08)60304-8. [DOI] [PubMed] [Google Scholar]
- Farnet CM, Bushman FD. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- Farnet CM, Haseltine WA. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassati A, Goff SP. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 2001;75:3626–3635. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 2005;33:977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YX, Campbell S, Harvin D, Ehresmann B, Ehresmann C, Rein A. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 1999;73:4251–4256. doi: 10.1128/jvi.73.5.4251-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YX, Copeland TD, Henderson LE, Gorelick RJ, Bosche WJ, Levin JG, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. U. S. A. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi A, Orthwein A, Mercier J, Cohen EA. Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. J. Virol. 2007;81:7476–7490. doi: 10.1128/JVI.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R, Fivash MJ, Stephen AG, Hagan NA, Shenoy SR, Medaglia MV, Smith LR, Worthy KM, Simpson JT, Shoemaker R, McNitt KL, Johnson DG, Hixson CV, Gorelick RJ, Fabris D, Henderson LE, Rein A. Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides. Nucleic Acids Res. 2006;34:472–484. doi: 10.1093/nar/gkj442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher TS, Darden T, Prasad VR. Substitutions at Phe61 in the [beta]3-[beta]4 hairpin of HIV-1 reverse transcriptase reveal a role for the fingers subdomain in strand displacement DNA synthesis. J. Mol. Biol. 2003;325:443–459. doi: 10.1016/s0022-2836(02)01225-1. [DOI] [PubMed] [Google Scholar]
- Fitzon T, Leschonsky B, Bieler K, Paulus C, Schröder J, Wolf H, Wagner R. Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology. 2000;268:294–307. doi: 10.1006/viro.1999.0178. [DOI] [PubMed] [Google Scholar]
- Forshey BM, Shi J, Aiken C. Structural requirements for recognition of the human immunodeficiency virus type 1 core during host restriction in Owl monkey cells. J. Virol. 2005;79:869–875. doi: 10.1128/JVI.79.2.869-875.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forshey BM, von Schwedler U, Sundquist WI, Aiken C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 2002;76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Meyer BE, Simon JH, Fischer U, Albright AV, González-Scarano F, Malim MH. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J. Virol. 1998;72:6004–6013. doi: 10.1128/jvi.72.7.6004-6013.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Meyer BE, Simon JH, Fischer U, Malim MH. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO, Englund G, Maldarelli F, Martin MA. Phosphorylation of residue 131 of HIV-1 matrix is not required for macrophage infection. Cell. 1997;88:171–173. doi: 10.1016/s0092-8674(00)81836-x. [DOI] [PubMed] [Google Scholar]
- Fu W, Gorelick RJ, Rein A. Characterization of human immunodeficiency virus type 1 diMéric RNA from wild-type and protease-defective virions. J. Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Ortiz-Conde BA, Gorelick RJ, Hughes SH, Rein A. Placement of tRNA primer on the primer-binding site requires pol gene expression in avian but not murine retroviruses. J. Virol. 1997;71:6940–6946. doi: 10.1128/jvi.71.9.6940-6946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Rein A. Maturation of diMéric viral RNA of Moloney murine leukemia virus. J. Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes GM, Palaniappan C, Fay PJ, Bambara RA. Strand displacement synthesis in the central polypurine tract region of HIV-1 promotes DNA to DNA strand transfer recombination. J. Biol. Chem. 1996a;271:29605–29611. doi: 10.1074/jbc.271.47.29605. [DOI] [PubMed] [Google Scholar]
- Fuentes GM, Rodriguez-Rodriguez L, Palaniappan C, Fay PJ, Bambara RA. Strand displacement synthesis of the long terminal repeats by HIV reverse transcriptase. J. Biol. Chem. 1996b;271:1966–1971. doi: 10.1074/jbc.271.4.1966. [DOI] [PubMed] [Google Scholar]
- Furuta RA, Shimano R, Ogasawara T, Inubushi R, Amano K, Akari H, Hatanaka M, Kawamura M, Adachi A. HIV-1 capsid mutants inhibit the replication of wild-type virus at both early and late infection phases. FEBS Lett. 1997;415:231–234. doi: 10.1016/s0014-5793(97)01132-0. [DOI] [PubMed] [Google Scholar]
- Gabus C, Auxilien S, Péchoux C, Dormont D, Swietnicki W, Morillas M, Surewicz W, Nandi P, Darlix JL. The prion protein has DNA strand transfer properties similar to retroviral nucleocapsid protein. J. Mol. Biol. 2001a;307:1011–1021. doi: 10.1006/jmbi.2001.4544. [DOI] [PubMed] [Google Scholar]
- Gabus C, Derrington E, Leblanc P, Chnaiderman J, Dormont D, Swietnicki W, Morillas M, Surewicz WK, Marc D, Nandi P, Darlix JL. The prion protein has RNA binding and chaperoning properities characteristic of nucleocapsid protein NCp7 of HIV-1. J. Biol. Chem. 2001b;276:19301–19309. doi: 10.1074/jbc.M009754200. [DOI] [PubMed] [Google Scholar]
- Gabus C, Ivanyi-Nagy R, Depollier J, Bucheton A, Pelisson A, Darlix JL. Characterization of a nucleocapsid-like region and of two distinct primer tRNALys,2 binding sites in the endogenous retrovirus Gypsy. Nucleic Acids Res. 2006;34:5764–5777. doi: 10.1093/nar/gkl722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. U. S. A. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallay P, Swingler S, Aiken C, Trono D. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell. 1995a;80:379–388. doi: 10.1016/0092-8674(95)90488-3. [DOI] [PubMed] [Google Scholar]
- Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995b;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- Gao K, Butler SL, Bushman F. Human immunodeficiency virus type 1 integrase: arrangement of protein domains in active cDNA complexes. EMBO J. 2001;20:3565–3576. doi: 10.1093/emboj/20.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K, Gorelick RJ, Johnson DG, Bushman F. Cofactors for human immunodeficiency virus type 1 cDNA integration in vitro. J. Virol. 2003;77:1598–1603. doi: 10.1128/JVI.77.2.1598-1603.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard PM, Bonnet-Mathonière B, Muriaux D, Paoletti J. A short autocomplementary sequence in the 5’ leader region is responsible for dimerization of MoMLV genomic RNA. Biochemistry. 1995;34:9785–9794. doi: 10.1021/bi00030a016. [DOI] [PubMed] [Google Scholar]
- Goel A, Mazur SJ, Fattah RJ, Hartman TL, Turpin JA, Huang M, Rice WG, Appella E, Inman JK. Benzamide-based thiolcarbamates: a new class of HIV-1 NCp7 inhibitors. Bioorg. Med. Chem. Lett. 2002;12:767–770. doi: 10.1016/s0960-894x(02)00007-0. [DOI] [PubMed] [Google Scholar]
- Gonsky J, Bacharach E, Goff SP. Identification of residues of the Moloney murine leukemia virus nucleocapsid critical for viral DNA synthesis in vivo. J. Virol. 2001;75:2616–2626. doi: 10.1128/JVI.75.6.2616-2626.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi G, Pursley M, Felock P, Witmer M, Hazuda D, Brackmann K, Grandgenett D. Efficiency and fidelity of full-site integration reactions using recombinant simian immunodeficiency virus integrase. J. Virol. 1999;73:8104–8111. doi: 10.1128/jvi.73.10.8104-8111.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick RJ, Benveniste RE, Gagliardi TD, Wiltrout TA, Busch LK, Bosche WJ, Coren LV, Lifson JD, Bradley PJ, Henderson LE, Arthur LO. Nucleocapsid protein zinc-finger mutants of simian immunodeficiency virus strain mne produce virions that are replication defective in vitro and in vivo. Virology. 1999a;253:259–270. doi: 10.1006/viro.1998.9513. [DOI] [PubMed] [Google Scholar]
- Gorelick RJ, Chabot DJ, Ott DE, Gagliardi TD, Rein A, Henderson LE, Arthur LO. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J. Virol. 1996;70:2593–2597. doi: 10.1128/jvi.70.4.2593-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick RJ, Chabot DJ, Rein A, Henderson LE, Arthur LO. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick RJ, Fu W, Gagliardi TD, Bosche WJ, Rein A, Henderson LE, Arthur LO. Characterization of the block in replication of nucleocapsid protein zinc finger mutants from moloney murine leukemia virus. J. Virol. 1999b;73:8185–8195. doi: 10.1128/jvi.73.10.8185-8195.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick RJ, Gagliardi TD, Bosche WJ, Wiltrout TA, Coren LV, Chabot DJ, Lifson JD, Henderson LE, Arthur LO. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology. 1999c;256:92–104. doi: 10.1006/viro.1999.9629. [DOI] [PubMed] [Google Scholar]
- Gorelick RJ, Henderson LE, Hanser JP, Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. U. S. A. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick RJ, Nigida SM, Jr, Bess JW, Jr, Arthur LO, Henderson LE, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttlinger HG, Sodroski JG, Haseltine WA. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U. S. A. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves BJ, Eisenman RN, McKnight SL. Delineation of transcriptional control signals within the Moloney murine sarcoma virus long terminal repeat. Mol. Cell. Biol. 1985;5:1948–1958. doi: 10.1128/mcb.5.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorov B, Décimo D, Smagulova F, Péchoux C, Mougel M, Muriaux D, Darlix JL. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology. 2007;4:54. doi: 10.1186/1742-4690-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Henderson LE, Bess J, Kane B, Levin JG. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J. Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wu T, Anderson J, Kane BF, Johnson DG, Gorelick RJ, Henderson LE, Levin JG. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 2000;74:8980–8988. doi: 10.1128/jvi.74.19.8980-8988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Kameoka M, Wei X, Roques B, Gotte M, Liang C, Wainberg MA. Suppression of an intrinsic strand transfer activity of HIV-1 Tat protein by its second-exon sequences. Virology. 2003;307:154–163. doi: 10.1016/s0042-6822(02)00068-5. [DOI] [PubMed] [Google Scholar]
- Hameau L, Jeusset J, Lafosse S, Coulaud D, Delain E, Unge T, Restle T, Le Cam E, Mirambeau G. Human immunodeficiency virus type 1 central DNA flap: dynamic terminal product of plus-strand displacement DNA synthesis catalyzed by reverse transcriptase assisted by nucleocapsid protein. J. Virol. 2001;75:3301–3313. doi: 10.1128/JVI.75.7.3301-3313.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LJ, Sandmeyer SB. Characterization of a transpositionally active Ty3 element and identification of the Ty3 integrase protein. J. Virol. 1990;64:2599–2607. doi: 10.1128/jvi.64.6.2599-2607.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargittai MR, Gorelick RJ, Rouzina I, Musier-Forsyth K. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J. Mol. Biol. 2004;337:951–968. doi: 10.1016/j.jmb.2004.01.054. [DOI] [PubMed] [Google Scholar]
- Hargittai MR, Mangla AT, Gorelick RJ, Musier-Forsyth K. HIV-1 nucleocapsid protein zinc finger structures induce tRNA(Lys,3) structural changes but are not critical for primer/template annealing. J. Mol. Biol. 2001;312:985–997. doi: 10.1006/jmbi.2001.5021. [DOI] [PubMed] [Google Scholar]
- Henderson LE, Bowers MA, Sowder RC, II, Serabyn SA, Johnson DG, Bess JW, Jr, Arthur LO, Bryant DK, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J. Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson LE, Copeland TD, Sowder RC, Smythers GW, Oroszlan S. Primary structure of the low molecular weight nucleic acid-binding proteins of murine leukemia viruses. J. Biol. Chem. 1981;256:8400–8406. [PubMed] [Google Scholar]
- Henriet S, Sinck L, Bec G, Gorelick RJ, Marquet R, Paillart JC. Vif is a RNA chaperone that could temporally regulate RNA dimerization and the early steps of HIV-1 reverse transcription. Nucleic Acids Res. 2007;35:5141–5153. doi: 10.1093/nar/gkm542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindmarsh P, Leis J. Retroviral DNA integration. Microbiol. Mol. Biol. Rev. 1999;63:836–843. doi: 10.1128/mmbr.63.4.836-843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombrouck A, De Rijck J, Hendrix J, Vandekerckhove L, Voet A, De Maeyer M, Witvrouw M, Engelborghs Y, Christ F, Gijsbers R, Debyser Z. Virus evolution reveals an exclusive role for LEDGF/p75 in chromosomal tethering of HIV. PLoS Pathog. 2007;3:e47. doi: 10.1371/journal.ppat.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WS, Temin HM. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. U. S. A. 1990a;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WS, Temin HM. Retroviral recombination and reverse transcription. Science. 1990b;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- Huang Y, Khorchid A, Gabor J, Wang J, Li X, Darlix JL, Wainberg MA, Kleiman L. The role of nucleocapsid and U5 stem/A-rich loop sequences in tRNA(3Lys) genomic placement and initiation of reverse transcription in human immunodeficiency virus type 1. J. Virol. 1998;72:3907–3915. doi: 10.1128/jvi.72.5.3907-3915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Stone JR, Maki JL, Collins T, Wagner G. Mammalian SCAN domain dimer is a domain-swapped homolog of the HIV capsid C-terminal domain. Mol. Cell. 2005;17:137–143. doi: 10.1016/j.molcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Jeanson L, Mouscadet JF. Ku represses the HIV-1 transcription: identification of a putative Ku binding site homologous to the mouse mammary tumor virus NRE1 sequence in the HIV-1 long terminal repeat. J. Biol. Chem. 2002;277:4918–4924. doi: 10.1074/jbc.M110830200. [DOI] [PubMed] [Google Scholar]
- Jeanson L, Subra F, Vaganay S, Hervy M, Marangoni E, Bourhis J, Mouscadet JF. Effect of Ku80 depletion on the preintegrative steps of HIV-1 replication in human cells. Virology. 2002;300:100–108. doi: 10.1006/viro.2002.1515. [DOI] [PubMed] [Google Scholar]
- Jeeninga RE, Huthoff HT, Gultyaev AP, Berkhout B. The mechanism of actinomycin D-mediated inhibition of HIV-1 reverse transcription. Nucleic Acids Res. 1998;26:5472–5479. doi: 10.1093/nar/26.23.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentoft JE, Smith LM, Fu XD, Johnson M, Leis J. Conserved cysteine and histidine residues of the avian myeloblastosis virus nucleocapsid protein are essential for viral replication but are not “zinc-binding fingers”. Proc. Natl. Acad. Sci. U. S. A. 1988;85:7094–7098. doi: 10.1073/pnas.85.19.7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Klarmann GJ, Preston BD. Effect of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein on HIV-1 reverse transcriptase activity in vitro. Biochemistry. 1996;35:132–143. doi: 10.1021/bi951707e. [DOI] [PubMed] [Google Scholar]
- Johnson MC, Scobie HM, Ma YM, Vogt VM. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 2002;76:11177–11185. doi: 10.1128/JVI.76.22.11177-11185.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PE, Turner RB, Wu ZR, Hairston L, Guo L, Levin JG, Summers MF. A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription. Biochemistry. 2000;39:9084–9091. doi: 10.1021/bi000841i. [DOI] [PubMed] [Google Scholar]
- Jolly C, Mitar I, Sattentau QJ. Requirement for an intact T-cell actin and tubulin cytoskeleton for efficient assembly and spread of human immunodeficiency virus type 1. J. Virol. 2007;81:5547–5560. doi: 10.1128/JVI.01469-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J. Virol. 2007;81:7873–7884. doi: 10.1128/JVI.01845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KS, Coleman J, Merkel GW, Laue TM, Skalka AM. Retroviral integrase functions as a multimer and can turn over catalytically. J. Biol. Chem. 1992;267:16037–16040. [PubMed] [Google Scholar]
- Jouve M, Sol-Foulon N, Watson S, Schwartz O, Benaroch P. HIV-1 buds and accumulates in “nonacidic” endosomes of macrophages. Cell Host Microbe. 2007;2:85–95. doi: 10.1016/j.chom.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJ, Bess C, Johnson MC, Virgen CA, Simon SM, Bieniasz PD. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2007;4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankia BI, Barany G, Musier-Forsyth K. Unfolding of DNA quadruplexes induced by HIV-1 nucleocapsid protein. Nucleic Acids Res. 2005;33:4395–4403. doi: 10.1093/nar/gki741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan AH, Krogstad P, Kempf DJ, Norbeck DW, Swanstrom R. Human immunodeficiency virus type 1 virions composed of unprocessed Gag and GagPol precursors are capable of reverse transcribing viral genomic RNA. Antimicrob. Agents Chemother. 1994a;38:2929–2933. doi: 10.1128/aac.38.12.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan AH, Manchester M, Swanstrom R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J. Virol. 1994b;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan AH, Zack JA, Knigge M, Paul DA, Kempf DJ, Norbeck DW, Swanstrom R. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of noninfectious particles. J. Virol. 1993;67:4050–4055. doi: 10.1128/jvi.67.7.4050-4055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz RA, Merkel G, Kulkosky J, Leis J, Skalka AM. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- Kaushik R, Ratner L. Role of human immunodeficiency virus type 1 matrix phosphorylation in an early postentry step of virus replication. J. Virol. 2004;78:2319–2326. doi: 10.1128/JVI.78.5.2319-2326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura M, Shimano R, Inubushi R, Akari H, Adachi A. Early function of HIV-1 Gag proteins is cell-dependent. Biochem. Biophys. Res. Commun. 1998;248:899–903. doi: 10.1006/bbrc.1998.9065. [DOI] [PubMed] [Google Scholar]
- Kelleher CD, Champoux JJ. Characterization of RNA strand displacement synthesis by Moloney murine leukemia virus reverse transcriptase. J. Biol. Chem. 1998;273:9976–9986. doi: 10.1074/jbc.273.16.9976. [DOI] [PubMed] [Google Scholar]
- Khan R, Giedroc DP. Recombinant human immunodeficiency virus type 1 nucleocapsid (NCp7) protein unwinds tRNA. J. Biol. Chem. 1992;267:6689–6695. [PubMed] [Google Scholar]
- Khan R, Giedroc DP. Nucleic acid binding properties of recombinant Zn2 HIV-1 nucleocapsid protein are modulated by COOH-terminal processing. J. Biol. Chem. 1994;269:22538–22546. [PubMed] [Google Scholar]
- Kiernan RE, Ono A, Englund G, Freed EO. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 1998;72:4116–4126. doi: 10.1128/jvi.72.5.4116-4126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilzer JM, Stracker T, Beitzel B, Meek K, Weitzman M, Bushman FD. Roles of host cell factors in circularization of retroviral DNA. Virology. 2003;314:460–467. doi: 10.1016/s0042-6822(03)00455-0. [DOI] [PubMed] [Google Scholar]
- Kirchner J, Sandmeyer S. Proteolytic processing of Ty3 proteins is required for transposition. J. Virol. 1993;67:19–28. doi: 10.1128/jvi.67.1.19-28.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasens BI, Huthoff HT, Das AT, Jeeninga RE, Berkhout B. The effect of template RNA structure on elongation by HIV-1 reverse transcriptase. Biochim. Biophys. Acta. 1999;1444:355–370. doi: 10.1016/s0167-4781(99)00011-1. [DOI] [PubMed] [Google Scholar]
- Koh K, Miyaura M, Yoshida A, Sakurai A, Fujita M, Adachi A. Cell-dependent gag mutants of HIV-1 are crucially defective at the stage of uncoating/reverse transcription in non-permissive cells. Microbes Infect. 2000;2:1419–1423. doi: 10.1016/s1286-4579(00)01295-8. [DOI] [PubMed] [Google Scholar]
- Kräusslich HG. Human immunodeficiency virus proteinase dimer as component of the viral polyprotein prevents particle assembly and viral infectivity. Proc. Natl. Acad. Sci. U. S. A. 1991;88:3213–3217. doi: 10.1073/pnas.88.8.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kräusslich HG, Fäcke M, Heuser AM, Konvalinka AM, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy G, Roques B, Darlix JL, Mély Y. DNA condensation by the nucleocapsid protein of HIV-1: a mechanism ensuring DNA protection. Nucleic Acids Res. 2003;31:5425–5432. doi: 10.1093/nar/gkg738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins LA, Tsichlis PN, Khoury G. Multiple enhancer domains in the 3’ terminus of the Prague strain of Rous sarcoma virus. Nucleic Acids Res. 1984;12:6427–6442. doi: 10.1093/nar/12.16.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix JL. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapadat-Tapolsky M, Gabus C, Rau M, Darlix JL. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J. Mol. Biol. 1997;268:250–260. doi: 10.1006/jmbi.1997.0978. [DOI] [PubMed] [Google Scholar]
- Laughrea M, Shen N, Jetté L, Darlix JL, Kleiman L, Wainberg MA. Role of distal zinc finger of nucleocapsid protein in genomic RNA dimerization of human immunodeficiency virus type 1; no role for the palindrome crowning the R-U5 hairpin. Virology. 2001;281:109–116. doi: 10.1006/viro.2000.0778. [DOI] [PubMed] [Google Scholar]
- Lee BM, De Guzman RN, Turner BG, Tjandra N, Summers MF. Dynamical behavior of the HIV-1 nucleocapsid protein. J. Mol. Biol. 1998;12:633–649. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- Lee EG, Linial ML. Basic residues of the retroviral nucleocapsid play different roles in gag-gag and Gag-Psi RNA interactions. J. Virol. 2004;78:8486–8495. doi: 10.1128/JVI.78.16.8486-8495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EG, Linial ML. Deletion of a Cys-His motif from the Alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects. Virology. 2006;347:226–233. doi: 10.1016/j.virol.2005.11.048. [DOI] [PubMed] [Google Scholar]
- Leis J, Baltimore D, Bishop JM, Coffin J, Fleissner E, Goff SP, Oroszlan S, Robinson H, Skalka AM, Temin HM, Vogt V. Standardized and simplified nomenclature for proteins common to all retroviruses. J. Virol. 1988;62:1808–1809. doi: 10.1128/jvi.62.5.1808-1809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner T, Foley B, Hahn B, Marx P, McCutchan F, Mellors J, Wolinksy S, Korber B, editors. HIV sequence compendium. Los Alamos, NM: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, LA-UR 07-7826; 20062007. [Google Scholar]
- Lener D, Tanchou V, Roques BP, Le Grice SF, Darlix JL. Involvement of HIV-1 nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J. Biol. Chem. 1998;273:33781–33786. doi: 10.1074/jbc.273.50.33781. [DOI] [PubMed] [Google Scholar]
- Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- Li F, Goila-Gaur R, Salzwedel K, Kilgore NR, Reddick M, Matallana C, Castillo A, Zoumplis D, Martin DE, Orenstein JM, Allaway GP, Freed EO, Wild CT. PA-457: A potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13555–13560. doi: 10.1073/pnas.2234683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Dou J, Ding L, Spearman P. Myristoylation is required for human immunodeficiency virus type 1 Gag-Gag multimerization in mammalian cells. J. Virol. 2007;81:12899–12910. doi: 10.1128/JVI.01280-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Olvera JM, Yoder KE, Mitchell RS, Butler SL, Lieber M, Martin SL, Bushman FD. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 2001;20:3272–3281. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Mizuuchi M, Burke TR, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limón A, Devroe E, Lu R, Ghory HZ, Silver PA, Engelman A. Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import. J. Virol. 2002a;76:10598–10607. doi: 10.1128/JVI.76.21.10598-10607.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limón A, Nakajima N, Lu R, Ghory HZ, Engelman A. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J. Virol. 2002b;76:12078–12086. doi: 10.1128/JVI.76.23.12078-12086.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial ML. Foamy viruses are unconventional retroviruses. J. Virol. 1999;73:1747–1755. doi: 10.1128/jvi.73.3.1747-1755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel LI, Murphy JE, Goff SP. The palindromic LTR-LTR junction of Moloney murine leukemia virus is not an efficient substrate for proviral integration. J. Virol. 1989;63:2629–2637. doi: 10.1128/jvi.63.6.2629-2637.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lori F, di Marzo Veronese F, de Vico AL, Lusso P, Reitz MS, Jr, Gallo RC. Viral DNA carried by human immunodeficiency virus type 1 virions. J. Virol. 1992;66:5067–5074. doi: 10.1128/jvi.66.8.5067-5074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyonnais S, Gorelick RJ, Mergny JL, Le Cam E, Mirambeau G. G-quartets direct assembly of HIV-1 nucleocapsid protein along single-stranded DNA. Nucleic Acids Res. 2003;31:5754–5763. doi: 10.1093/nar/gkg716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyonnais S, Hounsou C, Teulade-Fichou MP, Jeusset J, Le Cam E, Mirambeau G. G-quartets assembly within a G-rich DNA flap. A possible event at the center of the HIV-1 genome. Nucleic Acids Res. 2002;30:5276–5283. doi: 10.1093/nar/gkf644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrique ML, Rauddi ML, González SA, Affranchino JL. Functional domains in the feline immunodeficiency virus NC protein. Virology. 2004;327:83–92. doi: 10.1016/j.virol.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Marlor RL, Parkhurst SM, Corces VG. The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol. Cell. Biol. 1986;6:1129–1134. doi: 10.1128/mcb.6.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet R, Isel C, Ehresmann C, Ehresmann B. tRNAs as primer of reverse transcriptases. Biochimie. 1995;77:113–124. doi: 10.1016/0300-9084(96)88114-4. [DOI] [PubMed] [Google Scholar]
- Marsden MD, Zack JA. Human immunodeficiency virus bearing a disrupted central DNA flap is pathogenic in vivo. J. Virol. 2007;81:6146–6150. doi: 10.1128/JVI.00203-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Planelles V, Krogstad P, Chen IS. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J. Virol. 1995;69:6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méric C, Goff SP. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J. Virol. 1989;63:1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méric C, Gouilloud E, Spahr PF. Mutations in Rous sarcoma virus nucleocapsid protein p12 (NC): deletions of Cys-His boxes. J. Virol. 1988;62:3328–3333. doi: 10.1128/jvi.62.9.3328-3333.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méric C, Spahr PF. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J. Virol. 1986;60:450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkulov GV, Swiderek KM, Brachmann CB, Boeke JD. A critical proteolytic cleavage site near the C terminus of the yeast retrotransposon Ty1 Gag protein. J. Virol. 1996;70:5548–5556. doi: 10.1128/jvi.70.8.5548-5556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller Jenkins LM, Hara T, Durell SR, Hayashi R, Inman JK, Piquemal JP, Gresh N, Appella E. Specificity of acyl transfer from 2-mercaptobenzamide thioesters to the HIV-1 nucleocapsid protein. J. Am. Chem. Soc. 2007;129:11067–11078. doi: 10.1021/ja071254o. [DOI] [PubMed] [Google Scholar]
- Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Borde I, Reboud-Ravaux M, Restle T, Gorelick RJ, Le Cam E. HIV-1 protease and reverse transcriptase control the architecture of their nuclecapsid partner. PLoS One. 2007;2:e669. doi: 10.1371/journal.pone.0000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Justome A, Delain E, Gorelick RJ, Le Cam E. Transmission electron microscopy reveals an optimal HIV-1 nucleocapsid aggregation with single-stranded nucleic acids and the mature HIV-1 nucleocapsid protein. J. Mol. Biol. 2006;364:496–511. doi: 10.1016/j.jmb.2006.08.065. [DOI] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schröder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:e234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno A, Ido E, Goto T, Kuwata T, Nakai M, Hayami M. Mutational analysis of two zinc finger motifs in HIV type 1 nucleocapsid proteins: effects on proteolytic processing of Gag precursors and particle formation. AIDS Res. Hum. Retroviruses. 1996;12:793–800. doi: 10.1089/aid.1996.12.793. [DOI] [PubMed] [Google Scholar]
- Morcock DR, Thomas JA, Gagliardi TD, Gorelick RJ, Roser JD, Chertova EN, Bess JW, Jr, Ott DE, Sattentau QJ, Frank I, Pope M, Lifson JD, Henderson LE, Crise BJ. Elimination of retroviral infectivity by N-ethylmaleimide with preservation of functional envelope glycoproteins. J. Virol. 2005;79:1533–1542. doi: 10.1128/JVI.79.3.1533-1542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morellet N, de Rocquigny H, Mély Y, Jullian N, Déméné H, Ottmann M, Gérard D, Darlix JL, Fournié-Zaluski MC, Roques BP. Conformational behaviour of the active and inactive forms of the nucleocapsid NCp7 of HIV-1 studied by 1H NMR. J. Mol. Biol. 1994;7:287–301. doi: 10.1016/s0022-2836(05)80033-6. [DOI] [PubMed] [Google Scholar]
- Morellet N, Jullian N, De Rocquigny H, Maigret B, Darlix JL, Roques BP. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount SM, Rubin GM. Complete nucleotide sequence of the Drosophila transposable element copia: homology between copia and retroviral proteins. Mol. Cell Biol. 1985;5:1630–1638. doi: 10.1128/mcb.5.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux D, Costes S, Nagashima K, Mirro J, Cho E, Lockett S, Rein A. Role of murine leukemia virus nucleocapsid protein in virus assembly. J. Virol. 2004;78:12378–12385. doi: 10.1128/JVI.78.22.12378-12385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux D, de Rocquigny H, Roques BP, Paoletti J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J. Biol. Chem. 1996;271:33686–33692. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- Muriaux D, Mirro J, Harvin D, Rein A. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux D, Mirro J, Nagashima K, Harvin D, Rein A. Murine leukemia virus nucleocapsid mutant particles lacking viral RNA encapsidate ribosomes. J. Virol. 2002;76:11405–11413. doi: 10.1128/JVI.76.22.11405-11413.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy K, Young M, Baboonian C, Merson J, Whittle P, Oroszlan S. Antiviral activity of human immunodeficiency virus type 1 protease inhibitors in a single cycle of infection: evidence for a role of protease in the early phase. J. Virol. 1994;68:757–765. doi: 10.1128/jvi.68.2.757-765.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, Seeger C, Skalka AM, Katz RA. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negroni M, Buc H. Recombination during reverse transcription: an evaluation of the role of the nucleocapsid protein. J. Mol. Biol. 1999;286:15–31. doi: 10.1006/jmbi.1998.2460. [DOI] [PubMed] [Google Scholar]
- Neil S, Martin F, Ikeda Y, Collins M. Postentry restriction to human immunodeficiency virus-based vector transduction in human monocytes. J. Virol. 2001;75:5448–5456. doi: 10.1128/JVI.75.12.5448-5456.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nermut MV, Fassati A. Structural analyses of purified human immunodeficiency virus type 1 intracellular reverse transcription complexes. J. Virol. 2003;77:8196–8206. doi: 10.1128/JVI.77.15.8196-8206.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaitchik O, Rhodes TD, Ott D, Hu WS. Effects of mutations in the human immunodeficiency virus type 1 Gag gene on RNA packaging and recombination. J. Virol. 2006;80:4691–4697. doi: 10.1128/JVI.80.10.4691-4697.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Saïb A. Early steps of retrovirus replicative cycle. Retrovirology. 2004;1:9. doi: 10.1186/1742-4690-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nydegger S, Foti M, Derdowski A, Spearman P, Thali M. HIV-1 egress is gated through late endosomal membranes. Traffic. 2003;4:902–910. doi: 10.1046/j.1600-0854.2003.00145.x. [DOI] [PubMed] [Google Scholar]
- O’Brien WA, Namazi A, Kalhor H, Mao SH, Zack JA, Chen IS. Kinetics of human immunodeficiency virus type 1 reverse transcription in blood mononuclear phagocytes are slowed by limitations of nucleotide precursors. J. Virol. 1994;68:1258–1263. doi: 10.1128/jvi.68.2.1258-1263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott DE, Coren LV, Chertova EN, Gagliardi TD, Nagashima K, Sowder RC, 2nd, Poon DT, Gorelick RJ. Elimination of protease activity restores efficient virion production to a human immunodeficiency virus type 1 nucleocapsid deletion mutant. J. Virol. 2003;77:5547–5556. doi: 10.1128/JVI.77.10.5547-5556.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott DE, Coren LV, Gagliardi TD. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J. Virol. 2005;79:13839–13847. doi: 10.1128/JVI.79.22.13839-13847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann M, Gabus C, Darlix JL. The central globular domain of the NC protein of human immunodeficiency virus type 1 is critical for virion structure and infectivity. J. Virol. 1995;69:1778–1784. doi: 10.1128/jvi.69.3.1778-1784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson BA, Zhang M, Schultz SJ, Champoux JJ. Substitution of alanine for tyrosine-64 in the fingers subdomain of M-MLV reverse transcriptase impairs strand displacement synthesis and blocks viral replication in vivo. Virology. 2007;366:361–376. doi: 10.1016/j.virol.2007.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peliska JA, Balasubramanian S, Giedroc DP, Benkovic SJ. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- Peliska JA, Benkovic SJ. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258:1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- Perlman M, Resh MD. Identification of an intracellular trafficking and assembly pathway for HIV-1 Gag. Traffic. 2006;7:731–745. doi: 10.1111/j.1398-9219.2006.00428.x. [DOI] [PubMed] [Google Scholar]
- Petit C, Schwartz O, Mammano F. The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA. J. Virol. 2000;74:7119–7126. doi: 10.1128/jvi.74.15.7119-7126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit SC, Moody MD, Wehbie RS, Kaplan AH, Nantermet PV, Klein CA, Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit SC, Sheng N, Tritch R, Erickson-Viitanen S, Swanstrom R. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv. Exp. Med. Biol. 1998;436:15–25. doi: 10.1007/978-1-4615-5373-1_2. [DOI] [PubMed] [Google Scholar]
- Pettit SC, Simsic J, Loeb DD, Everitt L, Hutchison CA, 3rd, Swanstrom R. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J. Biol. Chem. 1991;266:14539–14547. [PubMed] [Google Scholar]
- Polge E, Darlix JL, Paoletti J, Fossé P. Characterization of loose and tight dimer forms of avian leukosis virus RNA. J. Mol. Biol. 2000;300:41–56. doi: 10.1006/jmbi.2000.3832. [DOI] [PubMed] [Google Scholar]
- Poljak L, Batson SM, Ficheux D, Roques BP, Darlix JL, Käs E. Analysis of NCp7-dependent activation of HIV-1 cDNA integration and its conservation among retroviral nucleocapsid proteins. J. Mol. Biol. 2003;329:411–421. doi: 10.1016/s0022-2836(03)00472-8. [DOI] [PubMed] [Google Scholar]
- Poon DT, Chertova EN, Ott DE. Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55(Gag): functional linkage between translation and RNA packaging. Virology. 2002;293:368–378. doi: 10.1006/viro.2001.1283. [DOI] [PubMed] [Google Scholar]
- Poon DT, Li G, Aldovini A. Nucleocapsid and matrix protein contributions to selective human immunodeficiency virus type 1 genomic RNA packaging. J. Virol. 1998;72:1983–1993. doi: 10.1128/jvi.72.3.1983-1993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon DT, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J. Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats AC, Housset V, de Billy G, Cornille F, Prats H, Roques B, Darlix JL. Viral RNA annealing activities of the nucleocapsid protein of Moloney murine leukemia virus are zinc independent. Nucleic Acids Res. 1991;11:3533–3541. doi: 10.1093/nar/19.13.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats AC, Sarih L, Gabus C, Litvak S, Keith G, Darlix JL. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988;7:1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja A, DeStefano JJ. Kinetic analysis of the effect of HIV nucleocapsid protein (NCp) on internal strand transfer reactions. Biochemistry. 1999;38:5178–5184. doi: 10.1021/bi9828019. [DOI] [PubMed] [Google Scholar]
- Raja C, Ferner J, Dietrich U, Avilov S, Ficheux D, Darlix JL, de Rocquigny H, Schwalbe H, Mély Y. A tryptophan-rich hexapeptide inhibits nucleic acid destabilization chaperoned by the HIV-1 nucleocapsid protein. Biochemistry. 2006;45:9254–9265. doi: 10.1021/bi052560m. [DOI] [PubMed] [Google Scholar]
- Ramirez BC, Simon-Loriere E, Galetto R, Negroni M. Implications of recombination for HIV viral diversity. Virus Res. 2008 doi: 10.1016/j.virusres.2008.01.007. in press. [DOI] [PubMed] [Google Scholar]
- Rascle JB, Ficheux D, Darlix JL. Possible roles of nucleocapsid protein of MoMuLV in the specificity of proviral DNA synthesis and in the genetic variability of the virus. J. Mol. Biol. 1998;280:215–225. doi: 10.1006/jmbi.1998.1873. [DOI] [PubMed] [Google Scholar]
- Rausch JW, Le Grice SF. ‘Binding, bending and bonding’: polypurine tract-primed initiation of plus-strand DNA synthesis in human immunodeficiency virus. Int. J. Biochem. Cell Biol. 2004;36:1752–1766. doi: 10.1016/j.biocel.2004.02.016. [DOI] [PubMed] [Google Scholar]
- Reil H, Bukovsky AA, Gelderblom HR, Göttlinger HG. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A, Harvin DP, Mirro J, Ernst SM, Gorelick RJ. Evidence that a central domain of NC protein is required for RNA packaging in murine leukemia virus. J. Virol. 1994;68:6124–6129. doi: 10.1128/jvi.68.9.6124-6129.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- Risco C, Menéndez-Arias L, Copeland TD, Pinto da Silva P, Oroszlan S. Intracellular transport of the murine leukemia virus during acute infection of NIH 3T3 cells: nuclear import of nucleocapsid protein and integrase. J. Cell Sci. 1995;108:3039–3050. doi: 10.1242/jcs.108.9.3039. [DOI] [PubMed] [Google Scholar]
- Roda RH, Balakrishnan M, Hanson MN, Wöhrl BM, Le Grice SF, Roques BP, Gorelick RJ, Bambara RA. Role of the reverse transcriptase, nucleocapsid protein, and template structure in the two-step transfer mechanism in retroviral recombination. J. Biol. Chem. 2003;278:31536–31546. doi: 10.1074/jbc.M304608200. [DOI] [PubMed] [Google Scholar]
- Roda RH, Balakrishnan M, Kim JK, Roques BP, Fay PJ, Bambara RA. Strand transfer occurs in retroviruses by a pause-initiated two-step mechanism. J. Biol. Chem. 2002;277:46900–46911. doi: 10.1074/jbc.M208638200. [DOI] [PubMed] [Google Scholar]
- Roe T, Reynolds TC, Yu G, Brown PO. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roebuck KA, Saifuddin M. Regulation of HIV-1 transcription. Gene Expr. 1999;8:67–84. [PMC free article] [PubMed] [Google Scholar]
- Rong L, Liang C, Hsu M, Guo X, Roques BP, Wainberg MA. HIV-1 nucleocapsid protein and the secondary structure of the binary complex formed between tRNA(Lys.3) and viral RNA template play different roles during initiation of (-) strand DNA reverse transcription. J. Biol. Chem. 2001;276:47725–47732. doi: 10.1074/jbc.M105124200. [DOI] [PubMed] [Google Scholar]
- Rossio JL, Esser MT, Suryanarayana K, Schneider DK, Bess JW, Jr, Vasquez GM, Wiltrout TA, Chertova E, Grimes MK, Sattentau Q, Arthur LO, Henderson LE, Lifson JD. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 1998;72:7992–8001. doi: 10.1128/jvi.72.10.7992-8001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner L, Nydegger S, Coren LV, Nagashima K, Thali M, Ott DE. Dynamic fluorescent imaging of human immunodeficiency virus type 1 gag in live cells by biarsenical labeling. J. Virol. 2005;79:4055–4065. doi: 10.1128/JVI.79.7.4055-4065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulli SJ, Jr, Hibbert CS, Mirro J, Pederson T, Biswal S, Rein A. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 2007;81:6623–6631. doi: 10.1128/JVI.02833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandefur S, Smith RM, Varthakavi V, Spearman P. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55(Gag) J. Virol. 2000;74:7238–7249. doi: 10.1128/jvi.74.16.7238-7249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandefur S, Varthakavi V, Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 1998;72:2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hot spots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Schultz SJ, Champoux JJ. RNase H activity: Structure, specificity, and function in reverse transcription. Virus Res. 2008 doi: 10.1016/j.virusres.2007.12.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehu-Xhilaga M, Crowe SM, Mak DP. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 2001a;75:1834–1841. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehu-Xhilaga M, Kraeusslich HG, Pettit S, Swanstrom R, Lee JY, Marshall JA, Crowe SM, Mak J. Proteolytic processing of the p2/nucleocapsid cleavage site is critical for human immunodeficiency virus type 1 RNA dimer maturation. J. Virol. 2001b;75:9156–9164. doi: 10.1128/JVI.75.19.9156-9164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng N, Pettit SC, Tritch RJ, Ozturk DH, Rayner MM, Swanstrom R, Erickson-Viitanen S. Determinants of the human immunodeficiency virus type 1 p15NC-RNA interaction that affect enhanced cleavage by the viral protease. J. Virol. 1997;71:5723–5732. doi: 10.1128/jvi.71.8.5723-5732.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer NM, Lehmann MJ, Jimenez-Soto LF, Ingmundson A, Horner SM, Cicchetti G, Allen PG, Pypaert M, Cunningham JM, Mothes W. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic. 2003;4:785–801. doi: 10.1034/j.1600-0854.2003.00135.x. [DOI] [PubMed] [Google Scholar]
- Sinha S, Grandgenett DP. Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. J. Virol. 2005;79:8208–8216. doi: 10.1128/JVI.79.13.8208-8216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Kafaie J, Yang L, Laughrea M. HIV-1 Viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization. J. Mol. Biol. 2007;371:1084–1098. doi: 10.1016/j.jmb.2007.06.010. [DOI] [PubMed] [Google Scholar]
- South TL, Summers MF. Zinc fingers. In: Marzilli LG, Eichorn G, editors. Metal Ion-induced Regulation of Gene Expression. Vol. 8. New York: Elsevier; 1990. pp. 199–248. [Google Scholar]
- Srivastava P, Schito M, Fattah RJ, Hara T, Hartman T, Buckheit RW, Jr, Turpin JA, Inman JK, Appella E. Optimization of unique, uncharged thioesters as inhibitors of HIV replication. Bioorg. Med. Chem. 2004;12:6437–6450. doi: 10.1016/j.bmc.2004.09.032. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Plainview, NY: Cold Spring Harbor Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- Takahashi K, Baba S, Koyanagi Y, Yamamoto N, Takaku H, Kawai G. Two basic regions of NCp7 are sufficient for conformational conversion of HIV-1 dimerization initiation site from kissing-loop dimer to extended-duplex dimer. J. Biol. Chem. 2001;276:31274–31278. doi: 10.1074/jbc.M104577200. [DOI] [PubMed] [Google Scholar]
- Tanchou V, Décimo D, Péchoux C, Lener D, Rogemond V, Berthoux L, Ottmann M, Darlix JL. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J. Virol. 1998;72:4442–4447. doi: 10.1128/jvi.72.5.4442-4447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanchou V, Gabus C, Rogemond V, Darlix JL. Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J. Mol. Biol. 1995;252:563–571. doi: 10.1006/jmbi.1995.0520. [DOI] [PubMed] [Google Scholar]
- Tang S, Murakami T, Agresta BE, Campbell S, Freed EO, Levin JG. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J. Virol. 2001;75:9357–9366. doi: 10.1128/JVI.75.19.9357-9366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesnitsky A, Goff SP. Reverse transcriptase and the generation of retroviral DNA. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Plainview, NY: Cold Spring Harbor Press; 1997. pp. 121–160. [PubMed] [Google Scholar]
- Thomas DC, Voronin YA, Nikolenko GN, Chen J, Hu W-S, Pathak VK. Determination of the ex vivo rates of human immunodeficiency virus type 1 reverse transcription by using novel strand-specific amplification analysis. J. Virol. 2007a;81:4798–4807. doi: 10.1128/JVI.02471-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JA, Gagliardi TD, Alvord WG, Lubomirski M, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid zinc-finger mutations cause defects in reverse transcription and integration. Virology. 2006a;353:41–51. doi: 10.1016/j.virol.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Thomas JA, Ott DE, Gorelick RJ. Efficiency of human immunodeficiency virus type 1 postentry infection processes: evidence against disproportionate numbers of defective virions. J. Virol. 2007b;81:4367–4370. doi: 10.1128/JVI.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JA, Shulenin S, Coren LV, Bosche WJ, Gagliardi TD, Gorelick RJ, Oroszlan S. Characterization of human immunodeficiency virus type 1 (HIV-1) containing mutations in the nucleocapsid protein at a putative HIV-1 protease cleavage site. Virology. 2006b;354:261–270. doi: 10.1016/j.virol.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Tisné C. Structural bases of the annealing of primer tRNA(3Lys) to the HIV-1 viral RNA. Curr. HIV Res. 2005;3:147–156. doi: 10.2174/1570162053506919. [DOI] [PubMed] [Google Scholar]
- Trono D. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J. Virol. 1992;66:4893–4900. doi: 10.1128/jvi.66.8.4893-4900.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trono D, Gallay P. In response to Freed et al. Cell. 1997;88:173–174. [Google Scholar]
- Tsuchihashi Z, Brown PO. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P, Doucas V, Craig E, Mangeat B, Klages N, Evans R, Kalpana G, Trono D. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: interference with early steps of viral replication. Mol. Cell. 2001;7:1245–1254. doi: 10.1016/s1097-2765(01)00255-6. [DOI] [PubMed] [Google Scholar]
- Turner BG, Summers MF. Structural biology of HIV. J. Mol. Biol. 1999;285:1–32. doi: 10.1006/jmbi.1998.2354. [DOI] [PubMed] [Google Scholar]
- Turpin JA, Song Y, Inman JK, Huang M, Wallqvist A, Maynard A, Covell DG, Rice WG, Appella E. Synthesis and biological properties of novel pyridinioalkanoyl thiolesters (PATE) as anti-HIV-1 agents that target the viral nucleocapsid protein zinc fingers. J. Med. Chem. 1999;42:67–86. doi: 10.1021/jm9802517. [DOI] [PubMed] [Google Scholar]
- Urbaneja MA, Kane BP, Johnson DG, Gorelick RJ, Henderson LE, Casas-Finet JR. Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J. Mol. Biol. 1999;287:59–75. doi: 10.1006/jmbi.1998.2521. [DOI] [PubMed] [Google Scholar]
- Urbaneja MA, Wu M, Casas-Finet JR, Karpel RL. HIV-1 nucleocapsid protein as a nucleic acid chaperone: spectroscopic study of its helix-destabilizing properties, structural binding specificity, and annealing activity. J. Mol. Biol. 2002;318:749–764. doi: 10.1016/S0022-2836(02)00043-8. [DOI] [PubMed] [Google Scholar]
- Van Maele B, De Rijck J, De Clercq E, Debyser Z. Impact of the central polypurine tract on the kinetics of human immunodeficiency virus type 1 vector transduction. J. Virol. 2003;77:4685–4694. doi: 10.1128/JVI.77.8.4685-4694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB, editors. Virus taxonomy classification and nomenclature of viruses: seventh report of the international committee on taxonomy of viruses. San Diego: Academic Press; 2000. [Google Scholar]
- Vink C, van Gent DC, Elgersma Y, Plasterk RH. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J. Virol. 1991;65:4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler U, Kornbluth RS, Trono D. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 1994;91:6992–6996. doi: 10.1073/pnas.91.15.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora A, Grandgenett DP. DNase protection analysis of retrovirus integrase at the viral DNA ends for full-site integration in vitro. J. Virol. 2001;75:3556–3567. doi: 10.1128/JVI.75.8.3556-3567.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapornin P, Lauhakirti D, Auewarakul P. The effect of capsid mutations on HIV-1 uncoating. Virology. 2006;358:48–54. doi: 10.1016/j.virol.2006.08.031. [DOI] [PubMed] [Google Scholar]
- Wang CT, Barklis E. Assembly, processing, and infectivity of human immunodeficiency virus type 1 gag mutants. J. Virol. 1993;67:4264–4273. doi: 10.1128/jvi.67.7.4264-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Noonan K, Aldovini A. Nucleocapsid-RNA interactions are essential to structural stability but not to assembly of retroviruses. J. Virol. 2004;78:716–723. doi: 10.1128/JVI.78.2.716-723.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow D, Harrich D. HIV-1 replication from after cell entry to the nuclear periphery. Curr. HIV Res. 2007;5:293–299. doi: 10.2174/157016207780636579. [DOI] [PubMed] [Google Scholar]
- Waters LC, Mullin BC. Transfer RNA into RNA tumor viruses. Prog. Nucleic Acid Res. Mol. Biol. 1977;20:131–160. doi: 10.1016/s0079-6603(08)60471-7. [DOI] [PubMed] [Google Scholar]
- Wei SQ, Mizuuchi K, Craigie R. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 1997;16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker R, Hohenberg H, Tessmer U, Huckhagel C, Kräusslich HG. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J. Virol. 2000;74:1168–1177. doi: 10.1128/jvi.74.3.1168-1177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S, Keppler OT, Habermann A, Allespach I, Krijnse-Locker J, Kräusslich HG. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog. 2007;3:e36. doi: 10.1371/journal.ppat.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiting SH, Champoux JJ. Properties of strand displacement synthesis by Moloney murine leukemia virus reverse transcriptase: mechanistic implications. J. Mol. Biol. 1998;278:559–577. doi: 10.1006/jmbi.1998.1720. [DOI] [PubMed] [Google Scholar]
- Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Kräusslich HG. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Guo J, Bess J, Henderson LE, Levin JG. Molecular requirements for human immunodeficiency virus type 1 plus-strand transfer: analysis in reconstituted and endogenous reverse transcription systems. J. Virol. 1999;73:4794–4805. doi: 10.1128/jvi.73.6.4794-4805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Heilman-Miller SL, Levin JG. Effects of nucleic acid local structure and magnesium ions on minus-strand transfer mediated by the nucleic acid chaperone activity of HIV-1 nucleocapsid protein. Nucleic Acids Res. 2007;35:3974–3987. doi: 10.1093/nar/gkm375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Henderson LE, Copeland TD, Gorelick RJ, Bosche WJ, Rein A, Levin JG. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J. Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Emerman M. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 2004;78:5670–5678. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Emerman M. The cell cycle independence of HIV infections is not determined by known karyophilic viral elements. PLoS Pathog. 2005;1:e18. doi: 10.1371/journal.ppat.0010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Emerman M. Retroviral infection of non-dividing cells: old and new perspectives. Virology. 2006;344:88–93. doi: 10.1016/j.virol.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Yoder KE, Bushman FD. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 2000;74:11191–11200. doi: 10.1128/jvi.74.23.11191-11200.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JC, McHenry CS. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J. Biol. Chem. 1994;269:31491–31495. [PubMed] [Google Scholar]
- Yu Q, Darlix JL. The zinc finger of nucleocapsid protein of Friend murine leukemia virus is critical for proviral DNA synthesis in vivo. J. Virol. 1996;70:5791–5798. doi: 10.1128/jvi.70.9.5791-5798.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- Zack JA, Haislip AM, Krogstad P, Chen IS. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J. Virol. 1992;66:1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- Zennou V, Serguera C, Sarkis C, Colin P, Perret E, Mallet J, Charneau P. The HIV-1 DNA flap stimulates HIV vector-mediated cell transduction in the brain. Nat. Biotechnol. 2001;19:446–450. doi: 10.1038/88115. [DOI] [PubMed] [Google Scholar]
- Zhang H, Dornadula G, Orenstein J, Pomerantz RJ. Morphologic changes in human immunodeficiency virus type 1 virions secondary to intravirion reverse transcription: evidence indicating that reverse transcription may not take place within the intact viral core. J. Hum. Virol. 2000a;3:165–172. [PubMed] [Google Scholar]
- Zhang H, Zhang Y, Spicer TP, Abbott LZ, Abbott M, Poiesz BJ. Reverse transcription takes place within extracellular HIV-1 virions: potential biological significance. AIDS Res. Hum. Retroviruses. 1993;9:1287–1296. doi: 10.1089/aid.1993.9.1287. [DOI] [PubMed] [Google Scholar]
- Zhang J, Crumpacker CS. Human immunodeficiency virus type 1 nucleocapsid protein nuclear localization mediates early viral mRNA expression. J. Virol. 2002;76:10444–10454. doi: 10.1128/JVI.76.20.10444-10454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JL, Sharma PL, Crumpacker CS. Enhancement of the basal-level activity of HIV-1 long terminal repeat by HIV-1 nucleocapsid protein. Virology. 2000b;268:251–263. doi: 10.1006/viro.2000.0194. [DOI] [PubMed] [Google Scholar]
- Zhang WH, Hwang CK, Hu WS, Gorelick RJ, Pathak VK. Zinc finger domain of murine leukemia virus nucleocapsid protein enhances the rate of viral DNA synthesis in vivo. J. Virol. 2002;76:7473–7484. doi: 10.1128/JVI.76.15.7473-7484.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J. Virol. 1995;69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Qian H, Love Z, Barklis E. Analysis of the assembly function of the human immunodeficiency virus type 1 gag protein nucleocapsid domain. J. Virol. 1998;72:1782–1789. doi: 10.1128/jvi.72.3.1782-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LJ, Mukherjee S, Narayan O. Biochemical mechanism of HIV-I Vpr function. Specific interaction with a cellular protein. J. Biol. Chem. 1994;269:15577–15582. [PubMed] [Google Scholar]