Preface
During metaphase, sister chromatids are positioned at the midpoint of the microtubule-based mitotic spindle in preparation for their segregation. The onset of anaphase triggers inactivation of the key mitotic kinase, cyclin dependent kinase 1 (CDK1), and the polewards movement of sister chromatids. During anaphase, the mitotic spindle reorganizes in preparation for cytokinesis. Kinesin motor proteins and microtubule associated proteins (MAPs) bundle the plus ends of interpolar microtubules and generate the central spindle, which regulates cleavage furrow initiation and completion of cytokinesis. Complementary approaches including cell biology, genetics, and computational modelling have provided new insights into the mechanism and regulation of central spindle assembly.
Introduction
Microtubules are cylindrical polymers assembled from dimers of α and β tubulin. They are polar filaments with a fast growing plus end and a slow growing minus end that is often capped by the γ-tubulin ring complex, a ring-shaped microtubule nucleator 1. Microtubules coordinate a diverse set of biological processes, which include chromosome segregation, spindle positioning and cytokinesis.
To orchestrate these diverse functions, microtubules self-organize into distinct structures. Chromosome segregation is driven by microtubule bundles termed kinetochore fibres in the bipolar mitotic spindle; spindle positioning is mediated by attachment of astral microtubules to cortical sites; and cytokinesis is coordinated, in large part, by the central spindle, an array of antiparallel microtubules that are bundled at their overlapping plus ends. The central spindle, emerges from the mitotic spindle as it elongates during anaphase. The mitotic and central spindles are both bipolar structures assembled from microtubules with overlapping plus ends 2. Despite their similar overall organization, these structures assemble at distinct times during the cell cycle. Are these structures independent from one another, or does the mitotic spindle template central spindle assembly? Are mitotic and central spindles organized by distinct microtubule motors and microtubule binding proteins? Accumulating evidence, reviewed herein, suggests that though the central spindle emerges from the mitotic spindle, distinct factors organize these two structures and they can even assemble a central spindle de novo.
Cytokinesis is mediated by an actomyosin-based contractile ring that assembles on the inner face of the plasma membrane 3,4. Myosin motor activity drives the sliding of actin filaments to constrict the ring and furrow the overlying plasma membrane. The site of contractile ring assembly has to be coordinated with the position of the mitotic spindle to ensure that the two sets of segregated chromosomes are sequestered into the two daughter cells. The central spindle has an important role in this coordination 5–8. In addition, the central spindle is required for the final step of cytokinesis, cell separation or abscission 9–12.
The contractile ring assembles through the coordinated activation of myosin motor activity and actin filament polymerization by the small GTPase RhoA 13–15. The central spindle contributes to the spatial regulation of contractile ring formation by concentrating a key activator of the small GTPase RhoA, namely its guanine nucleotide exchange factor (GEF) ECT2 16–19. However, the requirement for the central spindle for division plane positioning is not absolute as there is a second mechanism for division plane positioning that is controlled by astral microtubules 20–23. The aster dependent pathway involves biased accumulation of contractile components at sites of high microtubule density 22.
Here I will focus on assembly of the central spindle, as its function in cytokinesis has been the subject of several recent reviews 14,24. The structure of the central spindle and the individual and collective functions of the motors and microtubule associated proteins (MAPs) that contribute to the central spindle will be reviewed. I will also briefly summarize some insights into mitotic and central spindle assembly from computational modelling and conclude with a working model of central spindle assembly.
Organization of the central spindle
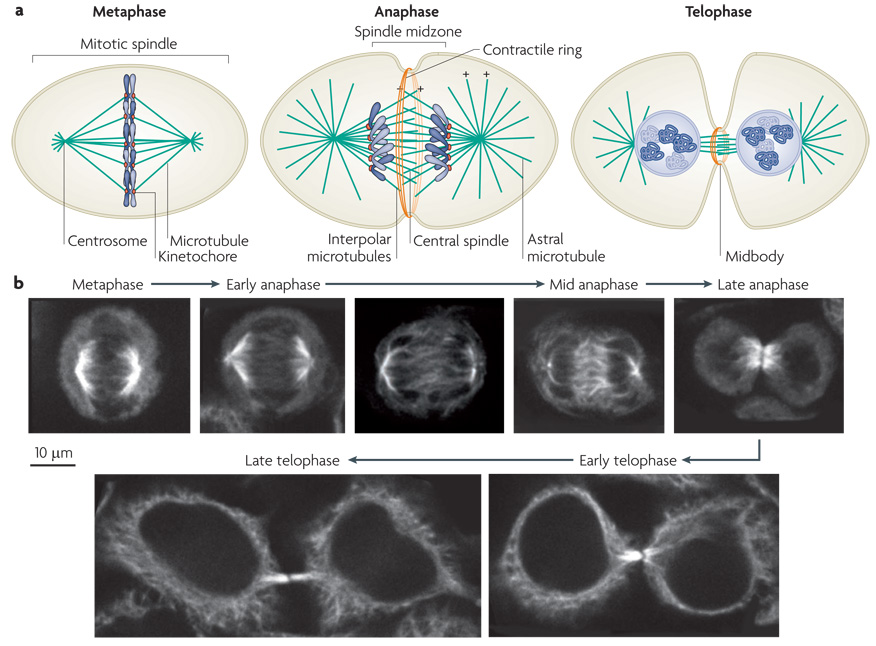
During metaphase, the mitotic spindle is comprised of kinetochore microtubule bundles, astral microtubules and interpolar microtubules (FIG. 1). To a first approximation, the fusiform shape of the spindle is generated by focusing microtubule minus ends at the poles and crosslinking interpolar microtubules in a region of overlap at the midzone. Pole focusing is mediated by the minus-end directed motor protein dynein and the two half spindles are crosslinked by a homotetrameric kinesin-5 motor protein named EG5 25,26.
Figure 1. Assembly of the central spindle.
a| Schematic diagrams of the distribution of microtubules and the chromosomes during cell division. In metaphase the chromosomes align on the metaphase plate. At anaphase, the chromosomes move polewards, the central spindle assembles and contractile ring assembly commences. In telophase, after cleavage furrow ingression, the contractile ring compresses the central spindle to form the midbody. Microtubule plus (+) ends are indicated (minus ends, which are positioned at the centrosomes, are not shown). b| Simulated time course of mitotic exit of a cultured human cell line with microtubules labelled by indirect immunofluorescence. At metaphase, the spindle microtubules position the chromosomes on the metaphase plate. In early anaphase, the chromosomes start to move polewards. At mid anaphase, the chromosomes lie at the poles, the spindle has elongated and spindle midzone microtubules are bundled at their overlapping plus ends. In late anaphase, the chromosomes start decondensing and the cleavage furrow has ingressed significantly. In early telophase, the furrow has fully ingressed and the central spindle is compacted into the midbody. In late telophase, the cytoplasmic bridge has narrowed and the cell is prepared for abscission.
On anaphase onset, the spindle reorganizes in a dramatic fashion. Kinetochore fibres shorten, delivering the sister chromatids toward the poles. Astral microtubules elongate 27,28, and several proteins that are crucial for central spindle assembly relocalize from the cytoplasm and initiate the bundling of antiparallel plus ends of microtubules (FIG. 1). The region between the two poles is called the spindle midzone and the microtubules that populate this region are called midzone microtubules (FIG 1). The term central spindle refers to the structure at the centre of the midzone, where the plus ends of the microtubules interdigitate. Although microtubule minus ends appear to emanate from the spindle pole during early anaphase, the microtubules of the central spindle ultimately lose their interaction with the spindle poles. Furthermore, the ends of the microtubules no longer cluster to a point, the poles are splayed.
As the cleavage furrow ingresses, the central spindle becomes compacted, forming a dense structure known as the midbody or flemming body 29. Electron microscopy indicates that microtubule plus ends overlap for ~2 µm 2; in tubulin immunofluorescence studies, this region of overlap is often obscured by epitope masking 30.The midbody concentrates proteins associated with vesicular transport leading to abscission at a site immediately adjacent to the dense midbody 31–39.
Stability of the central spindle
Mitotic and central spindles differ greatly in the stability of the microtubules contained therein. Precise measurements of microtubule dynamics requires visualization of individual microtubule ends over time so that their history can be tracked. It is not technically possible to make these measurements on bundled microtubules since their ends can not be tracked, moreover, the dynamics of free and bundled microtubules are likely to be quite different, even in the same cell.
The dynamics of bundled microtubules are therefore best assessed with bulk assays, using techniques such as fluorescence recovery after photobleaching (FRAP). Such measurement indicate that microtubules turn over far more rapidly in mitotic spindles during metaphase (t½= 10–20 sec) 40,41 as compared to microtubules in central spindles during anaphase, which turnover slowly (t½ > 2 min) 30. Central spindle microtubules are also resistant to doses of microtubule depolymerizing drugs sufficient to destabilize most astral microtubules, providing additional evidence for their stabilization 23. Although central spindle microtubules are stabilized relative to those of the mitotic spindle, they are not completely inert. There is some polymerization at the central spindle 30,42 and markers of plus end microtubule growth are detectable at this site 43. Since the structure does not grow appreciably, depolymerization must also take place at an equivalent rate so that it maintains a constant size.
Self organization of the central spindle
In an unperturbed anaphase, the central spindle forms through a rearrangement of the mitotic spindle, suggesting that the mitotic spindle may template the assembly of the central spindle. However, functional equivalents of the central spindle can assemble de novo as well as in the absence of prominent components of the mitotic spindle, such as chromosomes and centrosomes. For example, anucleate cells form normal central spindles that contain central spindle components 44, as do regions of overlap between neighbouring spindles 45. In some experimental situations, cell fragments that lack centrosomes and chromosomes or cells that have been treated with microtubule depolymerizing drugs during metaphase, can bundle microtubules into central spindle-like structures that have the capacity to induce furrowing 46,47; central spindle markers have not yet been localised on these bundles, but as in bonafide central spindles, anti-tubulin antibodies do not label the centre of these bundles, suggesting that central spindle components are likely present. These findings suggest that central spindles comprise a self-assembling structure that can arise independent of the bipolar cues normally provided by the preexisting mitotic spindle.
Motors and MAPs of the central spindle
Central spindle assembly is mediated by a set of MAPs, kinesin motor proteins and mitotic kinases. Chief among these components are the MAP protein regulating cytokinesis 1 (PRC1), the centralspindlin complex and the chromosome passenger complex (CPC) (Table 1, FIG. 2). Though these proteins are conserved in animal cells, the nomenclature has not been standardized. For clarity, species specific names have been avoided where possible. Additional components that contribute to central spindle assembly include mitotic kinesin-like protein 2 (MKLP2), M phase phosphoprotein 1(MPP1), orbit (also known as MAST or CLASP), abnormal spindle (Asp, known in humans as ASPM) and CEP55 (centrosome protein 55 kDa). Other factors that play important roles at the central spindle, but are not required for its assembly, include polo like kinase 1 (PLK1), FIP3 and ECT2 among many others.
Table 1.
List of conserved central spindle components
| H. sapiens | C. elegans | D. melanogaster | |
|---|---|---|---|
| Microtubule Associated Protein | |||
| PRC1 | PRC1 48,54 | SPD-1 89 | Fascetto 87 |
| KIF4 | KIF4 55 | KLP-19/KLP-12 120 | KLP3A 121 |
| Centralspindlin complex | |||
| CYK4 | MgcRacGAP/CYK4 57,122 | CYK-4 12 | RacGAP50C/Tumbleweed 16,123 |
| MKLP1 | MKLP159,124 | ZEN-410,11,57 | Pavarotti60 |
| Chromosomal Passenger Complex | |||
| Aurora B | Aurora B125 | AIR-291,126 | Aurora B |
| INCENP | INCENP127 | ICP-192 | Incenp128 |
| survivin | Survivin129 | BIR-1130 | Deterin 131 |
| borealin | Borealin132/Dasra B133 | CSC-1 134 | Borealin 132,135 |
| Additional Factors | |||
| MKLP2 74, MPP1 76, CEP55 34,86, PLK1 136,137, FIP3 138, ECT2 139, Asp 83, orbit 81 | |||
Table 1 Summary of the names of the proteins that have significant roles in central spindle assembly. Where applicable, the names in the first column were used throughout this review.
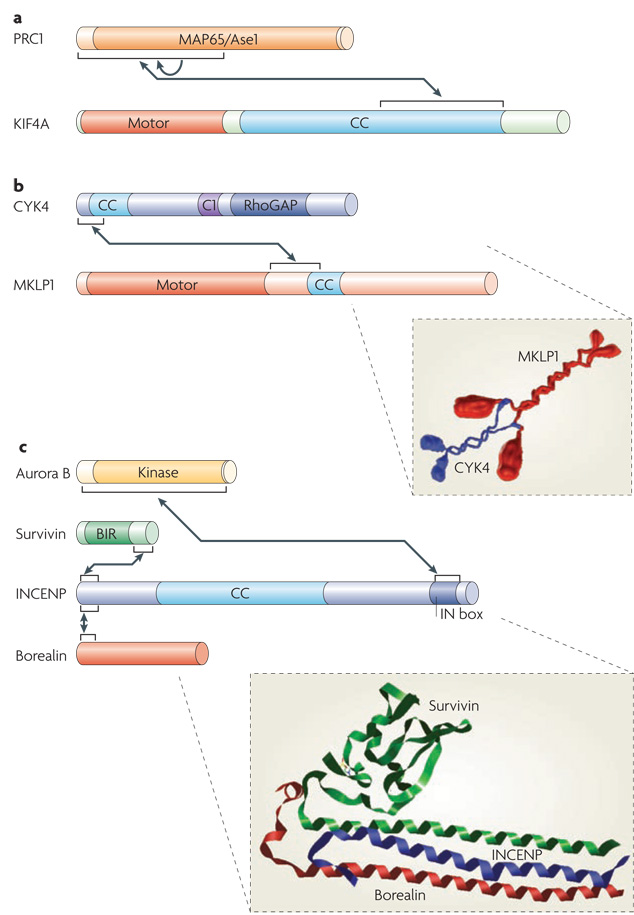
Figure 2. Structural features of central spindle components.
Box diagrams of featured central spindle components. Arrows indicate established protein-protein interactions. a| Protein regulating cytokinesis 1 (PRC1) contains a large central conserved domain ( MAP65/Ase1) that interacts with microtubules. The kinesin-4 motor KIF4 contains an N-terminal motor domain and a large coiled-coil region (CC). The N terminus of PRC1 is required for dimerization (indicated by the arrow) and for interacting with KIF4. b|Centralspindlin is a heterotetramer assembled from the Rho family GTPase activating protein (GAP) CYK4 and mitotic kinesin-like protein 1 (MKLP1) dimers. CYK4 consists of N-terminal coiled-coil, central C1 and C-terminal RhoGAP domains. MKLP1 consists of an N-terminal motor domain, an extended neck linker region and a short coiled-coil region. Both CYK4 and MKLP1 form parallel coiled-coils. Assembly of CYK4 and MKLP1 into centralspindlin is mediated by the N terminus of CYK4 binding to the neck linker region of MKLP1 (see inset). c| The chromosome passenger complex (CPC) is a heterotetramer comprised of Aurora B, survivin, INCENP and borealin. The N-terminal regions of survivin, borealin and INCENP form a three helical bundle. The BIR (Baculoviral inhibition of apoptosis protein repeat) domain of survivin is required for localization to the inner centromere but not the central spindle. The C-terminal IN box of INCENP binds to the kinase domain of Aurora B. Also shown is a structural model of the interacting regions of survivin-borealin-INCENP core complex (see inset) (http://dx.doi.org/10.2210/pdb2qfa/pdb). Protein box diagrams are drawn to scale.
PRC1: a conserved microtubule bundling protein
PRC1 is a highly conserved MAP. Found in metazoans, plants and yeast, PRC1 is involved in cell division in all organisms examined 48–52. PRC1 interacts with microtubules directly and localizes to the central spindle (FIG. 3). In vitro, purified PRC1 bundles microtubules 53. PRC1 contains a conserved central domain, which, when expressed alone induces microtubule bundling and accumulates over much of the spindle 54. However, a larger fragment containing the N-terminal region causes PRC1 to localize much more precisely to the central region of the central spindle 54. This region interacts with the kinesin-4 motor KIF4 and depletion of KIF4 causes PRC1 to localize to a broader region of the central spindle, however, the localization is more restricted as compared to the central domain of PRC1 alone 55. This suggests that the N terminus of PRC1 contains additional functionality beyond KIF4 binding. Indeed, the N terminus of PRC1 also contains a domain that mediates oligomerization 53. The budding and fission yeast orthologues of PRC1, Ase1, also localize to microtubules and promote their bundling with a preference for antiparallel microtubules 49,56. This activity is independent of a kinesin-4 motor, since this class of motors is not represented in either yeast genome.
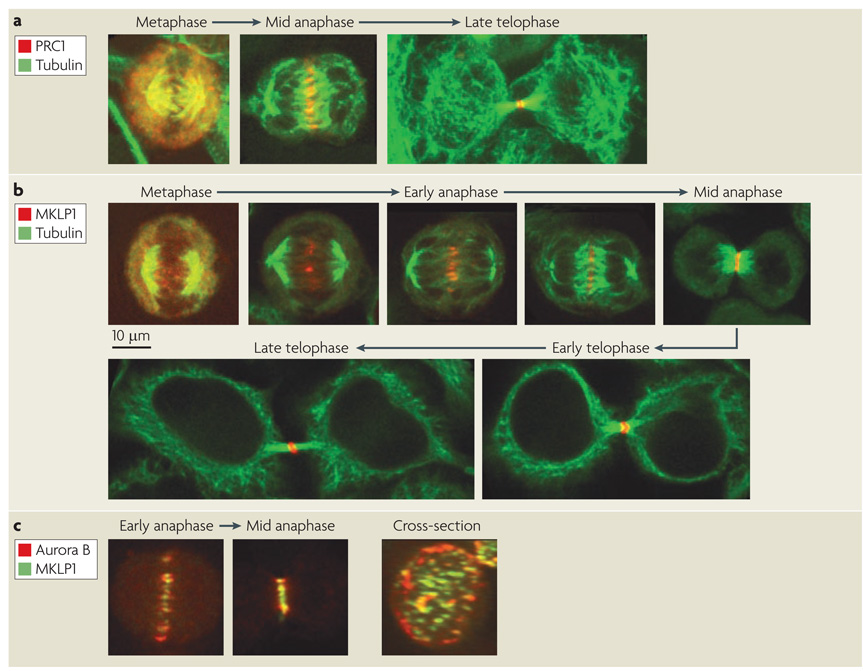
Figure 3. Localization of central spindle components.
Simulated time course of mitotic exit of a cultured human cell line with microtubules and central spindle components labelled by indirect immunofluorescence. a| Upon anaphase onset, protein regulating cytokinesis 1 (PRC1), localizes to the overlap zone on the ends of midzone microtubules and becomes compacted during furrow ingression to form the midbody. b | The localization of mitotic kinesin-like protein 1 (MKLP1), a subunit of centralspindlin, during mitotic exit. In metaphase, centralspindlin weakly associates with the spindle. Upon initiation of chromosome segregation, MKLP1 begins to concentrate where microtubule plus ends overlap. As anaphase proceeds, MKLP1 concentrates further and its localization becomes more restricted to the very center of the central spindle. In late telophase, centralspindlin reorganizes into a ring, surrounding the midbody. c| A comparison of the distribution of MKLP1 and the chromosome passenger complex (CPC) component Aurora B demonstrates that MKLP1 localizes more discretely on the central spindle as compared to Aurora B. The cross section through the central spindle shows the prominent cortical association of the CPC, with centralspindlin and CPC labelled microtubule bundles in the centre.
Centralspindlin: an unusual motor complex
A second important component for central spindle assembly is the centralspindlin complex. Centralspindlin is a tetrameric complex consisting of a dimer of the kinesin-6 motor protein MKLP1 bound to a dimer of the Rho family GTPase activating protein (GAP) CYK4 (also known as MgcRacGAP) 57,58. This complex localizes to the centre of the central spindle (Fig. 3) 10,11,57,59,60 where it promotes central spindle microtubule bundling, RhoA regulation and serves to recruit regulators of abscission 14,61. Neither CYK4 nor MKLP1 can localize in the absence of the other protein, only intact centralspindlin localizes 57. Similarly, the complex, but not the individual subunits are sufficient to promote microtubule bundling in vitro 57.
Centralspindlin function requires a high affinity interaction between CYK4 and MKLP1 58. The interface is created by an N-terminal domain in CYK4 and a ~85 residue interaction domain in MKLP1 that lies in the linker region C-terminal to the motor domain (FIG. 2). Although the interaction between these proteins is evolutionarily conserved, the sequences that mediate their interactions are not. Moreover, mutations that destabilize this interaction can be readily suppressed by a series of second site mutations, indicating a high degree of plasticity in this interaction interface 57,58. The 85 residue CYK4 interaction domain in MKLP1 starkly contrasts with the corresponding region in the majority of kinesin motor proteins. In most kinesins, the linker between the motor domain and the coiled-coil consists of 13–15 amino acids and is highly conserved 62. In kinesin-1 motors, this domain docks against the motor domain in a nucleotide sensitive manner and contributes to their plus end directed motility 63,64. CYK4 binding to this domain is essential for central spindle assembly 57,58, but the structural consequences are not yet understood in mechanistic detail. Attractive possibilities include a conformational change in MKLP1 that promotes binding to antiparallel microtubules and/or CYK4 directly participating in microtubule binding.
The CPC targets aurora B kinase to the central spindle
A third crucial component in central spindle assembly is the CPC that contains Aurora B kinase as a catalytic subunit (FIG. 2). The CPC is a multisubunit complex that consists of a triple helical bundle containing strands contributed by INCENP, survivin (also known as BIR1) and borealin (also known as CSC1) 65. A second domain in INCENP binds to and activates the kinase activity of Aurora B 66. This set of four proteins is active throughout mitosis, acting on chromosomes during metaphase and the central spindle during anaphase. The evocative name ‘chromosomal passenger complex’ derives from the fact that this complex concentrates at inner centromeres in the middle of the spindle during metaphase and then, during anaphase, it remains at a similar location in the cell - but at this time it associates with the central spindle and the cell cortex, as if it were delivered there by the chromosomes 67 (FIG. 3). However the CPC concentrates on the central spindle in cells that lack chromosomes 44. Furthermore, a specific survivin mutant can not localize to centromeres, but can localize to the central spindle 68. Thus, central spindle recruitment of the CPC is independent of its prior presence on chromosomes.
Although the CPC phosphorylates several central spindle components 69–71, it may also be directly involved in microtubule bundling. The N-terminal 42 residues of INCENP are required for its interaction with the central spindle 72, but this likely reflects a requirement for this region to bind to other subunits of the CPC 65. The localization of an N terminally deleted INCENP can be rescued by fusion of survivin to the remainder of INCENP, moreover this survivin-INCENP fusion can localize in the absence of borealin 68. Since neither INCENP nor survivin localize individually, these two factors must localize through a cooperative mechanism. INCENP also contains a tubulin binding domain 73, thus INCENP may be a structural component of the central spindle in addition to its role in activating and localizing Aurora B kinase.
Additional central spindle MAPs and motors
Whereas PRC1, centralspindlin and the CPC are the best characterized components of the central spindle, they are far from the only MAPs and motors that concentrate on this site. Other kinesins that are enriched on the central spindle include KIF4, MKLP2 and MPP1. As discussed above, KIF4 regulates PRC1 function 53,55. Like MKLP1, MKLP2 and MPP1 are kinesin-6 family members, and they are both required for late steps in cytokinesis 74–76. So far, only MKLP2 has defined functions at the central spindle, promoting the accumulation of Aurora B and PLK1 77,78. Interestingly, MKLP2 and MPP1 are not widely represented in sequenced genomes (MKLP2 being found in more genomes than MPP1). Caenorhabditis elegans has a sole kinesin-6 family member, an MKLP1 orthologue; Drosophila melanogaster has an MKLP2 orthologue, Subito, in addition to the MKLP1 orthologue Pavarotti 79. At this juncture, it is unclear why additional members of this kinesin family are required in some cell types but not all. Although MKLP1 and MKLP2 are highly related, MKLP2 is not known to have a stoichiometric binding partner comparable to CYK4 that binds to its neck linker region and facilitates its proper localization, thus the two motors are structurally and functionally distinct despite being paralogues.
Another MAP that appears to contribute to central spindle assembly is a microtubule plus-end binding protein, variously known as orbit, Clasp or Mast. This protein localizes to the centre of the central spindle and the midbody 80. Orbit has a crucial role in kinetochore-microtubule attachments and these earlier requirements make it difficult to study its role in cytokinesis. However, a hypomorphic mutation in orbit has been isolated in D. melanogaster. This allele does not severely compromise chromosome segregation, but it does cause penetrant defects in central spindle assembly 81. Further analysis of orbit’s role in central spindle assembly is clearly warranted.
As mentioned above, during telophase, central spindle microtubules appear to lose their attachment to the spindle poles. Little is known about what triggers this transition. However, there is some insight into how the released minus ends may be stabilized. An intriguing, evolutionarily conserved MAP, Asp, was first identified in D. melanogaster and named for its phenotype characterized by abnormal spindle poles 82. In D. melanogaster, Asp is highly concentrated at centrosomes for much of the cell cycle, but, during anaphase, it concentrates to the flanking regions of the central spindle 83. Although mutations in D. melanogaster are lethal, at least some humans lack the function of this gene, known as Aspm, and they are microcephalic 84. The viability of these individuals suggests that Asp is functionally redundant in most tissues except the brain. In D. melanogaster, loss of Asp causes disorganization of the central spindle and many central spindle factors are not properly localized 83. Asp may stabilize microtubule minus ends and could, additionally, contribute to nucleation of additional microtubules in the central spindle 85.
Finally, a direct interaction partner of MKLP1, CEP55, also concentrates on the central spindle and the midbody 34,86. CEP55 orthologues are readily identified in vertebrates but not in invertebrates. CEP55 is of significant interest because it directly mediates the recruitment of Tsg101, an endosomal sorting complex required for transport (ESCRT-I) subunit and the ESCRT-associated protein Alix. These factors are required for viral budding, which is topologically similar to membrane resolution during abscission 35,36. Their concentration at the midbody and their established function in regulating membrane topology suggests that they might have a similar role during cytokinesis. Indeed, CEP55, Alix and Tsg101 are required for abscission.
Complex interactions among central spindle components
Although several of the MAPs and motors described above are sufficient to bind and/or bundle microtubules in vitro, no single component is sufficient for central spindle assembly in vivo and the behaviour of many of these proteins are highly intertwined.
PRC1, centralspindlin and the CPC comprise a core set of interdependent factors involved in central spindle assembly. Absence of any of these factors significantly impacts the localization of the others, and, as a consequence, delocalization of the majority of peripheral components of the central spindle. Loss of PRC1 orthologues disturbs, but does not abolish, the localization of centralspindlin and the CPC. Centralspindlin still associates with the central spindle but fails to become highly concentrated at its centre 55,87. The CPC primarily associates with the cell cortex under these conditions and Asp also becomes delocalized 87,88. Loss of PRC1 in human cells causes the bipolar central spindle to split into two half spindles and often results in cytokinesis failure. However, in other cells such as in C. elegans embryos, inactivation of the PRC1 orthologue, SPD-1, does not invariably prevent cytokinesis 89. Although SPD-1 defective cells have highly disorganized central spindles, the residual structure is sufficient to permit completion of cytokinesis in many, but not all, cells of the embryo. As another example, some tissues in Xenopus laevis embryos express PRC1 at low levels and this causes their spindles to hyperelongate during anaphase and delays central spindle assembly 90.
Cells lacking centralspindlin or the CPC have profound defects in central spindle assembly. In C. elegans embryos and D. melanogaster cells, there is little if any microtubule bundling and PRC1 localization is greatly perturbed under such circumstances 10–12,60,89. Although both subunits of centralspindlin are CPC targets, there is no evidence yet that these phosphorylation events are required for central spindle assembly 69–71. In centralspindlin-depleted cells, the CPC associates with spindle microtubules at reduced levels 12. Conversely, centralspindlin does not stably localize in cells depleted of the CPC 91,92, perhaps explaining the requirement for the CPC in this process. Vertebrate cells depleted of MKLP1 retain the ability to recruit MKLP2 and the CPC 77, but numerous other central spindle factors fail to accumulate such as CEP55, the RhoGEF ECT2 and the endocytic protein FIP3, which is important for abscission 17–19,34,93. In summary, loss of centralspindlin or the CPC greatly inhibits central spindle assembly and prevents cytokinesis.
Other central spindle proteins are less crucial for the integrity of the structure itself, though many are essential for cytokinesis. For example, CEP55 is directly recruited by centralspindlin and serves to recruit additional factors for abscission 34. In CEP55 depleted cells, many midbody components (centralspindlin, PRC1, Aurora B, MKLP2) localize properly, at least initially. However, the prominent bulge in the cytoplasmic bridge, the Flemming body, is absent in CEP55-depleted cells and these cells are abscission defective. Likewise, the kinase PLK1, is an important regulator of cytokinesis that localizes to the central spindle, but it too is dispensable for central spindle assembly 94–97. PLK1 is recruited by PRC1 78 and, to a lesser extent, MKLP2 75. MKLP2 presents a puzzling case. Although MKLP2 depletion delocalizes the CPC, it does not dramatically affect PRC1 or MKLP1 localization, which are ordinarily CPC dependent for their localization 71. Thus, Aurora B must be functional without being highly localized, suggesting that the CPC acts catalytically at the central spindle, rather than structurally. As the CPC is recruited to the central spindle in organisms that lack a MKLP2 orthologue, there may be additional mechanisms for CPC recruitment. In sum, CEP55, PLK1 and MKLP2 have important roles at the central spindle, but they are not strictly required for its assembly.
Temporal regulation of spindle assembly
The mitotic spindle begins to assemble in prometaphase and it persists through metaphase, when the major mitotic kinase, cyclin dependent kinase 1 (CDK1) is highly active. In contrast, the central spindle assembles during anaphase, as CDK1 levels decline. Although these structures are related to one another, the proteins that control their assembly are largely non-overlapping. A number of these differences can be ascribed to the fact that the mitotic and central spindles are present during distinct cell cycle stages. For example, the tetrameric kinesin-5 motor EG5 has a crucial role in crosslinking microtubules during metaphase. Localization of EG5 to the mitotic spindle during metaphase requires phosphorylation of a single CDK1 site in the C terminus 98,99. Although this site would be predicted to be dephosphorylated during anaphase, this motor associates with the anaphase spindle, even after CDK1 has been inactivated 99. Perhaps spindle binding inhibits its dephosphorylation. Although EG5 remains associated with the spindle during anaphase and slows spindle elongation 100, inhibition of EG5 after anaphase onset does not perturb cytokinesis 26. Thus, one of the key factors that is required for mitotic spindle assembly is dispensable for central spindle assembly.
Conversely, the motors and MAPs that regulate central spindle assembly do not participate in mitotic spindle assembly. Many crucial central spindle components are inhibited prior to anaphase. For example, the centralspindlin complex is phosphorylated on a set of CDK1 sites that destabilize its interaction with microtubules and this complex is largely cytoplasmic during metaphase 101. Similarly, PRC1 is phosphorylated by CDK1 and this phosphorylation also reduces the efficiency with which it binds to the spindle and the extent to which it recruits PLK1 54,102. Finally, CDK1 inactivation is required for the CPC to bind to the central spindle, perhaps because it remains on chromosomes 103. Upon anaphase onset, Cyclins are degraded, CDK1 becomes inactive, and these inhibitory sites are dephosphorylated allowing central spindle assembly to commence. Thus, mitotic and central spindle assembly are mutually exclusive.
Modelling spindle assembly
How does a bipolar structure with antiparallel microtubules assemble? Computational modelling has emerged as an important discipline to answer questions of this ilk. Many individual aspects of spindle assembly have been modelled, including pole formation, establishing antiparallel microtubule overlap and the balancing of the mechanical forces generated by microtubule dynamics and microtubule motors within and external to the spindle 104. These models contribute to the overall understanding of spindle assembly, and they also shed light on the diversity of forms that microtubules, MAPs and motor proteins can create. However, a comprehensive model of the mitotic spindle remains unrealized. Nevertheless, these models are somewhat generic and many of their conclusions are also applicable to the central spindle. Progress made in this theoretical vein is useful for hypothesis development and to guide experimental design and interpretation.
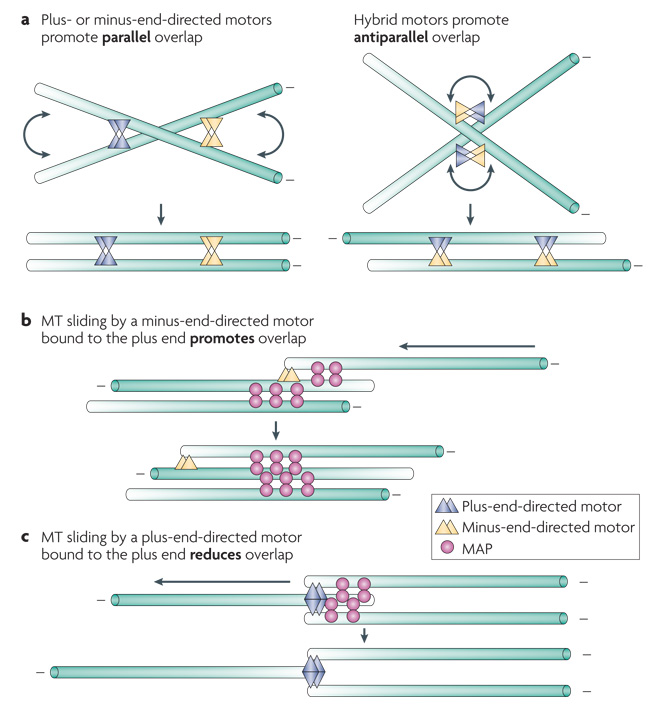
Computational modelling has been used to investigate how stable antiparallel microtubule overlap can arise. One approach is to model a minimal system consisting of dynamic microtubules and motors or MAPs. Specifically, multimeric motor proteins and motor protein/MAP fusions were compared for their ability to generate stable microtubule overlap (FIG. 4a). In this model, only a hybrid motor that contains both plus and minus end directed motors could give rise to stable antiparallel microtubule bundles (FIG. 4b) 105. The presence of motors of both directionalities was not sufficient: the two motors had to be physically connected. Although demonstrating that formation of a stable overlap zone requires coupling of microtubule motors that exert counter balancing forces is of significant interest, no motor with these properties is known to be involved in mitotic or central spindle assembly.
Figure 4. Microtubule bundling mechanisms.
a|Plus and minus end directed motor proteins primarily promote parallel microtubule bundling, but a hybrid motor can generate stable antiparallel overlap. b|Stable antiparallel overlap can be generated by a combination of a dimeric microtubule associated protein (MAP) and a minus end directed motor that stably associates with microtubule plus ends. c| In contrast, a plus end directed motor that stably associates with microtubule plus ends would reduce microtubule overlap. The polarity of microtubule indicated by the gradient.
One parameter that impacts these simulations is the residency time of the motor at the end of a microtubule. Hybrid motors that dissociate immediately from microtubule ends only create microtubule bundles that overlap for their entire length, whereas if the motors persist for a time with a microtubule end, the microtubule bundles can overlap less extensively. Interestingly, and more physiologically, antiparallel microtubule bundling could also be achieved through the combined action of a specialized microtubule motor and a MAP. Together with a MAP that binds preferentially to antiparallel microtubules, a motor that associates with the plus end of a microtubule and moves processively towards the minus end of a second microtubule can generate microtubule bundles with overlapping minus ends (FIG. 4c).
The properties of these molecules were selected to mimic the behaviour of the motors and MAPs that mediate the organization of microtubules in Schizosaccharomyces pombe (Box 1). Formation of microtubule bundles in S. pombe requires the PRC1 orthologue, Ase1, that preferentially bundles antiparallel microtubules and a kinesin-14 type motor that associates with the plus end of a microtubule, perhaps via a microtubule plus end tracking protein such as EB1 106 or a non catalytic, motor-like accessory protein 107, and moves processively towards the minus end of a second microtubule, allowing it to slide two microtubules relative to each other 56.
Box 1 Microtubule organization in Schizosaccharomyces pombe
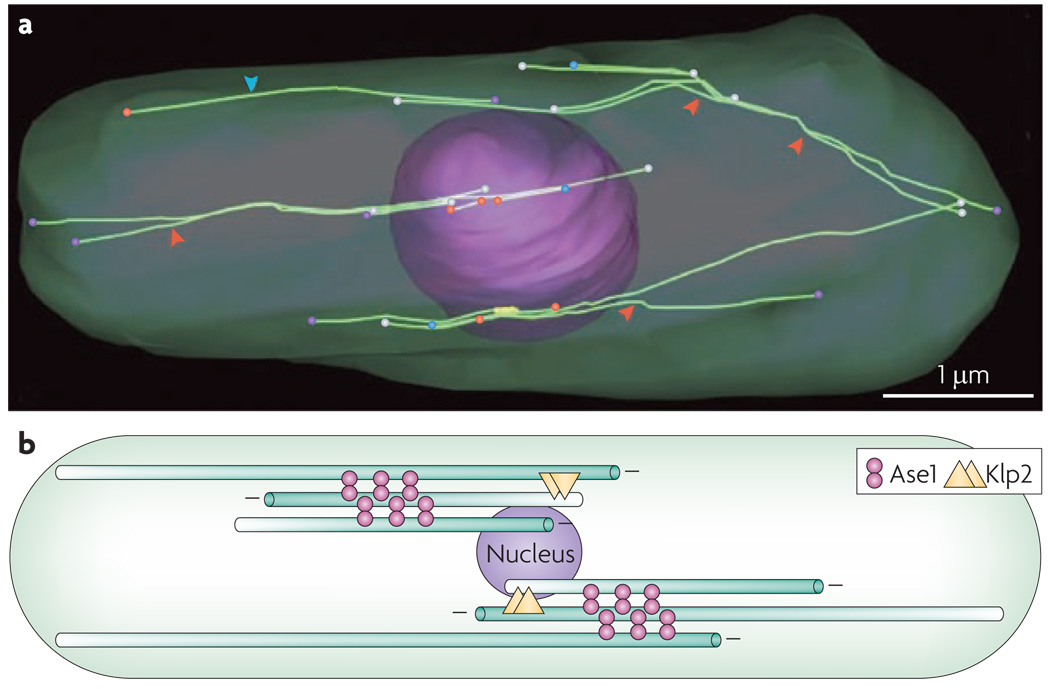
The fission yeast Schizosaccharomyces pombe provides an informative and complementary system to analyse organization of microtubule-based structures. This system is highly genetically and cytologically tractable, the main microtubule bundling proteins have been identified and entire cells have been reconstructed by electron tomography, revealing the detailed organization of its entire microtubule cytoskeleton 114. Interphase cells contain ~3–4 bundles of 4–5 microtubules 114–116. In this reconstruction, the nucleus is shown together with the entire complement of 16 microtubules (see figure panel a). The microtubules are extensively bundled, sites where bundles splay apart are indicate by red arrowheads. Microtubules have varied end structures, as indicated by coloured circles [capped (red); flared (turquoise); open sheet (yellow); blunt (blue); other (white)]. The arrangement of microtubules is schematized in panel b. The minus ends of the microtubules typically, but not invariably, lie near the nucleus (microtubules are shaded to indicate their polarity). The majority of microtubules are bundled and separated by crossbridges of 25–30 nm (double spheres), likely corresponding to the protein regulating cytokinesis 1 (PRC1) orthologue Ase1 114. Unlike the central spindle, the microtubules in these bundles overlap extensively near their minus ends. Although the bundles have a distinct organization from the central spindle, some of the components involved are orthologues, in particular Ase1. This organization also requires a kinesin motor which in this case is the kinesin-14 family member Klp2 117, which stably interacts with the plus end of one microtubule and slides it along an adjacent microtubule via its minus end-directed motor domain, thereby increasing the extent of microtubule overlap 56. These microtubule bundling and sliding activities are self organizing structures that can assemble in enucleated cells without the nucleus or the spindle pole body 118,119. The scale bar is 1 µm.
However, one can not generate the overlapping plus end organization of the central spindle simply by substituting a plus end-directed motor for the minus end directed motor in this system. This type of motor, located at a microtubule plus end, would induce an adjacent microtubule to slide past its end, thereby eliminating the overlap (FIG. 4d). However, factors that prevent the plus end directed motors from reaching the extreme plus end of the microtubule could, in principle, prevent complete separation and allow such motors to participate in microtubule bundling.
A second important consideration in modelling spindles is the dynamics of the constituent microtubules. Computational models permit facile exploration of how microtubule dynamics impact spindle assembly. For example, models reveal that more stable microtubules induce the assembly of longer spindles, even if the microtubules do not span the entire distance from the pole to the midzone; this has been confirmed experimentally 108,109. Not only will global changes in microtubule dynamics modulate the spindle, it is likely that microtubule dynamics are non-uniform within the spindle. Computational modelling indicates that non-uniform microtubule dynamics can alter spindle morphology 110. Significant differences in microtubule behaviour have been observed in D. melanogaster embryos before and after anaphase 111. These data could be recapitulated in a computational model involving spatial regulation of microtubule dynamics. In particular, increasing the length of microtubules is predicted to increase the extent of overlap 108. The observations converge with the experimental evidence indicating that central spindle microtubules are significantly more stable than the astral microtubules 23,30. Locally regulation of microtubule dynamics could even be a primary function of some central spindle components.
Working model for central spindle assembly
Based on the considerations above, a speculative outline of central spindle assembly can be proposed (FIG. 5). Upon anaphase onset, the mitotic factors that promote microtubule catastrophe and cross-linking of half spindles are down regulated, causing growth of astral and interpolar microtubules, which in turn induces spindle elongation. In parallel, central spindle assembly factors such as PRC1 and centralspindlin are relieved from mitotic inhibition and the CPC is released from centromeres. Antiparallel microtubules are bundled, primarily at their plus ends, by PRC1 dimers transported to the plus-end by the motor KIF4. Coordinately, the plus end directed motility of the kinesin subunit causes centralspindlin to concentrate at microtubule plus ends. Due to the presence of the CYK4 subunit, the centralspindlin complex may preferentially bind to antiparallel microtubules. The CPC could promote retention of centralspindlin on the microtubules, perhaps by stabilizing centralspindlin at microtubule plus ends. The combined presence of PRC1, centralspindlin and the CPC induces robust bundling of the microtubules, greatly stabilizing them. The high concentration of centralspindlin also serves as a direct docking site for additional cytokinetic regulators such as ECT2, which induces local activation of RhoA 17–19,112, and CEP55 and FIP3, which subsequently promote abscission 34,93. One important feature lacking from this model is an explanation for why the plus-end directed motility of centralspindlin would not drive the two half spindles apart. Is there a counteracting force that prevents the two half spindles from sliding apart? Are centralspindlin-mediated forces not sufficiently strong to disrupt the structure?
Figure 5. Working model for central spindle assembly.
A working model for conversion of a mitotic spindle to a central spindle. Overview is shown on the left, a detail of the overlap region on the right. During metaphase, short highly dynamic microtubules emanate from the centrosome. Factors such as phosphorylated homotetrameric kinesin-5 motor protein EG5 associates with spindle microtubules during metaphase and promote separation of spindle poles. Central spindle microtubule bundling factors are phosphorylated during metaphase and do not stably associate with the spindle. Upon anaphase onset, chromosomes move polewards and central spindle bundling factors become associated with the spindle. Due to plus end directed motility of the associated motor proteins (arrows), these factors move centrifugally and the cooperative action of these factors stabilize their association with overlapping microtubule plus ends. By late anaphase, the chromosomes have reached the poles and central spindle factors are highly concentrated at the central spindle. The presence of the bundling factors stabilizes the midzone microtubules. During telophase, chromosomes decondense, nuclear envelopes reform, the central spindle becomes increasingly ordered and the minus ends of central spindle microtubules lose their association with centrosomes.
One distinguishing feature of mitotic and central spindles is the shape of their poles. Mitotic spindles have focused poles whereas the poles of central spindles are largely frayed. Dynein inhibition causes fraying of the poles of the mitotic spindle 25. The findings suggest that dynein may not be active at central spindle poles. As dynein is not globally inactivated during anaphase, its activity could be spatially regulated. Whereas during metaphase, the minus ends of microtubules focus at the centrosome, during late anaphase, microtubules appear to dissociate from the centrosome. Several MAPs, which include Asp/Aspm, accumulate on these minus ends, presumably capping and stabilizing them; they may also inhibit dynein accessibility.
Informative perturbations
In unperturbed cells, central spindle components concentrate dramatically at the centre of the spindle where the microtubules are antiparallel. Some interesting experimental cases suggest that enrichment at overlapping antiparallel ends may reflect a binding site preference as opposed to an absolute requirement. Monopolar anaphase spindles can be generated by treating cells sequentially with a chemical inhibitor of EG5 followed by a CDK1 inhibitor 113. These cells accumulate MKLP1 and other central spindle components near clusters of plus ends of bundled microtubules 113. Interestingly, in these spindles or in the half spindles that result from PRC1 depletion 55, the CPC lies distal to MKLP1, suggesting that, perhaps, in unperturbed central spindles, the CPC that concentrates in each ‘half central spindle’ may in fact be positioned by microtubule plus ends in the opposite ‘half central spindle’. It is unclear why these components concentrate on a subset of microtubules of a monopolar spindle, nor whether their accumulation at this site reflects the same requirements and dynamics that allow them to concentrate in the central spindle. It is conceivable that these bundles may contain a few antiparallel microtubules. Alternatively, these components may bundle parallel microtubules, when antiparallel ones are absent.
Several possibilities, not mutually exclusive, could explain the preference of PRC1 and centralspindlin for binding to antiparallel microtubules. First, overlapping plus ends may preferentially exploit an intrinsic symmetry of these molecules. Second, the motor proteins in these complexes may travel along microtubules towards plus ends and rapidly fall off the ends, except at sites with overlapping plus ends. At these sites, they could cluster due to directed motility in both directions along the two sets of microtubules. Finally, these complexes could bind to a factor, as yet unidentified, that concentrates on these microtubules through one of these mechanisms.
Concluding remarks
The remarkable finding that clusters of beads coated with random DNA in concentrated mitotic cell extracts can nucleate the assembly of a beautiful mitotic spindle demonstrated that microtubules, motors and MAPs can self organize into complex supramolecular structures 25. Similar principles mediated by a different set of motors and MAPs organize the central spindle during anaphase. In an unperturbed dividing cell, this structure uniquely defines a plane that lies between the segregating chromosomes and is therefore the optimal position for the plane of cell division. Thus not only do these motors self assemble into a spectacular variant of a spindle-like structure, but they also create a signalling centre that initiates cytokinesis and subsequently mediates its completion.
Though central spindle assembly is understood at the conceptual level, large gaps in our understanding persist. In particular, further insight is needed to determine the structural and biophysical features that enable certain motors and MAPs to preferentially accumulate at sites of overlapping antiparallel microtubules. In addition, it will be important to determine whether central spindle motors continually generate force while concentrated at the central spindle and, if so, to identify the molecules that produce the counteracting forces that prevent spindle collapse. In addition, numerous biochemical questions remain, which include the mechanistic analysis of the roles of the CPC and CYK4 in central spindle assembly. Finally, it will be important to understand how the central spindle recruits accessory factors that regulate cytokinesis at appropriate times and how the entire structure ultimately disassembles upon completion of cytokinesis.
Online Summary
The central spindle consists of a set of microtubule bundles in anaphase cells that overlap for a short region at their plus ends.
The central spindle consists of a set of microtubule bundles in anaphase cells that overlap for a short region at their plus ends.
The central spindle regulates cleavage furrow formation and completion of cytokinesis.
The central spindle forms in anaphase as cells exit mitosis. In unperturbed cells, the central spindle forms from mitotic spindle microtubules.
Under appropriate conditions, a bipolar central spindle can form spontaneously from non-spindle microtubules, without a mitotic spindle template.
Central spindle microtubule bundles are highly stabilized.
Formation of the central spindle requires kinesin motor proteins, microtubule associated proteins (MAPs) and protein kinases. The central players include centralspindlin (a complex containing kinesin and RhoGAP subunits), the microtubule bundling protein PRC1 and the chromosome passenger complex (CPC).
Several of the proteins required for central spindle assembly are inactivated by phosphorylation during metaphase and activated during anaphase.
The precise mechanism of microtubule bundling that results in overlapping microtubule plus ends remains be determined.
Models of the interactions of motors, MAPs and microtubules provide useful insights into how stable microtubule overlap can be established and suggest that local regulation of microtubule dynamics may play and important role.
Box 1.
Acknowledgments
I would like to thank B. Wolfe, E. White and M. Mishima for comments on the manuscript and Z. Thakkar for assistance in producing the micrographs. I thank C. Antony and J. Höög for allowing the use of their figure. The author is supported by Award Number R01GM085087 from the National Institute of General Medical Sciences (M.G. is solely responsible for its content).
Glossary
- cytokinesis
The process by which a single cell divides into two physically distinct daughter cells.
- coiled-coil domain
A protein structural domain that mediates subunit oligomerization. Coiled-coils contain between two and five helices that twist around each other.
- kinetochore
The proteinaceous structure that serves as a physical link between microtubules and the chromatin during mitosis.
- mitotic spindle
The supramolecular structure comprised of microtubules, chromosomes, motor proteins, MAPs, etc. that is responsible for segregating chromosomes during mitosis.
- astral microtubule
Microtubules that emanate radially from the centrosome during metaphase and anaphase.
- abscission
The process that results in severing of the cytoplasmic bridge, finally separating the two daughter cells.
- interpolar microtubules
Microtubules that emanate from one spindle pole and bundle with microtubules emanating from the opposite pole.
- midbody
The highly compacted structure at the centre of the cytoplasmic bridge between two nascent daughter cells.
- FRAP (Fluorescence recovery after photobleaching)
An imaging technique in which a subset of fluorescent molecules are rendered non fluorescent by intense illumination. The time course of fluorescence recovery reflects the rate at which molecules exchange.
- centrosome
The structure enriched in gamma tubulin that nucleates and organizes microtubule minus ends. Often contains a pair of centrioles.
Biography
Biography Originally from Boston, Michael Glotzer began to study the cell cycle during his graduate studies with Marc Kirschner at UCSF. He began to focus on cytokinesis during a postdoc at the EMBL in Heidelberg. He started his lab at the IMP in Vienna, Austria and after eight years, moved to the University of Chicago, Chicago, IL. His major scientific interests are central spindle assembly, regulation of Rho family GTPases and the mechanism of division plane positioning.
Footnotes
Cytosim - Francois Nedelec's microtubule dynamics simulation program
References
- 1.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 2.Mastronarde DN, McDonald KL, Ding R, McIntosh JR. Interpolar spindle microtubules in PTK cells. J Cell Biol. 1993;123 doi: 10.1083/jcb.123.6.1475. 1475–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schroeder TE. Cytokinesis: filaments in the cleavage furrow. Exp Cell Res. 1968;53:272–276. doi: 10.1016/0014-4827(68)90373-x. [DOI] [PubMed] [Google Scholar]
- 4.Schroeder TE. Actin in dividing cells: contractile ring filaments bind heavy meromyosin. Proc Natl Acad Sci U S A. 1973;70 doi: 10.1073/pnas.70.6.1688. 1688–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mabuchi I, Okuno M. The effect of myosin antibody on the division of starfish blastomeres. J Cell Biol. 1977;74:251–263. doi: 10.1083/jcb.74.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knecht DA, Loomis WF. Antisense RNA inactivation of myosin heavy chain gene expression in Dictyostelium discoideum. Science. 1987;236:1081–1086. doi: 10.1126/science.3576221. [DOI] [PubMed] [Google Scholar]
- 7.De Lozanne A, Spudich JA. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- 8.Straight AF, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 9.Wheatley SP, Wang Y. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J Cell Biol. 1996;135:981–989. doi: 10.1083/jcb.135.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powers J, Bossinger O, Rose D, Strome S, Saxton W. A nematode kinesin required for cleavage furrow advancement. Curr Biol. 1998;8:1133–1136. doi: 10.1016/s0960-9822(98)70470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raich WB, Moran AN, Rothman JH, Hardin J. Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol Biol Cell. 1998;9:2037–2049. doi: 10.1091/mbc.9.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jantsch-Plunger V, et al. CYK-4: A Rho family GTPase activating protein (GAP) required for central spindle formation and cytokinesis. J Cell Biol. 2000;149:1391–1404. doi: 10.1083/jcb.149.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eggert US, Mitchison TJ, Field CM. Animal cytokinesis: from parts list to mechanisms. Annu Rev Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- 14.Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 15.Werner M, Glotzer M. Control of cortical contractility during cytokinesis. Biochem Soc Trans. 2008;36:371–377. doi: 10.1042/BST0360371. [DOI] [PubMed] [Google Scholar]
- 16.Somers WG, Saint RA. RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev Cell. 2003;4:29–39. doi: 10.1016/s1534-5807(02)00402-1. [DOI] [PubMed] [Google Scholar]
- 17.Yuce O, Piekny A, Glotzer M. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J Cell Biol. 2005;170:571–582. doi: 10.1083/jcb.200501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimura Y, Yonemura S. Centralspindlin regulates ECT2 and RhoA accumulation at the equatorial cortex during cytokinesis. J Cell Sci. 2006;119:104–114. doi: 10.1242/jcs.02737. [DOI] [PubMed] [Google Scholar]
- 19.Kamijo K, et al. Dissecting the role of Rho-mediated signaling in contractile ring formation. Mol Biol Cell. 2006;17:43–55. doi: 10.1091/mbc.E05-06-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dechant R, Glotzer M. Centrosome separation and central spindle assembly act in redundant pathways that regulate microtubule density and trigger cleavage furrow formation. Dev Cell. 2003;4:333–344. doi: 10.1016/s1534-5807(03)00057-1. [DOI] [PubMed] [Google Scholar]
- 21.Bringmann H, Hyman AA. A cytokinesis furrow is positioned by two consecutive signals. Nature. 2005;436:731–734. doi: 10.1038/nature03823. [DOI] [PubMed] [Google Scholar]
- 22.Werner M, Munro E, Glotzer M. Astral signals spatially bias cortical myosin recruitment to break symmetry and promote cytokinesis. Curr Biol. 2007;17:1286–1297. doi: 10.1016/j.cub.2007.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murthy K, Wadsworth P. Dual role for microtubules in regulating cortical contractility during cytokinesis. J Cell Sci. 2008 doi: 10.1242/jcs.027052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piekny A, Werner M, Glotzer M. Cytokinesis: welcome to the Rho zone. Trends Cell Biol. 2005;15:651–658. doi: 10.1016/j.tcb.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Heald R, et al. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–445. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 26.Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tournebize R, et al. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat Cell Biol. 2000;2:13–19. doi: 10.1038/71330. [DOI] [PubMed] [Google Scholar]
- 28.Rusan NM, Fagerstrom CJ, Yvon AM, Wadsworth P. Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Mol Biol Cell. 2001;12:971–980. doi: 10.1091/mbc.12.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paweletz N. Walther Flemming: pioneer of mitosis research. Nat Rev Mol Cell Biol. 2001;2:72–75. doi: 10.1038/35048077. [DOI] [PubMed] [Google Scholar]
- 30. Saxton WM, McIntosh JR. Interzone microtubule behavior in late anaphase and telophase spindles. J Cell Biol. 1987;105:875–886. doi: 10.1083/jcb.105.2.875. Provides early demonstration that central spindle microtubule bundles are unusually stable and that the sliding of bundles accompanies spindle elongation
- 31.Skop AR, Liu H, Yates J, 3rd, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305:61–66. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gromley A, et al. et al. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 33.Greenbaum MP, et al. TEX14 is essential for intercellular bridges and fertility in male mice. Proc Natl Acad Sci U S A. 2006;103:4982–4987. doi: 10.1073/pnas.0505123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao WM, Seki A, Fang G. Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol Biol Cell. 2006;17:3881–3896. doi: 10.1091/mbc.E06-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 36.Morita E, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goss JW, Toomre DK. Both daughter cells traffic and exocytose membrane at the cleavage furrow during mammalian cytokinesis. J Cell Biol. 2008;181:1047–1054. doi: 10.1083/jcb.200712137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pohl C, Jentsch S. Final Stages of Cytokinesis and Midbody Ring Formation Are Controlled by BRUCE. Cell. 2008;132:832–845. doi: 10.1016/j.cell.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 39.Durcan TM, et al. Tektin 2 is required for central spindle microtubule organization and the completion of cytokinesis. J Cell Biol. 2008;181:595–603. doi: 10.1083/jcb.200711160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saxton WM, et al. Tubulin dynamics in cultured mammalian cells. J Cell Biol. 1984;99:2175–2186. doi: 10.1083/jcb.99.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salmon ED, Leslie RJ, Saxton WM, Karow ML, McIntosh JR. Spindle microtubule dynamics in sea urchin embryos: analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. The Journal of cell biology. 1984;99:2165–2174. doi: 10.1083/jcb.99.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shelden E, Wadsworth P. Interzonal microtubules are dynamic during spindle elongation. J Cell Sci. 1990;97:273–281. doi: 10.1242/jcs.97.2.273. [DOI] [PubMed] [Google Scholar]
- 43.Rosa J, Canovas P, Islam A, Altieri DC, Doxsey SJ. Survivin modulates microtubule dynamics and nucleation throughout the cell cycle. Mol Biol Cell. 2006;17:1483–1493. doi: 10.1091/mbc.E05-08-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bucciarelli E, Giansanti MG, Bonaccorsi S, Gatti M. Spindle assembly and cytokinesis in the absence of chromosomes during Drosophila male meiosis. J Cell Biol. 2003;160:993–999. doi: 10.1083/jcb.200211029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Savoian MS, Earnshaw WC, Khodjakov A, Rieder CL. Cleavage furrows formed between centrosomes lacking an intervening spindle and chromosomes contain microtubule bundles, INCENP, and CHO1 but not CENP-E. Mol Biol Cell. 1999;10:297–311. doi: 10.1091/mbc.10.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Canman JC, Hoffman DB, Salmon ED. The role of pre- and post-anaphase microtubules in the cytokinesis phase of the cell cycle. Curr Biol. 2000;10:611–664. doi: 10.1016/s0960-9822(00)00490-5. [DOI] [PubMed] [Google Scholar]
- 47.Alsop GB, Zhang D. Microtubules are the only structural constituent of the spindle apparatus required for induction of cell cleavage. J Cell Biol. 2003;162:383–390. doi: 10.1083/jcb.200301073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jiang W, et al. PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol Cell. 1998;2:877–885. doi: 10.1016/s1097-2765(00)80302-0. The identification of a mammalian orthologue of Ase1, PRC1, and the initial indications for its involvement in cytokinesis
- 49.Schuyler SC, Liu JY, Pellman D. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J Cell Biol. 2003;160:517–528. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muller S, et al. The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr Biol. 2004;14:412–417. doi: 10.1016/j.cub.2004.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loiodice I, et al. Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol Biol Cell. 2005;16:1756–1768. doi: 10.1091/mbc.E04-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamashita A, Sato M, Fujita A, Yamamoto M, Toda T. The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol Biol Cell. 2005;16:1378–1395. doi: 10.1091/mbc.E04-10-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu C, Jiang W. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc Natl Acad Sci U S A. 2005;102:343–348. doi: 10.1073/pnas.0408438102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mollinari C, et al. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol. 2002;157:1175–1186. doi: 10.1083/jcb.200111052. Thorough domain analysis of PRC1 and depletion analysis demonstrating its role in central spindle assembly
- 55. Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. Embo J. 2004;23:3237–3248. doi: 10.1038/sj.emboj.7600347. Demonstration of biochemical and functional links between the kinesin KIF4 and PRC1
- 56. Janson ME, et al. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 2007;128:357–368. doi: 10.1016/j.cell.2006.12.030. Demonstration of how the combined activities of PRC1 and a plus end directed kinesin slide microtubules to generate bundles
- 57. Mishima M, Kaitna S, Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev Cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. Demonstration that Cyk-4 and Mklp1 form an evolutionarily conserved complex required for central spindle assembly
- 58.Pavicic-Kaltenbrunner V, Mishima M, Glotzer M. Cooperative assembly of CYK-4/MgcRacGAP and ZEN-4/MKLP1 to form the centralspindlin complex. Mol Biol Cell. 2007;18:4992–5003. doi: 10.1091/mbc.E07-05-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sellitto C, Kuriyama R. Distribution of a matrix component of the midbody during the cell cycle in Chinese hamster ovary cells. J Cell Biol. 1988;106:431–449. doi: 10.1083/jcb.106.2.431. Immunolocalization of a midbody component, subsequently identified as Mklp1, that colocalizes with the electron dense matrix
- 60. Adams RR, Tavares AA, Salzberg A, Bellen HJ, Glover DM. pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 1998;12:1483–1494. doi: 10.1101/gad.12.10.1483. Pioneering genetic analysis of the role of Mklp1 orthologues in cytokinesis
- 61.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 62.Vale RD, Fletterick RJ. The design plan of kinesin motors. Annu Rev Cell Dev Biol. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- 63.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 64.Case RB, Rice S, Hart CL, Ly B, Vale RD. Role of the kinesin neck linker and catalytic core in microtubule-based motility. Curr Biol. 2000;10:157–160. doi: 10.1016/s0960-9822(00)00316-x. [DOI] [PubMed] [Google Scholar]
- 65. Jeyaprakash AA, et al. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell. 2007;131:271–285. doi: 10.1016/j.cell.2007.07.045. Structural characterization of the CPC revealing that the three proteins co-assemble into a three stranded helix
- 66.Sessa F, et al. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol Cell. 2005;18:379–391. doi: 10.1016/j.molcel.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 67.Earnshaw WC, Cooke CA. Analysis of the distribution of the INCENPs throughout mitosis reveals the existence of a pathway of structural changes in the chromosomes during metaphase and early events in cleavage furrow formation. J Cell Sci. 1991;98:443–461. doi: 10.1242/jcs.98.4.443. [DOI] [PubMed] [Google Scholar]
- 68.Vader G, Kauw JJ, Medema RH, Lens SM. Survivin mediates targeting of the chromosomal passenger complex to the centromere and midbody. EMBO Rep. 2006;7:85–92. doi: 10.1038/sj.embor.7400562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ban R, Irino Y, Fukami K, Tanaka H. Human mitotic spindle-associated protein PRC1 inhibits MgcRacGAP activity toward Cdc42 during the metaphase. J Biol Chem. 2004;279:16394–16402. doi: 10.1074/jbc.M313257200. [DOI] [PubMed] [Google Scholar]
- 70.Guse A, Mishima M, Glotzer M. Phosphorylation of ZEN-4/MKLP1 by aurora B regulates completion of cytokinesis. Curr Biol. 2005;15:778–786. doi: 10.1016/j.cub.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 71.Neef R, Klein UR, Kopajtich R, Barr FA. Cooperation between mitotic kinesins controls the late stages of cytokinesis. Curr Biol. 2006;16:301–307. doi: 10.1016/j.cub.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 72.Mackay AM, Eckley DM, Chue C, Earnshaw WC. Molecular analysis of the INCENPs (inner centromere proteins): separate domains are required for association with microtubules during interphase and with the central spindle during anaphase. J Cell Biol. 1993;123:373–385. doi: 10.1083/jcb.123.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wheatley SP, Carvalho A, Vagnarelli P, Earnshaw WC. INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr Biol. 2001;11:886–890. doi: 10.1016/s0960-9822(01)00238-x. [DOI] [PubMed] [Google Scholar]
- 74.Hill E, Clarke M, Barr FA. The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. EMBO J. 2000;19:5711–5719. doi: 10.1093/emboj/19.21.5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neef R, et al. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J Cell Biol. 2003;162:863–875. doi: 10.1083/jcb.200306009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abaza A, et al. M phase phosphoprotein 1 is a human plus-end-directed kinesin-related protein required for cytokinesis. J Biol Chem. 2003;278:27844–27852. doi: 10.1074/jbc.M304522200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gruneberg U, Neef R, Honda R, Nigg EA, Barr FA. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J Cell Biol. 2004;166:167–172. doi: 10.1083/jcb.200403084. Demonstration that MKLP2 has a crucial role in mediating the localization of the CPC
- 78. Neef R, et al. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat Cell Biol. 2007;9:436–444. doi: 10.1038/ncb1557. Detailed analysis of PRC1 isoforms and the demonstration the mitotic phosphorylation of PRC1 inhibits the recruitment of Plk1
- 79.Jang JK, Rahman T, McKim KS. The kinesinlike protein Subito contributes to central spindle assembly and organization of the meiotic spindle in Drosophila oocytes. Molecular biology of the cell. 2005;16:4684–4694. doi: 10.1091/mbc.E04-11-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maiato H, et al. MAST/Orbit has a role in microtubule-kinetochore attachment and is essential for chromosome alignment and maintenance of spindle bipolarity. J Cell Biol. 2002;157:749–760. doi: 10.1083/jcb.200201101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Inoue YH, et al. Mutations in orbit/mast reveal that the central spindle is comprised of two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J Cell Biol. 2004;166:49–60. doi: 10.1083/jcb.200402052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gonzalez C, et al. Mutations at the asp locus of Drosophila lead to multiple free centrosomes in syncytial embryos, but restrict centrosome duplication in larval neuroblasts. J Cell Sci. 1990;96:605–616. doi: 10.1242/jcs.96.4.605. [DOI] [PubMed] [Google Scholar]
- 83. Wakefield JG, Bonaccorsi S, Gatti M. The drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. J Cell Biol. 2001;153:637–648. doi: 10.1083/jcb.153.4.637. One of the few papers with functional insight into factors that contribute to central spindle assembly by binding to the minus ends of the bundles
- 84.Bond J, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 85.do Carmo Avides M, Tavares A, Glover DM. Polo kinase and Asp are needed to promote the mitotic organizing activity of centrosomes. Nat Cell Biol. 2001;3:421–424. doi: 10.1038/35070110. [DOI] [PubMed] [Google Scholar]
- 86.Fabbro M, et al. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev Cell. 2005;9:477–488. doi: 10.1016/j.devcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 87.Verni F, et al. Feo, the Drosophila homolog of PRC1, is required for central-spindle formation and cytokinesis. Curr Biol. 2004;14:1569–1575. doi: 10.1016/j.cub.2004.08.054. [DOI] [PubMed] [Google Scholar]
- 88.Mollinari C, et al. Ablation of PRC1 by small interfering RNA demonstrates that cytokinetic abscission requires a central spindle bundle in mammalian cells, whereas completion of furrowing does not. Mol Biol Cell. 2005;16:1043–1055. doi: 10.1091/mbc.E04-04-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Verbrugghe KJ, White JG. SPD-1 Is Required for the Formation of the Spindle Midzone but Is Not Essential for the Completion of Cytokinesis in C. elegans Embryos. Curr Biol. 2004;14:1755–1760. doi: 10.1016/j.cub.2004.09.055. [DOI] [PubMed] [Google Scholar]
- 90.Kieserman EK, Glotzer M, Wallingford JB. Developmental regulation of central spindle assembly and cytokinesis during vertebrate embryogenesis. Curr Biol. 2008;18:116–123. doi: 10.1016/j.cub.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 91. Severson AF, Hamill DR, Carter JC, Schumacher J, Bowerman B. The aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr Biol. 2000;10:1162–1171. doi: 10.1016/s0960-9822(00)00715-6. An influential paper that used temperature sensitive alleles to demonstrate a functional interaction between the Mklp1 orthologue and Aurora B and to estimate when they act during cytokinesis
- 92.Kaitna S, Mendoza M, Jantsch-Plunger V, Glotzer M. Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr Biol. 2000;10:1172–1181. doi: 10.1016/s0960-9822(00)00721-1. [DOI] [PubMed] [Google Scholar]
- 93.Simon GC. Sequential Cyk-4 binding to ECT2 and FIP3 regulates cleavage furrow ingression and abscission during cytokinesis. EMBO J. 2008 doi: 10.1038/emboj.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Santamaria A, et al. Use of the novel Plk1 inhibitor ZK-thiazolidinone to elucidate functions of Plk1 in early and late stages of mitosis. Mol Biol Cell. 2007;18:4024–4036. doi: 10.1091/mbc.E07-05-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Petronczki M, Glotzer M, Kraut N, Peters JM. Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev Cell. 2007;12:713–725. doi: 10.1016/j.devcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 96.Burkard ME, et al. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc Natl Acad Sci U S A. 2007;104:4383–4388. doi: 10.1073/pnas.0701140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Johnson EF, Stewart KD, Woods KW, Giranda VL, Luo Y. Pharmacological and functional comparison of the polo-like kinase family: insight into inhibitor and substrate specificity. Biochemistry. 2007;46:9551–9563. doi: 10.1021/bi7008745. [DOI] [PubMed] [Google Scholar]
- 98.Blangy A, et al. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 99.Sawin KE, Mitchison TJ. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc Natl Acad Sci U S A. 1995;92:4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Saunders AM, Powers J, Strome S, Saxton WM. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr Biol. 2007;17:R453–R454. doi: 10.1016/j.cub.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mishima M, Pavicic V, Gruneberg U, Nigg EA, Glotzer M. Cell cycle regulation of central spindle assembly. Nature. 2004;430:908–913. [Google Scholar]
- 102. Zhu C, Lau E, Schwarzenbacher R, Bossy-Wetzel E, Jiang W. Spatiotemporal control of spindle midzone formation by PRC1 in human cells. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0506926103. 101 & 102 show that mitotic phosphorylation of Mklp1 and Prc1, respectively, inhibit central spindle assembly during mitosis
- 103.Murata-Hori M, Tatsuka M, Wang YL. Probing the Dynamics and Functions of Aurora B Kinase in Living Cells during Mitosis and Cytokinesis. Mol Biol Cell. 2002;13:1099–1108. doi: 10.1091/mbc.01-09-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Karsenti E, Nedelec F, Surrey T. Modelling microtubule patterns. Nat Cell Biol. 2006;8:1204–1211. doi: 10.1038/ncb1498. [DOI] [PubMed] [Google Scholar]
- 105. Nedelec F. Computer simulations reveal motor properties generating stable antiparallel microtubule interactions. J Cell Biol. 2002;158:1005–1015. doi: 10.1083/jcb.200202051. Computational exploration of mechanisms that could generate stable microtubule overlap
- 106.Goshima G, Nedelec F, Vale RD. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol. 2005;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sproul LR, Anderson DJ, Mackey AT, Saunders WS, Gilbert SP. Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus ends. Curr Biol. 2005;15:1420–1427. doi: 10.1016/j.cub.2005.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Burbank KS, Mitchison TJ, Fisher DS. Slide-and-cluster models for spindle assembly. Curr Biol. 2007;17:1373–1383. doi: 10.1016/j.cub.2007.07.058. [DOI] [PubMed] [Google Scholar]
- 109.Mitchison TJ, et al. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol Biol Cell. 2005;16:3064–3076. doi: 10.1091/mbc.E05-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Channels W, Nedelec F, Zheng Y, Iglesias P. Spatial regulation improves anti-parallel microtubule overlap during mitotic spindle assembly. Biophys J. 2007 doi: 10.1529/biophysj.107.117671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cheerambathur DK, et al. Quantitative analysis of an anaphase B switch: predicted role for a microtubule catastrophe gradient. J Cell Biol. 2007;177:995–1004. doi: 10.1083/jcb.200611113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chalamalasetty RB, Hummer S, Nigg EA, Sillje HH. Influence of human Ect2 depletion and overexpression on cleavage furrow formation and abscission. J Cell Sci. 2006;119:3008–3019. doi: 10.1242/jcs.03032. [DOI] [PubMed] [Google Scholar]
- 113.Hu CK, Coughlin M, Field CM, Mitchison TJ. Cell polarization during monopolar cytokinesis. J Cell Biol. 2008;181:195–202. doi: 10.1083/jcb.200711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hoog JL, et al. Organization of interphase microtubules in fission yeast analyzed by electron tomography. Dev Cell. 2007;12:349–361. doi: 10.1016/j.devcel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 115.Drummond DR, Cross RA. Dynamics of interphase microtubules in Schizosaccharomyces pombe. Curr Biol. 2000;10:766–775. doi: 10.1016/s0960-9822(00)00570-4. [DOI] [PubMed] [Google Scholar]
- 116.Tran PT, Marsh L, Doye V, Inoue S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Carazo-Salas RE, Antony C, Nurse P. The kinesin Klp2 mediates polarization of interphase microtubules in fission yeast. Science. 2005;309:297–300. doi: 10.1126/science.1113465. [DOI] [PubMed] [Google Scholar]
- 118.Carazo-Salas RE, Nurse P. Self-organization of interphase microtubule arrays in fission yeast. Nat Cell Biol. 2006;8:1102–1107. doi: 10.1038/ncb1479. [DOI] [PubMed] [Google Scholar]
- 119.Daga RR, Lee KG, Bratman S, Salas-Pino S, Chang F. Self-organization of microtubule bundles in anucleate fission yeast cells. Nat Cell Biol. 2006;8:1108–1113. doi: 10.1038/ncb1480. [DOI] [PubMed] [Google Scholar]
- 120.Powers J, et al. Loss of KLP-19 polar ejection force causes misorientation and missegregation of holocentric chromosomes. J Cell Biol. 2004;166:991–1001. doi: 10.1083/jcb.200403036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Williams BC, Riedy MF, Williams EV, Gatti M, Goldberg ML. The Drosophila kinesin-like protein KLP3A is a midbody component required for central spindle assembly and initiation of cytokinesis. J Cell Biol. 1995;129:709–723. doi: 10.1083/jcb.129.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Toure A, et al. MgcRacGAP, a new human GTPase-activating protein for Rac and Cdc42 similar to Drosophila rotundRacGAP gene product, is expressed in male germ cells. J Biol Chem. 1998;273:6019–6023. doi: 10.1074/jbc.273.11.6019. [DOI] [PubMed] [Google Scholar]
- 123.Goldstein AY, Jan YN, Luo L. Function and regulation of Tumbleweed (RacGAP50C) in neuroblast proliferation and neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:3834–3839. doi: 10.1073/pnas.0500748102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nislow C, Lombillo VA, Kuriyama R, McIntosh JR. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- 125.Terada Y, et al. AIM-1: a mammalian midbody-associated protein required for cytokinesis. Embo J. 1998;17:667–676. doi: 10.1093/emboj/17.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schumacher JM, Golden A, Donovan PJ. AIR-2: An Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J Cell Biol. 1998;143:1635–1646. doi: 10.1083/jcb.143.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cooke CA, Heck MM, Earnshaw WC. The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J Cell Biol. 1987;105:2053–2067. doi: 10.1083/jcb.105.5.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Adams RR, Maiato H, Earnshaw WC, Carmena M. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J Cell Biol. 2001;153:865–880. doi: 10.1083/jcb.153.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li F, et al. Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat Cell Biol. 1999;1:461–466. doi: 10.1038/70242. [DOI] [PubMed] [Google Scholar]
- 130.Fraser AG, James C, Evan GI, Hengartner MO. Caenorhabditis elegans inhibitor of apoptosis protein (IAP) homologue BIR-1 plays a conserved role in cytokinesis. Curr Biol. 1999;9:292–301. doi: 10.1016/s0960-9822(99)80137-7. [DOI] [PubMed] [Google Scholar]
- 131.Jones G, Jones D, Zhou L, Steller H, Chu Y. Deterin, a new inhibitor of apoptosis from Drosophila melanogaster. J Biol Chem. 2000;275:22157–22165. doi: 10.1074/jbc.M000369200. [DOI] [PubMed] [Google Scholar]
- 132.Gassmann R, et al. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. The Journal of cell biology. 2004;166:179–191. doi: 10.1083/jcb.200404001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sampath SC, et al. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 134.Romano A, et al. CSC-1: a subunit of the Aurora B kinase complex that binds to the survivin-like protein BIR-1 and the incenp-like protein ICP-1. J Cell Biol. 2003;161:229–236. doi: 10.1083/jcb.200207117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hanson KK, Kelley AC, Bienz M. Loss of Drosophila borealin causes polyploidy, delayed apoptosis and abnormal tissue development. Development. 2005;132:4777–4787. doi: 10.1242/dev.02057. [DOI] [PubMed] [Google Scholar]
- 136.Lee KS, Yuan YL, Kuriyama R, Erikson RL. Plk is an M-phase-specific protein kinase and interacts with a kinesin- like protein, CHO1/MKLP-1. Mol Cell Biol. 1995;15:7143–7151. doi: 10.1128/mcb.15.12.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mundt KE, Golsteyn RM, Lane HA, Nigg EA. On the regulation and function of human polo-like kinase 1 (PLK1): effects of overexpression on cell cycle progression. Biochem Biophys Res Commun. 1997;239:377–385. doi: 10.1006/bbrc.1997.7378. [DOI] [PubMed] [Google Scholar]
- 138.Wilson GM, et al. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol Biol Cell. 2005;16:849–860. doi: 10.1091/mbc.E04-10-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human ECT2 is an exchange factor for rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J Cell Biol. 1999;147:921–928. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]