SUMMARY
Asymmetric cell division is intensely studied because it can generate cellular diversity as well as maintain stem cell populations. Asymmetric cell division requires mitotic spindle alignment with intrinsic or extrinsic polarity cues, but mechanistic detail of this process is lacking. Here we develop a method to construct cortical polarity in a normally unpolarized cell line, and use this method to characterize Partner of Inscuteable (Pins; LGN/AGS3 in mammals)-dependent spindle orientation. We identify a previously unrecognized evolutionarily-conserved Pins domain (PinsLINKER) that requires Aurora-A phosphorylation to recruit Discs large (Dlg; PSD-95/hDlg in mammals) and promote partial spindle orientation. The well-characterized PinsTPR domain has no function alone, but placing the PinsTPR in cis to the PinsLINKER gives dynein-dependent precise spindle orientation. This "induced cortical polarity" assay is suitable for rapid identification of the proteins, domains, and amino acids regulating spindle orientation or cell polarity.
INTRODUCTION
Asymmetric cell division (ACD) occurs when one cell divides to generate two molecularly distinct daughter cells. ACD requires precise alignment of the mitotic spindle with the intrinsic or extrinsic polarity axis so that cellular components, such as fate determinants, are partitioned into different daughter cells (Knoblich, 2008). Recent work has highlighted the importance of ACD in generating cell diversity during early embryogenesis and in maintaining stem cell pool size (Doe, 2008). Thus, characterizing the mechanisms of ACD, including cell polarization and spindle orientation, is important for understanding many aspects of development and disease.
Drosophila neural progenitors, or neuroblasts, are an excellent model system for studying ACD. Neuroblasts have apical/basal polarity and they align their mitotic spindle with this polarity axis to generate a self-renewed apical neuroblast and a differentiating basal daughter cell. Cell fate determinants have been identified that partition into the apical neuroblast to maintain its fate (e.g. atypical protein kinase C, aPKC) and that partition into the basal daughter cell to induce differentiation (e.g. Miranda/Prospero)(reviewed in Doe, 2008). Progress has also been made on identifying proteins required for apical/basal spindle orientation. These include proteins that form an apical cortical crescent over one spindle pole, such as Inscuteable (Insc; mInsc in mammals), Partner of Inscuteable (Pins; LGN/AGS3 in mammals), Gαi/o, Discs large (Dlg), Scribble, and Mushroom body defect (Mud; NuMA in mammals); proteins enriched on centrosomes, such as Centrosomin, Sas-4 (CenpJ in mammals), Dynein, Dynactin, Lis1; and the mitotic kinases Aurora-A and Polo (Knoblich, 2008). Virtually all of these proteins are evolutionarily-conserved and have a similar function in regulating spindle alignment in yeast, C. elegans, and mammals (Siller and Doe, 2009).
Despite progress in identifying components involved in cortical polarity and spindle orientation, much remains unknown. Here we describe a system for generating cell polarity and spindle orientation in the normally unpolarized Drosophila S2 cell line, and use this system to test individual proteins, protein domains, and amino acids for their role in spindle orientation.
Results
Induced cell polarity in the Drosophila S2 cell line
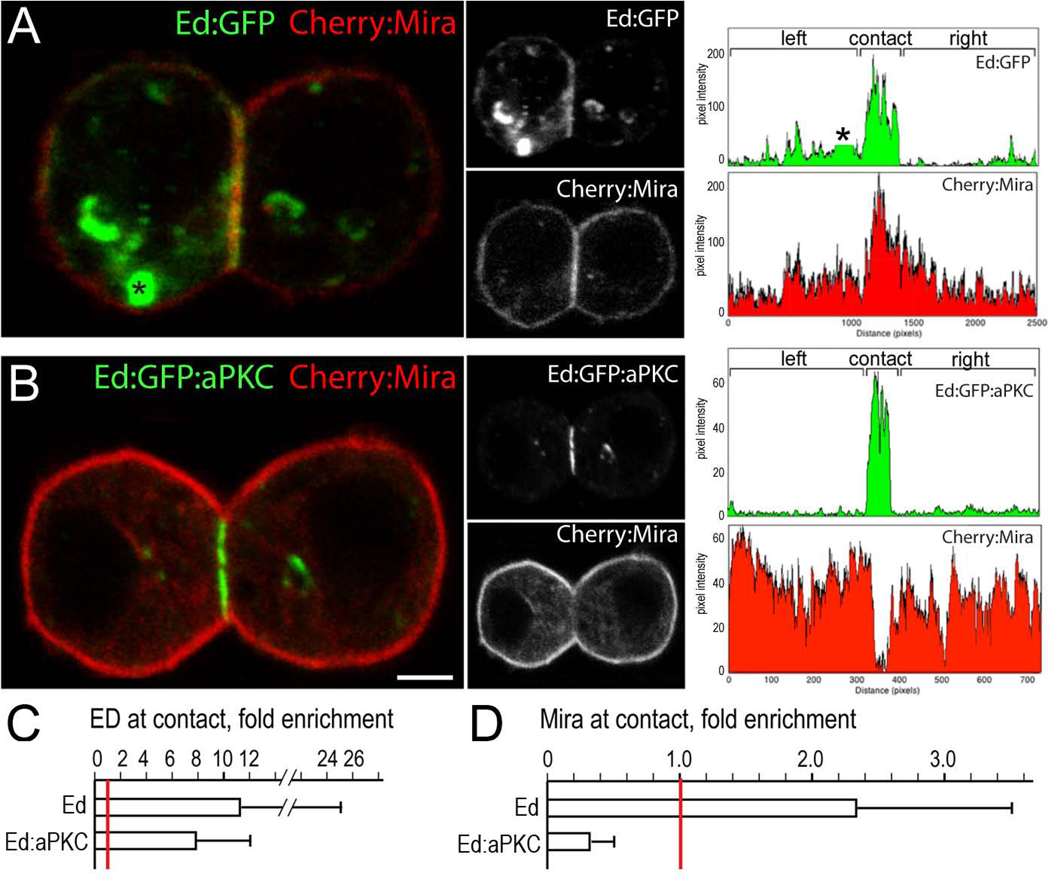
Transfection-induced expression of the homophilic cell adhesion molecule Echinoid (Ed) can induce cell-cell adhesion in S2 cells and restrict Ed protein to the site of cell-cell contact (Bai et al., 2001). We adapted this method to induce cortical polarity of a heterologous protein by fusing the protein of interest to the Ed cytoplasmic terminus. Ed:green fluorescent protein fusions (Ed:GFP) formed distinct cortical crescents containing the majority of the cortical protein (Figure 1A, C). Ed fusion proteins are also detected in cytoplasmic vesicles (Figure 1A, asterisk), as expected for transmembrane proteins, but the presence of vesicles had no effect on the cortical polarity or spindle orientation function of Ed fusion proteins (Supplemental Table 1). To test whether this system could be used to generate functional cortical polarity matching that of neuroblasts, we generated aPKC cortical polarity by expressing an Ed:GFP:aPKC fusion protein. We observed that cortical aPKC was necessary and sufficient to exclude Miranda from the cortex in S2 cells (Figure 1B,D), similar to larval neuroblasts (Rolls et al., 2003). We conclude that the Ed cell adhesion molecule can be used to induce functional cortical polarity in S2 cells.
Figure 1. Induced cell polarity in Drosophila S2 cells.
(A) The Echinoid (Ed) induced cortical polarity in S2 cells. Cortical polarization of Ed:GFP fusion protein (green); there is also cytoplasmic vesicle staining (asterisk); Cherry:Miranda protein (red) shows less cortical enrichment. Right: quantification of cortical pixel intensity (vesicle staining removed from plot).
(B) Induced functional cortical polarity in S2 cells. Polarized Ed:aPKC can exclude Cherry:Miranda (Mira) from the cortex, thereby inducing Cherry:Mira cortical polarization. Right: quantification of cortical pixel intensity. Scale bar 3 µm.
(C) Ed and Ed:aPKC are enriched at the site of cell-cell contact. Pixel intensity was measured for left (L), right (R), and contacting (C) cortical domains and fold enrichment at the contact site was calculated (C/L+R). Red line indicates a ratio of 1.0 (no enrichment or exclusion). n = 17 (Ed) and 18 (Ed:aPKC).
(D) aPKC excludes Miranda from the cortex. Cherry:Mira pixel intensity levels were measured as described in panel C. Red line indicates a ratio of 1.
Induced Pins cortical polarity promotes spindle orientation
Drosophila neuroblasts show tight alignment of the mitotic spindle with the apical cortical domain, and many apical proteins are required for proper spindle orientation (Knoblich, 2008). However, it is unknown which domain of each protein is essential for spindle orientation function, nor is it known which provide cortex-to-microtubule links or which act indirectly by regulating cortical polarity. Here we sought to test individual apical proteins for their ability to promote spindle orientation in Drosophila S2 cells, with the goal of understanding the function of individual proteins, protein domains, and specific amino acids.
We generated protein crescents as described above, and measured the angle of the mitotic spindle relative to the center of the Ed cortical crescent (Figure 2B). We measured the spindle axis using two different methods -- staining for the spindle marker α-tubulin or for the spindle pole marker Cnn -- and both gave identical results (Table 1). Precise alignment of the spindle with the crescent would generate a mean spindle angle near 0°, whereas random spindle orientation would give a mean spindle angle near 45° (see methods for details). Control experiments showed that polarization of Ed:FLAG or Ed:GFP resulted in nearly random spindle orientation with a mean spindle angle of 53° and a standard deviation of 23° (Figure 2C; Table 1). The large standard deviation results from the random distribution. We estimated the standard error of the spindle angle measurements to be ± 3° based on the analysis of multiple independent experiments (see methods). These control experiments allowed us to conclude that the spindle orientation is not affected by Ed, FLAG, GFP, or by potential cell shape changes due to cell-cell adhesive contacts. We next tested various proteins required for spindle orientation in neuroblasts for their ability to induce spindle orientation in S2 cells. Induced cortical crescents of Insc, EB1 or aPKC failed to orient the mitotic spindle (Table 1; although aPKC could efficiently displace Miranda from the cortex, see above). In contrast, full length Pins (Ed:PinsFL) could partially orient the mitotic spindle (27 ± 15°; Figure 2D; Table 1) consistent with its requirement for spindle orientation in Drosophila neuroblasts (Parmentier et al., 2000; Rebollo et al., 2007; Schaefer et al., 2000; Siegrist and Doe, 2005; Yu et al., 2000).
Figure 2. The Pins TPR and Linker domains synergistically orient the mitotic spindle in Drosophila S2 cells.
(A) N-terminal TPR domain (amino acids 42–398), central linker region (399–466), and 'GoLoco domain' containing three GoLoco motifs (467–658). Serine 436 (S436; black ball).
(B) Spindle angle assay: a vector perpendicular to the Ed crescent (red dashed line) and along the spindle axis (black dashed line) indicates spindle angle.
(C–K) Pins domains were fused in frame to Echinoid tagged with a FLAG epitope (Ed-FLAG) and transfected into S2 cells. Cells were stained for FLAG (red) and α-tubulin (green). FL, Pins full length; FL+Gαi, PinsFL co-transfected with Gαi (Gαi is colocalized with Pins, not shown); FL(R-F)+Gαi, PinsFL with a mutated Gαi binding domain co-transfected with Gαi (Gαi is delocalized, not shown); TPR, Pins TPR domain alone; GoLoco, Pins GoLoco domain alone; Linker, Pins Linker domain alone; ΔLinker, Pins full length protein lacking the linker domain; TPR+Linker, Pins TPR and Linker domains. Scale bar 3 µm.
(L) Quantification. Mean spindle angle and standard deviation is shown (sampling error is estimated at ± 3°; see methods). The Pins TPR, GoLoco, and ΔLinker proteins show no spindle orientation (30–60° spindle angles; Pins FL, FL(R-F)+Gαi, Linker proteins show partial spindle orientation (~15–30° spindle angles); Pins FL+Gαi and TPR+Linker proteins show good spindle orientation (~0–15° spindle angles). *, highly significant compared to Ed alone 'none' (p < 0.01); #, significant spindle orientation compared to Pins FL (p < 0.05), other proteins showed no difference from Ed alone (p > 0.05).
Table 1.
Echinoid fusion protein spindle orientation abilitya
| Echinoid:FLAG fusion partner | Average Angle |
Standard Deviation |
n | Pathway |
|---|---|---|---|---|
| PinsTPR+LINKER (in cis) | 10.7 | 7.1 | 26 | TPR+LINKER |
| bPinsTPR+LINKER (in cis) | 11.2 | 5.6 | 29 | " |
| Pins(1–446) | 13.4 | 13.3 | 25 | " |
| PinsTPR+LINKER (S436D) + aur-A RNAi | 13.6 | 12.9 | 35 | " |
| PinsTPR+LINKER (S436D) | 13.7 | 11.3 | 50 | " |
| PinsTPR+LINKER + baz RNAi | 13.7 | 9.3 | 24 | " |
| PinsFL + Gα | 17.2 | 15.4 | 55 | " |
| PinsFL(Arg-Phe) | 25.9 | 23.8 | 27 | LINKER |
| PinsFL | 27.2 | 14.8 | 40 | " |
| PinsTPR + PinsLINKER (in trans) | 27.2 | 18.1 | 34 | " |
| cPinsLINKER | 27.3 | 15.6 | 33 | " |
| PinsFL(Arg-Phe) + Gαi | 27.5 | 17.3 | 21 | " |
| DlgGK | 27.5 | 20.2 | 41 | " |
| PinsTPR+LINKER + dlc RNAi | 27.9 | 22.5 | 33 | " |
| PinsTPR+LINKER + mud RNAi | 29.1 | 18.1 | 28 | " |
| PinsLINKER | 29.2 | 20.5 | 41 | " |
| DlgGK + mud RNAi | 29.3 | 18 | 29 | " |
| PinsLINKER + dlc RNAi | 29.9 | 17.5 | 33 | " |
| DlgGK + dlg RNAi | 30.5 | 22.6 | 16 | " |
| PinsTPR+LINKER + lis1 RNAi | 31.1 | 25.9 | 22 | " |
| PinsTPR+LINKER (N226F) | 31.4 | 23.8 | 26 | " |
| dPinsLINKER | 32.4 | 17.0 | 30 | " |
| PinsTPR+LINKER + khc-73 RNAi | 40.5 | 21.9 | 41 | NONE |
| PinsFL + dlg RNAi | 40.8 | 27.1 | 48 | " |
| PinsTPR+LINKER + aur-A RNAi | 41.0 | 24.5 | 53 | " |
| Pins(1–426) | 41.1 | 23.1 | 21 | " |
| Pins(1–435) | 41.3 | 25.1 | 24 | " |
| Pins(1–406) | 41.7 | 24.1 | 28 | " |
| Pins(1–416) | 41.9 | 25.2 | 26 | " |
| PinsLINKER Deleted | 42.0 | 24.6 | 48 | " |
| PinsTPR+LINKER + dlg RNAi | 42.1 | 23.7 | 23 | " |
| PinsTPR+LINKER (S436A) | 43.0 | 20.8 | 44 | " |
| PinsTPR+LINKER + Nocodazole | 43.5 | 25 | 39 | " |
| PinsLINKER + dlg RNAi | 43.7 | 27.5 | 48 | " |
| EB1 | 44.5 | 29.7 | 27 | " |
| DlgGK + Nocodazole | 45.2 | 25 | 25 | " |
| PinsGoLOCO | 45.7 | 28.2 | 33 | " |
| PinsTPR | 46.1 | 25.2 | 57 | " |
| DlgGK + khc-73 RNAi | 46.7 | 30.5 | 21 | " |
| eInsc(252–583) | 48.9 | 27.6 | 15 | " |
| aPKC | 50.5 | 22.5 | 19 | " |
| PinsTPR+LINKER + Khc-73MBS | 51.9 | 23.5 | 11 | " |
| None | 52.8 | 23.3 | 31 | " |
| bNone | 46.5 | 27.5 | 33 | " |
all spindle angle measurements are made using tubulin stains except where noted;
spindle axis determined by Cnn staining;
Ed:GFP:PinsLINKER;
Ed:Cherry:PinsLINKER
this is the Insc domain necessary and sufficient for spindle orientation (Knoblich et al., 1999, Curr Biol., 9:155).
Full length Pins exists by default in a 'closed' or autoinhibited state due to intramolecular binding between its seven N-terminal tetratricopeptide repeat (TPR) domains and its three C-terminal Gαi-binding (GoLoco) motifs; binding of Gαi to the C-terminal GoLoco motif region can 'open' Pins (Du and Macara, 2004; Nipper et al., 2007). Gαi levels are undetectable in S2 cells (data not shown), raising the possibility that Ed:PinsFL is not fully active due to autoinhibition. To test this hypothesis we co-expressed Gαi and Ed:PinsFL, and observed a clear improvement in the mean spindle angle to 17 ± 15° (n = 55; Figure 2E; Table 1). Single amino acid mutations (Arg>Phe) to each GoLoco motif, which prevent Pins from binding Gαi (Willard et al., 2004), were found to block Gαi-induced improvement of spindle orientation (26 ± 24°; n=27; Figure 2F; Table 1). This mirrors the situation in wild type neuroblasts, where Gαi is required for tight alignment of the spindle with the Pins crescent (Nipper et al., 2007). We conclude that Gαi is required for Pins to accurately orient the mitotic spindle in S2 cells, similar to the role of Gαi/Pins in neuroblasts.
We next tested different fragments of Pins for their ability to promote spindle orientation: the N-terminal TPR region (residues 1–398), the central linker (residues 399–466), and the C-terminal GoLoco region (residues 467–658). The PinsTPR domain is the best candidate for providing Pins spindle orientation function, because it binds the microtubule-binding protein Mushroom body defect (Mud; NuMA in mammals; LIN-5 in C. elegans) which is required for spindle orientation in Drosophila and C. elegans (Bowman et al., 2006; Izumi et al., 2006; Lorson et al., 2000; Siller et al., 2006). Surprisingly, we found that the PinsTPR domain alone had no ability to orient the mitotic spindle in our S2 assay (46 ± 25°, Figure 2G, Table 1). The GoLoco region also failed to orient the spindle (46 ± 28°, Figure 2H; Table 1). The only region of Pins that promoted spindle orientation was the previously uncharacterized linker region (29 ± 20°, Figure 2I; Table 1), and deletion of the linker from full length Pins abolished spindle orientation activity (42 ± 25°, Figure 2J; Table 1).
PinsLINKER has spindle orientation activity that is similar to the 'closed' Pins full length protein, but not as good as the 'open' (Gαi-bound) Pins full length protein. This suggests that the PinsLINKER is active when Pins is in the 'closed' form, and that 'opening' of Pins reveals a second domain that can improve PinsLINKER-mediated spindle orientation. What is the second domain? PinsLINKER+GOLOCO did not improve on PinsLINKER spindle orientation, and PinsGOLOCO alone has no spindle orientation function (Table 1). In contrast, addition of the PinsTPR domain (PinsTPR+LINKER) showed striking improvement in spindle orientation over PinsLINKER alone (10.7 ± 7.1°, Figure 2K; Table 1). In fact, PinsTPR+LINKER spindle orientation was comparable to PinsFL + Gαi (17 ± 15°, Table 1) or neuroblast spindle orientation in vivo (Nipper et al., 2007; Siller et al., 2006; Siller and Doe, 2008), despite the absence of the GoLoco domain and endogenous Gαi. We draw two conclusions from these data. First, the PinsGOLOCO domain and Gαi are not essential for Pins-mediated spindle orientation; their function appears limited to Pins 'opening' and cortical tethering. Second, the PinsTPR can improve the spindle orientation ability of the PinsLINKER domain to a level matching Gαi-activated full length Pins.
PinsLINKER acts through Dlg and Khc-73 to promote spindle orientation
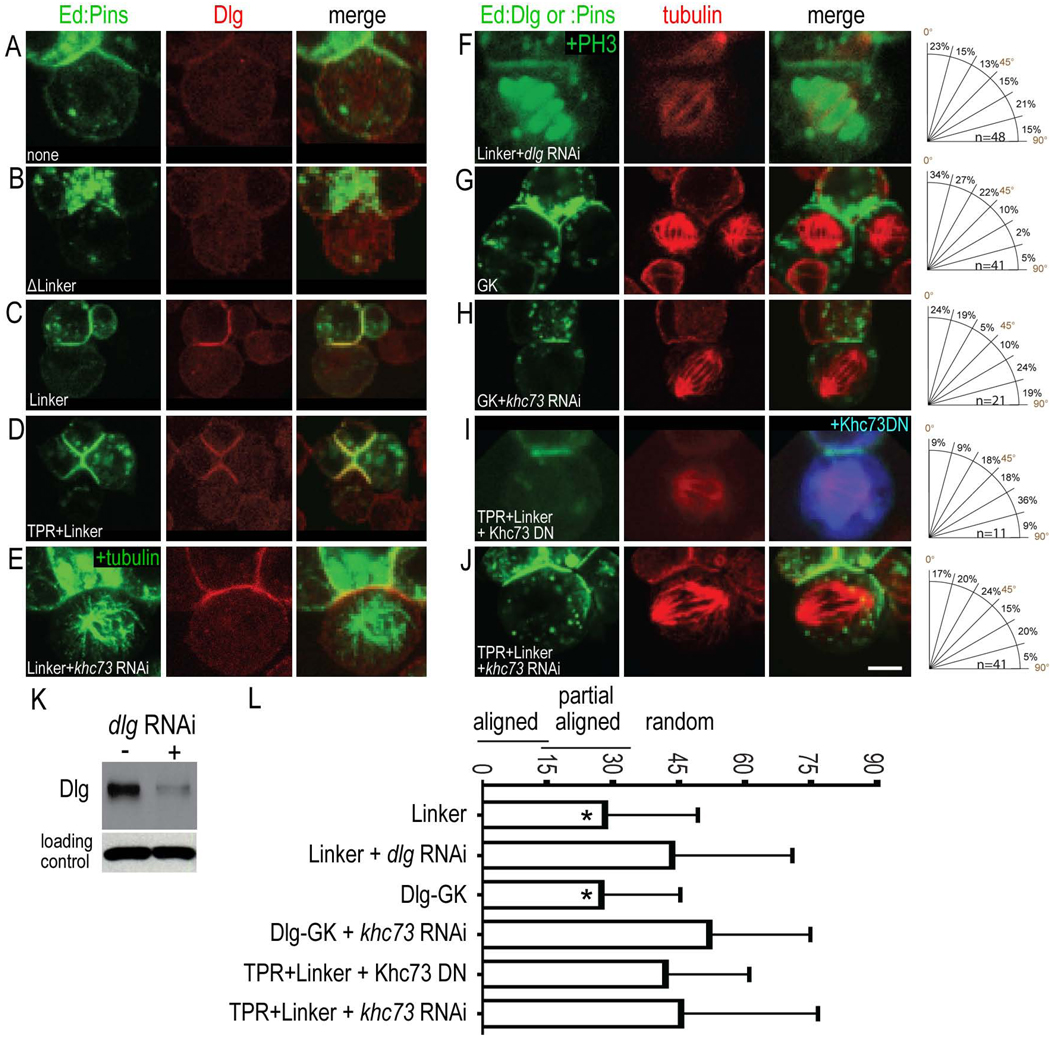
In this section we explore the mechanism of PinsLINKER-mediated spindle orientation, and in the following section we investigate how the PinsTPR domain improves PinsLINKER activity. Pins binds the tumor suppressor protein Dlg (Bellaiche et al., 2001; Sans et al., 2005) and Dlg is required for spindle orientation in Drosophila neuroblasts (Siegrist and Doe, 2005). Here we test whether Dlg is required for PinsLINKER-mediated spindle orientation. S2 cells transfected with Ed alone or with Ed-Pins lacking the linker domain localize endogenous Dlg primarily in the cytoplasm (Figure 3A,B). In contrast, we observed clear Dlg recruitment to PinsLINKER or PinsTPR+LINKER cortical crescents (Figure 3C,D). Moreover, dlg RNAi reduced Dlg levels and abolished PinsLINKER-mediated spindle orientation (42 ± 24°, Figure 3F; Table 1), as well as blocked spindle orientation conferred by full length Pins and PinsTPR+LINKER (Table 1); PinsTPR+LINKER cells treated with control RNAi constructs (bazooka or insc) had no effect on spindle orientation (Table 1). If PinsLINKER recruits Dlg to promote spindle orientation, we should be able to bypass Pins by inducing Dlg cortical polarity. We previously showed the Dlg guanylate kinase-like (DlgGK) domain was necessary for proper spindle orientation in neuroblasts (Siegrist and Doe, 2005); here we test whether the DlgGK domain is sufficient for spindle orientation in S2 cells. We used Ed to form a DlgGK cortical crescent, and found that it had spindle orientation ability similar to that of PinsLINKER (28 ± 20°; Figure 3G, Table 1). We conclude that the PinsLINKER recruits Dlg to the cortex, where the DlgGK domain promotes spindle orientation.
Figure 3. PinsLINKER requires Dlg and Khc73 for spindle orientation.
S2 cells were transfected with Ed:GFP alone or the indicated Ed:Pins domains (green) and immunostained for endogenous Dlg or Tubulin (red). RNAi knockdown was performed for the indicated genes.
(A–E) PinsLINKER recruits Dlg to the cortex.
(F–L) PinsLINKER requires Dlg and Khc-73 for spindle orientation. The mitotic marker PH3 is shown in F. Western blot analysis shows RNAi knockdown of Dlg; detection of α-tubulin demonstrates equal loading of lysates (K). Scale bar 3 µm for A–J.
(L) Quantification of spindle orientation (mean spindle angle ± standard deviation). *, significantly better spindle orientation compared to Ed alone control (p < 0.01); other proteins showed no difference from Ed alone control (p > 0.05).
How does the DlgGK interact with spindle microtubules? A good candidate is Khc-73 (Kif13a in mammals), a microtubule plus-end directed kinesin motor that can bind both microtubules and the DlgGK domain (s), and is required for proper spindle orientation in Drosophila neuroblasts (Siegrist and Doe, 2005). We found that expression of a dominant negative Khc-73 fragment prevents PinsLINKER-mediated spindle orientation (51.9 ±23.5°; Table 1; Figure 3I; Table 1). Similarly, RNAi directed against Khc-73 completely blocked PinsTPR+LINKER-mediated spindle orientation (40.5 ± 21.9°; Figure 3J; Table 1) as well as DlgGK-mediated spindle orientation (46.7 ± 30.5°; Figure 3H; Table 1). khc-73 RNAi does not block recruitment of Dlg to the PinsLINKER cortical crescent (Figure 3E), consistent with Khc-73 acting downstream of cortical Dlg, rather than transporting Dlg to the cortex as proposed for mammalian cells (Hanada et al., 2000). We conclude that Khc-73 acts downstream of Dlg to promote PinsLINKER/DlgGK-mediated spindle orientation.
PinsTPR recruits Mud and acts in cis to improve PinsLINKER-mediated spindle orientation
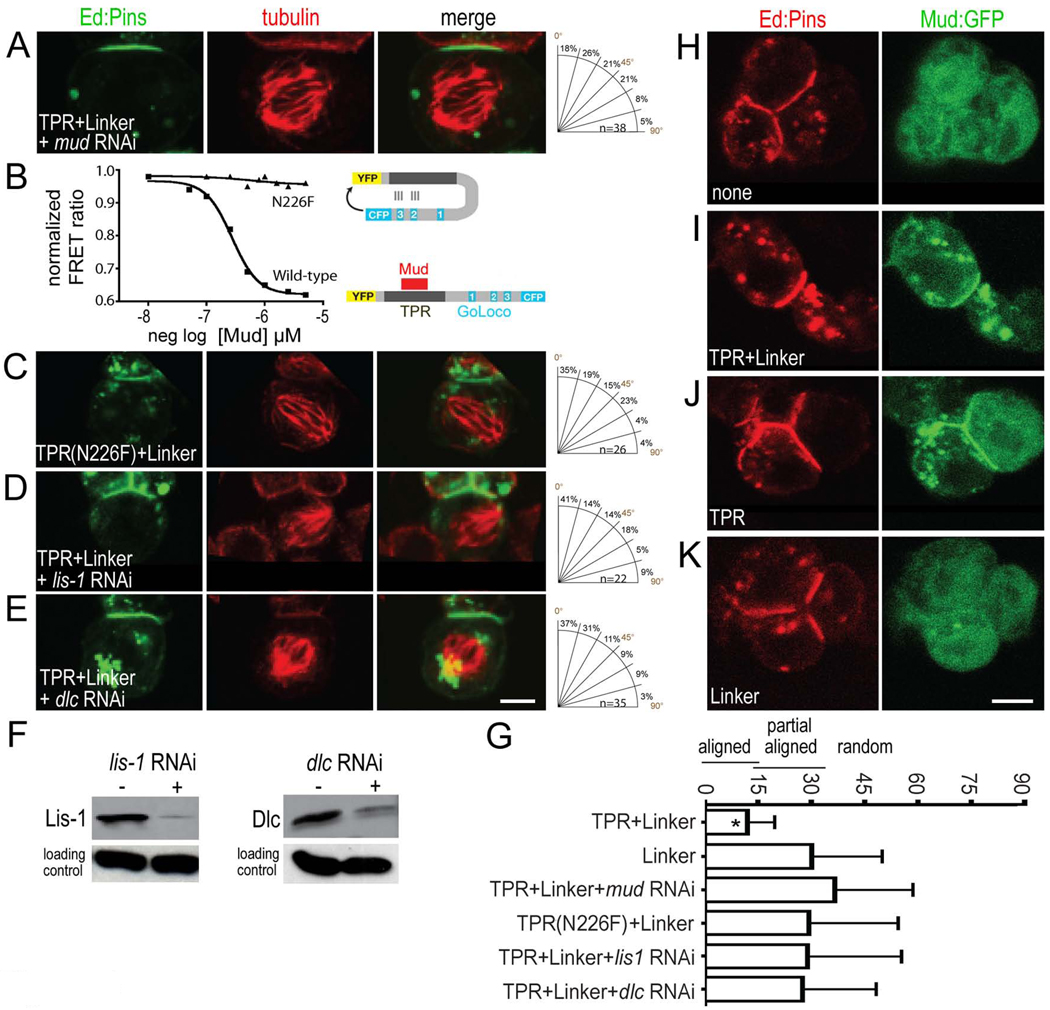
In this section we investigate how PinsTPR can significantly improve the spindle orientation ability of the PinsLINKER domain, despite having no spindle orientation function on its own. The PinsTPR binds Insc and Mud, which are both required for spindle orientation (Bowman et al., 2006; Izumi et al., 2006; Kraut et al., 1996; Siller et al., 2006). S2 cells express detectable levels of Mud but not Insc (Figure S1), and Ed:Insc has no spindle orientation ability in S2 cells (Table 1), so we tested the role of Mud in PinsTPR+LINKER spindle orientation. PinsTPR+LINKER was able to recruit Mud to the cortex (Figure 4I), and mud RNAi reduced PinsTPR+LINKER spindle orientation activity to match that of PinsLINKER alone (Figure 4A; Table 1). Thus, Mud is required for PinsTPR enhancement of the PinsLINKER spindle orientation. To confirm that Mud functions via binding the PinsTPR domain, we engineered a TPR(N226F) mutation based on previous structural studies that showed that this amino acid was likely to contact TPR ligands (Goebl and Yanagida, 1991). The N226F mutation blocked Pins-Mud binding in vitro (Figure 4B) without affecting stability of global TPR protein folding (Figure S2). When we placed the PinsTPR(N226F)+LINKER mutant into our S2 spindle assay, we observed a spindle orientation of 31 ± 24° (Figure 4C; Table 1), nearly identical to PinsLINKER or PinsTPR+LINKER plus mud RNAi (Table 1). Thus, the PinsTPR must bind Mud to improve PinsLINKER-mediated spindle orientation. However, PinsTPR - Mud interaction is not sufficient for spindle orientation: PinsTPR alone can recruit Mud (Figure 4J), but has no spindle orientation ability (Figure 2G; Table 1). We conclude that the PinsTPR can recruit Mud, which together with the PinsLINKER domain, promotes optimal spindle orientation.
Figure 4. PinsTPR+LINKER requires Mud and the dynein/dynactin complex to improve spindle orientation compared to PinsLINKER alone.
(A) mud RNAi reduces PinsTPR+LINKER spindle orientation to that of PinsLINKER alone. (B) The Pins N226F TPR domain mutation abolishes Mud-Pins interaction in a FRET assay. (C) The PinsTPR(N226F)+LINKER mutant has spindle orientation equal to that of PinsLINKER alone. (D) lis-1 RNAi reduces PinsTPR+LINKER spindle orientation to that of PinsLINKER alone. (E) dlc RNAi reduces PinsTPR+LINKER spindle orientation to that of PinsLINKER alone. (F) Western blots show RNAi knockdown of Lis-1 and Dlc; detection of α-tubulin demonstrates equal loading of lysates. Scale bar 3 µm for A–E.
(G) Quantification of spindle orientation (mean spindle angle ± standard deviation). *, significantly better spindle orientation compared to PinsLINKER (p < 0.01).
(H–K) The PinsTPR domain is sufficient to recruit Mud to the cortex. Transfected GFP:Mud (residues 1825–2016, the Pins-binding domain) is recruited to the cortex by PinsTPR+LINKER (I) or PinsTPR (J) but not by Ed alone (H) or PinsLINKER (K). Scale bar 3 µm for H–K.
We next asked whether the PinsTPR and PinsLINKER domains must be together in the same protein to provide full spindle orientation function. Whereas a single PinsTPR+LINKER protein gives excellent spindle orientation (10.7 ± 7.1°, Table 1), expression of the PinsTPR and PinsLINKER as separate Ed fusion proteins resulted in reduced spindle orientation equal to that of PinsLINKER alone (27.2 ± 18.1°, Table 1). We conclude that Mud binds the TPR domain within the PinsTPR+LINKER protein, and that PinsTPR/Mud acts in cis to stimulate the PinsLINKER/DlgGK/Khc-73 spindle orientation pathway.
PinsTPR+LINKER spindle orientation requires the Lis1/dynein complex
Lis1 and the dynein complex are evolutionarily-conserved proteins required for spindle orientation from yeast to mammals (reviewed in Siller and Doe, 2009). The dynein complex is a microtubule minus-end directed motor complex that, when anchored at the plasma membrane, can exert pulling force on spindle pole microtubules (reviewed in Siller and Doe, 2009). Moreover, dynein complex proteins are known to interact with the Mud-related proteins in C. elegans and mammals (Couwenbergs et al., 2007; Merdes et al., 2000; Nguyen-Ngoc et al., 2007). Here we test whether Lis1 or the dynein complex is required for PinsTPR+LINKER or PinsLINKER spindle orientation in S2 cells. We find that dynein light chain (dlc) RNAi reduced endogenous Dlc protein levels (Figure 4F) and decreased PinsTPR+LINKER spindle orientation to levels similar to PinsLINKER alone (Figure 4E; Table 1). Similarly, lis1 RNAi depleted Lis1 protein levels (Figure 4F) and reduced PinsTPR+LINKER spindle orientation to PinsLINKER levels (Figure 4D; Table 1). In contrast, dlc RNAi had no effect on PinsLINKER spindle orientation (Table 1), indicating that it acts 'downstream' of the PinsTPR domain. We conclude that PinsTPR+LINKER requires Lis1/dynein complex activity to achieve accurate spindle orientation; in the absence of Lis1 or the dynein complex only the PinsLINKER pathway appears to be functional.
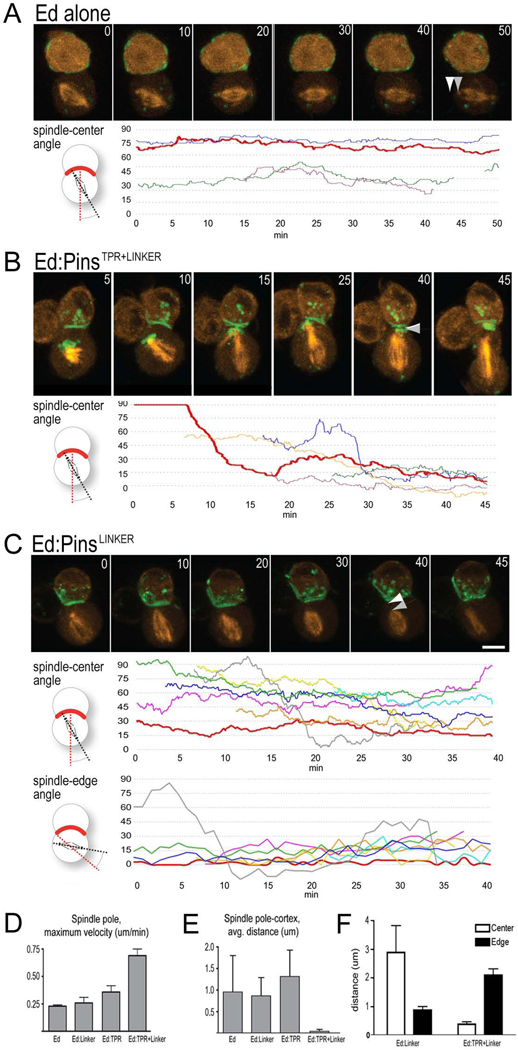
The PinsTPR+LINKER induces rapid, directional spindle alignment
We have shown that the PinsLINKER pathway provides partial spindle orientation, while the PinsTPR+LINKER pathway provides full spindle orientation. These differences could be due to many mechanisms that cannot be distinguished by fixed cell analysis, including timing of spindle orientation (e.g. the PinsLINKER pathway may only be active for part of mitosis), amplitude of spindle rocking (e.g. PinsLINKER could have larger amplitude), or directional spindle movement (e.g. only the PinsTPR+LINKER may induce movement towards the center of the crescent). To distinguish among these possibilities, we tagged Ed fusion proteins with GFP and the mitotic spindle with Cherry:α-tubulin and performed live imaging of spindle orientation in S2 cells expressing Ed alone, Ed:PinsLINKER, and Ed:PinsTPR+LINKER (Figure 5).
Figure 5. Live imaging of spindle orientation shows static spindles in PinsLINKER cells, but dynamic spindle movement in PinsTPR+LINKER cells.
To focus on the mechanism of spindle alignment, only Ed:Pins cells that started metaphase with misaligned spindles (>15°) were analyzed.
(A–C) Top row: A–C correspond to supplemental movie 1–supplemental movie 3. Time is in minutes; arrowheads, spindle pole-to-cortex distance. Each line is a different cell; red line, cell shown in still frames and supplemental movies. Bottom row: quantification of spindle orientation angle to crescent center or crescent edge (Y axis) over time (X axis). (A) Ed:GFP control cells have slowly drifting spindles (n=4). (B) Ed:PinsTPR+LINKER cells have dynamic spindle orientation (n=5). (C) Ed:PinsLINKER cells have poor spindle orientation to the crescent center (top tracing), but good spindle orientation to the crescent edge (bottom tracings; n = 8). Scale bar 2 µm.
(D) Ed:PinsTPR+LINKER cells have the greatest maximum spindle pole velocity.
(E) Ed:PinsTPR+LINKER cells have the closest spindle pole-to-cortex distance during the 10 min interval prior to anaphase onset.
(F) Ed:PinsLINKER cells show close spindle pole to edge of the crescent association, whereas Ed:PinsTPR+LINKER cells show close spindle pole to center of the crescent association.
We observed that Ed:GFP control cells had mitotic spindles that drifted relative to the Ed:GFP crescent (Figure 5A); the spindle never showed rapid movement (Figure 5D); and there was typically a gap between spindle poles and the cell cortex (Figure 5A, arrowheads; Figure 5E). This matches spindle movements in untransfected S2 cells (Echard et al., 2004; Goshima et al., 2007). A similar result was observed for Ed:PinsTPR cells (Figure 5D,E, and data not shown). In contrast, the Ed:PinsTPR+LINKER cells often showed rapid, directional spindle movement to align the mitotic spindle with the center of the Pins crescent (Figure 5B,F). Quantification of the maximum velocity of spindle movement confirms that the Ed:PinsTPR+LINKER induces a significantly greater spindle velocity compared to Ed alone, PinsLINKER, or PinsTPR (Figure 5D); similarly, the Ed:PinsTPR+LINKER cells showed a much tighter association with the spindle pole and the cell cortex than Ed alone, PinsLINKER, or PinsTPR (Figure 5E). We conclude that the PinsTPR+LINKER has the ability to induce rapid spindle movement towards the center of the cortical crescent.
Live imaging of the Ed:PinsLINKER cells revealed a novel phenotype that helped explain its partial spindle orientation phenotype. Ed:PinsLINKER cells showed no dynamic spindle movements (Figure 5D), no movement of the spindle pole towards the cortex (Figure 5E), and preferential alignment with the edge of the Pins crescent (Figure 5C, F). We conclude that slow spindle drift brings the initially misaligned spindle into contact with the edge of the crescent, where it becomes stabilized, leading to partial spindle orientation relative to the center of the crescent.
The Pins spindle orientation pathway is first active at prophase
To test for Pins function at interphase, we used the centriolar marker Sas-4, because mature centrosomal markers such as Centrosomin (Cnn) are not present during interphase in S2 cells (Goshima et al., 2007). We found that PinsTPR+LINKER has no effect on centriole positioning during interphase relative to Ed alone control (Figure 6A,C and Figure S3). We next determined whether PinsTPR+LINKER can anchor centrosomes during prophase by measuring centrosome position relative to the center of the Pins crescent (see methods). We found that the PinsTPR+LINKER showed excellent centrosome alignment at prophase (12 ± 7°; Figure 6B,C and Figure S3), indistinguishable from the angle of spindle orientation at metaphase (Figure 2K,L). Similarly, all other Pins constructs showed prophase centrosome alignment that matched metaphase spindle alignment: for example Ed alone showed no centrosome alignment (45 ± 22°) and PinsLINKER showed intermediate alignment (28.2 ± 16.2°; Figure S3). Thus, Pins can anchor centrosomes at prophase, and this foreshadows the spindle orientation observed at metaphase. This does not mean that one centrosome always maintains a position near the Pins crescent throughout mitosis, however, as movies show centrosome separation displacing both centrosomes from the PinsTPR+LINKER cortex, followed by rapid movement of the bipolar spindle to restore spindle alignment (Figure 5C,C'; data not shown). We conclude that the Pins spindle orientation pathway is first activated at the start of prophase.
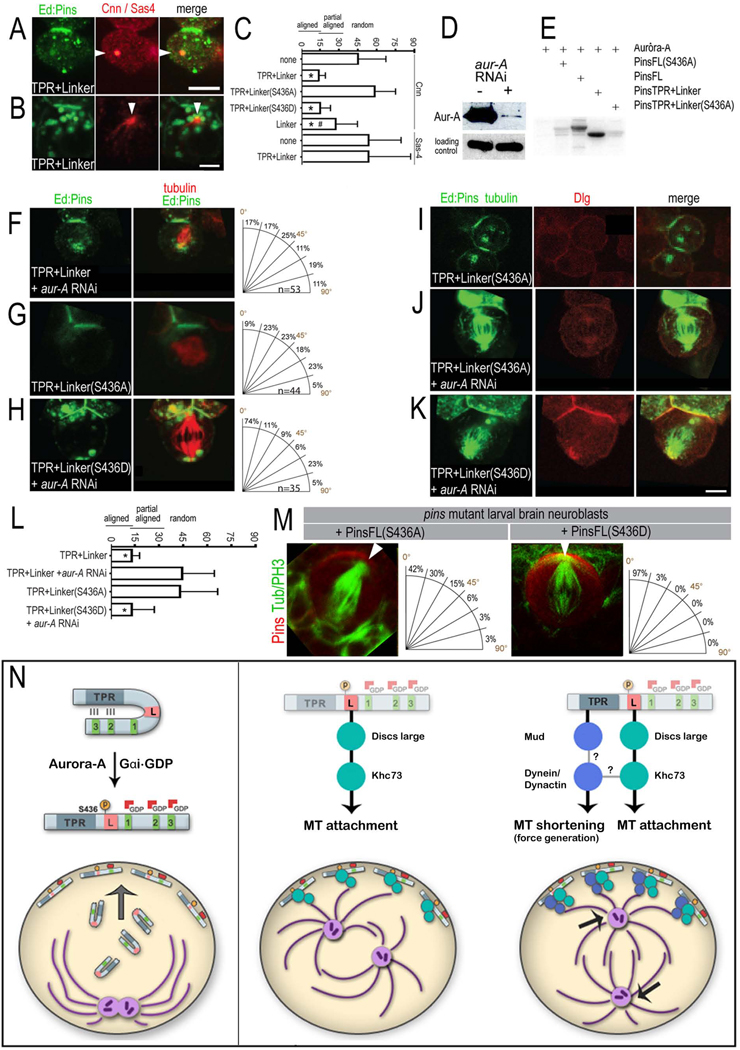
Figure 6. Aurora-A directly phosphorylates S436 in the Pins Linker to recruit the Dlg/Khc-73 complex and regulate spindle orientation.
(A) Interphase S2 cell transfected with Ed:FLAG:PinsTPR+LINKER stained for the centriolar marker Sas-4 showing no centriole anchoring to the crescent.
(B) Prophase S2 cell transfected with Ed:FLAG:PinsTPR+LINKER stained for the centrosomal marker Cnn showing centrosome anchoring to the crescent.
(C) Quantification of mean centrosome (Cnn) or centriole (Sas4) angle ± standard deviation. *, significant compared to Ed alone control (p < 0.01).
(D) Western blot demonstrates partial RNAi knockdown of Aurora-A; alpha-tubulin blot demonstrates equal loading of lysates.
(E) Aurora-A phosphorylates Pins on residue 436 in the Linker domain. Recombinant Aurora-A kinase and [γ-32P]-ATP were incubated alone or in the presence of wild-type or S436A versions of full-length or TPR+Linker Pins.
(F–K) Aurora-A phosphorylation of Pins S436 is required for Dlg recruitment and spindle orientation. Scale bar 3 µm for C–H.
(L) Quantification of spindle orientation (mean spindle angle ± standard deviation). The number of cells scored for each genotype are 26, 34, 28, 26, 8, 22, and 19 (from top to bottom). *, highly significant versus Ed alone (p < 0.01); #, significantly different from PinsTPR+LINKER (p < 0.05 when assayed by either Cnn or Sas-4).
(M) PinsLINKER/Aurora-A pathway is required in vivo for neuroblast spindle orientation. Larval neuroblasts in a pins62 homozygous mutant expressing the unphosphorylatable PinsFL(S436A) protein show defective spindle orientation (left panel; note alignment to edge of crescent), whereas the phosphomimetic PinsFL(S436D) protein show spindle pole alignment to the center of the Pins crescent (right panel). Pins (red), α-tubulin (green).
(N) A model for Pins-mediated spindle orientation. See text for details.
Aurora-A activates the Pins spindle orientation pathway by phosphorylating S436 within the PinsLINKER domain
Lack of Pins function during interphase could be due to immature centrosomes that nucleate few microtubules (Goshima et al., 2007), and/or due to activation of the Pins pathway at the onset of mitosis. The Aurora-A kinase is activated at the start of prophase (Hutterer et al., 2006) and is required for neuroblast spindle orientation (Lee et al., 2006; Wang et al., 2006); here we test whether it is required to activate the Pins spindle orientation pathway in S2 cells. We used aurora-A RNAi to reduce endogenous Aurora-A protein levels in S2 cells (Figure 6D), and found that this blocked PinsTPR+LINKER from orienting the mitotic spindle (41 ±24.5°; Figure 6F, Table 1). A second aurora-A RNAi construct gave the same effect (data not shown).
Aurora-A could be acting to promote centrosomal maturation (Berdnik and Knoblich, 2002), or more directly to phosphorylate a member of the Pins spindle orientation pathway. Pins contains three predicted Aurora-A consensus phosphorylation sites (Ferrari et al., 2005), so we performed a deletional analysis to identify which site, if any, warranted further characterization. The PinsTPR+LINKER protein extends to amino acid 466; truncation down to amino acid 446 (Pins1–446) retained excellent spindle orientation (13.4 ±13.3°), whereas truncations down to amino acid 435 (Pins1–435) completely blocked spindle orientation (41.3 ± 25.1°), as did all shorter Pins constructs (Table 1). We conclude that the Pins amino acids 436–446 are essential for its spindle orientation function. Interestingly, this small domain is evolutionarily conserved from sea urchin to mammals and contains a single predicted Aurora-A phosphorylation site (S436) (Figure S4). We used recombinant Aurora-A kinase to show that this site is a direct target of Aurora-A in vitro, and that mutation of this site (S436A) completely blocked Aurora-A phosphorylation of Pins (Figure 6E); additional predicted Aurora-A sites outside this essential domain (S479 and S571) were not phosphorylated (Figure 6E). Thus, Aurora-A specifically phosphorylates S436 within the PinsLINKER domain.
We next tested the role of S436 phosphorylation in spindle orientation, and found that a non-phosphorylatable mutant protein, PinsTPR+LINKER(S436A), failed to orient the mitotic spindle (43 ± 20.8°; Figure 6G), similar to the effect of aurora-A RNAi (41 ± 24.5°; Figure 6F). In contrast, the phosphomimetic S436D mutation resulted in excellent spindle orientation (13.7 ± 11.3°), similar to PinsTPR+LINKER (Table 1). Importantly, the phosphomimetic PinsTPR+LINKER(S436D) protein was able to orient the mitotic spindle even after nearly complete RNAi depletion of Aurora-A (Figure 6D,H), showing that the role of Aurora-A in spindle orientation is to phosphorylate Pins, rather than promoting centrosomal maturation.
How does S436 phosphorylation activate the Pins spindle orientation pathway? The similarity of aurora-A, dlg, and khc-73 RNAi phenotypes led us to test whether S436 phosphorylation was required to recruit Dlg to the PinsTPR+LINKER cortical crescent. Indeed, the non-phosphorylatable PinsTPR+LINKER(S436A) failed to recruit endogenous Dlg (Figure 6I). RNAi depletion of Aurora-A also blocked recruitment of Dlg to the wild type Pins cortical domain (Figure 6J), but did not have any effect on recruitment induced by the phosphomimetic PinsTPR+LINKER(S436D) protein (Figure 6K). We conclude that Aurora-A phosphorylates the PinsLINKER at S436, triggering recruitment of Dlg protein and the activation of the PinsLINKER/Dlg/Khc-73 spindle orientation pathway at the onset of mitosis.
The PinsLINKER pathway is required for spindle orientation in larval neuroblasts
We next tested whether the PinsLINKER/Aurora-A pathway is active in larval neuroblasts in vivo. We generated transgenic flies that allow inducible expression of full length Pins with either phosphorylation blocking or mimicking mutations -- PinsFL(S436A) and PinsFL(S436D) -- in a pins62 mutant background. We find that the PinsFL(S436D) phosphomimetic protein provides full spindle orientation function in larval brain neuroblasts lacking endogenous Pins (8.2 ± 4.0°, n=32; Figure 6M). In contrast, the non-phosphorylatable PinsFL(S436A) protein shows spindle orientation defects in larval brain neuroblasts (23.2 ± 17.8°, n=30; Figure 6M) that closely match the pins62 mutant alone (23.2 ± 23.1°, n=12; Siegrist and Doe, 2005). Thus, the single amino acid substitution in PinsFL(S436A) blocks most or all Pins-mediated spindle orientation function in larval brain neuroblasts, replicating and verifying our findings in the S2 assay.
We also assessed if PinsFL(S436A) protein has a dominant negative function. Indeed, expression of PinsFL(S436A) in a wild type background resulted in modest defects in spindle orientation (17.3 ± 13.6°, n=56), compared to wild type controls (6.2 ± 3.4°, n=48). In contrast, expression of the PinsFL(S436D) phosphomimetic protein did not significantly affect spindle orientation (6.3 ± 3.5°, n=13). Taken together, we conclude that the PinsLINKER/Aurora-A spindle orientation mechanism is active in vivo and required for proper spindle orientation in larval neuroblasts.
Discussion
We have developed an 'induced cell polarity' system using the Drosophila S2 cell line that permits rapid testing of individual proteins, domains, and amino acids for their role in cell polarity and spindle orientation. We use this system to dissect the role of the evolutionarily-conserved Pins protein in spindle orientation. We identify a previously unknown PinsLINKER pathway, show that it functions in larval neuroblasts, and show that the PinsTPR acts in cis to the PinsLINKER to improve spindle orientation. A model summarizing our results is shown in Figure 6N.
The PinsLINKER/Dlg/Khc-73 pathway
A surprising result of our studies is the importance of the PinsLINKER domain for spindle orientation in the S2 assay and within neuroblasts in vivo. Only this domain is sufficient for spindle orientation, and a single point mutation in the linker domain (S436A) results in spindle orientation defects in larval neuroblasts that closely mimic the pins null mutant phenotype. Based on domain mapping and epistasis analysis, we have identified a linear pathway from cortical PinsLINKER to the plus ends of astral microtubules: (1) Aurora-A phosphorylates PinsLINKER on a single amino acid, serine 436; (2) the phosphorylated PinsLINKER binds and recruits Dlg; (3) the kinesin Khc-73 moves to astral microtubule plus-ends using its motor domain and may be anchored at the plus ends by its Cap-Gly domain (Siegrist and Doe, 2005); (4) the Khc-73MBS domain binds the cortical DlgGK domain, thereby linking Khc-73+ astral microtubule plus ends to the Dlg cortical domain (Figure 6N). Interestingly, this pathway is active in both directions during mitosis. Cortical Pins acts through Dlg and Khc-73 to regulate spindle orientation (this work), and spindle associated Khc-73 acts through Dlg and Pins to induce Pins/Gαi functional cortical polarity in neuroblasts (Siegrist and Doe, 2005).
Why does the PinsLINKER pathway provide only partial spindle orientation function? Live imaging rules out several possible explanations, such as PinsLINKER-induced spindle rocking variability, or that PinsLINKER only functions during a narrow window during mitosis. Live imaging shows that in PinsLINKER cells, the spindle drifts until it is immobilized at the edge of the crescent (Figure 5B, D). This is consistent with the fact that Khc-73 is a plus-end directed motor protein, and thus unable to generate pulling forces to bring the centrosome closer to the cell cortex; at best it would provide a static link between astral microtubules and the cell cortex.
The PinsTPR/Mud/dynein pathway
The PinsTPR domain can improve the PinsLINKER spindle orientation to a level matching wild type neuroblasts. We propose that the PinsTPR domain directly binds Mud, and that Mud interacts with the dynein/dynactin/Lis1 complex to enhance PinsLINKER spindle orientation. This model is based on five observations. First, the PinsTPR domain binds Mud in vitro and the two proteins co-immunoprecipitate from in vivo lysates (Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006)(Figure 4B); this interaction is conserved in the related C. elegans and mammalian proteins (Couwenbergs et al., 2007; Merdes et al., 2000; Nguyen-Ngoc et al., 2007). Second, the PinsTPR and PinsTPR+LINKER but not the PinsLINKER can recruit Mud to the cortex of S2 cells. Third, PinsTPR+LINKER-mediated spindle orientation requires the dynein complex proteins Dlc and Lis1. Fourth, PinsTPR+LINKER-mediated spindle orientation exhibits rapid, directional spindle movement towards the center of the Pins cortical crescent, similar to dynein-dependent spindle orientation in Drosophila neuroblasts (Siller and Doe, 2008). Fifth, PinsTPR+LINKER-mediated spindle orientation leads to dynein-dependent movement of the spindle pole close to the cell cortex, consistent with dynein minus-end directed pulling of astral microtubules, as observed in other cell types (Siller and Doe, 2009).
If PinsTPR recruits Mud, and Mud recruits the dynein complex, then why doesn't PinsTPR have spindle orienting function on its own? The simplest model is that PinsTPR/Mud alone is unable to recruit or activate the dynein complex. Alternatively, the PinsLINKER pathway could be required for "presenting" microtubule plus ends to an active PinsTPR/Mud/Dynein complex, which fits with the requirement for PinsTPR and PinsLINKER acting in cis. In summary, our data show that the PinsTPR and PinsLINKER domains provide distinct functions, both of which are required for optimal spindle orientation (Figure 6N). Interestingly, spindle orientation in S2 cells does not show "telophase rescue" -- a phenomena whereby spindles that are partially oriented in metaphase/anaphase neuroblasts become aligned with the cell polarity axis by telophase (Siller and Doe, 2008) -- consistent with the absence of redundant spindle orientation pathways in this assay.
Regulation of Pins spindle orientation pathways
The PinsTPR pathway is regulated by Gαi binding to the GoLoco domain, relieving intramolecular TPR-GoLoco interactions, and making the TPR domain accessible for intermolecular interactions (Du and Macara, 2004; Nipper et al., 2007). In addition, Gαi is required to recruit Pins to the cell cortex where it can interact with regulator and effector proteins (Yu et al., 2003). In our S2 spindle orientation assay we can bypass a requirement for Gαi by simply deleting the GoLoco domain (thereby freeing the TPR for intermolecular interactions) and tethering the PinsTPR+LINKER protein to the cortex by fusion with the Ed transmembrane protein. Thus, Gαi is important to activate and localize full length Pins, but not as an effector of Pins-mediated spindle orientation.
In contrast, the PinsLINKER pathway is not regulated by Gαi, because full length Pins in the absence of Gαi provides equal spindle orientation to PinsLINKER, suggesting that the PinsLINKER is active when Pins is in the 'closed' form. The Khc-73 mammalian ortholog GAKIN transports hDlg to the cell cortex (Hanada et al., 2000), but there are several reasons to think that this mechanism does not activate the PinsLINKER pathway. First, cortically tethered DlgGK domain requires Khc-73 for spindle orientation, which rules out a role for Khc-73 in merely transporting Dlg to the cortex; second, khc-73 RNAi does not block the ability of PinsLINKER to recruit Dlg to the cortex.
We have shown that Aurora-A kinase activates the PinsLINKER spindle orientation pathway by phosphorylating S436 in the linker domain, and that this pathway is required for accurate spindle orientation in vivo for larval neuroblast asymmetric cell division. Neuroblasts expressing the non-phosphorylatable form of Pins (S436A) have a weaker spindle orientation phenotype than aurora-A null mutants (Lee et al., 2006; Wang et al., 2006), as expected due to Aurora-A regulating multiple Pins-independent processes required for spindle orientation, such as centrosome maturation, cell cycle progression, and cell polarity in flies (Barr and Gergely, 2007; Berdnik and Knoblich, 2002; Hutterer et al., 2006; Lee et al., 2006; Wang et al., 2006; Wirtz-Peitz et al., 2008). However, we show that a Pins phosphomimetic mutant (S436D) allows spindle orientation even after RNAi depletion of Aurora-A levels, suggesting that Aurora-A phosphorylation of PinsS436 is essential for Pins-dependent spindle orientation in the S2 cell assay. Furthermore, our finding that the PinsS436A protein has no spindle orientation activity in pins mutant larval neuroblasts, and has dominant negative activity in the presence of endogenous Pins, shows that the Aurora-A/PinsLINKER pathway is required for spindle orientation in larval neuroblasts as well.
The Pins spindle orientation pathway is cell cycle regulated: interphase S2 cells that have polarized PinsTPR+LINKER do not capture centriole/centrosomal microtubules. There are at least two reasons for the lack of Pins interphase activity. First, the level of the Aurora-A kinase is low during interphase, and we have shown that Aurora-A phosphorylation of Pins S436 is essential for Pins-mediated spindle orientation. Second, interphase centrosomes are immature, lacking Cnn and nucleating few microtubules (Goshima et al., 2007). Expression of the Pins S436D protein, which is fully functional during mitosis even after Aurora-A depletion, still has no ability to capture centrioles during interphase. Thus, both centrosome maturation and Aurora-A activation are required for Pins-mediated spindle orientation in S2 cells.
Concluding remarks
Here we have induced cell polarity and spindle orientation in a cultured cell line. We used this system to identify two pathways regulating spindle orientation, to establish molecular epistasis within each pathway, and to identify the target of the mitotic kinase Aurora-A that coordinates cell cycle progression with spindle orientation. In the future, this system should be useful for characterizing spindle orientation pathways from other Drosophila cell types or from other organisms, identifying the mechanisms that control centrosome or spindle asymmetry, and characterizing the establishment and maintenance of cortical polarity. In each of these cases, our induced polarity system should be useful for rapid protein structure/function studies and high-throughput drug or RNAi loss-of-function studies.
MATERIALS AND METHODS
Additional details are available in the supplemental methods.
Plasmid construction, cell culture, and RNAi
Ed:FLAG, Ed:GFP, and Ed:Cherry constructs were made in pMT-V5 (Invitrogen, Carlsbad, CA, USA) replacing the Ed cytoplasmic domain with the visualization tag and the protein of interest at the C-terminus (e.g. Ed:FLAG:Pins). Proteins with different visualization tags conferred similar spindle orientation (Table 1). Standard methods were used to grow S2 cells (Goshima et al., 2007). Cells were seeded at ~1–3 × 106 cells per well, transfected with 0.4-1 µg total DNA using Effectene® (Qiagen, Germantown, MD, USA), and gene expression was induced by 500 µM CuSO4 for 24–48 hours. Cell clustering was induced by rotation at ~175 RPM for 1–3 hours.
For RNAi treatment, primers that amplify ~400–600 base pairs were designed at http://www.dkfz.de/signaling2/e-rnai/ with T7 promoter tags. PCR-amplified sequences were reverse transcribed using the Megascript® T7 kit (Ambion, Austin, TX, USA). S2 cells were seeded at 1 × 106 cells per well in 1 ml of serum-free Schneider media and incubated with 100 µl (~10 µg). After 1 hour, 2 ml of serum-containing media was added and cells were incubated for an additional 3 days.
Live imaging, immunostaining, and western blots
For live imaging, cells were plated on poly-L-lysine coated chambered coverslip (Nalge). Three focal planes spaced 2 µm apart were collected every 5 seconds using Volocity software (Improvision) on a McBain spinning disc microscope with a 60× 1.4NA lens and a Hamamatsu EM-CCD camera. Volocity was used to XY crop, Z merge using maximum pixel intensity, and export as a tiff series; ImageJ was used to generate Quicktime movies.
For immunostaining, S2 cells were fixed for 20 minutes in 4% paraformaldehyde, stained (Goshima et al., 2007), and imaged on a Leica SP2 confocal microscope with a 60× 1.4 NA lens. For in vivo imaging of larval neuroblasts, we used neuroblast-specific worniugal4 to express UAS-pinsFL(S436A) or -pinsFL(S436D) in a pins62 homozygous mutant background (Nipper et al., 2007). For Westerns, 20 µg of S2 cell extracts were used per lane. The localization of endogenous polarity proteins in S2 cells is shown in Figure S1. Antibodies and dilutions: rat Lis1 1:1000 (Siller et al., 2005), rabbit Gαi 1:1000 (Nipper et al., 2007), mouse Dlg 1:250 (Developmental Studies Hybridoma Bank, Iowa), rabbit Sas-4 1:500 (Basto et al., 2006), rabbit Aurora-A (Berdnik and Knoblich, 2002), rabbit centrosomin 1:1000 (Megraw et al., 1999), rat Pins 1:500 (Yu et al., 2000), mouse FLAG (Sigma, 1:500), rat tubulin 1:1000 (Abcam), mouse tubulin 1:1000 (Sigma), rabbit phosphohistone-3 1:1000 (Upstate), and rabbit HA 1:1000 (Covance).
Measuring cortical polarity, spindle orientation, and centrosome alignment
Pixel intensity was measured in ImageJ. Ed crescents over 75° cell diameter were excluded. Spindle angles were measured using a vector perpendicular to the center of the Ed crescent and a vector matching the spindle or connecting the spindle poles. To estimate the spindle angle measurement error we compared the mean angle from multiple independent experiments of Ed-PinsLINKER and Ed-PinsFL + Gαi (three trials each). Based on this analysis, and assuming that the sources of error were consistent, we estimated a standard error of ± 3° in spindle angle measurement in our experiments.
In vitro kinase assays
Recombinant Aurora-A kinase was purchased from Millipore (Billerica, MA, USA). Pins constructs (10 µg) and Aurora-A (100 ng) were diluted in ice-cold assay buffer (20mM Tris, pH 7.4, 100mM NaCl, 1mM DTT, 10mM MgCl2, and 10 µM ATP). ATP-γ-32P (5 µCi) was added at 30 °C for 30 minutes. Reactions were quenched by addition of SDS loading buffer. Samples were resolved by SDS-PAGE and analyzed using a Storm 860 and Image Quant 5.1 (Molecular Dynamics).
FRET assays
Sensor proteins were diluted to 200 nM in 20 mM Tris, pH 7.5, 100 mM NaCl, 1 mM DTT with or without MudPBD, excited at 433 nm, and the YFP (525 nm) to CFP (475 nm) emission ratios were measured.
Supplementary Material
Acknowledgements
We thank Clemens Cabernard for Figure 6M; Taryn Gillies for Figure 6N; K; S. Liu, R. Vale, K. Siller, S. Spencer, T. Kaufman, J. Raff, J. Knoblich and the Bloomington Stock Center for providing fly, DNA, or antibody reagents; Brett Wee for technical assistance; and B. Bowerman, C. Cabernard, S. O'Rourke, S. Rogers and S. Atwood for comments. Funding: Damon Runyon postdoctoral fellowship (CAJ), NIH GM068032 (KEP), and HHMI (CQD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bai J, Chiu W, Wang J, Tzeng T, Perrimon N, Hsu J. The cell adhesion molecule Echinoid defines a new pathway that antagonizes the Drosophila EGF receptor signaling pathway. Development. 2001;128:591–601. doi: 10.1242/dev.128.4.591. [DOI] [PubMed] [Google Scholar]
- Barr AR, Gergely F. Aurora-A: the maker and breaker of spindle poles. J Cell Sci. 2007;120:2987–2996. doi: 10.1242/jcs.013136. [DOI] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Bellaiche Y, Radovic A, Woods DF, Hough CD, Parmentier ML, O'Kane CJ, Bryant PJ, Schweisguth F. The Partner of Inscuteable/Discs-large complex is required to establish planar polarity during asymmetric cell division in Drosophila. Cell. 2001;106:355–366. doi: 10.1016/s0092-8674(01)00444-5. [DOI] [PubMed] [Google Scholar]
- Berdnik D, Knoblich JA. Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr Biol. 2002;12:640–647. doi: 10.1016/s0960-9822(02)00766-2. [DOI] [PubMed] [Google Scholar]
- Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Couwenbergs C, Labbe JC, Goulding M, Marty T, Bowerman B, Gotta M. Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J Cell Biol. 2007;179:15–22. doi: 10.1083/jcb.200707085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Echard A, Hickson GR, Foley E, O'Farrell PH. Terminal cytokinesis events uncovered after an RNAi screen. Curr Biol. 2004;14:1685–1693. doi: 10.1016/j.cub.2004.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Marin O, Pagano MA, Meggio F, Hess D, El-Shemerly M, Krystyniak A, Pinna LA. Aurora-A site specificity: a study with synthetic peptide substrates. Biochem J. 2005;390:293–302. doi: 10.1042/BJ20050343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl M, Yanagida M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Lin L, Tibaldi EV, Reinherz EL, Chishti AH. GAKIN, a novel kinesin-like protein associates with the human homologue of the Drosophila discs large tumor suppressor in T lymphocytes. J Biol Chem. 2000;275:28774–28784. doi: 10.1074/jbc.M000715200. [DOI] [PubMed] [Google Scholar]
- Hutterer A, Berdnik D, Wirtz-Peitz F, Zigman M, Schleiffer A, Knoblich JA. Mitotic activation of the kinase Aurora-A requires its binding partner Bora. Dev Cell. 2006;11:147–157. doi: 10.1016/j.devcel.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat Cell Biol. 2006;8:586–593. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- Lee CY, Andersen RO, Cabernard C, Manning L, Tran KD, Lanskey MJ, Bashirullah A, Doe CQ. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 2006;20:3464–3474. doi: 10.1101/gad.1489406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson MA, Horvitz HR, van den Heuvel S. LIN-5 is a novel component of the spindle apparatus required for chromosome segregation and cleavage plane specification in Caenorhabditis elegans. J Cell Biol. 2000;148:73–86. doi: 10.1083/jcb.148.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw TL, Li K, Kao LR, Kaufman TC. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 1999;126:2829–2839. doi: 10.1242/dev.126.13.2829. [DOI] [PubMed] [Google Scholar]
- Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol. 2000;149:851–862. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc T, Afshar K, Gonczy P. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol. 2007;9:1294–1302. doi: 10.1038/ncb1649. [DOI] [PubMed] [Google Scholar]
- Nipper RW, Siller KH, Smith NR, Doe CQ, Prehoda KE. Galphai generates multiple Pins activation states to link cortical polarity and spindle orientation in Drosophila neuroblasts. Proc Natl Acad Sci U S A. 2007;104:14306–14311. doi: 10.1073/pnas.0701812104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier ML, Woods D, Greig S, Phan PG, Radovic A, Bryant P, O'Kane CJ. Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila. J Neurosci. 2000;20:RC84. doi: 10.1523/JNEUROSCI.20-14-j0003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo E, Sampaio P, Januschke J, Llamazares S, Varmark H, Gonzalez C. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell. 2007;12:467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Rolls MM, Albertson R, Shih HP, Lee CY, Doe CQ. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J Cell Biol. 2003;163:1089–1098. doi: 10.1083/jcb.200306079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans N, Wang PY, Du Q, Petralia RS, Wang YX, Nakka S, Blumer JB, Macara IG, Wenthold RJ. mPins modulates PSD-95 and SAP102 trafficking and influences NMDA receptor surface expression. Nat Cell Biol. 2005;7:1179–1190. doi: 10.1038/ncb1325. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Shevchenko A, Knoblich JA. A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr Biol. 2000;10:353–362. doi: 10.1016/s0960-9822(00)00401-2. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–1335. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Siller K, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009 doi: 10.1038/ncb0409-365. in press. [DOI] [PubMed] [Google Scholar]
- Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Lis1/dynactin regulates metaphase spindle orientation in Drosophila neuroblasts. Dev Biol. 2008;319:1–9. doi: 10.1016/j.ydbio.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller KH, Serr M, Steward R, Hays TS, Doe CQ. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol Biol Cell. 2005;16:5127–5140. doi: 10.1091/mbc.E05-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Somers GW, Bashirullah A, Heberlein U, Yu F, Chia W. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 2006;20:3453–3463. doi: 10.1101/gad.1487506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard FS, Kimple RJ, Siderovski DP. Return of the GDI: the GoLoco motif in cell division. Annu Rev Biochem. 2004;73:925–951. doi: 10.1146/annurev.biochem.73.011303.073756. [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Cai Y, Kaushik R, Yang X, Chia W. Distinct roles of Galphai and Gbeta13F subunits of the heterotrimeric G protein complex in the mediation of Drosophila neuroblast asymmetric divisions. J Cell Biol. 2003;162:623–633. doi: 10.1083/jcb.200303174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Morin X, Cai Y, Yang X, Chia W. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell. 2000;100:399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.