Abstract
Transmissible spongiform encephalopathies such as scrapie in sheep, Creutzfeldt-Jakob disease (CJD) in humans, and bovine sporadic encephalopathy in cattle are characterized by the accumulation of a misfolded protein: the pathological prion protein. Ever since bovine sporadic encephalopathy was discovered as the likely cause of the new variant of CJD in humans, parallels between human and animal transmissible spongiform encephalopathies must be viewed under the aspect of a disease risk for humans. In our study we have compared prion characteristics of different forms of sheep scrapie with those of different phenotypes of sporadic CJD. The disease characteristics of sporadic CJD depend considerably on the prion type 1 or 2. Our results show that there are obvious parallels between sporadic CJD type 1 and the so-called atypical/Nor98 scrapie. These parelleles apply to the deposition form of pathological prion protein in the brain, detected by the paraffin-embedded-tissue blot and the prion aggregate stability with regard to denaturation by the chaotropic salt guanidine hydrochloride. The same applies to sporadic CJD type 2 and classical scrapie. The observed parallels between types of sporadic CJD and types of sheep scrapie demonstrate that distinct groups of prion disease exist in different species. This should be taken into consideration when discussing interspecies transmission.
Transmissible spongiform encephalopathies (TSEs) are characterized by aggregates of a partly protease-resistant, self-replicating protein called a “proteinacous infectious particle” (hereafter referred to as “prion”) in the central nervous system (CNS). According to the prion hypothesis, the disease-associated prion protein (PrPSc) is the principal or only constituent of the infectious agent.1 The physiological isoform, a cell surface protein (PrPc), is expressed not only in the CNS, but also in a number of non-neuronal tissues.
A long incubation period followed by a short clinical disease course after experimental transmission to animals, led in the early 50s of the last century to the concept of a “slow virus disease.”2 Serial passages of scrapie isolates in inbred mice and hamsters revealed different incubation periods and vacuolation patterns defining strains, which was in line with the virus hypothesis,3 although a causative virus was not found.4 Prion strains are defined by their different incubation times and lesion profiles on inoculation in a new host species with an identical genetic background of the prion protein gene (PRNP).5,6 For human sporadic Creutzfeldt-Jakob disease (CJD) no animal model that propagates PrPSc characteristics of all known clinical phenotypes is currently available. The most promising attempt to find out whether clinical phenotypes of sporadic CJD represent strains in a new host seems to be the bank vole model.7 However, at least two biochemically distinguishable PrPSc types of sporadic CJD have been identified in the original species, termed type 1 and type 2. It has been shown in human sporadic CJD that, besides the methionine/valine polymorphism at codon 129 of the PRNP, the prion type is responsible for the clinical disease course as well as the pathological lesion profile.8 It is widely accepted that human prion types 1 and 2 result from different pathological protein conformations, leading to a difference in the proteinase K-cleavage sites despite identical primary sequences.9 If all clinically and neuropathologically distinguishable sporadic CJD phenotypes (resulting from the combination of PrPSc types and relevant PRNP polymorphisms) belong to different strains, each of the human prion types could contain several strains. Applying this notion to the numerous scrapie strains that have been isolated in inbred mouse lines after the transmission of ovine scrapie samples,10 classical sheep scrapie may represent only one prion type11 harboring a certain heterogeneity.12 The recently identified new forms in sheep scrapie13 and bovine spongiform encephalopathy (BSE)14,15 differ from the previously described “classical” forms in several aspects such as the Western blot profile, histopathological lesion profile, and epidemiology. Potential parallels between animal and human prion diseases can be detected15 which might be of some relevance regarding the transmissibility to humans. Parallels between human and animal TSEs have received increasing attention ever since classical BSE was discovered to be the likely cause of human variant CJD.16
This study compares the deposition characteristics and aggregate stability of naturally occurring atypical/Nor98 and classical sheep scrapie with those of human PrPSc type 1 and type 2 of sporadic CJD. We hypothesize that similarities between distinct prion diseases in sheep and groups of clinical phenotypes in sporadic CJD characterize interspecies groups of prion diseases. This concept suggests that PrPSc types are the major determinant of prion disease forms. On the basis of these prion types, prion strains may be caused by additional factors that manifest themselves as strain characteristics on inoculation into a new host. Consequently, we will use the term “prion strain” only if incubation time and lesion profile have been defined in a different host species. In contrast, the term “prion type” will apply to a group within a prion disease that is markedly set apart by biochemical characteristics such as the Western blot profile and the aggregate stability and distinct forms of PrPSc deposition. Thus, it is conceivable that within one prion type more than one strain may be found, but not vice versa.
Materials and Methods
Animals
CNS and lymphatic tissue were taken from 19 German sheep (15 ALRQ/ALRQ, two ALRQ/VLRQ, and two VLRQ/ALRH; letters represent amino acids at codon 136, 141, 154, and 171, respectively), five Norwegian sheep (four ALRQ/VLRQ and one VLRQ/ALRH), and nine French sheep (three ALRQ/ALRQ, three ALRQ/VLRQ, and three VLRQ/VLRQ) diagnosed with classical scrapie as well as 25 Norwegian Nor98 cases (one ALRQ/ALRQ, one ALHQ/ALRQ, six ALHQ/ALHQ, one ALHQ/ALRH, two ALHQ/AFRQ, five AFRQ/AFRQ, four ALHQ/ALRR, two ALRR/AFRQ, one ALRR/ALRR, and one unknown) and one German atypical case (ALHQ/ALRQ). Tissues from six clinically healthy sheep (four ALRR/ALRR, one ALRR/ALRH, and one ALRQ/ALRQ) originating from scrapie-free flocks were used as negative controls. The PrP genotypes were determined either by PCR and melting curve analysis17 or by automated sequencing as described previously.13
Sporadic CJD Cases
CNS tissues came from patients with a diagnosis of sporadic CJD, who were included in the National CJD Surveillance Study in Germany (approved by the ethics committee of the medical faculty of the University of Goettingen). The methionine (M)/valine (V) polymorphism at codon 129 of the PRNP was determined by using a standard protocol according to Windl et al.18 Tissues from frontal, temporal, and parietal cortices as well as cerebellum were used to determine the PrPSc type according to the Parchi classification, which was chosen due to its reproducibility in different laboratories. Alternative classifications do not take into account possible inconsistent pH-conditions during proteinase K-digestion, which result in band shifting during PrPSc typing.19 Cases with mixed PrPSc types or PRNP-mutations were excluded from this study. The final setting included tissue from 10 patients with PrPSc type 1 (four MM1, three MV1, and three VV1) and 10 patients with PrPSc type 2 (three MM2, three MV2, and four VV2).
Tissue Samples
CNS and lymphatic tissues were either snap frozen and stored at −80°C or fixed in 4% buffered formalin and embedded in paraffin. A decontamination step with formic acid (98%) was applied to the majority of fixed tissue blocks.
Paraffin-Embedded-Tissue Blot
The paraffin-embedded-tissue (PET) blot method was performed as described elsewhere.20,21 Primary antibodies were mAb L42, mAb P4 (both from R-Biopharm AG, Darmstadt, Germany), or F89/160.1.5 (Veterinary Medical Research and Development, Pullman, WA) for ovine tissues and mAb 12F10 (kindly provided by W. Bodemer and D. Motzkus, German Primate Center, Goettingen, Germany) for human sections (each at a dilution of 1:5000). An alkaline phosphatase-coupled goat anti-mouse antibody (Dako, Glostrup, Denmark) and NBT/BCIP subtrate were used to visualize the detection of PrPSc. Tissue sections of all samples were also put on glass slides and stained with H&E or Luxol Fast Blue/PAS to aid the identification of neuroanatomical structures in the PET blots. A total of 94 gray and white matter structures were assessed for intensity (quantified in 0.5 grades ranging from 0 to 4) and PrPSc deposition patterns, which were verified by immunohistochemistry. Intensity profiles using 17 representative neuroanatomic sites of the sheep brain were created by Sigma Plot software (Systat Software Inc., San Jose, CA).
Immunohistochemistry
Immunohistochemistry was performed as described before.21 The primary mAbs P4, L42, F89/160.1.5, and 12F10 were used 1:500 in combination with an alkaline phosphatase-coupled goat anti-mouse antibody (Dako) and neufuchsine-chromogen-subtrate. Alternatively, a commercially available kit from Dako (Envision AEC) was applied by using mAb F89/160.1.5 at a dilution of 1:2000 in combination with the mAb 2G11 (1:200).
Tissue Homogenates
Ten percent tissue homogenates (w/v) were prepared in PBS containing 0.5% deoxycholic acid sodium salt using glass grinding tubes and pestles or 20% homogenates were obtained by the standard sampling procedure of the TeSeE Western blot Kit (Bio-Rad, Hercules, CA).
Western Blot
Some ovine homogenates were processed by using the TeSeE sheep/goat Western blot Kit (Bio-Rad) according to the manufacturer’s instructions. P4 was added at a dilution of 1:1000 to the primary antibody of the kit. All other homogenates were subjected to a different protocol by using 15% acrylamid gels, a 0.45-μm nitrocellulose membrane (Bio-Rad) for semidry blotting, the primary antibodies P4 (1:2000) for ovine samples, and 3F4 (1:3000; kindly provided by M. Beekes, Robert-Koch Institute, Berlin, Germany) for human samples. Further primary antibodies used for the epitope mapping were L42, 12F10, and 3B5 (kindly provided by W. Bodemer and D. Motzkus). A horseradish peroxidase-conjugated goat anti-mouse (Dako, Carpinteria, CA) and Super Signal Femto West Maximum Sensitivity Substrate (Perbio, Erembodegem, Belgium) were used to visualize the result on X-ray film. The molecular size of PrPSc was compared within one system only. The membrane was treated with 4M guanidine isothiocyanate (GdnSCN, 30 minutes) and 0.2% Casein (30 minutes) before antibodies were applied. Deglycosylation was performed after homogenates were digested with proteinase K (20 μg/ml for ovine tissue and 50 μg/ml for human tissue), by using either 0.5U N-Glycosidase F (Roche, Indianapolis, IN) per mg brain wet weight (12 hours, 37°C) or a deglycosylation kit (New England Biolabs, Ipswich, NJ) according to the manufacturer’s instructions.
Membrane Adsorption Assay
Ten percent tissue homogenates (in PBS, 0.1% deoxycholic acid sodium salt) were incubated with DNase 1 (Applichem, Darmstadt, Germany) using 500 μg/ml for 30 minutes (37°C). On treatment with proteinase K (see above) samples were diluted in Tris-buffered saline and subjected to a commercially available slot blot/dot blot device (Bio-Rad), run by a diaphragm pump. Samples were drawn through a nitrocellulose membrane (0.45 μm, Bio-Rad), following the assay by Winklhofer et al22 with modifications. During suction, proteins adsorb to the membrane, which was prepared with 10% Roti Block (Roth, Karlsruhe, Germany). The slots/dots were rinsed with 200 μl 0.1% deoxycholic acid sodium salt before and after the samples were applied and the membrane was treated as described above with an additional blocking step (0.3% H2O2, 20 minutes).
Guanidine Hydrochloride Denaturation
Samples were incubated with suitable guanidine hydrochloride (GdnHCl) concentrations to achieve end concentrations of 1, 1.5, 2, 2.5, 3, 3.5, and 4M GdnHCl (Roth, Karlsruhe, Germany) for 1.5 hours (room temperature). Afterward, the samples were diluted in Tris-buffered saline containing 0.1% Brij (pH7.8) so that tissue concentrations of 1:1000 were obtained in all samples and proteinase K was added (see above). GdnHCl concentrations were only 0.1M in each sample in order not to interfere with proteinase K activity.23 Two hundred micrograms of brain wet weight were applied to each dot. The membrane was treated as described above.
Results
Biochemical Differences between Atypical/Nor98 and Classical Scrapie Are Consistent within Groups
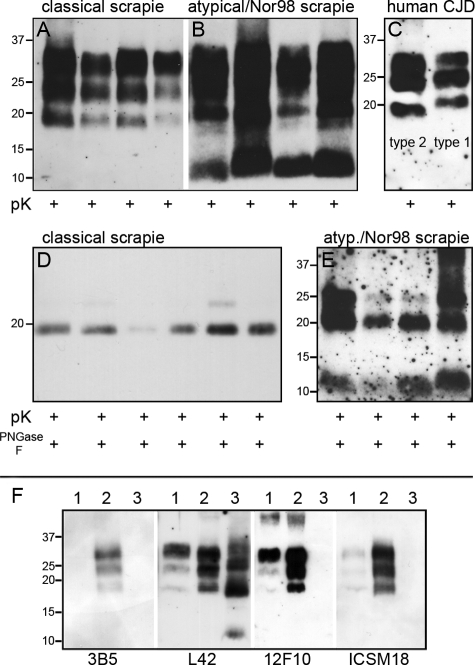
Proteinase K-digestion and Western blot analysis in all classical scrapie cases revealed the characteristic triplet band pattern of PrPSc comprising the di-, mono-, and unglycosylated band in the range of 18 to 30 kDa (Figure 1A). In contrast, all atypical/Nor98 scrapie cases displayed the same multiple band pattern with a small fragment at 11 to 12 kDa (Figure 1B),24 also characterized as migrating below 15 kDa.25
Figure 1.
PrPSc typing of sheep scrapie and human sporadic CJD. After treatment with proteinase K alone or combined with PNGase F, Western blot analysis of proteinase K digested classical scrapie cases (A) showed the typical triplet pattern of PrPSc, whereas atypical/Nor98 scrapie samples (B) comprised a multiple band pattern with the characteristic small fragment at 11 to 12 kDa. No differences were detectable in the size of the unglycosylated fragment after additional PNGase F digestion in classical scrapie cases (D). Minor variations of the small fragment of atypical/Nor98 scrapie samples (E) proved to be inconsistent. The differences in molecular size of human PrPSc type 1 (C; right) and type 2 (C; left) were highly reproducible. F: Epitope mapping of ovine scrapie types. Classical scrapie (lane 2) was detectable by antibodies against epitopes in the region of the octarepeats (mAb 3B5) and the helix 1 region (mAb L42, mAb 12F10, and ICSM18), whereas atypical/Nor98 scrapie (lane 3) was detectable by mAb L42 only. Lane 1 is a bovine classical BSE sample.
Epitope mapping (Figure 1F) demonstrated that classical scrapie samples were detected by antibodies against ovine epitopes in the region of the octarepeats (mAb 3B5 [AA 62 to 91]), the proteinase K cleavage site (mAb P4 [AA 89 to 104]; Figure 1A), and the helix 1 region (mAb L42 [AA 145 to 162], mAb 12F10 [AA 153 to 163], and ICSM18 [AA 146 to 156]). Atypical/Nor98 scrapie samples were only detected by mAb P4 and mAb L42, which is consistent with previous reports.26
No differences were detected in the size of the unglycosylated fragment of approximately 19 kDa after proteinase K-digestion and deglycosylation in classical scrapie cases of different genotypes (13 ALRQ/ALRQ, two ALRH/VLRQ, and two VLRQ/ALRQ) (Figure 1D). We are nevertheless aware that the analyzed samples might not be representative of the whole diversity found in classical scrapie isolates. Three fragments at approximately 25, 20, and 11 to 12 kDa were observed on digestion of atypical/Nor98 scrapie homogenates with proteinase K and PNGase F (Figure 1E). These are considered corresponding to the PrPSc fragments of approximately 23, 18, and 11 kDa, as described by Arsac et al12 since small differences in size may well be due to different protocols of sample preparation.
Atypical/Nor98 and Classical Sheep Scrapie Are Distinguishable by the Form and Neuroanatomical Distribution of PrPSc Deposits
The PET blot was used in this study since it is known to provide a high sensitivity and specificity in the topographic detection of disease-associated prion aggregates.20,21 PrPSc deposits were detected in all scrapie sheep but not in control animals. The observed PrPSc deposits were present in many forms including reticular/synaptic, granular, fine punctate, perivacuolar, intra- and perineuronal, and plaque-like and ramified (i.e., glia-associated27) deposits. The following results are based on the PET blot method, but PrPSc deposition patterns were also confirmed by conventional immunohistochemistry.
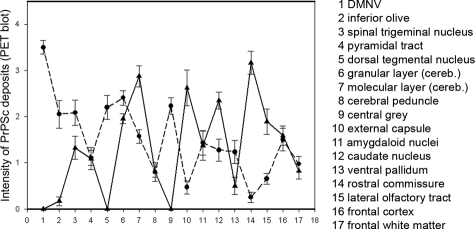
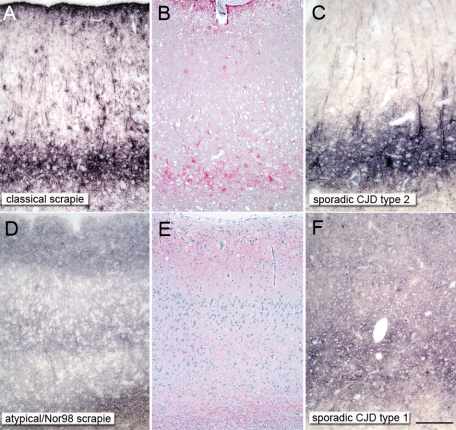
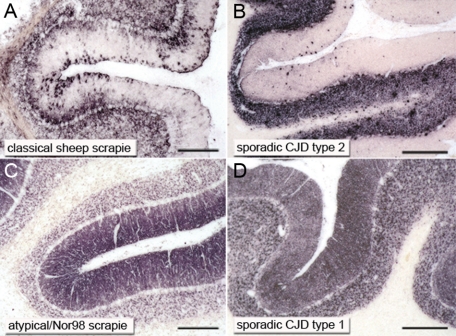
Classical and atypical/Nor98 scrapie were easily distinguished by their forms of PrPSc deposition. In atypical/Nor98 scrapie cases predominantly reticular/synaptic PrPSc deposits in the gray matter were observed, complemented by granular PrPSc deposits in a number of white matter tracts. In contrast, in sheep affected by classical scrapie a variety of PrPSc deposition forms were seen in the CNS, creating a complex deposition pattern that particularly involved intraneuronal, perineuronal, and glia-associated PrPSc deposits. Classical and atypical/Nor98 sheep scrapie also differed clearly in the neuroanatomical PrPSc distribution. The deposition of PrPSc in gray and white matter structures were scored28 on a scale from zero to four, as illustrated in Figure 2. A more rostral localization of PrPSc in atypical/Nor98 scrapie sheep was observed, contrasting strongly with the PrPSc profile of classical scrapie in sheep. In the latter, the spread from the brainstem to the cerebellum and cerebrum29 is mirrored by the more caudal accumulation of PrPSc. Classical scrapie sheep also showed large amounts of PrPSc in a number of gray matter structures that were devoid of PrPSc in all atypical/Nor98 scrapie sheep (Figure 2). These structures include the dorsal motor nucleus of the vagus nerve, the tegmental nucleus (rostral brainstem), and the central gray matter of the midbrain. The cerebral cortex of all atypical/Nor98 scrapie sheep displayed disease-associated PrPSc deposition in all cortical layers, resembling synaptic staining (Figure 3, D and E). Subcortical fibers also contained large amounts of PrPSc. In contrast, in classical sheep scrapie cortical deposits were mainly localized beneath the pia mater and in the deeper cortical layers (Figure 3, A and B), while staining of the white matter was usually glia-associated and never linked to subcortical fibers. Perivascular PrPSc deposition proved to be a prominent pattern in classical scrapie, but was not observed in atypical/Nor98 cases. Staining of the cerebellar cortex of classical scrapie showed that the majority of PrPSc accumulation was found as intra- and extracellular deposits in the granular layer and surrounding the Purkinje cells (Figure 4A). In most atypical/Nor98 cases, however, the molecular layer was most intensely stained (Figure 4C). The deposition form in all atypical/Nor98 cases displayed reticular/synaptic PrPSc aggregates in the molecular and granular layer.
Figure 2.
Classical (dots) and atypical/Nor98 scrapie sheep (triangles) differ clearly in their neuroanatomical distribution of PrPSc deposits. The amounts of PrPSc deposition in various brain structures were assessed on a scale from 0 to 4 by using the PET blot method.
Figure 3.
Obvious parallels in disease-associated PrPSc deposition forms between ovine and human prion types. A reticular/synaptic deposition pattern is observable in the cortices of atypical/Nor98 scrapie sheep (D and E) and sporadic CJD type 1 (F; codon 129 methionine homozygote), while a complex deposition pattern, which is mainly localized in the deep cortical layers, can be seen in classical sheep scrapie (A and B) and sporadic CJD type 2 (C; codon 129 valine homozygote). Ovine tissue: PET blot mAb P4 1:5000 (A and D) and immunohistochemistry mAb P4 1:500 (B and E); human tissue: PET blot 12F10 1:5000 (C and F); scale bar = 300 μm.
Figure 4.
Similarities in the cerebellar cortex between ovine and human prion types regarding the deposition pattern of PrPSc. Synaptically located PrPSc deposits were characteristic for sporadic CJD type 1 (D; codon 129 methionine homozygote) and atypical/Nor98 sheep scrapie (C). In sporadic CJD type 2 (B; codon 129 methione/valine heterozygote) and classical sheep scrapie (A) more complex PrPSc aggregates were visible. Human PET blots: 12F10 1:5000 (B and D); ovine PET blots: mAb L42 (C) and P4 1:5000 (A); scale bar = 300 μm.
In the majority of classical scrapie cases (29 out of 31) where lymphatic tissue was available, PrPSc deposits were visible in lymphatic tissue, predominantly restricted to the follicles. By contrast, no PrPSc was detected in the lymphatic tissue of 15 of the atypical/Nor98 scrapie animals of which tonsils and/or lymph nodes were available.
Pathological Prion Protein of Atypical/Nor98 and Classical Scrapie Differs in Its Stability against Denaturation
Using a membrane adsorption assay, PrPSc was reliably detected from an amount of 20-μg brain wet weight in the CNS from 15 examined classical scrapie cases (12 ALRQ/ALRQ, 2 VLRQ/ALRH, and 1 VLRQ/ALRQ) and six atypical/Nor98 scrapie cases (two ALHQ/ALHQ, one ALHQ/AFRQ, one ALHQ/ALRQ, one ALHQ/ALRH, and one AFRQ/AFRQ). To check the stability of the protein conformation, PrPSc of classical and atypical/Nor98, scrapie was first denatured by exposing brain tissue lysates to ascending concentrations of GdnHCl and then digested with proteinase K.23 To each dot 200 μg of brain wet weight were applied. PrPSc of atypical/Nor98 cases was only detectable after mild denaturation with GdnHCl: PrPSc was detected after denaturation with 1.5M GdnHCl but not 2M GdnHCl (Figure 5), although slightly different amounts of PrPSc were present in the samples. In the classical scrapie cases, PrPSc was still visible after denaturation with 3M and 4M GdnHCl. Even though displaying some variations in its stability at high amounts of GdnHCl, PrPSc of classical scrapie cases showed a greater stability with regard to denaturation by GdnHCl, clearly distinct from that of atypical/Nor98 scrapie (Figure 5). In this experimental setting there seemed to be no influence of genotypes, origin from different flocks or breeds on the PrPSc stability.
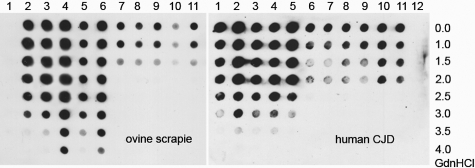
Figure 5.
Sheep scrapie and human sporadic CJD both exhibit a denaturation-resistant and a denaturation-sensitive PrPSc type. In classical scrapie samples (lanes 2 to 6, ALRQ/ALRQ, ALRQ/ALRQ, VLRQ/ALRH, VLRQ/ALRQ, ALRQ/ALRQ), PrPSc could still be seen at concentrations between 3M and 4M GdnHCl and in sporadic CJD type 2 (lanes 1 to 5, MM2, VV2, MV2, VV2, MM2) up to denaturation with concentrations of 3.5M and 4M GdnHCl. In contrast, in atypical/Nor98 sheep scrapie lanes (lanes 7 to 11, ALHQ/ALHQ, ALHQ/ALRQ, ALHQ/ALRH, ALHQ/ALHQ, ALHQ/AFRQ), PrPSc was only detectable up to 1.5M GdnHCl and sCJD type 1 (lanes 6 to 11, VV1, MV1, VV1, MM1, MV1, MM1) up to 2M GdnHCl. A negative control is present for ovine samples in lane 1 and for human samples in lane 12. Membrane adsorption assay: ovine tissue = mAb P4 1:2000; human tissue = mAb 3F4 1:3000.
In summary, classical and atypical/Nor98 scrapie show clear differences in their prion aggregate properties. This refers not only to the Western blot profile and the form and distribution of PrPSc deposition, but also to the aggregate stability regarding denaturation. It can thus be concluded that classical and atypical/Nor98 scrapie represent two different types of scrapie prion protein that elicit prion diseases in one species.
Different Scrapie Prion Types Show Similarities to Human Prion Types: PrPsc Deposition Pattern and Western Blot Results
After proteinase K-digestion and Western blot analysis, two different prion protein types were detectable in clinically distinct human Creutzfeldt-Jakob diseases.30 Depending on the PrPSc types 1 or 2 (Figure 1C) a difference in the form of PrPSc aggregates and the neuroanatomical distribution in the brain could be observed similar to differences identified in sheep scrapie. In patients with CJD that accumulate PrPSc type 1, reticular/synaptic were detected in cortical structures (Figure 3F), subcortical nuclei, and the cerebellar cortex (Figure 4D). By contrast, prion aggregates in patients accumulating PrPSc type 2 appeared to be complex as they displayed in particular perivacuolar, intra- and perineuronal, and/or plaque-like forms (Figures 3C and 5B). These differences concerning the deposition form of PrPSc aggregates were independent of the methionine/valine polymorphism at codon 129 of the PRNP. The topographical pattern of PrPSc distribution between these two prion types differed as follows: type 1 deposits were typically restricted to gray matter structures, while all type 2 patients showed deposits in the white matter. In patients with type 1 PrPSc the midbrain and brain stem structures were relatively spared, but in patients with type 2 PrPSc brain stem and midbrain were heavily affected. Although these prion type-related topographical differences are not completely identical to those in sheep scrapie, a comparable connection between prion type and deposition pattern is evident.
Aggregate Stability Regarding Denaturation
Similar to scrapie in sheep, the stability of PrPSc aggregates of human sporadic CJD against denaturation with GdnHCl showed two groups: denaturation-resistant and denaturation-sensitive PrPSc aggregates. This property correlated with the prion protein type according to Parchi et al8 and is independent from the physiologically occurring methionine/valine polymorphism at codon 129 of the PRNP. By membrane adsorption after GdnHCl denaturation and proteinase K-digestion, human PrPSc type 1 proved to be less stable than human PrPSc type 2. While human PrPSc type 2 was detectable up to GdnHCl concentrations between 3M and 4M, human PrPSc type 1 was stable up to 2M GdnHCl. Neither methionine nor valine at codon 129 in type 1 or type 2 seemed to alter the stability of the prion protein aggregates (Figure 5).
Summarizing the results, striking parallels between human PrPSc type 1 and atypical/Nor98 scrapie as well as human PrPSc type 2 and classical scrapie are observed with regard to PrPSc deposition and stability of the prion aggregates.
Discussion
In humans, different prion types are linked with clinically and neuropathologically distinct prion diseases.8 The present work emphasizes that the differences in deposition characteristics and stability with regard to denaturation between atypical/Nor98 and classical scrapie also account for different prion types. Moreover, the two scrapie types that have been characterized show a number of striking similarities with human PrPSc types in sporadic CJD. Hence, we propose that the existence of different PrPSc types might be a common denominator of prion diseases in humans and animals. Since these two prion types show an across-the-species comparability with similar biochemical and pathological characteristics, it is most likely that they exist due to a different conformational pattern of the disease-related prion protein.
Prion Types Depend on Conformation
The interpretation that the conformation of PrPSc accounts for prion types is supported by different proteinase K-cleavage sites of human prion types9 and the propagation of mutation-associated prion characteristics in human transgenic mice without PRNP-point mutation.31 However, differences in protein stability as they have been found in this study, provide direct evidence for a conformational distinction between these molecules.32 Further support for the relation between type and conformation is also given by experiments focusing on the size of prion protein aggregates. Using virus removal filters, Kobayashi et al33 were able to show differences in the size of CJD type 1 and type 2 aggregates: PrPSc type 2 forms larger aggregates than PrPSc type 1, independent of whether the disease was sporadic, iatrogenic or acquired. This difference is clearly reflected by the morphology of the PrPSc depositions we have found in sheep scrapie and human CJD. Sporadic CJD type 1 and atypical/Nor98 scrapie are characterized by fine (reticular) deposits, whereas CJD type 2 and classical scrapie display a complex aggregate pattern, regardless of the respective genotypes at the polymorphic positions of the PRNP that were investigated.
Prion Type Characteristics Versus Prion Strain Characteristics
Structural differences of the disease-associated protein have also been proposed as an explanation for the existence of strains. Partial digestion of the disease-associated protein with proteinase K as well as differences in antibody binding after the protein was partially denatured were used to identify structural characteristics in correlation with strain properties and different clinical TSE forms.23,34,35 It needs to be considered that the kinetics of proteinase K-digestion of PrPSc are markedly influenced by detergent effects in the buffer, demonstrating that the accessibility of the cleavage sites are variable.35 In contrast, differences in the stability against total unfolding of PrPSc seem to be a usable criterion to identify conformational differences or conformational motives. Whereas detergents affect the tertiary structure of a protein by interacting with hydrophilic and hydrophobic areas of protein molecules, chaotropic salts like GdnHCl destroy the hydrogen bonds in α-helices and β-sheets leading to an irregular coiled polypeptide chain.36 This is in line with the observation that detergents remove prion infectivity only partially, whereas chemicals that destroy secondary structures like chaotropic salts are highly effective.37 However, detectable differences regarding the stability against denaturation with GdnHCl shown for various prion strains in hamsters seem to be very small compared with the ones that can be shown here for the prion types of human and ovine prion diseases. Strains could thus correspond to structural differences that are less marked than those defining types and are probably constant only under defined conditions. Influences of polymorphisms or interactions with other genetic factors like the promotor region, species-specific factors like the recently detected incorporation of polyanionic molecules into prions,38 glycosaminoglycans or other yet unknown factors of the original host may also lead to different strains in a new host within the prion types of the original species.5,39 The existence of prion types does not exclude the existence of strains. The same variations that account for strains might be the reason for differences in the clinical disease course of the natural host.
Two Different Prion Types also in BSE?
Parallel to human sporadic CJD and our results in sheep scrapie, there is increasing evidence that two prion types also exist in cattle BSE. Two presumably sporadic forms of BSE known as H-type BSE14 and bovine amyloidotic spongiform encephalopathy, also called L-type BSE,15 have been described in cattle in addition to typical/classical BSE.40 The small variation in the apparent molecular weight of the unglycosylated band of bovine amyloidotic spongiform encephalopathy is considered to be well within the range of classical BSE,41,42 which would leave H-type BSE with a considerably larger unglycosylated fragment in Western blot analysis than the second BSE type. Interestingly, bovine amyloidotic spongiform encephalopathy converts into classical BSE after serial passages in bovine-transgenic mice,43 although displaying clinically different diseases in cattle.44 From the latter experiment the authors concluded that different strains were responsible for different phenotypes. Obviously the different clinical diseases were generated by agents that belong to a single prion type. These results together with our observations emphasize the need to differentiate strictly between prion types and prion strains and demonstrate that even in cattle BSE, one prion type may contain different prion strains.
Prion Type Displays Parallels in the Pathophysiology of Disease between Species
Biochemical and morphological similarities have been used to draw parallels between forms of BSE and human prion diseases.15 Parallels between species can also be observed with regard to the route of prion infection: in classical BSE, variant CJD, and classical scrapie, all of which presumably belong to one class of prion type (type 2 in humans) according to the observations made above, the oral route of infection has been identified. These TSEs use the dorsal motor nucleus of the vagus nerve as an entry site into the brain.29,45,46 This observation suggests that distinct prion types in human and animal TSEs possibly have an impact on the pathogenesis of prion diseases.
Conclusion
As the prion protein is a highly conserved protein in terms of evolution, parallels between characteristics of prion types in TSEs of different species are of interest. In the present study, we report previously unknown similarities between sheep scrapie forms and human sporadic CJD types. We propose that the observed similarities between sheep scrapie and sporadic CJD in humans justify new interspecies groups of prion diseases in which prion types, not prion strains, are the major determinant for prion disease forms. While epidemiology implies that classical scrapie is not related to human TSEs,47 the atypical/Nor98 scrapie risk for human transmission has not yet been elucidated. Currently there is no compelling evidence that sCJD has a different origin than sporadic genesis. However, the finding of prion types with an across-the-species comparability might provide further understanding of the pathogenesis in prion diseases.
Acknowledgments
We thank Tatjana Pfander, Nadine Rupprecht, and Kerstin Brekerbohm for their skillful technical assistance.
Footnotes
Address reprint requests to Walter J. Schulz-Schaeffer, Prion and Dementia Research Unit, Department of Neuropathology, University Medical Center Goettingen, Robert Koch Str. 40, 37075 Goettingen, Germany. E-mail: wjschulz@med.uni-goettingen.de.
Supported by the VolkswagenStiftung (grants ZN 1294 and ZN2168) to W.J.S.S.
References
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- Hadlow WJ. Scrapie and Kuru. Lancet. 1959;2:289–290. [Google Scholar]
- Fraser H, Dickinson AG. Scrapie in mice: agent-strain differences in the distribution and intensity of grey matter vacuolation. J Comp Pathol. 1973;83:29–40. doi: 10.1016/0021-9975(73)90024-8. [DOI] [PubMed] [Google Scholar]
- Kellings K, Meyer N, Mirenda C, Prusiner SB, Riesner D. Further analysis of nucleic acids in purified scrapie prion preparations by improved return refocusing gel electrophoresis. J Gen Virol. 1992;73:1025–1029. doi: 10.1099/0022-1317-73-4-1025. [DOI] [PubMed] [Google Scholar]
- Aguzzi A, Heikenwalder M, Polymenidou M. Insights into prion strains and neurotoxicity. Nat Rev Mol Cell Biol. 2007;8:552–561. doi: 10.1038/nrm2204. [DOI] [PubMed] [Google Scholar]
- Bessen RA, Marsh RF. Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J Gen Virol. 1992;73:329–334. doi: 10.1099/0022-1317-73-2-329. [DOI] [PubMed] [Google Scholar]
- Nonno R, Di Bari MA, Cardone F, Vaccari G, Fazzi P, Dell'Omo G, Cartoni C, Ingrosso L, Boyle A, Galeno R, Sbriccoli M, Lipp HP, Bruce M, Pocchiari M, Agrimi U. Efficient transmission and characterization of Creutzfeldt-Jakob disease strains in bank voles. PLoS Pathog. 2006;2:e12. doi: 10.1371/journal.ppat.0020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchi P, Giese A, Capellari S, Brown P, Schulz-Schaeffer W, Windl O, Zerr I, Budka H, Kopp N, Piccardo P, Poser S, Rojiani A, Streichemberger N, Julien J, Vital C, Ghetti B, Gambetti P, Kretzschmar H. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann Neurol. 1999;46:224–233. [PubMed] [Google Scholar]
- Parchi P, Zou W, Wang W, Brown P, Capellari S, Ghetti B, Kopp N, Schulz-Schaeffer WJ, Kretzschmar HA, Head MW, Ironside JW, Gambetti P, Chen SG. Genetic influence on the structural variations of the abnormal prion protein. Proc Natl Acad Sci USA. 2000;97:10168–10172. doi: 10.1073/pnas.97.18.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce ME, Boyle A, Cousens S, McConnell I, Foster J, Goldmann W, Fraser H. Strain characterization of natural sheep scrapie and comparison with BSE. J Gen Virol. 2002;83:695–704. doi: 10.1099/0022-1317-83-3-695. [DOI] [PubMed] [Google Scholar]
- Buschmann A, Biacabe AG, Ziegler U, Bencsik A, Madec JY, Erhardt G, Luhken G, Baron T, Groschup MH. Atypical scrapie cases in Germany and France are identified by discrepant reaction patterns in BSE rapid tests. J Virol Methods. 2004;117:27–36. doi: 10.1016/j.jviromet.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Arsac JN, Andreoletti O, Bilheude JM, Lacroux C, Benestad SL, Baron T. Similar biochemical signatures and prion protein genotypes in atypical scrapie and Nor98 cases: France and Norway. Emerg Infect Dis. 2007;13:58–65. doi: 10.3201/eid1301.060393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benestad SL, Sarradin P, Thu B, Schonheit J, Tranulis MA, Bratberg B. Cases of scrapie with unusual features in Norway and designation of a new type: Nor98. Vet Rec. 2003;153:202–208. doi: 10.1136/vr.153.7.202. [DOI] [PubMed] [Google Scholar]
- Biacabe AG, Laplanche JL, Ryder S, Baron T. Distinct molecular phenotypes in bovine prion diseases. EMBO Rep. 2004;5:110–115. doi: 10.1038/sj.embor.7400054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalone C, Zanusso G, Acutis P, Ferrari S, Capucci L, Tagliavini F, Monaco S, Caramelli M. Identification of a second bovine amyloidotic spongiform encephalopathy: molecular similarities with sporadic Creutzfeldt-Jakob disease. Proc Natl Acad Sci USA. 2004;101:3065–3070. doi: 10.1073/pnas.0305777101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce ME, Will RG, Ironside JW, McConnell I, Drummond D, Suttie A, McCardle L, Chree A, Hope J, Birkett C, Cousens S, Fraser H, Bostock CJ. Transmissions to mice indicate that “new variant” CJD is caused by the BSE agent. Nature. 1997;389:498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- Schutz E, Scharfenstein M, Brenig B. Genotyping of ovine prion protein gene (PRNP) variants by PCR with melting curve analysis. Clin Chem. 2006;52:1426–1429. doi: 10.1373/clinchem.2006.069666. [DOI] [PubMed] [Google Scholar]
- Windl O, Giese A, Schulz-Schaeffer W, Zerr I, Skworc K, Arendt S, Oberdieck C, Bodemer M, Poser S, Kretzschmar HA. Molecular genetics of human prion diseases in Germany. Hum Genet. 1999;105:244–252. doi: 10.1007/s004399900124. [DOI] [PubMed] [Google Scholar]
- Notari S, Capellari S, Giese A, Westner I, Baruzzi A, Ghetti B, Gambetti P, Kretzschmar HA, Parchi P. Effects of different experimental conditions on the PrPSc core generated by protease digestion: implications for strain typing and molecular classification of CJD. J Biol Chem. 2004;279:16797–16804. doi: 10.1074/jbc.M313220200. [DOI] [PubMed] [Google Scholar]
- Schulz-Schaeffer WJ, Tschoke S, Kranefuss N, Drose W, Hause-Reitner D, Giese A, Groschup MH, Kretzschmar HA. The paraffin-embedded tissue blot detects PrP(Sc) early in the incubation time in prion diseases. Am J Pathol. 2000;156:51–56. doi: 10.1016/S0002-9440(10)64705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemheuer WM, Benestad SL, Wrede A, Wemheuer WE, Brenig B, Bratberg B, Schulz-Schaeffer WJ. Detection of classical and atypical/Nor98 scrapie by the paraffin-embedded tissue blot method. Vet Rec. 2009;164:677–681. doi: 10.1136/vr.164.22.677. [DOI] [PubMed] [Google Scholar]
- Winklhofer KF, Hartl FU, Tatzelt J. A sensitive filter retention assay for the detection of PrP(Sc) and the screening of anti-prion compounds. FEBS Lett. 2001;503:41–45. doi: 10.1016/s0014-5793(01)02692-8. [DOI] [PubMed] [Google Scholar]
- Peretz D, Scott MR, Groth D, Williamson RA, Burton DR, Cohen FE, Prusiner SB. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci. 2001;10:854–863. doi: 10.1110/ps.39201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benestad SL, Arsac JN, Goldmann W, Noremark M. Atypical/Nor98 scrapie: properties of the agent, genetics, and epidemiology. Vet Res. 2008;39:19. doi: 10.1051/vetres:2007056. [DOI] [PubMed] [Google Scholar]
- EFSA Opinion of the Scientific Panel on Biological Hazards on classification of atypical transmissible spongiform encephalopathy (TSE) cases in small ruminants. EFSA J. 2005;276:1–30. [Google Scholar]
- Gretzschel A, Buschmann A, Langeveld J, Groschup MH. Immunological characterization of abnormal prion protein from atypical scrapie cases in sheep using a panel of monoclonal antibodies. J Gen Virol. 2006;87:3715–3722. doi: 10.1099/vir.0.81816-0. [DOI] [PubMed] [Google Scholar]
- Gonzalez L, Martin S, Begara-McGorum I, Hunter N, Houston F, Simmons M, Jeffrey M. Effects of agent strain and host genotype on PrP accumulation in the brain of sheep naturally and experimentally affected with scrapie. J Comp Pathol. 2002;126:17–29. doi: 10.1053/jcpa.2001.0516. [DOI] [PubMed] [Google Scholar]
- Gonzalez L, Martin S, Jeffrey M. Distinct profiles of PrP(d) immunoreactivity in the brain of scrapie- and BSE-infected sheep: implications for differential cell targeting and PrP processing. J Gen Virol. 2003;84:1339–1350. doi: 10.1099/vir.0.18800-0. [DOI] [PubMed] [Google Scholar]
- van Keulen LJ, Schreuder BE, Vromans ME, Langeveld JP, Smits MA. Pathogenesis of natural scrapie in sheep. Arch Virol Suppl. 2000:57–71. doi: 10.1007/978-3-7091-6308-5_5. [DOI] [PubMed] [Google Scholar]
- Parchi P, Castellani R, Capellari S, Ghetti B, Young K, Chen SG, Farlow M, Dickson DW, Sima AA, Trojanowski JQ, Petersen RB, Gambetti P. Molecular basis of phenotypic variability in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 1996;39:767–778. doi: 10.1002/ana.410390613. [DOI] [PubMed] [Google Scholar]
- Telling GC, Parchi P, DeArmond SJ, Cortelli P, Montagna P, Gabizon R, Mastrianni J, Lugaresi E, Gambetti P, Prusiner SB. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- Shamsir MS, Dalby AR. One gene, two diseases and three conformations: molecular dynamics simulations of mutants of human prion protein at room temperature and elevated temperatures. Proteins. 2005;59:275–290. doi: 10.1002/prot.20401. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Satoh S, Ironside JW, Mohri S, Kitamoto T. Type 1 and type 2 human PrPSc have different aggregation sizes in methionine homozygotes with sporadic, iatrogenic and variant Creutzfeldt-Jakob disease. J Gen Virol. 2005;86:237–240. doi: 10.1099/vir.0.80389-0. [DOI] [PubMed] [Google Scholar]
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- Uro-Coste E, Cassard H, Simon S, Lugan S, Bilheude JM, Perret-Liaudet A, Ironside JW, Haik S, Basset-Leobon C, Lacroux C, Peoch K, Streichenberger N, Langeveld J, Head MW, Grassi J, Hauw JJ, Schelcher F, Delisle MB, Andreoletti O. Beyond PrP res type 1/type 2 dichotomy in Creutzfeldt-Jakob disease. PLoS Pathog. 2008;4:e1000029. [PubMed] [Google Scholar]
- Tanford C. Protein denaturation. Anfinsen CB Jr, Edsall JT, editors. New York: Academic Press,; Advances in protein chemistry. 1968:pp. 121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- Prusiner SB, Groth DF, McKinley MP, Cochran SP, Bowman KA, Kasper KC. Thiocyanate and hydroxyl ions inactivate the scrapie agent. Proc Natl Acad Sci USA. 1981;78:4606–4610. doi: 10.1073/pnas.78.7.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JC, Valdes PA, Orem NR, Deleault NR, Williamson RA, Harris BT, Supattapone S. Selective incorporation of polyanionic molecules into hamster prions. J Biol Chem. 2007;282:36341–36353. doi: 10.1074/jbc.M704447200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales R, Abid K, Soto C. The prion strain phenomenon: molecular basis and unprecedented features. Biochim Biophys Acta. 2007;1772:681–691. doi: 10.1016/j.bbadis.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells GA, Scott AC, Johnson CT, Gunning RF, Hancock RD, Jeffrey M, Dawson M, Bradley R. A novel progressive spongiform encephalopathy in cattle. Vet Rec. 1987;121:419–420. doi: 10.1136/vr.121.18.419. [DOI] [PubMed] [Google Scholar]
- Baron T, Bencsik A, Biacabe AG, Morignat E, Bessen RA. Phenotypic similarity of transmissible mink encephalopathy in cattle and L-type bovine spongiform encephalopathy in a mouse model. Emerg Infect Dis. 2007;13:1887–1894. doi: 10.3201/eid13112.070635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Zheng M, Casalone C, Qing L, Huang S, Chakraborty B, Wang P, Chen F, Cali I, Corona C, Martucci F, Iulini B, Acutis P, Wang L, Liang J, Wang M, Li X, Monaco S, Zanusso G, Zou WQ, Caramelli M, Gambetti P. Evaluation of the human transmission risk of an atypical bovine spongiform encephalopathy prion strain. J Virol. 2008;82:3697–3701. doi: 10.1128/JVI.02561-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capobianco R, Casalone C, Suardi S, Mangieri M, Miccolo C, Limido L, Catania M, Rossi G, Fede GD, Giaccone G, Bruzzone MG, Minati L, Corona C, Acutis P, Gelmetti D, Lombardi G, Groschup MH, Buschmann A, Zanusso G, Monaco S, Caramelli M, Tagliavini F. Conversion of the BASE Prion Strain into the BSE Strain: the Origin of BSE? PLoS Pathog. 2007;3:e31. doi: 10.1371/journal.ppat.0030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi G, Casalone C, D'Angelo A, Gelmetti D, Torcoli G, Barbieri I, Corona C, Fasoli E, Farinazzo A, Fiorini M, Gelati M, Iulini B, Tagliavini F, Ferrari S, Caramelli M, Monaco S, Capucci L, Zanusso G. Intraspecies transmission of BASE induces clinical dullness and amyotrophic changes. PLoS Pathog. 2008;4:e1000075. doi: 10.1371/journal.ppat.1000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton DA. Pathogenesis and prevalence of variant Creutzfeldt-Jakob disease. J Pathol. 2006;208:134–141. doi: 10.1002/path.1880. [DOI] [PubMed] [Google Scholar]
- Schulz-Schaeffer WJ, Fatzer R, Vandevelde M, Kretzschmar HA. Detection of PrP(Sc) in subclinical BSE with the paraffin-embedded tissue (PET) blot. Arch Virol Suppl. 2000:173–180. doi: 10.1007/978-3-7091-6308-5_16. [DOI] [PubMed] [Google Scholar]
- Bosque PJ. Bovine spongiform encephalopathy, chronic wasting disease, scrapie, and the threat to humans from prion disease epizootics. Curr Neurol Neurosci Rep. 2002;2:488–495. doi: 10.1007/s11910-002-0034-1. [DOI] [PubMed] [Google Scholar]