Abstract
The presence of lymph node metastases is associated with poor prognosis in early stage cervical cancer. As of yet, no molecular markers predicting lymph node metastases have been identified. We examined single genetic markers and a composite marker, comprised of three fluorescence in situ hybridization (FISH) probes targeting the genes LAMP3, PROX1, and PRKAA1, in pretreatment cervical biopsies from 16 lymph node positive cases and 15 lymph node negative controls from women with stage IB and IIA cervical cancer. In addition, we determined clonal patterns by including CCND1 to compare the clonal constitution of primary tumors and associated lymph node metastases. The composite FISH marker allowed for classification of patients into those with and without lymph node metastases with a sensitivity and specificity of 75% and 87%, respectively (P = 0.001). The positive predictive value and negative predictive value were 86% and 76%, respectively. Clonal patterns varied among the tumors. In many cases, changes between the primary tumor and lymph node metastases in the most common clones may indicate that certain clones have a growth advantage for establishing metastases in lymph nodes. We conclude that the composite FISH marker may be useful for determining risk for subsequent development of lymph node metastases in patients with cervical cancer.
Cervical cancer is globally the second most common tumor in women, with 80% of the cases occurring in developing countries.1,2,3 Overall survival is generally high among patients with early stages of cervical cancer; however, the presence of lymph node metastases significantly reduces survival by a factor of four in stage Ib1 disease, therefore constituting the strongest prognostic factor for survival.4,5,6 Molecular markers as a means to detect lymph node metastases may therefore have therapeutic implications.
Genomic alterations in cervical cancer have been reported in many studies with consistent gains and amplifications found on chromosomes 1q, 3q, and 5p.7,8,9,10,11 Potential genes of interest in these regions of amplification include LAMP3, PROX1, and PRKAA1. LAMP3 resides on a region of high importance for cervical carcinogenesis, with high expression found to be associated with an enhanced metastatic potential in cervical cancer.12 The homeobox gene, PROX1, is a lymphatic endothelium specific marker involved in the developmental regulation of the lymphatic system.13,14,15 PRKAA1, a cellular metabolic stress regulator, may assist tumor cell growth under stress and is a potential cervical carcinogenesis marker.16 In addition to these genes, we included CCND1, a genetic marker of cellular proliferation. CCND1 is located on chromosome 11q13 and altered expression of this gene has been observed in many cancers. CCND1 overexpression is also associated with lymph node metastases in oral cancer.17
We analyzed genomic copy numbers of LAMP3, PROX1, PRKAA1, and CCND1 by fluorescence in situ hybridization (FISH) in pretreatment cervical biopsies from lymph node positive (cases) and lymph node negative (controls) stage IB and IIA patients with cervical cancer to explore their role as potential predicting factors for lymph node metastasis. In addition, we also analyzed the four markers in lymph node metastases from all cases with the objective to identify clonal patterns indicative of lymphatic spread.
Materials and Methods
Pretreatment formalin-fixed, paraffin-embedded biopsy specimens were obtained from 31 patients with cervical cancer at stages IB and IIA according to the International Federation of Gynecology and Obstetrics staging system, treated at the Department of Gynaecologic Oncology, Radiumhemmet (Stockholm, Sweden), from January 1994 to December 1997. Clinical information, including age, International Federation of Gynecology and Obstetrics stage, histology, tumor size, grading, lymph node metastasis, treatment modality, and follow-up status was retrieved from medical records. Radical hysterectomy and pelvic lymphadenectomy was performed in all patients. Preoperative intracavitary brachytherapy (remote after-loading technique with caesium) was given to 25 patients in the form of two uterovaginal insertions with a 3-week interval followed by surgery, 4 weeks after the second insertion. Of the 31 patients, 16 had histologically verified lymph node metastases (cases) and 15 were lymph node negative (controls). From all patients that presented during the observation time, we selected 31 patients that could be matched according to stage, grade, size, and lymph vascular invasion. In addition to the biopsy specimens already collected, we obtained samples from the positive lymph nodes. All histological samples were reviewed by a pathologist who was blinded to clinical outcome to confirm representative tumor specimens. The study was approved by the research ethical review board (Dnr: 01-269) at Karolinska Institutet. Pictorial presentation of the experimental design is shown in Figure 1.
Figure 1.
Pictorial representation of the experimental design.
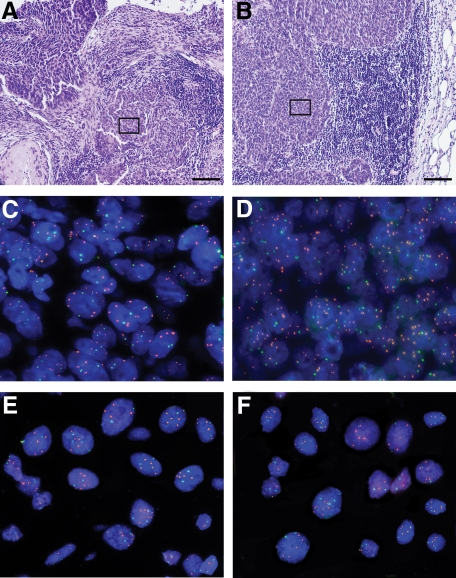
FISH was performed on 47 samples. Two 4-μm sections were cut before and after the FISH sections and H&E stained, to confirm tumor representation. Images for a primary tumor and its corresponding lymph node metastasis of the H&E stained sections, FISH on adjacent paraffin section, and FISH on cytospins prepared from adjacent paraffin sections are shown in Figure 2, A–F.
Figure 2.
Histological and FISH images of primary tumor C18 (A, C, and E) and its corresponding lymph node metastasis C19 (B, D, and F). A and B: Images of H&E stained sections showing areas representative for cervical cancer and cervical cancer lymph node metastasis, respectively. The length of the black bar in the bottom right corner is equivalent to 100 μm. The black frames indicate the tissue area shown in a larger magnification in the FISH images below. C and D: FISH probes PROX1 (green signals) and LAMP3 (orange signals) hybridized on sections corresponding to the H&E sections shown in A and B. The main signal numbers observed per nucleus were two signals for PROX1 and four to five signals for LAMP3. E and F: FISH probes PROX1 (green signals) and LAMP3 (orange signals) hybridized on cytospins prepared from sections corresponding to the H&E sections shown in A and B. The main signal numbers observed per nucleus were two signals for PROX1 and four to five signals for LAMP3. Note the improved signal countability in the cytospins compared with the sections (C and D). Most of the samples of this study had histological sections with highly overlapping nuclei, which made accurate enumeration impossible. We therefore disintegrated the sections and prepared monolayer cytospins from the resulting single cell suspensions.
Sample Preparation and FISH
FISH was performed by using a centromere-specific probe for chromosome 7 (CEP7; Abbott Molecular, Inc; Des Plaines, IL), and bacterial artificial chromosome (BAC) contigs for the following four probes: LAMP3 probe at chromosome band 3q26; PROX1 probe at chromosome band 1q41; PRKAA1 probe at chromosome band 5p19; and CCND1 probe at chromosome 11q13. Images of the individual FISH signals used are shown in Figure 3, A–F. The BAC contig of each probe comprised of four to five overlapping BAC sequences specific to the region of interest for each probe. The CEP7 probe (Abbott Molecular, Inc) was labeled in Spectrum Aqua. The BAC clone contigs were labeled by nick-translation with Spectrum Orange-dUTP (Abbott Molecular, Inc) for LAMP3, with Rhodamine Green-dUTP (Life Technologies; Carlsbad, CA) for PROX1, with Spectrum Orange-dUTP (Abbott Molecular, Inc) for PRKAA1, and with Rhodamine Green-dUTP (Life Technologies) for CCND1. The FISH markers were combined into two probe panels, with the first probe combination consisting of LAMP3, PROX1, and CEP7 (Abbott Molecular, Inc). The second probe combination contained PRKAA1 and CCND1. The two probe panels were subsequently hybridized to the samples, resulting in counts for all five FISH probes within the same nuclei.
Figure 3.
FISH images from a cytospin prepared from one of the primary biopsies (A) 4’,6’-diamidino-2-phenylindole hydrochloride stained cells, (B) CEP7 (Abbott Molecular, Inc) (Aqua), (C) LAMP3 signals (orange), (D) PROX1 signals (Green), (E) PRKAA1 signals (orange), and (F) CCND1 signals (Green).
For accurate signal number per nucleus enumeration, single nuclei preparations were prepared from 6-μm formalin-fixed, paraffin-embedded sections. The tissue was deparaffinized by using xylene (three times for 10 minutes) and rehydrated by using an ethanol series before centrifugation at 14,000 rpm. Excess 50% ethanol was removed before undergoing 20-minute incubation in sterile water at room temperature. The cells were then digested by adding 0.1% Protease (Type XXIV, Sigma, St. Louis, MO, USA) in 1 X PBS at 45°C for 45 to 60 minutes. Once an optimal single cell suspension was achieved, the reaction was stopped with 1 X PBS. The cells were then deposited onto slides by centrifugation, fixed in ethanol for 10 minutes, air dried, and stored at 4°C.
For hybridization, slides were pretreated with 0.05% pepsin for 20 to 25 minutes before undergoing fixation in 1% formaldehyde for 10 minutes followed by ethanol dehydration. After air drying, the slides were denatured in a 70% formamide/2 × saline sodium citrate (SSC) solution for 2 minutes. The slides were then put through an ice cold dehydrating ethanol series (70%, 90%, and 100%) and air dried. All probes were denatured for 5 minutes at 80°C followed by preannealing at 37°C for 2 hours except for CEP7 (Abbott Molecular, Inc), which requires no preannealing. Preannealed probes were mixed with the denatured CEP7 probe (Abbott Molecular, Inc) before being added to the denatured slides. The slides were then coverslipped and sealed with rubber cement before being placed in a humidified chamber for overnight hybridization at 37°C. After hybridization, slides were washed in 2 × SSC (three times for 3 minutes each time) followed by a dehydrating ethanol series (three times for 3 minutes each time). The slides were counterstained with a 4,6-diamidino-2-phenylindole (antifade) solution and mounted with a coverslip.
Hybridized FISH slides were viewed by using the Leica DM-RXA fluorescence microscope (Leica; Wetzlar, Germany) equipped with custom optical filters and a 40× objective. The Leica CW 4000 FISH software was used to acquire multifocal images for each filter. Fifteen to 25 images were taken in areas of optimal cell density with minimal cellular clumps and overlapping cells. Stage relocation was used to recapture the same cells hybridized with the second probe combination. Once imaging and relocation were completed for the first probe set, the probes were washed off with 50% formamide/SSC at 80°C for 2 to 3 minutes before undergoing ethanol dehydration. The air dried slides were then denatured in 70% formamide/2 × SSC at 80°C for 1 minute before being dehydrated in ice cold ethanol series. Once the slides air dried, the second probe combination (probes were denatured/preannealed according to the above protocol) was applied to the slide for overnight hybridization at 37°C. Probes were detected according to the protocol of the first probe combination. Images were acquired at the exact same regions as the first probe panel. Each hybridization set contained a lymphocyte control from a normal human donor.
FISH Analyses
FISH analyses were performed on all 47 samples with approximately 250 interphase cells counted in each case. The four gene probes and the CEP7 probe (Abbott Molecular, Inc) were scored within each of the 250 cells. The CEP7 probe (Abbott Molecular, Inc) was used to estimate the ploidy of the cell, since it was previously shown that chromosome 7 is the least involved chromosome in cervical carcinogenesis.7,10 Nuclei that could not be evaluated due to various reasons, including overlaps and insufficient hybridization, were excluded. Scoring of FISH markers was completed without prior knowledge of clinical outcome or histopathological evaluation.
FISH markers (LAMP3, PROX1, PRKAA1, and CCND1) were compared against the corresponding CEP7 probe (Abbott Molecular, Inc) in each cell and a ratio was calculated (FISH marker divided by CEP7 [Abbott Molecular, Inc]). A ratio per cell of greater than one was considered a gained signal. A ratio per cell of more than two was considered an amplified signal. Calculations were made according to the percentage of cells with gained or amplified signals within each case. The FISH composite marker status was based on the percentage of cells with amplified signals, relative to optimized thresholds established for each single FISH marker. For single FISH markers, cases with scores above the optimized threshold were considered a positive test. For the FISH composite marker, cases exhibiting a positive test for all three markers (LAMP3, PROX1, and PRKAA1) were considered a FISH positive test.
Statistical Analyses and Clonal Pattern Observations
Statistical analyses were performed with SAS version 9.1 (SAS Institute; Cary, NC), using the Mantel-Haenszel χ2 test to test patient characteristics with all FISH scores. The sensitivity (identifying specimens with lymph node metastasis) was compared with the specificity (identifying specimens without lymph node metastasis) by using the receiver operator characteristic (ROC) plot for the FISH composite marker. Sensitivities and specificities were calculated by independently varying the thresholds for all three probe signals over a wide range. Only the highest sensitivity value for each value of the specificity is plotted, with curves coming closest to 100% sensitivity and specificity (the ideal point, upper left corner of the graph) being optimal. The distance from ideal plot depicts the distance from the ideal point on the ROC plot as a function of threshold, with minimums identifying optimal threshold values, as previously described by Heselmeyer-Haddad et al.18
Clonal patterns were described as the association between four single FISH markers as evaluated in a single cell basis. The single FISH markers used in determining clonal patterns were compared against the CEP7 (Abbott Molecular, Inc) probe in each cell with ratios calculated (FISH marker divided by CEP7 [Abbott Molecular, Inc]). Gained (G) signals correspond to ratios greater than one and not gained (N) signals correspond to ratios of one or less than one. Clonal patterns, as seen in Figure 4, depict the percentages of cells with the observed gained (G) or not gained (N) signal combination for the four FISH markers (LAMP3, PROX1, PRKAA1, and CCND1). Patterns are listed by markers, with the first letter corresponding to LAMP3, then PROX1, PRKAA1, and CCND1. Example: GGGG = Gained signals for all four markers; NNGN = no gains for LAMP3, PROX1, and CCND1, but gain for PRKAA1.
Figure 4.
Clonal patterns observed in 16 primary tumor biopsies and their corresponding lymph node metastases. Each graph represents a comparison of signal patterns between the primary tumor (black bars) and the lymph node metastasis (gray bars) of one patient with the primary biopsy case number depicted in the right upper corner. For this analysis, each nucleus was enumerated for the control probe (CEP7 [Abbott Molecular, Inc]) and gene probe (LAMP3, PROX1, PRKAA1, and CCND1) signals. A gain (G) of a certain gene probe within a nucleus was defined by a signal ratio >1 of the respective gene probe divided by the control probe CEP7 (Abbott Molecular, Inc), while a not gained (N) signal number was reflected by a ratio of the gene probe divided by CEP7 (Abbott Molecular, Inc) of ≤1. Signal patterns observed in >10% of the cell population are displayed on the x axis of the graphs according to gained signals (G) or not gained (N) signals for the respective gene probes, with the first letter in the pattern corresponding to LAMP3, the second letter to PROX1, the third to PRKAA1, and the forth to CCND1. The y axis presents the percentage of nuclei observed with the respective pattern. Signals pattern examples: GGGG = Gained signals seen for all four markers; NNGN = No gains seen in LAMP3, PROX1, and CCND1. Gain is seen in PRKAA1.
Results
Clinical characteristics are presented in Table 1. Almost half (44%) of the lymph node positive patients died, while all of the lymph node negative patients remained alive during the 10-year follow-up period. Of the seven patients who died, lymph vascular invasion was observed in two patients.
Table 1.
Baseline Characteristics of Patients with Primary Cervical Cancer with (Cases) and without (Controls) Pelvic Lymph Node Metastases
| Characteristics | Cases, n = 16 | Controls, n = 15 |
|---|---|---|
| Age at diagnosis, yr | ||
| Median | 41.5 | 47 |
| Range | 29–66 | 34–69 |
| Stage* | ||
| IB | 12 | 13 |
| IIA | 4 | 2 |
| Tumor size | ||
| ≤4 cm | 8 | 7 |
| >4 cm | 8 | 8 |
| Histology | ||
| Squamous cell carcinoma | 10 | 10 |
| Adenocarcinoma | 2 | 2 |
| Adenosquamous carcinoma | 4 | 3 |
| Histopathologic grade† | ||
| 2 | 6 | 6 |
| 3 | 9 | 8 |
| Lymphvascular invasion | ||
| Present | 5 | 2 |
| Absent | 11 | 13 |
| Treatment | ||
| Preoperative brachytherapy | 14 | 11 |
| Radical hysterectomy and pelvic lymphadenectomy | 16 | 15 |
| Survival | ||
| Alive | 9 | 15 |
| Dead | 7 | 0 |
Tumor stage according to the International Federation of Gynecology and Obstetrics.
Tumor differentiation grade according to the World Health Organization international histologic classification of tumors.
FISH analyses were successfully performed on all 31 pretreatment cervical biopsies and 16 corresponding lymph node metastases. FISH results were analyzed in comparison with patient characteristics within the lymph node positive and negative cases, but none of these comparisons revealed differences that were statistically significant. Copy number gains were seen in all 31 biopsy specimens for all four markers. LAMP3 was the only marker where amplified cells were observed in all cases and controls, with an average of 17% of cells containing an amplified signal. The remaining three markers (PROX1, PRKAA1, and CCND1) each had no amplified signals in several cases. FISH marker comparisons of gained and amplified signals in lymph node positive and negative cases are depicted in Figure 5, A and B. When comparing amplified signals in lymph node positive and negative cases, we observed a larger percentage of amplified nuclei in samples with lymph node metastases, especially with the LAMP3 marker.
Figure 5.
Comparison between lymph node positive (black bars) and lymph node negative cases (gray bars) for the median percentage of cells with gained (gene probe divided by CEP7 [Abbott Molecular, Inc] signal ratio >1) signals for the respective gene probes (A) and the median percentage of cells with amplified (gene probe divided by CEP7 [Abbott Molecular, Inc] signal ratio >2) signals for the respective gene probes (B).
FISH Marker Combinations Predicts Lymph Node Metastases
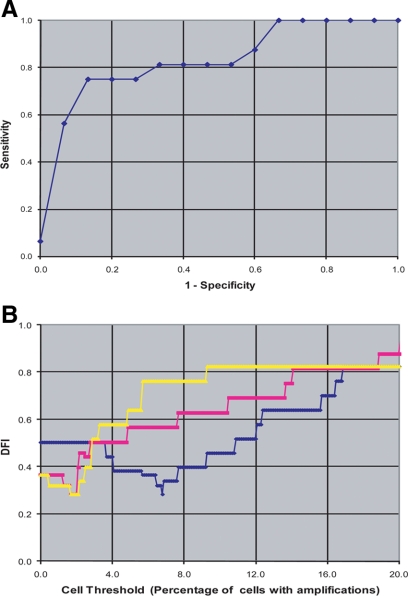
A combined FISH marker consisting of probes for LAMP3, PROX1, and PRKAA1 provided a significant (Ρ = 0.001) predictor for lymph node metastases (Table 2). The optimized threshold used in the composite marker was according to percentages of cells with amplified signals for each individual marker (LAMP: ≥6.8; PROX1: ≥2.0; PRKAA1: ≥2.0). CCND1, as a single marker, provided borderline significance (Ρ = 0.052), with most positive values having low percentages near zero. The specificity, sensitivity, positive predictive value, and negative predictive value of the composite FISH probes are shown in Table 3. These results indicate that 75% of lymph node positive patients were correctly identified with a positive test, while 87% of lymph node negative patients were correctly identified with a negative test. The positive predictive value represents the proportion of test positive patients (86%) who actually had the disease, while the negative predictive value at 76% represented the percentage of test negative patients who were correctly identified with a negative test result. The ROC curve illustrates the relationship between sensitivity and specificity of the FISH composite marker (Figure 6A). Figure 6B depicts the distance from ideal curves of all three single FISH markers used to determine the composite FISH marker and supports the selected threshold values (minimum in each curve). The composite FISH markers results were not related to tumor histology.
Table 2.
P-values to Distinguish between Lymph Node Positive Cases and Lymph Node Negative Controls by Combining Single FISH Markers into a Composite Marker in Pretreatment Cervical Biopsies
| Marker | Optimized thresholds of amplified nuclei, % | Lymph node positive cases,*n = 16 | Lymph node negative controls,*n = 15 | P† |
|---|---|---|---|---|
| LAMP3 | ≥6.8 | 12 | 8 | 0.215 |
| <6.8 | 4 | 7 | ||
| PROX1 | ≥2.0 | 12 | 9 | 0.379 |
| <2.0 | 4 | 6 | ||
| PRKAA1 | ≥2.0 | 14 | 9 | 0.085 |
| <2.0 | 2 | 6 | ||
| CCND1 | ≥1.3 | 11 | 5 | 0.052 |
| <1.3 | 5 | 10 | ||
| Composite | Positive test | 12 | 2 | 0.001 |
| Marker‡ | Negative test | 4 | 13 |
Cases with scores above optimized threshold for each single marker were considered a positive test.
Mantel-Haenszel χ2 test.
Composite marker includes LAMP3, PROX1, and PRKAA1, with each case requiring a positive test for all three markers to be considered a composite marker positive test.
Table 3.
Positive Predictive Value, Negative Predictive Value, and Sensitivity and Specificity Tests for Composite FISH Markers (LAMP3, PROX1, and PRKAA1)
| Lymph node positive cases, N | Lymph node negative controls, N | ||||
|---|---|---|---|---|---|
| Positive test | 12 | 2 | 14 | → | PPV = 0.86 |
| Negative test | 4 | 13 | 17 | → | NPV = 0.76 |
| 16 | 15 | ||||
| ↓ | ↓ | ||||
| Sensitivity = 0.75 | Specificity = 0.87 |
PPV, positive predictive value; NPV, negative predictive value.
Figure 6.
A: ROC plot depicting specificity vs 1− sensitivity curves for the composite FISH marker (B) distance from ideal curves for the three individual markers (LAMP3 in blue, PROX1 in pink, PRKAA1 in yellow) used in the FISH composite marker (see Materials and Methods).
Clonal Patterns
Examination of clonal patterns in tumor biopsies and the corresponding lymph node metastases provided evidence that in certain cases the proportions of different clones can vary between the primary tumor and the metastatic lymph node (Figure 4). Several cases (C7, C24, C30, C32, C37) showed similar clones and similar quantitative distribution between primary tumor and lymph node metastasis, indicating that the major clone(s) of the primary tumor were also the most successful clone(s) to establish lymph node metastasis. However, in many cases, a change in the frequency of specific clones was observed between the primary tumors and their associated lymph node metastases indicating that pre-existing, but minor clones in the primary tumors had a growth advantage in the lymph nodes. For example, in cases C1, C5, and C28, the major clonal change observed was the considerable increase of NNGN (gained signal seen in PRKAA1 only, see Materials and Methods) in the lymph node metastases. Although similarities in clonal patterns of certain cases were observed, these resemblances were not related to the patients’ clinical characteristics.
Discussion
For cervical cancer, we found that the enumeration of genomic copy numbers of the composite marker consisting of LAMP3, PROX1, and PRKAA1 allowed statistically significant prediction of lymph node metastases. We also observed that clonal patterns may differ between a primary tumor and its metastasis, indicating that certain clones may have a growth advantage in the lymph node environment.
To our knowledge, no other study has performed FISH analyses for the prediction of lymph node metastases in cervical cancer. Studies attempting to predict lymph node metastases have used techniques including immunohistochemistry, RT-PCR, and gene expression profiling. Immunohistochemistry is a cost-effective technique, but diverse epitopes (Cox-2, p16, CXCR4, CCR7, D2-40, etc) and controversial results were reported for the prediction of lymph node metastases in cervical cancer.19,20,21,22,23,24 Gene expression profiling studies also found divergent results with a study by Biewenga et al,25 which was not able to predict lymph node metastases in early stage cervical cancer, while a study by Kim et al26 did. Biewenga et al25 found five genes showing differential expression between patients with and without lymph node metastases. The prediction of lymph node positivity will assist in developing adequate treatment options for patients since histologically undetectable micrometastases in the lymphatic system may account for cervical cancer recurrence.27,28,29 Histological examinations are not infallible since examinations only comprise a small portion of lymph nodes, and estimates indicate that a pathologist has a 1% chance of detecting a micrometastatic focus within a three-tumor-cell diameter.28,30
Gene amplification is a common mechanism for oncogene activation in tumorigenesis and gains on chromosomes 1q, 3q, and 5p are frequently observed in cervical cancer.7,8,9,10,11 In our study, a gain of chromosome 3q was seen in all samples, regardless of histology, which confirms the dominant role of this chromosome in the development of cervical cancer.7,8,9,10,11,18,31 In the present study, LAMP3 is consistently gained and amplified, which supports the finding that 3q copy number gain is ubiquitous in cervical cancer. LAMP3, a potential gene of interest located on 3q26, was found to be associated with metastatic potential both in vitro and in vivo.12 Amplification of chromosome 3q has also been linked to lymph node metastases in two studies using comparative genomic hybridization, which exhibited an increase in 3q copy number gain in primary tumors with lymph node metastasis; however, the results by Allen et al10 were not statistically significant.10,32 Although the function of LAMP3 is relatively unknown, one study suggests that LAMP3 may be involved in tumor cell migration into surrounding lymph vessels.12 PROX1 has been found exclusively in lymphatic endothelial cells and is vital to embryonic lymphatic development.14,33,34 Decreased PROX1 expression has been found in several cancers, including hepatocellular carcinoma, biliary duct cancer, and breast cancer.35,36,37 However, in colon cancer, overexpression of PROX1 induces colon cancer progression by promoting a transition from a benign to a highly dysplastic phenotype.38 In cervical cancer, PROX1 lies within a region of amplification (chromosome 1q) which could suggest that PROX1 copy number gain may function similarly to that in colon cancer. Development of cervical cancer has also been linked to a copy number gain at chromosome 5p, with PRKAA1 being a potential gene of interest.16 A recent study found that high expression of PRKAA1 may help cancers establish their malignant nature through cellular resistance to cell death and overcoming their hypoxic condition.16,39 The three genes of interest, LAMP3, PROX1, and PRKAA1 have never been investigated in cervical cancer by using FISH. Previous studies on LAMP3, PROX1, and PRKAA1 have focused on the level of mRNA expression by using reverse transcription-PCR and protein expression using immunohistochemistry. An advantage of multicolor FISH is the ability to visualize several genes of interest simultaneously in each cell. In addition, formalin-fixed, archived samples can be used.
Clonal patterns varied among the primary cervical cancers. To our knowledge, this is the first study investigating the difference in FISH marker patterns between the cells of a primary cervical cancer and synchronous lymph node metastases. The approach to evaluate all four gene probes and the CEP7 (Abbott Molecular, Inc) ploidy control probe within the same nuclei, allows for comprehensive pattern comparison. In our study, several cases showed the exact same clones and their frequency between primary tumor and lymph node metastasis indicating that the major clone(s) of the primary tumor were also the most successful clone(s) to establish lymph node metastasis. Therefore, in these cases, at least on the genomic level tested, the majority of cells in the primary tumor could become metastatic cells. In that case metastatic potential would be already present in the bulk of the tumor. However, in some cases, changes in the clonal constitution between the primary tumor and lymph node metastasis indicate that a pre-existing but minor clone in the primary tumor had growth advantages for establishing metastases in the lymph nodes compared with the major clone of the primary tumor. This would be consistent with the hypothesis that a subset of cells in the primary tumor has the genetic makeup that eventually results in metastatic disease, which could indicate that additional mutations in a few cells are required for metastasis.40 This difference in patterns might reflect different mechanisms in the development of lymph node metastases in cervical cancer.
In summary, we found that a composite FISH marker, comprising LAMP3, PROX1, and PRKAA1, allowed statistically significant discernment between patients with and without risk for metastatic disease with a reasonable sensitivity and specificity. Further studies with additional patients are warranted to validate this molecular marker prospectively in the clinical setting.
Acknowledgments
We thank Margareta Rodensjö and Lisa Anfalk for excellent technical assistance. We also thank Catharine Beskow for her support and assistance.
Footnotes
Address reprint requests to Thomas Ried, Genetics Branch, Center for Cancer Research, National Cancer Institute/National Institutes of Health, 50 South Dr, Room 1306, Bethesda, MD 20892. E-mail: riedt@mail.nih.gov.
Supported by the Swedish Cancer Society (Cancerfonden), the Cancer Society of Stockholm (Cancerföreningen), Karolinska Institutet, the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and the National Library of Medicine.
References
- Alston RD, Geraci M, Eden TOB, Moran A, Rowan S, Birch JM. Changes in cancer incidence in teenagers and young adults (ages 13 to 24 years) in England 1979–2003. Cancer. 2008;113:2807–2815. doi: 10.1002/cncr.23901. [DOI] [PubMed] [Google Scholar]
- Parkin DM, Louie KS, Clifford G. Burden and trends of type-specific human papillomavirus infections and related diseases in the Asia Pacific region. Vaccine. 2008;26(Suppl 12):M1–M16. doi: 10.1016/j.vaccine.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Waggoner SE. Cervical cancer. Lancet. 2003;361:2217–2225. doi: 10.1016/S0140-6736(03)13778-6. [DOI] [PubMed] [Google Scholar]
- Buckley CH, Beards CS, Fox H. Pathological prognostic indicators in cervical cancer with particular reference to patients under the age of 40 years. Br J Obstet Gynaecol. 1988;95:47–56. doi: 10.1111/j.1471-0528.1988.tb06479.x. [DOI] [PubMed] [Google Scholar]
- Fuller AF, Jr, Elliott N, Kosloff C, Hoskins WJ, Lewis JL., Jr Determinants of increased risk for recurrence in patients undergoing radical hysterectomy for stage IB and IIA carcinoma of the cervix. Gynecol Oncol. 1989;33:34–39. doi: 10.1016/0090-8258(89)90598-2. [DOI] [PubMed] [Google Scholar]
- Quinn MA, Benedet JL, Odicino F, Maisonneuve P, Beller U, Creasman WT, Heintz APM, Ngan HY, Pecorelli S. Carcinoma of the cervix uteri. (Part of the FIGO 6th Annual Report on the Results of Treatment in Gynecological Cancer). Int J Gynaecol Obstet. 2006;95(Suppl 1):S43–S103. doi: 10.1016/S0020-7292(06)60030-1. [DOI] [PubMed] [Google Scholar]
- Heselmeyer K, Macville M, Schröck E, Blegen H, Hellström A-C, Shah K, Auer G, Ried T. Advanced-stage cervical carcinomas are defined by a recurrent pattern of chromosomal aberrations revealing high genetic instability and a consistent gain of chromosome arm 3q. Genes Chromosomes Cancer. 1997;19:233–240. [PubMed] [Google Scholar]
- Heselmeyer K, Schröck E, du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci USA. 1996;93:479–484. doi: 10.1073/pnas.93.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao PH, Arias-Pulido H, Lu XY, Harris CP, Vargas H, Zhang FF, Narayan G, Schneider A, Terry MB, Murty VVVS. Chromosomal amplifications, 3q gain and deletions of 2q33-q37 are the frequent genetic changes in cervical carcinoma. BMC Cancer. 2004;4:5. doi: 10.1186/1471-2407-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen DG, White DJ, Hutchins A-M, Scurry JP, Tabrizi SN, Garland SM, Armes JE. Progressive genetic aberrations detected by comparative genomic hybridization in squamous cell cervical cancer. Br J Cancer. 2000;83:1659–1663. doi: 10.1054/bjoc.2000.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff M, Rose H, Petersen BL, Maahr J, Gerdes T, Lundsteen C, Bryndorf T, Kryger-Baggesen N, Christensen L, Engelholm SA, Philip J. Comparative genomic hybridization reveals a recurrent pattern of chromosomal aberrations in severe dysplasia/carcinoma in situ of the cervix and in advanced-stage cervical carcinoma. Genes Chromosomes Cancer. 1999;24:144–150. doi: 10.1002/(sici)1098-2264(199902)24:2<144::aid-gcc7>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Kanao H, Enomoto T, Kimura T, Fujita M, Nakashima R, Ueda Y, Ueno Y, Miyatake T, Yoshizaki T, Buzard GS, Tanigami A, Yoshino K, Murata Y. Overexpression of LAMP3/TSC403/DC-LAMP promotes metastasis in uterine cervical cancer. Cancer Res. 2005;65:8640–8645. doi: 10.1158/0008-5472.CAN-04-4112. [DOI] [PubMed] [Google Scholar]
- Tervala T, Suominen E, Saaristo A. Targeted treatment for lymphedema and lymphatic metastasis. Ann NY Acad Sci. 2008;1131:215–224. doi: 10.1196/annals.1413.019. [DOI] [PubMed] [Google Scholar]
- Wigle JT, Oliver G. Prox1 function is required for the development of the murine lymphatic system. Cell. 1999;98:769–778. doi: 10.1016/s0092-8674(00)81511-1. [DOI] [PubMed] [Google Scholar]
- Witte MH, Jones K, Wilting J, Dictor M, Selg M, McHale N, Gershenwald JE, Jackson DG. Structure function relationships in the lymphatic system and implications for cancer biology. Cancer Metastasis Rev. 2006;25:159–184. doi: 10.1007/s10555-006-8496-2. [DOI] [PubMed] [Google Scholar]
- Huang FY, Chiu PM, Tam KF, Kwok YK, Lau ET, Tang MHY, Ng TY, Liu VWS, Cheung ANY, Ngan HYS. Semi-quantitative fluorescent PCR analysis identifies PRKAA1 on chromosome 5 as a potential candidate cancer gene of cervical cancer. Gynecol Oncol. 2006;103:219–225. doi: 10.1016/j.ygyno.2006.02.028. [DOI] [PubMed] [Google Scholar]
- Myo K, Uzawa N, Miyamoto R, Sonoda I, Yuki Y, Amagasa T. Cyclin D1 gene numerical aberration is a predictive marker for occult cervical lymph node metastasis in TNM Stage I and II squamous cell carcinoma of the oral cavity. Cancer. 2005;104:2709–2716. doi: 10.1002/cncr.21491. [DOI] [PubMed] [Google Scholar]
- Heselmeyer-Haddad K, Janz V, Castle PE, Chaudhri N, White N, Wilber K, Morrison LE, Auer G, Burroughs FH, Sherman ME, Ried T. Detection of genomic amplification of the human telomerase gene (TERC) in cytologic specimens as a genetic test for the diagnosis of cervical dysplasia. Am J Pathol. 2003;163:1405–1416. doi: 10.1016/S0002-9440(10)63498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dursun P, Yuce K, Usubutun A, Ayhan A. Cyclooxygenase-2 expression in cervical intraepithelial neoplasia III and squamous cell cervical carcinoma, and its correlation with clinicopathologic variables. Int J Gynecol Cancer. 2007;17:164–173. doi: 10.1111/j.1525-1438.2007.00798.x. [DOI] [PubMed] [Google Scholar]
- Kodama J, Hasengaowa , Kusumoto T, Seki N, Matsuo T, Ojima Y, Nakamura K, Hongo A, Hiramatsu Y. Association of CXCR4 and CCR7 chemokine receptor expression and lymph node metastasis in human cervical cancer. Ann Oncol. 2007;18:70–76. doi: 10.1093/annonc/mdl342. [DOI] [PubMed] [Google Scholar]
- Urabe A, Matsumoto T, Kimura M, Sonoue H, Kinoshita K. Grading system of lymphatic invasion according to D2–40 immunostaining is useful for the prediction of nodal metastasis in squamous cell carcinoma of the uterine cervix. Histopathology. 2006;49:493–497. doi: 10.1111/j.1365-2559.2006.02536.x. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Tomita S, Ichikawa K, Ono Y, Inaba F, Fukasawa I, Imai Y, Imura J, Fukui H, Fujimori T, Inaba N. P16-immunostaining pattern as a predictive marker of lymph node metastasis and recurrence in early uterine cervical cancer. Pathobiology. 2006;73:176–182. doi: 10.1159/000096018. [DOI] [PubMed] [Google Scholar]
- Mulvany NJ, Allen DG, Wilson SM. Diagnostic utility of p16INK4a: a reappraisal of its use in cervical biopsies. Pathology. 2008;40:335–344. doi: 10.1080/00313020802035907. [DOI] [PubMed] [Google Scholar]
- Baltazar F, Filho AL, Pinheiro C, Moreira MAR, Queiroz GS, Oton GJB, Júnior AF, Ribeiro LFJ, Schmitt FC. Cyclooxygenase-2 and epidermal growth factor receptor expressions in different histological subtypes of cervical carcinomas. Int J Gynecol Pathol. 2007;26:235–241. doi: 10.1097/pgp.0b013e31802f1996. [DOI] [PubMed] [Google Scholar]
- Biewenga P, Buist MR, Moerland PD, Ver Loren, van Themaat E, van Kampen AHC, ten Kate FJW, Baas F. Gene expression in early stage cervical cancer. Gynecol Oncol. 2008;108:520–526. doi: 10.1016/j.ygyno.2007.11.024. [DOI] [PubMed] [Google Scholar]
- Kim T-J, Choi J-J, Kim WY, Choi CH, Lee J-W, Bae D-S, Son D-S, Kim J, Park BK, Ahn G, Cho EY, Kim B-G. Gene expression profiling for the prediction of lymph node metastasis in patients with cervical cancer. Cancer Sci. 2008;99:31–38. doi: 10.1111/j.1349-7006.2007.00652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren L, O'Reilly MS, Folkman J. Dormancy of micrometastases: balanced proliferation and apoptosis in the presence of angiogenesis suppression. Nat Med. 1995;1:149–153. doi: 10.1038/nm0295-149. [DOI] [PubMed] [Google Scholar]
- Yuan S-H, Liang X-F, Jia W-H, Huang J-L, Wei M, Deng L, Liang L-Z, Wang X-Y, Zeng Y-X. Molecular diagnosis of sentinel lymph node metastases in cervical cancer using squamous cell carcinoma antigen. Clin Cancer Res. 2008;14:5571–5578. doi: 10.1158/1078-0432.CCR-08-0346. [DOI] [PubMed] [Google Scholar]
- Marchiolè P, Buènerd A, Scoazec J-Y, Dargent D, Mathevet P. Sentinel lymph node biopsy is not accurate in predicting lymph node status for patients with cervical carcinoma. Cancer. 2004;100:2154–2159. doi: 10.1002/cncr.20212. [DOI] [PubMed] [Google Scholar]
- Gusterson BA. The new TNM classification and micrometastases. Breast. 2003;12:387–390. doi: 10.1016/s0960-9776(03)00141-3. [DOI] [PubMed] [Google Scholar]
- Heselmeyer-Haddad K, Sommerfeld K, White NM, Chaudhri N, Morrison LE, Palanisamy N, Wang ZY, Auer G, Steinberg W, Ried T. Genomic amplification of the human telomerase gene (TERC) in pap smears predicts the development of cervical cancer. Am J Pathol. 2005;166:1229–1238. doi: 10.1016/S0002-9440(10)62341-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K-F, Lee W-Y, Huang SC, Lin YS, Kang CY, Liou CP, Tzeng CC. Chromosomal gain of 3q and loss of 11q often associated with nodal metastasis in early stage cervical squamous cell carcinoma. J Formos Med Assoc. 2007;106:894–902. doi: 10.1016/S0929-6646(08)60059-5. [DOI] [PubMed] [Google Scholar]
- Nathanson SD. Insights into the mechanisms of lymph node metastasis. Cancer. 2003;98:413–423. doi: 10.1002/cncr.11464. [DOI] [PubMed] [Google Scholar]
- Saharinen P, Tammela T, Karkkainen MJ, Alitalo K. Lymphatic vasculature: development, molecular regulation and role in tumor metastasis and inflammation. Trends Immunol. 2004;25:387–395. doi: 10.1016/j.it.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Shimoda M, Takahashi M, Yoshimoto T, Kono T, Ikai I, Kubo H. A homeobox protein, prox1, is involved in the differentiation, proliferation, and prognosis in hepatocellular carcinoma. Clin Cancer Res. 2006;12:6005–6011. doi: 10.1158/1078-0432.CCR-06-0712. [DOI] [PubMed] [Google Scholar]
- Van den Eynden GG, Van der Auwera I, Van Laere SJ, Trinh XB, Colpaert CG, van Dam P, Dirix LY, Vermeulen PB, Van Marck EA. Comparison of molecular determinants of angiogenesis and lymphangiogenesis in lymph node metastases and in primary tumours of patients with breast cancer. J Pathol. 2007;213:56–64. doi: 10.1002/path.2211. [DOI] [PubMed] [Google Scholar]
- Laerm A, Helmbold P, Goldberg M, Dammann R, Holzhausen H-J, Ballhausen WG. Prospero-related homeobox 1 (PROX1) is frequently inactivated by genomic deletions and epigenetic silencing in carcinomas of the bilary system. J Hepatol. 2007;46:89–97. doi: 10.1016/j.jhep.2006.07.033. [DOI] [PubMed] [Google Scholar]
- Petrova TV, Nykanen A, Norrmén C, Ivanov KI, Andersson LC, Haglund C, Puolakkainen P, Wempe F, von Melchner H, Gradwohl G, Vanharanta S, Aaltonen LA, Saharinen J, Gentile M, Clarke A, Taipale J, Oliver G, Alitalo K. Transcription factor PROX1 induces colon cancer progression by promoting the transition from benign to highly dysplastic phenotype. Cancer Cell. 2008;13:407–419. doi: 10.1016/j.ccr.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Kato K, Ogura T, Kishimoto A, Minegishi Y, Nakajima N, Miyazaki M, Esumi H. Critical roles of AMP-activated protein kinase in constitutive tolerance of cancer cells to nutrient deprivation and tumor formation. Oncogene. 2002;21:6082–6090. doi: 10.1038/sj.onc.1205737. [DOI] [PubMed] [Google Scholar]
- Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]