Abstract
FXYD3 is a FXYD-containing Na,K-ATPase ion channel regulator first identified as a protein overexpressed in murine breast tumors initiated by oncogenic ras or neu. However, our preliminary study revealed that FXYD3 expression was down-regulated in oncogenic KRAS-transduced airway epithelial cells. This contradiction led us to investigate the role of FXYD3 in carcinogenesis of the lung. FXYD3 mRNA and protein levels were lower in most of the lung cancer cell lines than in either the noncancerous lung tissue or airway epithelial cells. Protein levels were also lower in a considerable proportion of primary lung cancers than in nontumoral airway epithelia; FXYD3 expression levels decreased in parallel with the dedifferentiation process. Also, a somatic point mutation, g55c (D19H), was found in one cell line. Forced expression of the wild-type FXYD3, but not the mutant, restored the well-demarcated distribution of cortical actin in cancer cells that had lost FXYD3 expression, suggesting FXYD3 plays a role in the maintenance of cytoskeletal integrity. However, no association between FXYD3 expression and its promoter’s methylation status was observed. Therefore, inactivation of FXYD3 through a gene mutation or unknown mechanism could be one cause of the atypical shapes of cancer cells and play a potential role in the progression of lung cancer.
We recently proposed that potential tumor suppressors lie hidden downstream of oncogenic KRAS, based on the finding that a mutated KRAS (V12) induced severe growth suppression in primary and immortalized bronchial epithelial cells.1 We have identified several candidate tumor suppressors, including dual specificity phosphatase 6 (DUSP6)1 and insulin-like growth factor binding protein (IGFBP)-2/4,2 through comprehensive gene expression analyses. The results prompted us to further identify novel tumor suppressors from among the downstream targets of oncogenic KRAS.
Mammary tumor 8 kDa (MAT8)/FXDY domain-containing ion transport regulator 3 (FXYD3), a FXYD-containing Na,K-ATPase ion channel regulator, was first identified as a protein highly expressed in murine breast tumor initiated by oncogenic ras (v-Ha-ras) or neu (her2, egfr2)1,2,3,4,5,6,7,8,9,10,11,12,13 and later demonstrated to occur at (inner-) cellular membranes and to be involved in the regulation of chloride conductance.11,12,13 Recent studies found FXYD3 to be overexpressed breast, prostate, and pancreatic cancers,5,14,15 and suggested that it promotes carcinogenesis by providing a growth advantage.14,15 Moreover, its expression was reported to be up-regulated in response to stimuli inducing cellular differentiation16 and by treatment with an anticancer drug, 5-fluofouracil, in colon and/or breast cancer cells.17,18 Furthermore, contrary to initial observations, our reduced preliminary study revealed that its expression was rather in oncogenic KRAS-transduced immortalized human airway epithelial cells. Thus, FXYD3 could play diverse roles in different instances and its significance in carcinogenesis could vary in type of cancer.
To elucidate the potential role of FXYD3 in carcinogenesis of the lung, we here examined its expression, gene mutation, and biological function in lung cancer cell lines and/or primary human lung cancers, as well as analyzed the correlation between its expression levels and a variety of clinicopathologic parameters.
Materials and Methods
Cell Cultures
The immortalized human airway epithelial cell line (16HBE14o, Simian virus 40 [SV40]-transformed human bronchial epithelial cells) described by Cozens et al19 was kindly provided by D.C. Gruenert (California Pacific Medical Center Research Institute). A subclone of 16HBE14o cells, described as NHBE-T in this paper, was used for experiments. The immortalized airway epithelial cell lines (HPL1D and HPL1A, SV40-transformed human small airway epithelial cells) established by Masuda et al,20 was kindly provided by Takahashi T (Nagoya University School of Medicine). Human lung cancer cell lines (A549, H358, H1299, H2087, H526, H82, H1688, and H460), immortalized human lymphoblasts generated from the same person as the H2087 lung cancer cells, human lung fibroblasts (WI-38), and a human embryonic kidney cell line (HEK293T) were purchased from American type culture collection (ATCC, Manassas, VA). Human lung cancer cell lines, LC/MS, LCD, LC2/ad, Lu65, Lu134A, Lu135, Lu139, Lu140, ABC1, HLC1, and LCKJ were purchased form Riken cell bank (Tsukuba, Japan). Human lung cancer cell lines, PC1, PC3, PC9, PC13, and HARA were obtained form Riken cell bank (Tsukuba, Japan), and Immuno-Biological laboratories Co. (Gunma, Japan), respectively. Human lung cancer cell lines, TKB1, TKB2, TKB4, TKB5, TKB6, TKB7, TKB9, TKB12, TKB15, TKB16, TKB17, and TKB20, were kindly provided by Kamma (Kyorin University School of Medicine). These cells were cultured and grown in Dulbecco’s Modified Eagle Medium (Sigma Aldrich, St. Louis, MO) or RPMI1640 medium (Sigma) supplemented with 10% heat inactivated fetal bovine serum (FBS) (Sigma), 100 units/ml of penicillin (Sigma), and 100 μg/ml of streptomycin (Sigma). Primary small airway epithelial cells (SAEC) and bronchial epithelial cells (NHBE) were purchased from Sanko Kagaku (Tokyo, Japan), and grown in defined keratinocyte serum free medium (Invitrogen, Carlsbad, CA).
Plasmid Construction
Complementary DNA (cDNA) coding for the wild-type (KRAS/G12) and mutated KRAS (KRAS/V12) were PCR-amplified (PrimeSATR HS DNA polymerase, Takara Bio Inc., Kyoto, Japan) with primers having BamHI restriction sites, forward (F) 5′-CTCCGCGGATCCAAGCTTGCTGAAA-3′ and reverse (R) 5′-AGGGGCGGATCctcattacataa-3′ (BamHI restriction sites italicized), using plasmid vectors, pcDNA3.1-KRAS/G12 and -KRAS/V12 described elsewhere1 as templates. The amplified fragments were inserted into the BamHI site of pQCXIH retroviral vectors (BD Clontech, Palo Alto, CA). Enhanced green fluorescent protein (EGFP) cDNA was PCR-amplified (PrimeSATR HS DNA polymerase, Takara) using the pIRES2-EGFP vector (BD Clontech) as a template with mismatch primers having EcoRI restriction sites, F, 5′-ATGGGAATTCCCgggGTGAGCAAGGGCGAGGA-3′ (fist methionine [ATG] was converted to glycine [GGG], M1G) or 5′-ATGGaattcACCATGGTGAGCAAGGGCGAGGAG-3′; R, 5′-ATCTAGGAATTCGGCCGCTTTACTTGTA-3′ (EcoRI restriction sites italicized). It was inserted into the EcoRI site of pQCXIN (BD Clontech). FXYD3 variant 1 cDNA (GENE BANK ACCESSION# NM_005971) and a mutant (FXYD3 g55c [D19H]) were PCR-amplified (LA TaqDNA polymerase, Takara) with primers, F 5′-GGCCAGCGCTCTGACATGCAGAA-3′ and R 5′-CTGGGGACAGAGAACGGTCCTCC-3′, using cDNA reverse-transcribed (SuperScript III, Invitrogen, Carlsbad, CA) from total RNA of H2087 cells. After the adenylation of 3′ terminals with TaqDNA polymerase (Takara), the amplified fragment was subcloned into a pT7Blue vector (Novagen, Darmstadt, Germany). FXYD3 cDNA was PCR-amplified again (PrimeSATR HS DNA polymerase, Takara) using pT7Blue/FXYD3 as a template with mismatch primers having BamHI restriction sites, F 5′-ACCCGGGGATCCGATTGGCCAG-3′ and R 5′-CAGCTGGTGGATCCTCCTCCGCTTTG-3′ (stop codon [TGA] was converted into glycine [GGG]) (BamHI restriction sites italicized, mutant in bold). It was inserted into the BamHI site of pQCXIN/EGFP (M1G). A candidate promoter region (−2539 to −1615, −1615 to −5, −1159 to −5, −760 to −5, or −461 to −5 of the FXYD3 gene locus [NC_000019] taking the first base of the transcription start site as + 1), was PCR-amplified (PrimeSATR HS DNA polymerase, Takara) with primer sets: F-2539 5′-GGTGCTGAGACCACAGCGCTGTTC-3′ and R-1615, 5′-TGGGGGACCAGCAATTAGAGACTTC-3′; F-1615, 5′-TCTAATTGCTGGTCCCCCAGGTTGAAAC-3′ and R-4, 5′-GTCCCCTTCCAGGCCACATCTTGCCG-3′; F-1159, 5′-TGTACACGCCCCTACTCGCTCTCGG-3′ and R-5, 5′-GTCCCCTTCCAGGCCACATCTTGCCG-3′; F-760 5′-AGCGATGACAAGTGGGGGGTCCTTC-3′ and R-5 5′-GTCCCCTTCCAGGCCACATCTTGCCG-3′; F-461 5′-TGTCTAAGAAGGCGGAATTGAGGGG-3′ and R-5 5′-GTCCCCTTCCAGGCCACATCTTGCCG-3′, using genomic DNA extracted from nontumorous parts of surgically resected lung as a template. The products were subcloned into the pT7Blue vector (Novagen). They were cut out with HindIII (Takara) and KpnI (Takara), and inserted into pGL4.1 (Promega, Madison, WI). The accuracy of all of the constructs was verified by DNA sequencing (Dye-deoxy DNA sequencing kit, Amersham life science, Piscataway, NJ).
Retroviral-Mediated Gene Transfer
The pQCXIH or pQCXIH-based vectors and the pCL10A1 retroviral packaging vector (Imgenex, San Diego, CA) were cotransfected into HEK293T cells with Lipofectoamine 2000 reagent (Invitrogen). After 24 hours, the conditioned medium was recovered as a viral solution. Desired genes were introduced by incubating cells with the viral solution containing 10 μg/ml of polybren (Sigma) for 24 hours. Cells stably expressing desired genes were selected with 1000 μg/ml of neomycin (G418, Invitrogen) or 500 μg/ml of Hygromycin B (Invitrogen) for 7 days. Selected pooled clones were used for biological analyses.
Reverse Transcription-PCR
First-strand cDNA was synthesized from total RNA using the SuperScript First-Strand Synthesis System according to the protocol of the manufacturer (Invitrogen). Fragments of FXYD3 and ACTB (β-actin) were PCR-amplified with a LA TaqPCR kit using the following primers: for FXYD3 forward (F) 5′-GGCCAGCGCTCTGACATGCAGAA-3′ and reverse (R) 5′-CTGGGGACAGAGAACGGTCCTCC-3′, and for ACTB, F 5′-TGGCACCCAGCACAATGAA-3′ and R 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′.
Quantitative Reverse Transcription-PCR
First-strand cDNA was synthesized by the method described above. The cDNA generated was used as a template in real-time PCR with SYBR Premix EXTaq (Takara) and run on a Thermal Cycler DICE real-time PCR system (Takara). The primer set used for the detection of FXYD3 was forward (F) 5′-GGCCAGCGCTCTGACATGCAGAA-3′ and reverse (R) 5′-CTGGGGACAGAGAACGGTCCTCC-3′. That used for ACTB was F 5′-CTGGGGACAGAGAACGGTCCTCC-3′ and R 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′. The means and standard deviations of copy numbers of FXYD3 relative to ACTB mRNA were statistically obtained from triplicate reactions.
Direct DNA Sequencing
Genomic DNA was extracted from lung cancer cell lines and tumor cells microscopically dissected (PALM-MCB laser microdissection system, Carl Zeiss, Jena, Germany) from primary lung cancer tissues (90% ethanol-fixed materials) by a method described elsewhere.3 Coding regions of the FXYD3 gene were PCR-amplified (LA TaqDNA polymerase, Takara) using the genomic DNA as a template with the following sets of primers: for exon 3, forward (F) 5′-CAGCTGCGGCTTATCTCTCAGCCCA-3′ and reverse (R) 5′-AGATGCTGTTCTAACATTTACCACc-3′; for exons 4 and 5, F 5′-CCTGTCCCGCAGGAGACCCTTT-3′ and R 5′-TCTTTGTCCTGTGATGCTCACg-3′; for exons 6 and 7, F 5′-GTCTGTTTTCTTATGGCGGTGTC-3′ and R 5′-AGCCCTTTCACCCTGAAAAGCG-3′; and for exons 8 and 9, F 5′-ATGTCTGGGCAGGCTAAGAACCC-3′ and R 5′-TTCCATCCTGGAGTTCAAGTTTCT-3′. Surplus primers and nucleotides were removed enzymatically with EXO-SAP-IT (Amersham). The purified PCR products were subjected to a dye-terminator reaction with Big Dye version 3.1 (ABI, Foster, CA). For the dye-terminator reaction, the same primers as for the PCR-amplification were used. The DNA sequence was analyzed with an ABI 3100 DNA sequencer (ABI).
Western Blotting
Whole cell lysate was subjected to SDS-polyacrylamide gel electrophoresis, and transferred onto polyvinylidene difluoride membranes (Amersham). The membranes were incubated with nonfat dry milk in 0.01 M/L Tris-buffered saline containing 0.1% Tween-20 to block non-immunospecific protein binding, and then with 0.1 μg/ml of primary antibodies (EGFP, Santa Cruz, Santa Cruz, CA; FXYD3, ALTAS Antibodies, Stockholm, Sweden; β-actin, Sigma). After being washed with Tris-buffered saline containing 0.1% Tween-20, the membranes were incubated with animal-matched horseradish peroxidase-conjugated secondary antibodies (Amersham). Immunoreactivity was visualized with the enhanced chemiluminescence system (ECL, Amersham).
Treatment with 5-Azacytidine and Trichostatin A
Cells were treated either with 10 μmol/L of 5-azacytidine (AZA, Sigma) for 72 hours by exchanging the medium everyday or with 300 ng/ml of trichostatin A (Wako, Osaka, Japan) for 24 hours. In addition, cells were treated also with AZA for 48 hours and then with a combination of AZA and trichostatin A for an additional 24 hours.
Bisulfate Conversion-Based DNA Sequencing
Genomic DNA was extracted from cultured cells by a method described previously,3 and was subjected to bisulfate conversion treatment using a MethylEasy DNA bisulfate modification kit (Human Genetic Signatures, Macquarie Park, Australia) according to the manufacturer’s instructions. The regions from −1615 to −1159 and from −465 (or −175) to −5 of the FXYD3 gene were PCR-amplified (TaqHS DNA polymerase, Takara) using the bisulfate converted DNA as a template with the following sets of primers: forward (F)−1663 5′-TATTTTTAAGATGTAAAATTTAAAAA-3′ and reverse (R)−1140 5′-TGTTAATTTTTAAAAATATTTTATTTA-3′; F-465 5′-TGTTTAGGGTAGAGTTATTTTTTTT-3′ and R-5 5′-AAAACCTCCAAAACCATACAATAAA-3′; F-175 5′-AGGGTAGGGAGTAGGTTATGATTAg-3′ and R-5 5′-AAAACCTCCAAAACCATACAATAAA-3′, and were subcloned. Ten clones were randomly chosen. The conversion of nucleotide at CpG sites was examined for by DNA sequencing (Dye-deoxy DNA sequencing kit, Amersham).
Cell Growth and Migration of H1299 Cells
MOCK (EGFP) or FXYD3-EGFP was retrovirally transduced into H1299 cells. After the selection treatment, the surviving cells were harvested and counted, and 1.0 × 104 cells were re-seeded onto a 10 cm dish. After 12 days, the cells were fixed with methanol and stained with Giemsa. The selected cells (1.25 × 104) were cultured and grown in 1 ml of 0.3% soft agar in 3.5 cm culture dishes for 3 weeks, then fixed with a buffered 4% paraformaldehyde solution. The selected cells (2.5 × 105) were seeded onto 3.5 cm culture dishes and grown for 10 days. The cells were counted every 2 days.
Restriction Enzyme Mediated Selective-PCR Analysis for KRAS
Genomic DNA was extracted from fresh frozen lung cancer tissues or paraffin sections by methods described previously.21,22 For the detection of KRAS codon 12 mutations, the REMS PCR described by Ward et al21 was partially modified. Briefly, the reaction mixture, a final volume of 5 μl, containing 0.5 μl of 10 ng/μl DNA solution (1 ng/μl), 0.5 μl of 10×PCR buffer (1×), 0.4 μl of each 2.5 mmol/L dNTP (0.2 mmol/L), 0.04 μl of 50 μmol/L primers (three primer sets described by Ward R et al,21 each 0.4 μmol/L), 0.025 μl of TaqHS DNA polymerase (Takara), 0.4 μl of BstN1 (New England BioLabs, Northbrook, IL) (0.8 units/μl), and 2.935 μl of distilled water (final volume, 5 μl), was PCR-cycled on a Thermal Cycler Dice (Takara) with the following program; 94°C for 2 minutes (one cycle), 92°C for 20 seconds −60°C for 3 minutes (35 cycles), held at 4°C. The products were resolved by electrophoresis in 5% agar. Genomic DNA from A549 (mutated KRAS) and H1299 (wild-type KRAS) mixed indicated ratio (A549/H1299 = 1/0, 1/1, 1/10, 0/1), and from H358 (mutated KRAS), and pQCXIH vectors bearing wild-type (G12) and mutated KRAS (V12), served as experimental controls.
Luciferase Reporter Assays
The pGL4.1-based vectors constructed above and a pGL4.7-TK Renilla (Promega, Madison) were co-transfected into NHBE-T cells with Lipofectamine 2000 (Invitrogen). After 24 hours, the cells were solved and luminosity was measured using a Dual Luciferase Reporter Assay system (Promega) on a luminometer (TD-20/20, Turner BioSystems, Sunnyvale, CA). The luminosity derived from the pGL4.1-based vectors was corrected with that from the pGL4.7-TK Renilla, to normalize the transfection efficiency. Luminosity relative to that of the empty pGL4.1 vector was determined as promoter activity.
Primary Lung Cancers
All cases examined were of lung cancer patients who underwent surgical resection at Kanagawa Cardiovascular and Respiratory Disease Center Hospital during the period from 2001 to 2008. Informed consent for research use was obtained from all of the subjects providing materials. Sixty-five cases (adenocarcinomas [ADC, 36], squamous cell carcinomas [SQC, 21], adenosquamous carcinomas [ASC, 2], large cell carcinomas [LCC, 3], and small cell carcinomas [SCC, 3]), were analyzed for FXYD3 and Ki-67 expression by immunohistochemistry, and for KRAS codon 12 mutations by restriction endonuclease-mediated selective PCR (a representative result is shown: see Supplemental Figure S1 at http://ajp.amjpathol.org.). ADCs were classified into histological subtypes according to the criteria of the World Health Organization.3 If a case was of mixed type, the major component was described as the subtype in the present study.
Immunohistochemistry
Maximal tumor sections were subjected to immunohistochemistry. Four-micron–thick, formalin-fixed, paraffin-embedded tissue sections were deparaffinized and rehydrated, and boiled in citrated buffer (0.01 M/L, pH 6.0) to retrieve the masked epitope. Then, the sections were incubated with 3% hydrogen peroxide, followed by 5% goat serum to block endogenous peroxidase activities and nonimmunospecific protein binding. The sections were incubated with the primary antibody against either FXYD3 (ATLAS Antibodies) or Ki-67 (MIB1, DAKOcytomation, Dako, Ely, UK). Immunoreactivity was visualized with the Envision detection system (Dako), and the nuclei were counterstained with hematoxylin. The specificity of the anti-FXYD3 antibody was confirmed by an experiment of immunocytochemistry with sections cut out from paraffin-embedded H1299 cells transduced with the FXYD3-EGFP construct (see Supplemental Figure S2 at http://ajp.amjpathol.org.). Also, the specificity of an immunoreactive signal of FXYD3 in tissue sections was further confirmed by immunohistochemistry using nonimmunized rabbit serum instead of the primary specific antibody against FXYD3 (see Supplemental Figure S2 at http://ajp.amjpathol.org.). The expression of FXYD3 in lung cancer cells was categorized as, negative (0), weak (1), or strong (2). Weak expression was defined as a level similar to that in bronchial or alveolar epithelial cells. Strong expression was defined as a signal of unequivocally greater signal intensity than that in bronchial epithelial cells. The expression score was determined by averaging the degree of staining corresponding to the areal proportion. A labeling index of MIB1 was calculated as the proportion of positive cells by counting 500 to 1000 cancer cells. Those cases with a labeling index of less than 0.15 and of 0.15 or more were classified as lower and higher expressers, respectively.
Statistical Analysis
Differences in FXYD3 expression scores among clinicopathologic subgroups were analyzed with Student’s t-test. Correlations of FXYD3 expression levels with clinicopathologic parameters, KRAS gene mutations, and MIB index levels were analyzed with a χ2 test. P values of less than 0.05 were considered significant.
Cell Growth Assays
Cells (2.5 × 105) were seeded onto 3.5-cm culture dishes (Iwaki, Tokyo, Japan) and grown for 10 days. The cells were counted every 2 days without a change of medium until the finish of the experiments.
Colony Formation Assays
Cells (1.0 × 104) were seeded onto 10 cm culture dishes (Iwaki), and grown for 12 days. The cells were fixed with methanol and stained with Giemsa, and colonies more than 0.5 mm in diameter were counted.
Soft Agar Colony Formation Assays
Cells (1.25 × 104) were cultured and grown in 1 ml of Dalbecco’s Modified Eagle Medium-based 0.3% agar (agar noble; Becton Dickinson, Sparks, MD) containing 10% fetal bovine serum in 3.5-cm culture dishes (Iwaki) for 3 weeks. The agar was fixed with a buffered 4% paraformaldehyde solution, and colonies more than 0.5 mm in diameter were counted.
Cell Migration Assays
Trans-well chambers (24 wells, 8 μm pore size, Becton Dickinson, Franklin Lakes, NJ) were used for the assay. The lower chambers were filled with 0.7 ml of Dalbecco’s Modified Eagle Medium containing 10% fetal bovine serum, and the cells (5.0 × 104/0.2 ml of Dalbecco’s Modified Eagle Medium without fetal bovine serum) were seeded onto the filters of upper chambers. After 24 hours of incubation, nonmigrated cells on the upper side of the filter membranes were wiped away. The cells migrating through were fixed with methanol, and stained with Giemsa (Merck, Darmstadt, Germany). The cells in three fields of 1.0 mm2 on filter membranes were counted.
Results
FYXD3 mRNA Expression in Lung Cancer Cell Lines
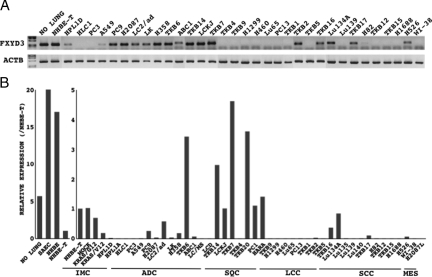
The semiquantitative reverse transcription (RT)-PCR analysis demonstrated that FXYD3 mRNA levels were unequivocally lower in some lung cancer cell lines than in normal lung tissue and primary or immortalized airway epithelial cells (nontumoral counterparts), as the PCR-amplified product could not be detected in HCL1, PC3, TKB5, TKB4, Lu130, Lu139, Lu65, H82, TKB12, TKB15, PC13, H1688, H1299, and H460 cells (Figure 1A). The quantitative RT-PCR analysis confirmed the result (Figure 1B). Interestingly, mutated KRAS (KRAS/V12)-transduced NHBE-T cells showed lower FXYD3 mRNA levels than the empty vector (mock)- and wild-type KRAS (KRAS/G12)-transduced NHEB-T cells (Figure 1B). Consistently, in lung cancer cell lines bearing KRAS mutations (A549 and H358), the levels were also lower than in the nontumoral counterparts (Figure 1B).
Figure 1.
Expression of FXYD3 mRNA in cell lines. Levels of FXYD3 mRNA were examined by RT-PCR. PCR products of FXYD3 and β-actin (ACTB) were resolved by electrophoresis in 1% agar and stained with ethidium bromide (A). Copy numbers of DUSP6 and β-actin (ACTB) mRNA were measured by quantitative RT-PCR. The mean and SD (error bars) of the mRNA levels of FXYD3 relative to that of ACTB in immortalized human airway cells and lung cancer cells from triplicate experiments are presented (B). NO LUNG, nontumoral lung tissue (cDNA was reverse transcribed from a mixture of total RNA extracted from 10 different nontumoral lung tissues); IMC, immortalized airway cells; MOCK, empty vector-transduced NHBE-T; G12, KRAS/G12-transduced NHBE-T; V12, KRAS/ V12-transduced NHBE-T; ADC, adenocarcinoma; SQC, squamous cell carcinoma; LCC, large cell carcinoma; SCC, small cell carcinoma; MES, mesenchymal cells.
Methylation in the FXYD3 Promoter
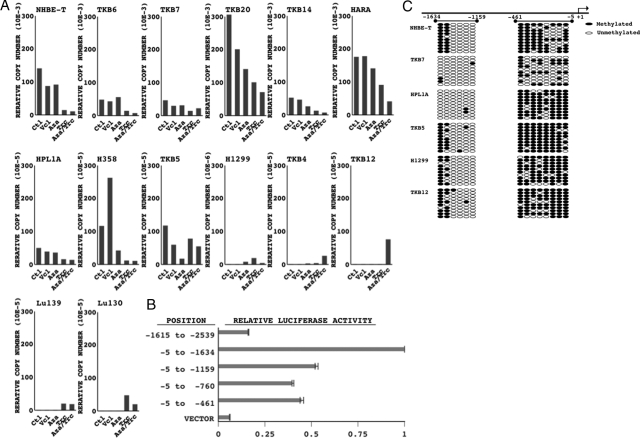
Treatment with an inhibitor for a DNA methyl-transferase (AZA)23 in combination with an inhibitor for histone deacetylase (trichostatin A, TSA)23 restored FXYD3 expression in H1299 and TKB12, but reduced it in the other cell lines examined (NHEB-T, TKB7, TKB6, TKB14, H358, HPL1A, and TKB5) (Figure 2A). The results suggested that epigenetic modifications could affect FXYD3 expression. To verify the possible involvement of DNA hypermethylation in the down-regulation, the promoter of FXYD3 was determined. The results of luciferase reporter assays suggested that the promoter lies between the position −461 and −5 or −1159, and −1634 of the FXYD3 gene locus, as luciferase reporter activity was elevated in these regions (Figure 2B). Thus, the methylation status of these loci was analyzed. The region from −461 to −5, especially −175 to −5, was highly methylated in the low expressers (HPL1A, TKB5, H1299, TKB12), although the other regions were randomly methylated, with no correlation between FXYD3 expression and methylation status (Figure 2B). We therefore focused on the region from −175 to −5 and further analyzed methylation status in additional cell lines. However, an association between hypermethylation of this region and the expression failed to be supported, as the site was methylated in high expresser (TKB20) and only faintly methylated in low expressers (Lu139) (see Supplemental Figure S3 at http://ajp.amjpathol.org.). Also, no association between the AZA/TSA-induced recovery status and methylation status of the promoter was found (Figure 2A and C, see Supplemental Figure S3 at http://ajp.amjpathol.org.).
Figure 2.
A: Methylation analyses of the FXYD3 promoter. The expression relative to the untreated controls (Ctl) is shown (note the difference in scale of the y axis). Luciferase reporter constructs, empty vector, −2539 to −1615, −1615 to −5, −1159 to −5, −760 to −5, and −461 to −5 (position of the first base of the transcription start was taken as + 1), were introduced into NHEB-T. Levels of luciferase activity were measured and normalized as described in the Materials and Methods. B: The means and standard deviations (error bars) of the ratio of the activities to those with empty vector (VECTOR) from duplicated experiments are presented C: The regions from −1615 to −1159 and −461 to −5 (or −175 to −5) in the FXYD3 gene locus were PCR-amplified using sodium bisulfate-modified genomic DNA as a template. Ten subclones of the PCR products were analyzed for methylation status by DNA sequencing. Open and filled circles indicate un-methylated and methylated cytosine at indicated CpG sites, respectively. Cells treated with vehicle (Vcl), 5-azacytidine (AZA), trichostatin A (TSA), or a combination of AZA and TSA, were examined for FXYD3 mRNA expression by quantitative RT-PCR. FXYD3 expression was normalized to β-actin expression.
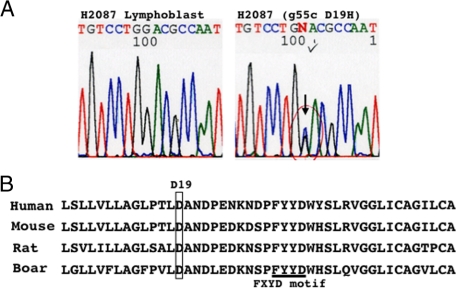
Mutation in FXYD3 Coding Regions
Among cell lines (44 lines, described in the cell culture section of the Materials and Methods) and primary lung cancers (72 tumors, 45 ADC, 19 SQC, 5 LCC, and 3 SCC) examined here, a missense mutation, g55c (D19H) at exon 5, was found in the H2087 cells that retained FXYD3 expression (Figure 3A). The absence of a base substitution at this position in H2087 lymphoblastic cells confirmed that the mutation was somatic, and not a rare polymorphism (Figure 3A). Asparate at the 19th amino acid (D19), part of the first transmembrane helix of FXYD3, is evolutionally conserved (Figure 3B),9,24,25 and likely to be essential to its function.
Figure 3.
A: Mutational analyses of coding regions of FXYD3. The results of direct sequencing analyses of H2087 lung cancer cells and H2087 lymphoblastic cells are shown. The position of a heterozygous mutation, g55c (D19H), is indicated with an arrow (also checked and circulated). B: Parts of protein sequences of FXYD3 in different species are shown. Asparate at the 19th amino acid (D19), which is evolutionally conserved, is enclosed. The FXYD3 motif is underlined.
Effect of FXYD3 on Cell Growth and Migration
The forced expression of EGFP-fused FXYD3 had no effect on colony-forming activity in culture dishes or in soft agar, on growth activity under ordinal and low serum culture conditions, or on the migration (see Supplemental Figure S4 at http://ajp.amjpathol.org.) of H1299 cells that expressed endogenous FXYD3 at a very low level (Figure 1). Similar results were observed in H1299 cells transduced with native FXYD3, excluding a critical influence of the EGFP tag on the biological function of FXYD3 and also confirming the former results (data not shown).
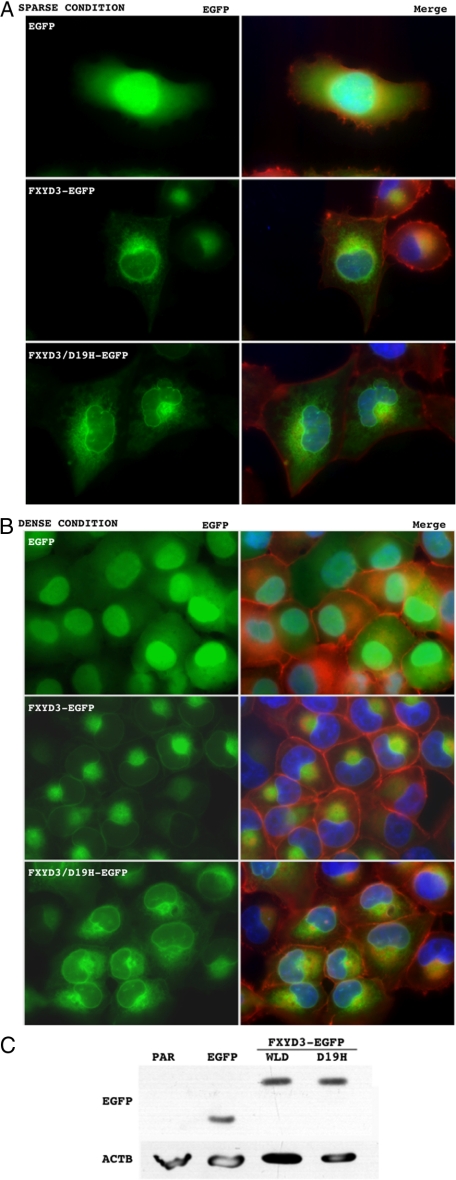
Effect of FXYD3 on Modulation of the Cytoskeleton
FXYD3 was concentrated in the nuclear envelope and radiated out from the perinuclear portion into the cytoplasm with a fine reticular pattern under sparse culture conditions (Figure 4A, middle panel), whereas EGFP was distributed diffusely throughout in the cell (Figure 4A, upper panel). FXYD3/D19H exhibited a similar distribution to wild-type FXYD3 (Figure 4A, middle and lower panels). These observations were consistent with those described by Arimochi et al.11 Noticeably, the FXYD3/D19H-transduced cells had nuclei with distorted outlines compared with the EGFP- and the wild-type FXYD3-transduced cells (Figure 4A). On the other hand, under dense culture conditions, the wild-type FXYD3 was distributed along with nuclear envelope and cell membranes, and also concentrated in perinuclear portions (Figure 4B, middle panel). In contrast, FXYD3/D19H was still distributed diffusely in the cytoplasm and was not localized to the cell membranes (Figure 4B, lower panel). The wild-type FXYD3-transduced cells had sharp and well-demarcated cortical actin where FXYD3 was co-localized, but the EGFP- and the FXYD3/D19H-transduced cells did not exhibit such changes (Figure 4B). The exogenous expression was confirmed by Western blotting (Figure 4C).
Figure 4.
Subcellular distribution of wild-type FXYD3 and the mutant (FXYD3/D19H), and their effects on morphology in H1299 cells. EGFP (upper panels in both A and B), FXYD3-EGFP (middle panels in both A and B), or FXYD3/D19H-EGFP (lower panels in both A and B) was retrovirally transduced into H1299 cells. After the selection treatment, the surviving cells were harvested and counted, and 1.25 × 104 (sparse condition, A) or 5.0 × 105 (dense condition, B) were re-seeded onto chamber slides (LAB-TEK 2-well chamber, Electron Microscopy Science) and cultured for 24 hours. The cells were fixed with a buffered 4% paraformaldehyde solution and stained with phalloidin-rhodamine (0.1 μg/ml, Sigma) and 4′,6-diamino-2-phenylindole (1.0 μg/ml, Sigma). Green, red, and blue fluorescence indicate the distribution of FXYD3-EGFP (or EGFP alone), filamentous actin, and nuclei, respectively. Left panels in both A and B show representative images of the green fluorescence alone (EGFP). Right panels in both A and B show images layered with green, red and blue fluorescence (MERGE). Expression of the transduced EGFP, the wild-type FXYD3-EGFP (WLD) and the mutant FXYD3/D19H-EGFP (D19H) was confirmed by Western blotting using antibody against EGFP (C, upper panel). Equal loading of protein samples was confirmed by Western blotting using antibody against β-actin (ACTB) (C, lower panel). Par, parental H1299 cells.
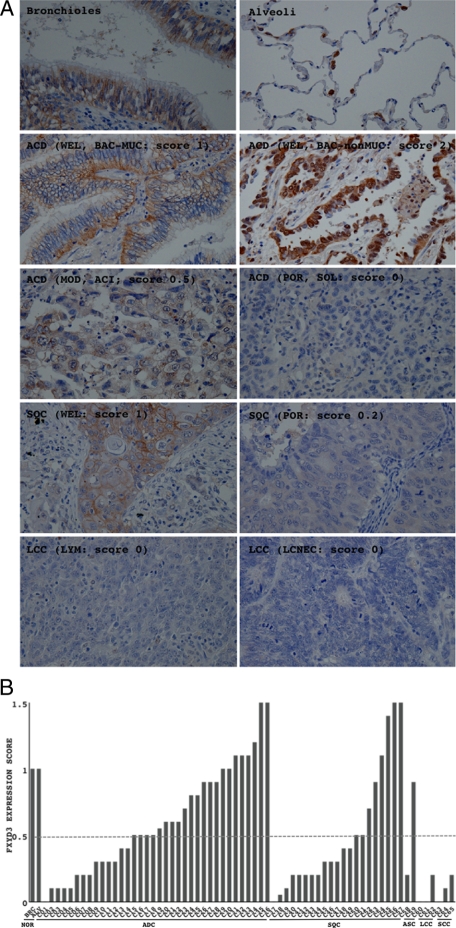
FYXD3 Protein Expression in Primary Lung Cancers
For 65 cases of primary lung cancers, FXYD3 expression was examined by immunohistochemistry. Representative results are shown in Figure 5A. Immunoreactivity for FXYD3 was observed in epithelial cells, but not in mesenchymal cells (Figure 5A). FXYD3 was extensively expressed in cellular membranes mainly on the basolateral sides of bronchial epithelial cells (Figure 5A, bronchioles), and in cytoplasm where it was concentrated at the perinuclear site of alveolar epithelial cells (Figure 5A, alveoli), but was not expressed in the interstitial cells (fibroblasts, lymphocytes, vascular endothelial cells, and smooth muscle cells). In lung cancers, particularly in poorly differentiated cancers, its expression was faint or seen in the cytoplasm of only a small proportion of neoplastic cells (Figure 5A, third to fifth panels). The immunoreactivity was semiquantified according to the scoring system described in the Materials and Methods. Scores of less than 0.5 were taken to indicate down-regulation. FXYD3 expression was found to be down-regulated in 38.9% (14/36) of ADC, 61.9% (13/21) of SQC, 50% (1/2) of ASC, 100% (3/3) of LCC, and 100% (3/3) of SCC (Figure 5, A and B, Table 1). Statistical analyses demonstrated that its expression was significantly weaker in LCC and SCC than in ADC (Table 1), and decreased significantly as the histological grade of SQC progressed from well to poorly differentiated (Table 1). No significant association between the expression score and growth activities evaluated by the MIB1 index was found, which supported the result in the in vitro experiment of no effect of exogenous expression of FXYD3 on the growth activity of H1299 cells (see Supplemental Figure S4 at http://ajp.amjpathol.org.). On the other hand, no significant association between the expression score and KRAS mutations was found (Table 1), suggesting there could be potential mechanisms other than the oncogenic activation of KRAS.
Figure 5.
Expression of FXYD3 in normal lung and lung cancers. A: Expression of FXYD3 in bronchial epithelia (bronchiole), alveolar epithelia (alveoli), and primary lung cancers was examined by immunohistochemistry. Levels of FXYD3 expression were evaluated according to the scoring system described in the Materials and Methods. Representative photographs are shown. It is noticeable that the expression of FXYD3 was restricted to the epithelial cells (A, upper two panels). Magnification is × 400 for each. B: Expression scores of all cases examined (65 cases) are presented in a graph. Scores lower than 0.5 (bordered with a dashed line) were taken to indicate down-regulation. ADC, adenocarcinoma; SQC, squamous cell carcinoma; ASC, adenosquamous carcinoma; SCC, small cell carcinoma; LCC, large cell carcinoma; WEL, well differentiated; MOD, moderately differentiated; BAC, bronchioloalveolar carcinoma; ACI, acinar carcinoma; SOL, solid carcinoma with mucin production; LYM, lymphoepithelioma-like large cell carcinoma; LCNEC, large cell neuroendocrine carcinoma.
Table 1.
Correlation between FXYD3 Expression and Clinicopathologic Status in Primary Lung Cancers
| FXYD3 Score < 0.5 | FXYD3 Score ≥ 0.5 | Score (Mean ± SD) | |
|---|---|---|---|
| MIB1 labeling index (65) | |||
| Low level (<15%) (20) | 7 (35.0%) | 13 (65.0%) | 0.655 ± 0.371 |
| High level (≥15%) (45) | 27 (60.0%) | 18 (40.0%) | 0.471 ± 0.448 |
| Chi square test (P = 0.0625) | Student’s t-test (P = 0.1131) | ||
| KRAS Codon12 (65) | |||
| Mutant-type (8) | 2 (25.0%) | 6 (75.0%) | 0.750 ± 0.378 |
| Wild-type (57) | 32 (56.1%) | 25 (43.9%) | 0.496 ± 0.432 |
| Chi square test (P = 0.0987) | Student’s t-test (P = 0.1202) | ||
| Histology (65) | |||
| ADC (36) | 14 (38.9%) | 22 (61.1%) | 0.604 ± 0.401*† |
| Grade | |||
| WEL (20) | 6 (30.0%) | 13 (70.0%) | 0.642 ± 0.364 |
| MOD (7) | 3 (42.9%) | 4 (57.1%) | 0.657 ± 0.469 |
| POR (9) | 5 (55.6%) | 4 (44.4%) | 0.417 ± 0.391 |
| Chi square test (P = 0.4741) | |||
| Subtype | |||
| BAC (15) | 4 (26.7%) | 11 (73.3%) | 0.693 ± 0.561 |
| ACI (5) | 4 (80.0%) | 1 (20.0%) | 0.500 ± 0.361 |
| PAP (7) | 2 (28.6%) | 5 (71.4%) | 0.686 ± 0.398 |
| SOL (9) | 4 (55.6%) | 5 (44.4%) | 0.450 ± 0.416 |
| Chi square test (P = 0.1029) | |||
| SQC (21) | 13 (61.9%) | 8 (38.1%) | 0.521 ± 0.478 |
| Grade | |||
| WEL (3) | 1 (33.3%) | 2 (66.7%) | 0.600 ± 0.265‡ |
| MOD (15) | 9 (60.0%) | 6 (40.0%) | 0.593 ± 0.516 |
| POR (3) | 3 (100.0%) | 0 (0.0%) | 0.083 ± 0.104‡ |
| Chi square test (P = 0.2337) | |||
| ASC (2) | 1 (50.0%) | 1 (50.0%) | 0.550 ± 0.495 |
| LCC (3) | 3 (100.0%) | 0 (0.0%) | 0.067 ± 0.115*† |
| SCC (3) | 3 (100.0%) | 0 (0.0%) | 0.100 ± 0.100 |
| Chi square test (P = 0.0650) | |||
| T factor (65) | |||
| T1 (24) | 12 (50.0%) | 12 (50.0%) | 0.517 ± 0.449 |
| T2 (27) | 12 (32.4%) | 15 (67.6%) | 0.565 ± 0.414 |
| T3 (5) | 5 (100.0%) | 0 (0.0%) | 0.270 ± 0.130 |
| T4 (9) | 5 (55.6%) | 4 (44.4%) | 0.589 ± 0.291 |
| Chi square test (P = 0.1500) | |||
| N factor (65) | |||
| N0 (40) | 22 (55.0%) | 18 (45.0%) | 0.542 ± 0.427 |
| N1 (8) | 2 (25.0%) | 6 (75.0%) | 0.688 ± 0.557 |
| N2 (14) | 7 (55.6%) | 7 (44.4%) | 0.479 ± 0.379 |
| N3 (3) | 3 (100.0%) | 0 (0.0%) | 0.133 ± 0.153 |
| Chi square test (P = 0.1529) | |||
| Stage (65) | |||
| 1A (20) | 9 (45.0%) | 11 (55.0%) | 0.590 ± 0.448 |
| 1B (14) | 8 (57.1%) | 6 (42.9%) | 0.518 ± 0.429 |
| 2A (2) | 1 (50.0%) | 1 (50.0%) | 0.250 ± 0.354 |
| 2B (7) | 4 (57.1%) | 3 (42.9%) | 0.536 ± 0.437 |
| 3A (10) | 5 (50.0%) | 5 (50.0%) | 0.460 ± 0.389 |
| 3B (10) | 6 (60.0%) | 4 (40.0%) | 0.530 ± 0.542 |
| 4 (2) | 1 (50.0%) | 1 (50.0%) | 0.550 ± 0.354 |
| Chi square test (P = 0.9894) | |||
ADC, adenocarcinoma; SQC, squamous cell carcinoma; ASC, adenosquamous carcinoma; LCC, large cell carcinoma; SCC, small cell carcinoma; WEL, well differentiated; MOD, moderately differentiated; POR, poorly differentiated carcinomas; BAC, bronchioloalveolar carcinoma; ACI, aciner adenocarcinoma; PAP, papillary adenocarcinoma; SOL, solid adenocarcinoma with mucin production.
P = 0.0281;
P = 0.0386;
P = 0.0346, student’s t-test.
Discussion
FXYD3 mRNA levels were lower in the majority of lung cancer cell lines than in either normal lung tissue or an noncancerous airway epithelial cell line (SAEC, NHBE, and NHEB-T) (Figure 1), and the protein expression was also down-regulated in a considerable proportion of primary lung cancers (Figure 5, Table 1). The protein’s expression decreased with the histological grade of SQC (Table 1) and was significantly reduced in highly malignant subtypes such as LCC and SCC (Table 1). These results suggested that FXYD3 could be a potential tumor suppressor and its down-regulation could play some role particularly in the progression of lung cancers. Consistently, a previous study also demonstrated that FXYD3 expression was down-regulated in lung cancers.5,14,15 On the other hand, among our results, a discrepancy was noticed. That is, FXYD3 protein levels were found to be lower in SQC than in ADC among primary lung cancers, although the mRNA levels were lower in ADC than in SQC among the cell lines. To verify the possibility of a lag between the mRNA and protein levels, the lung cancer cell lines were examined further. The protein levels were found to be not necessarily associated with the mRNA levels. Protein levels in SQC cell lines, TKB7 and TKB20, were lower than those in NHBE-T and PC9 (ADC cell line), despite their high level mRNA expressions (see Supplemental Figure S5 at http://ajp.amjpathol.org.). There might be a potential mechanism involved in the down-regulation of FXYD3 expression at the posttranscriptional level, especially in SQC cells.
In contrast to our results, FXYD3 expression was reported to be up-regulated in breast, prostate, and pancreatic cancers.5,14,15 Also, our finding that the forced expression of oncogenic KRAS reduced the expression of FXYD3 in NHBE-T cells was in contrast to the initial observation that FXYD3 was highly expressed in oncogenic ras-initiated murine breast tumors.5 The regulation of FXYD3 expression and its roles in carcinogenesis are assumed to differ among the type of cancer, species, or subtype of the RAS oncogene, and the disordered expression itself, whether strong or weak, might affect cellular homeostasis resulting in the transformation.
The mutational analysis demonstrated a somatic mutation (g55t, D19H) in a lung cancer cell line (H2087) (Figure 3). This is a novel finding, although the mutation is very rare. Exogenous expression of the wild-type FXYD3, but not the mutant (FXYD3/D19H), restored the well-demarcated distribution of cortical actin in H1299 cells (Figure 4). FXYD3/D19H actually distorted the outline of nuclear envelope in H1299 cells (Figure 4). These results implied that loss of FXYD3 expression or loss of function resulting from a gene mutation could attenuate the integrity of the nuclear envelope and the cytoskeleton. In other words, loss of FXYD3 function might be one of the causes of morphological atypism, a common feature of cancer cells. A recent study on intestinal epithelia suggested that FXYD3 could regulate indirectly the development of tight junctions through its interaction with Na,K-ATPase, to promote cellular polarization and differentiation,16,18,26,27 which seems consistent with our observations. Considering that FXYD3 was expressed exclusively in epithelial cells (see the result for WI-38 lung fibroblastic cells and H2087 lymphoblastic cells [H2087L] in Figure 1A, and its negative staining in mesenchymal cells in Figure 5A), the down-regulation of FXYD3 expression might be involved in the epithelial–mesenchymal transition, as is the down-regulation of the expression of E-cadherin, an epithelial cell–specific molecule. Epithelial–mesenchymal transition is linked with the vigorous migration and invasiveness of cancer cells.26 The restoration of FXYD3 expression, however, did not affect the migration of H1299 cells (see Supplemental Figure S5 at http://ajp.amjpathol.org.). The instability of cortical actin alone, possibly caused by FXYD3’s inactivation, alone might be not enough to provide cells with actual migrating activity. Additional events such as the complete disruption of intercellular junctions28 and active enhancement of cytoskeletal dynamism29 were considered to be required.
The present study also determined the promoter region of FXYD3 and examined the methylation status in the region to elucidate the mechanism of its down-regulation (Figure 2). The result suggested that hypermethylation of the promoter was not a primary cause of the down-regulation of FXYD3 expression, since no significant association between the expression level and methylation status was found. An in silico analysis using computer software (Genomatix, Munich, Germany) estimated that the region from −261 to −5 in the FXYD3 gene locus contained consensus-binding sites for several transcription factors including PAX3, SOX17, and INSM1. The expression of PAX3 and SOX17 was reportedly severely down-regulated in some types of human cancers including small cell lung carcinomas.30,31 INSM1, a transcriptional repressor, was reportedly expressed at high levels in neuroendocrine carcinomas, such as small cell lung carcinomas.32 Thus, we have speculated the involvement of such factors in the down-regulation of FXYD3 expression in the lung cancer cells.
In summary, FXYD3 expression was down-regulated in a considerable proportion of lung cancer cell lines and/or primary lung cancers through a mechanism other than hypermethylation of the promoter. One cell line had a somatic mutation resulting in a loss of function of FXYD3, and restoration of FXYD3 expression resulted in a sharp and well-demarcated distribution of cortical actin in the lung cancer cells. The results suggested FXYD3 to be a potential tumor suppressor and its inactivation to be a cause of morphological atypism. To our knowledge, no study has investigated in such detail the involvement of FXYD3 in carcinogenesis of the lung. We hope our efforts will advance understanding of the molecular characteristics of lung cancers.
Supplementary Material
Acknowledgments
We especially thank Masaichi Ikeda (Department of Pathology, Yokohama City University Graduate School of Medicine, Yokohama, Japan), Sigeko Iwanade, and Emi Honda (Division of Pathology, Kanagawa Prefectural Cardiovascular and Respiratory Disease Center Hospital, Yokohama, Japan) for excellent technical assistance, and Reika Iwakawa (Tokyo University of Science, Tokyo, Japan) for encouragement.
Footnotes
Address reprint requests to Hitoshi Kitamura, Department of Pathology, Yokohama City University Graduate School of Medicine, 3-9 Fukuura, Kanazawa-Ku, 236-0004 Yokohama, Japan, E-mail: pathola@med.yokohama-cu.ac.jp.
Supported by the Japanese Ministry of Education, Culture, Sports, and Science (Tokyo Japan), and by grants from the Yokohama Medical Facility (Yokohama, Japan) and the Smoking Research Foundation (Tokyo, Japan).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Okudela K, Yazawa T, Woo T, Sakaeda M, Ishii J, Mitsui H, Shimoyamada H, Sato H, Tajiri M, Ogawa N, Masuda M, Takahashi T, Sugimura H, Kitamura H. Down-regulation of DUSP6 expression in lung cancers: its mechanism and potential role in carcinogenesis. Am J Pathol. 2009;175:867-881. doi: 10.2353/ajpath.2009.080489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Yazawa T, Suzuki T, Shimoyamada H, Okudela K, Ikeda M, Hamada K, Yamada-Okabe H, Yao M, Kubota Y, Takahashi T, Kamma H, Kitamura H. Growth regulation via insulin-like growth factor binding protein-4 and -2 in association with mutant K-ras in lung epithelia. Am J Pathol. 2006;169:1550–1566. doi: 10.2353/ajpath.2006.051068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC, editors. Lyon: IARC Press,; World Health Organization classification of tumors. Pathology and genetics of the lung, pleura, thymus and heart. 2004 [Google Scholar]
- Kitamura H, Kameda Y, Ito T, Hayashi H. Atypical adenomatous hyperplasia of the lung. Implications for the pathogenesis of peripheral lung adenocarcinoma. Am J Clin Pathol. 2000;113:593–594. doi: 10.1093/ajcp/111.5.610. [DOI] [PubMed] [Google Scholar]
- Morrison BW, Leder P. Neu and ras initiate murine mammary tumors that share genetic markers generally absent in c-myc and int-2-initiated tumors. Oncogene. 1994;9:3417–3426. [PubMed] [Google Scholar]
- Crambert G, Geering K. FXYD proteins: new tissue-specific regulators of the ubiquitous Na, K-ATPase. Sci STKE. 2003;166:RE1. doi: 10.1126/stke.2003.166.re1. [DOI] [PubMed] [Google Scholar]
- Crambert G, Li C, Claeys D, Geering K. FXYD3 (Mat-8), a new regulator of Na, K-ATPase. Mol Biol Cell. 2005;16:2363–2371. doi: 10.1091/mbc.E04-10-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geering K. Function of FXYD proteins, regulators of Na,K-ATPase. J Bioenerg Biomembr. 2005;37:387–392. doi: 10.1007/s10863-005-9476-x. [DOI] [PubMed] [Google Scholar]
- Geering K. FXYD proteins: new regulators of Na-K-ATPase. Am J Physiol Renal Physiol. 2006;290:241–250. doi: 10.1152/ajprenal.00126.2005. [DOI] [PubMed] [Google Scholar]
- Franzin CM, Gong XM, Teriete P, Marassi FM. Structures of the FXYD regulatory proteins in lipid micelles and membranes. J Bioenerg Biomembr. 2007;39:379–383. doi: 10.1007/s10863-007-9105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimochi J, Kobayashi A, Maeda M. Stable expression and visualization of Mat-8 (FXYD-3) tagged with a fluorescent protein in Chinese hamster ovary (CHO)-K1 cells. Biotechnol Lett. 2005;27:1017–1024. doi: 10.1007/s10529-005-7870-4. [DOI] [PubMed] [Google Scholar]
- Arimochi J, Ohashi-Kobayashi A, Maeda M. Interaction of Mat-8 (FXYD-3) with Na+/K+-ATPase in colorectal cancer cells. Biol Pharm Bull. 2007;30:648–654. doi: 10.1248/bpb.30.648. [DOI] [PubMed] [Google Scholar]
- Morrison BW, Moorman JR, Kowdley GC, Kobayashi YM, Jones LR, Leder P. Mat-8, a novel phospholemman-like protein expressed in human breast tumors, induces a chloride conductance in Xenopus oocytes. J Biol Chem. 1995;270:2176–2182. doi: 10.1074/jbc.270.5.2176. [DOI] [PubMed] [Google Scholar]
- Kayed H, Kleeff J, Kolb A, Ketterer K, Keleg S, Felix K, Giese T, Penzel R, Zentgraf H, Büchler MW, Korc M, Friess H. FXYD3 is overexpressed in pancreatic ductal adenocarcinoma and influences pancreatic cancer cell growth. Int J Cancer. 2006;118:43–54. doi: 10.1002/ijc.21257. [DOI] [PubMed] [Google Scholar]
- Grzmil M, Voigt S, Thelen P, Hemmerlein B, Helmke K, Burfeind P. Up-regulated expression of the MAT-8 gene in prostate cancer and its siRNA-mediated inhibition of expression induces a decrease in proliferation of human prostate carcinoma cells. Int J Oncol. 2004;24:97–105. [PubMed] [Google Scholar]
- Anderle P, Rakhmanova V, Woodford K, Zerangue N, Sadée W. Messenger RNA expression of transporter and ion channel genes in undifferentiated and differentiated Caco-2 cells compared to human intestines. Pharm Res. 2003;20:3–15. doi: 10.1023/a:1022282221530. [DOI] [PubMed] [Google Scholar]
- Maxwell PJ, Longley DB, Latif T, Boyer J, Allen W, Lynch M, McDermott U, Harkin DP, Allegra CJ, Johnston PG. Identification of 5-fluorouracil-inducible target genes using cDNA microarray profiling. Cancer Res. 2003;63:4602–4606. [PubMed] [Google Scholar]
- Bibert S, Roy S, Schaer D, Felley-Bosco E, Geering K. Structural and functional properties of two human FXYD3 (Mat-8) isoforms. J Biol Chem. 2006;281:39142–39151. doi: 10.1074/jbc.M605221200. [DOI] [PubMed] [Google Scholar]
- Cozens AL, Yezzi MJ, Kunzelmann K, Ohrui T, Chin L, Eng K, Finkbeiner WE, Widdicombe JH, Gruenert DC. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am J Respir Cell Mol Biol. 1994;10:38–47. doi: 10.1165/ajrcmb.10.1.7507342. [DOI] [PubMed] [Google Scholar]
- Masuda A, Kondo M, Saito T, Yatabe Y, Kobayashi T, Okamoto M, Suyama M, Takahashi T, Takahashi T. Establishment of human peripheral lung epithelial cell lines (HPL1) retaining differentiated characteristics and responsiveness to epidermal growth factor, hepatocyte growth factor, and transforming growth factor beta1. Cancer Res. 1997;57:4898–4904. [PubMed] [Google Scholar]
- Ward R, Hawkins N, O'Grady R, Sheehan C, O'Connor T, Impey H, Roberts N, Fuery C, Todd A. Restriction endonuclease-mediated selective polymerase chain reaction: a novel assay for the detection of K-ras mutations in clinical samples. Am J Pathol. 1998;153:373–379. doi: 10.1016/S0002-9440(10)65581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okudela K, Hayashi H, Ito T, Yazawa T, Suzuki T, Nakane Y, Sato H, Ishi H, KeQin X, Masuda A, Takahashi T, Kitamura H. K-ras gene mutation enhances motility of immortalized airway cells and lung adenocarcinoma cells via Akt activation: possible contribution to non-invasive expansion of lung adenocarcinoma. Am J Pathol. 2004;164:91–100. doi: 10.1016/S0002-9440(10)63100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16:R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- Franzin CM, Gong XM, Thai K, Yu J, Marassi FM. NMR of membrane proteins in micelles and bilayers: the FXYD family proteins. Methods. 2007;41:398–408. doi: 10.1016/j.ymeth.2006.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzin CM, Yu J, Thai K, Choi J, Marassi FM. Correlation of gene and protein structures in the FXYD family proteins. J Mol Biol. 2005;354:743–750. doi: 10.1016/j.jmb.2005.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekaran AK, Rajasekaran SA. Role of Na-K-ATPase in the assembly of tight junctions. Am J Physiol Renal Physiol. 2003;285:F388–F396. doi: 10.1152/ajprenal.00439.2002. [DOI] [PubMed] [Google Scholar]
- Contreras RG, Shoshani L, Flores-Maldonado C, Lázaro A, Cereijido M. Relationship between Na(+), K(+)-ATPase and cell attachment. J Cell Sci. 1999;112:4223–4232. doi: 10.1242/jcs.112.23.4223. [DOI] [PubMed] [Google Scholar]
- Guarino M. Epithelial-mesenchymal transition and tumour invasion. Int J Biochem Cell Biol. 2007;39:2153–2160. doi: 10.1016/j.biocel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Vignjevic D, Montagnac G. Reorganisation of the dendritic actin network during cancer cell migration and invasion. Semin Cancer Biol. 2008;18:12–22. doi: 10.1016/j.semcancer.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Fukasawa M, Kimura M, Morita S, Matsubara K, Yamanaka S, Endo C, Sakurada A, Sato M, Kondo T, Horii A, Sasaki H, Hatada I. Microarray analysis of promoter methylation in lung cancers. J Hum Genet. 2006;51:368–374. doi: 10.1007/s10038-005-0355-4. [DOI] [PubMed] [Google Scholar]
- Zhang W, Glöckner SC, Guo M, Machida EO, Wang DH, Easwaran H, Van Neste L, Herman JG, Schuebel KE, Watkins DN, Ahuja N, Baylin SB. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res. 2008;68:2764–2772. doi: 10.1158/0008-5472.CAN-07-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan MS, Russell EK, Lu J, Johnson BE, Notkins AL. IA-1, a new marker for neuroendocrine differentiation in human lung cancer cell lines. Cancer Res. 1993;53:4169–4171. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.