Abstract
In less than a decade after discovery, RNA interference-mediated gene silencing is already being tested as potential therapy in clinical trials for a number of diseases. Lentiviral vectors provide a means to express short hairpin RNA (shRNA) to induce stable and long-term gene silencing in both dividing and non-dividing cells and thus, are being intensively investigated for this purpose. However, induction of long-term shRNA expression can also cause toxicities by inducing off target effects and interference with the endogenous micro RNA (miRNA) pathway that regulates cellular gene expression. Recently, several advances have been made in the shRNA vector design to mimic cellular miRNA processing and to express multiplex siRNAs in a tightly regulated and reversible manner to overcome toxicities. In this review we describe some of these advances, focusing on the progress made in the development of lentiviral shRNA delivery strategies to combat viral infections.
Keywords: RNAi, shRNA, miRNA, viral infection, lentivirus
1. Introduction
RNA interference (RNAi) was first recognized as a natural antiviral defense mechanism in plants wherein long double-stranded RNAs generated during viral replication are cleaved by the enzyme Dicer into short 21–23 nucleotide double-stranded RNA molecules called small interfering RNAs (siRNAs) that mediate sequence-specific gene silencing [1, 2]. The siRNA associates with a multiprotein complex called the RNA-induced silencing complex (RISC), during which the “passenger” sense strand is cleaved by the enzyme, Argonaute 2 (Ago2) [3]. The antisense “guide” strand contained in the activated RISC then guides the RISC to the corresponding mRNA because of sequence homology and the same Ago2 nuclease cuts the target mRNA, resulting in specific gene silencing. Although RNAi is a natural phenomenon in plants and worms, long dsRNA induces an interferon response in mammalian cells resulting in non-specific global suppression of protein synthesis and cell death [4]. However, introduction of double stranded small synthetic RNA resembling the Dicer-processed siRNA into mammalian cells induces sequence-specific gene silencing without evoking the interferon response [5]. Since this discovery, RNAi has been widely used as a quick reverse genetics approach for gene function analysis as well as for ablating specific genes for therapeutic purpose. What makes RNAi an attractive gene silencing approach is its exquisite sequence specificity and high potency [2]. This is probably due to the autocatalytic effect of RNAi whereby a single siRNA molecule gets reused for the cleavage of many target mRNA molecules [5, 6].
RNAi can be induced by the introduction of synthetic siRNA or by intracellular generation of siRNA from vector driven expression of the precursor small hairpin (sh) RNAs. In the latter method, an oligonucleotide containing the siRNA sequence followed by a ~9 nt loop and a reverse complement of the siRNA sequence is cloned in plasmid or viral vectors to endogenously express shRNA which is subsequently processed in the cytoplasm to siRNA. While the synthetic siRNA effects are short lived (a few days) probably because of siRNA dilution with cell division and also degradation, the shRNA effects are long lasting because they are continually produced within the cells. Although shRNAs can be produced in plasmid-based systems, because they are easy to deliver, non-replicating recombinant viral vectors are commonly used for shRNA expression. Adeno, adeno-associated and lentiviruses are generally used because they infect actively dividing as well as resting and differentiated cells such as the stem cells, macrophages and neurons. Lentiviruses may be particularly suited for long-term shRNA expression and gene silencing since the viral DNA gets incorporated in the host genome. Although viral vectors efficiently deliver shRNA leading to persistent siRNA expression [7], they may also have potential toxicities associated with unregulated expression of large amounts siRNA (see later) and thus, much emphasis has been laid in recent years to achieve temporally and spatially regulated shRNA expression.
Endogenously expressed small non-coding regulatory RNAs called microRNAs (miRNA) constitutes an important arm of RNAi. In fact, the existence of miRNA machinery is what makes the si/shRNAs function in the cells and thus, understanding of miRNA biogenesis would provide insights to designing better RNAi strategies for application. MiRNAs are genomically encoded and are transcribed as long primary transcripts (pri-miRNAs). The pri-miRNA is processed in the nucleus into ~65 nt hairpin shaped precursor miRNAs (pre-miRNAs) by a microprocessor complex consisting of the RNase III enzyme Drosha and its cofactor DGCR8 (Pasha in invertebrates) [8–13]. The pre-miRNA is exported out of the nucleus by exportin 5 [14, 15]. The pre-miRNAs are further processed in the cytoplasm by another RNase III enzyme, Dicer that cuts the pre-miRNA ~22 bp from the end set by Drosha cleavage to generate the double-stranded miRNA duplex [16–19]. For most miRNAs, only one strand (the guide strand) of the double-stranded miRNA duplex is loaded into RISC while the other (*) strand is destroyed rapidly [3, 20–22]. However, in some cases such as miR-142, miR-125, miR-126, miR-342, both strands (5p and 3p) are selected [23]. Schwarz et al proposed that the relative instability of the miRNA duplex termini determines which strand will be loaded into RISC. When both termini of the miRNA have the similar stability, both strands might be selected [20, 24].
Unlike siRNAs, which are designed to be homologous to the coding regions of mRNA and suppress gene expression by mRNA degradation, miRNAs predominantly regulate gene expression by translational repression, but can sometimes also mediate mRNA degradation [25, 26]. Also unlike siRNAs, the two strands of the miRNA duplex are imperfectly homologous and forms internal bulges and the guide strand is also imperfectly complementary to the 3′ UTR of the target mRNA, with only 2–7 nt at the 5′ end being perfectly complementary [27, 28]. The 5′ 2–7 nt (“seed”) region defines the target specificity of the miRNA [29–31]. Probably because of the small complementary sequence needed for target recognition, a single miRNA can regulate many target mRNAs containing the miRNA seed binding sites[32]. This also imposes restriction on using artificial si/shRNAs in that, although the antisense strand of si/shRNA is usually designed to be completely complimentary to the target gene, it can also act like miRNA and repress unintended genes that contain the target sequences recognized by the first 2–7 nt of the selected si/shRNA antisense strand. If the unintended sense strand of si/shRNA is also loaded into RISC, the off-target effect will presumably be doubled. Thus, precaution should be taken to design si/shRNA to avoid seed matches in the 3′UTR of cellular genes as well as to ensure proper strand selection by RISC by engineering the termini with distinct thermodynamic stability.
2. Generation of non-replicating transducing lentivirus
2.1. Lentiviral vectors and packaging constructs
As a part of their life cycle, retro- and lentiviruses integrate into the host genome and thus their genomic backbone provides a means for life-long expression of a transgene of interest. Because the native viruses can cause serious diseases, non-replicating viruses are used for transgene expression. The basic idea here is to generate a virus that after a single round of infection ensures stable integration of the transgene with only a minimal component of the viral genome, so that the transgene is continually expressed but infectious virus is not generated. This is generally achieved by introducing the different components of the viral genome in 3–4 separate plasmid vectors to generate the transducing virus (Fig. 1). Unlike retroviruses, which do not infect non-dividing cells, lentiviruses infect actively dividing as well as resting and differentiated cells and thus, lentiviral vectors are generally preferred for long-term expression of transgenes. They have proven effective in transducing non-dividing cells in a variety of organs in vivo including the brain, eye, liver and hematopoietic cells to induce long-term transgene expression [33]. Another advantage is that relatively large sequences of transgenes can be accommodated. Although several lentiviruses including the equine infectious anemia virus and feline as well as bovine immunodeficiency virus have also been used as gene therapy vehicles, HIV-based vectors are predominantly used because the basic biology of HIV replication has been extensively characterized over a number of years, leading to steady improvements in the vector design (reviewed in [34]). During a retroviral gene therapy clinical trial for x-linked immunodeficiency, 4 patients developed leukemia due to proviral integration near proto-oncogenes [35–37], highlighting the potential risks of using retroviral vectors. HIV-1 also preferentially integrates within the transcribed genes [38]. However, HIV infection itself has not been associated with insertional mutagenesis and several preclinical studies in animal models also indicate that HIV-based lentiviral vectors may not have a predilection for insertional oncogenesis [34]. In fact, a recent study comparing the integration sites of both retro- and lentiviruses in human hematopoietic cells showed that retro- but not lentiviral integration occurs at high frequency (20%) at genomic locations (hot spots) that are significantly enriched in protooncogenes and genes involved in the control of cell proliferation [39]. Also, a recent phase I clinical trial using a HIV-based vector encoding antisense RNA targeting the HIV envelope gene was found to be safe and with out the risks of mutagenesis or recombination-mediated activation of the virus [40]. However, it must be cautioned that lentiviral gene therapy is still in its infancy to draw definitive conclusions. Considering the potential risks of using a vector derived from a dangerous virus like HIV, biosafety to obviate all potential risks has been the core issue in their development. Thus, steady progress has been made in designing replication-deficient HIV-based lentiviral vectors to avoid accidental generation of replication competent virus arising from recombination between the different vector components or following HIV infection. These include deleting the cis elements from the packaging cassette, removing accessory genes, partitioning packaging genes into separate expression cassettes and minimizing homology between the different elements.
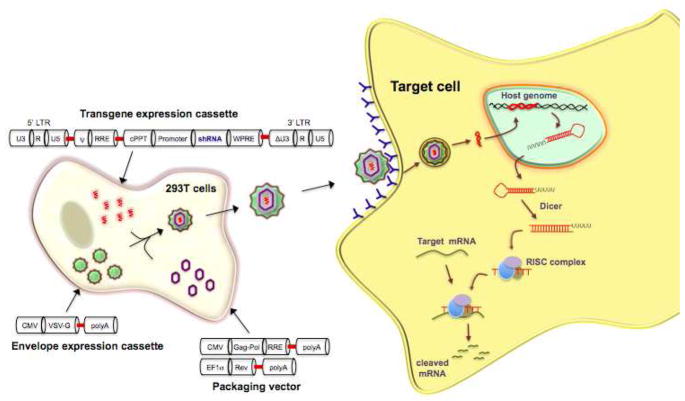
Fig. 1. Schematic of non-replicating lentiviral vector for stable shRNA expression.
Suitable host cells (such as 293T cells) are trasfected with a mixture of plasmids consisting of i) an shRNA expression cassette, ii) a packaging cassette and iii) a heterologous (VSV-G) viral envelope expression cassette. The generated lentivirus is then used to transduce the desired cell type for shRNA expression. Because only the vector containing the shRNA expression cassette (devoid of the viral structural genes) integrates into the host cell genome in the transduced cells, shRNA is continually expressed but infectious virus is not produced.
The generation transducing lentivirus production generally involves transfection of cells (such as the SV-40 large T antigen expressing HEK 293 T cells which are highly transfectable) with 3–4 different plasmids (see Fig. 1): one or two packaging plasmids containing the viral structural genes, a vector plasmid containing the transgene and cis elements needed for integration and packaging and 3) a heterologous viral (generally, VSV-G) envelope encoding plasmid. The replication incompetent virus released is then used to transduce cells for transgene expression.
The packaging cassettes, essentially derived from the HIV genome, have evolved by a sequential removal of cis elements needed for viral replication but retains the trans elements such as structural gag protein and enzymatic pol protein coding region. In the first generation packaging cassettes, only the envelope gene was deleted. In order to enhance biosfety, several groups systematically deleted the accessory genes and deletion of all accessory genes except Tat and Rev led to the second-generation packaging vector. In the third generation cassette, even the Tat gene is deleted and the gag/pol and rev genes are placed on separate expression plasmids [34]. Thus, in the currently used third generation packaging palsmid, only the cis-element rev response element (RRV) and the parental splice donor site necessary for post-transcriptional mRNA processing are retained and instead of the LTR, a heterologous promoter (generally CMV) and polyadenylation signal (SV40 or bovine growth hormone gene) is used to drive the full length packaging cassette mRNA. Remarkably, the extensive modification introduced in the packaging vector does not reduce the derived transducing viral titers. The packaging cassette has been further modified by codon optimization (humanization) that renders the gag/pol expression rev-independent, thus allowing deletion of RRE [41]. This modification is particularly important in the context of expressing anti-HIV shRNAs, in that shRNAs to target the gag and pol genes of wild type HIV-1 can be expressed without targeting of the packaging cassette mRNA itself [42]. To further reduce the chances of generating replication competent virus by non-homologous recombination events, a translentiviral packaging construct in which the gag/pol coding region is split into two separate expression cassettes has also been described [43]. Similarly, the integrase-deficient episomally replicating lentiviral vectors have also been used for stable transgene (although not shRNA) expression in non-dividing cells in vivo [43, 44]. The vector cassette contains the cis elements such as the primer binding site, packaging signal, RRE, central polypurine tract (cPPT), splice donor and acceptor sites, in addition to the transgene expression cassette and a modified woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) to increase the transgene expression. Although the overlapping sequences in WPRE containing the C-terminally truncated version of the WHV X protein might be potentially carcinogenic [45], this risk can be minimized by modifying the WPRE to delete the X protein promoter and all ORFs larger than 25 amino acids by mutating residual ATGs [46, 47]. A significant improvement in the so-called “self-inactivating” (SIN) vector that is currently used is the deletion of the U3 region in the LTR (that makes it Tat-independent) and the use of a heterologous (such as CMV) promoter to drive the entire vector mRNA transcription [48]. Several modifications can also be incorporated in the vector cassette to make the shRNA expression conditional (see later).
Several packaging cell lines stably expressing the packaging and envelope genes have also been developed [34]. Because of the toxicity of constitutive expression of VSV-G, generally all the packaging and envelope genes are expressed in a drug regulated manner. Although these cell lines provide a method to induce virus production by a single transfection with the vector cassette, generally the resulting transducable lentiviral titers are lower by many folds and thus, multi-plasmid transfection remains the preferred method.
2.2. Pseudotyping with heterologous viral envelopes
A major breakthrough in lentiviral vector development was the substitution of HIV-1 envelope with the vesicular stomatitis virus glycoprotein (VSV-G). Since VSV-G infects almost all cell types, this enabled broadening of the host tropism. Moreover VSV-G also directs viral entry to the endocytic pathway thereby reducing the viral accessory proteins needed for infectivity [49]. One more added benefit is that they are resistant to freeze thawing and ultracentrifugation, making it easy to concentrate the viral particles. However one constraint of VSV-G pseudotyping for systemic delivery is its rapid elimination from the circulation by complement and antibody. Although this can be reduced by conjugation with PEG [50], preexisting anti-VSV antibody still remains a problem. One way around this is to use the VSV-G derived from a different strain (Chandipura strain to substitute the commonly used Indiana strain). However, VSV-G is well suited for in vitro transduction as well as for local expression of transgenes in tissues. Many investigators including us have used VSV-G psuedotyped lentivirus to achieve shRNA expression in select tissues in vivo to silence genes for a variety of purposes (reviewed in [51]).
Apart from VSV-G, several other heterologous viral glycoproteins have also been used to psudotype lentiviral vectors expressing a variety of transgenes (reviewed in [52]). Pseudotyping with alternate envelopes have generally been used to restrict the viral tropism for delivery in a cell and tissue-specific manner. For eample., psudotyping the lentiviral vector with the neurotropic rabies virus glycoprotein (RVG) allows retro-axonal and trans-synoptic spread, thereby enhancing the transgene expression within the brain [53]. We have used this system to express antiviral shRNA against West Nile and Japanese encephalitis virus in the mouse brain [54]. RVG psudotyping restricted shRNA expression to neuronal cells and resulted in a much enhanced protection compared to VSV-G pseudotyped vector expressing the same shRNA, presumably due to a more extensive spread of the injected virus.
The pseudotype viral envelope glycoprotein can also be engineered to display specific targeting moieties. The E2 glycoprotein of Sindbis virus and HA of influenza have been generally used for this purpose. An interesting construct is the E2 protein of Sindbis virus fused to the Ig binding ZZ domain of Protein A. Because the ZZ domain binds potentially any immunoglobulin, this envelope can be made cell type specific just by incubating with a cell-specific mAb. In fact, gene delivery could be retargeted by this psudotyping with the specificity of the applied antibody [55]. An optimized version of this envelope pseudotyped lentivirus, coated with an antibody specific to metastatic malignant melanoma cells effectively targeted the melanoma cells in vivo in mice [56]. Notably the viral titers were not reduced by this pseudotyping and the background non-specific delivery to liver and spleen was much lower compared to VSV-G pseudotyped virus. To further reduce background, this pseudotyping/antibody targeting has been combined with a tissue-specific promoter for transgene expression. A Tie-2 promoter was used for endothelial cell-specific expression of a transgene [57] and more recently a prostrate cancer cell-specific promoter was used to express a luciferase reporter. The latter construct, pseudotyped with the modified Sinbis envelope and coated with a monoclonal antibody to prostrate stem cell antigen was highly effective in specifically targeting the xenotransplanted human prostrate cancer cells in mice with very little non-specific delivery [58]. Another group has used a chimeric Sindbis glycoprotein displaying CD20 antigen for targeting the lentiviral delivery to B cells [59]. The Sindbis envelope contains a binding site for the ubiquitously expressed cellular receptor heparan sulfate as well as a site to bind the dendritic cell-expressed DC-SIGN. Yang et al recently showed that just by mutating the heparan sulfate binding site, the delivery can be made dendritic cell-specific [60]. Remarkably, when a lentiviral vector expressing a nominal ovalbumin transgene was pseudotyped with this envelope and injected subcutaneously in mice, the mice developed a substantial ova-specific immune response that could protect the animals from ova-expressing tumor challenge. Although the above mentioned exciting studies suggest that shRNAs expressing lentivirus could also similarly pseudotyped for cell-specific delivery in vivo, this has so far not been shown except for our study using RVG pseuotyping mentioned earlier.
3. shRNA-induced toxicities
We highlight the possible undesirable effects of shRNA expression upfront, so that efforts to overcome this can be better understood. Similar to the externally introduced synthetic siRNA, endogenous expression of siRNAs can also cause side effects such as activation of innate immunity via induction of an interferon response as well as off-target gene silencing. Because they are continually expressed, these risks are enhanced in the case of shRNAs. In addition, because they use some of the miRNA machinery for their generation and export, shRNAs also have the risk of competition with cellular miRNAs. Si/sh RNAs can potentially elicit interferon responses either through the cytosolic dsRNA-activated protein kinase PKR or binding to toll-like receptors 3 and 7 that recognize RNA on the cell surface or in endosomes [61, 62]. Certain nucleotide motifs such as 5′-UGUGU-3′ or 5′-GUCCUUCAA-3′ within siRNAs appear be responsible for the induction interferon and interleukin production by plasmacytoid dendritic cells [62, 63]. Thus, it is important to avoid such motifs in the shRNA design. Apart from immunostimulation, si/shRNAs can also induce off-target effects. Since the majority of off-target effects are caused by small regions of seed sequence homology to 3′ untranslated region (UTR) of cellular miRNAs, it is important to scan for such homologies in the guide strand as well as to ensure that the passenger strand is not loaded to RISC. In the latter context, designing shRNAs to ensure thermodynamically less stable 5′ end and more stable 3′ end of the hairpin after shRNA processing by Drosha and Dicer would be important. High-level expression of shRNAs can also cause toxicity unrelated to off target effects by possible dysregulation of the cellular miRNAs. Long-term and sustained expression of shRNA under the control of a RNA polymerase III (Pol III) promoter via adeno-associated viral (AAV) vector in the mouse liver resulted in a dose-dependent liver injury and led to mortality at higher doses of expression [64]. This morbidity has been attributed to the saturation of exportin-5 (the factor required for export of miRNA from the nucleus) by shRNA since the liver-derived miRNAs were significantly downregulated in these mice. Consistent with this, Yi et al also showed that over expression of exportin-5 could relieve competition with cellular miRNAs [65]. Another study by Boudreau et al also showed that the Pol III U6 promoter-processed transcripts accumulate both in the cytosol and nucleus, indicating that both exportin-5 and Dicer might be saturated [66]. Similarly, lentiviral expression of large amounts shRNA generated from the U6 promoter resulted in toxicity in primary human T lymphocytes in vitro that could be mitigated by lower level of expression under the control of H1 promoter [67]. In summary, although endogenous shRNA expression has tremendous potential, it also possesses several intrinsic risks that could be dangerous and thus, the importance of carefully designing shRNA constructs to optimize the dose and the exact sequence of siRNA expression cannot be overemphasized.
4. shRNA design considerations
4.1 Conventional and miRNA-based shRNAs
Soon after the discovery that synthetic siRNAs induce gene silencing in mammalian cells, it was also shown that siRNAs also can be endogenously produced by expressing a short hairpin RNA (shRNA) from a DNA construct consisting of a siRNA sequence of 21–29 nt, a short loop region, and the reverse complement of the siRNA sequence and a short terminator (5–6 T residues) sequence (reviewed in [68, 69]. We refer to this type of shRNAs as conventional shRNAs (Fig. 2). When transcribed in vivo, this short transcript folds back on itself to form a hairpin structure, which is converted by cellular nucleases into siRNAs. Because the expressed RNA is short, generally Pol III promoters such as U6 or H1 are used. These promoters are strong and usually generate a huge amount of transcription products (although H1 promoter is weaker than U6) that are presumably processed by Dicer directly. Another advantage is that the sequence of the generated siRNA can be controlled because the transcription starts from the +1 position of the promoter transcription unit and termination occurs within a stretch of uracils in the terminator sequence, facilitating the generation of double stranded shRNA with 3′ overhangs that is essential for dicer processing. However, unlike the typical Pol II transcripts, the Pol III transcripts lack the 5′ cap and 3′ Poly (A) tail. These conventional shRNA transcripts are not processed by Drosha but instead are exported to the cytoplasm by exportin-5. They are thought to be further processed in the cytoplasm by Dicer into double stranded siRNAs. Because of the ease of vector construction, many investigators have used the conventional shRNAs in a variety of contexts to knockdown different genes since it was first described in 2002. However, the potential of this system for inducing toxicities was highlighted in 2006, when it was shown that overexpression of U6 driven shRNA could interfere with cellular miRNA expression, leading to lethality in mice [64]. Moreover, the shRNAs generated by these promoters generally have triphosphate at their 5′ ends and a variable number of overhangs at the 3′ end and thus, are not the optimal substrate of Exportin-5 and Dicer, although the huge amount of the transcription products can compensate for the low efficiency and generate a good silencing effect. The triphosphate 5′ end of the siRNAs might also activate RIG1 to induce an interferon response, resulting in the activation of PKR and subsequent shut down of cellular translation [70–72]. The recently described tRNA promoter-driven tRNA-shRNA chimeras might mitigate some of these problems [73]. The chimeras will be processed by endogenous endonuclease to release the shRNA that has a 5′ monophosphate. Also the expression level of shRNA driven by tRNA promoter is much lower compared to the commonly used Pol III promoters such as U6 and H1, which might reduce the nonspecific side effects [73].
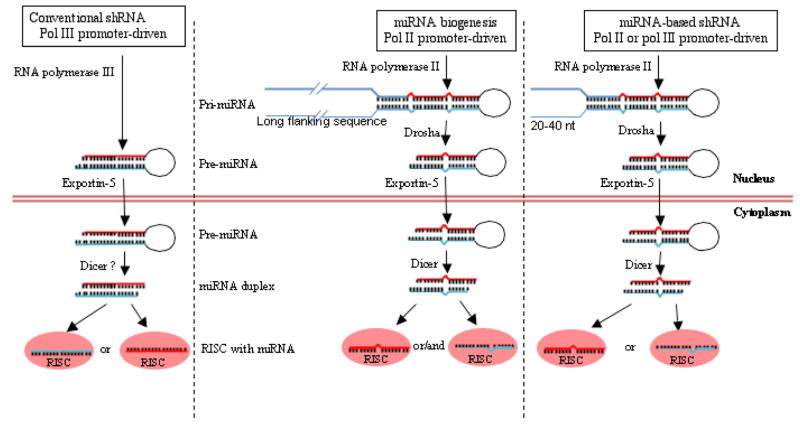
Fig. 2. Biogenesis of conventional and miRNA-based shRNA.
The left panel shows the biogenesis of the Pol III promoter-driven conventional shRNAs (originally developed without the knowledge of miRNA biogenesis). Here the transcript only contains the hairpin RNA sequence that is directly exported to the cytoplasm. The middle panel indicates the natural miRNA biogenesis. The primary miRNA transcript contains the mature miRNA hairpin as well as a long flanking sequence and is processed first by Drosha into pre-miRNA that is then exported to the cytoplasm for Dicer processing. The right panel shows miRNA-based shRNA expression. Here the shRNA construct (controlled by either Pol III or Pol II promoter) is designed to mimic the pri-miRNA by including the miRNA flanking sequence into to shRNA stem. The resulting transcript is processed like miRNA by Drosha, followed by Dicer into siRNA. Although during miRNA biogenesis, both strands are sometimes selected, selection of the passenger strand should be avoided in the shRNA design to reduce off-target effects.
Probably the best way to induce effective gene silencing without toxicities is to use the naturally occurring miRNA pathway to generate the desired shRNA and thus, understanding of miRNA biogenesis appears to be key to the rational design of shRNAs (Fig. 2). Although the above described conventional shRNA somewhat resembles the pre-miRNA and is probably processed by Dicer, it is not processed by Drosha. Processing by Drosha is the first critical step in miRNA biogenesis that extracts the miRNA out from the long sequence within the pri-miRNA [13]. Drosha cleaves the pri-miRNA to yield a pre-miRNA with a 3′ 2 nt overhang and Dicer just recognizes this overhang and cuts the hairpin 22 nt away to produce the miRNA duplex [74, 75]. Form this duplex generally one strand of the mature miRNA is then selected for incorporation to RISC according to the 5′ end base-pairing rule [20, 24]. Thus, the initial cleavage sites chosen by Drosha largely dictates where Dicer will cleave and hence, which miRNA strand enters the RISC [26]. Because of these reasons, there has been a tremendous interest in mimicking the pri-miRNA processing to generate shRNAs. However, the critical issue for miRNA-based shRNA design is what exactly determines the Drosha cleavage site. Without this knowledge, earlier attempts to design shRNA mimicking a mature miRNA duplex were largely unsuccessful. In fact, after testing a large number shRNA variants, Paddison et al concluded that the conventional shRNAs appears to be much better than miRNA-based shRNAs that were mimicking let-7 [76].
The first glimpse of pri-miRNA processing was provided by a study that showed that an authentic human miRNA, mir-30 could be correctly excised from within a long irrelevant Pol II transcript, if it contained the 71nt precursor, but not if the transcript contained only the ~22 nt mature mir-30 sequence [77]. Moreover, the generated mature miRNA was capable of repressing its target gene and more importantly, substitution of miR-30 stem in the construct with an artificial shRNA also led to specific gene silencing. Thus this study, for the first time showed that the precursor might contain all cis-acting elements required for miRNA processing. However, there were some unusual features in that, whereas only one strand of endogenous mir-30 is selected by RISC, both strands were selected in this artificial system. Also, although mir-30 and mir-21 precursors were correctly processed, several other precursors including mir-27, let-7a3 and lin-4 precursors in the same context failed to generate mature miRNAs, indicating more needs to be learned about miRNA biogenesis [77]. In another study, inclusion of ~125 bp flanking sequence from the miRNA stem loop was found to generate mature miRNAs uniformly for several tested miRNAs in murine hematopeoitic cells, indicating that flanking sequence might be important for pri-miRNA processing [78]. Mutational analysis of mir-30 processing by Zeng et al showed that a large (~10 nt) terminal loop is important and that Drosha cleaves the RNA hairpin approximately two helical RNA turns (~22nt) from the loop into the stem to produce the pre-miRNA [79]. In addition, a few nucleotides 5′ to precursor miRNA cleavage sites was also found to be essential for efficient processing. A caveat in this study however, is that the pre-miRNAs were expressed within a long irrelevant RNA and thus do not have their natural flanking sequences beyond cleavage site except for the first 4 nts and thus, the importance of the flanking sequence could not be fully evaluated. However, a later systematic mechanistic study by Han et al showed that the terminal loop itself is irrelevant for Drosha processing and that the microprocessor component DGCR8 recognizes both the single-stranded (ss) flanking segment and the double-stranded (ds) RNA stem following which, Drosha cuts the pri-miRNA ~11nt from the ss-dsRNA junction [13]. This is also consistent with an earlier study showing that varying either the loop size from 4 nt to 23 nt or the loop sequence did not change shRNA generation efficiency [76]. Moreover, the miRNA-based shRNAs optimized by including the single-stranded flanking sequences of pri-miRNA showed consistent repression effect [80–82]. Thus as a general rule, it may be advisable to include ~15–20 nt flanks in the design of shRNA. Although most studies of miR-based shRNA expression demonstrate the functionality of the generated siRNAs in terms of target gene silencing, they use stems with varying number of nts (from 19–23) and unfortunately the actual sequence of siRNAs generated has not been elucidated. Also whether the passenger strand is also selected remains unknown. Thus if the Han’s model is correct, the siRNAs in some cases are also likely to include irrelevant sequences from the stem-loop and/or select the passenger strand that might cause unintended effects. Thus, it appears to be important to keep the stem length to ~22 nt from the Drosha cleavage site as well as to ensure that the thermodynamic properties of the ends favor selection of the anti-sense strand. Han’s model also indicates that most miRNA stem loops have mismatches around 12 bp from the 5′ end of mature miRNA that might avoid abortive Drosha processing (if the loop is large, it can also be considered ss-ds junction and Drosha can possibly cleave from this end). Thus, it may also be profitable to design shRNA to contain mismatches ~12 nt from the 5′ end of mature miRNA. The design of an ideal shRNA based on the present understanding of miRNA biogenesis is depicted in Fig. 3.
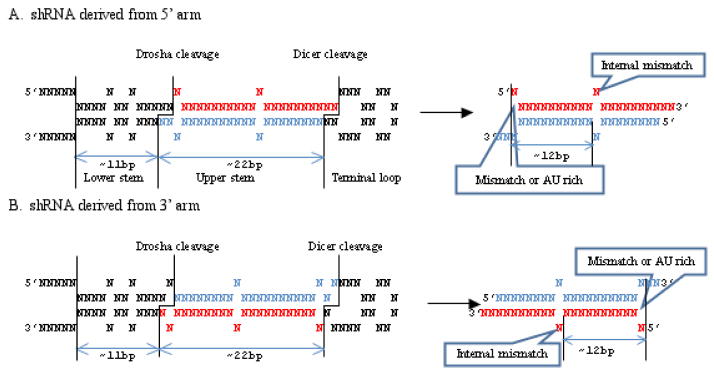
Fig. 3. Rational design of shRNA based on the current knowledge about Drosha and Dicer processing.
Drosha cleaves the pri-miRNA ~11bp from the ss-ds RNA junction irrespective of whether the mature miRNA (marked in red) is derived from the 5′ (A) or the 3′ (B) arm of the hairpin. The miRNA stem also generally contains internal mismatches ~12nt from the 5′ end of mature miRNA that might help avoid abortive processing. Thus for the design of shRNA depicted on the right, 20–40 nt flanking sequence and an internal bulge as depicted should be included to ensure accurate Drosha processing. Also the shRNA terminus should be designed to favor the selection of guide vs passenger strand by incorporating mismatches or AU at the 5′ end as depicted.
Although the Han’s model has provided tremendous insights into rational design of shRNA, it should be pointed out that this model of pri-miRNA processing is mainly based on in vitro assays on miR-16 biogenesis and computational analysis of other miRNAs [13] and whether it holds true in vivo and for all pri-miRNA processing is not known. For example, using this model, it is difficult to reconcile with some of the findings with miR-30 processing described earlier. Moreover, the exact cutting site of Drosha/Dicer is mainly deduced from the annotated mature miRNA sequences. However, recent studies indicate that the ends of mature miRNA are highly varied [83–85] and it is speculated that the variations might be a consequence of imprecision in the Drosha/Dicer processing [83, 84] or modifications introduced after Drosha/Dicer processing [85]. Also, although according to Han et al’s model, the terminal loop is irrelevant for pri-miRNA processing, a recent study showed that the terminal loop of let-7 mediates negative regulation the miRNA processing by binding to Lin 28 [86–88]. Thus, although great progress has been made in the rational shRNA design, further insights from miRNA biogenesis will undoubtedly continue to aid efforts in further optimizing the shRNA design.
Although miRNA-based shRNA can be expressed both under the control of Pol III or Pol II promoters, pol II promoters may be preferable. With the latter, the transcript level is similar to the low levels of the endogenously expressed miRNAs and thus, the toxicity associated with overexpression can be avoided. Moreover even though a much lower level of shRNA is produced with miRNA-based expression, the shRNA works equally well as compared to overexpression in a conventional system. For example, expression in a lentiviral vector of a U6 driven shRNA embedded within the stem of a miRNA mitigated the brain toxicity associated with the same shRNA not in a miRNA context, expressed under the control of the same U6 promoter [89]. Thus, sequential processing by Drosha and Dicer in a physiologic way might somehow render the shRNA functionally more competent. Also since hundreds of miRNA are naturally expressed in a cell, expression of a few more similarly processed shRNAs is unlikely to compete with endogenous miRNA pathway. These features of miRNA-based shRNA expression will be a big plus for in vivo application.
4.2. Multiplex shRNA
As discussed earlier, one of the major concerns for using shRNA to repress target gene expression is the unintended effects, such as saturation of the endogenous miRNA machinery, interferon response and off-target effect. Reducing the intracellular concentration of the mature siRNAs can decrease all these side effects. Multiple shRNAs targeting different region of the same gene has been used to reduce the effective concentration of individual siRNA, yet maintaining the overall silencing effect. Moreover, expression of single siRNA to treat viral infection rapidly results in the emergence of escape mutants and the use of multiplex siRNAs is imperative to reduce the chances of generation of viral escape mutants, particularly for chronic infection with viruses having propensity for high degree of mutations like HIV-1 [90–92].
Because lentiviral vectors can accommodate up to 7 kb foreign sequences, several strategies have been used to express multiple shRNAs in these vectors. In one approach, complementary single stranded RNAs or long hairpin RNAs expressed under the control of Pol II promoters in lentiviral constructs have been tried, but these were not very effective probably due to problems in the nuclear export of long RNAs as well as the difficulty in viral propagation [93, 94]. However, unlike transfection with long double stranded RNAs, expressing long dsRNAs intracellularly did not induce an interferon response. In another approach, longer hairpins encoding 2–3 anti-HIV shRNAs, harboring multiple C to U (or A to G) mutations within the sense strand, have also been expressed under the control of a single U6 promoter [95]. This strategy has also been used to suppress HCV and HBV replication [96, 97]. However in general, the processivity declines progressively from the proximal end and results in unequal silencing by different shRNAs. Because the Pol III promoter system is small (200–400bp), another strategy is to use multiple shRNA expression cassettes, which can be conveniently cloned into the lentiviral vector. One of the common methods is to insert multiple shRNAs cassettes, each driven by the same Pol III promoter [42, 98, 99]. However as a recent study by Brake et al showed, although the initial cotransfection experiments using plasmid DNA constructs suggest that each of the individual shRNAs can be expressed that can silence the corresponding target, repeated promoter sequence within the same lentiviral construct induces genetic instability during the production of transducible lentivirus [100]. In fact, several shRNA cassettes were frequently deleted from the virus because of recombination and only less than 15% of the cell clones transduced with a triple shRNA lentivirus contained an intact provirus. To overcome this, the authors constructed a lentiviral vector harboring multiple shRNAs driven by different pol III promoters. A cassette containing different anti-HIV shRNAs driven by three different human pol III promoters U6, H1 and 7SK (with no sequence homology between them) was inserted into a lentiviral vector. Cells transduced with the lentivirus generated from this construct stably expressed all shRNAs and effectively prevented the generation of viral escape mutants. In a similar strategy, a vector to express shRNAs bidirectionally using 4 different pol III (mouse U6, human U6, mouse H1 and 7SH) promoters has been found to effectively silence 4 different targets [101]. Another multiplex strategy is to use shRNA in combination with other antiviral proteins. Notably, a lentiviral vector expressing a U6 driven anti-HIV shRNA, U6-driven TAR RNA decoy and a chimeric adenoviral VA1 CCR5 ribozyme has been used to inhibit HIV replication by multiple mechanisms. This vector stably suppressed HIV replication in vitro and was safe in humanized mice [102, 103] and is currently under Phase I clinical trial.
Perhaps the most ideal strategy to express multiplex shRNA is to use the template of miRNA polycistrons. Several miRNAs are encoded in genomic clusters and transcribed as a single polycistron, such as the miR-17-92 cluster, that result in the generation of multiple miRNAs from a single transcription unit. Thus, multiple shRNAs could be easily expressed in equimolar amounts from a single miRNA polycistron driven by a Pol II promoter [104]. The advantage of this approach is that the expression level of shRNAs will be similar to endogenously expressed miRNAs and the shRNAs are processed by the cellular miRNA machinery. The human miR-17-92 cluster on chromosome 13 encodes a ~1kb pri-miRNA that is processed into 6 different pre-miRNAs. In an elegant recent study, Liu et al cloned and used this template to express multiple anti-HIV shRNAs [104]. The individual pre-miRNAs with at least 40 nt flanks on each side of the hairpin were subcloned and the miRNA sequences were systematically replaced with anti-HIV shRNA sequences. Individual shRNA sequences were optimized to resemble the natural miRNA by designing the passenger strand to mimic the structural features (mismatches, bulges and thermodynamic stability) of the natural pre-miRNAs. The optimized hairpins were found to be as efficient functionally as the conventional U6-driven shRNAs, although the amounts of siRNAs generated were much smaller than conventional shRNAs used as positive control, indicating a better biofunctionality of the Dorsha and Dicer processed shRNAs. When these shRNAs were chained in different permutations and combinations, it was found that the individual siRNA amounts increased by linkage to another hairpin and that the position of the shRNA within the polycistron did not affect siRNA production or efficacy. The chained polycistron encoding 4 different anti-HIV shRNAs under the control of CMV promoter was then cloned into a lentiviral vector. The anti-HIV efficacy of this lentivirus was more robust than individual shRNAs, attesting to the feasibility of using this for clinical application. However, it is worth noting that although the shRNAs containing either 19 or 23 nt stems were both equally functional in terms of silencing HIV replication, the actual sequence of the mature siRNA produced was not determined and thus the siRNA generated from the shorter hairpins might contain additional vector-derived sequences that might not be desirable.
In a similar approach, the miR-106 cluster has also been used to express multiplex shRNA [105]. Located within an intron in the human chromosome 13, this cluster encodes 3 miRNAs (miRs-106b, 93 and 25) from a single transcription unit that is ~800 bp long. Three anti-HIV shRNAs were inserted into the miR-106 cluster scaffold. This study also found that the shRNA flanking sequences are important for processing and that the passenger strand selection can be avoided by mimicking the natural miRNA stems by introducing mismatches and bulges rather than as two perfectly complimentary siRNA strands. In addition to shRNAs, a U6 driven TAR decoy was also expressed in the intronic region. This construct, when expressed in a lentiviral vector, enhanced the anti-HIV activity seen with shRNAs alone.
4.3 Conditional Expression of shRNAs
Regulated RNAi systems may be desirable in many circumstances. For example, conditional expression would be essential to generate knockdown mice for genes whose inactivation would be embryonically lethal. Such a system would also allow loss of function analysis under induced and non-induced conditions in the same cells. It would also provide a means to study the kinetics of gene silencing and determine the concentration of shRNA needed for silencing. Moreover, given that long-term shRNA expression in vivo could be toxic, it would provide a way to express shRNA only when needed or only in certain tissues or virally infected cells for therapeutic application. Essentially two general strategies are used. In the first strategy, a Pol II or Pol III promoter controlled by drug (generally tetracycline or ecdysone)-responsive repressors or transactivators or the HIV-1 Tat protein is used for regulated expression. In this case, expression can be turned on or off at will simply by using or withdrawing the drug. In the second strategy that is mainly used for transgenic purposes, a cre-lox recombination system is used to induce shRNA expression. In this case, the expression is irreversible after induction.
Generally in the tetracyclin based-regulated RNAi systems, the Pol II or III promoter driving the shRNA is modified to contain a single or multiple copies of the tetracyclin operator (TetO) sequence and a Tet repressor (TetR) is also separately expressed. The TetR binding to TetO represses transcription in the absence of tetracycline and in the presence of tetracycline or its analog doxycyclin, which binds with high affinity to TetR, the repression is relieved. Since the inhibition mediated by steric hindrance is generally not complete, there is some leakiness in this simple system. Hence, to provide more tightly controlled inducible expression, several versions of the repressor system including using different transactivators and transcription factor domains that mediate epigenetic repression, fused to tetR components that either result in turning the shRNA transcription on or off with doxycyclin have been developed (reviewed in [106]). In some of the earlier versions, two separate vectors (one containing the shRNA expression cassette and another expressing the transactivator/repressor element) and high multiplicities of infection had to be used for shRNA expression generally under the control of pol III promoters [82, 107–112]. However, in the last 2 years, significant improvements in the vector design to conditionally express either Pol II or Pol III driven shRNA in a single lentiviral vectors have been made. Stegmeier et al developed a lentivirus system for regulated expression of miRNA-based shRNAs in a Pol II CMV-based promoter [82]. Here the miR-30 based shRNAs were cloned in front of the CMV promoter containing TetO sequences in a lentiviral vector which was then used to infect a cell line stably expressing the reverse tet-controlled transcriptional activator (rtTA), which activates transcription from the promoter in response to doxycyclin. Based on this vector, a second generation, Tet-inducible shRNA library that allows genome wide shRNA screen has also been developed [80]. However, this system is not self-contained in that, it relies on rTA expression from another source. Recently, several investigators have also developed “all in one” single lentiviral vectors that contain all the elements needed for regulated expression of shRNAs [108, 109, 113, 114]. Notably in an elegant approach, Szulc et al developed a system where virtually any transcription unit (expressing transgenes, shRNAs or miRNAs under the control of either Pol III or Pol II promoter) can be accommodated into a single lentiviral vector design and subjected to epigenetic regulation [109]. Here, the DNA binding domain of a transcriptional silencer (derived from the Kruppel-associated box (KRAB) domain found in many transcriptional regulators), promiscuous for RNA II and -III promoters that silences gene expression by triggering locally confined epigenetic changes around their binding sites, is fused to tetR. This (tTRKRAB) cassette is bicistronically expressed along with a transgene (such as GFP) under the control of a Pol II promoter (different promoters such as CMV, EF-1α, Ubiquitin c were all shown to be effective) in a lentiviral vector. In addition, a pol III shRNA cassette is cloned downstream of TetO sequence in the LTR. Since the epigenetic regulation mediated by tTRKRAB extends to ~2–3 kb, the conditional repression effect also extends to the transgene and the KRAB control element itself, thus providing a way for autoregulation, whereby its expression is at basal levels in the drug “off” mode. The versatility of this compact single virus system for tightly regulated and readily reversible doxycyclin-dependent expression was shown in a variety of situations in vitro and in vivo including for the expression of a GNDF transgene in the mouse brain, conditional knockdown of GATA 1 expression in human hematopoietic progenitors, TP53 and GFP knockdown in xenotransplanted tumor cells in mice as well as in the generation of conditional GFP knockdown mice. Shin et al also developed a single lentiviral vector for conditional RNAi [113]. Here, a minimal TRE promoter is used to drive miR-based shRNAs followed by a human ubi-c promoter driving the constitutive expression of a bicistronic transcript containing the tet-dependent transcativator rTA or rtTA3 and a GFP derivative, venous. Using this system called pSLIK, the authors effectively achieved conditional expression of multiple shRNAs for simultaneous knockdown of Gα12 as well as Gα13 in MEF cells. Similarly, using a steric hindrance-based single lentiviral system consisting of a U6 driven shRNA and CMV-driven tetR, Aagaard et al were able to conditionally knockdown Drosha or protein knase R (PKR) in HEK293 cells, although there appeared to be ~25% leakiness in the shRNA expression under basal uninduced conditions [114]. Recently a single lentiviral vector expressing shRNA under TetO/H1 promoter followed by tetR/T2A/GFP under the control of a ubiquitin promoter has also been used to generate transgenic rats for doxycyclin regulated reversible knockdown of insulin receptor [115].
In another approach, Unwalla et al demonstrated that HIV-TAT can be used for conditional expression of shRNA from a lentiviral vector [116]. This study was based on the concept that similar to HIV-1 LTR promoter, the drosophila hsp70 promoter can also be transactivated by TAT. Thus, the authors placed an anti-HIV rev shRNA sequence and a minimal poly A sequence in front of the core hsp70 promoter and this cassette was then cloned into a lentiviral vector downstream of the TAR element in the LTR. In the absence of TAT-mediated transactivation, the transcription from the LTR is stalled at the TAR site and thus no shRNA is made. When TAT is induced, it binds to the TAR element and recruits the transcription elongation factor P-TEFb, which in turn phosphorylates the RNA polymerase assembled at the downstream hsp promoter to relieve the transcription block and initiate shRNA expression. This conditional rev shRNA expression using the plasmid construct resulted in 90% reduction of HIV replication when cotransfected with pNL4-3, a molecular clone of HIV-1. Moreover, the conditional shRNA cassette cloned in the lentiviral vector pHIV-7 was also effective in a gene therapy setting in that, after transduction of hematopoietic stem cells with pHIV-7 shRNA encoding lentivirus, the cells were remarkably resistant to infection with different strains of HIV-1 including HIV IIIb and JRFL. The main advantage of this system is that the shRNA expression is only induced in HIV infected cells, thus avoiding non-specific toxic effects. In another study, a doxycyclin-dependent conditionally replicating HIV vector has been used to express anti-HIV nef shRNA [117]. Here, the nef gene was deleted and replaced with rtTA and TetO sites incorporated into the 5′LTR to make the virus replication dependent on doxycyclin and a Pol III shRNA expression cassette was also cloned in the U3 region of the 3′LTR. The idea was to start a spreading infection with this “therapeutic virus” in all susceptible cells with transient dox treatment ensuring uniform integration of the provirus, so that after dox withdrawal, although virus replication is inhibited, shRNA is constitutively expressed that would inhibit a nef expressing wild type HIV. Although some inhibition was seen in SupT1 cells with low levels of wild type HIV infection, breakthrough replication occurred at higher multiplicities of infection and moreover, the shRNA expression cassette was deleted after several passages and thus, this strategy appears too dangerous for any clinical application.
The cre/loxP recombination method to obtain conditional shRNA-mediated gene knockdown is primary used to generate transgenic animals for genes (such as Dicer, Drosha and many transcription factors) whose constitutive silencing might result in embryonic lethality. This also allows the study of temporal and tissue-specific effects of gene expression and the availability of a plethora of cell- and tissue-specific cre expressing mice for crossing provide a valuable asset for this purpose. Essentially two loxP sites flanking a sequence (such as a stop signal or a stuffer sequence) that hinders constitutive shRNA expression are inserted within the promoter driving the shRNA [118–120] to generate transgenic mice. After crossing these mice with a cell or tissue-specific cre-expressing mice, the shRNA is activated because of cre-mediated deletion of the hindering sequence. Although most studies have used a U6 or H1 Pol III promoters for regulated shRNA design, one study has also used a Pol II U1 promoter by inserting a loxP-stuffer-loxP cassette just upstream the transcription initiation site [121]. Some investigators have also inserted the loxP-stop-loxP sequence within the shRNA loop between the sense and antisense strand of shRNA to generate conditional shRNA knockout mice [122–124]. However in this system, a 34 bp loxP site remains within the shRNA loop after cre recombination that might affect shRNA cytoplasmic transport. However in an interesting recent study, this system was used to generate multiple gene mutant mice using wild type and a mutant loxP site that are recombined by cre independently. The two loxP sites flanking a stop signal was inserted within the loops (intervening between the sense and antisense strands of shRNAs) of two shRNA expression cassettes to generate simultaneous conditional knockout of the redundant genes ERK1 and ERK2 or Gsk3α and Gsk3b [125]. Two sets of mutant loxP sites have also been used to invert (flip) a DNA segment containing a mir-based shRNA from antisense to sense orientation after cre recombination [126]. Stern et al showed the versatility of this “Filp” vectors to conditionally express multiple miR-based shRNAs, various combinations of reporter genes and oncogenes in transgenic mice as well as in cell culture and hematopoietic stem cells.
5. Lentiviral shRNA delivery for viral infections
Although lentivirally expressed shRNAs have also been evaluated for potential therapeutic application in cancer [127–129] and neurodegenerative diseases (reviewed in [130–132]) in this review, we will restrict our discussion to viral infections.
5.1. HIV-1
Stable expression of siRNA would be particularly valuable in chronic infections such as HIV-1, where durable antiviral effects are needed. Moreover, the siRNAs need to be expressed in the hard to transfect primary T cells and macrophages, which are the primary targets of HIV-1. Thus, many investigators have used lentiviral vectors for shRNA expression. However, HIV-1 also poses numerous challenges for RNAi-based therapies and in fact, these challenges have been the driving force in moving the technology forward, resulting in incremental improvements in lentiviral shRNA design and delivery.
One constraint in using HIV-based lentiviral vectors to express anti-HIV shRNAs is that a potential target sequence might also be present in the vector or the packaging construct [133]. Although the sequence of any shRNA cassette is also present in the genomic vector-derived transcript and theoretically might be subject to “self-targeting” by the mature siRNA generated, this obviously does not happen (or else production of any shRNA would become impossible). The reason for this may be the inaccessibility of the target site within genomic transcript for RNAi, due to the secondary structure assumed [133]. However, the presence of gag/pol and rev sequences within the packaging construct can interfere with efforts to express shRNAs targeting these HIV genes and in fact, attempts to express gag/pol shRNA significantly reduced the generated transducing viral titers. However, this constraint could be effectively overcome by using human codon optimized gag/pol sequence (where the vector sequences differ from that in the wild type HIV) in the packaging vector [133]. Similar problem might also arise with shRNAs targeting highly conserved regulatory sequence elements that are also likely to be present in the lentiviral vector, which could be overcome by introducing small mutations in the vector sequence. These issues may become particularly important when trying to express multiplex anti-HIV shRNAs and thus, care should be taken in the design of such vectors.
Theoretically, targeting any region within the primary viral transcript should lead to degradation of the whole transcript and thus mediate antiviral activity. In fact, several proof of principle studies using shRNAs to target several different HIV-1 genes showed effectiveness in cell lines [134–136]. However, the exquisite sequence specificity of RNAi can also become a handicap for RNAi-based therapy for HIV-1. For example, the usefulness of any selected siRNA target would be limited because of the existence of multiple genetic subtypes or clades (clades A, B, C, D, E, F1, F2, G, H, J, and K) of HIV-1 with differing sequences [137]. Moreover, many different viral quasispecies are present at any given time in an infected individual, the number of which increases with disease progression [138, 139]. Further compounding the problem is the ability of the virus to readily generate escapes mutations (with single or multiple nucleotide changes within the shRNA recognition sequence) that resist shRNA interference. In fact, several studies have documented the propensity of HIV-1 to generate RNAi escape mutants in long-term cultures [90–92, 140]. This problem can be circumvented to some extent by limiting the choice of RNAi targets to highly conserved regions, where the virus is averse to mutate because of their essential structural and/or functional roles [140–143]. We were able to show that a lentiviral shRNA targeting one such well-conserved target in the viral vif sequence could effectively protect primary CD4 T cells from all HIV-1 clades, including multiple isolates of clade B, prevalent in the West [144]. In a more extensive analysis, the Berkhout group screened 86 different highly conserved viral sequences to identify shRNA targets that could inhibit viral replication, of which 21 were found to be potent HIV-1 inhibitors [42]. However, it seems unlikely that just selecting an shRNA that targets conserved region will be enough to prevent the emergence of escape mutants. A recent study using shRNAs targeting highly conserved viral genomic sequences in Gag, protease, integrase, and Tat-Rev regions of HIV-1 could not identify any shRNA inhibitors that that did not allow viral escape [140]. A comprehensive target sequence analyses of 500 escape viruses revealed that the viruses had acquired single point mutations, predominantly at target positions 6, 8, 9, 14, and 15 but not in positions 1, 2, 5, 18, and 19.
Another approach to prevent viral escape is to use shRNAs to target host genes that are important in the viral life cycle. A good example is the viral co-receptor CCR5, where a natural 32 bp homozygous deletion of the coding gene sequence abrogates its function but has no deleterious immunological consequences and even provides protection from HIV-1 infection [145, 146]. In fact, several studies have shown that HIV replication could be blocked in primary CD4 T cells or macrophages by delivery of CCR5 shRNA [147–149]. As HIV-1 has a large dependency on host factors for its replication, many more such druggable host targets are likely to be identified. In fact, a recent genome wide RNAi screen has identified several host genes necessary for viral replication [150]. Some of these HIV-dependency factors (HDFs) were also found to be not essential for host cell viability. Thus, potentially some of these host genes that participate at different stages in the viral life cycle could be used as targets for suppressing HIV infection. Using a combination of siRNAs targeting conserved viral sequences and host genes important in the viral life cycle may be the optimal therapeutic approach, akin to antiretroviral drug cocktails.
To restrict the viral evolutionary possibilities further, many groups have designed lentiviral vectors that express more than one conserved shRNAs using several strategies described earlier. Yee et al have used a combination of pol III and pol II promoters to express antiviral siRNAs in the miR-30a backbone to show that HIV escape can be prevented when two shRNAs are simultaneously expressed in the cells [151]. As discussed earlier, multiple anti-HIV shRNAs have also been expressed in polycistronic miRNA scafolds [104] [105]. Long-term analysis of these multiplex shRNAs in future should reveal how effective they actually are in preventing viral escape.
Currently the most practical use of shRNA for HIV disease is to derive virus-resistant cells of all key lineages, primarily progeny T cells and macrophages from lentivrally transduced CD34+ hematopoietic stem cells (HSC) [103, 152]. The feasibility of this approach was shown by transplanting human CD34+ hematopoietic stem cells transduced with a lentivirus expressing an anti-rev shRNA into thy/liv grafts in SCID-hu mice. The progeny T cells and macrophages isolated from these grafts were shown to resist HIV challenge [102, 153, 154]. In vitro cultured CD34+ HSCs similarly transduced with single or bispecific lentiviral constructs expressing CCR-5 and CXCR-4 shRNA have also been shown to give rise to progeny macrophages that are HIV resistant, which also attests to the feasibility of the approach [99]. Significantly, stable reduction of CCR-5 in progeny T cells has also been demonstrated in non-human primates transplanted with CD34+ HSC expressing the CCR-5 shRNA [155]. Once again, the antiviral efficacy was shown in ex vivo experiments with T cells sorted on the basis of GFP marker gene expression. Interestingly, a strong but toxic U6 promoter had to be replaced with a weaker H1 promoter to derive progeny cells in vivo, probably due to the toxicity in HSC mediated by high-level expression of shRNA preventing effective engraftment. This study, showing sustained (over a year) shRNA transgene expression in primates in vivo augurs well for future translational studies. However, considering that only a small proportion of progeny cells expressed the transgene, improvements to achieve a better level of gene marked cells in vivo is an issue that will need attention. This may entail reducing the time in culture during transduction of HSC to avoid lineage commitment associated with loss of self-renewal capacity and/or developing better targeted delivery approaches.
As mentioned earlier, RNAi has also been combined with other gene therapy approaches in a single lentiviral vector. The lentivirus engineered to encode an shRNA targeting HIV-1 rev/tat mRNA, a short RNA homologous to the viral transactivation response region (TAR) to act as a decoy for TAT binding and a ribozyme targeting the host CCR5 gene [103] is already undergoing Phase I clinical trial. The result from this trial is likely to have a significant impact on the future lentiviral-based therapies in general.
5.2 Hepatitis viruses
Hepatitis B (HBV) and Hepatitis C (HCV) viruses also cause chronic infections and thus stable expression of shRNAs to suppress these viruses would be desirable. Many investigators have used both synthetic siRNA and shRNA to suppress these viruses (reviewed in [156–158]). Both the viruses lack a convenient animal model although mice transfected with plasmids containing the HBV viral genome have been used to test the efficacy of RNAi. However for long-term expression of shRNA to target HBV, adeno and adeno-associated viral vectors rather than lentiviral vectors have been used for in vivo delivery because of their tropism for liver cells. In case of HCV, the effectiveness of RNAi has mostly been tested in vitro in cell lines transfected with HCV replicons using synthetic siRNAs or plasmid expressed shRNAs. One recent study however, has used a lentiviral vector to express multiple shRNAs. Two antiviral shRNAs targeting the viral IRES and NS5b sequences as well an shRNA targeting the cellular receptor, CD81 were cloned in tandem under the control of the Pol III H1 promoter. These were shown to be effective in reducing viral replication as well as inhibiting CD81 in HCV replicon-transfected Huh-7 cells for at least up to 17 days of culture [98].
5.3 Human Papilloma virus
Infection with certain types of HPV (most commonly HPV16, 18) can cause cervical and other types of cancers in humans via the HPV E6 and E7-mediated inactivation of the tumor suppressor proteins p53 and pRB. Since E6 and E7 proteins are selectively expressed in caner cells, several investigators have used RNAi to suppress these proteins in HPV transformed cell lines [159–162]. In one recent study, a lentiviral vector was used to express shRNA under U6 promoter to target both E6 and E7 proteins of HPV 18 [160]. Transduction of HeLa cells in vitro with this lentivirus resulted in a dose-dependent reduction of E7 protein level associated with upregulation of its target proteins, p53 and p21, leading to the induction of senescence or cell death (at high high-dose transduction). The tumorigenicity of the E7-silenced HeLa cell xenotransplants in Rag-/- mice was abolished and moreover, systemic treatment with the lentivirus after the tumor metastasis was already established, also significantly reduced the tumor progression in the lungs. It was also shown in another study that suppression of E6, E7 enhances the chemotherapeutic effect of cisplatin in HeLa cells [162]. Thus, specific suppression of viral oncogenes in conjunction with chemotherapy appears to be promising for human therapy.
5.4 Coxsackie virus
Coxsackie B enteroviral infection that can cause viral cardiomyopathy with a high degree of fatality in children and young adults. RNAi has therefore also been evaluated for suppressing this virus in vitro and in vivo [163–166]. In a recent study, an shRNA targeting the viral conserved cis-acting replication element was expressed in a lentiviral vector. Transduction with this lentivirus effectively suppressed coxsackie B3 viral replication in cell lines. Moreover, mice treated with this lentivirus intraperitoneally and challenged with coxsackie B3 developed a markedly reduced cardiac viral titer, cardiac inflammation and mortality compared to control mice [164]. In another study, iv treatment with a AAV9 vector encoding a shRNA targeting the coxsackie viral RdRp also dramatically reduced the cardiac inflammation [163]. However in both cases, the shRNA was administered before the viral challenge and thus, whether it can be used as a treatment after the infection is diagnosed remains unclear.
5.5 Encephalitogenic Flaviviruses
Japanese encephalitis (JE) and West Nile (WN) viruses can cause a devastating neurological illness. We have used a lentiviral vector to express shRNAs targeting conserved regions in the viral envelope genes of JEV and WNV [167]. A single intracranial treatment with the lentiviral vector was enough to provide near 100% protection from fatality induced by WNV or JEV in mice. Furthermore, pseudotyping the lentivirus with RVG instead of VSV-G enabled neuronal cell-specific delivery and significantly reduced the dose of lentivirus required for protection [54]. Moreover, targeting a sequence that is highly conserved in both JEV and WNV, protection could be obtained against either viral challenge, showing the broad-spectrum utility of this approach [167].
5.6 Prion Disease
Spongiform encephalopathy or Prion disease is a chronic and fatal neurodegenerative disease characterized by accumulation of an infectious and protease-resistant form of the cellular protein PrpC. Lentiviruses have been used to knockdown PrPC in scrapie prion infected cells in vitro as well as in vivo in mice [168, 169]. One study used PrPC shRNA encoding lentivirus to generate transgenic mice via transduction of embryonic stem cells. The PrPC levels were correspondingly reduced in the brains of transgenic animals and when infected with scrapie prion agent, their survival was more prolonged compared to control mice [168, 169]. In another interesting study, the shRNA expressing lentivirus was used as treatment several weeks after prion infection in mice. A single hypothalamic injection of the lentivirus was enough to significantly reduce the neurobehavioral changes and resulted in prolongation of survival by 25% [168, 169].
6. Summary and Future Perspectives
The field of RNA interference is moving forward with a remarkable pace and several clinical trials are being conducted to assess the safety and efficacy of this approach. Because of its ability to induce stable long-term RNAi, shRNA expression via viral vectors has received considerable attention for potential clinical use, particularly in chronic diseases. Although the therapeutic application of viral vectors suffered a set back following the occurrence of malignancies in clinical trials using oncoretroviral vectors, lentiviral vectors have been generally thought to be safer and are being tried in several clinical trials [34, 39]. Particularly heartening is the apparent lack of toxicity in a Phase I trial to express anti-sense RNA [40]. In fact, a recent ongoing Phase I trial to express a combination of shRNAs and other anti-HIV agents also appears to be going well [170]. Apart from hazards of vector integration, stable endogenous shRNA expression could also be potentially harmful because of activation of innate immunity, off-target effects as well as interference with the endogenous miRNA pathway. In addition, the shRNA could be rendered ineffective by generation of escape mutants in viral infections. In a short span of time, great progress has been made in addressing some of these issues such as the improvement in shRNA design, development of multiplex shRNAs and inducible expression systems. Despite these commendable achievements, further improvements in safety, efficacy and reliability will be required before the lentiviral RNAi-based therapies become clinically viable. Some of the possible improvements likely in the future are briefly discussed below.
With an increasing understanding of the miRNA biogenesis, it has become easier to predict the processing of lentivirally expressed shRNA precursors. To minimize off target effects, it is important to make sure that the lentivirally expressed shRNA is processed to accurately generate the intended siRNA sequence and also to make sure that only the guide strand but not the passenger strand is loaded into RISC. To insure Drosha processing, the shRNAs should be expressed within a miRNA backbone preferably under the control of a Pol II promoter. This would also avoid interference with the miRNA and generate a more effective siRNA that works at lower concentrations. Even with these precautions, since the miRNA biogenesis is still not perfectly understood, it would perhaps be beneficial to actually sequence the mature siRNAs generated in the lentivirally transduced cells. In addition to ensuring that the guide strand does not contain any vector-derived sequences, this would also make sure that the passenger strand is destroyed, thereby greatly reducing the chances of off target effects. With the advent of newer technologies for easy sequencing and genome wide proteomic analysis, it would also be an advantage to perform a combined microarray and genome-wide proteomic analysis in transduced cells to validate potential candidates before clinical trials. This would be particularly important when trying to express multiplex shRNAs because of increased risks of unintended effects.
Currently the most practical approach for lentiviral shRNA therapy appears to be reinfusion into patients of hematopoietic stem cells that have been transduced with lentiviral vectors in vitro. However, the transduction efficiencies are still rather low (10% or less) and methods to enhance this are urgently needed. Moreover, even selection of transduced cells before transfusion does not guarantee uniform expression in the progeny cells generated in vivo. One possible way to increase the transduction efficacy is to develop methods for targeted delivery using cell surface receptors.
Recent advances in gene therapy approaches also indicate that systemic treatment with letiviral shRNA vectors may be feasible. Some form of targeted delivery approach may be needed for this. In this regard, pseudotyping the lentiviral vector with the modified Sindbis virus envelope appears to be promising. Sindbis envelope with the ZZ domain appears to be particularly interesting because the delivery can be targeted to any cell type just by using a cell type-specific mAb. Although these methods have not so far been used to deliver shRNA, one could easily envisage such approaches. For example, we have recently used a single chain antibody directed to CD7 for targeted siRNA delivery to T cells as treatment for HIV infection in humanized mice [171]. Thus, conceivably CD7 scFv coating after Sindbis ZZ envelope psudotyping would also allow delivery of lentivirus encoding any shRNA specifically to T cells. A similar approach might also be used to enhance transduction of primary cells such as hematopoietic stem cells. Moreover, another level of regulation can be superimposed by using a tissue-specific promoter in addition to cell-specific targeting.
Finally, the future studies will undoubtedly concentrate on combining several elements to enhance safety and efficacy while reducing toxicities at the same time. An ideal designer shRNA lentiviral therapeutic vector would therefore be aimed at 1) expression of multiplex shRNA in a miRNA background that have been proven to be efficiently and accurately processed by Drosha and Dicer 2) achieve regulated and reversible expression using a conditional and/or tissue-specific expression system 3) targeting the delivery to appropriate cell type using psuodotyping and/or other approaches.
Acknowledgments
This work was supported by NIH grants AI071882 to PS and AI075419 to (NM)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 2.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 3.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 4.Geiss G, Jin G, Guo J, Bumgarner R, Katze MG, Sen GC. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J Biol Chem. 2001;276:30178–30182. doi: 10.1074/jbc.c100137200. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 6.Plasterk RH. RNA silencing: the genome’s immune system. Science. 2002;296:1263–1265. doi: 10.1126/science.1072148. [DOI] [PubMed] [Google Scholar]
- 7.Van den Haute C, Eggermont K, Nuttin B, Debyser Z, Baekelandt V. Lentiviral vector-mediated delivery of short hairpin RNA results in persistent knockdown of gene expression in mouse brain. Hum Gene Ther. 2003;14:1799–1807. doi: 10.1089/104303403322611809. [DOI] [PubMed] [Google Scholar]
- 8.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. Embo J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 10.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 11.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 12.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 14.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 16.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 17.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 18.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 20.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 21.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 23.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 25.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 26.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 27.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 28.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 29.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 30.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 31.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 33.Wiznerowicz M, Trono D. Harnessing HIV for therapy, basic research and biotechnology. Trends Biotechnol. 2005;23:42–47. doi: 10.1016/j.tibtech.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Cockrell AS, Kafri T. Gene delivery by lentivirus vectors. Mol Biotechnol. 2007;36:184–204. doi: 10.1007/s12033-007-0010-8. [DOI] [PubMed] [Google Scholar]
- 35.Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, Radford I, Villeval JL, Fraser CC, Cavazzana-Calvo M, Fischer A. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 36.Check E. Gene therapy put on hold as third child develops cancer. Nature. 2005;433:561. doi: 10.1038/433561a. [DOI] [PubMed] [Google Scholar]
- 37.Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, Clappier E, Caccavelli L, Delabesse E, Beldjord K, Asnafi V, MacIntyre E, Dal Cortivo L, Radford I, Brousse N, Sigaux F, Moshous D, Hauer J, Borkhardt A, Belohradsky BH, Wintergerst U, Velez MC, Leiva L, Sorensen R, Wulffraat N, Blanche S, Bushman FD, Fischer A, Cavazzana-Calvo M. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 39.Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, Cassani B, Schmidt M, von Kalle C, Howe S, Thrasher AJ, Aiuti A, Ferrari G, Recchia A, Mavilio F. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007;110:1770–1778. doi: 10.1182/blood-2007-01-068759. [DOI] [PubMed] [Google Scholar]
- 40.Levine BL, Humeau LM, Boyer J, MacGregor RR, Rebello T, Lu X, Binder GK, Slepushkin V, Lemiale F, Mascola JR, Bushman FD, Dropulic B, June CH. Gene transfer in humans using a conditionally replicating lentiviral vector. Proc Natl Acad Sci U S A. 2006;103:17372–17377. doi: 10.1073/pnas.0608138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kotsopoulou E, Kim VN, Kingsman AJ, Kingsman SM, Mitrophanous KA. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J Virol. 2000;74:4839–4852. doi: 10.1128/jvi.74.10.4839-4852.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Kappes JC, Wu X, Wakefield JK. Production of trans-lentiviral vector with predictable safety. Methods Mol Med. 2003;76:449–465. doi: 10.1385/1-59259-304-6:449. [DOI] [PubMed] [Google Scholar]
- 44.Philpott NJ, Thrasher AJ. Use of nonintegrating lentiviral vectors for gene therapy. Hum Gene Ther. 2007;18:483–489. doi: 10.1089/hum.2007.013. [DOI] [PubMed] [Google Scholar]
- 45.Themis M, Waddington SN, Schmidt M, von Kalle C, Wang Y, Al-Allaf F, Gregory LG, Nivsarkar M, Holder MV, Buckley SM, Dighe N, Ruthe AT, Mistry A, Bigger B, Rahim A, Nguyen TH, Trono D, Thrasher AJ, Coutelle C. Oncogenesis following delivery of a nonprimate lentiviral gene therapy vector to fetal and neonatal mice. Mol Ther. 2005;12:763–771. doi: 10.1016/j.ymthe.2005.07.358. [DOI] [PubMed] [Google Scholar]
- 46.Kingsman SM, Mitrophanous K, Olsen JC. Potential oncogene activity of the woodchuck hepatitis post-transcriptional regulatory element (WPRE) Gene Ther. 2005;12:3–4. doi: 10.1038/sj.gt.3302417. [DOI] [PubMed] [Google Scholar]
- 47.Schambach A, Bohne J, Baum C, Hermann FG, Egerer L, von Laer D, Giroglou T. Woodchuck hepatitis virus post-transcriptional regulatory element deleted from X protein and promoter sequences enhances retroviral vector titer and expression. Gene Ther. 2006;13:641–645. doi: 10.1038/sj.gt.3302698. [DOI] [PubMed] [Google Scholar]
- 48.Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aiken C. Pseudotyping human immunodeficiency virus type 1. HIV-1 by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J Virol. 1997;71:5871–5877. doi: 10.1128/jvi.71.8.5871-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Croyle MA, Callahan SM, Auricchio A, Schumer G, Linse KD, Wilson JM, Brunner LJ, Kobinger GP. PEGylation of a vesicular stomatitis virus G pseudotyped lentivirus vector prevents inactivation in serum. J Virol. 2004;78:912–921. doi: 10.1128/JVI.78.2.912-921.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manjunath N, Kumar P, Lee SK, Shankar P. Interfering antiviral immunity: application, subversion, hope? Trends in Immunology. 2006;27:328–335. doi: 10.1016/j.it.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cronin J, Zhang XY, Reiser J. Altering the tropism of lentiviral vectors through pseudotyping. Curr Gene Ther. 2005;5:387–398. doi: 10.2174/1566523054546224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong LF, Azzouz M, Walmsley LE, Askham Z, Wilkes FJ, Mitrophanous KA, Kingsman SM, Mazarakis ND. Transduction patterns of pseudotyped lentiviral vectors in the nervous system. Mol Ther. 2004;9:101–111. doi: 10.1016/j.ymthe.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 54.Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL, Lee SK, Shankar P, Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 55.Morizono K, Bristol G, Xie YM, Kung SK, Chen IS. Antibody-directed targeting of retroviral vectors via cell surface antigens. J Virol. 2001;75:8016–8020. doi: 10.1128/JVI.75.17.8016-8020.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morizono K, Xie Y, Ringpis GE, Johnson M, Nassanian H, Lee B, Wu L, Chen IS. Lentiviral vector retargeting to P-glycoprotein on metastatic melanoma through intravenous injection. Nat Med. 2005;11:346–352. doi: 10.1038/nm1192. [DOI] [PubMed] [Google Scholar]
- 57.Pariente N, Mao SH, Morizono K, Chen IS. Efficient targeted transduction of primary human endothelial cells with dual-targeted lentiviral vectors. J Gene Med. 2008;10:242–248. doi: 10.1002/jgm.1151. [DOI] [PubMed] [Google Scholar]
- 58.Pariente N, Morizono K, Virk MS, Petrigliano FA, Reiter RE, Lieberman JR, Chen IS. A novel dual-targeted lentiviral vector leads to specific transduction of prostate cancer bone metastases in vivo after systemic administration. Mol Ther. 2007;15:1973–1981. doi: 10.1038/sj.mt.6300271. [DOI] [PubMed] [Google Scholar]
- 59.Ziegler L, Yang L, Joo K, Yang H, Baltimore D, Wang P. Targeting lentiviral vectors to antigen-specific immunoglobulins. Hum Gene Ther. 2008;19:861–872. doi: 10.1089/hum.2007.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang L, Yang H, Rideout K, Cho T, Joo KI, Ziegler L, Elliot A, Walls A, Yu D, Baltimore D, Wang P. Engineered lentivector targeting of dendritic cells for in vivo immunization. Nat Biotechnol. 2008;26:326–334. doi: 10.1038/nbt1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sioud M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J Mol Biol. 2005;348:1079–1090. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 62.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, Endres S, Hartmann G. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 63.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 64.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 65.Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. Rna. 2005;11:220–226. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boudreau RL, Monteys AM, Davidson BL. Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs. Rna. 2008;14:1834–1844. doi: 10.1261/rna.1062908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.An DS, Qin FX, Auyeung VC, Mao SH, Kung SK, Baltimore D, Chen IS. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol Ther. 2006;14:494–504. doi: 10.1016/j.ymthe.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 69.Mittal V. Improving the efficiency of RNA interference in mammals. Nat Rev Genet. 2004;5:355–365. doi: 10.1038/nrg1323. [DOI] [PubMed] [Google Scholar]
- 70.Kim DH, Rossi JJ. Coupling of RNAi-mediated target downregulation with gene replacement. Antisense Nucleic Acid Drug Dev. 2003;13:151–155. doi: 10.1089/108729003768247619. [DOI] [PubMed] [Google Scholar]
- 71.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 72.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 73.Scherer LJ, Frank R, Rossi JJ. Optimization and characterization of tRNA-shRNA expression constructs. Nucleic Acids Res. 2007;35:2620–2628. doi: 10.1093/nar/gkm103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. Embo J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 76.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 78.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 79.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. Embo J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: microRNA-based shRNA libraries. Nat Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- 81.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G, Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 82.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 84.Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5′ precision of both MicroRNAs and their miRNA strands in flies. Curr Biol. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA Profiling of Naive, Effector and Memory CD8 T Cells. PLoS ONE. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Piskounova E, Viswanathan SR, Janas M, Lapierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008 doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 87.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. Rna. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, Carter BJ, Davidson BL. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boden D, Pusch O, Lee F, Tucker L, Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, Berkhout B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Akashi H, Miyagishi M, Yokota T, Watanabe T, Hino T, Nishina K, Kohara M, Taira K. Escape from the interferon response associated with RNA interference using vectors that encode long modified hairpin-RNA. Mol Biosyst. 2005;1:382–390. doi: 10.1039/b510159j. [DOI] [PubMed] [Google Scholar]
- 94.Konstantinova P, ter Brake O, Haasnoot J, de Haan P, Berkhout B. Trans-inhibition of HIV-1 by a long hairpin RNA expressed within the viral genome. Retrovirology. 2007;4:15. doi: 10.1186/1742-4690-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sano M, Li H, Nakanishi M, Rossi JJ. Expression of long anti-HIV-1 hairpin RNAs for the generation of multiple siRNAs: advantages and limitations. Mol Ther. 2008;16:170–177. doi: 10.1038/sj.mt.6300298. [DOI] [PubMed] [Google Scholar]
- 96.Watanabe T, Sudoh M, Miyagishi M, Akashi H, Arai M, Inoue K, Taira K, Yoshiba M, Kohara M. Intracellular-diced dsRNA has enhanced efficacy for silencing HCV RNA and overcomes variation in the viral genotype. Gene Ther. 2006;13:883–892. doi: 10.1038/sj.gt.3302734. [DOI] [PubMed] [Google Scholar]
- 97.Weinberg MS, Ely A, Barichievy S, Crowther C, Mufamadi S, Carmona S, Arbuthnot P. Specific inhibition of HBV replication in vitro and in vivo with expressed long hairpin RNA. Mol Ther. 2007;15:534–541. doi: 10.1038/sj.mt.6300077. [DOI] [PubMed] [Google Scholar]
- 98.Henry SD, van der Wegen P, Metselaar HJ, Tilanus HW, Scholte BJ, van der Laan LJ. Simultaneous targeting of HCV replication and viral binding with a single lentiviral vector containing multiple RNA interference expression cassettes. Mol Ther. 2006;14:485–493. doi: 10.1016/j.ymthe.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 99.Anderson J, Akkina R. CXCR4 and CCR5 shRNA transgenic CD34+ cell derived macrophages are functionally normal and resist HIV-1 infection. Retrovirology. 2005;2:53. doi: 10.1186/1742-4690-2-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.ter Brake O, Hooft Kt, Liu YP, Centlivre M, von Eije KJ, Berkhout B. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol Ther. 2008;16:557–564. doi: 10.1038/sj.mt.6300382. [DOI] [PubMed] [Google Scholar]
- 101.Gou D, Weng T, Wang Y, Wang Z, Zhang H, Gao L, Chen Z, Wang P, Liu L. A novel approach for the construction of multiple shRNA expression vectors. J Gene Med. 2007;9:751–763. doi: 10.1002/jgm.1080. [DOI] [PubMed] [Google Scholar]
- 102.Li MJ, Kim J, Li S, Zaia J, Yee JK, Anderson J, Akkina R, Rossi JJ. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- 103.Anderson J, Li MJ, Palmer B, Remling L, Li S, Yam P, Yee JK, Rossi J, Zaia J, Akkina R. Safety and efficacy of a lentiviral vector containing three anti-HIV genes--CCR5 ribozyme, tat-rev siRNA, and TAR decoy--in SCID-hu mouse-derived T cells. Mol Ther. 2007;15:1182–1188. doi: 10.1038/sj.mt.6300157. [DOI] [PubMed] [Google Scholar]
- 104.Liu YP, Haasnoot J, ter Brake O, Berkhout B, Konstantinova P. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008;36:2811–2824. doi: 10.1093/nar/gkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aagaard LA, Zhang J, von Eije KJ, Li H, Saetrom P, Amarzguioui M, Rossi JJ. Engineering and optimization of the miR-106b cluster for ectopic expression of multiplexed anti-HIV RNAs. Gene Ther. 2008;15:1536–1549. doi: 10.1038/gt.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wiznerowicz M, Szulc J, Trono D. Tuning silence: conditional systems for RNA interference. Nat Methods. 2006;3:682–688. doi: 10.1038/nmeth914. [DOI] [PubMed] [Google Scholar]
- 107.Pluta K, Diehl W, Zhang XY, Kutner R, Bialkowska A, Reiser J. Lentiviral vectors encoding tetracycline-dependent repressors and transactivators for reversible knockdown of gene expression: a comparative study. BMC Biotechnol. 2007;7:41. doi: 10.1186/1472-6750-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Amar L, Desclaux M, Faucon-Biguet N, Mallet J, Vogel R. Control of small inhibitory RNA levels and RNA interference by doxycycline induced activation of a minimal RNA polymerase III promoter. Nucleic Acids Res. 2006;34:e37. doi: 10.1093/nar/gkl034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Szulc J, Wiznerowicz M, Sauvain MO, Trono D, Aebischer P. A versatile tool for conditional gene expression and knockdown. Nat Methods. 2006;3:109–116. doi: 10.1038/nmeth846. [DOI] [PubMed] [Google Scholar]
- 110.Miyagishi M, Sumimoto H, Miyoshi H, Kawakami Y, Taira K. Optimization of an siRNA-expression system with an improved hairpin and its significant suppressive effects in mammalian cells. J Gene Med. 2004;6:715–723. doi: 10.1002/jgm.556. [DOI] [PubMed] [Google Scholar]
- 111.Gupta S, Schoer RA, Egan JE, Hannon GJ, Mittal V. Inducible, reversible, and stable RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:1927–1932. doi: 10.1073/pnas.0306111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI, Rebres R, Roach T, Seaman W, Simon MI, Fraser ID. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc Natl Acad Sci U S A. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Aagaard L, Amarzguioui M, Sun G, Santos LC, Ehsani A, Prydz H, Rossi JJ. A facile lentiviral vector system for expression of doxycycline-inducible shRNAs: knockdown of the pre-miRNA processing enzyme Drosha. Mol Ther. 2007;15:938–945. doi: 10.1038/sj.mt.6300118. [DOI] [PubMed] [Google Scholar]
- 115.Herold MJ, van den Brandt J, Seibler J, Reichardt HM. Inducible and reversible gene silencing by stable integration of an shRNA-encoding lentivirus in transgenic rats. Proc Natl Acad Sci U S A. 2008;105:18507–18512. doi: 10.1073/pnas.0806213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Unwalla HJ, Li MJ, Kim JD, Li HT, Ehsani A, Alluin J, Rossi JJ. Negative feedback inhibition of HIV-1 by TAT-inducible expression of siRNA. Nat Biotechnol. 2004;22:1573–1578. doi: 10.1038/nbt1040. [DOI] [PubMed] [Google Scholar]
- 117.Westerhout EM, Vink M, Haasnoot PC, Das AT, Berkhout B. A conditionally replicating HIV-based vector that stably expresses an antiviral shRNA against HIV-1 replication. Mol Ther. 2006;14:268–275. doi: 10.1016/j.ymthe.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 118.Shukla V, Coumoul X, Deng CX. RNAi-based conditional gene knockdown in mice using a U6 promoter driven vector. Int J Biol Sci. 2007;3:91–99. doi: 10.7150/ijbs.3.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yu J, McMahon AP. Reproducible and inducible knockdown of gene expression in mice. Genesis. 2006;44:252–261. doi: 10.1002/dvg.20213. [DOI] [PubMed] [Google Scholar]
- 120.Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Iovino N, Denti MA, Bozzoni I, Cortese R. A loxP-containing pol II promoter for RNA interference is reversibly regulated by Cre recombinase. RNA Biol. 2005;2:86–92. doi: 10.4161/rna.2.3.2045. [DOI] [PubMed] [Google Scholar]
- 122.Hitz C, Wurst W, Kuhn R. Conditional brain-specific knockdown of MAPK using Cre/loxP regulated RNA interference. Nucleic Acids Res. 2007;35:e90. doi: 10.1093/nar/gkm475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kasim V, Miyagishi M, Taira K. Control of siRNA expression using the Cre-loxP recombination system. Nucleic Acids Res. 2004;32:e66. doi: 10.1093/nar/gnh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fritsch L, Martinez LA, Sekhri R, Naguibneva I, Gerard M, Vandromme M, Schaeffer L, Harel-Bellan A. Conditional gene knock-down by CRE-dependent short interfering RNAs. EMBO Rep. 2004;5:178–182. doi: 10.1038/sj.embor.7400064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Steuber-Buchberger P, Wurst W, Kuhn R. Simultaneous Cre-mediated conditional knockdown of two genes in mice. Genesis. 2008;46:144–151. doi: 10.1002/dvg.20376. [DOI] [PubMed] [Google Scholar]
- 126.Stern P, Astrof S, Erkeland SJ, Schustak J, Sharp PA, Hynes RO. A system for Cre-regulated RNA interference in vivo. Proc Natl Acad Sci U S A. 2008;105:13895–13900. doi: 10.1073/pnas.0806907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dykxhoorn DM, Chowdhury D, Lieberman J. RNA interference and cancer: endogenous pathways and therapeutic approaches. Adv Exp Med Biol. 2008;615:299–329. doi: 10.1007/978-1-4020-6554-5_14. [DOI] [PubMed] [Google Scholar]
- 128.Sumimoto H, Kawakami Y. Lentiviral vector-mediated RNAi and its use for cancer research. Future Oncol. 2007;3:655–664. doi: 10.2217/14796694.3.6.655. [DOI] [PubMed] [Google Scholar]
- 129.Putral LN, Gu W, McMillan NA. RNA interference for the treatment of cancer. Drug News Perspect. 2006;19:317–324. doi: 10.1358/dnp.2006.19.6.985937. [DOI] [PubMed] [Google Scholar]
- 130.Farah MH. RNAi silencing in mouse models of neurodegenerative diseases. Curr Drug Deliv. 2007;4:161–167. doi: 10.2174/156720107780362276. [DOI] [PubMed] [Google Scholar]
- 131.Boudreau RL, Davidson BL. RNAi therapy for neurodegenerative diseases. Curr Top Dev Biol. 2006;75:73–92. doi: 10.1016/S0070-2153(06)75003-7. [DOI] [PubMed] [Google Scholar]
- 132.Wong LF, Goodhead L, Prat C, Mitrophanous KA, Kingsman SM, Mazarakis ND. Lentivirus-mediated gene transfer to the central nervous system: therapeutic and research applications. Hum Gene Ther. 2006;17:1–9. doi: 10.1089/hum.2006.17.1. [DOI] [PubMed] [Google Scholar]
- 133.ter Brake O, Berkhout B. Lentiviral vectors that carry anti-HIV shRNAs: problems and solutions. J Gene Med. 2007;9:743–750. doi: 10.1002/jgm.1078. [DOI] [PubMed] [Google Scholar]
- 134.Capodici J, Kariko K, Weissman D. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J Immunol. 2002;169:5196–5201. doi: 10.4049/jimmunol.169.9.5196. [DOI] [PubMed] [Google Scholar]
- 135.Hu WY, Myers CP, Kilzer JM, Pfaff SL, Bushman FD. Inhibition of retroviral pathogenesis by RNA interference. Curr Biol. 2002;12:1301–1311. doi: 10.1016/s0960-9822(02)00975-2. [DOI] [PubMed] [Google Scholar]
- 136.Surabhi RM, Gaynor RB. RNA interference directed against viral and cellular targets inhibits human immunodeficiency Virus Type 1 replication. J Virol. 2002;76:12963–12973. doi: 10.1128/JVI.76.24.12963-12973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stebbing J, Moyle G. The clades of HIV: their origins and clinical significance. AIDS Rev 5. 2003:205–213. [PubMed] [Google Scholar]
- 138.Bello G, Casado C, Garcia S, Rodriguez C, del Romero J, Lopez-Galindez C. Co-existence of recent and ancestral nucleotide sequences in viral quasispecies of human immunodeficiency virus type 1 patients. J Gen Virol. 2004;85:399–407. doi: 10.1099/vir.0.19365-0. [DOI] [PubMed] [Google Scholar]
- 139.Ahumada-Ruiz S, Casado C, Toala-Gonzalez I, Flores-Figueroa D, Rodriguez-French A, Lopez-Galindez C. High divergence within the major HIV type 1 subtype B epidemic in Panama. AIDS Res Hum Retroviruses. 2008;24:1461–1466. doi: 10.1089/aid.2008.0153. [DOI] [PubMed] [Google Scholar]
- 140.von Eije KJ, ter Brake O, Berkhout B. Human immunodeficiency virus type 1 escape is restricted when conserved genome sequences are targeted by RNA interference. J Virol. 2008;82:2895–2903. doi: 10.1128/JVI.02035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chang LJ, Liu X, He J. Lentiviral siRNAs targeting multiple highly conserved RNA sequences of human immunodeficiency virus type 1. Gene Ther. 2005;12:1133–1144. doi: 10.1038/sj.gt.3302509. [DOI] [PubMed] [Google Scholar]
- 142.Han W, Wind-Rotolo M, Kirkman RL, Morrow CD. Inhibition of human immunodeficiency virus type 1 replication by siRNA targeted to the highly conserved primer binding site. Virology. 2004;330:221–232. doi: 10.1016/j.virol.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 143.Dave RS, Pomerantz RJ. Antiviral effects of human immunodeficiency virus type 1-specific small interfering RNAs against targets conserved in select neurotropic viral strains. J Virol. 2004;78:13687–13696. doi: 10.1128/JVI.78.24.13687-13696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lee SK, Dykxhoorn DM, Kumar P, Ranjbar S, Song E, Maliszewski LE, Francois-Bongarcon V, Goldfeld A, Swamy NM, Lieberman J, Shankar P. Lentiviral delivery of short hairpin RNAs protects CD4 T cells from multiple clades and primary isolates of HIV. Blood. 2005;106:818–826. doi: 10.1182/blood-2004-10-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth RJ, Collman RG, Doms RW, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 146.Huang Y, Paxton WA, Wolinsky SM, Neumann AU, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau NR, Phair J, Ho DD, Koup RA. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 147.Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci U S A. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lee MT, Coburn GA, McClure MO, Cullen BR. Inhibition of human immunodeficiency virus type 1 replication in primary macrophages by using Tat- or CCR5-specific small interfering RNAs expressed from a lentivirus vector. J Virol. 2003;77:11964–11972. doi: 10.1128/JVI.77.22.11964-11972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Butticaz C, Ciuffi A, Munoz M, Thomas J, Bridge A, Pebernard S, Iggo R, Meylan P, Telenti A. Protection from HIV-1 infection of primary CD4 T cells by CCR5 silencing is effective for the full spectrum of CCR5 expression. Antivir Ther. 2003;8:373–377. [PubMed] [Google Scholar]
- 150.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 151.Lo HL, Chang T, Yam P, Marcovecchio PM, Li S, Zaia JA, Yee JK. Inhibition of HIV-1 replication with designed miRNAs expressed from RNA polymerase II promoters. Gene Ther. 2007;14:1503–1512. doi: 10.1038/sj.gt.3303011. [DOI] [PubMed] [Google Scholar]
- 152.Strayer DS, Akkina R, Bunnell BA, Dropulic B, Planelles V, Pomerantz RJ, Rossi JJ, Zaia JA. Current status of gene therapy strategies to treat HIV/AIDS. Mol Ther. 2005;11:823–842. doi: 10.1016/j.ymthe.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 153.Banerjea A, Li MJ, Bauer G, Remling L, Lee NS, Rossi J, Akkina R. Inhibition of HIV-1 by lentiviral vector-transduced siRNAs in T lymphocytes differentiated in SCID-hu mice and CD34+ progenitor cell-derived macrophages. Mol Ther. 2003;8:62–71. doi: 10.1016/s1525-0016(03)00140-0. [DOI] [PubMed] [Google Scholar]
- 154.Li M, Rossi JJ. Lentiviral vector delivery of siRNA and shRNA encoding genes into cultured and primary hematopoietic cells. Methods Mol Biol. 2005;309:261–272. doi: 10.1385/1-59259-935-4:261. [DOI] [PubMed] [Google Scholar]
- 155.An DS, Donahue RE, Kamata M, Poon B, Metzger M, Mao SH, Bonifacino A, Krouse AE, Darlix JL, Baltimore D, Qin FX, Chen IS. Stable reduction of CCR5 by RNAi through hematopoietic stem cell transplant in non-human primates. Proc Natl Acad Sci U S A. 2007;104:13110–13115. doi: 10.1073/pnas.0705474104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Arbuthnot P, Thompson LJ. Harnessing the RNA interference pathway to advance treatment and prevention of hepatocellular carcinoma. World J Gastroenterol. 2008;14:1670–1681. doi: 10.3748/wjg.14.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen Y, Cheng G, Mahato RI. RNAi for treating hepatitis B viral infection. Pharm Res. 2008;25:72–86. doi: 10.1007/s11095-007-9504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ma Y, Chan CY, He ML. RNA interference and antiviral therapy. World J Gastroenterol. 2007;13:5169–5179. doi: 10.3748/wjg.v13.i39.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gu W, Putral L, McMillan N. siRNA and shRNA as anticancer agents in a cervical cancer model. Methods Mol Biol. 2008;442:159–172. doi: 10.1007/978-1-59745-191-8_12. [DOI] [PubMed] [Google Scholar]
- 160.Gu W, Putral L, Hengst K, Minto K, Saunders NA, Leggatt G, McMillan NA. Inhibition of cervical cancer cell growth in vitro and in vivo with lentiviral-vector delivered short hairpin RNA targeting human papillomavirus E6 and E7 oncogenes. Cancer Gene Ther. 2006;13:1023–1032. doi: 10.1038/sj.cgt.7700971. [DOI] [PubMed] [Google Scholar]
- 161.Bai L, Wei L, Wang J, Li X, He P. Extended effects of human papillomavirus 16 E6-specific short hairpin RNA on cervical carcinoma cells. Int J Gynecol Cancer. 2006;16:718–729. doi: 10.1111/j.1525-1438.2006.00380.x. [DOI] [PubMed] [Google Scholar]
- 162.Putral LN, Bywater MJ, Gu W, Saunders NA, Gabrielli BG, Leggatt GR, McMillan NA. RNA interference against human papillomavirus oncogenes in cervical cancer cells results in increased sensitivity to cisplatin. Mol Pharmacol. 2005;68:1311–1319. doi: 10.1124/mol.105.014191. [DOI] [PubMed] [Google Scholar]
- 163.Fechner H, Sipo I, Westermann D, Pinkert S, Wang X, Suckau L, Kurreck J, Zeichhardt H, Muller O, Vetter R, Erdmann V, Tschope C, Poller W. Cardiac-targeted RNA interference mediated by an AAV9 vector improves cardiac function in coxsackievirus B3 cardiomyopathy. J Mol Med. 2008;86:987–997. doi: 10.1007/s00109-008-0363-x. [DOI] [PubMed] [Google Scholar]
- 164.Kim YJ, Ahn J, Jeung SY, Kim DS, Na HN, Cho YJ, Yun SH, Jee Y, Jeon ES, Lee H, Nam JH. Recombinant lentivirus-delivered short hairpin RNAs targeted to conserved coxsackievirus sequences protect against viral myocarditis and improve survival rate in an animal model. Virus Genes. 2008;36:141–146. doi: 10.1007/s11262-007-0192-y. [DOI] [PubMed] [Google Scholar]
- 165.Fechner H, Pinkert S, Wang X, Sipo I, Suckau L, Kurreck J, Dorner A, Sollerbrant K, Zeichhardt H, Grunert HP, Vetter R, Schultheiss HP, Poller W. Coxsackievirus B3 and adenovirus infections of cardiac cells are efficiently inhibited by vector-mediated RNA interference targeting their common receptor. Gene Ther. 2007;14:960–971. doi: 10.1038/sj.gt.3302948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kim JY, Chung SK, Hwang HY, Kim H, Kim JH, Nam JH, Park SI. Expression of short hairpin RNAs against the coxsackievirus B3 exerts potential antiviral effects in Cos-7 cells and in mice. Virus Res. 2007;125:9–13. doi: 10.1016/j.virusres.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 167.Kumar P, Lee SK, Shankar P, Manjunath N. A single siRNA suppresses fatal encephalitis induced by two different flaviviruses. PLoS Med. 2006;3:e96. doi: 10.1371/journal.pmed.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.White MD, Farmer M, Mirabile I, Brandner S, Collinge J, Mallucci GR. Single treatment with RNAi against prion protein rescues early neuronal dysfunction and prolongs survival in mice with prion disease. Proc Natl Acad Sci U S A. 2008;105:10238–10243. doi: 10.1073/pnas.0802759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pfeifer A, Eigenbrod S, Al-Khadra S, Hofmann A, Mitteregger G, Moser M, Bertsch U, Kretzschmar H. Lentivector-mediated RNAi efficiently suppresses prion protein and prolongs survival of scrapie-infected mice. J Clin Invest. 2006;116:3204–3210. doi: 10.1172/JCI29236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL, Laouar A, Yao J, Haridas V, Habiro K, Yang YG, Jeong JH, Lee KY, Kim YH, Kim SW, Peipp M, Fey GH, Manjunath N, Shultz LD, Lee SK, Shankar P. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]