Abstract
Culex pipiens, the mosquito that vectors West Nile virus in North America, overwinters in an adult diapause (dormancy) that is programmed by the short day length and low temperatures of autumn. In response to these environmental signals, females cease feeding on blood and instead seek sources of nectar used to generate the huge lipid reserves required for winter survival. To identify regulatory networks that regulate fat accumulation and fat consumption during diapause, we compared expression of fat-related genes from nondiapausing females with expression of those same genes in early and late diapause and at diapause termination. Among the 31 genes we examined, 4 were expressed more highly in early diapause than in nondiapause, while 14 genes were downregulated in early diapause. In the transition from early to late diapause, 19 genes related to fat metabolism were upregulated. As reported previously, fatty acid synthase, identified as fas-1 in this study, was upregulated in early diapause. Numerous fat metabolism genes, including multiple kinetic classes and genes involved in β-oxidation, an energy-generation step, were suppressed in early diapause but were highly expressed in late diapause and at diapause termination. RNA interference (RNAi) analysis revealed that the fas-1 gene and others (fas-3 and fabp) have important roles in fat storage during early diapause. When expression of these genes is suppressed, female mosquitoes fail to sequester the lipids needed for overwintering.
Keywords: diapause, RNA interference, lipid metabolism
in late summer and early autumn, the mosquito Culex pipiens, like many other temperate insect species, responds to short day length and decreasing temperatures by entering an overwintering adult diapause (16). Diapause-programmed females do not seek blood meals but instead feed on nectar and other sugar sources (13) that are converted into lipid stores used as the energy source for survival during the long winter diapause (24). Thus fat storage in early diapause and regulation of its utilization are critical features for successful overwintering.
Insulin signaling appears to be a key component of the cascade regulating diapause in C. pipiens (17, 18). The insulin signaling pathway is not activated in diapausing females, thus lifting suppression of a forkhead transcriptional factor (FOXO). This, in turn, leads to induction of target genes associated with the diapause syndrome, including lipid accumulation (17). Concurrently, the key genes encoding enzymes needed to digest a blood meal (trypsin and a chymotrypsin-like protease) are repressed in diapause-destined females, while a gene associated with accumulation of lipid reserves (fatty acid synthase) is highly induced (15). These observations underscore the presence of a molecular switch in early diapause that exclusively directs mosquito metabolism toward lipid synthesis and sequestration. Later, when the mosquitoes have consumed their glycogen energy sources (after the first month of diapause), the energy demands are fueled from lipid stores (24).
In this study we exploited a genomics approach to elucidate the lipid metabolic pathways utilized by C. pipiens during diapause. The near completion of the Culex genome project has provided the needed resources to probe modulation of lipid metabolism genes in association with diapause and to develop functional assays using RNA interference (RNAi). Our approach is based on a priori knowledge of lipid metabolism pathways, including fatty acid synthesis, lipolysis, β-oxidation, desaturation, fatty acid binding, and transport. Using two genome databases, Vectorbase and Kyoto Encyclopedia of Genes and Genomes (KEGG), we identified putative genes in multiple kinetic classes of lipid metabolism and compared expression levels of these genes in nondiapausing females and in females representing early and late diapause, as well as diapause termination. RNAi was then used to determine which genes are most important for converting free fatty acids into the triacylglycerides that are stored in early diapause.
MATERIALS AND METHODS
Insect rearing.
The stock colony of C. pipiens (Buckeye strain) was reared at 25°C and 75% relative humidity (RH) under a 15:9-h light-dark (L:D) photoperiod as previously described (15). When larvae reached the second instar, rearing containers were placed under one of two environmental conditions: nondiapausing adults were generated by rearing at 18°C, 75% RH, and 15:9 L:D; to induce diapause mosquitoes were reared at 18°C, 75% RH, and 9:15 L:D. To confirm diapause status, the stage of ovarian development was determined according to methods described previously (5). To generate three biologically independent samples, three batches of 15 mosquitoes were collected from each of the following developmental stages: nondiapausing (ND) and early diapausing (ED) female adults, both collected 1 wk after eclosion; late diapausing (LD) females sampled 2 mo after eclosion; and females that had broken diapause (BD), collected 2 wk after females were transferred from diapausing conditions to 25°C, 75% RH, and 15:9 L:D.
Identification and bioinformatic analysis of fatty acid metabolic genes.
To retrieve sequences of fatty acid metabolism genes in C. pipiens, sequences of fatty acid metabolism genes in Drosophila melanogaster were identified with the KEGG pathway database (www.genome.ad.jp/kegg/pathway.html); those sequences were then used for discontinuous MEGA-BLAST searches on trace archives of genome data from the NCBI database (http://www.ncbi.nlm.nih.gov/blast/tracemb.shtml). RT-PCR was used to clone the putative genes related to fatty acid metabolism in C. pipiens. Total RNA samples were extracted with TRIzol (Invitrogen). To remove genomic DNA contamination, RNA samples were treated with 1.0 μl of DNase I according to the manufacturer's instructions (50–375 U/μl; Invitrogen). For reverse transcription, 5 μg of total RNA was reverse transcribed with SuperScript III RNase H-reverse transcriptase (Invitrogen). Single-stranded cDNAs of different dilutions were amplified by PCR using recombinant Taq DNA polymerase (Invitrogen). Identities of PCR products were confirmed by performing BLASTN searches against the vectorbase (http://cpipiens.vectorbase.org/Tools/BLAST/).
Transcript levels determined by quantitative RT-PCR.
To compare transcript levels of the fat metabolism genes, total RNA samples were extracted with TRIzol, genomic DNA contamination was removed, and reverse transcription was performed as described above. Quantitative RT-PCR (qRT-PCR) was performed with an iQ5 real-time PCR detection system (Bio-Rad, Hercules, CA), as described previously (19, 20). To select appropriate qRT-PCR primer sets, primers were selected only if the amplification efficiency was >0.99 and <1.01 and only if the dissociation curve of each primer set yielded a single peak. The standard curves were generated for each transcript tested with 10-fold serial dilutions of plasmids that included fragments of the fat metabolism genes, ranging from 100 to 0.01 pg per reaction. These curves, based on linear regression, were generated with plasmid input transformed by natural logarithm as the dependent variable and threshold cycle (Ct) numbers as the independent variable. Ct numbers from cDNA samples were then used in the linear regressions to estimate transcript levels for each fat-related gene.
All reactions were performed in triplicate in a total volume of 20 μl containing 10 μl of SYBR Green PCR Master Mix (Bio-Rad) and 300 nmol of each primer under the following conditions: 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 15 s, annealing at 55°C for 30 s, and extension at 72°C for 30s. 28S large-subunit ribosomal RNA (28S) and ribosomal protein large-subunit 19 (RpL19) were used as internal controls. The geometric averages of transcript levels of the internal controls were used to normalize transcript levels of each fat-related gene. Statistical significance of differences in transcript levels was then determined with a Student's t-test between the relative transcript values of ND vs. ED, LD, and BD samples, using three biologically independent replicates for each gene. A P value <0.05 was considered to be a significant transcript level change. Sequences of gene-specific qRT-PCR primer sets are listed in Supplemental Table S1.1
Double-stranded RNA preparation and injection into adult female mosquitoes.
Double-stranded RNA (dsRNA) for putative C. pipiens fas-1 (dsFAS-1), fas-3 (dsFAS-3), fabp (dsFABP), fad-1 (dsFAD-1), and β-galactosidase (dsβ-gal) genes were prepared with the MEGAscript T7 transcription kit (Ambion, Austin, TX), as previously described (19), and T7 primers are listed accordingly (5′-3′): dsFAS-1, TAATACGACTCACTATAGGGTCCTACAGCAGCCAAGTCCT and TAATACGACTCACTATAGGGAGACCGGCACTGTCAGAGTT; dsFAS-3, TAATACGACTCACTATAGGGGGAATGGCCAACTCTGTGAT and TAATACGACTCACTATAGGGAACATTGCTACTCCGGTTGG; dsFABP, TAATACGACTCACTATAGGGGTCAAGGAGGGCGATGAGTA and TAATACGACTCACTATAGGGCGGTAAACTCCCGGATGAT; dsFAD-1, TAATACGACTCACTATAGGGGACAAAAGCTCCACCAAGGA and TAATACGACTCACTATAGGGACGGATCCAGTCGTACAAGG; dsβ-gal, TAATACGACTCACTATAGGGGTCGCCAGCGGCACCGCGCCTTTC and TAATACGACTCACTATAGGGCCGGTAGCCAGCGCGGATCATCGG. Each PCR-derived fragment was sequenced and blasted against the C. pipiens genome database (http://cpipiens.vectorbase.org/Tools/BLAST/) to validate redundancy of the sequence and to confirm unique sequences. For knockdown experiments, ∼0.7 μl of dsFAS-1, dsFAS-3, dsFABP, dsFAD-1 (1.5 μg/μl), or dsβ-gal (control, 1.5 μg/μl) was injected into the thorax of 3-day-old, cold-anesthetized females with a microinjector (Tritech Research, Los Angeles, CA). Thus all adult females were injected with ∼1.0 μg of the ds construct.
RNAi efficiency evaluation with qRT-PCR.
qRT-PCR of the dsRNA-injected mosquitoes was carried out as above and as previously described (17) to evaluate RNAi efficiency; primers were used to amplify endogenous putative fas-1, fas-3, fabp, and fad-1. Ribosomal protein large subunit 19 (RpL19) from C. pipiens was used as a loading control.
Lipid assay after dsRNA of fat metabolism genes.
Diapausing females injected with the ds constructs were provided a 10% sucrose solution, and lipid levels were measured 5 and 10 days after dsRNA injection with a slight modification of an assay previously described (23). Each mosquito was placed in a 2.0-ml tube, homogenized in 500 μl of chloroform-methanol (1:1), and centrifuged. The supernatant was transferred and placed in a 90°C incubator to evaporate the solvent. After 1 h, 0.2 ml of sulfuric acid was added and the sample was again heated for 10 min. After cooling, 5 ml of vanillin reagent (600 mg vanillin, 100 ml hot water, and 400 ml 85% phosphoric acid) was added and mixed for 5 min. Samples were read directly in a spectrophotometer at 490 nm.
RESULTS AND DISCUSSION
Overview of fat metabolism during diapause in C. pipiens.
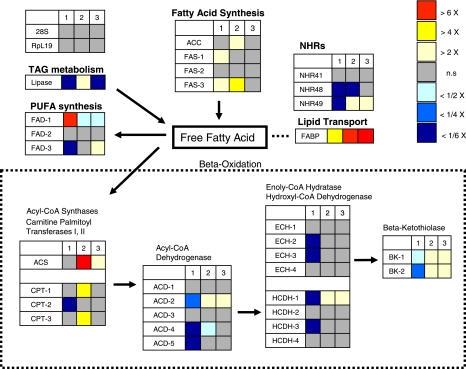
Using a comparative genomics approach and the Vectorbase and KEGG database, we identified 31 putative fat-related genes having putative functions in fatty acid synthesis, lipolysis, β-oxidation, desaturation, fatty acid binding, and transport. We measured transcript levels in early ND, ED, LD, and BD females. By comparison to nondiapausing females of the same age, four genes were significantly upregulated during fat accumulation in early diapause and 14 genes were downregulated in early diapause. The upregulated genes were involved in fatty acid synthesis, and the downregulated genes were mostly involved in β-oxidation. By contrast, in late diapause, genes involved in β-oxidation were upregulated while the genes involved in fat accumulation that were highly expressed in early diapause were downregulated. At diapause termination transcript levels of some genes involved in fatty acid synthesis, fatty acid binding, and lipolysis were downregulated, but the expression pattern at that developmental stage differed little from the pattern observed in late diapause.
The fact that fat metabolism genes in the diapausing and nondiapausing female mosquitoes displayed different transcript profiles suggests that these genes may be regulated by distinct energetic needs, nutritional influx, or deprivation. Because the genes of β-oxidation were strongly suppressed during fat accumulation in early diapause, these genes are likely responding to an enhancement of fatty acid catabolism, and eventually ATP production, as an energy source. By contrast, in late diapause, after completing a period of intense sugar feeding and sequestration of fat reserves, the females start to utilize their triacylglyceride stores, convert stored fat to free fatty acids, and transport these fatty acids to mitochondria or peroxisomes to meet their energetic needs (ATP) through β-oxidation. Because glucose persists in the diapausing mosquito only for the first month of overwintering (24), glucose deprivation may provide a cue to initiate lipolysis of stored fat and eventually to induce the β-oxidation genes, leading to energy production.
Upregulation of genes involved in fatty acid synthesis in early diapause.
Two of the four putative fatty acid synthesis genes were significantly upregulated (>2-fold) in early diapause compared with nondiapause (Table 1 and Fig. 1): both were putative fatty acid synthase (FAS) genes (fas-1 and fas-3). FAS enzymes are large, multifunctional polypeptides with seven functional domains required for fatty acid synthesis from acetyl-CoA and malonyl-CoA (12). Thus the significant upregulation of putative fas-1 (2.2-fold up) and putative fas-3 (3.7-fold up) in early diapause suggests that fat accumulation is induced by these genes involved in fatty acid synthesis. A higher transcript level of putative fas-1 was also observed in diapausing females by Northern blot analysis (15). This evidence is consistent with the observation that C. pipiens switches from blood feeding to sugar feeding as a component of the diapause program and more than doubles its lipid reserves compared with nondiapausing females after adult eclosion. Recently, we showed (17) that storage of lipid reserves in fat body cells can be inhibited by using RNAi targeted to a forkhead transcriptional factor (FOXO). FOXO is normally activated by suppression of insulin signaling; thus FOXO may be involved in increasing transcript levels of the genes in fatty acid synthesis, as observed in early diapausing females. The transcriptional regulation of the putative fas-1 and -3 genes, however, remains to be elucidated.
Table 1.
Fat metabolism genes and expression in nondiapausing females of Culex pipiens and females in early diapause, in late diapause, and after diapause was broken
| Putative Gene ID | C. pipiens Gene ID | Accession No. | Putative Physiological Process (Homology, BlastN <E−6) | Average Fold Changes (SE) |
||||
|---|---|---|---|---|---|---|---|---|
| ED/ND | LD/ND | BD/ND | ||||||
| Fatty acid synthesis | ||||||||
| acc | CPIJ005524 | GQ232408 | Acetyl-CoA carboxylase | 1.1 (0.8) | 4.1 (0.3) | −7.9 (6.7) | ||
| fas-1 | CPIJ005595 | GQ232409 | Fatty acid synthase S-acetyltransferase | 2.2 (0.4) | 1.3 (0.1) | 1.9 (0.4) | ||
| fas-2 | CPIJ003494 | GQ232410 | Fatty acid synthase S-acetyltransferase | 3.7 (2.1) | 4.1 (2.0) | 2.0 (0.7) | ||
| fas-3 | CPIJ008367 | GQ232411 | Fatty acid synthase S-acetyltransferase | 3.7 (0.8) | 5.1 (1.0) | 2.0 (0.8) | ||
| β-Oxidation | ||||||||
| acs | CPIJ000426 | GQ232412 | Acetyl-CoA synthetase | 1.4 (0.2) | 6.7 (0.2) | 3.0 (0.2) | ||
| cpt-1 | CPIJ005329 | GQ232413 | Carnitine O-octanoyltransferase | 1.1 (0.1) | 5.1 (0.1) | 4.1 (2.0) | ||
| cpt-2 | CPIJ010031 | GQ232414 | Carnitine O-palmitoyltransferase | −111.9 (98.0) | 0.9 (0.1) | 1.4 (0.2) | ||
| cpt-3 | CPIJ019087 | GQ232415 | Carnitine O-acetyltransferase | 1.3 (0.2) | 4.5 (1.0) | 1.7 (0.3) | ||
| acd-1 | CPIJ008217 | GQ232416 | Acyl-CoA dehydrogenase | 2.2 (0.5) | 2.4 (0.6) | 1.9 (0.5) | ||
| acd-2 | CPIJ016454 | GQ232417 | Acyl-CoA dehydrogenase | −5.0 (1.3) | 3.5 (0.1) | 3.1 (0.3) | ||
| acd-3 | CPIJ016451 | GQ232418 | Crotonobetainyl-CoA dehydrogenase | 1.6 (0.2) | 0.6 (0.1) | 0.6 (0.4) | ||
| acd-4 | CPIJ009148 | GQ232419 | Acyl-CoA dehydrogenase | −52.0 (33.0) | −2.2 (0.4) | 0.8 (0.2) | ||
| acd-5 | CPIJ014783 | GQ232420 | Isovaleryl-CoA dehydrogenase | −29.7 (2.0) | 0.6 (0.2) | 1.2 (0.1) | ||
| ech-1 | CPIJ006455 | GQ232425 | Enoyl-CoA hydratase | 0.6 (0.2) | 1.1 (0.1) | 1.6 (0.2) | ||
| ech-2 | CPIJ013796 | GQ232426 | 3,2-Trans-enoyl-CoA isomerase | −8.1 (2.0) | 1.2 (0.2) | 2.0 (0.3) | ||
| ech-3 | CPIJ006158 | GQ232427 | Cyclohex-1-ene-1-carboxyl-CoA hydratase | −9.4 (0.8) | 1.2 (0.1) | 1.4 (0.1) | ||
| ech-4 | CPIJ002685 | GQ232428 | Enoyl-CoA hydratase | 1.5 (0.8) | 2.8 (1.7) | 4.8 (2.5) | ||
| hcdh-1 | CPIJ006479 | GQ232421 | 3-Hydroxyacyl-CoA dehydrogenase | −9.6 (8.7) | 2.4 (0.2) | 2.4 (0.4) | ||
| hcdh-2 | CPIJ006478 | GQ232422 | 3-Hydroxyacyl-CoA dehydrogenase | 0.7 (0.1) | −3.4 (1.1) | −3.0 (2.3) | ||
| hcdh-3 | CPIJ020263 | GQ232423 | 3-Hydroxyacyl-CoA dehydrogenase | −10.1 (1.2) | 0.9 (0.1) | 1.0 (0.1) | ||
| hcdh-4 | CPIJ012030 | GQ232424 | 3-Hydroxyisobutyryl-CoA hydrolase | 0.8 (0.1) | 1.2 (0.2) | 1.7 (0.2) | ||
| bk-1 | CPIJ002342 | GQ232429 | β-Ketoacyl-CoA thiolase | −3.0 (0.2) | 2.7 (0.3) | 2.9 (0.2) | ||
| bk-2 | CPIJ018065 | GQ232430 | β-Ketoacyl-CoA thiolase | −3.9 (1.1) | 3.8 (0.4) | 3.0 (0.5) | ||
| Lipid transport | ||||||||
| fabp | CPIJ013698 | GQ232431 | Fatty acid binding protein, allergen | 5.1 (1.9) | 42.2 (8.1) | 22.9 (10.7) | ||
| Lipolysis | ||||||||
| lipase | CPIJ015981 | GQ232432 | Hormone-sensitive lipase | −6.7 (2.4) | 2.4 (0.2) | −6.3 (1.2) | ||
| Polyunsaturated fatty acid synthesis | ||||||||
| fad-1 | CPIJ013749 | GQ232433 | Δ(9)-desaturase | 8.1 (0.4) | −2.3 (0.3) | −2.5 (0.4) | ||
| fad-2 | CPIJ013748 | GQ232434 | Δ(9)-desaturase | 0.6 (0.1) | 1.8 (0.1) | 1.6 (0.1) | ||
| fad-3 | CPIJ013752 | GQ232435 | Acyl-CoA Δ(11) desaturase | −7.5 (0.2) | 1.8 (0.1) | 2.4 (0.1) | ||
| Nuclear hormone receptor | ||||||||
| nhr-41 | CPIJ001878 | GQ232436 | Nuclear hormone receptor nhr-41 | 1.2 (0.1) | 1.3 (0.2) | 1.9 (0.3) | ||
| nhr-48 | CPIJ016038 | GQ232437 | Nuclear receptor nhr-48 | −7.3 (0.7) | −6.7 (1.5) | −2.0 (0.8) | ||
| nhr-49 | CPIJ010249 | GQ232438 | Retinoid X receptor α | −7.8 (1.4) | 3.4 (0.1) | 3.5 (0.2) | ||
| Control gene | ||||||||
| 28S | CPIJ016534 | DQ401446 | 28S large-subunit ribosomal RNA | 0.7 (0.1) | 0.8 (0.3) | 1.3 (0.2) | ||
| rpl19 | CPIJ014540 | FJ266017 | ribosomal protein large subunit 19 | 0.6 (0.2) | 0.7 (0.3) | 0.9 (0.2) | ||
Bold numbers are used to designate changes that are significantly up or down; numbers in parenthesis are SE, each n = 3. If a fold change is <0.5, the ratio value was negatively inversed, e.g., if value was 0.2, value is stated as −5.0. ND, nondiapausing; ED, early diapause; LD, late diapause; BD, broken diapause.
Fig. 1.
Putative Culex pipiens fatty acid metabolism pathway. Each box represents a gene predicted by sequence homology to encode for the enzyme indicated. Quantitative RT-PCR (qRT-PCR) was used to measure the expression ratios of these 31 genes (1, early diapause/nondiapause; 2, late diapause/nondiapause; 3, diapause termination/nondiapause). TAG, triacylglyceride; PUFA, polyunsaturated fatty acid; FAD, fatty acid desaturase; ACC, acetyl-coA carboxylase; FAS, fatty acid synthase; NHRs, nuclear hormone receptors; FABP, fatty acid binding protein; 28S, 28S large-subunit ribosomal RNA; RpL19, ribosomal protein large subunit 19 (loading controls). Data are presented as means of 3 independent samples. n.s., Not significant.
RNA interference of upregulated genes reveals their functional role in fat storage during early diapause.
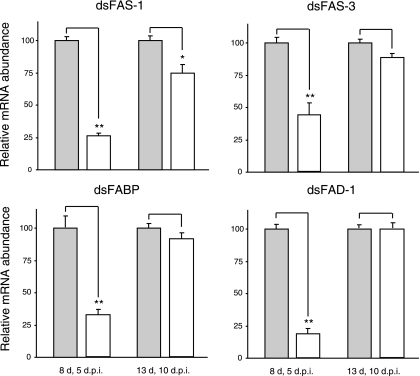
The RNAi efficiency targeting putative fas-1, fas-3, fabp, and fad-1 genes was first assessed by qRT-PCR. In contrast to the relatively high induction of putative fas-1, fas-3, fabp, and fad-1 in dsβ-gal-injected diapausing females, only 18–43% of these four transcripts were detected 5 days after dsRNA injection compared with control females (Fig. 2), thus indicating that injection of dsRNA successfully suppressed the mRNAs of each gene. However, these knockdown effects faded within 10 days after dsRNA injection. There were no significant differences in transcript levels of any of the genes 10 days after injection, except for putative fas-1, but even in that case the gene recovered to >70% of the control level within 10 days (Fig. 2). Thus we conclude that by 10 days after a dsRNA injection (1 μg/female), the mosquitoes recovered the functions of the putative fas-1, fas-3, fabp, and fad-1 gene products. RpL19, the loading control, showed no differences in expression 5 and 10 days after injection (data not shown), thus indicating that the low expression levels observed for the putative fas-1, fas-3, fabp, and fad-1 genes were related to the knockdown effect of dsRNAi rather than variation in sample loading.
Fig. 2.
RNA interference (RNAi) efficacy in knocking down expression of genes that were upregulated in early diapausing mosquitoes. Double-stranded RNA (dsRNA) of each gene (∼1.0 μg/female) was injected into the thorax of cold-anesthetized mosquitoes 3 days after eclosion (control, dsβ-gal), and expression was measured 5 and 10 days later [days postinjection (dpi)]. Gray bars represent mRNA abundance after injection of dsβ-gal; open bars represent mRNA abundance after injection of dsRNA. Bars represent means ± SE, each n = 3. *P < 0.05, **P < 0.01, compared with control, dsβ-gal by Student's t-test.
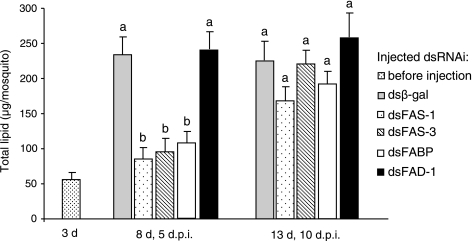
Prior to injection 3 days after eclosion females contained 56.4 ± 10.5 μg lipid/female (mean ± SE, n = 10), and 5 days later the lipid level had increased more than fourfold in the dsβ-gal control females (Fig. 3). Although there was no significant difference in lipid levels in response to the dsFAD-1 injection, in response to an injection of dsFAS-1, dsFAS-3, and dsFABP, diapausing females synthesized and stored half the amount of lipid stored by dsβ-gal control females 5 days after injection (Fig. 3). Interestingly, by 10 days after injection, the lipid levels in mosquitoes injected with dsRNA of all the candidate genes recovered, along with the recovery of the transcript levels (Fig. 3). These rescues of lipid levels 10 days after injection are interpreted as pseudo-gains of function because the RNAi effects remained for at least 5 days but were lost by 10 days. Thus our results indicate that the putative fas-1, fas-3, and fabp genes play a significant role in fat storage during early diapause.
Fig. 3.
Lipid levels in females 5 and 10 dpi compared with dsβ-gal (control), dsFAS-1, dsFAS-3, dsFABP, or dsFAD-1. Baseline represents lipid level 3 days after eclosion. Bars (means ± SE, each n = 10) with the same superscript letters are not significantly different at P = 0.05 (ANOVA).
Suppression of fatty acid β-oxidation in early diapause.
Ten of the nineteen putative β-oxidation genes were downregulated in early diapause compared with nondiapause (Fig. 1 and Table 1). Four were strongly downregulated (>10-fold down): one putative carnitine O-palmitoyltransferase (CPT) gene (cpt-2), two putative acyl-CoA dehydrogenase (ACD) genes (acd-4 and acd-5), and one putative 3-hydroxyacyl-CoA dehydrogenase (HCDH) gene (hcdh-3). CPT is an enzyme that transports fatty acids into mitochondria or peroxisomes (6); thus downregulation of putative cpt-2 may indicate suppression in the transport of fatty acids into the mitochondria matrix or peroxisomes for β-oxidation in early diapause. ACD, ECH, HCDH, and β-ketoacyl-CoA thiolase (BK) are a series of enzymes that shorten the acetyl groups of fatty acids, subsequently producing ATPs by β-oxidation of acetyl-CoA. This strong downregulation of putative acd-4, acd-5, and hcdh-3 (all >10-fold down) likely reflects the high level of fat accumulation in early diapause, the concurrent suppression of fatty acid catabolism, and induction of triacylglyceride synthesis from free fatty acids during this early phase of diapause.
Upregulation of genes involved in fatty acid β-oxidation in late diapause.
In late diapause, 7 of the 19 putative β-oxidation genes were upregulated compared with expression levels in early diapause (Fig. 1 and Table 1): one putative acetyl-CoA synthetase (acs), two putative carnitine O-octanoyltransferase (cpt-1 and -3), one putative acyl-CoA dehydrogenase (acd-2), one putative 3-hydroxyacyl-CoA dehydrogenase (hcdh-1), and two putative β-ketoacyl-CoA thiolase (bk-1 and -2) genes. The energy contained within fatty acid molecules is made available by β-oxidation of fatty acids (3, 11). The induction of putative acs, cpt, acd, hcdh, and bk in this study indicates enhanced catabolism of fatty acids and energy expenditure through β-oxidation and lipolysis in late diapausing females. These results are similar to observations made in fasted Caenorhabditis elegans, which also showed increased transcript levels of acs, cpt, and hcdh. Products of these induced genes stimulate fat breakdown to meet nutritional and energy needs (22). A recent study of metabolic changes during diapause in C. pipiens also indicated a steady decrease in glycogen during the first month of diapause, but lipid levels did not significantly decrease until after day 35 of diapause (24). Together, these observations imply that diapausing females store fat reserves during the first month of winter and then, when they deplete other energy sources such as glycogen, they utilize their fat reserves to meet their energy needs. This scenario raises interesting questions about how they sense deficits in their nutritional condition, how they convert fat reserves to energy, and how they minimize their energy expenditure to survive the remaining 3–4 mo of diapause. Although we now know that diapausing females regulate fatty acid synthesis, lipolysis, lipid transport, and β-oxidation genes, all of which are important regulatory steps in utilizing fat reserves, the details of the molecular sensing of energy deficits and mechanisms regulating the fat reserves remain unknown.
Transcript levels of fatty acid-binding protein are influenced by the diapause program.
The diapause-responsive gene, putative fabp, shares significant homology with mammalian fatty acid-binding proteins (FABPs) [both chimpanzee (Pan troglodytes) and human (Homo sapiens) E values = 7E−08 by BLASTN]. Fatty acid transport to the mitochondria for β-oxidation is a major requirement for energy production in mammals and insects (8, 10). Interestingly, putative fabp expression was upregulated in early diapausing females, but expression was even higher in late diapause. This expression pattern in association with diapause is unique among the fat metabolism genes that we investigated: putative fabp was the only gene that was highly upregulated in early and late diapause, as well as at diapause termination. (5.1-fold up in ED, 42.2-fold up in LD, and 22.9 fold up in BD; Table 1). In early diapause FABP may be involved in mobilizing triacylglycerides for storage in the fat body, and in late diapause FABP may help to mobilize and distribute fatty acids for use in generating energy. The high upregulation of the putative lipase gene (Fig. 1) also supports the hypothesis that FABP mobilizes fatty acids in late diapause. The fact that lipid levels decrease in later stages of diapause (24) is consistent with a function for lipase in utilization of fat reserves in late diapause.
NHR-49 has expression profile similar to genes associated with β-oxidation.
Because of their ability to interact with fatty acids and other lipids, nuclear hormone receptors (NHRs) are thought to be important regulators of fat metabolism (21). However, most of the fat-regulating NHRs known from mammals are not present in flies or nematodes; only a few of the NHRs are conserved between vertebrates and invertebrates (4). Among the NHRs in C. elegans, NHR-41, -48, and -49 were identified as potential transcriptional factors regulating fat metabolism in dauer larvae (1, 2, 9). Screening for deletions in the nhr-49 gene also revealed suppression of transcript levels of the genes involved in fatty acid β-oxidation (21). In this study, we compared expression levels of the candidate nuclear hormone receptors (nhr-41, -48, and -49). Only the transcript profile of the putative nhr-49 gene showed an expression profile similar to the genes of β-oxidation (Fig. 1). Like the genes involved in β-oxidation, putative nhr-49 expression was suppressed in early diapause but enhanced in late diapause. This correlation raises the possibility that putative mosquito nhr-49 may be a regulator of fat metabolism in diapausing mosquitoes, but this relationship remains tentative until a functional analysis is completed. Putative nhr-48 is also suppressed in early diapause, but this suppression is not lifted in late diapause as seen for putative nhr-49. Suppression of putative nhr-48 is lifted only after the mosquitoes have broken diapause. Thus the expression of putative nhr-48 does not correlate with the pattern of fat metabolism, but its elevation at diapause termination could imply a link to the physiological response involved in postdiapause activities such as blood feeding and the subsequent utilization of amino acids.
Conclusions.
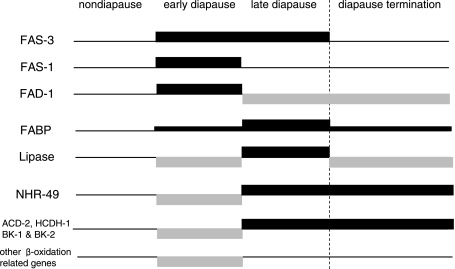
Fat accumulation is a key feature of diapause in C. pipiens, and in this study we began to examine how fat reserves are accumulated and utilized during diapause by examining transcript profiles of fat-related genes. A summary of the temporal patterns of expression in select genes related to fat storage and utilization in relation to diapause is depicted in Fig. 4. The genes of interest fall into several discrete categories. First is a category of genes that are upregulated in early diapause by comparison to nondiapausing females of the same age. This broad category included putative fas-1, -3, fad-1, and fabp, genes that would be expected to be involved in fat accumulation. Within this broad category are genes such as putative fas-1 and fad-1 that are highly expressed only in early diapause and are then turned off in late diapause. By contrast, putative fas-3 remains highly expressed throughout diapause. The putative fabp expression pattern is also distinct. Although it is already upregulated in early diapause, it is more highly expressed in late diapause and expression then again declines at diapause termination. Another major category includes genes that are suppressed in early diapause and are then upregulated in late diapause or revert to the expression levels observed in nondiapause. This category includes putative lipase, nhr-49, acd-2, hcdh-1, bk-1, bk-2, and a number of other genes involved in β-oxidation. Again, there are distinctions within this category: while most of these genes continue to be highly expressed even after diapause has been terminated, putative lipase expression is not highly expressed after diapause termination. One of the dominant patterns that thus emerges is the upregulation of genes involved in fat accumulation in early diapause and the concurrent suppression of genes involved in fat utilization (β-oxidation). The opposite pattern dominates in late diapause: several genes involved in fat accumulation are turned off and those involved in utilization are turned on.
Fig. 4.
Representative expression patterns of genes related to fat metabolism in the mosquito C. pipiens in relation to adult reproductive diapause. Proteins encoded by the genes are indicated on left. Categories include genes that are upregulated throughout diapause (putative fas-3 and fabp), early diapause genes (putative fas-1 and fad-1), late diapause genes (putative lipase, nhr-49, acd-2, hcdh-1, bk-1, and bk-2) and genes that are downregulated only in early diapause (putative cpt-2, acd-5, ech-2, ech-3, and hcdh-3).
Consistent results from mammals and nematodes suggest that fasting mediates induction of genes involved in β-oxidation and genes that encode FABPs (14, 22). Those genes produce proteins used to convert stored fat into energy, a feature that is similar to the requirement for mosquito fat metabolism in late diapause. However, not all genes involved in β-oxidation and FABP are upregulated in nematodes, and in fact some are repressed or not significantly changed during fasting (22). In this study, we also observed no changes in the expression levels of some genes involved in β-oxidation in early diapause. This is a time when energetic needs are less demanding, but even in early diapause lipid catabolism persists at a basal level.
RNAi targeted against putative fas and fabp genes demonstrated the functional importance of these genes in fat synthesis and storage at the onset of diapause. Although putative fas-1 and fas-3 were induced in early diapause, fas-2 was not. Recovery from knockdown also differed between putative fas-1 and fas-3; recovery was much faster for putative fas-1 than for fas-3 (Fig. 2). These differences may reflect differences in tissue specificity for these two genes or may have some other basis. Blood feeding is, of course, another important component of mosquito digestive physiology, an event associated with a huge nutritional influx. Although blood feeding does not occur in association with diapause, mosquitoes must be able to exploit this resource after the termination of diapause. Thus the female mosquito must have the ability to utilize glucose, amino acids, and lipids present in a blood meal, and this resource will require different metabolic cascades that possibly elicit responses from different fas genes. Our current working hypothesis is that different sensing mechanisms are involved in these two feeding options: blood feeding that leads to egg laying in nondiapause and postdiapause stages and nectar feeding leading to fat hypertrophy during diapause. Perhaps putative fas-1, -2, and -3 are uniquely induced in response to these different developmental states. If so, it is not yet clear what sensing mechanisms may operate in early and late diapause, or in response to blood digestion. Answering these questions may be a first step toward understanding the general regulatory mechanism of lipid catabolism and synthesis in hematophagous arthropods.
In addition, we also report that the transcript profile of putative nhr-49 is similar to that of seven β-oxidation genes. In late diapause, when stored fat is converted to energy, putative nhr-49 is highly induced compared with early diapause, along with the β-oxidation genes. The nematode homolog of nhr-49 is a key component of the fasting response, converting stored fat to energy and eliciting fasting-dependent induction of β-oxidation genes (21). Thus this expression pattern in C. elegans is similar to the expression of putative nhr-49 in late diapause of C. pipiens. The ligand binding domain of NHR-49 shares similarity to the mammalian HNF4 receptor, which directly binds fatty acids (7, 22). It will thus be interesting to determine whether NHR-49 binds and senses free fatty acids and thus eventually regulates transcription of the β-oxidation genes.
The signaling network that activates fat metabolism in diapause is yet to be determined, but we suspect it may involve insulin signaling and FOXO (17). How does this regulatory network enable overwintering mosquitoes to survive food deprivation and perhaps the cold? This study describing the profiles of key fat-related genes provides a first step toward understanding the gene regulatory network of fat metabolism in diapausing mosquitoes, a major goal for deciphering the molecular underpinning of diapause energetics.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by National Institute of Allergy and Infectious Diseases Grant R01-AI-058279.
Footnotes
1The online version of this article contains supplemental material.
REFERENCES
- 1.Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev 14: 1512–1527, 2000 [PMC free article] [PubMed] [Google Scholar]
- 2.Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421: 268–272, 2003 [DOI] [PubMed] [Google Scholar]
- 3.Bartlett K, Eaton S. Mitochondrial beta-oxidation. Eur J Biochem 271: 462–469, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Bertrand S, Brunet FG, Escriva H, Parmentier G, Laudet V, Robinson-Rechavi M. Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Mol Biol Evol 21: 1923–1937, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Christophers SR. The development of the egg follicle in anophelines. Paludism 2: 73–88, 1911 [Google Scholar]
- 6.Coleman RA, Lewin TM, Van Horn CG, Gonzalez-Baro MR. Do long-chain acyl-CoA synthetases regulate fatty acid entry into synthetic versus degradative pathways? J Nutr 132: 2123–2126, 2002 [DOI] [PubMed] [Google Scholar]
- 7.Dhe-Paganon S, Duda K, Iwamoto M, Chi YI, Shoelson SE. Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand. J Biol Chem 277: 37973–37976, 2002 [DOI] [PubMed] [Google Scholar]
- 8.Erol E, Kumar LS, Cline GW, Shulman GI, Kelly DP, Binas B. Liver fatty acid binding protein is required for high rates of hepatic fatty acid oxidation but not for the action of PPARalpha in fasting mice. FASEB J 18: 347–349, 2004 [DOI] [PubMed] [Google Scholar]
- 9.Gissendanner CR, Crossgrove K, Kraus KA, Maina CV, Sluder AE. Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans. Dev Biol 266: 399–416, 2004 [DOI] [PubMed] [Google Scholar]
- 10.Haunerland NH, Andolfatto P, Chisholm JM, Wang Z, Chen X. Fatty-acid-binding protein in locust flight muscle. Developmental changes of expression, concentration and intracellular distribution. Eur J Biochem 210: 1045–1051, 1992 [DOI] [PubMed] [Google Scholar]
- 11.Kim JJ, Battaile KP. Burning fat: the structural basis of fatty acid beta-oxidation. Curr Opin Struct Biol 12: 721–728, 2002 [DOI] [PubMed] [Google Scholar]
- 12.Maier T, Jenni S, Ban N. Architecture of mammalian fatty acid synthase at 4.5 Å resolution. Science 311: 1258–1262, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Mitchell C, Briegel H. Inability of diapausing Culex pipiens (Diptera: Culicidae) to use blood for producing lipid reserves for overwinter survival. J Med Entomol 26: 318–326, 1989 [DOI] [PubMed] [Google Scholar]
- 14.Nagao M, Parimoo B, Tanaka K. Developmental, nutritional, and hormonal regulation of tissue-specific expression of the genes encoding various acyl-CoA dehydrogenases and alpha-subunit of electron transfer flavoprotein in rat. J Biol Chem 268: 24114–24124, 1993 [PubMed] [Google Scholar]
- 15.Robich RM, Denlinger DL. Diapause in the mosquito Culex pipiens evokes a metabolic switch from blood feeding to sugar gluttony. Proc Natl Acad Sci USA 102: 15912–15917, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanburg LL, Larsen JR. Effect of photoperiod and temperature on ovarian development in Culex pipiens pipiens. J Insect Physiol 19: 1173–1190, 1973 [DOI] [PubMed] [Google Scholar]
- 17.Sim C, Denlinger DL. Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex pipiens. Proc Natl Acad Sci USA 105: 6777–6781, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sim C, Denlinger DL. A shut-down in expression of an insulin-like peptide, ILP-1, halts ovarian maturation during the overwintering diapause of the mosquito Culex pipiens. Insect Mol Biol 18: 325–332, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sim C, Hong Y, Tsetsarkin K, Vanlandingham D, Higgs S, Collins F. Anopheles gambiae heat shock protein cognate 70B impedes o'nyong-nyong virus replication. BMC Genomics 8: 231, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sim C, Hong YS, Vanlandingham DL, Harker BW, Christophides GK, Kafatos FC, Higgs S, Collins FH. Modulation of Anopheles gambiae gene expression in response to o'nyong-nyong virus infection. Insect Mol Biol 14: 475–481, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol 3: e53, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Gilst MR, Hadjivassiliou H, Yamamoto KR. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc Natl Acad Sci USA 102: 13496–13501, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Handel E. Rapid determination of total lipids in mosquitoes. J Am Mosq Control Assoc 1: 302–304, 1985 [PubMed] [Google Scholar]
- 24.Zhou G, Miesfeld RL. Energy metabolism during diapause in Culex pipiens mosquitoes. J Insect Physiol 55: 40–46, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.