Abstract
Diets high in fat and cholesterol are associated with increased obesity and metabolic disease in mice and humans. To study the molecular basis of the metabolic response to dietary fat, 10 inbred strains of mice were fed atherogenic high-fat and control low-fat diets. Liver gene expression and whole animal phenotypes were measured and analyzed in both sexes. The effects of diet, strain, and sex on gene expression were determined irrespective of complex processes, such as feedback mechanisms, that could have mediated the genomic responses. Global gene expression analyses demonstrated that animals of the same strain and sex have similar transcriptional profiles on a low-fat diet, but strains may show considerable variability in response to high-fat diet. Functional profiling indicated that high-fat feeding induced genes in the immune response, indicating liver damage, and repressed cholesterol biosynthesis. The physiological significance of the transcriptional changes was confirmed by a correlation analysis of transcript levels with whole animal phenotypes. The results found here were used to confirm a previously identified quantitative trait locus on chromosome 17 identified in males fed a high-fat diet in two crosses, PERA × DBA/2 and PERA × I/Ln. The gene expression data and phenotype data have been made publicly available as an online tool for exploring the effects of atherogenic diet in inbred mouse strains (http://cgd-array.jax.org/DietStrainSurvey).
Keywords: dietary fat, liver, microarray analysis, obesity, strain survey
high levels of dietary fat intake have been associated with obesity (3). While obesity is becoming increasingly prevalent (28, 39), individuals with a genetic predisposition to obesity are more likely than control subjects to gain weight on a high-fat diet (2). Obesity, in turn, is associated with a number of health problems, including hypertension (5), diabetes mellitus (8, 10), dyslipidemia (5), coronary heart disease (13), stroke (31, 38), gallstones (35), osteoarthritis (7, 15), and various forms of cancer (9, 16, 19).
Mice that are fed an atherogenic high-fat diet containing 1% cholesterol experience many of the same health conditions related to obesity and heart disease as humans (30). These responses depend on genetic factors. For instance, the often-studied inbred mouse strain C57BL/6J (B6) develops severe obesity, hyperglycemia, and hyperinsulinemia in response to high-fat diet where the A/J strain remains resistant to these conditions despite a similar caloric intake on the same diet (37, 26). The observation that adiposity levels vary substantially even among genetically identical mice suggests that epigenetic mechanisms may also be involved in the determination of individual susceptibility to obesity (24).
As with humans, different inbred strains of mice respond differently to high-fat diet (18). Therefore, understanding the metabolic mechanisms underlying diet-induced obesity in mice may lead to a better understanding of human obesity. Here, we examine gene expression profiles in liver tissues of ten inbred strains of mice fed either high-fat or standard low-fat diets. The strains were selected based on previously published genetic mapping studies used to find genomic regions associated with high density lipoprotein (HDL) cholesterol levels. Our results indicate that the atherogenic diet represses cholesterol biosynthesis and induces an immune response that may be related to liver damage. Processed gene expression data and precalculated test statistics generated from these analyses are accessible through a web-based query tool at http://cgd-array.jax.org/DietStrainSurvey. The phenotype data can be found at the same location. The raw data are deposited in the National Center for Biotechnology Information's Gene Expression Omnibus with accession number GSE10493.
MATERIALS AND METHODS
Experimental design.
Ten male and 10 female mice from 12 inbred strains (129S1/SvImJ, A/J, BALB/cJ, C3H/HeJ, C57BL/6J, CAST/EiJ, DBA/2J, I/LnJ, MRL/MpJ-Tnfrsf6lpr/J, NZB/BINJ, PERA/Ei, and SM/J) were fed a standard diet (6% fat, product 5K52 from LabDiets, St. Louis, MO) until at least 6 wk of age. The mice were subsequently divided into two dietary groups for a 4 wk period. Five animals from each strain and sex were fed an atherogenic high-fat diet (30% Kcal from dairy fat) containing by weight 1% cholesterol and 0.5% cholic acid to increase cholesterol uptake, while the remaining animals were kept on the standard diet. Nine of the 12 strains were housed in clean animal rooms, which were found to be free of 15 viruses, 17 bacterial species (including Helicobacter spp., Pasteurella pneumotropica, and two Mycoplasma spp.), ecto- and endoparasites, and the microsporidium, Encephalitozoon cuniculi. CAST/EiJ, PERA/Ei, and I/LnJ were maintained in an animal room known to harbor Helicobacter spp., P. pneumotropica, and Klebsiella spp. (other than K. pneumoniae). For acclimation, all animals were group-housed for 3 wk after acquisition, with no more than 5 mice per pen, in duplex polycarbonate cages measuring 31 × 31 × 214 cm. Autoclaved white pine shavings were used as bedding, and cages were changed every 2 wk. All animals had ad libitum access to acidified water (pH 2.8–3.1) and pelleted diet.
Most mice were bled and killed at 10–13 wk old. However, difficulties associated with obtaining CAST/EiJ and PERA/Ei mice resulted in ages at bleeding and death ranging from 20 to 55 wk. Due to the chance that this age difference would result in unwanted variability, we removed these strains from our primary analysis. However, the data are available in the web tool, and PERA/Ei was used in the QTL examination presented here. All mice were housed singly for 3 days prior to bleeding and death and were placed in clean boxes without food at 6 AM on the day of death. Retro-orbital bleeding followed by euthanasia by cervical dislocation and tissue harvesting were consistently conducted between 11 AM and noon to minimize circadian variation in mRNA expression. Mice were perfused with saline before dissection. The median lobe of the liver was removed, cut into <1 cm2 sections, placed in RNAlater (Ambien, Austin, TX) at room temperature for 24 h, and then stored at −80°C until RNA extraction.
Body weight and blood chemistry data were obtained for all mice resulting in phenotype data for five replicate animals per experimental group. Specifically, we provide data on blood urine nitrogen, calcium, total cholesterol (CHOL), glutamate dehydrogenase (GLDH), glucose (GLU), HDL, nonesterified fatty acids (NEFA), thyroid hormone (T4), triglycerides (TG), and body weight. These data were measured using a SYNCHRON CX5 Delta Clinical System chemistry analyzer from Beckman Coulter and are available online at www.cgd.org.
Microarray procedures.
Three animals per experimental group were assayed for liver mRNA expression. Samples were extracted in batches randomized with respect to strain and collection date. RNA was extracted from the entire median lobe of the liver homogenized in TRIzol (Invitrogen, Carlsbad, CA); quality was assessed using a 2100 Bioanalyzer instrument and an RNA 6000 Nano LabChip Assay (Agilent Technologies, Palo Alto, CA). As described previously (34), total RNA was reverse transcribed with oligo(dT)-T7 primers before double-stranded cDNA was generated with the Superscript double-stranded cDNA synthesis custom kit (Invitrogen). The cDNA was linearly amplified with biotinylated nucleotides (Enzo Diagnostics, Farmingdale, NY) through an in vitro transcription reaction with T7 RNA polymerase. We then hybridized 15 μg of biotin-labeled and fragmented cRNA onto MOE430v2.0 GeneChip arrays for 16 h at 45°C. Posthybridization staining and washing were performed according to the manufacturer's protocols using the Fluidics Station 450 instrument (Affymetrix, Santa Clara, CA). Arrays were scanned with a GeneChip Scanner 3000 laser confocal slide scanner and quantified using GeneChip Operating Software v1.2 (Affymetrix).
Microarray data preprocessing.
Probe intensity data from Affymetrix GeneChip arrays containing 45,101 probe sets were read into the R software environment (http://www.R-project.org) directly from .CEL files using the R/affy package (17). Data quality was assessed using image reconstruction, histograms of raw signal intensities, and MA plots. Normalization was carried out using the robust multiarray average (RMA) method (22) to form one expression measure per probe set per array. Briefly, the RMA method was used to adjust the background of perfect match (PM) probes, apply a quantile normalization of the corrected PM values and calculate final expression measures using the Tukey median polish algorithm.
Probe sets were mapped to Entrez Gene identifiers using the R/mouse4302 package. A gene universe was defined based on Entrez Gene identifiers exhibiting variation across the experimental groups. Specifically, we used Fs, a modified F-statistic incorporating shrinkage variance components (11), to test for variation among any of the 40 groups (2 sexes × 2 diets × 10 strains) using the general linear model
| (Eq.1) | (Eq. 1) |
where Yi represents log2 transformed expression data, μ is the mean for each array, GROUP is a 40 level factor identifying each experimental group, and εi is random error. Using a one-way analysis of variance (ANOVA), we found a total of 26,564 probe sets to have significant variation at the nominal P < 0.01 level based on permutation of group labels. For cases in which multiple probe sets mapped to a single Entrez gene ID, the probe set with the highest Fs statistic was chosen to represent the gene. Overall, ANOVA selection and filtering reduced the 45,101 probe sets on the array to 20,831 probe sets representing 20,831 unique genes.
Differential gene expression.
A database containing the results of 532 statistical tests for differences between strains, diets, sexes and strain-by-diet interactions was created (see Supplemental Table S1).1 P values were calculated by permuting model residuals 1,000 times using the R/maanova package (41, 43). All statistical tests were performed using the Fs statistic. The false discovery rate (FDR) was estimated using the q-value method (36). Unless otherwise noted, significant changes were determined at the FDR threshold of 0.01.
Computation of gene expression profiles.
Global gene expression was analyzed using multidimensional scaling and clustering approaches. K-means and hierarchical cluster analyses were used to identify groups of genes with similar expression patterns across the 40 sex-strain-diet groups. In the bootstrapped k-means algorithm, a gene was assigned to a cluster if it appears in 80% of 1,000 bootstrap iterations (23). The bootstrapped k-means procedure was repeated for different values of k to find the minimum number of clusters explaining >90% of the variance in the data after bootstrapping. For hierarchical clustering, Pearson correlation coefficients were calculated between group-averaged phenotype data and group-averaged expression data. The P values resulting from the correlation tests were used to calculate an FDR for each gene (36). Hierarchical biclustering of group mean data based on Euclidean distance and Ward's method was applied to the group-averaged expression data for all significant genes (FDR < 0.01). Heat maps of clustering results were visualized in TreeView (32). Supplemental Tables S2 and S3 contain correlations between gene expression and HDL or TG, respectively.
Eigengene analysis.
A linear mixed-effects model was fit to data from each gene to quantify variation associated with main effects and interactions among experimental factors. The model used to calculate variances is given as follows
| (Eq.2) | (Eq. 2) |
where μ is a fixed effect. Random effects were assumed to be independently normally distributed with a mean of 0. Singular value decomposition (SVD) applied directly to the matrix of calculated variances was used to separate the eight sources of variation in Eq. 2 into three orthogonal vectors (i.e., “eigengenes”) accounting for >98% of the variation in the data. A permutation test based on the multivariate Hotelling T2 statistic was used to uncover genes with significant scores. Results for all genes are given in Supplemental Table S4.
Enrichment of gene sets.
The Gene Ontology (GO) Consortium provides relationships between genes and annotation terms using a restricted vocabulary (4). Hypergeometric tests for overrepresentation of “biological process” GO terms in selected gene lists were performed using the R/GOstats package (14) or R/allez (29). GO leaves were found by reconstructing the GO tree corresponding to all significantly overrepresented GO terms (P < 0.01) and extracting the leaves from the induced tree structure.
The Ingenuity Pathways Knowledge Base consists of data from >400 journals with known biological relationships between genes and gene products (www.ingenuity.com). Overrepresentation of Ingenuity Canonical Pathway members in gene lists was found using a right-tailed Fisher's exact test for 2 × 2 contingency tables.
RESULTS
Global gene expression analysis.
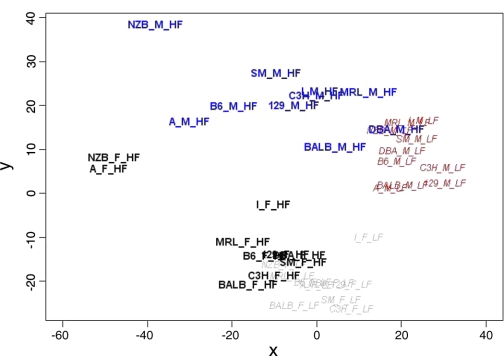
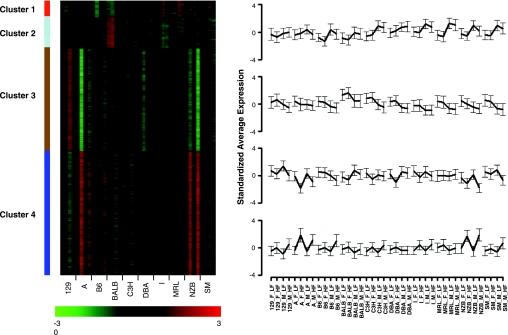
A total of 120 mice from 40 different experimental groups (diet-sex-strain) were used for global expression profiling. Multidimensional scaling was used to display differences in summarized expression profiles between any pair of groups as the distances in a two-dimensional plane. As shown in Fig. 1, groups of strains could be reasonably well separated based on differences in diet or sex. There were relatively few transcriptional differences between strains when mice of the same sex were fed a low-fat diet. By contrast, the atherogenic diet led to pronounced disparity among the strain-specific profiles. Bootstrapped k-means clustering applied to group mean expression data produced four stable clusters representing ∼27% of the mapped genes in the data set (Fig. 2). With the single exception of strain A/J, the first two clusters reflect sex-specific alterations; 355 genes showed higher expression in males and 630 genes show higher expression in females. When fed a low-fat diet, A/J males show similar expression as the other female strains (cluster 1) and A/J females show expression levels similar to the other male strains (cluster 2). GO terms enriched in cluster 1 are associated with a higher transport and processing of proteins (see Supplemental Table S5). Important GO terms in cluster 2 include “electron transport” and acyl-CoA metabolism (Supplemental Table S5), suggesting an increased production of energy from fat in most female strains.
Fig. 1.
Multidimensional scaling of summarized gene expression samples. A 2-dimensional projection shows each sex-strain-diet group plotted onto an arbitrarily scaled x/y-plane. The distance between any 2 groups best represents the total difference computed between their summarized complete gene expression profiles considering all 20,831 genes. Low-fat groups (LF) are italicized, while high-fat groups (HF) are shown in bold. Different colors separate each sex-diet combination.
Fig. 2.
Gene expression profiles generated by k-means clustering. Expression measures were averaged within groups, standardized across groups, and analyzed using a bootstrapping k-means cluster analysis. Left: the heat map shows standardized average expression for each gene along the vertical axis for the 40 groups along the horizontal axis. The levels of expression are indicated in the color bar at bottom. Right: k-mean cluster averages and standard deviations are shown with each strain connected by a solid line. The order in each strain is given for female (F), male (M), low-fat (L), and high-fat (H) conditions.
The two remaining clusters in Fig. 2 reflect gene expression differences due to diet. A total of 2,085 genes (cluster 3) are repressed, and 2,527 genes (cluster 4) are induced, with the high-fat diet. Here, strains A/J and NZB show the largest differences. No diet-specific changes are apparent in females from strains C57BL/6J and MRL (cluster 3) or males from DBA (cluster 4). Genes repressed under high-fat diet are enriched for ion transport and cell signaling, while genes induced under high-fat diet (cluster 4) are enriched for terms related to immune system and cell signaling (Supplemental Table S5).
Most of the expression differences between groups within the four clusters in Fig. 2 represent sex or diet effects, but the scale of these changes varies considerably across strains. Variance distributions derived from global ANOVA modeling were bimodal (Supplemental Fig. S2), with genes showing no variance (peak at zero) or some variation associated with a model term (peak greater than zero). SVD was used to explore these distributions to find the prevailing dimensions of the data referred to as “eigengenes.” Here, three eigengenes explained >98% of the variation in the data set (Supplemental Fig. S3). Genes correlated with the first eigengene (e.g., Xist and Sult2a2) vary between sexes, but not by strain, diet, or higher-order interactions between factors (Supplemental Table S6). Similarly, genes correlated with the second eigengene (e.g., H2-K1, Camk2b, and Zfp236) and third eigengene (e.g., Idi1 and Sqle) only vary between strains or diets, respectively.
Response to atherogenic diet.
Improving our understanding of the molecular basis of the metabolic response to the high-fat diet was central to this investigation. We tested for transcriptional differences between animals fed the high-fat atherogenic and low-fat diets within each of the 20 strain-sex groupings. In a similar manner, we looked for sex-specific differences within each of the 20 strain-diet groupings. A large number of diet and sex effects were uncovered by this pair-wise ANOVA approach. While there is substantial overlap between these lists of differentially expressed genes, most of these alterations were not shared across strains (Table 1). GO terms significantly repressed or induced with atherogenic diet were determined for each strain. Only two biological process GO term leaves were repressed in all strains (GO:0006695:cholesterol biosynthesis and GO:0006720:isoprenoid metabolism), and no GO terms were found to be induced in all strains on the atherogenic diet. However, an analysis based on Ingenuity Pathways Knowledge Base uncovered a small list of pathways repressed or induced across most strain-sex groupings (Table 2). Notably, every gene in the cholesterol biosynthesis pathway was repressed with atherogenic diet in every strain and in both sexes (Supplemental Fig. S4). The atherogenic diet induced genes associated with liver damage, especially genes associated with X receptor activation (LXR/RXR, FXR/RXR, and VDR/RXR), interleukin signaling (IL-6 and IL-10), and pentose/glucuronate interconversions. However, the evidence for induction of interleukin signaling and the repression of other categories (e.g., “metabolism of xenobiotics by cytochrome P450”, “linoleic acid metabolism,” “glycine, serine and threonine metabolism,” “glycolysis/gluconeogenesis”) varied somewhat between strains and sexes.
Table 1.
Numbers of differentially expressed genes in sex and diet ANOVA tests
| Strain | Sex Effects |
Diet Effects |
||||
|---|---|---|---|---|---|---|
| Low Fat | High Fat | Overlap (LF/HF) | Females | Males | Overlap (F/M) | |
| 129 | 762 | 584 | 207 | 510 | 970 | 172 |
| A | 299 | 480 | 78 | 2,600 | 1,455 | 986 |
| B6 | 660 | 546 | 184 | 347 | 1,203 | 166 |
| BALB | 1,852 | 1,091 | 603 | 766 | 644 | 200 |
| C3H | 900 | 1,195 | 443 | 818 | 1,483 | 473 |
| DBA | 648 | 780 | 303 | 286 | 481 | 115 |
| I | 377 | 319 | 159 | 835 | 718 | 375 |
| MRL | 1,180 | 1,471 | 663 | 717 | 609 | 208 |
| NZB | 996 | 473 | 232 | 2,054 | 2,298 | 1,249 |
| SM | 823 | 1,193 | 394 | 314 | 1,444 | 150 |
| Overlap | 66 | 33 | 27 | 26 | 21 | 15 |
The data were subset by strain and either diet (to find significant sex effects in mice fed low-fat or high-fat atherogenic diets) or sex (to find significant diet effects in male or female strains). The number of overlapping genes differentially expressed when mice were fed low-fat and high-fat diets, or between males and females, is given in the 3rd column for each effect. The overlap between strains is given in the last row. Differential expression is determined based on a false discovery threshold of 0.01. LF, low fat; HF, high fat; F, female; M, male.
Table 2.
Ingenuity pathway analysis describing strain-specific response to high-fat diet
| Pathway | 129 |
A |
B6 |
BALB |
C3H |
DBA |
I |
MRL |
NZB |
SM |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F | M | F | M | F | M | F | M | F | M | F | M | F | M | F | M | F | M | F | M | |
| Repressed with high fat | ||||||||||||||||||||
| Biosynthesis of steroids | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** |
| Metabolism of xenobiotics by cytochrome P450 | ** | * | * | * | * | ** | * | * | ** | ** | ** | * | * | * | * | ** | ** | ** | ** | |
| Stilbene, coumarine, and lignin biosynthesis | ** | ** | * | ** | ** | ** | ** | ** | ** | * | ** | * | ** | ** | ** | ** | ** | ** | ** | ** |
| Tryptophan metabolism | ** | * | ** | ** | * | ** | * | * | * | * | * | * | * | * | * | * | ** | ** | * | ** |
| Linoleic acid metabolism | ** | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||
| Androgen and estrogen metabolism | * | * | ** | ** | * | ** | * | * | * | * | ** | * | * | * | * | * | * | ** | ** | * |
| Glycine, serine, and threonine metabolism | * | ** | * | * | * | * | * | * | * | * | * | * | * | * | ||||||
| Fatty acid metabolism | * | * | * | * | * | * | * | * | * | * | ** | * | * | * | ||||||
| C21-steroid hormone metabolism | * | * | * | ** | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * |
| Glycolysis/gluconeogenesis | * | * | ** | * | * | * | * | * | * | * | * | * | ||||||||
| Induced with high fat | ||||||||||||||||||||
| LXR/RXR activation | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** |
| FXR/RXR activation | ** | ** | * | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** |
| VDR/RXR activation | * | ** | ** | ** | ** | ** | * | ** | ** | ** | * | * | ** | ** | * | * | ** | * | ** | ** |
| Hepatic fibrosis/hepatic stellate cell activation | * | * | ** | * | * | * | * | * | ** | ** | * | * | * | * | ** | ** | * | ** | * | |
| Pentose and glucuronate interconversions | * | * | ** | ** | ** | * | * | ** | ** | ** | ** | ** | * | ** | ** | ** | ** | * | ** | ** |
| IL-10 signaling | * | ** | * | ** | ** | ** | * | * | ** | * | ** | * | ** | * | ** | * | ** | |||
| IL-6 signaling | * | * | * | ** | ** | * | ** | * | ** | ** | ** | * | ** | * | * | ** | ** | * | * | ** |
Significant Ingenuity Canonical Pathways members exclusively induced or repressed with HF feeding (P < 0.001). Pathway significance is represented with asterisks: ****P < 10−12, **P < 10−3, or *P < 0.05 significance thresholds.
We performed an ANOVA-based analysis including all strains and both sexes to detect global diet effects with greater power. We also tested for diet effects that varied between strains (i.e., DIET-by-STRAIN effects) or varied differentially among strains according to sex (i.e., DIET-by-STRAIN-by-SEX effects). A total of 11,312 changes (∼56.6% of the genes examined) can be attributed to diet, while 2,638 alterations show strain-specific changes in diet and 293 genes exhibit both sex- and strain-specific diet alterations (see Fig. S5). However, 442 genes out of the 2,638 DIET-by-STRAIN effects were not explained by diet alone. In addition, 40 genes out of the 293 DIET-by-STRAIN-by-SEX effects were not found when we considered only diet- or strain-specific diet effects. Some of these genes are part of overrepresented GO terms associated with diet effects (Supplemental Fig. S5) and described in greater detail below.
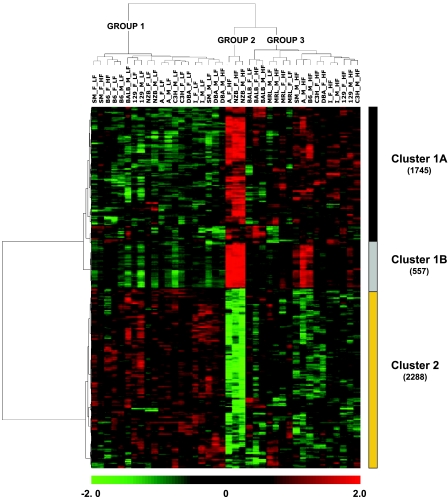
Correlations between gene expression profiles and metabolic phenotypes were calculated to extend the biological significance of the transcriptional analyses. Six phenotypes were examined (TG, NEFA, GLU, GLDH, CHOL, and HDL). A total of 3,936 genes had a significant correlation for at least one phenotype (Table 3). Hierarchical biclustering of the gene expression levels of these 3,936 genes gave rise to three groups (Fig. 3). Group 1 strains have a similar correlation between transcripts and phenotypes when fed the low-fat diet. However, two females (strains SM and B6) and one male (strain DBA) fed the atherogenic diet also appear in group 1. Group 2 strains consist of NZB (females and males) and A/J (females) fed the atherogenic diet. Group 3 has strains fed the atherogenic diet but also includes two females (BALB, MRL) and one male (MRL) fed the low-fat diet.
Table 3.
Number of genes correlated with different physiological phenotypes
| CHOL | HDL | WGT | GLU | TG | T4 | CA | GLDH | BUN | NEFA | |
|---|---|---|---|---|---|---|---|---|---|---|
| CHOL | 3,609 | 177 | 29 | 1 | 15 | 1 | 0 | 0 | 0 | 0 |
| HDL | 229 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| WGT | 197 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| GLU | 68 | 3 | 0 | 0 | 0 | 0 | 0 | |||
| TG | 40 | 0 | 0 | 0 | 0 | 0 | ||||
| T4 | 16 | 0 | 0 | 0 | 0 | |||||
| CA | 9 | 0 | 0 | 0 | ||||||
| GLDH | 1 | 0 | 0 | |||||||
| BUN | 1 | 0 | ||||||||
| NEFA | 0 |
Pearson correlation threshold is based on false discovery rate <0.01. CHOL, total cholesterol; HDL, high density lipoprotein; WGT, weight; GLU, glucose; TG, triglycerides; T4, thyroid hormone; CA, calcium; GLDH, glutamate dehydrogenase; BUN, blood urine nitrogen; NEFA, nonesterified fatty acids.
Fig. 3.
Biclustering for genes with correlations between gene expression and phenotype. A gene was included in the analysis if it was significantly correlated with at least 1 phenotype [Pearson correlation, false discovery rate (FDR) < 0.01]. The color bar at bottom indicates relative expression level. The total number of genes in each cluster is shown in parentheses. Total cholesterol (CHOL), glutamate dehydrogenase (GLDH), glucose (GLU), high density lipoprotein (HDL), nonesterified fatty acids (NEFA), and triglyceride levels (TG) were included in the analysis.
This biclustering approach divided the expression profiles across experimental groups into two major clusters, where cluster 1 can be further divided into two subclusters (Fig. 3). Genes in clusters 1A and 1B were induced with the high-fat atherogenic diet. Strains NZB and A and males from B6 and SM showed noticeably higher expression levels when fed the atherogenic diet than other experimental groups. Cluster 2 genes are negatively correlated with cluster 1 and were mostly repressed with high-fat atherogenic diet. A GO enrichment analysis corresponding to the expression profiles in Clusters 1A, 1B, and 2 is summarized in Table 4. Cluster 1A was enriched for GO terms associated with endocytosis, regulation of cell differentiation (osteoclast and myeloid), and angiogenesis, while cluster 1B was related to the immune response. Overrepresented GO terms in cluster 2 are strongly associated with cholesterol biosynthesis and also show enrichment in acetyl-CoA metabolism and eye development.
Table 4.
Top GO term leaves in phenotype correlation clusters
| Cluster | GOTERM | Description | Counts* | Category† | P Value |
|---|---|---|---|---|---|
| 1A | GO:0006897 | endocytosis | 33 | 163 | 2.46×10−4 |
| 1A | GO:0007015 | actin filament organization | 14 | 48 | 3.71×10−4 |
| 1A | GO:0015031 | protein transport | 95 | 635 | 4.89×10−4 |
| 1A | GO:0045670 | regulation of osteoclast differentiation | 7 | 16 | 7.83×10−4 |
| 1A | GO:0001525 | angiogenesis | 27 | 133 | 8.32×10−4 |
| 1A | GO:0045638 | negative regulation of myeloid cell differentiation | 8 | 21 | 9.91×10−4 |
| 1B | GO:0019886 | antigen processing, exogenous peptide antigen via MHC class II | 11 | 16 | 1.79×10−13 |
| 1B | GO:0030593 | neutrophil chemotaxis | 8 | 14 | 3.49×10−9 |
| 1B | GO:0042535 | positive regulation of tumor necrosis factor-α biosynthetic process | 5 | 8 | 2.03×10−6 |
| 1B | GO:0030048 | actin filament-based movement | 6 | 15 | 5.02×10−6 |
| 1B | GO:0045582 | positive regulation of T cell differentiation | 5 | 11 | 1.54×10−5 |
| 1B | GO:0006911 | phagocytosis, engulfment | 5 | 12 | 2.57×10−5 |
| 1B | GO:0051085 | chaperone cofactor-dependent protein folding | 5 | 12 | 2.57×10−5 |
| 1B | GO:0045059 | positive thymic T cell selection | 4 | 7 | 3.88×10−5 |
| 1B | GO:0019221 | cytokine and chemokine-mediated signaling pathway | 8 | 42 | 6.05×10−5 |
| 1B | GO:0030032 | lamellipodium biogenesis | 5 | 15 | 8.99×10−5 |
| 1B | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway | 3 | 4 | 1.42×10−4 |
| 1B | GO:0009968 | negative regulation of signal transduction | 11 | 89 | 1.68×10−4 |
| 1B | GO:0050766 | positive regulation of phagocytosis | 5 | 17 | 1.75×10−4 |
| 1B | GO:0050852 | T cell receptor signaling pathway | 5 | 18 | 2.36×10−4 |
| 1B | GO:0002573 | myeloid leukocyte differentiation | 7 | 45 | 6.45×10−4 |
| 1B | GO:0050798 | activated T cell proliferation | 4 | 13 | 6.75×10−4 |
| 1B | GO:0008360 | regulation of cell shape | 7 | 47 | 8.44×10−4 |
| 1B | GO:0007249 | I-κB kinase/NF-κB cascade | 9 | 77 | 9.84×10−4 |
| 2 | GO:0006695 | cholesterol biosynthetic process | 18 | 22 | 1.31×10−14 |
| 2 | GO:0006084 | acetyl-CoA metabolic process | 12 | 34 | 1.17×10−4 |
| 2 | GO:0001747 | eye development (sensu Mammalia) | 14 | 51 | 6.46×10−4 |
Gene Ontology (GO) term leaves were generated based on the induced GO graph resulting from all significant GO terms (P < 0.01).
Number of differentially expressed genes in a GO term category (false discovery rate <0.01);
Total number of genes in a GO term category.
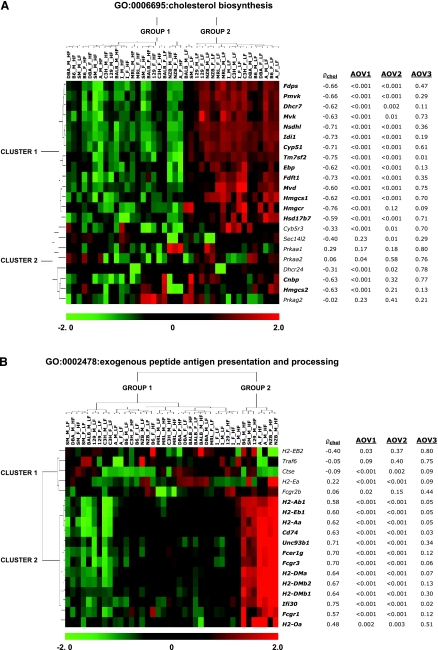
The most statistically significant biological processes uncovered here involve the repression of cholesterol biosynthesis and induction of the antigen presentation (see Tables 2 and 4, Fig. 3, and Supplemental Fig. S5). Therefore, we explored these categories in greater detail. Gene expression patterns and results of the ANOVA and correlation analyses are shown in Fig. 4 for every gene in the GO classes “cholesterol biosynthesis” (Fig. 4A) and “exogenous antigen peptide presentation and processing” (Fig. 4B). In Fig. 4A, strains separate into two groups based on diet (low-fat strains vs. high-fat strains), with the exception that SM males and BALB females group with the high-fat strains. Most genes in the “cholesterol biosynthesis” category show repression of expression with the high-fat diet that correlate with cholesterol levels, but a second group of genes show more diverse expression with varying degrees of correlation to the phenotype. The “antigen presentation” category (Fig. 4B) separates the strains into two groups: group 1 contains strains A (females and males), NZB (females and males), B6, SM, and 129 (all males), and group 2 contains the rest of the experimental strains. The genes in Fig. 4B are also divided into two clusters. Genes in cluster 1 have expression patterns distinct from other genes in the grouping and are not correlated with cholesterol levels. By contrast, expression levels for genes in cluster 2 are notably higher for the group 2 strains than the group 1 strains and are correlated with cholesterol.
Fig. 4.
Biological processes enriched for genes with correlations between gene expression and cholesterol levels. The expression of genes involved in cholesterol biosynthesis (A) is inversely correlated with cholesterol levels (Z = −6.84), while genes involved in antigen presentation (B) are positively correlated with cholesterol (Z = 6.07). Correlation between gene expression and cholesterol (ρchol) is given for each gene. The ANOVA-based significance levels for each test is shown for main effects of diet (AOV1), 2-way interactions between diet and strain (AOV2) and 3-way interactions between diet, strain, and sex (AOV3).
Both GO classes in Fig. 4 contain diet effects that were only found when testing for diet differences (main effect ANOVA), diet × strain interactions (two-way interaction ANOVA) or diet × strain × sex interactions (three-way ANOVA). For instance, like most genes involved in cholesterol biosynthesis in Fig. 4A, Hmgcr, Cnbp, and Hmgcs2 show significant differences between diet regiments when tested across strains (P < 0.001 in each case). However, unlike most genes with significant diet changes, the evidence for strain-specific diet effects in these genes does not reach statistical threshold levels (P = 0.12, 0.32, and 0.21, respectively). Also, correlation with phenotype can distinguish genes possessing greater biological weight than data based on gene expression analyses alone. For instance, Cyb5r3, Prkaa2, and Dhcr24 in Fig. 4A and H2-EB2, Ctse, H2-Ea, and Fcgr2b in Fig. 4B are downregulated with high-fat diet, but unlike other genes supported with significant gene expression differences, these genes are not significantly correlated with phenotypes.
Strain survey database.
We provide a publicly available database that can be used to explore differentially expressed transcripts between experimental factors for any of 532 different statistical tests (see Supplemental Table S1). Users can query the data by selecting an ANOVA model, tested term, and subset of the data for a list of candidate genes. Selected genes can be explored graphically within the Java interface. F statistics (classical F and Fs), permutation P values, FDR, fold changes, and error variances can be obtained. Results from fixed and mixed effects models are also available. In the analysis presented above, all genes differentially expressed in mixed-effects model tests were also found to be differentially expressed in corresponding fixed-effects tests (Supplemental Table S7).
To illustrate the utility of the information that can be extracted from the database, graphical representations for genes exhibiting strong effects in our study are presented in Fig. 5. The gene encoding the X (inactive)-specific transcript (Xist) varies significantly across sexes (Fig. 5A). The genes encoding the major histocompatibility complex, class I, C protein (H2-K1), and apolipoprotein C-IV (Apoc4) vary across strains (Fig. 5, B and C, respectively). The hepatocanalicular cholesterol transporter (Abcg5) expression increases significantly on high-fat diet in all strains and both sexes (Fig. 5D). This gene is particularly interesting due to its role in response to dietary fat. In humans, it plays a role in reducing absorption of cholesterol in the intestine and encourages secretion of sterols in the bile. Mutations in this gene have been linked to atherosclerosis, sterol accumulation, and sitosterolemia (27, 20). With this in mind, we speculate that Abcg5 expression is upregulated in response to accumulating lipids in the high-fat diet. Other diet-induced changes include a large decrease in expression with the atherogenic diet for the cholesterol biosynthesis genes isopentenyl-diphosphate delta isomerase 1 (Idi1) and squalene epoxidase (Sqle) (Fig. 5, E and F, respectively).
Fig. 5.
Selected effects plots for strong main and interaction effects. A: Xist exhibits a strong sex effect. Both H2-K1 (B) and Apoc4 (C) exhibit strain effects. Diet effects include the induction of Abcg5 (D) and the repression of Idi1 (E) and Sqle (F) after atherogenic diet feeding. Strain-by-diet interactions are observed in Soat1 (G) and Cd207 (H). Differences in HDL levels (I) are observed across strains.
Sterol O-acyltransferase 1 (Soat1) produces cholesterol esters from cholesterol (27) and shows a strong strain-by-diet effect here (Fig. 5G). Its expression is increased on high-fat diet in NZB and A, with a less pronounced effect in 129, B6, BALB, C3H, I, and MRL. However, in females of DBA and SM and males from DBA, the expression doesn't change across diets. Strong strain-by-diet interactions are also seen between multiple strains for Cd207 (Fig. 5H). The database may be used to examine phenotypes. For instance, NZB also has the highest HDL levels among these strains and increases substantially on high fat diet (Fig. 5I). Conversely, BALB shows little change in HDL in response to diet (Fig. 5I).
Using the strain survey database to find quantitative trait locus genes.
There are two ways in which the strain survey database can be useful in the search for genes within a quantitative trait locus (QTL). First, one can use the database to identify all genes in a QTL region that are differentially expressed between the two strains that gave rise to the QTL. This step can become even more powerful if the QTL has been found in multiple crosses. Second, one can then ask whether gene expression is correlated with the phenotype across all strains, sexes, and diets. Neither of these criteria is an absolute requirement for a QTL gene. For example, a gene may cause a QTL through a mutation in the coding region that changes function; such changes can be searched for in single nucleotide polymorphism (SNP) databases or by sequencing. Furthermore, it is not necessary that a gene causing a QTL through an expression difference be correlated with the phenotype in all strains. Nevertheless, such a correlation provides additional evidence that a candidate gene is related to the basic functions of the phenotype.
As an example, we selected a QTL on chromosome 17 from 72 to 90 Mb that was previously identified in males on a high-fat diet in two crosses, PERA × DBA/2 and PERA × I/Ln (40). Using BioMart (www.biomart.org), we found 91 genes within this region that were represented on our microarray. This list of genes was used as input first for the PERA × DBA/2 males fed high-fat diet and then for the PERA × I males on high fat. Only 15 genes out of the 91 were differentially expressed in both crosses. We then asked whether all 15 of these genes had an expression difference that was consistent with the phenotype in both crosses. For example, if gene expression was low in DBA/2, then it also had to be low in I/Ln. This step eliminated two genes. Next, we used the Mouse Phenome Database (http://phenome.jax.org/pub-cgi/phenome/mpdcgi?rtn=snps/door) to obtain SNPs in the region and conducted a block haplotype analysis (6, 12). We required that the gene and its regulatory regions differ between PERA and DBA/2 as well as between PERA and I. PERA carried the allele for high HDL in both cases, while DBA/2 and I both carried the allele for low HDL. Block haplotyping was used to eliminate three genes with regulatory and coding regions that were identical between PERA and one of the other strains. This left 10 potential candidate genes (Abcg5, Abcg8, Ehd3, Galm, Haao, Mta3, Prkce, Slc3a1, Srbd1, Thada). However, only three of these genes were correlated with HDL levels, and these three remained as the most likely QTL genes (Abcg5, Abcg8, Thada).
Here, the database helped us to reduce a list of 91 potential candidates to only three. The next bioinformatic step in QTL gene identification is to add knowledge about tissue distribution and gene function. Immediately the genes encoding Abcg5 and Abcg8 stand out as genes highly expressed in the liver and known to have a role in lipid metabolism. In contrast, Thada (thyroid ademona associated) is expressed at relatively low levels in the liver and has no obvious connection to lipid metabolism. These results are consistent with previously published studies. We have previously demonstrated that the differential expression of Abcg5/8 is controlled by cis elements within the QTL region (40), and recently it has been shown that Abcg5/8 polymorphisms are associated with HDL levels in humans (1).
DISCUSSION
The ratio of between-groups variance to within-groups variance (the F-ratio) is often used to make inferences about differential gene expression. However, nearly all of the tested genes investigated in our study (i.e., 17,215 genes out of 20,831 genes on the microarray) show significant differences between at least two of the 40 diet-sex-strain experimental groups using this criterion (FDR < 0.01). Nevertheless, strategic exploration of the data revealed meaningful global expression patterns and demonstrated that diet, sex, and strain can each have substantial influence on gene expression response in laboratory mice. For instance, predominant patterns in the data reveal considerable variation in gene expression levels during high-fat feeding compared with low-fat feeding (Fig. 1) and strain-specific differences due to diet or sex (Fig. 2). Functional annotations associated with eigengenes provided additional evidence of the influence of each factor and external validation of our experiment. For example, the gene for the noncoding Xist RNA was associated with differences between sexes (the first eigengene) and is known to be expressed in females where it plays a key role maintaining gene dose equality between sexes (42).
Some strains had notable gene expression differences compared with most of the strains studied. Strains A/J and NZB showed greater gene expression differences between diets than most other mice (Fig. 4), and BALB had a relatively large number of sex-specific differences compared with other strains (Table 1). The importance of using multiple experimental factors to examine the response to dietary fat can be illustrated by strain DBA. Female DBA mice show a response to high-fat diet similar to other strains, but male DBA mice are different from most of the other strains (Fig. 2). In both sexes, strain 129 shows a less pronounced response to high-fat feeding, while strains A/J and NZB have strong responses to diet compared with other strains. These relationships are supported by similarities and differences based on the global gene expression response to diet.
The high-fat atherogenic diet repressed cholesterol biosynthesis and induced immune-related responses in all strains examined (Table 2). The correlation patterns between transcripts and phenotypes confirm that the repression of cholesterol biosynthesis and induction of the immune response are the major responses to atherogenic diet. Levels of TG, NEFA, and GLU are negatively correlated with GLDH, CHOL, and HDL. The transcription of genes involved in cholesterol biosynthesis is repressed during high-fat feeding, when total cholesterol levels are high (Fig. 3, clusters 1A and 1B). Under these conditions, GLDH and HDL levels are also high but levels of TG, NEFA, and GLU are low.
Cholesterol is essential for life and is primarily synthesized from smaller molecules within the body. However, the mice fed the high-fat diet do not need to make cholesterol; it is available in abundance in the diet, and cholic acid aids in the uptake of the steroid. Even so, strains may have noticeable differences with respect to the list of genes that accomplish these responses and the degree to which they achieve such alterations (see Fig. 4A). The induction of the immune response could be due to inflammation, which is known to accompany obesity in adipose tissue (21). However, enrichment analysis of Ingenuity Canonical Pathway members provides evidence that the high-fat diet used here damages the liver (see Table 2). This atherogenic diet-linked damage could promote inflammation and explain the release of GLDH with atherogenic diet (see accompanying phenotype data), which is an indicator of liver damage (33). The atherogenic diet induced particularly large gene expression changes in the immune response in NZB mice in our study (Figs. 2–4), which is consistent with a previous report in which a high-fat diet increased the severity of autoimmune disease in NZB × NZW mice (25).
The statistical framework underlying this study ensures that the effects of diet, strain, and sex on hepatic gene expression are casually upstream of the genomic responses to these factors. However, the transcriptional responses could have been mediated by a number of complex processes, including feedback-driven mechanisms. For example, the introduction of inflammatory cells, followed by inflammation and tissue damage, can occur in response to high-fat diet. Accordingly, liver tissue is likely to respond to dietary conditions by remodeling. Changes in gene expression may also reflect changes in tissue composition or changes in the expression patterns of individual cell types.
The data generated in this strain survey can be mined to further investigate the gene expression response to high-fat diet in mice. We used the expression database to sort rapidly through a large list of potential candidate genes for a QTL and to select a smaller number of genes for further study. It is important to keep in mind that a causal QTL gene may be due to a coding region polymorphism that changes function rather than RNA (and protein) abundance. Such cases should be searched for in the SNP databases. Also, not all genes are tested in a microarray hybridization. In our example, we found 44 genes in the investigated QTL region that had no probe in the microarray. Most, but not all, of these genes were noncoding RNA genes, predicted genes, or pseudogenes. Finally, once the list of genes has been reduced to a small number, they require further testing to demonstrate causal activity. Despite the limitations, our database will enable researchers to search for genes associated with cholesterol production, select candidate model systems to study metabolic traits, and investigate novel questions surrounding diet-induced metabolism.
GRANTS
This work was supported by Novartis Institutes for Biomedical Research (Boston, MA) and by U.S. National Institutes of Health (NIH) Grants GM-076468 (G. A. Churchill) and HL-77796 and HL-74086 (B. J. Paigen). K. R. Shockley acknowledges NIH/National Human Genome Research Institute Ruth L. Kirchstein Postdoctoral Fellowship HG-003968. D. Witmer acknowledges the Howard Hughes Medical Institute, the Betterment Fund Scholarship Endowment, the Clarence Cook Little Scholarship Fund, the L. G. Balfour Foundation, and The Horace W. Goldsmith Foundation.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Keith Sheppard, Dave Walton, and Nazira Bektassova (The Jackson Laboratory, Bar Harbor, ME) for assistance with constructing the public database.
Current addresses: K. R. Shockley, Biostatistics Branch, National Institute of Environmental Health Sciences/National Toxicology Program, 111 TW Alexander Dr., Research Triangle Park, NC 27709; D. Witmer, Dept. of Electrical Engineering and Computer Science, and MIT Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA 02139; S. L. Burgess-Herbert, Zoological Society of San Diego, Conservation Research for Endangered Species, 15600 San Pasqual Valley Rd., Escondido, CA 92027.
Footnotes
1The online version of this article contains supplemental material.
REFERENCES
- 1.Acalovschi M, Ciocan A, Mostean O, Tirziu S, Chiorean E, Keppeler H, Schirin-Sokhan R, Lammert F. Are plasma lipid levels related to ABCG5/ABCG8 polymorphisms? A preliminary study in siblings with gallstones. Eur J Intern Med 17: 490–494, 2006 [DOI] [PubMed] [Google Scholar]
- 2.Allison DB, Kaprio J, Korkeila M, Koskenvuo M, Neale MC, Hayakawa K. The heritability of body mass index among an international sample of monozygotic twins reared apart. Int J Obes Relat Metab Disord 20: 501–506, 1996 [PubMed] [Google Scholar]
- 3.Astrup A, Buemann B, Western P, Toubro S, Raben A, Christenson NJ. Obesity as an adaptation to a high-fat diet: evidence from a cross-sectional study. Am J Clin Nutr 59: 350–355, 1994 [DOI] [PubMed] [Google Scholar]
- 4.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown CD, Higgins M, Donato KA, Rohde FC, Garrison R, Obarzanek E, Ernst ND, Horan M. Body mass index and the prevalence of hypertension and dyslipidemia. Obes Res 8: 605–619, 2000 [DOI] [PubMed] [Google Scholar]
- 6.Burgess-Herbert SL, Cox A, Paigen B. Practical applications of the bioinformatics toolbox for narrowing quantitative trait loci. Genetics 180: 107–113, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carman WJ, Sowers M, Hawthorne VM, Weissfeld LA. Obesity as a risk factor for osteoarthritis of the hand and wrist: a prospective study. Am J Epidemiol 139: 119–129, 1994 [DOI] [PubMed] [Google Scholar]
- 8.Chan JM, Rimm EB, Colditz GA, Stampfer MJ, Willett WC. Obesity, fat distribution, and weight gain as risk factors for clinical diabetes in men. Diabetes Care 17: 961–969, 1994 [DOI] [PubMed] [Google Scholar]
- 9.Chu SY, Lee NC, Wingo PA, Senie RT, Greenberg RS, Peterson HB. The relationship between body mass and breast cancer among women enrolled in the Cancer and Steroid Hormone Study. J Clin Epidemiol 44: 1197–1206, 1991 [DOI] [PubMed] [Google Scholar]
- 10.Colditz GA, Willett WC, Stampfer MJ, Manson JE, Hennekens CH, Arky RA, Speizer FE. Weight as a risk factor for clinical diabetes in women. Am J Epidemiol 132, 501–513, 1990 [DOI] [PubMed] [Google Scholar]
- 11.Cui X, Hwang JT, Qui J, Blades NJ, Churchill GA. Improved statistical tests for differential gene expression by shrinking variance component estimates. Biostatistics 6: 59–75, 2005 [DOI] [PubMed] [Google Scholar]
- 12.DiPetrillo K, Wang X, Stylianou IM, Paigen B. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet 21: 683–692, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Donahue RP, Abbott RD, Bloom E, Reed DM, Yano K. Central obesity and coronary heart disease in men. Lancet 1: 821–824, 1987 [DOI] [PubMed] [Google Scholar]
- 14.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics 23: 257–258, 2007 [DOI] [PubMed] [Google Scholar]
- 15.Felson DT, Anderson JJ, Naimark A, Walker AM, Meenan RF. Obesity and knee osteoarthritis. The Framingham Study. Ann Intern Med 109: 18–24, 1988 [DOI] [PubMed] [Google Scholar]
- 16.Garfinkel L. Overweight and mortality. Cancer 58: 1826–1829, 1986 [DOI] [PubMed] [Google Scholar]
- 17.Gautier L, Cope LM, Bolstad BM, Irizarry RA. Affy-Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315, 2004 [DOI] [PubMed] [Google Scholar]
- 18.Ghazalpour A, Doss S, Zhang B, Wang S, Plaisier C, Costellanos R, Brozell A, Schadt EE, Drake TA, Lusis AJ, Horvath S. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet 2: e130, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giovannucci E, Colditz GA, Stampfer MJ, Willett WC. Physical activity, obesity, and risk of colorectal adenoma in women (United States). Cancer Causes Control 7: 253–263, 1996 [DOI] [PubMed] [Google Scholar]
- 20.Grunhage F, Acalovschi M, Tirziu S, Walier M, Wienker TF, Ciocan A, Mosteanu O, Sauerbruch T, Lammert F. Increased gallstone risk in humans conferred by common variant of hepatic ATP-binding cassette transporter for cholesterol. Hepatology 46: 793–801, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Guzik TJ, Mangalat D, Korbut R. Adipocytokines-novel link between inflammation and vascular function? J Physiol Pharmacol 57: 505–528, 2006 [PubMed] [Google Scholar]
- 22.Irizarry RA, Bolstad BM, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264, 2003 [DOI] [PubMed] [Google Scholar]
- 23.Kerr MK, Churchill GA. Bootstrapping cluster analysis: assessing the reliability of conclusions from microarray experiments. Proc Natl Acad Sci USA 98: 8961–8965, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koza RA, Nikonova L, Hogan J, Rim JS, Mendoza T, Faulk C, Skaf J, Kozak LP. Changes in gene expression foreshadow diet-induced obesity in genetically identical mice. PLoS Genet 2: e81, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy JA, Ibrahim AB, Shirai T, Ohta K, Nagasawa R, Yoshida H, Estes J, Gardner M. Dietary fat affects immune response, production of antiviral factors, and immune complex disease in NZB/NZW mice. Proc Natl Acad Sci USA 79: 1974–1978, 1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin S, Thomas TC, Storlien LH, Huang XF. Development of high fat diet-induced obesity and leptin resistance in C57Bl/6J mice. Int J Obes 24: 639–646, 2000 [DOI] [PubMed] [Google Scholar]
- 27.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 33: D54–D58, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mokdad AH, Ford ES, Bowman BA, Dietz WH, Vinicor F, Bales VS, Marks JS. Prevalence of obesity, diabetes, and obesity-related health risk factors, 2001. JAMA 289: 76–79, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Newton MA, Quintana FA, den Boon JA, Sengupta S, Ahlquist P. Random-set methods identify distinct aspects of the enrichment signal in gene-set analysis. Ann Appl Stat 1: 85–106, 2007 [Google Scholar]
- 30.Paigen B, Morrow A, Brandon C, Mitchell D, Holmes PA. Variation in susceptibility to atherosclerosis among inbred strains of mice. Atherosclerosis 57: 65–73, 1985 [DOI] [PubMed] [Google Scholar]
- 31.Rexrode KM, Hennekens CH, Willett WC, Colditz GA, Stampfer MJ, Rich-Edwards JW, Speizer FE, Manson JE. A prospective study of body mass index, weight change, and risk of stroke in women. JAMA 277: 1539–45, 1997 [DOI] [PubMed] [Google Scholar]
- 32.Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics 20: 3246–3248, 2004 [DOI] [PubMed] [Google Scholar]
- 33.Schmidt ES, Schmidt FW. Glutamate dehydrogenase: biochemical and clinical aspects of an interesting enzyme. Clin Chim Acta 173: 43–55, 1988 [DOI] [PubMed] [Google Scholar]
- 34.Shockley KR, Churchill GA. Gene expression in mouse chromosome substitution strains. Mamm Genome 17: 598–614, 2006 [DOI] [PubMed] [Google Scholar]
- 35.Stampfer MJ, Maclure KM, Colditz GA, Manson JE, Willett WC. Risk of symptomatic gallstones in women with severe obesity. Am J Clin Nutr 55: 652–658, 1992 [DOI] [PubMed] [Google Scholar]
- 36.Storey JD. A direct approach to false discovery rates. J R Stat Soc B 64: 479–498, 2002 [Google Scholar]
- 37.Surwit RS, Feinglos MN, Rodin J, Sutherland A, Petro AE, Opara EC, Kuhn CM, Rebuffé-Scrive M. Differential effects of fat and sucrose on the development of obesity and diabetes in C57BL/6J and A/J mice. Metabolism 44: 645–51, 1995 [DOI] [PubMed] [Google Scholar]
- 38.Walker SP, Rimm EB, Ascherio A, Kawachi I, Stampfer MJ, Willett WC. Body size and fat distribution as predictors of stroke among US men. Am J Epidemiol 144: 1143–1150, 1996 [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Beydoun MA. The obesity epidemic in the United States-gender, age, socioeconomic, racial/ethnic, and geographic characteristics: a systematic review and meta-regression analysis. Epidemiol Rev 29: 6–28, 2007 [DOI] [PubMed] [Google Scholar]
- 40.Wittenburg H, Lyons MA, Li R, Churchill GA, Carey MC, Paigen B. FXR and ABCG5/ABCG8 as determinants of cholesterol gallstone formation from quantitative trait locus mapping in mice. Gastroenterology 125: 868–881, 2003 [DOI] [PubMed] [Google Scholar]
- 41.Wu H, Kerr MK, Cui XQ, Churchill GA. MAANOVA: a software package for the analysis of spotted cDNA microarray experiments. In: The Analysis of Gene Expression Data: an Overview of Methods and Software, edited by Parmgiani G, Garrett ES, Irizarry RA, Zeger SL. New York: Springer, 2003, p. 313–431 [Google Scholar]
- 42.Wust A. Xist function: bridging chromatin and stem cells. Trends Genet 23: 457–464, 2007 [DOI] [PubMed] [Google Scholar]
- 43.Yang H, Churchill GA. Estimating p-values in small microarray experiments. Bioinformatics 23: 38–43, 2007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.