Abstract
The Postn gene encodes protein periostin. During embryonic development, it is highly expressed in the outflow tract (OFT) endocardial cushions of the developing heart, which give rise to several structures of the mature heart including the aortic valve. Periostin was previously implicated in osteoblast differentiation, cancer metastasis, and tooth and bone development, but its role in cardiac OFT development is unclear. To elucidate the role that periostin plays in the developing heart we analyzed cardiac OFT phenotype in mice after deletion of the Postn gene. We found that lack of periostin in the embryonic OFT leads to ectopic expression of the proosteogenic growth factor pleiotrophin (Ptn) and overexpression of delta-like 1 homolog (Dlk1), a negative regulator of Notch1, in the distal (prevalvular) cushions of the OFT. This resulted in suppression of Notch1 signaling, strong induction of the central transcriptional regulator of osteoblast cell fate Runx2, upregulation of osteopontin and osteocalcin expression, and subsequent calcification of the aortic valve. Our data suggest that periostin represses a default osteogenic program in the OFT cushion mesenchyme and promotes differentiation along a fibrogenic lineage. Lack of periostin causes derepression of the osteogenic potential of OFT mesenchymal cells, calcium deposition, and calcific aortic valve disease. These results establish periostin as a key regulator of OFT endocardial cushion mesenchymal cell fate during embryonic development.
Keywords: development, outflow tract, endocardial cushions
a family of notch receptors controls a highly conserved signaling pathway that regulates a multitude of biological processes, including cell fate specification, progenitor cell maintenance, tissue boundary formation, proliferation, and apoptosis. Notch pathway plays a crucial role in cardiac development as evidenced by the wide range of cardiac phenotypes in mice with targeted disruption of genes encoding various components of the Notch signaling pathway (15, 28). These data indicate that Notch is involved in cardiac mesoderm differentiation during early embryonic development, atrioventricular canal development, epithelial-to-mesenchymal transition and valve development, ventricular trabeculation, and outflow tract (OFT) development. Human mutations in several Notch signaling components result in congenital cardiovascular malformations such as Alagille syndrome and tetralogy of Fallot (15, 28). Loss-of-function mutations in NOTCH1 cause aortic valve disease (14). Although a consensus has emerged that the Notch1 signaling cascade plays a key role in aortic valve disease (13), two recent genomewide linkage studies for valve calcification susceptibility loci suggested that NOTCH1 mutations do not account for all cases of calcific valve disease and found that other genes are involved (4, 25).
Periostin, a 90-kDa secreted protein, was first isolated from the mesenchymal cell line MC3T3-E1 (16). Subsequent studies revealed that periostin plays an important regulatory role in osteoblast differentiation (37), cancer metastasis (3), and embryonic development of teeth and skeleton (41). We have shown (20, 21, 30) that periostin is also expressed in the embryonic heart. Its expression first appears in the mouse heart at embryonic day (E) 10.5 and is localized to the prevalvular endocardial cushions of the OFT and atrioventricular canal (21). However, the role of periostin in cardiac development is unclear. In this study, we analyzed the role of periostin in OFT development, using periostin-knockout (Postn−/−) mice. Our data suggest that by modulating Notch1 signaling periostin promotes OFT mesenchymal cell differentiation along a fibrogenic lineage while repressing a default osteogenic program. Lack of periostin causes derepression of the osteogenic potential of OFT cushion mesenchyme and calcific aortic valve disease.
MATERIALS AND METHODS
Periostin-knockout mice.
All animals received water and food ad libitum and were maintained in accordance with a protocol approved by the Medical University of South Carolina's Institutional Animal Care and Use Committee. The colony of Postn−/− mice was maintained on the C57BL/6J background from original stock described by Oka et al. (35). A total of 73 wild-type, 137 Postn+/−, and 48 Postn−/− animals were analyzed. Genotype was determined by PCR analysis of genomic DNA from either tails (adult animals) or placentas (embryos). Genomic DNA was isolated and amplified by PCR using the following primers: 5′-CCTTGCCAGTCTCAATGAAGG-3′ (forward primer) and 5′-TGACAGAGTGAACACATGCC-3′ (reverse primer directed against wild-type allele) or 5′-GGAAGACAATAGCAGGCATG-3′ (reverse primer corresponding to mutant allele).
Morphology.
Gross anatomic examination of the Postn−/− hearts was carried out under a Leica MZ 7.5 stereomicroscope, and images were captured with a Leica EC3 digital camera (Leica Microsystems). Masson's trichrome staining was performed with a Masson's trichrome stain kit (DakoCytomation). Calcium deposits were identified with a calcium phosphate-specific von Kossa stain kit (Diagnostic BioSystems) and alizarin red S reagent (Sigma-Aldrich). Sections were viewed with a Leitz Ortholux II microscope.
Echocardiographic analysis of cardiac function.
Echocardiography was performed with a Vevo 660 ultrasound system equipped with a 40-MHz scanhead (VisualSonics). Two-dimensional imaging and Doppler interrogation of blood flow was done as previously described (55). Mice were anesthetized with 4% isoflurane, and anesthesia was maintained with 2% isoflurane. Hair was removed from the ventral thorax with a hair removal cream (Nair, Carter-Horner). Body temperature was monitored continuously with a rectal probe (Indus Instruments) and maintained at 36–38°C with a heated platform and a small heat lamp.
Differential screening.
Screening for differentially expressed genes in the hearts of Postn−/− mice was performed with a suppression-subtractive-hybridization-based technique essentially as previously described (50). Poly(A)+ RNA was isolated from either 10 wild-type or 10 Postn−/− E12.5 hearts. Subtracted/normalized cDNA libraries were generated with a PCR-Select cDNA Subtraction Kit (Clontech). Secondary PCR products were cloned into pCRII-TOPO vector with a TOPO TA cloning kit (Invitrogen), and resulting libraries were organized in 384-well plates. Bacterial clones from these plates were manually printed onto Hybond-XL nylon membranes (GE Healthcare) by using a 384-pin tool (V&P Scientific) to prepare two identical sets of bacterial colony filters. Six sets of duplicate filters each containing 384 randomly picked clones of either library (total of 2,304 clones) were hybridized with 32P-labeled subtracted cDNA probes, and differential clones were selected as previously described (50).
In situ hybridizations.
In situ hybridizations were performed essentially as previously described (53). E12.5 C57BL and Postn−/− embryos were dissected in ice-cold PBS, fixed in 4% paraformaldehyde in PBS for 30 min, cryoprotected in 30% sucrose in PBS, and sectioned in a cryostat at a thickness of 10 μm. Sections were hybridized with digoxigenin (DIG)-labeled cDNA probes followed by incubation with anti-DIG antibodies conjugated with horseradish peroxidase (HRP) (Roche). The peroxidase activity was localized and amplified with biotinyl tyramide amplification reagent (PerkinElmer) followed by alkaline phosphatase-conjugated streptavidin. The signal was detected with nitro blue tetrazolium (NBT; 0.25 mg/ml) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP; 0.125 mg/ml) (Roche) as substrates. For the double fluorescent in situ hybridizations, the first probe was labeled with DIG while the second probe was labeled with fluorescein. Slides were incubated with either anti-DIG HRP-conjugated antibodies (Roche) or anti-fluorescein HRP-conjugated antibodies (Roche). Detection of the signal was carried out with the Fluorophore Tyramide Amplification System (Molecular Probes) according to the manufacturer's protocol. Periostin was visualized with Alexa Fluor 488 (green), and the second gene was visualized with Alexa Fluor 546 (red).
Real-time PCR.
cDNA synthesized with 1 μg of mRNA isolated from either 10 wild-type or 10 Postn−/− E12.5 hearts was used for PCR reactions, which were carried out with AmpliTaqGold thermostable DNA polymerase (Applied Biosystems). SuperROX (Biosearch Technologies) was used as a passive reference at a final concentration of 300 nM. The level of gene expression in the hearts was estimated by multiplex real-time PCR using gene-specific primers and TaqMan probes: Ptn: 5′-CCATGAAGACTCAGAGATGTAAGATCC-3′ (forward primer), 5′-CAAGGCGGTATTGAGGTCACATT-3′ (reverse primer), and 6FAM-AGCCTGGAACTGGTACTTGCACTCAGCTC-BHQ1 (TaqMan probe); Dlk1: 5′-CCCTCTGTGACAAGTGTGTAACTG-3′ (forward primer), 5′-GGAGCATTCGTACTGGCCTTTC-3′ (reverse primer), and 6FAM-CCAGGTCCACGCAAGTTCCATTGTTGGC-BHQ1 (TaqMan probe); Notch1: 5′-CCAACTGAGGACAGACGGACT-3′ (forward primer), 5′-GTGAGCACAGCCCTGAACCA-3′ (reverse primer), and 6FAM-CCCAGAAGCAGGCTCCACCCACATTCCAG-BHQ1 (TaqMan probe); Hes1: 5′-GTCCTAACGCAGTGTCACCTTC-3′ (forward primer), 5′-ATGGTCAGTCACTTAATACAGCTCTC-3′ (reverse primer), and 6FAM-TGGCTCCTCGCTCACTTCGGACTCCA-BHQ1 (TaqMan probe); Hey1: 5′-GAGGTGAAGGGAGAAAGGTGTCT-3′ (forward primer), 5′-CAAGTGCAGGCAAGGTCTACAT-3′ (reverse primer), and 6FAM-CTGTCTTGCTTGCTGCCAACGCTCCAA-BHQ1 (TaqMan probe); Hey2: 5′-AGCAATCAGCCCACCCTTGT-3′ (forward primer), 5′-CCGAAATGGACGACATCAGTTACT-3′ (reverse primer), and 6FAM-TCCGCAGCCTCCAGTCCTCAGCAGA-BHQ1 (TaqMan probe); Runx2: 5′-ACTCTTCTGGAGCCGTTTATGTG-3′ (forward primer), 5′-CCTTTGAGAACCGTGTGCATCT-3′ (reverse primer), and 6FAM-ACTAACCAACCCTTCCCTCCACTTCCCTG-BHQ1 (TaqMan probe); Bglap1/2: 5′-ATGTCCAAGCAGGAGGGCAATA-3′ (forward primer), 5′-CTCGTCACAAGCAGGGTTAAGC-3′ (reverse primer), and 6FAM-AGTGAACAGACTCCGGCGCTACCTTGG-BHQ1 (TaqMan probe); SPP1: 5′-CACATGAAGAGCGGTGAGTCTAAG-3′ (forward primer), 5′-GCTTTGGAACTTGCTTGACTATCG-3′ (reverse primer), and 6FAM-AGTCCCTCGATGTCATCCCTGTTGCCCA-BHQ1 (TaqMan probe). Gapdh was used as an internal control for normalization: 5′-CCAATGTGTCCGTCGTGGATC-3′ (forward primer), 5′-TGTAGCCCAAGATGCCCTTCA-3′ (reverse primer), and VIC-CCCTCAGATGCCTGCTTCACCACCTTCTT-MGBNFQ (TaqMan probe). PCRs were carried out on the 7300 Real-Time PCR System (Applied Biosystems), and data were processed with the manufacturer's software.
RESULTS
Loss of periostin leads to calcific aortic valve disease.
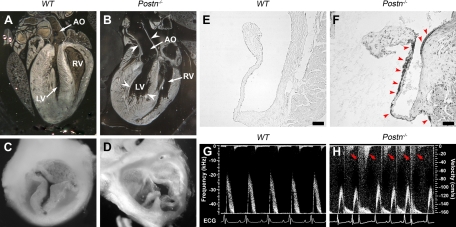
To explore the role of periostin in heart development, we analyzed the cardiac phenotype in mutant mice with targeted deletion of Postn gene (Postn−/− mice). Gross anatomic examination of adult hearts revealed that the hearts of the 10-mo-old Postn−/− mice had a pronounced dilation of the left ventricle and the aortic root (Fig. 1, A and B). A closer examination revealed that aortic valves in Postn−/− mice were severely deformed and had a bicuspid-like morphology (Fig. 1, C and D). These pathological features, which often accompany severe cases of calcific aortic valve disease (5), were fully penetrant. Therefore, we investigated whether aortic valve leaflets of Postn−/− mice undergo calcification. The calcium-specific von Kossa staining revealed that the leaflets of the aortic valve in 6-mo-old Postn−/− mice have extensive calcium deposits, while no such deposits were observed in the age-matched wild-type animals (Fig. 1, E and F; Fig. 2). Occasional small calcium deposits were observed in 3-mo-old Postn−/− animals (data not shown), while no calcifications were found in neonatal Postn−/− mice. Since backflow of the blood through a leaky calcified aortic valve (aortic regurgitation) represents one of the main pathological features of calcific aortic valve disease (5), we analyzed cardiac blood flow in Postn−/− mice, using Doppler echocardiography that revealed aortic regurgitation in 6-mo-old Postn−/− animals (Fig. 1, G and H). One of the hallmarks of pediatric aortic valve disease is a reduced survival rate; therefore, we analyzed the survival rate of the offspring from Postn+/− × Postn+/− matings and found that the survival rate of the Postn−/− animals was significantly reduced compared with both Postn+/+ and Postn+/− animals (Table 1). We did not detect any pathological changes in the Postn+/− hearts. Thus only Postn−/− mice exhibited pathological features characteristic of aortic valve disease.
Fig. 1.
Postn−/− mice exhibit pathological features reminiscent of calcific aortic valve disease. A and B: left ventricle (LV) and aortic root were severely dilated (white arrowheads) in 10-mo-old Postn−/− mice (B) compared with age-matched wild-type (WT) control mice (A). C and D: aortic valve in 10-mo-old Postn−/− mice was severely deformed and had a bicuspid-like morphology (D) compared with age-matched WT control mice (C). E and F: calcification of the aortic valve leaflets (black precipitates, red arrowheads) in 6-mo-old Postn−/− mice (F) revealed by von Kossa stain; no calcification was observed in age-matched WT control mice (E). G and H: Doppler echocardiographic examination of the cardiac blood flow in 6-mo-old Postn−/− mice (H) revealed aortic regurgitation (red arrows) compared with WT control mice (G). AO, aorta; bpm, beats/minute; ECG, electrocardiogram; RV, right ventricle. Scale bars, 50 μm.
Fig. 2.
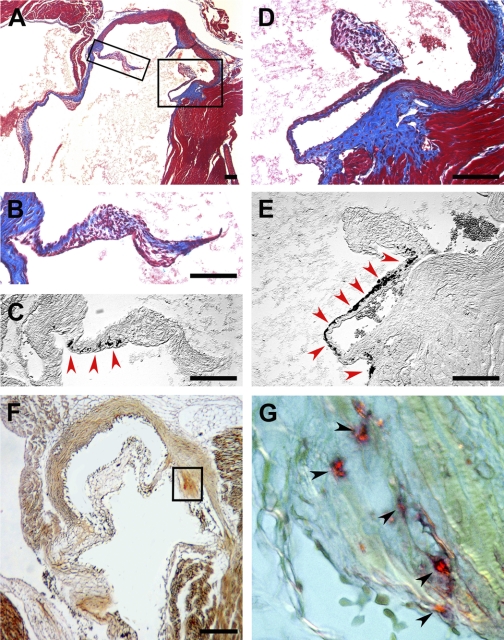
Calcification of the aortic valve leaflets in 6-mo-old Postn−/− mice. A: Masson's trichrome staining of the aortic valve from 6-mo-old Postn−/− mouse. B–E: high magnification of insets in A depicting aortic valve leaflet stained with Masson's trichrome stain (B and D) and adjacent section stained for calcium deposits with von Kossa stain (black precipitates, red arrowheads; C and E). F: aortic valve leaflet stained for calcium deposits with alizarin red (orange-red precipitates). G: high magnification of inset in F depicting aortic valve leaflet stained with alizarin red (calcium deposits, orange-red precipitates, black arrowheads). Scale bars, 100 μm.
Table 1.
Analysis of Postn−/− offspring survival rate
| Mating (n) | Offspring |
Litter Size (means ± SD) | ||
|---|---|---|---|---|
| Postn−/− | Postn+/− | WT | ||
| WT × WT (7) | 0 | 0 | 36 | 5.1±1.5 |
| Postn+/− × Postn+/− (69) | 48 (19%/25%) | 137 (53%/50%) | 73 (28%/25%) | 3.7±1.9 |
| Postn−/− × Postn−/− (3) | 8 | 0 | 0 | 2.7±0.6 |
Number of offspring with Postn−/− genotype from Postn+/− × Postn+/− matings was significantly reduced compared with normal Mendelian distribution [P < 0.022, Postn−/− vs. wild type (WT), n = 69]. As evidenced by the litter size, the survival rate of Postn−/− mice was significantly lower compared with both Postn+/− (P < 0.05) and WT (P < 0.005) animals. n, No. of matings. Percentages of offspring per genotype are shown at postnatal day 1 compared with expected frequency from Mendelian distribution (bold).
Ptn and Dlk1 are overexpressed in Postn−/− hearts.
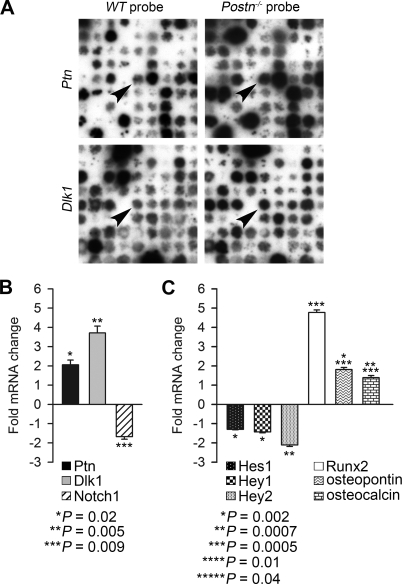
Since we have shown (21) that periostin is highly expressed in the prevalvular endocardial cushions of the OFT during embryonic development as early as E10.5, we hypothesized that adult valve calcific disease can have its roots in embryonic development. To determine whether the adult aortic valve disease has a developmental basis and to unravel the molecular mechanism of aortic valve calcification in Postn−/− mice, we analyzed alterations of gene expression in the hearts of Postn−/− E12.5 embryos (vs. wild-type embryos), i.e., 2 days after the onset of Postn gene expression in the heart. To do so, we used a strategy for a gene expression profiling based on a combination of suppression subtractive hybridization and reverse Northern analysis (50). Two subtracted cDNA libraries were generated, one enriched in clones upregulated and the other enriched in clones downregulated in the hearts of Postn−/− embryos. One thousand one hundred and fifty-two clones from each library were organized on nylon filters. After these filters were hybridized with subtracted cDNA probes, we identified 69 genes with a difference in expression between the hearts of Postn−/− mice and wild-type animals (Table 2). Our data suggested that two genes, pleiotrophin (Ptn) and delta-like 1 homolog (Dlk1), among others, were upregulated in the heart of Postn−/− mice (Fig. 3A).
Table 2.
Genes differentially expressed in heart of Postn−/− mouse
| Clone Name | GB_ACC | Gene Name | Gene Symbol | UniGene ID | Expression Postn−/−/WT |
|---|---|---|---|---|---|
| NTA2A14 | EF437324 | Expressed in nonmetastatic cells 2, protein | Nme2 | Mm.1260 | ↓ |
| NTA2B16 | EF437325 | Phosphoglycerate kinase 1 | Pgk1 | Mm.336205 | ↓ |
| NTA2B18 | EF437326 | Eukaryotic translation elongation factor 1 ε1 | Eef1e1 | Mm.36683 | ↓ |
| NTA2C11 | EF437327 | Leucine rich repeat containing 41 | Lrrc41 | Mm.260786 | ↓ |
| NTA2C12 | EF437328 | Iron responsive element binding protein 2 | Ireb2 | Mm.208991 | ↓ |
| NTA2C17 | EF437329 | Integrin β3 binding protein (β3-endonexin) | Itgb3bp | Mm.257094 | ↓ |
| NTA2D10 | EF437330 | Pleiomorphic adenoma gene-like 1 | Plagl1 | Mm.287857 | ↓ |
| NTA2D14 | EF437331 | Unknown | ↓ | ||
| NTA2D15 | EF437332 | RD RNA-binding protein | Rdbp | Mm.279907 | ↓ |
| NTA2D16 | EF437333 | Solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), 5 | Slc25a5 | Mm.371544 | ↓ |
| NTA2E1 | EF437334 | ATPase, Na+/K+ transporting, β1 polypeptide | Atp1b1 | Mm.4550 | ↓ |
| NTA2E20 | EF437335 | Coenzyme Q2 homolog, prenyltransferase (yeast) | Coq2 | Mm.260661 | ↓ |
| NTA2F8 | EF437336 | RIKEN cDNA 4921517L17 gene | 4921517L17Rik | Mm.435448 | ↓ |
| NTA2G11 | EF437337 | Septin 2 | Sept2 | Mm.428652 | ↓ |
| NTA2H11 | EF437339 | Ribosomal protein L14 | Rpl14 | Mm.289810 | ↓ |
| NTA2H12 | EF437340 | Nuclear autoantigenic sperm protein (histone-binding) | Nasp | Mm.257181 | ↓ |
| NTA2H2 | EF437338 | RIKEN cDNA 1810026J23 gene | 1810026J23Rik | Mm.39704 | ↓ |
| NTA2J19 | EF437341 | Aldolase 1, A isoform | Aldoa | Mm.275831 | ↓ |
| NTA2K23 | EF437342 | Selenoprotein | Sep15 | Mm.29812 | ↓ |
| NTA2L11 | EF437343 | Phosphoglucomutase 2 | Pgm2 | Mm.217764 | ↓ |
| NTA2L12 | EF437344 | Coiled-coil domain containing 132 | Ccdc132 | Mm.45539 | ↓ |
| NTA2L13 | EF437345 | NOP17 | Nop17 | Mm.181689 | ↓ |
| NTA2L17 | EF437347 | Ubiquitin A-52 residue ribosomal protein fusion product 1 | Uba52 | Mm.297372 | ↓ |
| NTA2L18 | EF437348 | A disintegrin and metallopeptidase domain 19 (meltrin β) | Adam19 | Mm.89940 | ↓ |
| NTA2M16 | EF437351 | RNA binding motif protein, X chromosome retrogene | Rbmxrt | Mm.24718 | ↓ |
| NTA2M18 | EF437352 | F-box protein 11 | Fbxo11 | Mm.386857 | ↓ |
| NTA2M6 | EF437349 | Myeloblastosis oncogene-like 2 | Mybl2 | Mm.4594 | ↓ |
| NTA2M7 | EF437350 | Expressed sequence AI506816 (AI506816) | AI506816 | Mm.205125 | ↓ |
| NTA2N13 | EF437353 | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase | Atic | Mm.38010 | ↓ |
| NTA3B12 | EF437354 | Zinc finger protein 289 | Zfp289 | Mm.43636 | ↓ |
| NTA3B13 | EF437355 | Myosin, light polypeptide 6, alkali, smooth muscle and nonmuscle | Myl6 | Mm.337074 | ↓ |
| NTA3C8 | EF437356 | ATPase, H+ transporting, lysosomal V1 subunit B2 | Atp6v1b2 | Mm.249096 | ↓ |
| NTA3I8 | EF437357 | Transmembrane protein 126A | Tmem126a | Mm.28963 | ↓ |
| NTA3J20 | EF437358 | KRIT1, ankyrin repeat containing | Krit1 | Mm.32368 | ↓ |
| NTA3J21 | EF437359 | Exocyst complex component 7 | Exoc7 | Mm.22530 | ↓ |
| NTA3K14 | EF437360 | Eukaryotic translation initiation factor 3, subunit 5 (ε) | Eif3 s5 | Mm.182962 | ↓ |
| NTA3K20 | EF437361 | Lysosomal-associated protein transmembrane 4A | Laptm4a | Mm.30071 | ↓ |
| NTA3K21 | EF437362 | Zinc finger, AN1-type domain 5 | Zfand5 | Mm.292405 | ↓ |
| NTA3K23 | EF437363 | Farnesyltransferase, CAAX box, β | Fntb | Mm.151174 | ↓ |
| NTA3K24 | EF437364 | Ras homolog gene family, member U | Rhou | Mm.168257 | ↓ |
| NTA3L6 | EF437365 | Diazepam binding inhibitor | Dbi | Mm.2785 | ↓ |
| NTA3L7 | EF437366 | Pyrophosphatase (inorganic) 1 | Ppa1 | Mm.28897 | ↓ |
| NTA3L8 | EF437367 | Glutamate dehydrogenase 1 | Glud1 | Mm.10600 | ↓ |
| NTA3O4 | EF437368 | Eukaryotic translation elongation factor 1 α1 | Eef1a1 | Mm.435662 | ↓ |
| PTA4A4 | EF437369 | Myeloid/lymphoid or mixed lineage-leukemia translocation to 3 homolog (Drosophila) | Mllt3 | Mm.288898 | ↑ |
| PTA4B19 | EF437370 | Mitochondrial cytb gene for cytochrome b | Cytb | ↑ | |
| PTA4B20 | EF437371 | Zinc finger, AN1-type domain 3 | Zfand3 | Mm.220972 | ↑ |
| PTA4B21 | EF437372 | Low-density lipoprotein receptor-related protein 10 | Lrp10 | Mm.28465 | ↑ |
| PTA4B22 | EF524068 | Stathmin 1 | Stmn1 | Mm.378957 | ↑ |
| PTA4C2 | EF437373 | Insulin-like growth factor 2 (Igf2) | Igf2 | Mm.3862 | ↑ |
| PTA4C6 | EF437374 | Procollagen, type VI, α1 | Col6a1 | Mm.2509 | ↑ |
| PTA4D10 | EF437375 | SCY1-like 1 (Saccharomyces cerevisiae) | Scyl1 | Mm.276063 | ↑ |
| PTA4D22 | EF437376 | Nucleoporin 205 | Nup205 | Mm.261208 | ↑ |
| PTA4F21 | EF437377 | Integrator complex subunit 2 | Ints2 | Mm.171304 | ↑ |
| PTA4G24 | EF437378 | Potassium voltage gated channel, Shab-related subfamily, member 1 | Kcnb1 | Mm.387390 | ↑ |
| PTA4I7 | EF437379 | Pleiotrophin | Ptn | Mm.279690 | ↑ |
| PTA4J20 | EF437380 | ADP-ribosylation factor-like 6 interacting protein 1 | Arl6ip1 | Mm.29924 | ↑ |
| PTA4K23 | EF437381 | UDP-glucose ceramide glucosyltransferase-like 1 | Ugcgl1 | Mm.261022 | ↑ |
| PTA4L14 | EF437382 | Zinc finger protein 422, related sequence 1 (Zfp422-rs1) | Zfp422-rs1 | Mm.276296 | ↑ |
| PTA4N24 | EF437383 | SAC1 (suppressor of actin mutations 1, homolog)-like (S. cerevisiae) | Sacm1l | Mm.273671 | ↑ |
| PTA4P11 | EF437384 | Metastasis associated lung adenocarcinoma transcript 1 | Malat1 | Mm.392400 | ↑ |
| PTA5H10 | EF437386 | Troponin T2, cardiac | Tnnt2 | Mm.247470 | ↑ |
| PTA5H6 | EF437385 | Purine rich element binding protein B | Purb | Mm.296150 | ↑ |
| PTA5K10 | EF437387 | Neuroguidin, EIF4E binding protein | Ngdn | Mm.21214 | ↑ |
| PTA5M16 | EF437388 | Hexosaminidase (glycosyl hydrolase family 20, catalytic domain) containing | Hexdc | Mm.275130 | ↑ |
| PTA5N12 | EF437389 | Dehydrodolichyl diphosphate synthase | Dhdds | Mm.100290 | ↑ |
| PXL2L15 | EF437346 | Delta-like 1 homolog (Drosophila) | Dlk1 | Mm.157069 | ↑ |
| PXL3J19 | EF524069 | Tafazzin | Taz | Mm.268483 | ↑ |
| PXL3N17 | EF524070 | Ras homolog gene family, member G | Rhog | Mm.259795 | ↑ |
Reverse Northern analysis using subtracted probes resulted in identification of 141 differential clones that gave a strong hybridization signal with 1 of the probes and at least 10-fold weaker signal with the probe subtracted in the opposite direction. Sequencing analysis of the corresponding cDNAs identified a total of 69 differentially expressed genes, thus revealing an average redundancy of 2.0 that indicates a very good coverage of genes differentially expressed in embryonic day 12.5 Postn−/− hearts.
Fig. 3.
Analysis of gene expression in embryonic day (E) 12.5 Postn−/− hearts. A: differential gene expression analysis identified Ptn and Dlk1 as genes overexpressed (arrowheads) in E12.5 Postn−/− hearts. B: real-time PCR confirmed significant increase in expression of both Ptn and Dlk1 and revealed downregulation of Notch1 gene expression in E12.5 Postn−/− hearts. C: real-time PCR analysis of the expression of Notch1 downstream target genes revealed that expression of Hes1, Hey1, and Hey2 (direct downstream targets of Notch1) were diminished in E12.5 Postn−/− hearts. Conversely, expression of Runx2 and 2 main effectors of osteoblast mineralization, osteopontin and osteocalcin, were strongly upregulated in E12.5 Postn−/− hearts. Bars represent means ± SD (n = 9).
Ptn and Dlk1/Notch1 signaling cascades have been implicated in Runx2-mediated osteoblast differentiation (2, 14, 49, 54). Therefore, we analyzed the expression of Ptn, Dlk1, and Notch1 in E12.5 Postn−/− and wild-type hearts with real-time quantitative PCR. Both Ptn and Dlk1 expression were significantly upregulated in Postn−/− hearts, while Notch1 expression was reduced (Fig. 3B; Table 3). Together, these data suggested that Ptn and Dlk1 upregulation in Postn−/− mice may trigger changes in the Notch1 signaling cascade causing activation of osteoblast-specific gene expression program in the mesenchymal cells of the endocardial cushions of the OFT and calcification of the aortic valve.
Table 3.
Real-time PCR data
| Gene | Ratio Postn−/−/WT | SD | P Value (n = 9) |
|---|---|---|---|
| Ptn | 2.1 | 0.25 | 0.02283 |
| Dlk1 | 3.7 | 0.35 | 0.00455 |
| Notch1 | −1.7 | 0.13 | 0.00879 |
| Hes1 | −1.3 | 0.03 | 0.00198 |
| Hey1 | −1.4 | 0.04 | 0.00181 |
| Hey2 | −2.1 | 0.08 | 0.00066 |
| Runx2 | 4.8 | 0.14 | 0.00055 |
| Osteocalcin (Bglap1/2) | 1.4 | 0.11 | 0.03879 |
| Osteopontin (SPP1) | 1.8 | 0.11 | 0.00954 |
n, No. of samples per group.
Periostin functions upstream of Ptn, Dlk1, and Notch1.
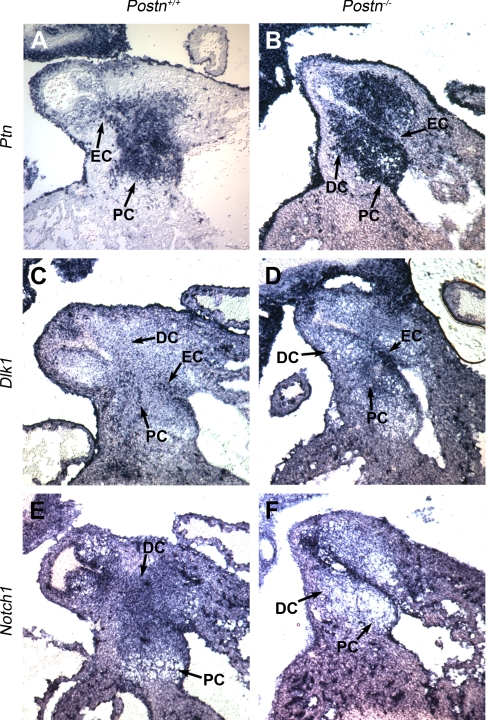
To test the hypothesis that lack of periostin may activate Notch1-mediated calcification of the aortic valve, we determined the patterns of expression of Ptn, Dlk1, and Notch1 in the heart during embryonic development and investigated how they are affected by the absence of periostin. We analyzed expression of these genes in E12.5 wild-type and Postn−/− embryos, using in situ hybridization focusing on the OFT. All three genes were expressed in the endocardial cushions of the OFT (Fig. 4). Specifically, in wild-type embryos, Ptn expression was restricted to the mesenchyme and the endocardial cells of the proximal (preseptal) endocardial cushions (Fig. 4A). Dlk1 was expressed in the OFT endocardium and the mesenchyme of both the proximal and distal cushions (Fig. 4C). Notch1 mRNA transcripts were mostly restricted to the mesenchyme of the distal portion of the OFT (Fig. 4E). Consistent with the real-time PCR data, we observed an increase in the abundance of Ptn and Dlk1 mRNA transcripts and a decrease in the abundance of Notch1 mRNA transcripts in the OFT of Postn−/− embryos, with the biggest difference in the distal endocardial cushions, which give rise to the leaflets of the outlet (arterial) valves (Fig. 4). Remarkably, we also observed that in the Postn−/− embryos the Ptn expression domain, normally restricted to the proximal (nonvalvular) OFT cushions, extended into the distal (valvular) cushions (Fig. 4, A and B). These findings further suggested that Ptn, Dlk1, and Notch1 expression are developmentally regulated by periostin.
Fig. 4.
In situ hybridizations revealed abnormal expression of Ptn, Dlk1, and Notch1 in E12.5 Postn−/− outflow tract (OFT). Nonradioactive in situ hybridization on frontal sections of E12.5 OFT is shown. A and B: Ptn expression pattern. Ptn expression in WT embryos (A) was restricted to the proximal endocardial cushion (PC) and endocardium (EC). In Postn−/− embryos (B), Ptn expression was markedly increased and the expression domain extended into the distal endocardial cushion (DC). C and D: Dlk1 expression pattern. Dlk1 mRNA transcripts were found throughout the OFT cushion mesenchyme and endocardium in both WT (C) and Postn−/− (D) embryos. Note a significant increase of Dlk1 expression level in Postn−/− embryos. E and F: Notch1 expression pattern. Notch1 expression was reduced in Postn−/− embryos (F) and was mostly restricted to the mesenchyme of the distal cushion (E).
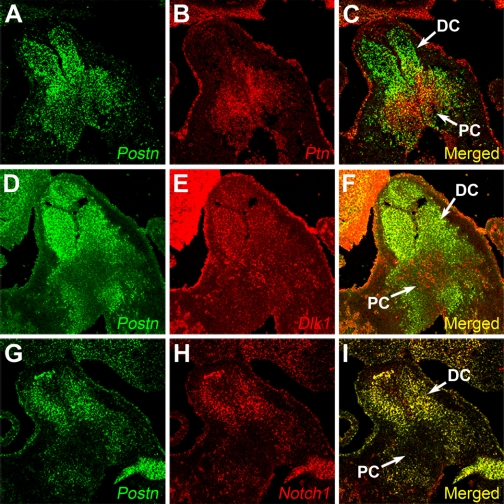
Since the in situ hybridization data and real-time PCR data clearly suggested that there was a link between Postn expression and expression of Ptn, Dlk1, and Notch1, we performed double fluorescent in situ hybridizations to determine expression domains of these genes in relation to Postn in the wild-type E12.5 OFT. Consistent with the in situ data (above), Ptn and Postn were expressed in mutually exclusive domains (Fig. 5, A–C). The Dlk1 gene was expressed throughout the OFT mesenchyme and coexpressed with Postn in the distal cushions (Fig. 5, D–F). The Notch1 expression domain completely overlapped that of Postn except for the endocardium, where no Postn expression was detected (Fig. 5, G–I). Ptn expression was restricted to the proximal cushion mesenchyme, where it was coexpressed with Dlk1, marking a mesenchymal cell population that appears to be distinct from the valvulogenic mesenchymal cells of the distal cushions and lacks both Notch1 and Postn expression (Fig. 5).
Fig. 5.
Colocalization of Postn with Ptn, Dlk1, and Notch1. Double fluorescent in situ hybridization on frontal sections of WT E12.5 OFT is shown. A–C: colocalization of Ptn and Postn gene expression; Ptn and Postn have mutually exclusive expression domains. D–F: colocalization of Dlk1 and Postn gene expression; Dlk1 was expressed in both distal and proximal endocardial cushions and was coexpressed with Postn in the distal cushion. G–I: colocalization of Notch1 and Postn gene expression; Notch1 and Postn expression domains completely overlapped.
Thus Dlk1 and Notch1 are coexpressed with periostin in the distal cushions of the OFT (Fig. 5, D–I). Absence of periostin expression in the E12.5 cushions, i.e., 48 h after the onset of periostin expression in the heart, leads to a 3.7-fold increase in Dlk1 expression (Fig. 3B, Fig. 4, C and D; Table 3). Since Dlk1 acts as a negative regulator of Notch1 (2), overexpression of Dlk1 combined with diminished expression of Notch1 should lead to a strong repression of Notch1 signaling in Postn−/− hearts. Indeed, real-time quantitative PCR analysis revealed that the expression of Hes1, Hey1, and Hey2, direct downstream targets of Notch1 (18, 27), were downregulated in Postn−/− hearts (Fig. 3C; Table 3). This is a direct indication that Notch1 signaling is repressed in Postn−/− hearts.
Postn and Ptn are expressed in mutually exclusive domains in E12.5 OFT (Fig. 5, A–C). Absence of periostin expression in the Postn−/− E12.5 OFT, i.e., 48 h after the onset of periostin expression in the heart, results in expansion of Ptn expression domain into the distal OFT cushions (Fig. 4, A and B) (i.e., where periostin is normally expressed in WT mice) and a 2.1-fold increase in its overall expression (Fig. 3B; Table 3), which indicates that periostin has a suppressive effect on Ptn expression. Thus these data suggest that periostin functions upstream of Ptn, Dlk1, and Notch1 and its absence leads to suppression of Notch1 signaling.
Suppression of Notch1 signaling in Postn−/− hearts leads to mesenchymal cell differentiation along an osteogenic lineage.
Our data strongly suggested that lack of periostin causes repression of Notch1 signaling in E12.5 hearts. Therefore, we analyzed expression of the central transcriptional regulator of osteoblast cell fate, Runx2, which was implicated in aortic valve calcification downstream of Notch1 (14) and was found to be upregulated in patients with valvular calcification (39). Real-time PCR analysis revealed that Runx2 was strongly upregulated in the hearts of E12.5 Postn−/− embryos (Fig. 3C; Table 3). Moreover, we also found that the expression of two main effectors of osteoblast mineralization, osteopontin and osteocalcin, were also significantly increased (Fig. 3C; Table 3). This suggests that even at this early stage of embryonic development a lack of periostin leads to activation of an osteogenic program in prevalvular endocardial cushions that is manifested as calcific aortic valve disease in early adult life.
DISCUSSION
Origin and formation of aortic valve.
During cardiogenesis, aortic valve leaflets develop from the endocardial cushions of the distal OFT as a result of a highly coordinated process, which involves an endothelial-to-mesenchymal transformation, remodeling accompanied by differentiation of cushion mesenchyme into cells of a fibrogenic lineage, and cushion maturation (8, 17). Several signaling pathways and transcription factors that control early stages of semilunar valve development, i.e., cushion formation, have been identified. Loss-of-function mutations in various components of the bone morphogenetic protein (BMP)2 signaling pathway in mice result in anomalies of semilunar valves, suggesting that the BMP2 signaling pathway plays an important role in development of the OFT endocardial cushions (6, 10, 12). The transforming growth factor (TGF)-β2 signaling pathway also seems to be critical for this process, since deletion of TGFβ2 in mice leads to the loss of semilunar valves (44). This is highlighted by the observation of severe OFT defects in embryos deficient for Pinch1, an adaptor protein acting upstream of TGFβ2 (22). In addition to BMP2 and TGF-β2 signaling pathways, several transcription regulators, including Sox4 (45), Nfatc1 (9, 40), Foxp1 (52), and Tbx20 (48), were shown to be essential for early stages of semilunar valve development. However, the genetic network guiding late stages of cushion remodeling and maturation is largely unknown.
Genetic basis of calcific aortic valve disease.
Calcific aortic valve disease is identified by thickening and calcification of the aortic valve leaflets and often leads to aortic stenosis (47), which represents the second most common indication for cardiac surgery (43). It affects up to 25% of the population over 65 yr of age (47) and up to 2% of pediatric patients (42).
Age-related aortic valve calcification has many features of an active pathobiological process, including inflammation (36, 38), lipoprotein deposition (34), and calcification (33, 39). Certain alleles of APOE and ESR1 were found to be associated with increased risk of age-related aortic valve calcification in adults, indicating that genetic factors influence disease development (29, 32).
Calcification of the aortic valve in pediatric patients is commonly associated with bicuspid aortic valve (BAV) (24). The heritability of BAV was estimated at 89%, suggesting that pediatric aortic stenosis is almost entirely determined by genetic factors (7). Loss-of-function mutations in NOTCH1 were shown to be associated with both BAV and age-related aortic valve calcification (14). However, two recent genomewide linkage studies for valve calcification susceptibility loci have identified multiple other chromosomal loci linked to calcific aortic valve disease, in addition to 9q harboring NOTCH1. Strong linkage was found to chromosomes 1q42, 2q37, 5q21, 13q34, 16q22, 17q23-24, 18q22, and 19q13, suggesting that multiple other genes are involved in the development of aortic valve disease (4, 25).
Although the upstream events may differ in different forms of aortic valve disease, they all lead to the activation of Runx2 (1, 14, 39), a central transcriptional activator of osteoblast differentiation that directly activates expression of several osteoblast-specific genes (11). Overexpression of Runx2 and its direct downstream targets osteopontin and osteocalcin was found to be associated with calcification of the aortic valve in patients with aortic stenosis (14, 39) and in a mouse model of the aortic valve disease (1).
Periostin controls OFT mesenchymal cell fate by modulating Notch1 signaling.
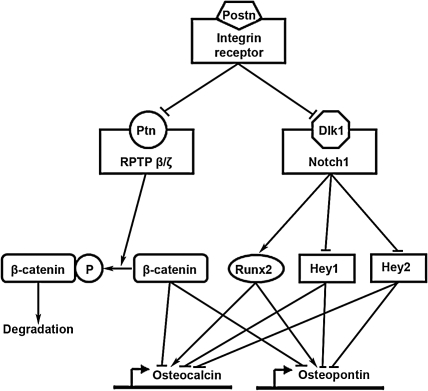
Periostin has recently become a popular target of investigation for its role in cell migration, cancer metastasis, myocardial remodeling, and atrioventricular valve maturation (23, 31, 35), but the discovery that periostin plays a key role in aortic valve development and OFT mesenchymal cell fate determination represents a new and very intriguing avenue of research. Our data suggest that periostin plays a critical role in the morphogenetic remodeling of the OFT endocardial cushions, i.e., the process that ultimately leads to the formation of muscular septal tissue from proximal cushions and mature aortic valve from distal cushions (8, 51). Our data indicate that periostin regulates Dlk1/Notch1 and Ptn signaling cascades in the OFT during embryonic development. Dlk1 is a negative regulator of Notch1 (2). Deactivating mutations in Notch1 can lead to Runx2-dependent overexpression of osteocalcin and calcification of the aortic valve (14). Ptn is a growth/differentiation factor that has been implicated in early osteoblast differentiation (54). Overexpression of Ptn resulted in an increase in bone thickness and mineralization (49). Binding of Ptn to its receptor, the receptor-type protein tyrosine phosphatase β/ζ (RPTPβ/ζ), increases phosphorylation and subsequent degradation of β-catenin (26). Activated (dephosphorylated) β-catenin was shown to inhibit Runx2-mediated transcriptional activation of the osteocalcin promoter (19). Therefore, Ptn-mediated β-catenin degradation can lead to activation of osteocalcin transcription. Under normal circumstances, periostin appears to negatively regulate Ptn and Dlk1 expression in the distal OFT cushions, while it seems to be necessary for maintaining adequate levels of Notch1 in these cushions. Our data suggest that this suppresses the default mesenchymal cell potential to differentiate into osteoblast-like cells and promotes their differentiation along a fibrogenic lineage necessary for normal valvular development. Thus we propose that the lack of periostin causes derepression of the osteogenic program by causing overexpression of Dlk1 and misexpression of Ptn in the distal OFT cushions, which in turn leads to suppression of Notch1 signaling and upregulation of the central regulator of osteoblast cell fate, Runx2. As summarized in Fig. 6, activation of Runx2 combined with suppression of its corepressors Hey1 and Hey2 would lead to activation of osteopontin and osteocalcin transcription and ultimate calcification of the aortic valve leaflets in early adulthood.
Fig. 6.
Periostin signaling cascade: model for proposed periostin signaling cascade in the cardiac OFT. We propose that periostin binds to an integrin receptor. This leads to suppression of Ptn and Dlk1 expression that results in activation of receptor-type protein tyrosine phosphatase β/ζ (RPTPβ/ζ) and Notch1. This causes activation of Runx2 corepressors β-catenin, Hey1, and Hey2 and, combined with suppression of Runx2 transcription, leads to suppression of osteogenic program in the mesenchymal cells of the distal OFT cushions and their subsequent differentiation along a fibrogenic lineage. Lack of periostin triggers a reversal of the cascade, activation of osteopontin and osteocalcin transcription, and ultimate calcification of the aortic valve leaflets (see text).
Recently, a marked reduction in periostin expression was observed in pediatric patients with aortic stenosis (46), suggesting that our mouse developmental findings may extend to the human disease. Thus delivering periostin to calcific valves at the early stages of the disease may have a therapeutic potential because it may prevent disease progression. The intriguing possibility of β-catenin phosphorylation (and degradation) as a result of RPTPβ/ζ inhibition by Ptn and subsequent further enhancement of Runx2-mediated activation of osteopontin and osteocalcin promoters needs to be further investigated. Thus periostin has emerged as an important regulator of cardiac valve development and a potential therapeutic target.
GRANTS
This work was supported by National Institutes of Health Grants HL-077342 (to S. J. Conway) and HL-33756, NCRR C06, and COBRE 2P20-RR-016434 (to R. R. Markwald).
ACKNOWLEDGMENTS
We thank Debi Turner for technical assistance and Tim C. McQuinn for help with echocardiography.
REFERENCES
- 1.Aikawa E, Nahrendorf M, Sosnovik D, Lok VM, Jaffer FA, Aikawa M, Weissleder R. Multimodality molecular imaging identifies proteolytic and osteogenic activities in early aortic valve disease. Circulation 115: 377–386, 2007 [DOI] [PubMed] [Google Scholar]
- 2.Baladron V, Ruiz-Hidalgo MJ, Nueda ML, Diaz-Guerra MJ, Garcia-Ramirez JJ, Bonvini E, Gubina E, Laborda J. Dlk acts as a negative regulator of Notch1 activation through interactions with specific EGF-like repeats. Exp Cell Res 303: 343–359, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Bao S, Ouyang G, Bai X, Huang Z, Ma C, Liu M, Shao R, Anderson RM, Rich JN, Wang XF. Periostin potently promotes metastatic growth of colon cancer by augmenting cell survival via the Akt/PKB pathway. Cancer Cell 5: 329–339, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Bella JN, Tang W, Kraja A, Rao DC, Hunt SC, Miller MB, Palmieri V, Roman MJ, Kitzman DW, Oberman A, Devereux RB, Arnett DK. Genome-wide linkage mapping for valve calcification susceptibility loci in hypertensive sibships: the Hypertension Genetic Epidemiology Network Study. Hypertension 49: 453–460, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Braverman AC, Guven H, Beardslee MA, Makan M, Kates AM, Moon MR. The bicuspid aortic valve. Curr Probl Cardiol 30: 470–522, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Clark TG, Conway SJ, Scott IC, Labosky PA, Winnier G, Bundy J, Hogan BL, Greenspan DS. The mammalian Tolloid-like 1 gene, Tll1, is necessary for normal septation and positioning of the heart. Development 126: 2631–2642, 1999 [DOI] [PubMed] [Google Scholar]
- 7.Cripe L, Andelfinger G, Martin LJ, Shooner K, Benson DW. Bicuspid aortic valve is heritable. J Am Coll Cardiol 44: 138–143, 2004 [DOI] [PubMed] [Google Scholar]
- 8.de la Cruz MV, Markwald RR. Living Morphogenesis of the Heart Boston: Birkhäuser, 1997, p. xviii [Google Scholar]
- 9.de la Pompa JL, Timmerman LA, Takimoto H, Yoshida H, Elia AJ, Samper E, Potter J, Wakeham A, Marengere L, Langille BL, Crabtree GR, Mak TW. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature 392: 182–186, 1998 [DOI] [PubMed] [Google Scholar]
- 10.Delot EC, Bahamonde ME, Zhao M, Lyons KM. BMP signaling is required for septation of the outflow tract of the mammalian heart. Development 130: 209–220, 2003 [DOI] [PubMed] [Google Scholar]
- 11.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89: 747–754, 1997 [DOI] [PubMed] [Google Scholar]
- 12.Galvin KM, Donovan MJ, Lynch CA, Meyer RI, Paul RJ, Lorenz JN, Fairchild-Huntress V, Dixon KL, Dunmore JH, Gimbrone MA, Jr, Falb D, Huszar D. A role for smad6 in development and homeostasis of the cardiovascular system. Nat Genet 24: 171–174, 2000 [DOI] [PubMed] [Google Scholar]
- 13.Garg V. Molecular genetics of aortic valve disease. Curr Opin Cardiol 21: 180–184, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D . Mutations in NOTCH1 cause aortic valve disease. Nature 437: 270–274, 2005 [DOI] [PubMed] [Google Scholar]
- 15.High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat Rev Genet 9: 49–61, 2008 [DOI] [PubMed] [Google Scholar]
- 16.Horiuchi K, Amizuka N, Takeshita S, Takamatsu H, Katsuura M, Ozawa H, Toyama Y, Bonewald LF, Kudo A. Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor beta. J Bone Miner Res 14: 1239–1249, 1999 [DOI] [PubMed] [Google Scholar]
- 17.Hurle JM, Colvee E, Blanco AM. Development of mouse semilunar valves. Anat Embryol (Berl) 160: 83–91, 1980 [DOI] [PubMed] [Google Scholar]
- 18.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature 377: 355–358, 1995 [DOI] [PubMed] [Google Scholar]
- 19.Kahler RA, Westendorf JJ. Lymphoid enhancer factor-1 and beta-catenin inhibit Runx2-dependent transcriptional activation of the osteocalcin promoter. J Biol Chem 278: 11937–11944, 2003 [DOI] [PubMed] [Google Scholar]
- 20.Kern CB, Hoffman S, Moreno R, Damon BJ, Norris RA, Krug EL, Markwald RR, Mjaatvedt CH. Immunolocalization of chick periostin protein in the developing heart. Anat Rec A Discov Mol Cell Evol Biol 284: 415–423, 2005 [DOI] [PubMed] [Google Scholar]
- 21.Kruzynska-Frejtag A, Machnicki M, Rogers R, Markwald RR, Conway SJ. Periostin (an osteoblast-specific factor) is expressed within the embryonic mouse heart during valve formation. Mech Dev 103: 183–188, 2001 [DOI] [PubMed] [Google Scholar]
- 22.Liang X, Sun Y, Schneider J, Ding JH, Cheng H, Ye M, Bhattacharya S, Rearden A, Evans S, Chen J. Pinch1 is required for normal development of cranial and cardiac neural crest-derived structures. Circ Res 100: 527–535, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litvin J, Zhu S, Norris R, Markwald R. Periostin family of proteins: therapeutic targets for heart disease. Anat Rec A Discov Mol Cell Evol Biol 287: 1205–1212, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Mack G, Silberbach M. Aortic and pulmonary stenosis. Pediatr Rev 21: 79–85, 2000 [DOI] [PubMed] [Google Scholar]
- 25.Martin LJ, Ramachandran V, Cripe LH, Hinton RB, Andelfinger G, Tabangin M, Shooner K, Keddache M, Benson DW. Evidence in favor of linkage to human chromosomal regions 18q, 5q and 13q for bicuspid aortic valve and associated cardiovascular malformations. Hum Genet 121: 275–284, 2007 [DOI] [PubMed] [Google Scholar]
- 26.Meng K, Rodriguez-Pena A, Dimitrov T, Chen W, Yamin M, Noda M, Deuel TF. Pleiotrophin signals increased tyrosine phosphorylation of beta-catenin through inactivation of the intrinsic catalytic activity of the receptor-type protein tyrosine phosphatase beta/zeta. Proc Natl Acad Sci USA 97: 2603–2608, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakagawa O, McFadden DG, Nakagawa M, Yanagisawa H, Hu T, Srivastava D, Olson EN. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proc Natl Acad Sci USA 97: 13655–13660, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niessen K, Karsan A. Notch signaling in cardiac development. Circ Res 102: 1169–1181, 2008 [DOI] [PubMed] [Google Scholar]
- 29.Nordstrom P, Glader CA, Dahlen G, Birgander LS, Lorentzon R, Waldenstrom A, Lorentzon M. Oestrogen receptor alpha gene polymorphism is related to aortic valve sclerosis in postmenopausal women. J Intern Med 254: 140–146, 2003 [DOI] [PubMed] [Google Scholar]
- 30.Norris RA, Kern CB, Wessels A, Moralez EI, Markwald RR, Mjaatvedt CH. Identification and detection of the periostin gene in cardiac development. Anat Rec A Discov Mol Cell Evol Biol 281: 1227–1233, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Norris RA, Moreno-Rodriguez RA, Sugi Y, Hoffman S, Amos J, Hart MM, Potts JD, Goodwin RL, Markwald RR. Periostin regulates atrioventricular valve maturation. Dev Biol 316: 200–213, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novaro GM, Sachar R, Pearce GL, Sprecher DL, Griffin BP. Association between apolipoprotein E alleles and calcific valvular heart disease. Circulation 108: 1804–1808, 2003 [DOI] [PubMed] [Google Scholar]
- 33.O'Brien KD, Kuusisto J, Reichenbach DD, Ferguson M, Giachelli C, Alpers CE, Otto CM. Osteopontin is expressed in human aortic valvular lesions. Circulation 92: 2163–2168, 1995 [DOI] [PubMed] [Google Scholar]
- 34.O'Brien KD, Reichenbach DD, Marcovina SM, Kuusisto J, Alpers CE, Otto CM . Apolipoproteins B, (a), and E accumulate in the morphologically early lesion of “degenerative” valvular aortic stenosis. Arterioscler Thromb Vasc Biol 16: 523–532, 1996 [DOI] [PubMed] [Google Scholar]
- 35.Oka T, Xu J, Kaiser RA, Melendez J, Hambleton M, Sargent MA, Lorts A, Brunskill EW, Dorn GW, 2nd, Conway SJ, Aronow BJ, Robbins J, Molkentin JD. Genetic manipulation of periostin expression reveals a role in cardiac hypertrophy and ventricular remodeling. Circ Res 101: 313–321, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsson M, Dalsgaard CJ, Haegerstrand A, Rosenqvist M, Ryden L, Nilsson J. Accumulation of T lymphocytes and expression of interleukin-2 receptors in nonrheumatic stenotic aortic valves. J Am Coll Cardiol 23: 1162–1170, 1994 [DOI] [PubMed] [Google Scholar]
- 37.Oshima A, Tanabe H, Yan T, Lowe GN, Glackin CA, Kudo A. A novel mechanism for the regulation of osteoblast differentiation: transcription of periostin, a member of the fasciclin I family, is regulated by the bHLH transcription factor, twist. J Cell Biochem 86: 792–804, 2002 [DOI] [PubMed] [Google Scholar]
- 38.Otto CM, Kuusisto J, Reichenbach DD, Gown AM, O'Brien KD. Characterization of the early lesion of “degenerative” valvular aortic stenosis. Histological and immunohistochemical studies. Circulation 90: 844–853, 1994 [DOI] [PubMed] [Google Scholar]
- 39.Rajamannan NM, Subramaniam M, Rickard D, Stock SR, Donovan J, Springett M, Orszulak T, Fullerton DA, Tajik AJ, Bonow RO, Spelsberg T. Human aortic valve calcification is associated with an osteoblast phenotype. Circulation 107: 2181–2184, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranger AM, Grusby MJ, Hodge MR, Gravallese EM, de la Brousse FC, Hoey T, Mickanin C, Baldwin HS, Glimcher LH. The transcription factor NF-ATc is essential for cardiac valve formation. Nature 392: 186–190, 1998 [DOI] [PubMed] [Google Scholar]
- 41.Rios H, Koushik SV, Wang H, Wang J, Zhou HM, Lindsley A, Rogers R, Chen Z, Maeda M, Kruzynska-Frejtag A, Feng JQ, Conway SJ. Periostin null mice exhibit dwarfism, incisor enamel defects, and an early-onset periodontal disease-like phenotype. Mol Cell Biol 25: 11131–11144, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts WC. The congenitally bicuspid aortic valve. A study of 85 autopsy cases. Am J Cardiol 26: 72–83, 1970 [DOI] [PubMed] [Google Scholar]
- 43.Roberts WC, Ko JM. Frequency by decades of unicuspid, bicuspid, and tricuspid aortic valves in adults having isolated aortic valve replacement for aortic stenosis, with or without associated aortic regurgitation. Circulation 111: 920–925, 2005 [DOI] [PubMed] [Google Scholar]
- 44.Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschman T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development 124: 2659–2670, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, Clevers H. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature 380: 711–714, 1996 [DOI] [PubMed] [Google Scholar]
- 46.Snider P, Hinton RB, Moreno-Rodriguez RA, Wang J, Rogers R, Lindsley A, Li F, Ingram DA, Menick D, Field L, Firulli AB, Molkentin JD, Markwald R, Conway SJ. Periostin is required for maturation and extracellular matrix stabilization of noncardiomyocyte lineages of the heart. Circ Res 102: 752–760, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart BF, Siscovick D, Lind BK, Gardin JM, Gottdiener JS, Smith VE, Kitzman DW, Otto CM. Clinical factors associated with calcific aortic valve disease. Cardiovascular Health Study. J Am Coll Cardiol 29: 630–634, 1997 [DOI] [PubMed] [Google Scholar]
- 48.Takeuchi JK, Mileikovskaia M, Koshiba-Takeuchi K, Heidt AB, Mori AD, Arruda EP, Gertsenstein M, Georges R, Davidson L, Mo R, Hui CC, Henkelman RM, Nemer M, Black BL, Nagy A, Bruneau BG. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development 132: 2463–2474, 2005 [DOI] [PubMed] [Google Scholar]
- 49.Tare RS, Oreffo RO, Sato K, Rauvala H, Clarke NM, Roach HI. Effects of targeted overexpression of pleiotrophin on postnatal bone development. Biochem Biophys Res Commun 298: 324–332, 2002 [DOI] [PubMed] [Google Scholar]
- 50.Tkatchenko AV, Le Cam G, Leger JJ, Dechesne CA. Large-scale analysis of differential gene expression in the hindlimb muscles and diaphragm of mdx mouse. Biochim Biophys Acta 1500: 17–30, 2000 [DOI] [PubMed] [Google Scholar]
- 51.van den Hoff MJ, Kruithof BP, Moorman AF, Markwald RR, Wessels A. Formation of myocardium after the initial development of the linear heart tube. Dev Biol 240: 61–76, 2001 [DOI] [PubMed] [Google Scholar]
- 52.Wang B, Weidenfeld J, Lu MM, Maika S, Kuziel WA, Morrisey EE, Tucker PW. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development 131: 4477–4487, 2004 [DOI] [PubMed] [Google Scholar]
- 53.Yang H, Wanner IB, Roper SD, Chaudhari N. An optimized method for in situ hybridization with signal amplification that allows the detection of rare mRNAs. J Histochem Cytochem 47: 431–446, 1999 [DOI] [PubMed] [Google Scholar]
- 54.Yang X, Tare RS, Partridge KA, Roach HI, Clarke NM, Howdle SM, Shakesheff KM, Oreffo RO. Induction of human osteoprogenitor chemotaxis, proliferation, differentiation, and bone formation by osteoblast stimulating factor-1/pleiotrophin: osteoconductive biomimetic scaffolds for tissue engineering. J Bone Miner Res 18: 47–57, 2003 [DOI] [PubMed] [Google Scholar]
- 55.Zhou YQ, Foster FS, Nieman BJ, Davidson L, Chen XJ, Henkelman RM. Comprehensive transthoracic cardiac imaging in mice using ultrasound biomicroscopy with anatomical confirmation by magnetic resonance imaging. Physiol Genomics 18: 232–244, 2004 [DOI] [PubMed] [Google Scholar]