Summary
The Notch receptor and its ligands are involved in many developmental processes. They are highly expressed in the thymus and have been implicated in the CD4 versus CD8 lineage decision. We identified the constitutively active intracellular fragment of murine Notch-1 as capable of rendering thymomas resistant to glucocorticoid-induced apoptosis. This effect was confirmed in other T cell lines and in CD4+CD8+ DP thymocytes. Activation of the Notch signaling pathway also upregulated a number of other markers that, like steroid resistance, correlate with DP maturation into both the CD4 and CD8 lineages. These results suggest that Notch signaling is critically involved in the maturation of DP thymocytes into both CD4+ and CD8+ SP thymocytes.
Introduction
The differentiation of immature DP thymocytes into mature CD4+ and CD8+ SP thymocytes is a highly regulated process. DP thymocytes, which express T cell receptor (TCR) and both CD4 and CD8 coreceptor molecules, are evaluated for their ability to recognize self-peptide/MHC ligands expressed on thymic epithelial cells. DP thymocytes that express a TCR with too-low affinity for self-peptide/MHC undergo apoptosis by a process termed “death by neglect.” DP thymocytes that express a TCR with too-high affinity for self-peptide/MHC undergo apoptosis by a process termed “negative selection.” In contrast, DP thymocytes that express TCR with an intermediate affinity for self-peptide/MHC are rescued from apoptosis and differentiate into mature SP thymocytes by a process termed “positive selection” (for review see Kisielow and von Boehmer, 1995; Jameson and Bevan, 1998). This step in thymocyte maturation also involves a choice between differentiation into either the CD4+ or CD8+ SP lineages. DP thymocytes that recognize MHC class I ligands during positive selection downregulate CD4 expression and acquire the ability to mediate cytotoxic effector functions, whereas DP thymocytes that recognize MHC class II ligands downregulate CD8 expression and acquire the ability to mediate helper effector functions.
Our understanding of positive selection and lineage commitment is based primarily on the characterization of requisite TCR and CD4 and CD8 coreceptor signals and the description of phenotypic changes that occur during maturation. Relatively little is known, however, regarding transcriptional mechanisms responsible for regulating these processes. One interesting event that occurs during the differentiation of DP thymocytes into both the CD4+ and CD8+ SP lineages is the acquisition of resistance to glucocorticoids. DP thymocytes are exquisitely sensitive to glucocorticoid-induced apoptosis, whereas SP thymocytes are relatively resistant (Scollay et al., 1984; Screpanti et al., 1989; Cohen, 1992). The molecular basis for this difference in glucocorticoid sensitivity is not known. Glucocorticoids, produced both by the adrenal gland and endogenously within the thymus, have complex effects on the development of thymocytes (Vacchio et al., 1998). Since circulating levels of corticosteroids approach that which causes apoptosis in vitro, it is hypothesized that glucocorticoids may contribute to the process of death by neglect of DP thymocytes and that the acquisition of resistance to glucocorticoids is a key event that rescues thymocytes from apoptosis during positive selection (Sprent et al., 1988; Cohen, 1992).
To further our understanding of molecular events associated with thymocyte maturation, we sought to identify molecules that regulate thymocyte sensitivity to glucocorticoids. For this purpose, we screened a thymocyte cDNA library for genes that confer glucocorticoid resistance in a glucocorticoid-sensitive DP thymoma cell line. This approach led to the repeated identification of the intracellular domain of mNotch-1. Notch is an evolutionarily conserved cell surface receptor that regulates changes in gene expression during development (Artavanis et al., 1995). Following productive interaction with ligands such as Jagged, Notch undergoes cleavage within the transmembrane domain, allowing the intracellular portion to traffic to the nucleus (Schroeter et al., 1998; Struhl and Adachi, 1998). In many systems, ectopic expression of the intracellular domain of Notch (NotchIC) results in constitutive activation of the Notch signaling pathway (Lieber et al., 1993; Rebay et al., 1993; Struhl et al., 1993). Mice expressing NotchIC as a transgene in the thymus develop an excess of CD8+ SP thymocytes relative to CD4+ SP thymocytes (Robey et al., 1996). This led to the suggestion that the CD4 versus CD8 lineage choice following a positively selecting interaction with MHC ligands is determined by Notch signaling. According to this idea, MHC class I recognition leads to Notch signaling while MHC class II recognition allows DP thymocytes to avoid Notch signaling.
We show here that activation of the Notch signaling pathway in T cell lines and in thymocytes induces multiple events that are associated with the maturation of DP thymocytes into SP thymocytes. These include the acquisition of resistance to glucocorticoids and the upregulation of Bcl-2, TCR, and Deltex expression. Importantly, these phenotypic changes occur during the differentiation of DP thymocytes into both the CD4 and CD8 lineages. These results lead us to propose that activation of the Notch signaling pathway is a crucial event in the maturation of both CD4+ and CD8+ SP thymocytes.
Results
Expression of the Intracellular Domain of mNotch-1 Confers Resistance to Glucocorticoids in T Cells
We employed an expression cloning strategy to identify genes that regulate thymocyte sensitivity to glucocorticoids. A thymocyte cDNA library was constructed in a retroviral vector (Onishi et al., 1996) and transferred into the glucocorticoid-sensitive DP thymoma cell line, AKR1010, followed by selection for clones that proliferate in the presence of dexamethasone. Retrovirally transferred cDNAs from glucocorticoid-resistant clones were sequenced, and multiple independent clones were found to express cDNAs derived from the mNotch-1 gene (Figures 1A and 1B). Conceptual translation of the cDNAs identified by expression cloning indicates that they encode polypeptides corresponding to the intracellular domain of mNotch-1. Since ectopic expression of the intracellular domain of Notch results in constitutive activation of the Notch signaling pathway (Lieber et al., 1993; Rebay et al., 1993; Struhl et al., 1993), this suggested that Notch signaling confers resistance to glucocorticoids in T cells. To confirm this, the intracellular domain of mNotch-1 was expressed in AKR1010 and in the glucocorticoid-sensitive T cell hybridoma, 2B4.11 (Figure 1C). The pMI retroviral vector used in these experiments allows identification of transgene-expressing cells by cell-surface expression of human CD2 (see Experimental Procedures). Infection of AKR1010 and 2B4.11 with retrovirus prepared from the pMI/NotchIC vector resulted in 20%–30% transgene-expressing cells (Figure 2A, top panels). These populations were cultured in medium alone or in medium containing dexamethasone for 7 days. When cultured in medium alone, there was a gradual loss of transgene-expressing cells, suggesting that expression of NotchIC may confer a growth disadvantage to these T cells (Figure 2A, middle panels). In contrast, when cultured in the presence of dexamethasone, there was a dramatic loss of uninfected cells, while transgene-expressing cells selectively survived (Figure 2A, bottom panels). In control experiments using cells infected with the empty pMI vector, there was no loss of hCD2-expressing cells in medium alone and few viable cells remaining in cultures containing dexamethasone (data not shown). This confirms that expression of NotchIC confers resistance to glucocorticoids in multiple T cell lines.
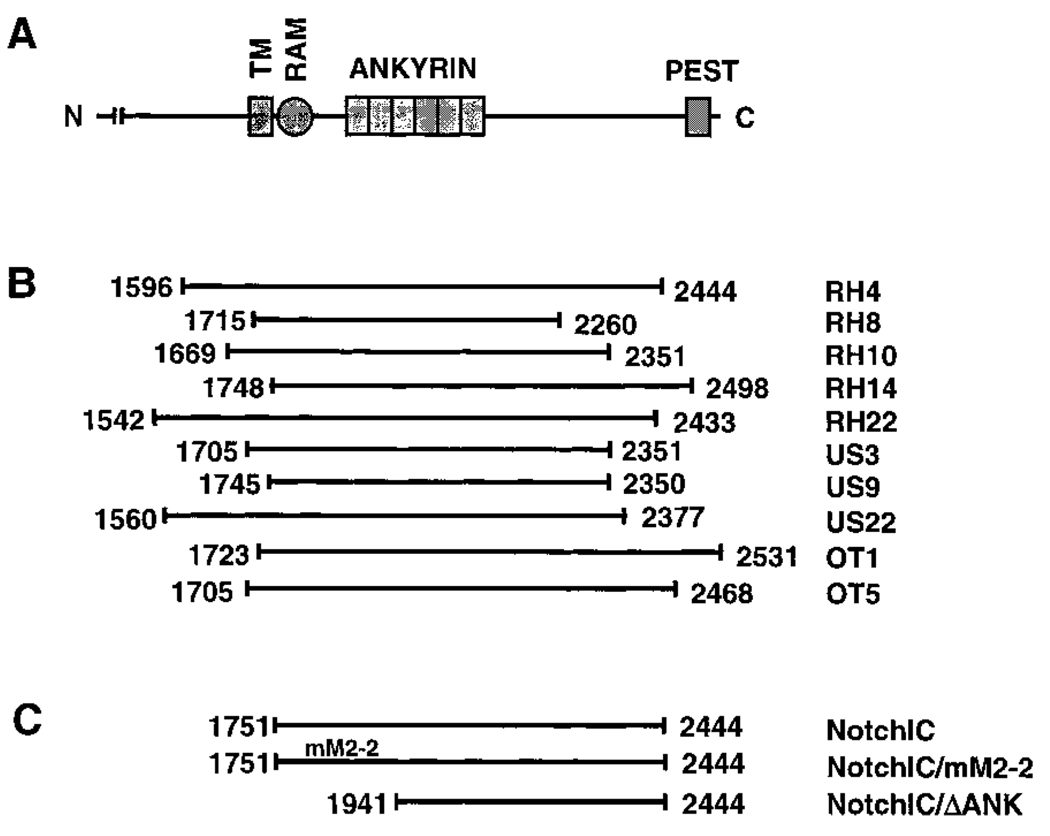
Figure 1.
Identification of NotchIC as a Mediator of Resistance to Glucocorticoids in T Cells
(A) Schematic representation of the mNotch-1 amino acid sequence. Labeled are the transmembrane domain (TM), residues 1724–1746; the RAM domain, residues 1751–1806; the ankyrin domain, residues 1865–2075; and the PEST domain, residues 2481–2503 (del Amo et al., 1993; Tamura et al., 1995). The majority of the extracellular domain has been omitted.
(B) Schematic representation of the coding regions of mNotch-1 cDNAs identified by expression cloning. Numbers indicating the amino- and carboxyl-terminal amino acid residues are based on conceptual translation of the cDNA sequences. The initiation methionine for these cDNAs has not been determined. RH, US, and OT refer to clones derived from these respective thymocyte cDNA libraries.
(C) Schematic representation of the regions of mNotch-1 cloned into the pMI retroviral vector and transferred into the AKR1010 and 2B4.11 T cell lines for functional assays. NotchIC begins at the first amino acid of the RAM domain. NotchIC/mM2-2 contains a mutation in the RAM domain that disrupts RBP/Su(H) binding (Tamura et al., 1995). NotchIC/ΔAnk begins immediately amino terminal to the third ankyrin repeat of Notch.
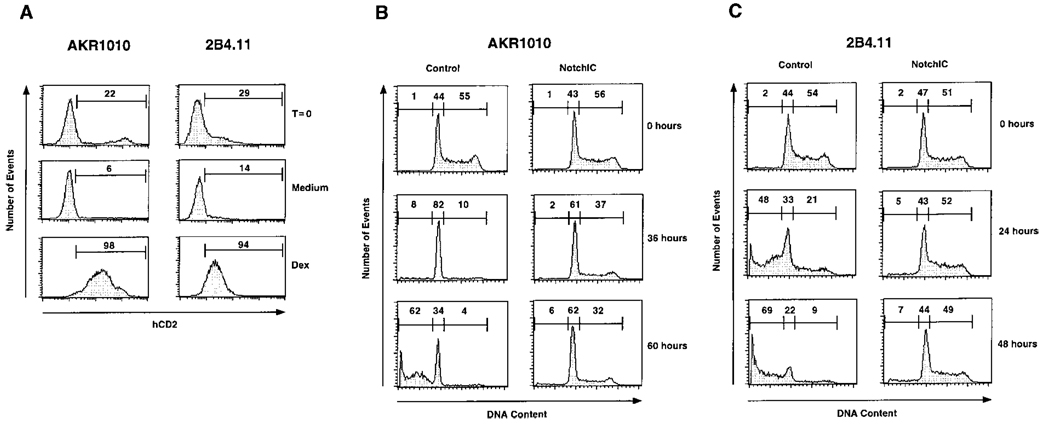
Figure 2.
Expression of NotchIC Confers Resistance to Glucocorticoid-Induced Apoptosis and Cell Cycle Arrest in T Cells
(A) Expression of NotchIC in AKR1010 and 2B4.11 confers resistance to dexamethasone. AKR1010 and 2B4.11 were infected with retrovirus derived from the pMI/NotchIC vector. Following infection, transgene-expressing cells were identified by FACS analysis for IRES-driven hCD2 expression. The cells were then cultured in medium alone or in medium containing 10−7 M dexamethasone for 7 days followed by staining for hCD2 expression. Numbers represent the percentage of transgene-expressing cells in the indicated gates.
(B) Expression of NotchIC confers resistance to glucocorticoid-induced apoptosis and cell-cycle arrest in AKR1010. Control cells and polyclonal AKR1010 cells expressing NotchIC were cultured in the presence of 10−7 M dexamethasone for the indicated times and analyzed for DNA content by FACS. Numbers represent the percentage of cells in the indicated gates.
(C) Expression of NotchIC confers resistance to glucocorticoid-induced apoptosis and cell cycle arrest in 2B4.11. Control cells and polyclonal 2B4.11 cells expressing NotchIC were assayed for sensitivity to glucocorticoid-induced apoptosis and cell cycle arrest as in (B).
Glucocorticoids induce both cell cycle arrest and apoptosis in susceptible T cells (Zacharchuk et al., 1990; Miyashita and Reed, 1992). The survival of AKR1010 and 2B4.11 cells in the presence of dexamethasone suggests that expression of NotchIC inhibits both of these effects. To confirm this, we generated polyclonal cell lines derived from AKR1010 and 2B4.11 that express NotchIC by infecting with retrovirus prepared from the pMI/NotchIC vector and panning for cells that express hCD2. Cell cycle analysis of these and control cells was performed following exposure to dexamethasone (Figures 2B and 2C). Dexamethasone causes AKR1010 cells to arrest in the G1 phase of the cell cycle as indicated by the accumulation of cells with a diploid DNA content. This is followed by the gradual emergence of apoptotic cells with a subdiploid DNA content (Figure 2B). In contrast, AKR1010 cells expressing NotchIC do not undergo cell cycle arrest or apoptosis in response to dexamethasone. Similarly, dexamethasone induces 2B4.11 cells to undergo apoptosis, and this is inhibited by expression of NotchIC (Figure 2C). These results confirm that expression of NotchIC inhibits both glucocorticoid-induced apoptosis and cell cycle arrest in T cells. In contrast, expression of NotchIC has no effect on apoptosis induced by staurosporine, ceramide, or etoposide (data not shown). Additional experiments indicated that NotchIC has no effect on expression of the glucocorticoid receptor or the induction of multiple glucocorticoid-regulated genes in these cell lines (data not shown).
Resistance to Glucocorticoids Correlates with Activation of the Notch Signaling Pathway and Maps to the RAM Domain of mNotch-1
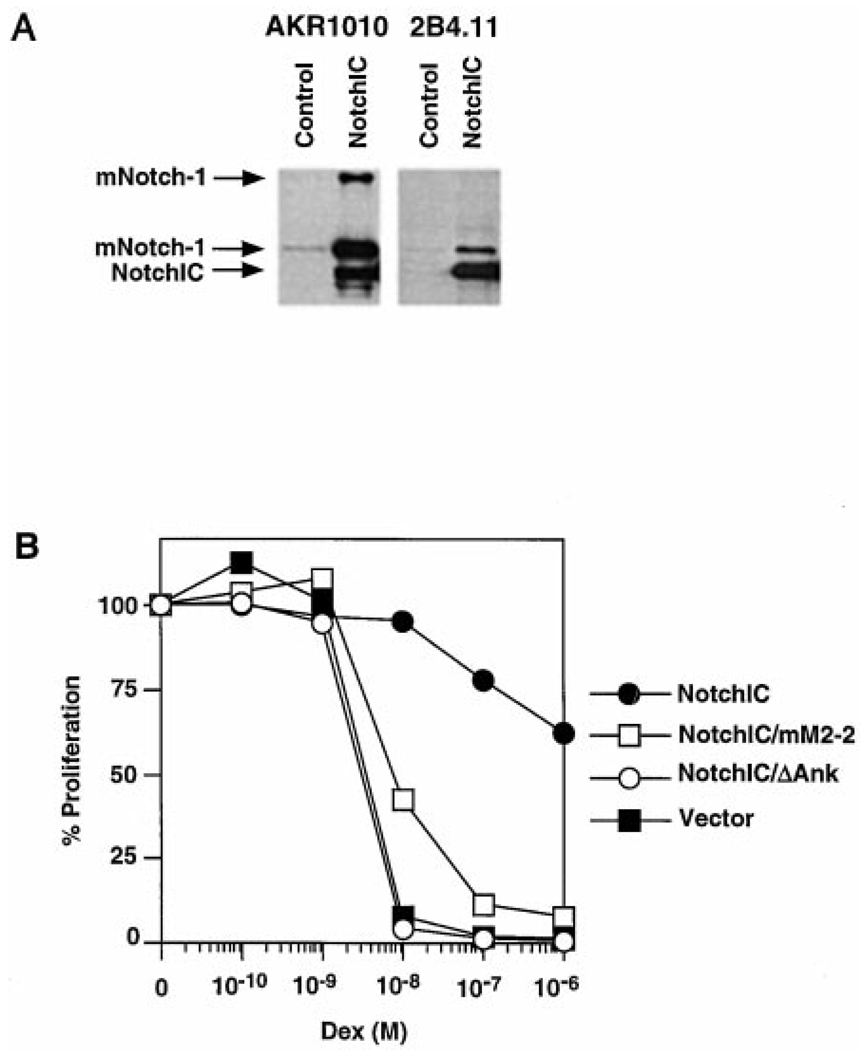
In many cell lineages, activation of the Notch signaling pathway results in the upregulation of Notch expression in a positive feedback loop (Wilkinson et al., 1994; Luo et al., 1997; Weinmaster, 1997). We performed Western blot analysis to determine whether expression of NotchIC induces this downstream effect of Notch signaling in T cells (Figure 3A). Both AKR1010 and 2B4.11 express low levels of mNotch-1. In contrast, polyclonal AKR1010 and 2B4.11 cells expressing NotchIC express high levels of endogenous mNotch-1. The two mNotch-1 species that are upregulated represent the full-length ~300 kDa and the proteolytically processed ~120 kDa forms of mNotch-1 (Kopan et al., 1996; Blaumueller et al., 1997). This suggests that expression of NotchIC activates downstream events of the Notch signaling pathway in these T cells. Together with the ability of NotchIC expression to inhibit glucocorticoid-induced apoptosis and cell cycle arrest, these data suggest that activation of the Notch signaling pathway confers resistance to glucocorticoids in T cells.
Figure 3.
NotchIC-Mediated Resistance to Glucocorticoids Correlates with Activation of the Notch Signaling Pathway and Maps to the RAM Domain of Notch
(A) Expression of NotchIC upregulates endogenous mNotch-1 expression in AKR1010 and 2B4.11. Vector control cells and polyclonal AKR1010 and 2B4.11 cells expressing NotchIC were analyzed for endogenous mNotch-1 expression by Western blot analysis.
(B) Resistance to glucocorticoids in T cells is mediated by the RAM and ankyrin domains of mNotch-1. Polyclonal 2B4.11 cell lines expressing the different NotchIC constructs diagrammed in Figure 1C were incubated in increasing concentrations of dexamethasone for 24 hr and their proliferation measured by 3H thymidine incorporation during an additional 12 hr of culture. Expressed is the percentage of proliferation relative to cells cultured in the absence of dexamethasone. Counts incorporated in the absence of dexamethasone are 62158 ± 2686 cpm for NotchIC; 65778 ± 3759 cpm for NotchIC/mM2-2; 72343 ± 4864 cpm for NotchIC/ΔAnk; and 89784 ± 2942 cpm for vector alone.
Notch signaling regulates gene expression by activating the RBP/Su(H) transcription factor (Artavanis et al., 1995). This requires the RAM and ankyrin domains of Notch (Kato et al., 1997). In some cases, however, downstream effects of Notch signaling may be mediated by RBP/Su(H)-independent mechanisms (Shawber et al., 1996; Matsuno et al., 1997). To address whether resistance to glucocorticoids is mediated by RBP/Su(H), we assayed Notch constructs that lack functional RAM and ankyrin domains (Figure 1C) for their ability to confer resistance to glucocorticoids. To quantitate this effect, the proliferative rate of 2B4.11 cells following culture in the presence of dexamethasone was measured (Figure 3B). Expression of NotchIC, which contains functional RAM and ankyrin domains, allows the proliferation of 2B4.11 in the presence of dexamethasone. Expression of NotchIC/mM2-2, which contains point mutations in the RAM domain that inhibit RBP/Su(H) interaction, also allows proliferation in the presence of dexamethasone but at reduced levels relative to wild-type NotchIC. In other experimental systems, deletion or mutation of the RAM domain similarly reduces, but does not eliminate, RBP/Su(H)-mediated transcriptional activation (Kato et al., 1997; Wettstein et al., 1997). In contrast, expression of NotchIC/ΔAnk, which contains a deletion of the RAM domain and the first two ankyrin repeats, does not confer on 2B4.11 the ability to proliferate in the presence of dexamethasone. Similar results were obtained with the AKR1010 cell line (data not shown). These results suggest that downstream events of the Notch signaling pathway leading to glucocorticoid-resistance are mediated by RBP/Su(H).
Activation of the Notch Signaling Pathway Upregulates Bcl-2 Expression in T Cells
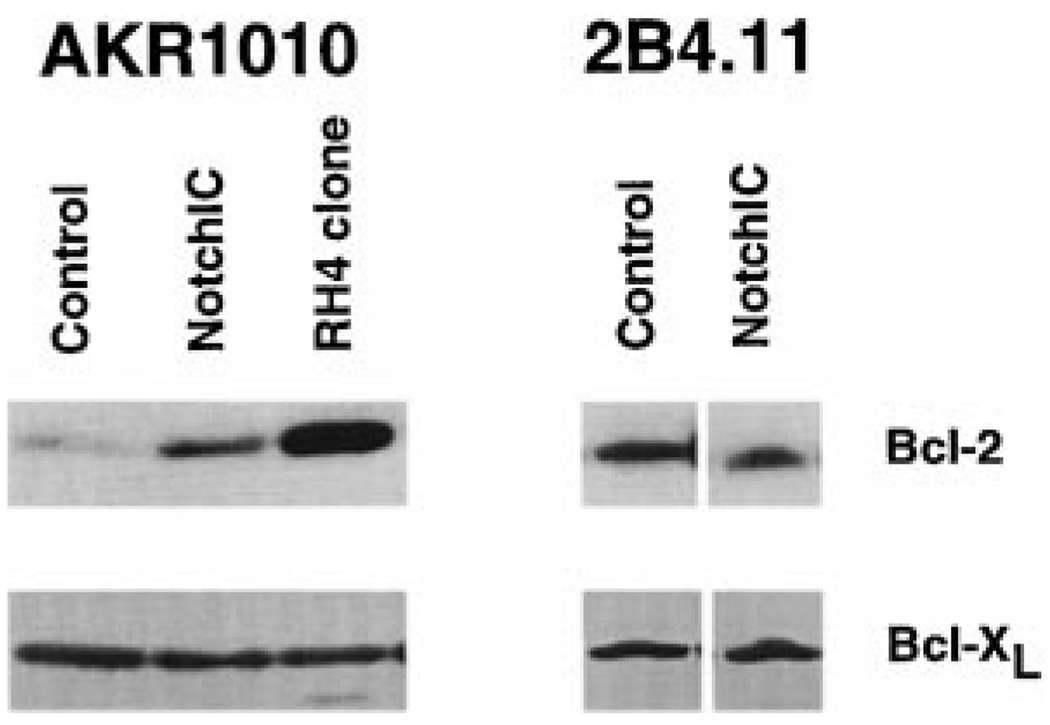
The mechanism by which glucocorticoids induce apoptosis in thymocytes is not understood. Glucocorticoids bind to the glucocorticoid receptor (GR) and either activate or inhibit gene expression (Beato et al., 1995). Glucocorticoid-induced apoptosis requires de novo RNA and protein synthesis and is dependent on the DNA-binding domain of the GR (Cohen and Duke, 1984; Wyllie et al., 1984; Reichardt et al., 1998). This suggests that glucocorticoids induce the expression of genes that activate the apoptotic pathway. One determinant of susceptibility to glucocorticoids is the Bcl-2 family of proteins. Overexpression of Bcl-2 or Bcl-XL in T cells inhibits glucocorticoid-induced apoptosis but not cell cycle arrest (Sentman et al., 1991; Strasser et al., 1991; Siegel et al., 1992; Miyashita and Reed, 1992; Chao et al., 1995; Grillot et al., 1995). We performed Western blot analysis to determine whether expression of NotchIC affects the expression of Bcl-2 or Bcl-2 family members (Figure 4). Polyclonal AKR1010 cell lines expressing NotchIC upregulate Bcl-2 expression 3-fold. Furthermore, in dexamethasone-resistant AKR1010 clones generated by expression cloning, Bcl-2 is upregulated 10-fold. The difference in the level of Bcl-2 upregulation may be due to differences in the level of transgene expression. In contrast, expression of NotchIC does not upregulate Bcl-2 expression in 2B4.11 cells. No change in the expression of Bcl-XL or Bax is observed in either cell line (Figure 4). These data suggest that Notch signaling can upregulate expression of Bcl-2 in some T cell lines but not in others. Furthermore, the ability of NotchIC to confer resistance to glucocorticoids in 2B4.11 without upregulating Bcl-2 expression indicates that this may occur by a Bcl-2-independent mechanism.
Figure 4.
Western Blot Analysis of Bcl-2 and Bcl-XL Expression in AKR1010 and 2B4.11 Cells Expressing NotchIC
Lysates from the indicated cell lines were analyzed by Western blot analysis for Bcl-2 expression. The blots were then stripped and reprobed for Bcl-XL. Analysis of two additional dexamethasone-resistant AKR1010 clones isolated by expressing cloning gave identical results to the RH4 clone.
Activation of the Notch Signaling Pathway Upregulates TCR Expression
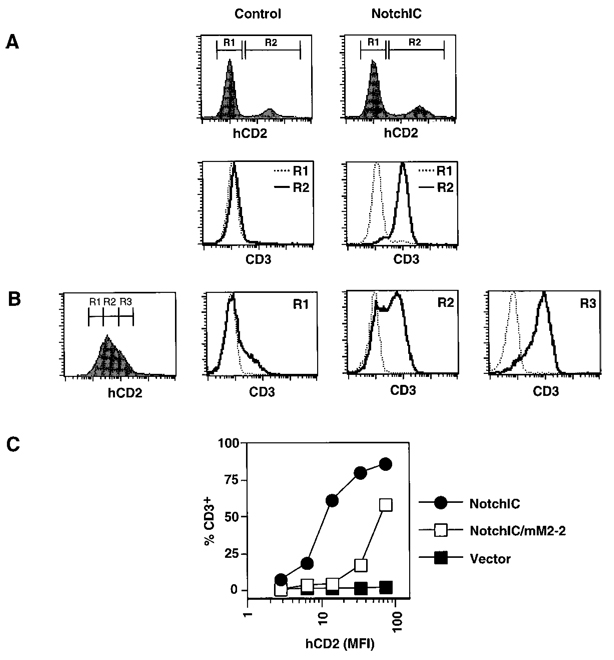
NotchIC expression confers resistance to glucocorticoids and upregulates Bcl-2 expression in the AKR1010 DP thymoma cell line. Both of these events occur during the differentiation of DP thymocytes into SP thymocytes. This suggests that activation of the Notch signaling pathway may induce multiple events associated with thymocyte maturation. We therefore sought to identify additional phenotypic changes induced by Notch signaling in AKR1010. One important event that occurs during the differentiation of DP thymocytes into SP thymocytes is the upregulation of TCR expression. To determine whether expression of NotchIC upregulates TCR expression, we infected AKR1010 with retrovirus prepared from the pMI/NotchIC or control vector and assayed transgene-expressing cells for CD3 and αβTCR expression by FACS. Uninfected AKR1010 and AKR1010 infected with retrovirus prepared from control vector do not express detectable TCR on their surface (Figure 5A, left panels). Remarkably, AKR1010 cells infected with retrovirus prepared from the pMI/NotchIC vector express high levels of TCR of their surface (Figure 5A, right panels). To determine the dose-response relationship between NotchIC expression and TCR upregulation, we gated on different levels of hCD2 expression in polyclonal AKR1010 cells expressing NotchIC and analyzed these for TCR expression (Figure 5B). Interestingly, increasing the level of NotchIC expression leads to a greater percentage of cells expressing TCR rather than an increase in the level of TCR expression. Furthermore, TCR surface upregulation is partially dependent on the RAM domain of mNotch-1, since mutation of the RAM domain results in 5- to 10-fold fewer cells that express TCR at the same level of transgene expression (Figure 5C). We also performed similar experiments to determine whether NotchIC expression affects CD4 or CD8 coreceptor expression in AKR1010. While there was no effect on CD4 expression, NotchIC downregulated CD8 expression approximately 5-fold (data not shown). Interestingly, a similar level of CD8 coreceptor downregulation also occurs during the transition of DP thymocytes into both the CD4+ and CD8+ SP lineages (Lucas and Germain, 1996).
Figure 5.
Expression of NotchIC Upregulates TCR Expression
(A) Expression of NotchIC in AKR1010 induces TCR surface expression. AKR1010 was infected with retrovirus prepared from control vector or the pMI/NotchIC vector. Two days following infection, the cells were assayed for IRES-driven hCD2 expression and CD3 expression by FACS analysis. Gates on uninfected and infected cells were established based on hCD2 expression (top panels). Cells falling in these gates were then analyzed for CD3 expression (lower panels). Identical results were obtained using the H57-597 MAb directed against the αβTCR.
(B) Dose-dependent relationship between the level of NotchIC expression and TCR expression. Vector control and polyclonal AKR1010 cell lines expressing NotchIC were stained simultaneously for hCD2 expression and CD3 expression. Gates for different levels of hCD2 expression were established, and cells within these were analyzed for CD3 expression. The dotted line represents background staining levels.
(C) Upregulation of TCR expression maps to the RAM domain of Notch. Polyclonal AKR1010 cell lines expressing NotchIC, NotchIC/mM2-2, or control vector were analyzed for CD3 expression as in (B). The graph represents the percentage of cells expressing CD3 relative to the level of transgene expression.
Notch Signaling Regulates Deltex Expression
To further characterize downstream effects of Notch signaling in T cells, we sought to identify novel genes that are regulated by Notch signaling. For this purpose, we used the representation difference analysis (RDA) technique (Hubank and Schatz, 1994) to identify genes upregulated by expression of NotchIC in AKR1010. This approach yielded multiple cDNA fragments derived from the Deltex gene. Northern blot analysis confirmed that expression of NotchIC results in the upregulation of Deltex expression in AKR1010 (Figure 6A). Deltex was originally identified in Drosophila melanogaster as a gene that interacts with the Notch signaling pathway (Xu and Artavanis, 1990). Homologs of Deltex have subsequently been cloned from mice and humans (Pampeno and Meruelo, 1996; Matsuno et al., 1998). Genetic and biochemical studies suggest that Deltex functions as a positive regulator of the Notch signaling pathway (Matsuno et al., 1995). Our finding identifies Deltex as a novel target of the Notch signaling pathway in T cells.
Figure 6.
Deltex Is Positively Regulated by Notch Signaling and Is Expressed at High levels in DN and CD4+ and CD8+ SP Thymocytes and Low Levels in DP Thymocytes
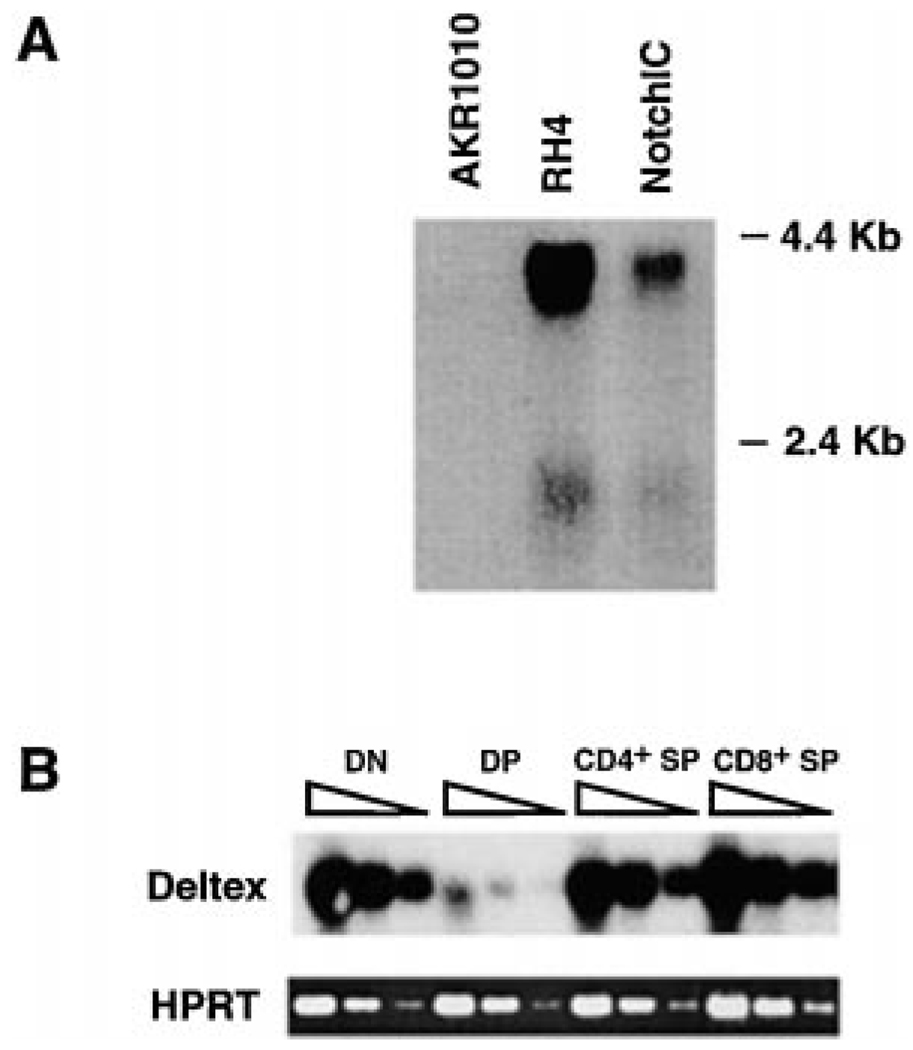
(A) NotchIC expression upregulates Deltex expression. AKR1010, the RH4 dexamethasone-resistant clone, and polyclonal AKR1010 expressing NotchIC were analyzed for Deltex expression by Northern blot analysis.
(B) Expression of Deltex in thymocyte subpopulations. cDNA prepared from FACS-sorted thymocyte subpopulations was serially diluted at 3-fold and amplified by PCR with primers specific for Deltex. The PCR products were analyzed by Southern blot using a Deltex-specific probe. Amplification of the same cDNA using primers specific for HPRT was performed to control for template quality.
Since Deltex is positively regulated by Notch signaling in T cells, we reasoned that thymocytes that have active Notch signaling would express Deltex whereas thymocytes that are not receiving Notch signals would lack Deltex expression. Therefore, we analyzed FACS-sorted DN, DP, CD4+ SP, and CD8+ SP thymocytes for Deltex expression by RT-PCR (Figure 6B). Relatively high levels of Deltex expression were detected in DN, CD4+ SP, and CD8+ SP thymocytes, whereas much lower levels were detected in DP thymocytes. This pattern of Deltex expression suggests that the Notch signaling pathway is active in DN and CD4+ and CD8+ SP thymocytes and inactive in DP thymocytes. Importantly, this agrees with our data indicating that glucocorticoid resistance and Bcl-2 are positively regulated by Notch signaling since DN and CD4+ and CD8+ SP thymocytes are resistant to glucocorticoids and express high levels of Bcl-2, whereas DP thymocytes are sensitive to glucocorticoids and express low levels of Bcl-2 (Scollay et al., 1984; Screpanti et al., 1989; Veis et al., 1993; Linette et al., 1994). This also agrees with the expression pattern of mNotch-1 in thymocyte subsets: Notch signaling positively regulates mNotch-1 expression in T cells, and mNotch-1 is expressed at higher levels in DN and CD4+ and CD8+ SP thymocytes relative to DP thymocytes (Hasserjian et al., 1996).
NotchIC Expression Confers Resistance to Glucocorticoids in DP Thymocytes
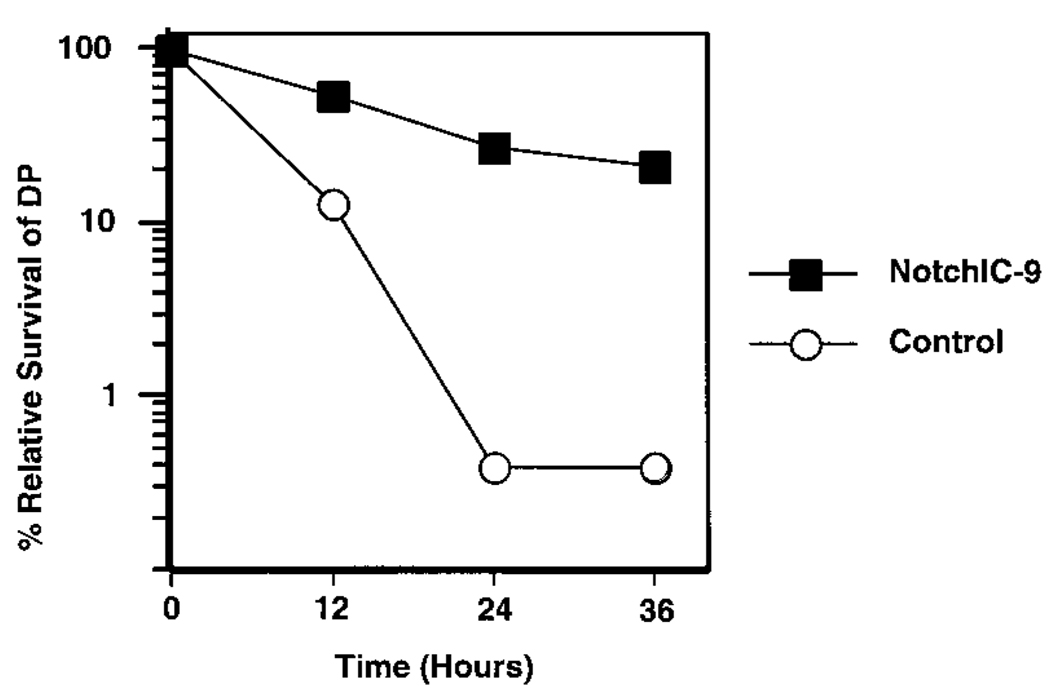
Our data indicate that activation of the Notch signaling pathway in a DP thymoma cell line induces multiple phenotypic changes that are associated with the maturation of DP thymocytes into SP thymocytes. To confirm that similar effects occur in vivo, we analyzed DP thymocytes from the NotchIC-9 mice, which express NotchIC under the control of the lck proximal promoter (Robey et al., 1996). Thymocytes from these mice and littermate controls were cultured in the presence of dexamethasone and analyzed by FACS to determine their sensitivity to glucocorticoids (Figure 7). As expected, DP thymocytes from nontransgenic littermate control mice are exquisitely sensitive to glucocorticoids. In contrast, DP thymocytes from transgene-expressing NotchIC-9 mice are relatively resistant to glucocorticoid-induced apoptosis. Therefore, activation of the Notch signaling pathway confers resistance to glucocorticoids in both transformed T cell lines and thymocytes.
Figure 7.
Expression of NotchIC Confers Resistance to Glucocorticoids in DP Thymocytes from Transgenic Mice
Thymocytes from NotchIC-9 and littermate control mice were incubated in medium alone or medium supplemented with 10−8 M dexamethasone for the indicated times and analyzed by FACS for viability and CD4 and CD8 expression. The graph shows the percentage of viable DP thymocytes in cultures containing dexamethasone relative to viable DP thymocytes in cultures containing medium alone. Similar results were obtained in three independent experiments.
Discussion
Notch is an evolutionary conserved cell surface receptor that plays a role in many developmental processes. Upon binding to its ligand, Notch is proteolytically processed within the transmembrane domain and the intracellular domain is released (Schroeter etal., 1998; Struhl and Adachi, 1998). Following transport to the nucleus, the intracellular domain of Notch physically interacts with the RBP/Su(H) transcription factor and changes RBP/Su(H) from a repressor to an activator of transcription (Hsieh et al., 1996; Kao et al., 1998). Notch thereby functions in a signal transduction pathway that directly links activation of a cell surface receptor to changes in gene expression.
Notch is expressed at high levels in developing thymocytes (Hasserjian et al., 1996), and a role for Notch signaling in regulating the CD4 versus CD8 cell fate choice has been proposed (Robey et al., 1996). We have shown that activation of the Notch signaling pathway in a DP thymoma cell line and in thymocytes results in the induction of multiple phenotypic changes that are associated with the differentiation of DP thymocytes into SP thymocytes. These changes include the acquisition of resistance to glucocorticoid-induced apoptosis and the upregulation of Bcl-2, TCR, and Deltex expression. Furthermore, phenotypes that are associated with active Notch signaling are present in DN and CD4+ and CD8+ SP thymocytes and absent from DP thymocytes in normal mice. Thus, both DN and CD4 + and CD8+ SP thymocytes are resistant to glucocorticoids and express high levels of Bcl-2 and Deltex, whereas DP thymocytes are sensitive to glucocorticoids and express low levels of Bcl-2 and Deltex. Similarly, Notch signaling upregulates mNotch-1 expression and DN thymocytes, and CD4+ and CD8+ SP thymocytes express higher levels of mNotch-1 than DP thymocytes. Based on these observations, we propose that the activity of the Notch signaling pathway is high in DN and CD4+ and CD8+ SP thymocytes and low in DP thymocytes. Furthermore, we suggest that Notch signaling regulates the expression of multiple genes that are differentially expressed within these thymocyte subsets.
What regulates activation of Notch signaling in developing thymocytes? In many developmental processes, Notch signaling is regulated by cell–cell interactions between Notch and Notch ligand-expressing cells. Jagged-2, which is a ligand for mNotch-1, is expressed at high levels in the thymus (Luo et al., 1997), suggesting that cell–cell interactions between thymocytes and Jagged-2-expressing cells may play a crucial role in regulating Notch signaling. If interaction with Jagged-2-expressing cells directly regulates Notch signaling in thymocytes, our data suggests that DN and SP thymocytes, but not DP thymocytes, productively interact with Jagged-2-expressing cells. A characterization of the pattern of expression of Jagged-2 and other Notch ligands in the thymus is necessary to test this hypothesis. An alternative possibility is that Jagged-2 or other Notch ligands may be ubiquitously expressed in the thymus and additional factors may regulate activation of the Notch signaling pathway. It is intriguing to speculate that Notch signaling may be regulated, directly or indirectly, by signals delivered through the pre-TCR and TCR. These cell surface receptors play a critical role in regulating the development of thymocytes. Signals from the pre-TCR are required for the maturation of DN thymocytes into DP thymocytes, and our data suggest that this is accompanied by the inhibition of Notch signaling. Similarly, the maturation of DP thymocytes into SP thymocytes is dependent upon appropriate signals from the TCR and may be accompanied by activation of the Notch signaling pathway. Changes in the activity of the Notch signaling pathway that occur concomitant with thymocyte maturation may be a secondary consequence of maturation or may play an active role in inducing thymocyte maturation. If Notch plays an active role in driving the maturation of DP thymocytes into SP thymocytes, this suggests the possibility that positively selecting TCR signals may facilitate activation of the Notch signaling pathway. One important event in this process would be the rescue of DP thymocytes from glucocorticoid-induced apoptosis. Interestingly, we have identified Deltex as a gene whose expression is upregulated by Notch signaling in T cells. Deltex was originally identified in Drosophila as a gene that genetically interacts with the Notch signaling pathway (Xu and Artavanis, 1990). Ectopic expression of Deltex in Drosophila results in phenotypes similar to those that result from expression of constitutively active Notch (Matsuno et al., 1995). This suggests that Deltex functions to positively regulate the Notch signaling pathway. If murine Deltex functions similarly, this suggests a positive feedback loop in which initial activation of Notch signaling is positively reinforced by the upregulation of Deltex expression.
Our model differs from a previous model suggesting that the Notch signaling pathway functions to regulate the CD4 versus CD8 lineage choice (Robey et al., 1996). This model was based on the observation that expression of NotchIC under control of the lck proximal promoter in transgenic mice leads to the development of an excess of CD8+ SP thymocytes over CD4+ SP thymocytes. This led to the hypothesis that positively selected DP thymocytes that receive a Notch signal develop into CD8+ SP thymocytes, whereas, in the absence of Notch signals, DP thymocytes adopt the CD4+ cell fate. We note, however, that mice expressing Bcl-2 as a trans-gene under control of the lck proximal promoter have a very similar phenotype to the NotchIC transgenic mice. Both types of transgenic mice develop an excess of CD8+ SP thymocytes that express high levels of TCR but do not survive in the periphery, and in both cases the development in these CD8+ SP thymocytes is independent of MHC class I expression but does depend on some MHC expression, either class I or class II (Sentman et al., 1991; Linette et al., 1994; Robey et al., 1996). Why do both the Bcl-2 and NotchIC transgenic mice develop excess CD8+ SP thymocytes? As we have shown for the NotchIC transgene, the Bcl-2 transgene also renders DP thymocytes resistant to the apoptotic effects of glucocorticoids (Sentman et al., 1991; Strasser et al., 1991; Siegel et al., 1992), and the preferential effect of NotchIC and Bcl-2 on CD8+ SP thymocyte development may be secondary to this effect. Enhanced survival in the presence of endogenous glucocorticoids may allow DP thymocytes a greater chance to interact with positively selecting MHC ligands. Since CD8 lineage commitment may require weaker signals from the TCR compared to the signal strength needed for CD4 lineage commitment (Itano et al., 1996; Matechak et al., 1996), then cells which have an extremely low affinity interaction with self-MHC, and which would normally die by neglect, may be rescued preferentially to the CD8 lineage in the Bcl-2 and NotchIC mice. Alternatively, Notch signaling may play an active role in driving the maturation of DP thymocytes into both the CD4+ and CD8+ SP lineages. Expression of NotchIC under control of the lck proximal promoter may induce maturation of DP thymocytes that have not received appropriate TCR and coreceptor signals. Again, since less stringent TCR signals may be required for commitment to the CD8 lineage relative to the CD4 lineage, these cells may preferentially adopt the CD8 cell fate. According to this model, Notch signaling plays a role in the positive selection of DP thymocytes, but different factors, most likely signals derived from the interaction of the TCR and CD4 or CD8 coreceptor molecules with MHC ligands, drive lineage commitment. The excess CD8+ SP cells that appear in the thymus of Bcl-2 and NotchIC transgenic mice are not seen in the lymphoid periphery (Sentman et al., 1991; Robey et al., 1996). Either they die within the thymus or shortly after leaving. Both models we have suggested above imply that these cells have not received appropriate positively selecting signals for maturation and may be only partially along the pathway to full maturity. In line with this idea, the excess CD8+ SP thymocytes in the NotchIC-9 mice express lower levels of CD8 and TCR than wild-type CD8+ SP thymocytes (data not shown).
The ability of NotchIC to induce multiple phenotypic changes in the AKR1010 cell line that are associated with the maturation of DP thymocytes into SP thymocytes provides a useful system for studying molecular events that occur during thymocyte development. Our data suggest that the Notch signaling pathway regulates the expression of genes that confer resistance to glucocorticoids and upregulate TCR surface expression in thymocytes. Mechanisms regulating glucocorticoid-sensitivity and TCR surface expression in developing thymocytes are not known (Cohen, 1992; Kearse et al., 1995). The identification of genes regulated by Notch signaling in AKR1010 and other T cell lines may provide a productive approach toward elucidating the molecular basis for these and other events of thymocyte maturation.
Experimental Procedures
Cell Lines and Tissue Culture
AKR1010 is a spontaneous CD4+CD8+ thymoma cell line derived from an AKR/J mouse (Richie et al., 1988). 2B4.11 is a T cell hybridoma specific for pigeon cytochrome C (Ashwell et al., 1987). ФNX-A is a retrovirus packaging cell line that produces high-titer amphotropic retrovirus upon transient transfection (Pear, 1996). All cells were cultured in DMEM containing 10% FCS, 2 mM glutamine, 25 mM HEPES, 50 µM β-mercaptoethanol, 100 U/ml penicillin, and 100 µg/ml streptomycin. Dexamethasone was purchased from Sigma.
Thymocyte cDNA Library Construction
PolyA+ RNA was prepared from thymocytes from 6- to 8-week-old female C57BL/6 mice using the FastTrack 2.0 kit (Invitrogen). Double-stranded cDNA was synthesized using random hexamer or oligo dT primers, size selected, and ligated into the BstXI site of the pMX vector (Onishi et al., 1996). The ligation reaction was transformed into ElectroMAX DH10B E. coli (GIBCO-BRL) and plated onto multiple LB plates containing 100 µg/ml ampicillin. Approximately 1 × 107 independent clones were obtained from 2 µg of polyA+ RNA. These clones were pooled and plasmid DNA prepared using the Qiagen Plasmid Maxi Kit. Two random hexamer primed libraries (RH and US) and one oligo-dT primed library (OT) were constructed.
Retrovirus Production and Infection of T Cell Lines
Plasmid DNA was transfected into the ФNX-A packaging cell line using CaPO4 precipitation as described (Pear, 1996). Retrovirus-containing supernatant was collected following overnight incubation of the transfected packaging cells at 32°C. AKR1010 and 2B4.11 cells were infected by resuspending mid-log phase cells at 5 × 105 cells/ml in retrovirus-containing supernatant supplemented with 8 µg/ml polybrene followed by centrifugation for 90 min at 2500 rpm and overnight incubation at 32°C.
Expression Cloning
Retrovirus-containing supernatant was prepared from the RH, US, and OT thymocyte cDNA libraries and used to infect 2 × 107 AKR1010 cells. Two days following infection, dexamethasone was added to the cultures at a final concentration of 10−6 M and the cells were plated at 1 × 104 cells per well in 96-well plates. Culture for 2–4 weeks in the presence of dexamethasone resulted in the isolation of numerous dexamethasone-resistant clones. RT-PCR using vector-specific primers that flank the polycloning site of pMX was used to amplify cDNA prepared from the dexamethasone-resistant clones. PCR products were purified and directly sequenced with nested vector-specific primers on an ABI automated sequencer.
Construction of NotchIC Plasmids and Generation of Transgene-Expressing Polyclonal Cell Lines
The pMI vector was constructed by cloning the internal ribosome entry site (IRES) from the encephalomyocarditis virus and a cDNA encoding the extracellular and transmembrane domains of human CD2 into the pMX vector. The resulting vector contains a polycloning site upstream of an IRES-hCD2 cassette. cDNA cloned into this vector is transcribed into bicistronic mRNA that concomitantly directs translation of the cDNA and hCD2. pMI-based plasmids used in this work include the following: pMI/NotchIC contains a Kozak consensus initiation methionine followed by the coding region of mNotch-1 extending from amino acids 1751 to 2444; pMI/NotchIC/mM2-2 is identical to pMI/NotchIC except that it contains the mM2-2 point mutations in the RAM domain (Tamura et al., 1995); pMI/NotchIC/ΔAnk contains a Kozak consensus initiation methionine followed by the coding region of mNotch-1 extending from amino acids 1941 to 2444. The mNotch-1 cDNA used to generate these constructs was obtained by RT-PCR from the RH4 dexamethasone-resistant clone using Pfu polymerase (Stratagene). Polyclonal cell lines expressing various Notch polypeptides were generated by infecting AKR1010 and 2B4.11 with retrovirus produced from the pMI/NotchIC, pMI/NotchIC/mM2-2, and pMI/NotchIC/ΔAnk plasmids. Following infection, transgene-expressing cells were isolated by multiple rounds of panning on tissue culture plates coated with 50 µg/ml anti-hCD2 antibody (MAb 35.1). Control cells were similarly generated by using retrovirus produced from the control pMI vector.
Flow Cytometric Analysis
Cells were incubated with an excess of labeled antibodies on ice for 30 min and washed with PBS containing 1% FCS. The anti-hCD2 MAb 35.1 was purified and labeled in our laboratory. Labeled anti-CD3, anti-CD4, and anti-CD8 MAb were obtained from Pharmingen. For cell cycle analysis, cells were washed in PBS, fixed overnight at 4°C in 70% ethanol, and stained for DNA content using 50 µg/ml propidium iodide in the presence of 100 µg/ml RNAseA. Data were collected on a FACScan flow cytometer (Becton Dickinson) using CellQuest software.
Proliferation Assay
Cells expressing various NotchIC constructs or hCD2 alone were incubated in triplicate in 96-well plates at 1 × 104 cells/well in media or media containing various concentrations (1 × 10−10 M to 1 × 10−6 M) of dexamethasone for 24 hr followed by the addition of 1 mCi of 3H thymidine. After an additional 12 hr, the cells were harvested and incorporated thymidine measured.
Western Blot Analysis
Cells were lysed at 1 × 108 cells/ml in lysis buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 1 mM EDTA, 1% NP-40 containing 1 mM PMSF, 10 µg/ml aprotinin, and 10 µg/ml pepstatin) for 30 min and cleared of debris by centrifugation. 2 × 106 cell equivalents were separated on polyacrylamide gels, transferred to nitrocellulose, and probed with the antibodies to mNotch-1 (C-20, Santa Cruz), Bcl-2 (3F11, Pharmingen), or Bcl-XL (S-18, Santa Cruz). Bound antibodies were detected with HRP-conjugated secondary antibodies and enhanced chemiluminesence according to the manufacturer’s instructions (Amersham).
Representation Difference Analysis and Northern Blot Analysis
RDA was performed as described (Hubank and Schatz, 1994) using cDNA prepared from AKR1010 as “driver” and cDNA prepared from the RH4 dexamethasone-resistant clone as “tester.” Multiple distinct products were observed by agarose gel electrophoresis following two rounds of subtractive PCR. Sequence analysis of these products revealed multiple DpnII fragments derived from the murine Deltex gene (Pampeno and Meruelo, 1996). Northern blot analysis was performed according to standard protocols (Sambrook et al., 1989). 10 µg of total RNA from each cell line was analyzed. Equivalent loading and transfer was verified by visual inspection of ribo-somal RNA bands. The probe consisted of a cloned fragment of murine Deltex corresponding to nucleotides 1028 to 1938.
RT-PCR Analysis of Thymocyte Subsets
Thymocytes from 6- to 8-week-old C57BL/6 mice were sorted for DN, DP, CD4+ SP, andCD8+ SP subsets by FACS. Following sorting, each subset contained less than 0.5% contamination with other sorted subsets. RNA prepared from sorted cells was converted into cDNA and used as a template for PCR. Sequences of primers used for Deltex amplification are 5′CACTGGCCCTGTCCACCCAGCCTTGGCAGG3′ and 5′ATGCGAATTCGGGAAGGCGGGCAACTCAGG3′. PCR reactions were prepared according to manufacturer instructions (Perkin Elmer) and consisted of 30 cycles (45 sec 94°C, 45 sec 55°C, and 60 sec 72°C). The PCR products were analyzed by Southern blot using standard protocols (Sambrook et al., 1989). The probe consisted of a cloned fragment of murine Deltex corresponding to nucleotides 1028 to 1938.
Analysis of Thymocyte Sensitivity to Glucocorticoids
Thymocytes from NotchIC-9 transgenic and littermate control mice were incubated at 3 × 106 cell/ml in medium alone or in medium supplemented with 10−8 M dexamethasone. After the indicated times, the cells were harvested, stained with antibodies specific for CD4 and CD8, and 50,000 events analyzed by FACS. A viable cell gate was established based on FSC and SSC, and the number of CD4+CD8+ events falling in the viable gate was used to quantitate the number of viable DP cells. The percentage of relative survival was calculated by dividing the number of viable DP thymocytes in cultures containing dexamethasone by the number of viable DP thymocytes in cultures containing medium alone.
Acknowledgments
We thank Ellen Richie for the AKR1010 thymoma cell line, Garry Nolan for the ФNX-A retrovirus packaging cell line, and Ellen Robey for providing the NotchIC-9 mice. We also thank Ananda Goldrath for FACS sorting of thymocyte subsets and Jacqueline Kirchnerfor help with RDA. This work was supported by National Institutes of Health grants CA09537 and AI29802, the Howard Hughes Medical Institute, and Medical Scientist Training Program Fellowship NIGMS GM07266.
References
- Artavanis TS, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- Ashwell JD, Cunningham RE, Noguchi PD, Hernandez D. Cell growth cycle block of T cell hybridomas upon activation with antigen. J. Exp. Med. 1987;165:173–194. doi: 10.1084/jem.165.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M, Herrlich P, Schutz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- Blaumueller CM, Qi H, Zagouras P, Artavanis TS. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- Chao DT, Linette GP, Boise LH, White LS, Thompson CB, Korsmeyer SJ. Bcl-XL and Bcl-2 repress a common pathway of cell death. J. Exp. Med. 1995;182:821–828. doi: 10.1084/jem.182.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JJ. Glucocorticoid-induced apoptosis in the thymus. Semin. Immunol. 1992;4:363–369. [PubMed] [Google Scholar]
- Cohen JJ, Duke RC. Glucocorticoid activation of a calcium-dependent endonuclease in thymocyte nuclei leads to cell death. J. Immunol. 1984;132:38–42. [PubMed] [Google Scholar]
- del Amo FF, Gendron MM, Swiatek PJ, Jenkins NA, Copeland NG, Gridley T. Cloning, analysis, and chromosomal localization of Notch-1, a mouse homolog of Drosophila Notch. Genomics. 1993;15:259–264. doi: 10.1006/geno.1993.1055. [DOI] [PubMed] [Google Scholar]
- Grillot DA, Merino R, Nunez G. Bcl-XL displays restricted distribution during T cell development and inhibits multiple forms of apoptosis but not clonal deletion in transgenic mice. J. Exp. Med. 1995;182:1973–1983. doi: 10.1084/jem.182.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasserjian RP, Aster JC, Davi F, Weinberg DS, Sklar J. Modulated expression of notch1 during thymocyte development. Blood. 1996;88:970–976. [PubMed] [Google Scholar]
- Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubank M, Schatz DG. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itano A, Salmon P, Kioussis D, Tolaini M, Corbella P, Robey E. The cytoplasmic domain of CD4 promotes the development of CD4 lineage T cells. J. Exp. Med. 1996;183:731–741. doi: 10.1084/jem.183.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson S, Bevan M. T-cell selection. Curr. Opin. Immunol. 1998;10:214–219. doi: 10.1016/s0952-7915(98)80251-3. [DOI] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano NN, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura OS, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- Kearse KP, Roberts JP, Wiest DL, Singer A. Developmental regulation of alpha beta T cell antigen receptor assembly in immature CD4 + CD8+ thymocytes. Bioessays. 1995;17:1049–1054. doi: 10.1002/bies.950171209. [DOI] [PubMed] [Google Scholar]
- Kisielow P, von Boehmer H. Development and selection of T cells: facts and puzzles. Adv. Immunol. 1995;58:87–209. doi: 10.1016/s0065-2776(08)60620-3. [DOI] [PubMed] [Google Scholar]
- Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl. Acad. Sci. USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber T, Kidd S, Alcamo E, Corbin V, Young MW. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- Linette GP, Grusby MJ, Hedrick SM, Hansen TH, Glimcher LH, Korsmeyer SJ. Bcl-2 is upregulated at the CD4+ CD8+ stage during positive selection and promotes thymocyte differentiation at several control points. Immunity. 1994;1:197–205. doi: 10.1016/1074-7613(94)90098-1. [DOI] [PubMed] [Google Scholar]
- Lucas B, Germain RN. Unexpectedly complex regulation of CD4/CD8 coreceptor expression supports a revised model for CD4+CD8+ thymocyte differentiation. Immunity. 1996;5:461–477. doi: 10.1016/s1074-7613(00)80502-6. [DOI] [PubMed] [Google Scholar]
- Luo B, Aster JC, Hasserjian RP, Kuo F, Sklar J. Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol. Cell. Biol. 1997;17:6057–6067. doi: 10.1128/mcb.17.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matechak EO, Killeen N, Hedrick SM, Fowlkes BJ. MHC class II-specific T cells can develop in the CD8 lineage when CD4 is absent. Immunity. 1996;4:337–347. doi: 10.1016/s1074-7613(00)80247-2. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Diederich RJ, Go MJ, Blaumueller CM, Artavanis TS. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 1995;121:2633–2644. doi: 10.1242/dev.121.8.2633. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Go MJ, Sun X, Eastman DS, Artavanis TS. Suppressor of Hairless-independent events in Notch signaling imply novel pathway elements. Development. 1997;124:4265–4273. doi: 10.1242/dev.124.21.4265. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Eastman D, Mitsiades T, Quinn AM, Carcanciu ML, Ordentlich P, Kadesch T, Artavanis TS. Human deltex is a conserved regulator of Notch signaling. Nat. Genet. 1998;19:74–78. doi: 10.1038/ng0598-74. [DOI] [PubMed] [Google Scholar]
- Miyashita T, Reed JC. bcl-2 gene transfer increases relative resistance of S49.1 and WEHI7.2 lymphoid cells to cell death and DNA fragmentation induced by glucocorticoids and multiple chemotherapeutic drugs. Cancer Res. 1992;52:5407–5411. [PubMed] [Google Scholar]
- Onishi M, Kinoshita S, Morikawa Y, Shibuya A, Phillips J, Lanier LL, Gorman DM, Nolan GP, Miyajima A, Kitamura T. Applications of retrovirus-mediated expression cloning. Exp. Hematol. 1996;24:324–329. [PubMed] [Google Scholar]
- Pampeno CL, Meruelo D. A novel cDNA transcript expressed in fractionated X-irradiation-induced murine thymomas. Cell Growth Differ. 1996;7:1113–1123. [PubMed] [Google Scholar]
- Pear W. Transient transfection methods for preparation of high-titer retroviral supernatants. In: Auaubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley and Sons, Inc.; 1996. pp. 9–11. [DOI] [PubMed] [Google Scholar]
- Rebay I, Fehon RG, Artavanis TS. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993;74:319–329. doi: 10.1016/0092-8674(93)90423-n. [DOI] [PubMed] [Google Scholar]
- Reichardt H, Kaestner K, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, Schutz G. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- Richie ER, McEntire B, Crispe N, Kimura J, Lanier LL, Allison JP. Alpha/beta T-cell antigen receptor gene and protein expression occurs at early stages of thymocyte differentiation. Proc. Natl. Acad. Sci. USA. 1988;85:1174–1178. doi: 10.1073/pnas.85.4.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey E, Chang D, Itano A, Cado D, Alexander H, Lans D, Weinmaster G, Salmon P. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell. 1996;87:483–492. doi: 10.1016/s0092-8674(00)81368-9. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Second Edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signaling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Scollay R, Bartlett P, Shortman K. T cell development in the adult murine thymus: changes in the expression of the surface antigens Ly2, L3T4 and B2A2 during development from early precursor cells to emigrants. Immunol. Rev. 1984;82:79–103. doi: 10.1111/j.1600-065x.1984.tb01118.x. [DOI] [PubMed] [Google Scholar]
- Screpanti I, Morrone S, Meco D, Santoni A, Gulino A, Paolini R, Crisanti A, Mathieson BJ, Frati L. Steroid sensitivity of thymocyte subpopulations during intrathymic differentiation. Effects of 17 beta-estradiol and dexamethasone on subsets expressing T cell antigen receptor or IL-2 receptor. J. Immunol. 1989;142:3378–3383. [PubMed] [Google Scholar]
- Sentman CL, Shutter JR, Hockenbery D, Kanagawa O, Korsmeyer SJ. bcl-2 inhibits multiple forms of apoptosis but not negative selection in thymocytes. Cell. 1991;67:879–888. doi: 10.1016/0092-8674(91)90361-2. [DOI] [PubMed] [Google Scholar]
- Shawber C, Nofziger D, Hsieh JJ, Lindsell C, Bogler O, Hayward D, Weinmaster G. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- Siegel RM, Katsumata M, Miyashita T, Louie DC, Greene MI, Reed JC. Inhibition of thymocyte apoptosis and negative antigenic selection in bcl-2 transgenic mice. Proc. Natl. Acad. Sci. USA. 1992;89:7003–7007. doi: 10.1073/pnas.89.15.7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent J, Lo D, Gao EK, Ron Y. T cell selection in the thymus. Immunol. Rev. 1988;101:173–190. doi: 10.1111/j.1600-065x.1988.tb00737.x. [DOI] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Cory S. bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell. 1991;67:889–899. doi: 10.1016/0092-8674(91)90362-3. [DOI] [PubMed] [Google Scholar]
- Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Curr. Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- Vacchio MS, Ashwell JD, King LB. A positive role for thymus-derived steroids in formation of the T-cell repertoire. Ann. NY Acad. Sci. 1998;840:317–327. doi: 10.1111/j.1749-6632.1998.tb09571.x. [DOI] [PubMed] [Google Scholar]
- Veis DJ, Sentman CL, Bach EA, Korsmeyer SJ. Expression of the Bcl-2 protein in murine and human thymocytes and in peripheral T lymphocytes. J. Immunol. 1997;151:2546–2554. [PubMed] [Google Scholar]
- Weinmaster G. The ins and outs of notch signaling. Mol. Cell. Neurosci. 1997;9:91–102. doi: 10.1006/mcne.1997.0612. [DOI] [PubMed] [Google Scholar]
- Wettstein DA, Turner DL, Kintner C. The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development. 1997;124:693–702. doi: 10.1242/dev.124.3.693. [DOI] [PubMed] [Google Scholar]
- Wilkinson HA, Fitzgerald K, Greenwald I. Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. elegans cell fate decision. Cell. 1994;79:1187–1198. doi: 10.1016/0092-8674(94)90010-8. [DOI] [PubMed] [Google Scholar]
- Wyllie AH, Morris RG, Smith AL, Dunlop D. Chromatin cleavage in apoptosis: association with condensed chromatin morphology and dependence on macromolecular synthesis. J. Pathol. 1984;142:67–77. doi: 10.1002/path.1711420112. [DOI] [PubMed] [Google Scholar]
- Xu T, Artavanis TS. deltex, a locus interacting with the neurogenic genes, Notch, Delta and mastermind in Drosophila melanogaster. Genetics. 1990;126:665–677. doi: 10.1093/genetics/126.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharchuk CM, Mer’cep M, Chakraborti PK, Simons SSJ, Ashwell JD. Programmed T lymphocyte death. Cell activation- and steroid-induced pathways are mutually antagonistic. J. Immunol. 1990;145:4037–4045. [PubMed] [Google Scholar]