Abstract
A Binding Ensemble PROfiling with (F)photoaffinity Labeling (BEProFL) approach that utilizes photolabeling of HDAC8 with a probe containing a UV-activated aromatic azide, mapping the covalent modifications by liquid chromatography-tandem mass-spectrometry, and a computational method to characterize the multiple binding poses of the probe is described. Using the BEProFL approach two distinct binding poses of the HDAC8 probe were identified. The data also suggest that an “upside-down” pose with the surface binding group of the probe bound in an alternative pocket near the catalytic site may contribute to the binding.
Keywords: histone deacetylases, HDAC, binding ensemble profiling with photoaffinity labeling, proteomics, drug design
Introduction
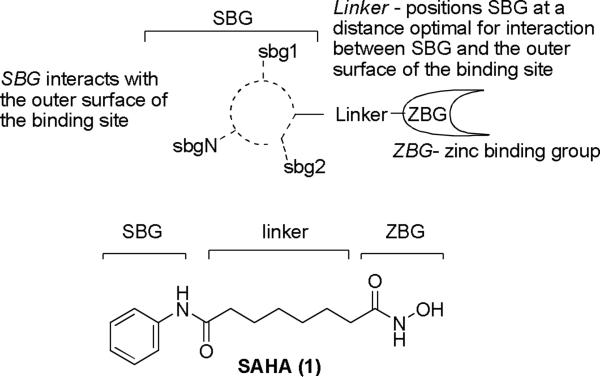
Histone deacetylases (HDACs) comprise a family of enzymes that regulate chromatin remodeling, gene transcription and activity of partner proteins. They control critical cellular processes, including cell growth, cell cycle regulation, DNA repair, differentiation, proliferation, and apoptosis.1 Chemical inhibitors of HDACs (Figure 1) have been shown to inhibit tumor cell growth and induce differentiation and cell death.2 SAHA (1, Figure 1) has been recently approved by FDA for use against cutaneous T-cell lymphoma. The HDACs can be divided into four classes based on structure, sequence homology, and domain organization.3 It is hypothesized that depending on isoform selectivity HDAC ligands can be either cytotoxic or neuroprotective.4 The development of isoform-selective HDAC inhibitors would be a significant step in reducing off-target effects of HDAC-based therapeutics.
Figure 1.
General structure of HDAC inhibitors.
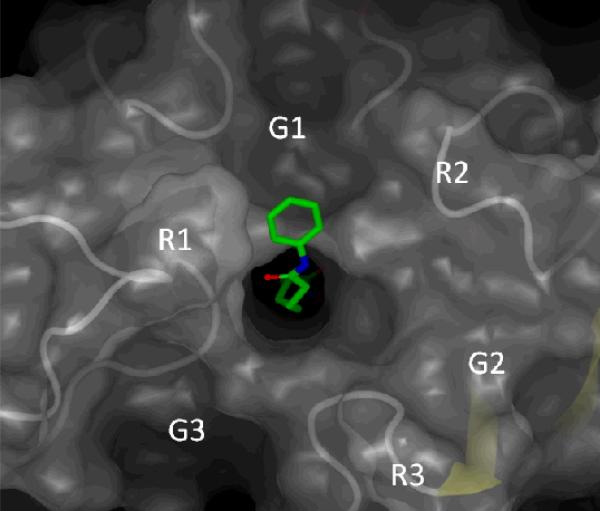
One of the key challenges in designing isoform-selective HDAC inhibitors is a poor understanding of the binding modes (poses) available to the highly solvent exposed surface binding group (SBG) of HDAC inhibitors targeting the grooves and ridges on the protein surface directly adjacent to the catalytic well of HDACs (Figure 2). It has been hypothesized by us in our preliminary experiments and a recent publication by Wiest and colleagues5 that the SBG groups may have more than one preferred position on the surface and each of them contributes to the overall binding affinity. Although the available HDAC X-ray data provide information on structure of proteins and binding of ligands, its use is limited due to high solvent exposure of the SBG of the ligands and additional copies of the same protein in the crystallographic cell that interfere with the binding of the co-crystallized ligands.
Figure 2.
Description of the grooves (G1-G3) and ridges (R1-R3) formed by the protein surface of HDAC8 (PDB:1T69).
Photoaffinity labeling is a strategy offering methods for the identification of drug target proteins of biologically active compounds and mapping their binding sites.6-8 Among several photosensitive groups, aryl azide is perhaps one of the most widely used due to its universality, reactivity, and relative simplicity in implementation.6 One of the recent advances in the photoaffinity labeling probe development is application of “click-chemistry” and bioorthogonal probes for activity-based proteomics profiling by Cravatt et al.9-15 and its recent modification by Suzuki et al.16-18 This latter study is based on a concept in which a bifunctional ligand is connected to a target protein by activation of a photoreactive group, such as an aromatic azido or 3-trifluoromethyl-3H-diazirin-3-yl group, and identification of the ligand product is achieved by anchoring a detectable tag to an alkyl azido moiety, which survives photolysis, using the Staudinger-Bertozzi ligation.19-24 A similar approach was recently utilized to discover a new binding site in HIV-1 integrase.25 In this case a different photoactivated moiety - benzophenone – facilitated by mass spectrometry and docking analysis was used to determine the exact ligand binding position. The recent study by Cravatt et al.15 presents a novel proteomics probe for histone deacetylases and is intended to discover new proteins interacting with the histone deacetylases. It does not address, however, the problem of multiple binding poses of HDAC inhibitors.
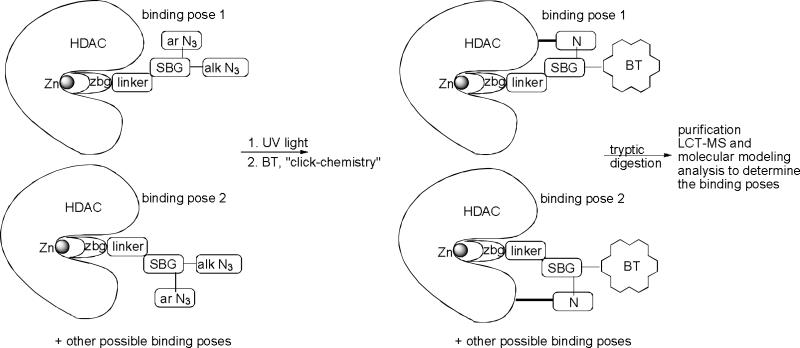
Here we introduced for the first time the use of diazide-based photoaffinity labeling probes, liquid chromatography-tandem mass-spectrometry, and molecular dynamics simulations to map the ensemble of the poses of HDAC8 inhibitors upon binding (Figure 3) or BEProFL – (B)inding (E)nsemble (Pro)filing with (F)photoaffinity (L)abeling.
Figure 3.
Application of photoaffinity probes for detection of binding poses of an HDAC ligand.
Materials and methods
Synthesis
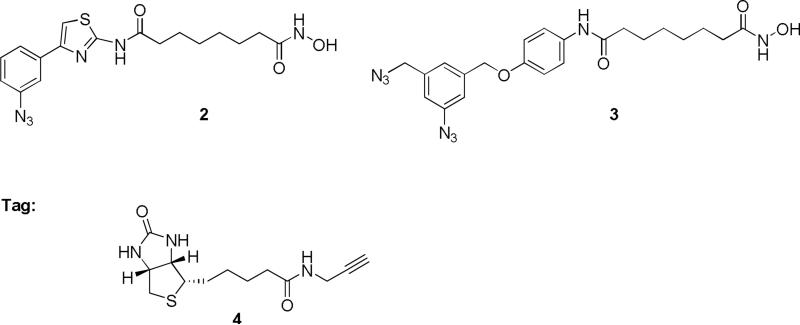
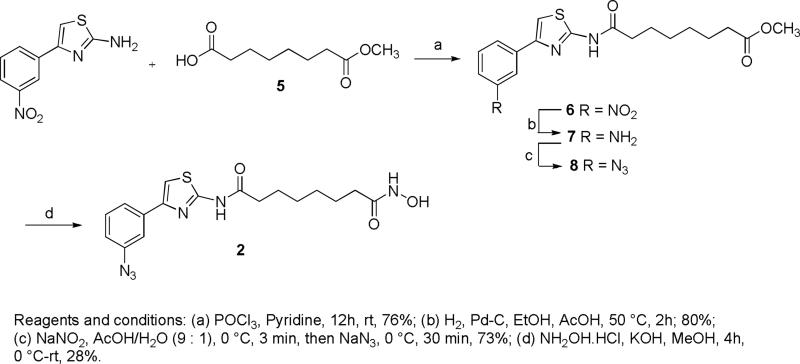
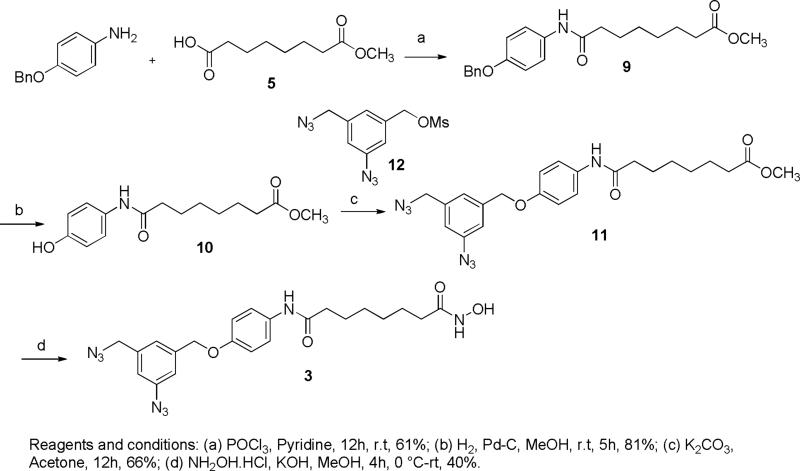
The synthesis of probes 2 and 3 (Figure 4) is outlined in schemes 1 and 2. Commercially available subericacid monomethyl ester 5 was coupled with 4-(3-nitrophenyl)-thiazol-2-ylamine in presence pyridine and POCl3 to give ester 6.26 The nitro group of ester 6 was reduced resulting in amine 7 that was converted to azide 8 by diazotization and nucleophilic substitution with sodium azide. Further treatment of 8 with hydroxylamine resulted in hydroxamic acid 2. The condensation of 5 with an equivalent amount of 4-benzyloxyaniline in presence of POCl3 and pyridine gave amide 9. The benzylhydroxy group of amide 9 was deprotected by catalytic hydrogenation and coupled with mesylate 1227 resulting in diazide 11 that was then converted to the corresponding hydroxamic acid 3 in a 40% yield. BT 4 was synthesized following the procedure described earlier.28
Figure 4.
Chemical structures of the probes 2 and 3 and the biotin tag 4.
Scheme 1.
Synthesis of probe 2.
Scheme 2.
Synthesis of probe 3.
HDAC activity assay
The inhibition of HDAC8 was measured as recommended by the supplier BIOMOL International using the fluorescent acetylated HDAC substrate Fluor de Lys (BIOMOL, KI178) and commercially available recombinant human HDAC8 (BIOMOL). The activity data are summarized in Table 1.
Table 1.
IC50 and KI values of inhibition of HDAC8 with SAHA and probes 2 and 3.
| Compound | HDAC8 | |
|---|---|---|
| |
IC50 μ M |
Ki μM |
| SAHA |
0.44 ±0.021 |
0.396 |
|
2 |
2.71±0.14 |
2.43 |
| 3 | 7.34±0.22 | 6.59 |
Photolabeling and visualization of probes
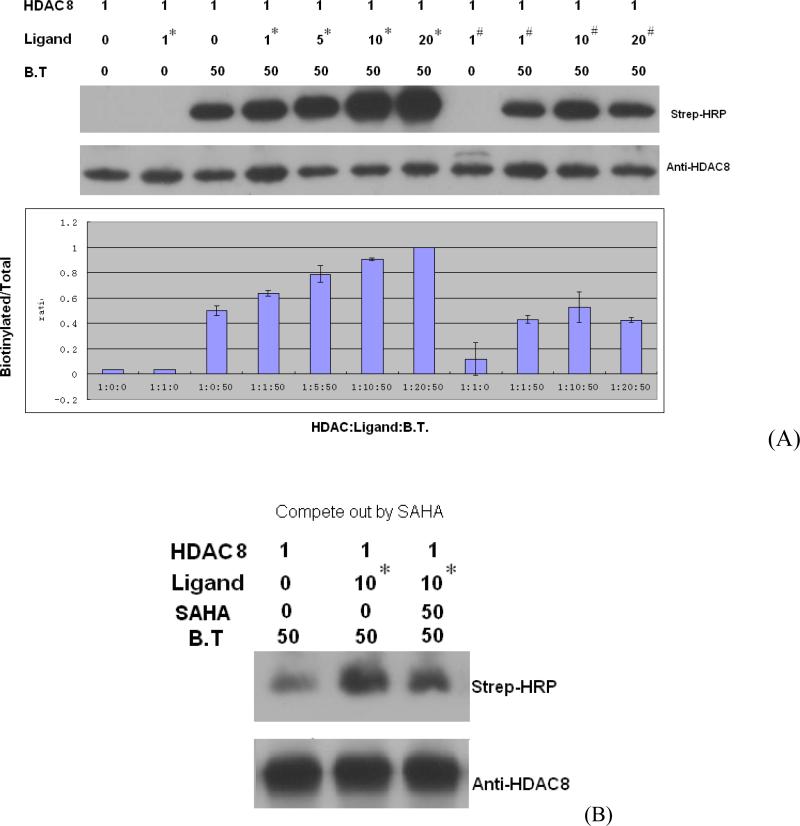
The probes 2 and 3 or probe 3-SAHA mixture (in the competition experiment) were incubated with the HDAC8 protein, exposed to 254 nM UV light 3×1 min with 1 min resting. In case of 3, BT 4 was attached to HDAC8-3 adduct using [3+2]-cycloligation catalyzed by TBTA and Cu(I) produced in situ from CuSO4 with TCEP. The tagged HDAC8 protein was visualized with Western blotting. The procedures were in general similar to those recently described by Cravatt et al.15 and Suzuki et al.16-18 The results are shown in Figure 5.
Figure 5.
Characterization of biotinylated HDAC proteins and total HDAC proteins. (A) Western blot analyses of probe 2 (labeled as #) and probe 3 (labeled as *) binding to HDAC8 probed by strep-HRP and anti-HDAC8 antibodies. All the numbers stand for the concentration in μM. (B) Same as A. Competition of probe 3 with SAHA.
MALDI-ToF MS Analysis of intact HDAC8 protein
The four HDAC8 samples (three of them were treated with 2 at different concentrations) were purified and subjected to MALDI-ToF MS positive ion analysis. The results are shown in Table 2.
Table 2.
MALDI MS of intact HDAC8 and treated with 2.
| AVG MW, DA [M+H]+ | Std Deviation | Mass Shift Δ CTRL | ||
|---|---|---|---|---|
| Sample | N | |||
| HDAC8-CTRL | 6.0 | 45620 | 57.5 | 0.0 |
| HDAC8-(1:−1) | 6.0 | 45624 | 41.3 | 4.0 |
| HDAC8-(1:−5) | 6.0 | 45733 | 37.1 | 113.5 |
| HDAC8-(1:−10) | 4.0 | 45877 | 37.1 | 256.8 |
Tryptic digestion and 1-D LC-MS/MS Analysis
HDAC8 protein was incubated with the photoaffinity probe 3, exposed to UV light to form HDAC8-3 covalent adduct, and tagged with BT 4. Next, the modified protein was purified using avidin-agarose chromatography. Unmodified HDAC8 was used as a negative control. To facilitate the analysis of the modified protein, tryptic digestion was carried out followed by mass spectrometric liquid chromatography-tandem mass spectrometry (LC-MS/MS) and proteomics database analyses using SEQUEST proteomics software. Two proteomics data sets were generated; one used an accurate mass peptide mass tolerance of 5 ppm between the theoretical and measured peptide masses, and second data set used a mass tolerance of 10 ppm. It should be noted that both 5 and 10 ppm are considered accurate mass measurements. The results are provided in Table 3 and Figures 6-8.
Table 3.
Summary of identified HDAC8 peptides modified with probe 3:BT 4 adduct.
| Entry No. | Peptide | Sequence (AA#s) | [M+H]+ | m/z | Charge state (z) | # of Mods | Mass tolerance ppm |
|---|---|---|---|---|---|---|---|
| 1 | IPKRASM*VHSLIEAYALHKQM*Ra,b | 034-055 | 3330.72571 | 1109.57524 | 3 | 1 | 5 |
| 2 | RASMVHSLIEAYALHKQM*RIVKPK | 037-060 | 3541.88969 | 886.22242 | 4 | 1 | 10 |
| 3 | ASM*VHSLIEAYALHKQMRIVKPK | 038-060 | 4105.10938 | 1367.70313 | 3 | 2 | 5 |
| 4 | ASM*VHSLIEAYALHKQMRIVKPK | 038-060 | 4105.10938 | 1367.70313 | 3 | 2 | 5 |
| 5 | QM*RIVKPK. | 053-060 | 1734.93338 | 867.96669 | 2 | 1 | 10 |
| 6 | VAINWSGGWHHAKK | 133-145 | 2901.37544 | 1450.18772 | 2 | 2 | 5 |
| 7 | DEASGFCYLNDAVLGILRLRRKFER | 147-171 | 3660.86731 | 609.31122 | 6 | 1 | 5 |
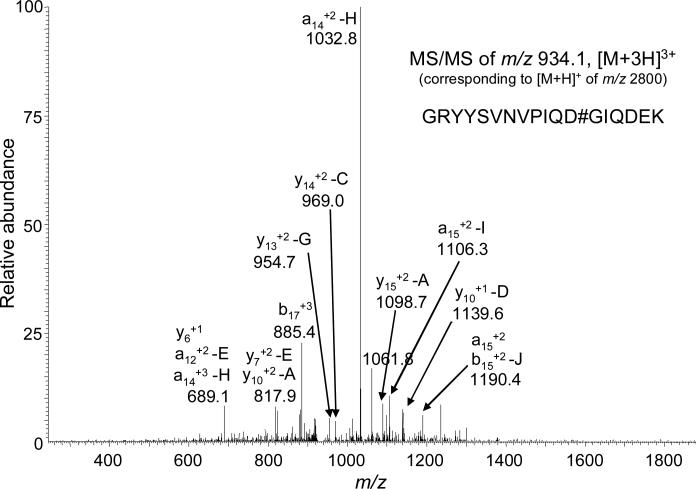
| 8 | GRYYSVNVPIQDGIQDEK | 222-239 | 2800.35061 | 934.11687 | 3 | 1 | 10 |
| 9 | YYQICESVLKEVYQAFNPK | 240-258 | 3041.46828 | 1521.23414 | 2 | 1 | 10 |
| 10 | AVVLQLGADTIAGDPMCSFNMTPVGIGK | 259-286 | 4244.03997 | 1060.25999 | 4 | 2 | 5 |
| 11 | AVVLQLGADTIAGDPMCSFNMTPVGIGK | 259-286 | 4244.03997 | 849.60799 | 5 | 2 | 10 |
* indicates an oxidized methionine
The underlined residues correspond to the region of the HDAC8 protein where modification by probe 3 occurred.
Figure 6.
MS/MS spectrum of HDAC8 tryptic peptide 222-239, GRYYSVNVPIQDGIQDEK (see Table 3, entry 8) showing alkylation at Asp233. The peaks are annotated using the conventional proteomics MS/MS nomenclature (e.g., y6+1). Upper case letters denote the additional side chain cleavages of ligand 3 modified by biotin tag 4 (the structures of possible fragmentation of the resulting construct are shown in Figure 7).
Figure 8.
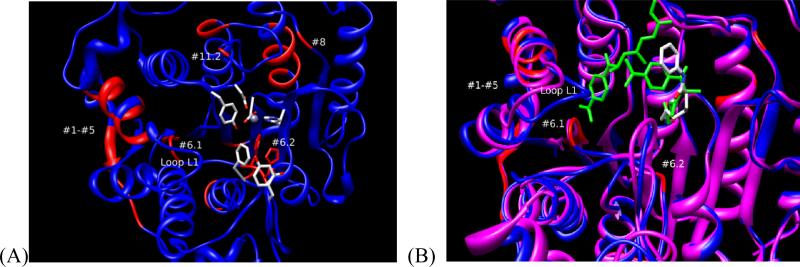
(A) Location of the peptides (red) modified with probe 3 mapped on HDAC8 PDB:1T69 (blue model). The numbers # correspond to the entries in Table 3. The residues rendered as “sticks” correspond to the residues in the immediate proximity to SAHA in 1T69. (B) Same as panel A. Overlay of HDAC8 1T69 (blue) and 1T64 (magenta). SAHA and TSA are rendered as “stick” models colored either by atom types (SAHA) or green color (TSA).
Molecular Dynamics Simulations
Molecular Dynamics Simulations of SAHA and ligand 3 in complex with HDAC8 were performed using the Gromacs software package (version 3.3.1)29-32 and the Amber03 force field33 as ported to Gromacs 34. Ligands were parameterized using the General Amber force field (GAFF)35, 36 augmented with custom azide parameters37and AM1BCC charges.38, 39 The structure of HDAC8 and the starting conformation of SAHA were taken from PDB:1T69. The starting structures for 3 were obtained from the docking with Gold 3.2.40 Root Mean Square Deviation (RMSD) matrices of the ligands were built for each simulation trajectory by calculating the RMSD of each structure of the ligand with all other structures of the same ligand in the trajectory. All protein structures in the trajectory were aligned using the least square fitting of the protein backbone. The final RMSD calculations of the ligands were performed on their heavy atoms using the atoms corresponding to the scaffold of SAHA. The results of the RMSD calculations are given in Supplemental material together with the analysis of the distances between the SBGs of SAHA and ligand 3 and representative amino acids in G1-G3. The most representative binding poses are shown in Figure 9.
Figure 9.
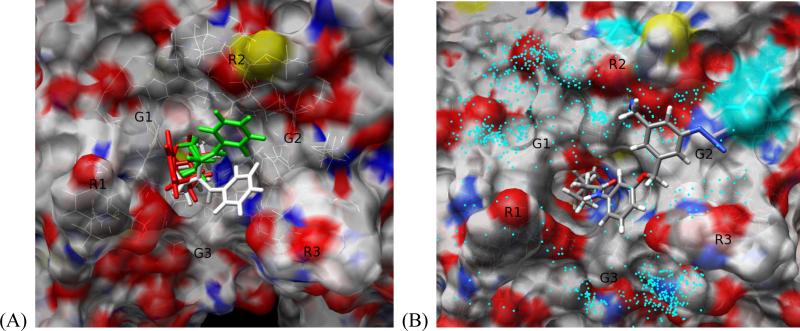
(A) Binding mode 1, 2 and 3 of SAHA indicated in white, green and red, respectively. The surface is colored according to atom types using standard atom color convention. (B) The residues Asp233 and Asp272 modified by photoactivation of the probe 3 are marked by a cyan surface. The locations of the aromatic azide throughout the simulations are projected on the surface and are marked by cyan dots. Probe 3 is represented in a binding mode similar to the binding mode 2 observed for SAHA. G1-G3 and R1-R3 are grooves and ridges on the protein surface, respectively.
Results and discussion
A general idea for capturing the binding poses of the HDAC probe using BEProFL approach is outlined in Figure 3. Among other reactive group typically used for covalent modification of the protein we selected an aromatic azide because (1) it is hydrophobic and, therefore, is likely to stay near the protein surface, minimizing the reaction with the solvent and maximizing the amount of the covalently modified protein, and (2) upon UV-photoactivation it generates a nitrene moiety known to be very reactive and non-specific, which is expected to be essential for capturing a “snapshot” of the bound probe in a close proximity to where this reactive moiety was generated. In addition there are several critical requirements for photoaffinity probes in general. First, the probes should exhibit a respectable level of HDAC binding to maximize the yield of the modified HDAC protein. Second, the procedures for UV-activated crosslinking should be efficient enough so that the UV damage of the protein is minimized while the yield of the crosslinked adduct is maximized. Third, the attachment procedure of the biotin probe should be fast and efficient to minimize possible aggregation and precipitation of the protein.
The probes used in this study were designed to mimic the scaffold of SAHA (1) and a phenylthiazole-based HDAC inhibitor.41 The synthesis of probes 2 and 3 is shown in Schemes 1 and 2. It was found that probe 2 is 6.1-fold and probe 3 is 16.6-fold less active in inhibiting HDAC8 compared to SAHA (Table 1). The moderate HDAC8 activity of the ligands with this scaffold is consistent with other reports.42 The decrease in activity of the probes is not surprising since introduction of the azide moieties in probes 2 and 3 increased the lipophilic nature of the solvent exposed SBGs.
The success of the procedure shown in Figure 3 depends on the ability of the probes to modify the protein rather than the solvent surrounding the ligand and the protein. Since the SBG of HDAC inhibitors is highly solvent exposed there was a chance that the aromatic azide moiety would not be able to reach the protein surface and instead would prefer to react with the solvent.
As the first step, we confirmed that an aromatic azide group can indeed be used to covalently attach the probes to the HDAC8 protein by detecting a mass shift in a MALDI MS experiment. The results of the MALDI MS analysis are summarized in Table 2. The average mass of the untreated HDAC8 protein was 45620 Da. After incubation with ligand 2, the mass of the HDAC8 protein increased in dose-dependent manner. A maximum mass shift of 256.8 Da was observed at an enzyme to reagent ratio of 1:10. As the amount of protein modified approaches 100%, the observed average mass shift would be expected to approach the mass of the reagent (360 Da). We attribute relatively high concentration of probe 2 needed to its relatively poor potency against HDAC8 and high solvent exposure of the SBG. The results of the MALDI-MS analysis of the intact protein supported the conclusion that the protein was successfully modified by the UV activated nitrene group generated from the probe 2 and we moved to the next stage – cross-linking followed by attachment of BT 4 to the alkyl azide moiety of probe 3.
To evaluate the selectivity of photolabeling in vitro, purified recombinant HDAC8 was incubated with various concentrations of ligands 2 and 3. The conditions for UV crosslinking were optimized to increase signal-to-noise ratio. We found that in case of HDAC8 3×1 min exposure to UV light with 1 min rest gives the best signal to noise ratio. The “click-chemistry” conditions with catalysis by Cu(I) generated in situ were successfully used to attach BT 4 to the adduct between HDAC8 and probe 3. In our preliminary studies we also tried the Staudinger-Bertozzi ligation19-24 as an attachment procedure of the biotin tag and determined that the “click-chemistry” approach is much faster and gives better yield as determined by Western blotting. This is consistent with the observation made by Cravatt et al.9
As expected, the amount of biotinylated HDAC increases in a concentration-dependent manner in the presence of ligand 3 but not with ligand 2 (Figure 5A) that lacks the alkyl azide group designed to bind the biotin tag; only non-specific, background level signals are observed. A comparison of the background levels observed in lanes 3, 9-11 shows that the background levels of biotinylation are due to non-specific modification of the HDAC8 protein with BT 4 and do not depend on the presence of probes 2 and 3. It appears that the non-specific biotinylation with BT 4 may be specific to the HDAC8 protein as in a similar experiment HDAC3 protein showed negligible levels of non-specific biotinylation with BT 4 (data not shown). In the competition experiment shown in Figure 5B, the level of the covalent modifications with probe 3 drops by 50% when SAHA is present during the photolabeling, indicating that the cross-linking of the probe 3 to HDAC8 is likely to happen when 3 is bound at the catalytic site.
A total of 11 modified HDAC8 peptides were identified from the avidin-agarose purified HDAC8-3-4 adduct based on peptide mapping using high resolution accurate mass measurement (within 10 ppm) and peptide sequencing using MS/MS (Table 3). No modified peptides were detected in the negative control using unmodified HDAC8 (data not shown). The amino acids underlined in Table 3 correspond to the residues modified by 3 based on MS/MS analysis. Due to occasional missed trypsin cleavage sites, which are common in trypsinization reactions, there are some amino acid sequence overlaps among several peptides. For example, entries 1 and 2 correspond to overlapping amino acid sequences 35-55 and 37-60, respectively. Also, due to the high reactivity of the photoaffinity ligand, several amino acid residues became modified in entries # 3,4,6,10, and 11 (Table 3).
Among the sites modified by probe 3 (Table 3, Figure 8A), the two nearest to the binding site amine acids located on the surface of the protein were Asp233 and Asp272. Asp272 maps to the R2/G1 region of HDAC8, and Asp233 neighbors with the R2/G2 (Figure 2). The two other areas near the binding site corresponding to His142-His143 (entry #6, Table 3) and Ile135-Asn136-Trp137-Ser138 (entry #6, Table 3) are not on the surface of the protein. Photolabeling of these residues may be explained if the binding site of HDAC8 adopts a conformation similar to that found in 1T64 X-ray structure where the second molecule of TSA is bound upside down.43 The access to these residues in other crystal structures is blocked by the loop L1 (Figure 8B) that was shown to shift its position to accommodate the second molecule of TSA. If the loop L1 swings far enough it may expose the residues in entries #1-#6 for modification by the SBG of probe 3. The other labeled peptides correspond to sites on the outer periphery of HDAC8 and were likely modified by excess unbound 3 upon activation by UV light. An analysis of these areas shows that they are either hydrophobic or in close proximity to hydrophobic areas and, thus, may participate in aggregation with probe 3.
Modified Asp233 was contained in peptide GRYYSVNVPIQDGIQDEK (222-239), and the MS/MS spectrum of this modified peptide is shown in Figure 6. Based on Sequest proteomics analysis, the standard peptide fragment ions such as y6+1 and a15+2 were identified, and manual inspection of the data was used to identify additional fragment ions formed by side chain fragmentation of 3. By accounting for these side chain fragments, it was possible to identify and assign the other peaks present in the MS/MS spectra.
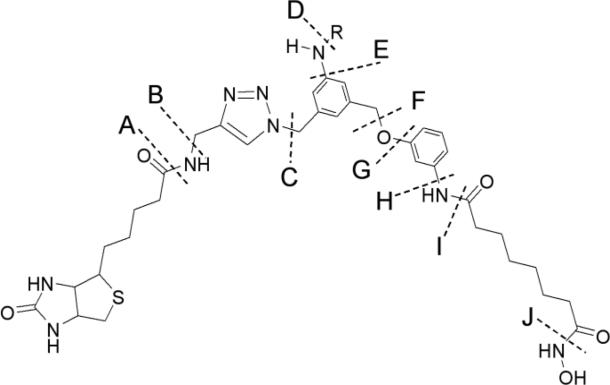
During MS/MS analysis of peptides from HDAC8 that had been modified by 3, fragmentation of probe 3 was observed at the sites indicated in Figure 7. These types of cleavages (e.g., y15+2-A) are indicated in the tandem mass spectra of modified HDAC8 peptides. There were a total of three fragment ions m/z 689.1 (y6+1), 885.4 (b17+3), and 1190.4 (a15+2), that could be directly assigned to peptide backbone bond cleavages. Of those three, only the y6+1 fragment corresponds to a region of the peptide not containing probe 3. The remaining fragment ions were assigned to peptide backbone cleavages plus fragmentation of 3. For three of the fragment ions, more than one type of fragment ion could be assigned. The overall HDAC8 sequence analyses ranged between 61 – 68 % for the control HDAC8 samples and 52 – 58 % for modified HDAC8 protein samples.
Figure 7.
Structure and ten possible bond cleavages of ligand 3 modified with biotin tag 4. Bond cleavages are labeled A through J, and R denotes the amino acid side chain.
To explore if there are more than one binding mode for HDAC ligands we performed extensive MD simulations of ligands 1 and 3 bound to the HDAC8 and compared them with the outcomes of the photolabeling experiments described above for ligand 3. The binding modes of the ligands are characterized by the conformation of the ligand inside the binding site and the resulting interactions between the SBG of the ligands and the various structural elements of the HDAC8 surface, i.e. R1-R3 and G1-G3 (Figure 2). To determine the binding modes available to the ligands, identical conformations sampled during the course of an MD simulation were identified using the matrix RMSD and confirmed with an analysis of the distances between the SBGs of SAHA and ligand 3 and representative amino acids in G1-G3 (Supplemental Material).
In ligand 1 the binding mode 1 is represented by the areas in the RMSD matrix at 1 ns, 5 ns, 8.5-12 ns and 19.5-20 ns, the binding mode 2 at 2-4.5 ns, 5.5 ns, 13 ns, 14-17 ns and 17.5-19.5 ns, and the binding mode 3 at 2 ns, 5.5-8.5 ns, 12 ns, 13.5 ns and 17 ns. The RMSD matrix (Supplemental Material) shows that ligand 1 changed from one binding mode to another multiple times during the course of the simulation, suggesting that they are the only ones available to the ligand. The fact that no new conformations were found after 2 ns in the remaining 18 ns of the simulation suggests that the conformational space of the bound ligand was fully sampled.
Characteristic interactions of each binding mode are determined by visual inspection of the conformation of the ligand inside the binding site of HDAC8. In the first binding mode, the phenyl ring interacts with the area G2/R3 of the protein, whereas in the second binding mode the phenyl ring interacts with G2/R2 (Figure 9A). The third binding mode is similar to the binding mode of 1 observed in X-ray as they are both characterized by strong electrostatic interaction between the amide group of 1 and Asp101 and weak interaction between the phenyl ring of 1 and the protein surface.
The analysis indicates that the second binding mode dominates, contributing approximately 35% of the total conformations. The first and third binding modes follow with an approximately equal weight, 22% and 21% respectively. It is, therefore, expected that all the three major binding modes of SAHA collectively contribute to inhibition of HDAC8. Only the binding mode 3 is observed in the crystal structure PDB: 1T69. The only difference between 1T69 and the binding mode 3 is the conformation of the amide group of SAHA. In 1T69 the carbonyl points toward the carboxylate of Asp101, one of the residues conserved in class I and II HDAC isoforms, whereas in the binding mode 3 the amide nitrogen of SAHA forms hydrogen bond with Asp101. Although recent crystal structures44 of HDAC8 indicate the presence of hydrogen bond between Asp101 and the amide nitrogen of the inhibitors, the protonation state of Asp101 remains a matter of debate.45 Interestingly, with very few exceptions,45 the majority of the HDAC X-rays contain an additional copy of the HDAC protein that not only shields the ligand from the solvent but also directly affects the binding of the ligand. It is, therefore, unclear how relevant the binding pose observed in the X-ray is to the actual pose of the co-crystallized ligands in the solution.
Similar exploration of all conformational space available to probe 3 would require running simulations for unrealistically long time due to the increased number of degrees of freedom. To avoid it we decided to use multiple binding poses of ligand 3 as starting conformations and perform several simulations simultaneously, a well accepted approach for exploring large conformational spaces. Probe 3 was docked to HDAC8, and ten diverse binding poses were selected for further MD simulations. The total cumulated simulation time was more than 160 ns. The RMSD matrix of ligand 3 (SI) shows that similarly to ligand 1 ligand 3 has three main binding modes. The binding mode 1 corresponds to 0-3 ns, 28-32 ns, 60-65 ns, and 135-160 ns, the binding mode 2 to 20-28 ns, 32-40 ns, 50-60 ns, 85-90 ns, 120-135 ns, and 165-168 ns, and the binding mode 3 to 3-20 ns, 40-50 ns, 65-85 ns, 90-120 ns, 120-135 ns, 160-165 ns and 168-180 ns. As expected for a larger and more flexible molecule, the transition time from one binding mode to another is now longer compared to that observed for the binding modes of SAHA (SI). The binding modes 1, 2 and 3 have a weight of approximately 19%, 12% and 46%, respectively, with the binding mode 3 is now predominant. Whereas these binding modes are not present in the same proportion as in ligand 1, more than one major binding mode is available for ligand 3 as well.
Figure 9B shows the spatial distribution of the aromatic azide group throughout the simulations. The cyan dots represent all the locations occupied by the last nitrogen atom of the azide group of ligand 3. The coordinates of this atom were taken from every frame of the simulation trajectory. The main density areas are located in the grooves and ridges G1/R2, R2 and G2/R2. The spatial distribution of the aromatic azide group during the simulation is consistent with the modification of the residues Asp233 and Asp272 observed by LC-MS/MS analysis, as it is mostly located in the vicinity of these residues. Importantly, the modification of these residues cannot be accomplished if the ligand exists only in a single binding pose as the residues modified are located too far away from each other to be reacting with any single binding pose. The absence of modification corresponding to the azido group oriented toward G3 in the pose 1 of probe 3 is consistent with its predominant solvent exposure and the fact that G3 is relatively deeper compared to G1 and G2. The sharper curvature of the surface near G3 prevents the remotely located aromatic azide moiety of probe 3 to contact with the surface. These observations suggest that to improve the surface contact with G3 the introduction of a more flexible moiety between the linker and the SBG portions of the HDAC ligands may be necessary.
Conclusions
This is the first study where the binding poses available to the ligands in HDAC8 were determined experimentally by BEProFL approach. The tagging of the probe 3 attached to HDAC8 with BT 4 resulted in a concentration dependent increase in biotinylation of the protein with the increase of the concentration of probe 3 but not with probe 2 that lacks the second azido group. The selectivity of probe 3 for the active site of HDAC8 was demonstrated by blocking it with SAHA. Using the photoaffinity experiments we show that probe 3 modifies the HDAC8 binding site at or near Asp233 and Asp272– the interface areas between G1 and R2 and R2 and G2 that can only be reached if probe 3 adopts at least two different binding poses. The results of MD simulations are consistent with these observations. Sharper curvature of the HDAC8 protein surface in G3 explains the lack of covalent modifications in it. The modifications observed at His142-His143 and Ile135-Asn136-Trp137-Ser138 are likely attributed to the “upside-down” binding pose of probe 3 analogous to that of TSA. The other few modifications of the HDAC8 protein on the side opposite to the binding site appear to be a result of aggregation with probe 3. Although the three major poses available to SAHA and probe 3 are similar, they are not equally populated as determined by MD analysis. Overall, the findings demonstrated that multiple binding poses of the HDAC ligands are likely to contribute to the binding. Future studies are needed to learn how this observation can be used in HDAC drug design to improve the activity and selectivity of the ligands.
The study also highlights further directions in refinement of the BEProFL approach. Minimization of the fragmentation of the probe is highly desirable to facilitate the analysis of MS/MS data. The source of non-specific tagging of the HDAC8 protein upon incubation with biotin tag 4 remains unclear, warranting further investigation.
The methodology supplies a unique power in analysis of the binding of ligands to their macromolecular targets by expanding the data typically obtained in the protein x-ray crystallography or providing an alternative tool when co-crystallization of the protein with the ligand of interest has failed. Most importantly, the binding poses determined by BEProFL are determined in solution and, thus, the captured “snapshots” reflect the dynamic nature of the ligand and its macromolecular target conformations. Extension of the BEProFL approach to study the binding poses of the ligands in cells is in progress in our laboratory. The approach has the potential not only to guide ligand optimization but also to become a new tool in disciplines such as molecular modeling, development, validation and application of computer-aided drug design methods, especially those for rapid prediction of the protein-ligand interactions such as docking and scoring.
Experimental Procedures
General synthetic procedures
1H NMR and 13C NMR spectra were recorded on Bruker spectrometer at 300/400 MHz and 75/100 MHz, respectively, with TMS as an internal standard. Standard abbreviations indicating multiplicity were used as follows: s = singlet, bs = broad singlet, d = doublet, t = triplet, q = quadruplet, and m = multiplet. HRMS experiment was performed on Q-TOF-2TM (Micromass). TLC was performed with Merck 60F254 silica gel plates. Column chromatography was performed using Merck silica gel (40-60 mesh). Analytical HPLC was carried out on an Ace 5AQ column (100 mm × 4.6 mm), with a Shimadzu 10 VP Series HPLC with a diode array detector with detection at λ = 254 nm; Method A: H2O/MeCN (0.05% TFA), 70/30 to 0/100 in 25 min, flow rate of 1.3 mL/min; Method B: H2O/MeCN (0.05% TFA), 90/10 to 0/100 in 25 min, flow rate of 1.3 mL/min. The purity of compounds determined by HPLC (methods A, B) was ≥95% for all the synthesized compounds.
Synthesis of probes 2 and 3
7-[4-(3-Azido-phenyl)-thiazol-2-ylcarbamoyl]-heptanoic acid methyl ester (8)
To a solution of 7 (0.350 g, 0.96 mmol) in a 9:1 mixture of acetic acid and water (15 mL) was added NaNO2 (0.100 g, 1.45 mmol) at 0 °C and the mixture was stirred for 5 min. To this was added NaN3 (0.094 g, 1.45 mmol) at the same temperature and stirring was continued for 30 min. To this was added saturated aqueous NaHCO3 solution followed by NaHCO3 powder until the mixture reached pH 7. The mixture was extracted with EtOAc and the combined organic extracts were washed with brine, dried (Na2SO4), filtered, and concentrated under reduced pressure to afford 8 (0.273 g, 73%); 1H NMR (400 MHz, CD3OD) δ 7.70 (d, J = 8.0 Hz, 1H), 7.66 (s, 1H), 7.44 (s, 1H), 7.41 (t, J = 7.6 Hz, 1H), 7.00 (dd, J = 7.6, 1.6 Hz, 1H), 3.65 (3H, s), 2.49 (t, J = 7.2 Hz, 2H), 2.34 (t, J = 7.6 Hz, 2H),1.73 (t, J = 7.2 Hz, 2H), 1.64 (t, J = 6.8 Hz, 2H), 1.41(brs, 4H); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 174.1, 170.2, 158.5, 148.5, 146.7, 136.0, 130.2, 122.5, 122.5, 117.5, 116.7, 108.7, 51.5, 36.2, 33.9, 28.6, 24.6, 24.5.
Octanedioic acid [4-(3-azido-phenyl)-thiazol-2-yl]-amide hydroxyamide (2)
KOH (0.607 g, 10.85 mmol) was added at 40 °C for 10 min to a solution of hydroxylamine hydrochloride (0.753 g, 10.85 mmol) in MeOH. The reaction mixture was cooled to 0 °C and filtered. Compound 8 (0.210 g, 0.54 mmol) was added to the filtrate followed by KOH (36 mg, 0.65 mmol) at room temperature for 30 min and stirred for 4h. The reaction mixture was extracted with EtOAc, and the organic layer was washed with a saturated NH4Cl aqueous solution and brine, dried over Na2SO4, filtered, and concentrated. The crude solid was purified by preparative HPLC to give compound 2 (0.058 g, 28%); 1H NMR (400 MHz, DMSO-d6) δ (ppm) 12.25 (s, 1H), 10.32 (s, 1H), 8.64 (S, 1H), 7.74-7.62 (m, 3H), 7.05 (brs, 1H), 6.67 (brs, 1H), 2.49 (m, 2H), 1.93 (m. 2H), 1.26 (m, 2H), 1.10 (brs, 4H); 13C NMR (100 MHz, DMSO-d6): δ (ppm) 172.0, 169.5, 158.4, 148.0, 140.3, 136.5, 130.8, 122.8, 118.8, 116.3, 109.5, 35.2, 32.6, 25.4, 25.0; MS (ESI) m/z 389.1 [M+H]+; HRMS (ESI) calculated for [C17H20N6O3S1+H]+ 389.1390; found 389.1383.
7-(4-Benzyloxy-phenylcarbamoyl)-heptanoic acid methyl ester (9)
A stirring solution of 4-Benzyloxyaniline hydrochloride (2.01 g, 10.10 mmol) and suberic acid monomethyl ester 5 (1.90 g, 10.10 mmol) in dry pyridine (20 ml) was cooled to −15 °C and POCl3 (1.2 mL, 13.13 mmol ) was added dropwise over 30 minutes and stirred for 12h at room temperature. The reaction mixture was diluted with EtOAc and washed thoroughly with saturated KHSO4 solution and brine. The organic phase was dried over Na2SO4. The solvent was removed by rotary evaporation. The crude solid was washed with EtOAc to give compound 9 (2.27 g, 61%); 1H NMR (400 MHz, CD3OD): δ 7.42-7.26 (m, 7H), 7.11 (s, 1H), 6.93 (d, J = 7.8 Hz, 2H), 5.22 (s, 2H), 3.66 (s, 3H), 2.34-2.31 (m, 4H), 1.77-1.71 (m, 2H), 1.70-1.62 (m, 2H), 1.40-1.36 (m, 4H); 13C NMR (100 MHz, CD3OD): δ 174.2, 171.0, 155.5, 136.9, 131.3, 128.5, 127.9, 127.4, 121.6, 115.2, 70.3, 51.5, 37.4, 33.9, 28.7, 28.7, 25.4, 24.6.
7-(4-Hydroxy-phenylcarbamoyl)-heptanoic acid methyl ester (10)
A suspension of compound 9 (1.20 g, 3.24 mmol) and Pd/C (10% wt, 0.120 g) in MeOH (20 mL) was reacted under hydrogen atmosphere at room temperature for 5h. The catalyst was removed by filtration through a pad of celite. The solvent was evaporated. The crude material was purified by flash chromatography (EtOAc/hexane, 2:1) to give compound 10 (0.742 g, 81%); 1H NMR (400 MHz, CD3OD): δ 7.31 (d, J = 8.8 Hz, 2H), 6.74 (d, J = 8.8 Hz, 2H), 3.65 (s, 3H), 2.34-2.31 (m, 4H), 1.71-1.61 (m, 4H), 1.39-1.34 (m, 4H); 13C NMR (100 MHz, CD3OD) δ: 174.5, 172.8, 153.9, 130.2, 121.9, 114.7, 50.5, 36.2, 33.2, 28.4, 25.4, 24.2.
7-[4-(3-Azido-5-azidomethyl-benzyloxy)-phenylcarbamoyl]-heptanoic acid methyl ester (11)
To a solution of 12 (0.250 g, 0.886 mmol) and 7-(4-Hydroxy-phenylcarbamoyl)-heptanoic acid methyl ester 10 (0.247 g, 0.886 mmol) in dry acetone (8 mL) was added K2CO3 (0.367 g, 2.65 mmol) at ambient temperature, and the reaction mixture was stirred at the same temperature for 12 h. The mixture was poured into water and extracted with EtOAc. The organic layer was washed with water three times, dried over Na2SO4, and evaporated in vacuo. The residue was purified by flash chromatography (EtOAc/hexane, 2:1) to afford 11 (0.272 g, 66%); 1H NMR (400 MHz, CDCl3): δ 7.43 (d, J = 8.8 Hz, 2H), 7.15 (s, 1H), 7.11 (s, 1H), 7.08 (s, 1H), 6.92 (d, J = 9.2 Hz, 2H), 5.04 (s, 1H), 4.37 (s, 1H), 3.69 (s, 3H), 2.36-2.31 (m, 4H), 1.74 (t, J = 8.0 Hz, 2H), 1.65 (t, J = 7.2 Hz, 2H), 1.40 (brs, 4H); 13C NMR (100 MHz, CDCl3): δ 174.2, 171.0155.0, 141.0, 139.8, 137.8, 131.6, 124.2, 123.1, 121.6, 119.1, 118.7, 117.9, 117.5, 115.1, 69.3, 54.1, 51.4, 37.4, 33.9, 28.6, 25.3, 24.6.
Octanedioic acid [4-(3-azido-5-azidomethyl-benzyloxy)-phenyl]-amide hydroxyamide (3)
KOH (0.481 g, 8.60 mmol) was added at 40 °C for 10 min to a solution of hydroxylamine hydrochloride (0.597 g, 8.60 mmol) in MeOH. The reaction mixture was cooled to 0 °C and filtered. Compound 11 (0.200 g, 0.43 mmol) was added to the filtrate followed by KOH (28 mg, 0.51 mmol) at room temperature for 30 min and stirred for 4h. The reaction mixture was extracted with EtOAc, and the organic layer was washed with a saturated NH4Cl aqueous solution and brine, dried over Na2SO4, filtered, and concentrated. The crude solid was purified by preparative HPLC to give compound 3 (0.080 g, 40%); 1H NMR (400 MHz, DMSO-d6): δ 10.32 (s, 1H), 9.72 (s, 1H), 8.65 (s, 1H), 7.48 (d, J = 7.3 Hz, 2H), 7.26 (s, 1H), 7.16 (s, 1H), 7.09 (s, 1H), 6.94 (d, J = 8.6 Hz, 2H), 5.09 (s, 1H), 4.50 (s, 1H), 2.24 (t, J = 7.2 Hz, 2H), 1.93 (t, J = 7.0 Hz, 2H), 1.55-1.46 (m, 4H), 1.26 (brs, 4H); 13C NMR (100 MHz, DMSO-d6) δ 171.1, 169.52, 154.19, 140.48, 140.38, 138.56, 133.37, 124.36, 120.96, 118.61, 118.0, 115.2, 68.9, 53.3, 36.6, 32.6, 28.8, 25.52, 25.47; MS (ESI) m/z 467.2 [M+H]+; HRMS (ESI) calculated for [C22H26N8O4+H]+ 467.2140; found 467.2149.
Biological materials
The HDAC8 fluorescent assay kit and the human recombinant HDAC8 protein were purchased from Biomol International (Plymouth Meeting, PA). Cell culture media MEM/EBSS, Ham's F12 as well as fetal bovine serum (FBS) and non-essential amino acids (NEAA) were purchased from Hyclone (Logan, UT). Cell culture dishes were purchased from BD Falcon (BD Biosciences, San Jose, CA). The primary anti-HDAC8 antibody was from Santa Cruz (Santa Cruz, CA) respectively. The anti-rabbit secondary antibody conjugated to horse radish peroxidase (HRP) was from GE (Piscataway, NJ). Streptavidin and the colorimetric HRP substrate o-phenylenediamine (OPD) were from Pierce (Rockford, IL) and the 96-well plates from Nunc (Rochester, NY). Tris(2-carboxyethyl)phosphine) (TCEP) was from (Alfa Aesar chemicals, Ward Hill, MA), CuSO4, dimethyl sulfoxide (DMSO), tris ((1-benzy l-1H-1,2,3- triazol-4-yl) methyl) amine (TBTA) and all other chemicals were purchased from Sigma (St. Louis, MO) if not stated otherwise.
HDAC activity assay
The assay proceeded through a simple two-step procedure that can be carried out in half-volume microtitration plates. The in vitro fluoro HDAC assay was performed in HDAC assay buffer (25 mM Tris (PH 8.0), 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2) supplemented with 0.5 mg/mL BSA. After 30 min, 50 μL of developer, supplemented with 5 μM Trichostatin A to inactivate the enzyme were added. The fluorescence intensity was measured at λexcit = 360 nm and λemis = 460 nm using a POLARStar Optima microtitration plate reader (BMG Labtech, Durham, NC). The IC50 values were determined using the GraphPad Prism 5 software and KI were calculated using Cheng-Prusoff equation46 and a Km for the substrate equal to 222μM for HDAC8.
Visualization of probe 3 attached to HDAC 8 using biotinylated tag 4
Purified recombinant human HDAC8 (0.5 μg in 5 μL of assay buffer) was incubated with various concentrations of compounds 2, 3 and SAHA for three hours in the dark, after which the formation of a covalent bond between the azide group present in the ligand and reactive side chains of the HDAC was initiated by UV irradiation (λ = 254 nm) for 3×1 min with 1 min rest. Then “click chemistry” was used to attach BT 4 to probe 3 attached covalently to the protein. The biotin tag 4 was added to all the tubes at a final concentration of 50 μM, and the chemical reaction was initiated by addition of TCEP (0.5 mM final concentration), TBTA 0.1 mM ) and CuSO4 (1 mM). After 90 min incubation at room temperature, protein samples were analyzed by SDS-PAGE and western blot using an anti-biotin primary antibody, streptavidin conjugated to horse radish peroxidase (Pierce (Rockford, IL) and ECL (Pierce (Rockford, IL). The membranes were then stripped in 0.2M glycine, pH 2.6 for 10 minutes, then in 0.2M glycine, pH 2.3 for another 10 minutes before being reblocked and redecorated with an anti-HDAC primary antibody and an anti-rabbit-HRP secondary antibody.
Western Blotting
Western blot was performed using 10 μg of total protein from cell lysate or 0.5 μg of purified protein with 5· loading buffer containing 10% SDS, 0.05% bromophenol blue, 50% glycerol and β-mercaptoethanol, protein samples were boiled for 5 min and allowed to cool before loading on a denaturing 10% polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, protein was transferred to a polyvinylidiene difluoride membrane (Imobilon-Millipore, Bedford, MA, USA). The membrane was incubated for 1 h with 1% Albumin Fraction V (USB, OH, USA) and washed three times with 1× phosphate buffer saline supplemented with 0.05% of Tween 20 (PBS-T). The membrane was then incubated with an anti-HDAC8 antibody (1:3000) for 2 h under room temperature with slight agitation. After three washes in PBS-T, the membranes were incubated with a secondary antibody anti-rabbit-HRP for 1 h at room temperature. The signals were detected using the enhanced chemiluminescence (ECL) kit from Pierce (Pierce Biotechnology, Rockford, IL, USA). Densitometry scanning of the films was performed using the ImageJ software provided by NIH.47, 48 Data obtained from at least three independent experiments are presented as the mean of the relative intensity of the biotinylated bands over the total immunoreactive HDAC bands +/− standard deviation (SD).
MALDI-ToF MS Analysis of intact HDAC8 protein
The samples were purified and concentrated using Centricon 10,000 molecular weight cutoff filter filters (Millipore) according to the manufacturer's instructions. Prior to use, the filters were washed three times with 400 μL of 50 mM ammonium bicarbonate, pH 7.8. Aliquots of 25 μL of each HDAC8 sample were then diluted to a total volume of 250 μL with the ammonium bicarbonate buffer and applied to the pre-rinsed filters. The samples were centrifuged at 13,500×g at 4 °C for 60 min. After centrifugation, the filter was transferred to a second 1.5 mL microcentrifuge tube inverted and centrifuged at 1000×g for 6 min. The recovered protein (approx 3 - 5 μL) was mixed with 3 μL of MALDI matrix and spotted on a MALDI plate for analysis. The MALDI matrix was saturated with sinapinic acid in 50:50 (v/v) H2O/ACN + 0.1% TFA. Positive ion MALDI mass spectra were acquired using an Applied Biosystems Voyager DE Pro MALDI-ToF MS.
Tryptic digestion of HDAC8 and modified HDAC8
HDAC8 (~10 μg) was incubated with the five equivalents of probe 3, irradiated with UV light to activate the probe, and tagged with BT 4. Next, the modified protein was purified using avidin-agarose chromatography Unmodified HDAC8 was used as a negative control. Each sample of modified or unmodified HDAC8 was diluted to a final volume of 100 μL with an aqueous buffer containing 50 mM ammonium bicarbonate (AMBIC) and 1 mM TCEP (pH 7.8). Prior to digestion, all samples were tested to ensure that their pH was between 7.0 and 8.0. Control samples containing 10 μg of HDAC8 were treated with 200 ng of sequence grade modified trypsin (Promega) while the modified HDAC8 samples (≤10 μg each) were treated with 100 mg of trypsin. Digestions were carried out for 15 h and then analyzed using LC-MS/MS.
1-D LC-MS/MS Analysis of Tryptic Digests
Tryptic digests were analyzed using an Ultimate 3000 capillary HPLC system (Dionex) interfaced with a LTQ-FT hybrid linear ion trap FT ICR mass spectrometer (Thermo-Fisher Scientific) that was operated using positive ion nanoelectrospray with data-dependent MS/MS. The resolving power of the FT ICR mass spectrometer was ~100,000. Charge state screening was used to exclude singly charged ions. Samples were diluted to a concentration of ~0.1 pmol/μL of total protein using 50 mM AMBIC. The LC-MS/MS injection volume was 5 μL. The mobile phases consisted of A: 95/5 (v/v) H2O/CH3CN + 0.1% formic acid and B: 5/95 (v/v) H2O/CH3CN + 0.1% formic acid. The trapping column (0.3 mm i.d. × 5 mm Dionex C18 PepMap column) was operated at a flow rate of 15 μL/min using 100% solvent A during sample loading. The analytical column (75 μm × 150 mm Agilent Zorbax 300-SB C18 column) was used to separate the peptide digests of HDAC and modified HDAC was operated at a flow rate of 190 nL/min and was equipped with a PEEK sleeve fitting containing a 75 μm i.d. pulled fused silica nanoelectrospray emitter (New Objective) with a tip i.d. of 15 μm. An 85 min linear gradient was used from 3 to 95% solvent B followed by a 10 min re-equilibration prior to the next analysis. Following LC-MS/MS analysis, the data were searched using SEQUEST (Bio Works 3.2) proteomics software.
Graphics
The protein surface in Figure 2 is rendered in Vida3.0.49 The images in Figures 8 and 9 were rendered in Chimera.50
Molecular Dynamics Simulations
Molecular Dynamics Simulations were performed using the Gromacs software package (version 3.3.1) 29-32, the Amber03 force field 33 and the General Amber force field (GAFF) 35, 36 together with AM1BCC charges 38, 39. The structure of HDAC8 and the starting conformation of SAHA were taken from the x-ray structure of HDAC8 forming a complex with SAHA (pdb structure 1T69). Since no crystal structure is available for ligand 3, the starting structures were obtained from the docking of ligand 3 to HDAC8 using the docking software Gold 3.2.40 TIP3P explicit solvent was used 51. All simulations were run in a dodecahedral box containing approximately 9100 water molecules. The bonds of the protein and ligands were constrained using the LINCS algorithm 52 while the bonds and angle of the water molecules were constrained using the SETTLE algorithm 53. A time step of 2 fs was used for integrating the equations of motion. The system was simulated in an NTP ensemble at a fixed temperature of 300 K and a fixed pressure of 1 bar. Temperature was kept constant using a Berendsen thermostat 54 with a coupling time of 0.1 ps and pressure was controlled by a weak coupling to a reference pressure 55 with a coupling time of 1 ps and an isothermal compressibility of 4.6 10−5 bar−1. Long-range coulombic interactions were evaluated using Particle Mesh Ewald (PME) algorithm 56 with an interpolation order of 4 and Fourier spacing of 1.2 Å. The system was energy minimized and then equilibrated for 200 ps in total. Finally the simulations were run for 20 ns each.
Root Mean Square Deviation (RMSD) matrices of the ligands were built for each simulation trajectory by calculating the RMSD of each structure of the ligand with all other structures of the same ligand in the trajectory. All protein structures in the trajectory were aligned using the least square fitting of the protein backbone. The final RMSD calculations of the ligands were performed on their heavy atoms using the atoms corresponding to the scaffold of SAHA.
Any two conformations of a ligand were considered different if their RMSD was more than 2 Å. By using this cut-off value as a similarity criterion for the RMSD between two conformations, identical conformations were identified and represented by a white point on the matrix, whereas different conformations were represented by a black point. As a consequence a white block on the matrix diagonal represents a group of identical conformations and a white block off diagonal indicates that the corresponding two groups on the diagonal contain identical conformations. By using this technique different sets of identical conformations corresponding to the different binding modes of the ligand can be readily identified. The (RMSD) matrices of the ensemble of conformations sampled for ligands 1 and 3 are shown in the Supplemental material.
The binding modes of 1 and 3 determined using the RMSD matrix were confirmed by monitoring characteristic distances between the interacting groups. The residues were selected in such a way that a distance less than 5 Å between the center of mass of the phenyl ring of 1 and Phe208 would indicate that ligand 1 is in G2/R3. Similarly a distance less than 5 Å between the center of mass of the phenyl ring of 1 and the center of mass of Met274 would be indicative of ligand 1 in G2/R2, a distance less than 5 Å between the amide NH of the SBG and the center of mass of Asp101 would be indicative of ligand 1 near R1. The same distance characteristics of the three binding modes of 1 have been monitored for 3. The results of this analysis are shown in the supplemental material.
Supplementary Material
Acknowledgments
This study was in part funded by the Breast Cancer Congressionally Directed Research Program of Department of Defense Idea Award BC051554 and by the National Cancer Institute/NIH grant 1R01 CA131970-01A1. We also thank Chicago Biomedical Consortium and Searle family for the support of FT ICR MS and the National Center for Research Resources/NIH grant 1S10 RR14686 for the MALDI ToF mass spectrometer. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081) and OpenEye Scientific Software, Santa Fe, NM.
Abbreviations
- HDAC
histone deacetylase
- BEProFL
(B)inding (E)nsemble (Pro)filing with (F)photoaffinity (L)abeling
- BT
biotin tag
- ArN3
aromatic azide group
- AlkN3
aliphatic azide group
- ZBG
zinc binding group
- SBG
surface binding group
- linker
a portion of the molecule connecting the ZBG and SBG
- SAHA
suberoyl anilide hydroxamic acid
- TSA
Trichostatin A
Footnotes
Supporting Information Available: The RMSD matrixes and distance analysis figures and synthetic procedures for BT 4. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Korner M, Tibes U. Histone deacetylase inhibitors: a novel class of anti-cancer agents on its way to the market. Prog. Med. Chem. 2008;46:205–280. doi: 10.1016/S0079-6468(07)00005-7. [DOI] [PubMed] [Google Scholar]
- 2.Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J. Natl. Cancer Inst. 2000;92:1210–1216. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 3.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hahnen E, Hauke J, Trankle C, Eyupoglu IY, Wirth B, Blumcke I. Histone deacetylase inhibitors: possible implications for neurodegenerative disorders. Expert Opin. Investig. Drugs. 2008;17:169–184. doi: 10.1517/13543784.17.2.169. [DOI] [PubMed] [Google Scholar]
- 5.Wang DF, Helquist P, Wiech NL, Wiest O. Toward selective histone deacetylase inhibitor design: Homology modeling, docking studies, and molecular dynamics simulations of human class I histone deacetylases. J. Med. Chem. 2005;48:6936–6947. doi: 10.1021/jm0505011. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen PE. Photochemical Probes in Biochemistry. Vol. 272. Kluwer Academic Publishers; Dordrecht / Boston / London: 1989. [Google Scholar]
- 7.Rajagopalan K, Watt DS, Haley BE. Orientation of GTP and ADP within their respective binding sites in glutamate dehydrogenase. Eur. J. Biochem. 1999;265:564–571. doi: 10.1046/j.1432-1327.1999.00736.x. [DOI] [PubMed] [Google Scholar]
- 8.Olcott MC, Bradley ML, Haley BE. Photoaffinity labeling of creatine kinase with 2-azido- and 8-azidoadenosine triphosphate: identification of two peptides from the ATP-binding domain. Biochemistry. 1994;33:11935–11941. doi: 10.1021/bi00205a032. [DOI] [PubMed] [Google Scholar]
- 9.Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 10.Adam GC, Burbaum J, Kozarich JW, Patricelli MP, Cravatt BF. Mapping enzyme active sites in complex proteomes. J. Am. Chem. Soc. 2004;126:1363–1368. doi: 10.1021/ja038441g. [DOI] [PubMed] [Google Scholar]
- 11.Sieber SA, Mondala TS, Head SR, Cravatt BF. Microarray platform for profiling enzyme activities in complex proteomes. J. Am. Chem. Soc. 2004;126:15640–15641. doi: 10.1021/ja044286+. [DOI] [PubMed] [Google Scholar]
- 12.Speers AE, Cravatt BF. A tandem orthogonal proteolysis strategy for high-content chemical proteomics. J. Am. Chem. Soc. 2005;127:10018–10019. doi: 10.1021/ja0532842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanson SR, Hsu TL, Weerapana E, Kishikawa K, Simon GM, Cravatt BF, Wong CH. Tailored glycoproteomics and glycan site mapping using saccharide-selective bioorthogonal probes. J. Am. Chem. Soc. 2007;129:7266–7267. doi: 10.1021/ja0724083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Blankman JL, Cravatt BF. A functional proteomic strategy to discover inhibitors for uncharacterized hydrolases. J. Am. Chem. Soc. 2007;129:9594–9595. doi: 10.1021/ja073650c. [DOI] [PubMed] [Google Scholar]
- 15.Salisbury CM, Cravatt BF. Activity-based probes for proteomic profiling of histone deacetylase complexes. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1171–1176. doi: 10.1073/pnas.0608659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosoya T, Hiramatsu T, Ikemoto T, Nakanishi M, Aoyama H, Hosoya A, Iwata T, Maruyama K, Endo M, Suzuki M. Novel bifunctional probe for radioisotope-free photoaffinity labeling: compact structure comprised of photospecific ligand ligation and detectable tag anchoring units. Org. Biomol. Chem. 2004;2:637–641. doi: 10.1039/b316221d. [DOI] [PubMed] [Google Scholar]
- 17.Sun P, Wang GX, Furuta K, Suzuki M. Synthesis of a bis-azido analogue of acromelic acid for radioisotope-free photoaffinity labeling and biochemical studies. Bioorg. Med. Chem. Lett. 2006;16:2433–2436. doi: 10.1016/j.bmcl.2006.01.083. [DOI] [PubMed] [Google Scholar]
- 18.Hosoya T, Hiramatsu T, Ikemoto T, Aoyama H, Ohmae T, Endo M, Suzuki M. Design of dantrolene-derived probes for radioisotope-free photoaffinity labeling of proteins involved in the physiological Ca2+ release from sarcoplasmic reticulum of skeletal muscle. Bioorg. Med. Chem. Lett. 2005;15:1289–1294. doi: 10.1016/j.bmcl.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 19.Saxon E, Armstrong JI, Bertozzi CR. A “traceless” Staudinger ligation for the chemoselective synthesis of amide bonds. Org. Lett. 2000;2:2141–2143. doi: 10.1021/ol006054v. [DOI] [PubMed] [Google Scholar]
- 20.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 21.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc. Natl. Acad. Sci. U. S. A. 2002;99(1):19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saxon E, Luchansky SJ, Hang HC, Yu C, Lee SC, Bertozzi CR. Investigating cellular metabolism of synthetic azidosugars with the Staudinger ligation. J. Am. Chem. Soc. 2002;124:14893–14902. doi: 10.1021/ja027748x. [DOI] [PubMed] [Google Scholar]
- 23.Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vocadlo DJ, Bertozzi CR. A strategy for functional proteomic analysis of glycosidase activity from cell lysates. Angew. Chem. Int. Ed. Engl. 2004;43:5338–5342. doi: 10.1002/anie.200454235. [DOI] [PubMed] [Google Scholar]
- 25.Al-Mawsawi LQ, Fikkert V, Dayam R, Witvrouw M, Burke TR, Jr., Borchers CH, Neamati N. Discovery of a small-molecule HIV-1 integrase inhibitor-binding site. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10080–10085. doi: 10.1073/pnas.0511254103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozikowski AP, Chen Y, Gaysin A, Chen B, D'Annibale MA, Suto CM, C LB. Functional differences in epigenetic modulators-superiority of mercaptoacetamide based-HDAC inhibitors relative to hydroxamates in cortical neuron neuroprotection studies. J. Med. Chem. 2007;50:3054–3061. doi: 10.1021/jm070178x. [DOI] [PubMed] [Google Scholar]
- 27.Hosoya T, Hiramatsu T, Ikemoto T, Aoyama H, Ohmae T, Endo M, Suzuki M. Design of dantrolene-derived probes for radioisotope-free photoaffinity labeling of proteins involved in the physiological Ca2+ release from sarcoplasmic reticulum of skeletal muscle. Bioorg. Med. Chem. Lett. 2005;15:1289–1294. doi: 10.1016/j.bmcl.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 28.Lin PC, Ueng SH, Yu SC, Jan MD, Adak AK, Yu CC, Lin CC. Surface Modification of Magnetic Nanoparticle via Cu(I)-Catalyzed Alkyne-azide [2+3] Cycloaddition. Org. Lett. 2007;9:2131–2134. doi: 10.1021/ol070588f. [DOI] [PubMed] [Google Scholar]
- 29.Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comp. Phys. Comm. 1995;91:43–56. [Google Scholar]
- 30.Lindal E, Hess B, van der Spoel D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Mod. 2001;7:306–317. [Google Scholar]
- 31.Van Der Spoel D, Van Buuren AR, Apol E, Meulenhoff PJ, Tieleman DP, Sijbers ALTM, Hess B, Feenstra KA, Lindahl E, Drunen R. v., Berendsen HJC. Gromacs User Manual version 3.0. In Nijenborgh 4, 9747 AG Groningen. The Netherlands: 2001. Internet: http://www.gromacs.org. [Google Scholar]
- 32.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE. GROMACS: Fast, flexible and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 33.Cornell WD, Cieplak P, Barly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A Second Generation Force Field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 34.Sorin EJ, Rhee YM, Pande VS. Does water play a structural role in the folding of small nucleic acids? Biophys. J. 2005;88:2516–2524. doi: 10.1529/biophysj.104.055087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 37.Pieffet G, Petukhov PA. Parameterization of aromatic azido groups: application as photoaffinity probes in molecular dynamics studies. J. Mol. Model. 2009;15:1291–1297. doi: 10.1007/s00894-009-0488-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakalian A, Bush BL, Jack DB, Bayly CI. Fast, Efficient Generation of High-Quality Atomic Charges. AM1-BCC Model: I: Method. J. Comput. Chem. 2000;21:132–146. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 39.Jakalian A, Jack DB, Bayly CI. Fast, Efficient Generation of High-Quality Atomic Charges. AM1-BCC Model: II: Parameterization and Validation. J. Comput. Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 40.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Bio. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki T, Kouketsu A, Matsuura A, Kohara A, Ninomiya S, Kohda K, Miyata N. Thiol-based SAHA analogues as potent histone deacetylase inhibitors. Bioorg. Med. Chem. Lett. 2004;14:3313–3317. doi: 10.1016/j.bmcl.2004.03.063. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Lopez-Sanchez M, Savoy DN, Billadeau DD, Dow GS, Kozikowski AP. A series of potent and selective, triazolylphenyl-based histone deacetylases inhibitors with activity against pancreatic cancer cells and Plasmodium falciparum. J. Med. Chem. 2008;51:3437–3448. doi: 10.1021/jm701606b. [DOI] [PubMed] [Google Scholar]
- 43.Somoza JR, Skene RJ, Katz BA, Mol C, Ho JD, Jennings AJ, Luong C, Arvai A, Buggy JJ, Chi E, Tang J, Sang BC, Verner E, Wynands R, Leahy EM, Dougan DR, Snell G, Navre M, Knuth MW, Swanson RV, McRee DE, Tari LW. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure (Camb) 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 44.Vannini A, Volpari C, Gallinari P, Jones P, Mattu M, Carfi A, De Francesco R, Steinkuhler C, Di Marco S. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8-substrate complex. Embo. Rep. 2007;8:879–884. doi: 10.1038/sj.embor.7401047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dowling DP, Gantt SL, Gattis SG, Fierke CA, Christianson DW. Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors. Biochemistry. 2008;47:13554–13563. doi: 10.1021/bi801610c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng YC, Prusoff WH. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50% inhibition (IC50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 47.Girish V, Vijayalakshmi A. Affordable image analysis using NIH Image/ImageJ. Indian J. Cancer. 2004;41:47. [PubMed] [Google Scholar]
- 48.Eliceiri KW, Rueden C. Tools for visualizing multidimensional images from living specimens. Photochem. Photobio. 2005;81:1116–1122. doi: 10.1562/2004-11-22-IR-377. [DOI] [PubMed] [Google Scholar]
- 49.Vida, 3.0; Openeye Scientific Software LLC. Santa Fe, NM: 2008. [Google Scholar]
- 50.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 52.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 53.Miyamoto S, Kollman PA. SETTLE: An Analytical Version of the SHAKE and RATTLE Algorithms for Rigid Water Models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 54.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 55.Hoover WG, editor. In Molecular-Dynamics Simulations of Statistical-Mechanical Systems. North-Holland; Amsterdam: 1986. Amsterdam, 1986. [Google Scholar]
- 56.Darden T, York D, Pedersen L. Particle mesh Ewald: An N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.