Abstract
Fatty acid desaturases (FADS) play an important role in the formation of omega-6 and omega-3 highly unsaturated fatty acids (HUFAs). The composition of HUFAs in the human metabolome is important for membrane fluidity and for the modulation of essential physiological functions such as inflammation processes and brain development. Several recent studies reported significant associations of single nucleotide polymorphisms (SNPs) in the human FADS gene cluster with HUFA levels and composition. The presence of the minor allele correlated with a decrease of desaturase reaction products and an accumulation of substrates. We performed functional studies with two of the associated polymorphisms (rs3834458 and rs968567) and showed an influence of polymorphism rs968567 on FADS2 promoter activity by luciferase reporter gene assays. Electrophoretic mobility shift assays proved allele-dependent DNA-binding ability of at least two protein complexes to the region containing SNP rs968567. One of the proteins binding to this region in an allele-specific manner was shown to be the transcription factor ELK1 (a member of ETS domain transcription factor family). These results indicate that rs968567 influences FADS2 transcription and offer first insights into the modulation of complex regulation mechanisms of FADS2 gene transcription by SNPs.
Keywords: delta-6 desaturase, fatty acid metabolism, desaturation, single nucleotide polymorphism
Fatty acids are among other metabolites essential components of the human metabolome. In cells, phospholipids containing highly unsaturated fatty acids (HUFAs) such as arachidonic acid (all-cis-5,8,11,14-eicosatetraenoic acid or C20:4n-6) and docosahexaenoic acid [22:6(ω-3), all-cis-docosa-4,7,10,13,16,19-hexaenoic acid or C22:6n-3] have a positive effect on the fluidity of cell membranes. On the molecular level, HUFAs fulfill several other central functions like acting as second messengers in intracellular signaling pathways or regulating transcription. On the physiological level, HUFAs are important for brain development, acquisition of cognitive behaviors, and development of visual functions in early life. In addition, HUFAs are precursors for eicosanoids (leukotriens and prostaglandins), which play an important role in inflammatory processes (1).
The production of HUFAs from dietary fatty acids includes several desaturation and elongation steps. The desaturases involved in this reaction cascade, delta-6 desaturase and delta-5 desaturase, are the rate-limiting enzymes. Both are expressed in the majority of human tissues, with highest levels in liver and to a smaller amount in brain, heart, and lung (2, 3). Delta-6 desaturase inserts a double bond at position 6 and after an elongation step, delta-5 desaturase inserts an additional double bond at position 5 of the elongated fatty acid chain. These conversions result in the formation of either arachidonic acid (C20:4n-6) in the omega-6 pathway or of eicosapentaenoic acid (C20:5n-3) in the omega-3 pathway. These molecules are either converted into eicosanoids or further elongated and desaturated, again with the help of the delta-6 desaturase (1). The importance of delta-6 desaturase for the formation of HUFAs and their influence on membrane integrity and fluidity was shown in a recent study by Stoffel et al. (4) who generated a fads2 −/− (fatty acid desaturase) mouse. In this animal model, membrane polarity of Sertoli and ovarian follicle cells was completely disturbed due to the lack of HUFAs in knockout mice caused by the delta-6 desaturase deficiency. Furthermore, both male and female mice were infertile and eicosanoid synthesis was disturbed. However, the administration of a HUFA-rich diet (either C20:4n-6 or C20:5n-3/C22:6n-3) enabled the fads2 −/− mice to overcome the genetic defect, restored the fatty acid pattern in membrane lipids, and rescued spermatogenesis as well as normal follicle development. Similarly, eicosanoid synthesis was restored by administration of arachidonic acid. Similar effects were observed in another fads2 −/− mouse by Stroud et al. (5).
These studies showed that the level and composition of HUFAs in the body highly depends on the conversion rate of the delta-6 desaturase, which is in turn regulated by supply with dietary fatty acids and hormone signaling. The effect of dietary fatty acids on desaturase transcription regulation is mediated by two transcription factors, sterol regulatory element binding protein (SREBP1) and peroxisome proliferator activated receptor (PPARA) (6). The feedback regulation mechanisms by which dietary fatty acids act on SREBP1 processing and stability, which in turn influence FADS2 gene expression, have been investigated intensively (7–11). The induction of desaturases by PPARA was shown to occur both by indirect and direct mechanisms (12–15). Besides the mediation of fatty acid effects, SREBP1 may also mediate the insulin effect on FADS2 gene expression, as was observed in experimentally induced diabetic rats (16, 17).
Although dietary and hormonal influences seem to play an important role in transcription regulation of delta-6 desaturase, genetic factors are important as well for influencing the level and composition of HUFAs in human tissues. Of special interest is the FADS gene cluster on chromosome 11, with a head-to-head orientation of the FADS1 and FADS2 genes, which encode the delta-5 and delta-6 desaturase, respectively. A third putative desaturase gene, FADS3, is located in the 6.0 kb telomeric side from the FADS2 gene in a tail-to-tail orientation (18). Several candidate gene studies reported an association of a number of single nucleotide polymorphisms (SNPs) in the FADS gene cluster with fatty acid composition in human tissues (19–22). These results were strengthened recently by our study, which for the first time compared genome-wide SNP data with metabolomics data and replicated the previous findings by this new approach (23). Additionally, several genome-wide association studies meanwhile reported an association of FADS polymorphisms with polyunsaturated fatty acids (24) and more complex lipid traits like low-density lipoprotein, high-density lipoprotein, and triglycerides (25–27).
In the first association study (19), the minor alleles of 11 SNPs located in and around the FADS1 and FADS2 genes were associated with enhanced levels of desaturase substrates in serum phospholipids. In contrast, levels of desaturase products (especially arachidonic acid, with a genetically explained variance of 28%) were lower. The same significant associations were found for haplotype analyses. This observation speaks for a strong influence of the genetic variants on the activity of the desaturases. Until now, functional data on the described polymorphisms were not available. The aim of this study was to identify causative SNPs within the FADS1/FADS2 haplotype, and we therefore performed functional analyses of polymorphisms in the FADS2 promoter region to gain insight into regulatory mechanisms of the FADS2 gene resulting from the presence of these polymorphisms on the transcriptional level. Based on their close proximity to the translation start site of FADS2, we selected the one base pair deletion/insertion polymorphism (DIP) rs3834458 (position −942) and the SNP rs968567 (−299). In addition, both polymorphisms are located in a CpG-rich region predicted to contain interesting binding sites for transcription factors known to be involved in fatty acid metabolism such as SREBP1 and PPARA.
MATERIALS AND METHODS
Bioinformatic analysis of transcription factor binding sites
The prediction of transcription factor binding sites (TFBS) in promoter sequences was performed using the Genomatix MatInspector software with standard settings for the highest matrix similarity (28). This program uses a large library of weight matrices based on known in vivo binding sites to predict TFBS in nucleotide sequences.
Plasmid constructions
To obtain constructs for luciferase assays, the FADS2 promoter sequence from position −1014 to −1 relative to the translation start site was amplified by PCR from human genomic DNA. The PCR product was first cloned into the vector pGEM T-Easy (Promega) and then subcloned into the reporter vector pGL4.12 (Promega). Constructs containing all possible combinations of major and minor alleles of rs3834458 (T/Del, position −942) and rs968567 (C/T, position −299) were obtained by PCR mutagenesis. Truncated constructs (containing region −414 to −1 and −214 to −1) were generated by PCR from the original respective plasmids and subsequent cloning into pGL4.12. All constructs were verified by sequencing.
Luciferase reporter assays
HeLa, HEK293, and HepG2 cells were seeded at a density of 1 × 105 cells/well in 12-well plates in MEM or DMEM medium with stable l-glutamine (PAA Laboratories), respectively, containing 10% FBS (PAA Laboratories) and 1% penicillin/streptomycin (Gibco) and incubated overnight. All cell lines were transfected with 500 ng of the promoter construct per assay using FuGene6 (Roche Diagnostics) according to the manufacturer's instructions in an appropriate ratio of FuGene-DNA. For normalization, 50 ng of the pGL4.74 vector (Promega), which constitutively expresses Renilla luciferase, was cotransfected. Transfected cells were incubated for 32 h at 37°C in a 5% CO2 atmosphere. Cells were then washed once in PBS buffer before 200 µl of 1× passive lysis buffer (Promega) was added. After gentle shaking for 30 min, the plate containing the lysed cells was frozen at −80°C overnight. After thawing, luciferase activity was measured. For this, 50 µl of both Dual Luciferase Reporter Assay System reagents (Promega) was added successively to 20 µl of the lysate according to the manufacturer's instructions. Measurements were done in a Tecan GeniosPro microplate reader. Calculation of the intensity ratios of Firefly-Renilla luciferase activity resulted in the relative promoter activity of the constructs. The significance of difference in promoter activity between the constructs was tested by independent-samples t-test using the SPSS 16.0 software.
Nuclear protein extraction and electrophoretic mobility shift assays
Confluent HeLa cells grown in T75 flasks were harvested and nuclear proteins were extracted with the NE-PER® Nuclear and Cytoplasmic Extraction reagents (Pierce) according to the manufacturer's instructions. For electrophoretic mobility shift assays (EMSAs), oligonucleotides containing predicted TFBSs surrounding the DIP rs3834458 and the SNP rs968567 were designed and purchased from the company Metabion. The oligo sequences are summarized in Table 1. A total of 20 pmol of double-stranded oligos containing either the major or the minor allele were 5′-end labeled with γ-32P-ATP (Hartmann Analytic) and T4 polynucleotide kinase (Fermentas) according to the manufacturer's protocol. Unincorporated label was separated from labeled DNA by gel filtration on G-25 columns (GE Healthcare). Binding reaction was carried out with or without different concentrations of unlabeled competitor oligonucleotides using 15 µg of nuclear extract in 1× binding buffer (20 mM Tris/HCl, pH 7.9, 50 mM NaCl, 1 mM EDTA, 10% glycerol, 0.05% NP40, 2.5 mM DTT) with 1 µg poly dI-dC (Roche Diagnostics) and 20 fmol of labeled probe in a total volume of 20 µl for 30 min at room temperature. Protein-DNA complexes were separated on 10% nondenaturing polyacrylamide gels by electrophoresis in 1× tris-borate-EDTA (TBE) buffer. The gels were dried and radioactivity was visualized by autoradiography on Kodak films.
TABLE 1.
Oligonucleotides representing the putative TFBSs used for EMSAs
| Polymorphism | Allele | Strand | Sequence (5′→3′) | Size (bp) |
|---|---|---|---|---|
| rs3834458 | T | for | ATTCTTTTCTAAGATTGTC | 19 |
| T | rev | GACAATCTTAGAAAAGAAT | 19 | |
| Del | for | ATTCTTTTCAAGATTGTC | 18 | |
| Del | rev | GACAATCTTGAAAAGAAT | 18 | |
| rs968567 | C | for | GAGGCCCTGAGCTCCCGGGGAGTTTTTACT | 30 |
| C | rev | AGTAAAAACTCCCCGGGAGCTCAGGGCCTC | 30 | |
| T | for | GAGGCCCTGAGCTTCCGGGGAGTTTTTACT | 30 | |
| T | rev | AGTAAAAACTCCCCGGAAGCTCAGGGCCTC | 30 |
Nucleotides representing the polymorphisms are underlined.
Gene expression analysis
Correlation analysis of peripheral blood gene expression was performed in 322 KORA F3 samples with whole-genome expression profiles available. A detailed description of the KORA F3 study, which is a population-based study comprising individuals living in the region of Augsburg, has been given elsewhere (29). Gene expression analysis was performed with the Illumina Human-6 v2 Expression BeadChip as described earlier (30). Raw data from the Illumina ‘Beadstudio’ software were exported to R software. Data were logarithmized and normalized using the LOWESS method (31). Associations between the expressions of two genes were computed with a linear regression model. Correlations were determined using the Pearson correlation coefficient.
DNA affinity purification and immunoblotting
Oligonucleotides for DNA affinity purification contained four repeats of the predicted TFBSs surrounding SNP rs968567 to ensure maximal binding efficiency (see Table 2). Double-stranded oligonucleotide binding sites were constructed by annealing 26 pmol of complementary single-stranded 5′-end biotinylated oligonucleotides containing the −299 major C allele or the −299 minor T allele in annealing buffer (89.6 mM Tris-HCl, pH 9.0, 448.2 mM KCl, and 13.4 mM MgCl2). Oligonucleotides with four repeats of an experimentally verified ELK1 (member of ETS domain transcription factor family) binding site were generated accordingly as positive control. Proteins binding to the oligonucleotides were purified using streptavidin-coated Dynabeads M-280 (Invitrogen). Briefly, 26 pmol of double-stranded biotinylated oligonucleotides were coupled to 250 µg (25 µl) of the streptavidin magnetic beads according to the manufacturer's protocol. A total of 50 µg of HeLa nuclear extract was applied to the DNA-magnetic beads complex and incubated in protein binding buffer (4.6 mM Tris-HCl, pH 8.0, 18.4 mM KCl, 0.02% NP-40, 0.37% glycerol, 4.8 mM DTT, 22.9 µM ZnSO4 with 9.7 mM MgCl2) for 10 min at room temperature. Nonspecific DNA binding was inhibited by the subsequent addition of 2.5 µg poly[d(I-C)] (Roche) and incubation for an additional 20 min. Afterwards, the supernatant containing unbound proteins was removed by use of a magnetic separator and the beads with the DNA-protein complexes were washed three times with wash buffer (9.9 mM Tris-HCl, pH 8.0, 39.6 mM KCl, 0.05% NP-40, 0.8% glycerol, 10 mM DTT, 49.5 µM ZnSO4). Bound proteins were eluted from the magnetic beads by use of a high ionic strength elution buffer (9.5 mM Tris-HCl, pH 8.0, 1.9 M KCl, 0.048% NP-40, 0.76% glycerol, 10 mM DTT, 47.5 µM ZnSO4, and 10 mM MgCl2), separated on a 10% Tris-tricine SDS-PAGE gel, and subsequently blotted onto a polyvinylidene fluoride membrane (Pall). Incubation with ELK1 antibody (SC-355 X, Santa Cruz, 1:500 in PBS containing 0.5% milk powder) was carried out at 4°C overnight. As secondary antibody, a peroxidase-conjugated goat anti-rabbit IgG (A-6154, Sigma, 1:5000 in PBS containing 0.5% milk powder) was used with an incubation time of 1 h at room temperature. Peroxidase reaction was carried out using the Western Lightning Chemiluminescence Reagent Plus (PerkinElmer) and specific ELK1 bands were visualized by exposing Kodak films.
TABLE 2.
Oligonucleotides used for DNA affinity purification of ELK1
| Primer | Strand | Sequence (5′ → 3′) | Size (bp) |
|---|---|---|---|
| ELK1_control | for | *GAATAACCGGAAGTAACCGAATAACCGGAAGTAA CCGAATAACCGGAAGTAACCGAATAACCGGAAGTAACC | 72 |
| rev | *GGTTACTTCCGGTTATTCGGTTACTTCCGGTTAT TCGGTTACTTCCGGTTATTCGGTTACTTCCGGTTATTC | 72 | |
| rs968567_C | for | *TGAGCTCCCGGGGAGTTGAGCTCCCGGGGA GTTGAGCTCCCGGGGAGTTGAGCTCCCGGGGAGT | 71 |
| rev | *ACTCCCCGGGAGCTCAACTCCCCGGGAGCT CAACTCCCCGGGAGCTCAACTCCCCGGGAGCTCA | 71 | |
| rs968567_T | for | *TGAGCTTCCGGGGAGTTGAGCTTCCGGGGA GTTGAGCTTCCGGGGAGTTGAGCTTCCGGGGAGT | 71 |
| rev | *ACTCCCCGGAAGCTCAACTCCCCGGAAGCT CAACTCCCCGGAAGCTCAACTCCCCGGAAGCTCA | 71 |
Nucleotides representing the rs968567 polymorphism are underlined. The first nucleotide of each repetitive element is written in bold. An asterisk (*) represents the position of the biotinylation site.
RESULTS
Bioinformatic analyses predict the allele-dependent presence of different TFBS in the SNP-containing FADS2 promoter regions
Bioinformatic analyses using the Genomatix software predicted transcription factors with the highest core matrix similarities and revealed that our DIP of interest (rs3834458, position −942) was located in close proximity to predicted SREBP1 and PPARA binding sites, with the SREBP binding element being 48 bp and the PPAR/RXR binding element 12 bp away. Several other binding sites for transcription factors were predicted for the region containing the DIP rs3834458: C/EBP-β in 6 bp distance from the DIP, and PAX4/PAX6 and BCL6 directly spanning the −942 position. Interestingly, the BCL6 binding site is present only when the sequence contains the −942 major T allele and is lost when the deletion mutation is present, because the −942 major T allele is part of the binding site core sequence of BCL6 (Fig. 1A, C).
Fig. 1.
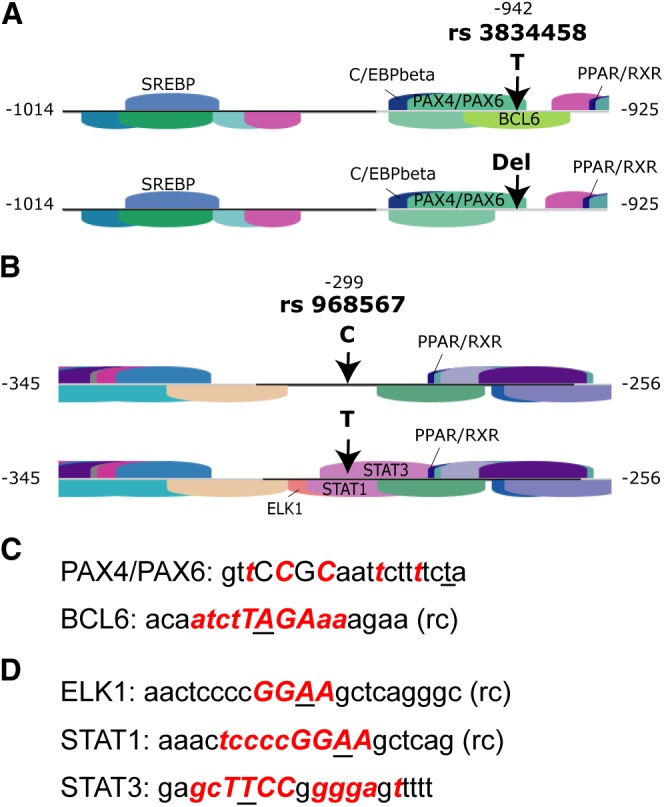
Schematic presentation of location of analyzed polymorphisms in the human FADS2 gene promoter and surrounding TFBSs modified from Genomatix MatInspector software output. A: A BCL6 binding site is predicted when the major T allele of rs3834458 is present but lacking when the deletion is present. B: The minor T allele of rs968567 leads to prediction of binding sites for ELK1, STAT1, and STAT3. In contrast, no binding sites are predicted when the major C allele is present. C and D: Sequence that was recognized by Genomatix MatInspector to contain a TFBS and also rs3834458 (C) or rs968567 (D), respectively. Nucleotides marked in bold red show a high degree of matrix conservation at this position, i.e., information content is very high. Nucleotides in capitals denote the core sequence used by MatInspector. Underlined nucleotides highlight the position of the respective polymorphism; rc means that a TFBS sequence was found for the opposite strand.
The promoter region surrounding SNP rs968567 (position −299) is also predicted to contain several TFBS. Once more, a PPAR/RXR binding site is located in the neighborhood of the SNP only 12 bp away. Three additional binding sites are predicted for the sequence containing the −299 minor T allele: ELK1, STAT1, and STAT3, which are not present for the −299 major C allele (Fig. 1B). Again, the −299 minor T allele is part of the matrix core sequences of all three TFBSs (Fig. 1D).
Luciferase reporter gene assays reveal an influence of SNP rs968567 on promoter activity
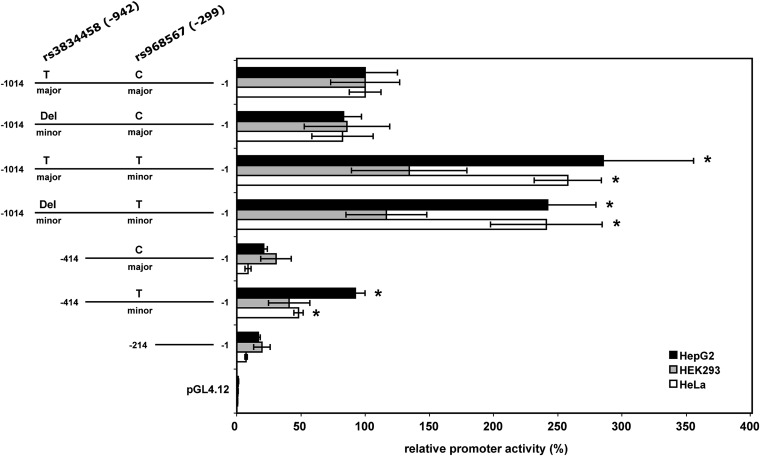
To determine the functional effects of the two polymorphisms (rs3834458, T/Del, −942 and rs968567, C/T, −299) on transcriptional regulation, luciferase reporter gene assays were conducted to measure promoter activity (Fig. 2). Three different human cell lines (HepG2, HEK293, and HeLa) were transiently transfected with the promoter constructs or the empty reporter vector pGL4.12 as control. Three individual experiments for each construct and cell line were performed and promoter activity was measured in triplicates for each construct and experiment. Luciferase activity was slightly lower for all constructs containing the −942 minor deletion mutation compared with the constructs containing the −942 major T allele. This was a modestly not significant effect, however, with a decrease in luciferase activity of around 20% averaged over all tested cell lines and constructs. The replacement of the −299 major C allele of rs968567 by the −299 minor T allele resulted in a 2- to 3-fold increase of luciferase activity in HeLa and HepG2 cells in full-length as well as truncated constructs. This effect was statistically significant in HepG2 (P < 2.0E-05) and HeLa (P < 1.0E-6) cells but not in HEK293 cells. Altogether, the results indicate a strong regulatory function of polymorphism rs968567 in different cell lines.
Fig. 2.
Relative promoter activity of different constructs of human FADS2 gene promoter in three different human cell lines. Values represent the mean of three independent experiments performed in triplicate. Promoter activity of the major/major construct was used as reference and set at 100%. Asterisks denote statistically significant results calculated by t-test. Numbers indicate the relative position to the FADS2 translation start site.
EMSA demonstrates altered DNA-binding ability of nuclear proteins to the FADS2 promoter due to SNP rs968567
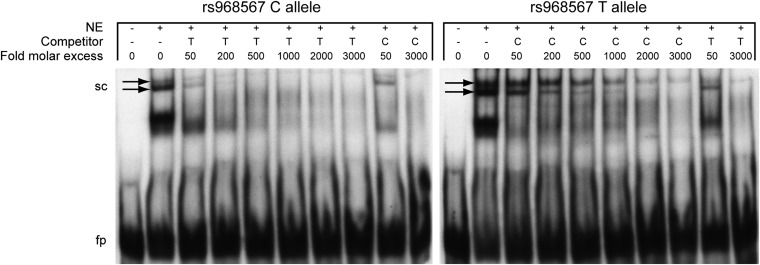
Next we asked if the polymorphisms effect the DNA-binding ability of nuclear proteins. HeLa nuclear protein extracts were subjected to binding to oligonucleotides representing the region surrounding SNP rs968567 with either the −299 major C allele or the −299 minor T allele, and DNA-protein complexes were analyzed by EMSA (Fig. 3). Specific binding of nuclear protein to the respective oligonucleotide was tested by adding increasing amounts of competing unlabeled oligonucleotide probe, containing the respective other allele. Two bands corresponding to shifted complexes showed different intensity, depending on which allele was present. Both bands showed weaker intensity when the labeled oligonucleotide with the C allele was present, whereas a higher intensity was achieved when the labeled oligonucleotide contained the T allele. Competition of labeled C allele with unlabeled T allele resulted in a significant decrease of band intensities already at low concentrations of competitor. The upper band was still visible at very high competitor concentrations, whereas the lower band vanished completely. In contrast, competition for protein binding of labeled T allele with unlabeled C allele resulted in slightly decreased band intensities only at high concentrations of competitor. At the highest competitor concentration, the lower band vanished as well, but the upper band was much stronger than in the vice versa competition experiment. These effects were observed in two independent experiments. The results indicated that the −299 T allele increased binding affinity of the tested promoter region for at least two protein complexes. The same experiment was conducted with oligonucleotides containing the major and minor alleles of the rs3834458 polymorphism. Only very weak band intensities, hinting to very weak binding of two nuclear proteins, could be observed and no significant difference of competing effects between oligonucleotides was found (data not shown).
Fig. 3.
Verification of molecular interactions in the human FADS2 promoter region. Autoradiographs of competitive EMSA gels of the rs968567 polymorphism using allele-specific 32P-labeled oligonucleotides. NE: nuclear extract, sc: shifted complexes, fp: free probe. Arrows indicate bands corresponding to shifted complexes and having different intensity dependent on the tested allele.
Gene expression analysis shows statistically significant association between expression levels of FADS2 and ELK1
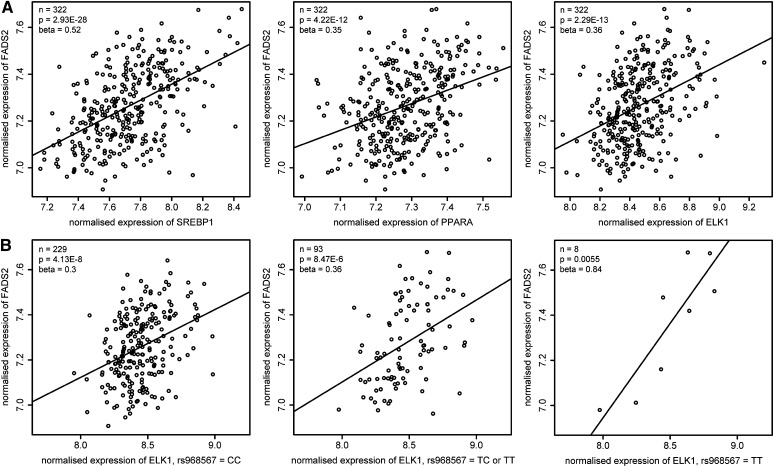
Because we have found a significant impact on promoter activity and binding of nuclear protein complexes only for SNP rs968567 and not for DIP rs3834458, we focused on the region surrounding SNP rs968567 for further characterization. Prediction of TFBSs in this region resulted in three binding sites when the rs968567 minor T allele was present in the sequence: ELK1, STAT1, and STAT3. Regression analysis between FADS2 whole blood mRNA expression levels and expression levels of these three transcription factors in 322 subjects revealed a statistically significant association between mRNA expression levels of FADS2 and ELK1 with a P-value of 2.29E-13 and an effect size of 0.36 (Fig. 4A). No significant P-values were obtained for the correlation of FADS2 with STAT1 and STAT3 expression levels. To test the plausibility of this approach, regression analyses of FADS2 expression levels with PPARA and SREBP1, two transcription factors already known to be involved in FADS2 transcription regulation, were performed as positive controls. The expression levels of both transcription factors were significantly associated with FADS2 expression (PPARA: P = 4.22E-12, effect size = 0.35 and SREBP1: P = 2.93E-28, effect size = 0.52) and by this proved the reliability of our expression data. We furthermore tested the association between FADS2 and ELK1 gene expression dependent on the rs968567 genotype. The effect size of association between FADS2 and ELK1 in homozygous carriers of the rs968567 major C allele (n = 229) was 0.3 (P = 4.13E-8). In heterozygous (CT) and homozygous minor T allele carriers (n = 93), it reached 0.36 (P = 8.47E-6), and in homozygous minor T allele carriers alone (n = 8) the effect size increased to 0.84 (P = 0.0055) (Fig. 4B). These results strongly point to ELK1 as a newly identified regulator of FADS2 gene expression with a higher impact of ELK1 in carriers of the rs968567 minor T allele.
Fig. 4.
Correlation analysis of FADS2 and transcription factor mRNA expression in human whole blood samples. Axes represent normalized mRNA expression levels of the respective genes. Numbers of subjects (n), P-value, and effect size (β) of the association are indicated for each graph. A: Normalized expression level of FADS2 plotted against SREBP1 and PPARA as positive control and against ELK1. B: Normalized expression level of FADS2 plotted against ELK1 dependent on rs968567 genotype.
DNA affinity purification with immunoblotting reveals allele-specific binding of ELK1 to the region surrounding SNP rs968567
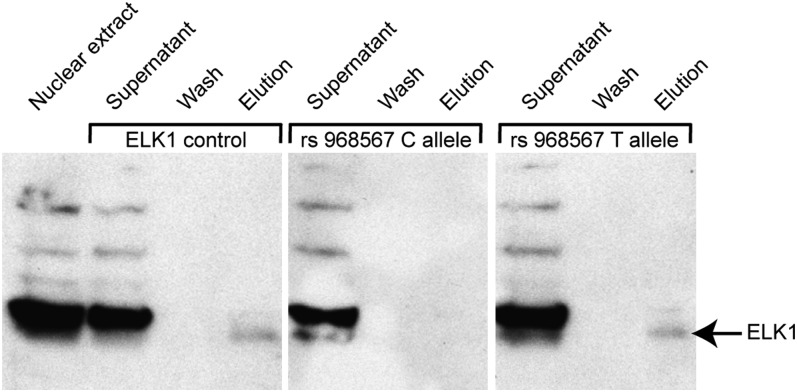
Our gene expression analyses in a population-based study revealed a significant association between expression levels of FADS2 mRNA and ELK1 mRNA in whole blood, with a higher effect size in carriers of the rs968567 minor T allele. Additionally, the Genomatix MatInspector software predicted allele-specific binding of ELK1 to the region surrounding SNP rs968567. We therefore tested the binding of ELK1 protein to the respective sequence by performing DNA affinity purification of nuclear proteins from HeLa nuclear extract using biotinylated oligonucleotides representing the region surrounding SNP rs968567 with either the −299 major C allele or the −299 minor T allele. An oligonucleotide containing an experimentally verified ELK1 binding site (32) was used as positive control. The supernatant and wash fractions containing unbound proteins as well as the elution fraction with the bound proteins were immunoblotted and a specific antibody against human ELK1 was used to detect presence of ELK1 protein in the fractions (Fig. 5). A specific band corresponding to ELK1 was present in the elution fraction of the positive control, showing that ELK1 from HeLa nuclear extract is able to bind to its consensus sequence under the used buffer conditions and experimental setup. The appearance of ELK1 in the elution fraction of the −299 minor T allele, which was lacking in the elution fraction of the −299 major C allele, confirms binding of ELK1 to the FADS2 promoter sequence exclusively when the minor T allele is present.
Fig. 5.
Detection of human ELK1 protein in DNA affinity purification fractions by immunoblotting. The first lane represents crude HeLa nuclear extract and verifies the presence of ELK1 protein in HeLa cells. The supernatant and wash fractions contain all unbound proteins, whereas the elution fraction contains all oligonucleotide binding proteins. A specific band at 62 kDa corresponds to the ELK1 protein (indicated by an arrow).
DISCUSSION
Disorders of delta-6 desaturase activity affect essential physiological functions
Recent association studies showed an association of delta-6 and delta-5 desaturase gene polymorphisms with HUFA level and composition in different human tissues accompanied by an accumulation of desaturase substrates and a decline in desaturase products (19–23). This suggests that the desaturase activity is not only regulated by nutritional and hormonal influences but also by genetic factors. The observed change of HUFA levels and composition in different human tissues due to the polymorphisms might alter several important physiological processes and is thought to modulate the development of complex diseases. The effect of FADS polymorphisms on brain development has been shown by Caspi et al. (33), who reported a modulation of the positive effect of breastfeeding on development of intelligence by polymorphisms in the FADS gene cluster in two independent birth cohorts. The importance of an intact delta-6 desaturase function on eicosanoid synthesis and membrane lipid composition was underlined by previous reports of two different fads2 knockout mice (4, 5). The assumption that there is a direct effect of FADS polymorphisms on the outcome of fatty acid-related diseases has been supported by Schaeffer et al. (19), who reported an association of the FADS gene cluster with allergic rhinitis and atopic eczema, though without statistical significance after correction for multiple testing. Another study recently reported an association of FADS genotypes with inflammation and coronary artery disease (34). All these observations hint at a strong role of delta-6 desaturase in regulating fatty acid composition in human tissues to maintain health. Approaches to investigate the influence of genetic polymorphisms on the regulation of the human enzyme activity are therefore needed to understand the role of delta-6/delta-5 desaturases in the development of fatty acid-related complex diseases.
Detection of a critical polymorphism-containing region that influences delta-6 desaturase activity
Many studies have reported associations of several SNPs in the FADS gene cluster with HUFA levels and composition in different human tissues and have contributed to the understanding of the influence of SNPs on the regulation of fatty acid synthesis (19–23). However, the causative functional variant(s) are not known up to date. The analysis of linkage disequilibrium (LD) structures in the FADS gene cluster suggests that all polymorphisms in this region are in very high LD and most of them are highly correlated. The real functional variant(s) could therefore cause associations of all other SNPs being in high LD and cannot be directly identified by association studies for this reason. Functional approaches are needed to determine the effect of the associated SNPs on the molecular level and by this identify the causative variant(s).
By performing luciferase reporter gene assays, we showed that one of the two analyzed FADS2 promoter polymorphisms (rs968567) is located in a region that seems to be important for transcription regulation. While the minor deletion mutation of rs3834458 had only a little, statistically not significant effect on promoter activity, the minor T allele of rs968567 highly increased promoter activity compared with the construct containing both major alleles. The effect was the same in all three tested cell lines; however, the response in HEK293 cells was lower and not statistically significant for both polymorphisms. Because transcription regulation is tissue dependent (35), this is likely due to the tissue-specific expression pattern of involved transcription factors. To investigate if altered binding of transcription factors to the polymorphism-containing regions is the cause for the observed effects in the luciferase assays, molecular interactions were analyzed by EMSA. Bioinformatic analyses predicted several putative binding sites in the polymorphism-containing regions, and we consequently checked their functionality for protein binding. Indeed, several protein complexes were shown to bind to the regions of interest by EMSA. In the case of rs968567, a clear allele-specific binding affinity of at least two protein complexes was shown by using a competitive method. The minor T allele of rs968567 facilitated the binding in comparison to the major C allele. No differential binding affinity could be shown for the region containing rs3834458. All these observations speak for a strong influence of the rs968567 polymorphism on transcription regulation of the FADS2 gene.
Identification of ELK1 as a potential new regulator of FADS2 gene transcription
In this study, it was shown that the FADS2 promoter region surrounding SNP rs968567 exhibits promoter activity, which increases when the major C allele of SNP rs968567 is replaced by the minor T allele. We assumed that this effect could be caused by allele-specific differential binding affinity of transcription factors. An in silico analysis of TFBSs predicted three additional binding sites (ELK1, STAT1, and STAT3) in the sequence when the major C allele of rs968567 was replaced by the minor T allele. This was substantiated by EMSA experiments that revealed allele-specific binding of at least two nuclear protein complexes to this promoter region. Linear regression analysis of whole blood mRNA levels of the predicted transcription factors and expression levels of FADS2 mRNA resulted in a highly significant association between ELK1 and FADS2, with a much higher effect size in subjects being homozygous for the rs968567 minor T allele. We used the correlation of PPARA and SREBP1 with FADS2 as positive control, because these two transcription factors are known to activate FADS2 transcription (1). The significant association results of our positive controls approve reliability of the expression data and substantiate the significant association between ELK1 and FADS2. ELK1 is a member of the ETS domain family of transcription factors, was first cloned in 1989 (36), and is primarily known for its role in the transcriptional regulation of immediate early genes, including c-fos (37) and egr-1 (38), by forming ternary complexes with serum response factor on the serum response elements of gene promoters (39). To our knowledge, a role of ELK1 in lipid metabolism has not been reported until now. We tested binding of ELK1 protein to the predicted binding site in the FADS2 gene promoter by DNA affinity purification and subsequent immunoblotting. Specific ELK1 bands in the elution fraction were only present when the major C allele of SNP rs968567 was replaced by the minor T allele. This effect is in clear accordance with the Genomatix MatInspector prediction and identifies ELK1 as a putative new regulator of FADS2 gene transcription in an allele-specific manner. The fact that correlation analysis between FADS2 and ELK1 mRNA expression gives significant results for both alleles (however, with lower effect size for the major C allele) suggests that ELK1 also binds to the FADS2 promoter in the presence of the −299 major C allele, but with lower affinity so that we were not able to detect ELK1 protein in that case in our immunoblotting experiment. Another possibility would be an additional functional ELK1 binding site in another region of the FADS2 gene, of which several are predicted by Genomatix MatInspector.
Controversial impact of the rs3834458 deletion polymorphism on promoter activity
Nwankwo et al. (40) published a study in 2003 that already dealt with functional investigations of the rs3834458 polymorphism. The authors aimed to identify the molecular mechanism of FADS2 deficiency in skin fibroblasts from a patient with severe symptoms like corneal ulceration, growth failure, skin abnormalities, and photophobia previously shown to be caused by a deficiency of delta-6 desaturase (41). By sequencing the FADS2 promoter region of DNA derived from patient fibroblasts and comparing the sequence to DNA from three healthy controls, they identified a thymidine insertion in the patient DNA, which corresponded to the T allele of rs3834458. Luciferase reporter gene assays in a mouse fibroblast cell line (NIH/3T3) with promoter sequences derived from patient (T allele present) and healthy control (T deletion) fibroblasts resulted in significantly decreased promoter activity when the T allele was present. This result could not be replicated in any of our three tested human cell lines. Possible explanations could be that Nwankwo et al. (40) used a mouse fibroblast cell line (NIH/3T3) for their assays, which might express a different set of transcription factors compared with our human cell lines or that another unrecognized polymorphism in the tested sequences caused the effect in the study of Nwankwo et al. (40).
Conclusion and outlook
In this study, we showed that polymorphism rs968567 influences FADS2 gene promoter activity and alters DNA-binding affinity of nuclear proteins. One of the proteins binding to this region in an allele-specific manner was shown to be the transcription factor ELK1. Further experiments are required to completely characterize the interaction of ELK1 with the FADS2 gene promoter or other functional elements in the gene and its impact on FADS2 gene expression in vivo.
Acknowledgments
We thank K. Schwerdtner for excellent technical assistance and F. Haller for critical reading of the manuscript.
Footnotes
This study was partly funded by the German Federal Ministry of Education, Science and Technology (National Genome Research Net-2, NGFNplus 01GS0823) and has received funding from the European Community's 7° Framework Programme (FP7/2008-2013) under grant agreement no 212652 (NUTRIMENTHE Project “The Effect of Diet on the Mental Performance of Children”).
Abbreviations:
- C20:4n-6 or arachidonic acid
- all-cis-5,8,11,14-eicosatetraenoic acid
- C22:6n-3 or docosahexaenoic acid, 22:6(ω-3)
- all-cis-docosa-4,7,10,13,16,19-hexaenoic acid
- DIP
- deletion/insertion polymorphism
- ELK1
- member of ETS domain transcription factor family
- EMSA
- electrophoretic mobility shift assay
- FADS
- fatty acid desaturase
- HUFA
- highly unsaturated fatty acid
- LD
- linkage disequilibrium
- PPAR
- peroxisome proliferator activated receptor
- SNP
- single nucleotide polymorphism
- SREBP
- sterol regulatory element binding protein
- TFBS
- transcription factor binding site
REFERENCES
- 1.Nakamura M. T., Nara T. Y. 2004. Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu. Rev. Nutr. 24: 345–376. [DOI] [PubMed] [Google Scholar]
- 2.Cho H. P., Nakamura M., Clarke S. D. 1999. Cloning, expression, and fatty acid regulation of the human delta-5 desaturase. J. Biol. Chem. 274: 37335–37339. [DOI] [PubMed] [Google Scholar]
- 3.Cho H. P., Nakamura M. T., Clarke S. D. 1999. Cloning, expression, and nutritional regulation of the mammalian Delta-6 desaturase. J. Biol. Chem. 274: 471–477. [DOI] [PubMed] [Google Scholar]
- 4.Stoffel W., Holz B., Jenke B., Binczek E., Gunter R. H., Kiss C., Karakesisoglou I., Thevis M., Weber A. A., Arnhold S., et al. 2008. Delta6-desaturase (FADS2) deficiency unveils the role of omega3- and omega6-polyunsaturated fatty acids. EMBO J. 27: 2281–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stroud C. K., Nara T. Y., Roqueta-Rivera M., Radlowski E. C., Lawrence P., Zhang Y., Cho B. H., Segre M., Hess R. A., Brenna J. T., et al. Disruption of FADS2 gene in mice impairs male reproduction and causes dermal and intestinal ulceration. J Lipid Res. Epub ahead of print. April 7, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuzaka T., Shimano H., Yahagi N., Amemiya-Kudo M., Yoshikawa T., Hasty A. H., Tamura Y., Osuga J., Okazaki H., Iizuka Y., et al. 2002. Dual regulation of mouse Delta(5)- and Delta(6)-desaturase gene expression by SREBP-1 and PPARalpha. J. Lipid Res. 43: 107–114. [PubMed] [Google Scholar]
- 7.Hannah V. C., Ou J., Luong A., Goldstein J. L., Brown M. S. 2001. Unsaturated fatty acids down-regulate srebp isoforms 1a and 1c by two mechanisms in HEK-293 cells. J. Biol. Chem. 276: 4365–4372. [DOI] [PubMed] [Google Scholar]
- 8.Mater M. K., Thelen A. P., Pan D. A., Jump D. B. 1999. Sterol response element-binding protein 1c (SREBP1c) is involved in the polyunsaturated fatty acid suppression of hepatic S14 gene transcription. J. Biol. Chem. 274: 32725–32732. [DOI] [PubMed] [Google Scholar]
- 9.Xu J., Nakamura M. T., Cho H. P., Clarke S. D. 1999. Sterol regulatory element binding protein-1 expression is suppressed by dietary polyunsaturated fatty acids. A mechanism for the coordinate suppression of lipogenic genes by polyunsaturated fats. J. Biol. Chem. 274: 23577–23583. [DOI] [PubMed] [Google Scholar]
- 10.Xu J., Teran-Garcia M., Park J. H., Nakamura M. T., Clarke S. D. 2001. Polyunsaturated fatty acids suppress hepatic sterol regulatory element-binding protein-1 expression by accelerating transcript decay. J. Biol. Chem. 276: 9800–9807. [DOI] [PubMed] [Google Scholar]
- 11.Yoshikawa T., Shimano H., Yahagi N., Ide T., Amemiya-Kudo M., Matsuzaka T., Nakakuki M., Tomita S., Okazaki H., Tamura Y., et al. 2002. Polyunsaturated fatty acids suppress sterol regulatory element-binding protein 1c promoter activity by inhibition of liver X receptor (LXR) binding to LXR response elements. J. Biol. Chem. 277: 1705–1711. [DOI] [PubMed] [Google Scholar]
- 12.Kawashima Y., Musoh K., Kozuka H. 1990. Peroxisome proliferators enhance linoleic acid metabolism in rat liver. Increased biosynthesis of omega 6 polyunsaturated fatty acids. J. Biol. Chem. 265: 9170–9175. [PubMed] [Google Scholar]
- 13.Reddy J. K., Hashimoto T. 2001. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu. Rev. Nutr. 21: 193–230. [DOI] [PubMed] [Google Scholar]
- 14.Song He, W., Nara T. Y., Nakamura M. T. 2002. Delayed induction of delta-6 and delta-5 desaturases by a peroxisome proliferator. Biochem. Biophys. Res. Commun. 299: 832–838. [DOI] [PubMed] [Google Scholar]
- 15.Li Y., Nara T. Y., Nakamura M. T. 2005. Peroxisome proliferator-activated receptor alpha is required for feedback regulation of highly unsaturated fatty acid synthesis. J. Lipid Res. 46: 2432–2440. [DOI] [PubMed] [Google Scholar]
- 16.Shimomura I., Bashmakov Y., Ikemoto S., Horton J. D., Brown M. S., Goldstein J. L. 1999. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. USA. 96: 13656–13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rimoldi O. J., Finarelli G. S., Brenner R. R. 2001. Effects of diabetes and insulin on hepatic delta6 desaturase gene expression. Biochem. Biophys. Res. Commun. 283: 323–326. [DOI] [PubMed] [Google Scholar]
- 18.Marquardt A., Stohr H., White K., Weber B. H. 2000. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 66: 175–183. [DOI] [PubMed] [Google Scholar]
- 19.Schaeffer L., Gohlke H., Muller M., Heid I. M., Palmer L. J., Kompauer I., Demmelmair H., Illig T., Koletzko B., Heinrich J. 2006. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum. Mol. Genet. 15: 1745–1756. [DOI] [PubMed] [Google Scholar]
- 20.Malerba G., Schaeffer L., Xumerle L., Klopp N., Trabetti E., Biscuola M., Cavallari U., Galavotti R., Martinelli N., Guarini P., et al. 2008. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids. 43: 289–299. [DOI] [PubMed] [Google Scholar]
- 21.Rzehak P., Heinrich J., Klopp N., Schaeffer L., Hoff S., Wolfram G., Illig T., Linseisen J. 2009. Evidence for an association between genetic variants of the fatty acid desaturase 1 fatty acid desaturase 2 (FADS1 FADS2) gene cluster and the fatty acid composition of erythrocyte membranes. Br. J. Nutr. 101: 20–26. [DOI] [PubMed] [Google Scholar]
- 22.Baylin A., Ruiz-Narvaez E., Kraft P., Campos H. 2007. alpha-Linolenic acid, Delta6-desaturase gene polymorphism, and the risk of nonfatal myocardial infarction. Am. J. Clin. Nutr. 85: 554–560. [DOI] [PubMed] [Google Scholar]
- 23.Gieger C., Geistlinger L., Altmaier E., Hrabé de Angelis M., Kronenberg F., Meitinger T., Mewes H. W., Wichmann H. E., Weinberger K. M., Adamski J., et al. 2008. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 4: e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka T., Shen J., Abecasis G. R., Kisialiou A., Ordovas J. M., Guralnik J. M., Singleton A., Bandinelli S., Cherubini A., Arnett D., et al. 2009. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 5: e1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aulchenko Y. S., Ripatti S., Lindqvist I., Boomsma D., Heid I. M., Pramstaller P. P., Penninx B. W., Janssens A. C., Wilson J. F., Spector T., et al. 2009. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 41: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kathiresan S., Willer C. J., Peloso G. M., Demissie S., Musunuru K., Schadt E. E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. 2009. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabatti C., Service S. K., Hartikainen A. L., Pouta A., Ripatti S., Brodsky J., Jones C. G., Zaitlen N. A., Varilo T., Kaakinen M., et al. 2009. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 41: 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. 2005. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 21: 2933–2942. [DOI] [PubMed] [Google Scholar]
- 29.Doring A., Gieger C., Mehta D., Gohlke H., Prokisch H., Coassin S., Fischer G., Henke K., Klopp N., Kronenberg F., et al. 2008. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat. Genet. 40: 430–436. [DOI] [PubMed] [Google Scholar]
- 30.Meisinger C., Prokisch H., Gieger C., Soranzo N., Mehta D., Rosskopf D., Lichtner P., Klopp N., Stephens J., Watkins N. A., et al. 2009. A genome-wide association study identifies three loci associated with mean platelet volume. Am. J. Hum. Genet. 84: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y. H., Dudoit S., Luu P., Lin D. M., Peng V., Ngai J., Speed T. P. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shore P., Bisset L., Lakey J., Waltho J. P., Virden R., Sharrocks A. D. 1995. Characterization of the Elk-1 ETS DNA-binding domain. J. Biol. Chem. 270: 5805–5811. [DOI] [PubMed] [Google Scholar]
- 33.Caspi A., Williams B., Kim-Cohen J., Craig I. W., Milne B. J., Poulton R., Schalkwyk L. C., Taylor A., Werts H., Moffitt T. E. 2007. Moderation of breastfeeding effects on the IQ by genetic variation in fatty acid metabolism. Proc. Natl. Acad. Sci. USA. 104: 18860–18865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinelli N., Girelli D., Malerba G., Guarini P., Illig T., Trabetti E., Sandri M., Friso S., Pizzolo F., Schaeffer L., et al. 2008. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. Am. J. Clin. Nutr. 88: 941–949. [DOI] [PubMed] [Google Scholar]
- 35.Ohnesorg T., Keller B., Hrabé de Angelis M., Adamski J. 2006. Transcriptional regulation of human and murine 17beta-hydroxysteroid dehydrogenase type-7 confers its participation in cholesterol biosynthesis. J. Mol. Endocrinol. 37: 185–197. [DOI] [PubMed] [Google Scholar]
- 36.Rao V. N., Huebner K., Isobe M., ar-Rushdi A., Croce C. M., Reddy E. S. 1989. elk, tissue-specific ets-related genes on chromosomes X and 14 near translocation breakpoints. Science 244: 66–70. [DOI] [PubMed] [Google Scholar]
- 37.Dalton S., Treisman R. 1992. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 68: 597–612. [DOI] [PubMed] [Google Scholar]
- 38.Clarkson R. W., Shang C. A., Levitt L. K., Howard T., Waters M. J. 1999. Ternary complex factors Elk-1 and Sap-1a mediate growth hormone-induced transcription of egr-1 (early growth response factor-1) in 3T3–F442A preadipocytes. Mol. Endocrinol. 13: 619–631. [DOI] [PubMed] [Google Scholar]
- 39.Hipskind R. A., Rao V. N., Mueller C. G., Reddy E. S., Nordheim A. 1991. Ets-related protein Elk-1 is homologous to the c-fos regulatory factor p62TCF. Nature. 354: 531–534. [DOI] [PubMed] [Google Scholar]
- 40.Nwankwo J. O., Spector A. A., Domann F. E. 2003. A nucleotide insertion in the transcriptional regulatory region of FADS2 gives rise to human fatty acid delta-6-desaturase deficiency. J. Lipid Res. 44: 2311–2319. [DOI] [PubMed] [Google Scholar]
- 41.Williard D. E., Nwankwo J. O., Kaduce T. L., Harmon S. D., Irons M., Moser H. W., Raymond G. V., Spector A. A. 2001. Identification of a fatty acid delta6-desaturase deficiency in human skin fibroblasts. J. Lipid Res. 42: 501–508. [PubMed] [Google Scholar]