Abstract
The APOA1/C3/A4/A5 gene cluster encodes important regulators of fasting lipids, but the majority of lipid metabolism takes place in the postprandial state and knowledge about gene regulation in this state is scarce. With the aim of characterizing possible regulators of lipid metabolism, we studied the effects of nine single nucleotide polymorphisms (SNPs) during postprandial lipid metabolism. Eighty-eight healthy young men were genotyped for APOA1 -2630 (rs613808), APOA1 -2803 (rs2727784), APOA1 -3012 (rs11216158), APOC3 -640 (rs2542052), APOC3 -2886 (rs2542051), APOC3 G34G (rs4520), APOA4 N147S (rs5104), APOA4 T29T (rs5092), and A4A5_inter (rs1263177) and were fed a saturated fatty acid-rich meal (1g fat/kg of weight with 60% fat, 15% protein and 25% carbohydrate). Serial blood samples were extracted for 11 h after the meal. Total cholesterol and fractions [HDL-cholesterol, LDL-cholesterol, trifacylglycerols (TGs) in plasma, TG-rich lipoproteins (TRLs) (large TRLs and small TRLs), apolipoprotein A-I and apolipoprotein B] were determined. APOA1 -2803 homozygotes for the minor allele and A4A5_inter carriers showed a limited degree of postprandial lipemia. Carriers of the rare alleles of APOA4 N147S and APOA4 T29T had lower APOA1 plasma concentration during this state. APOC3 -640 was associated with altered TG kinetics but not its magnitude. We have identified new associations between SNPs in the APOA1/C3/A4/A5 gene cluster and altered postprandial lipid metabolism.
Keywords: apolipoproteins, diet, gene-nutrient interaction, nutrigenomics, postprandial
Pronounced postprandial hypertriglyceridemia is proatherogenic (1). The extent of this phenomenon depends on several factors, both intrinsic and extrinsic. Diet is the main external determinant of postprandial lipemia magnitude. It has been stated that carbohydrate intake increases the plasma concentration of postprandial lipid particles [triacylglycerols (TGs) and VLDL] when replacing fat in the diet (2, 3). Dietary fat type also influences postprandial lipemia. Saturated fats induce a prolonged lipemia compared with other types of fat (4). Diets rich in monounsaturated fat provoke a faster postprandial TG-rich lipoprotein (TRL) clearance when compared with diets pronounced in saturated fat, thus, shortening the lipemia, which may be mediated by postprandial apolipoproteins (5) and TRL metabolism (6). Diets rich in N3 PUFA (>2.7–4 g/d) can lower the postprandial TG response (7). The effects of diet on postprandial metabolism were detailed elsewhere (4).
In addition to external determinants, others that are specific to the individual may modulate the response to dietary interventions. A number of genes have been identified as responsible for triglyceride concentration in both the fasting and postprandial states. The most studied genetic region with regard to lipid metabolism is that encoding the apolipoprotein genes APOA1, APOC3, APOA4, and APOA5, also known as the APOA1/C3/A4/A5 gene cluster (8).
Apolipoprotein A1 (APOA1), the main protein included in HDL cholesterol particles, is an essential element of reverse cholesterol transport (9), but it has been published that a genetic variation (−276 base pairs G/A) in the promoter region of this apolipoprotein is associated with altered postprandial lipid metabolism (10). Three other single nucleotide polymorphisms (SNPs) (APOA1 -2630G/A, APOA1 -2803G/A, and -3012A/G) in the promoter region were previously tested for their influence in fasting lipids and response to hypolipidemic medication (11). Although these variants were not associated with lipid concentrations, we decided to investigate their potential influence in postprandial lipids.
Apolipoprotein C-III (APOC3) is a component of TRLs and HDL, whose major function in lipid metabolism is to inhibit LPL and, thereby, plasma APOC3 concentrations are positively associated with TG concentrations (12). Several SNPs within APOC3 coding or promoter regions have been associated with altered triglycerides (4, 13–16). The three SNPs analyzed here were also previously studied: APOC3 -2886T/G, reported as −2854T/G, has been associated with altered fasting triglycerides (17). APOC3 -640, rs2542052, has been described as −641C/A and associated with longevity and HDL levels (18, 19), although other authors did not replicate the findings (18). The minor allele of the APOC3 synonymous G34G variant (rs4520, also known as 1100C>T) has been widely studied, first reported to associate with elevated triglyceride levels (20). Although these three SNPs were previously reported as putative functional SNPs, their influence in the postprandial situation has not been tested.
Apolipoprotein A-IV (APOA4) influences dietary fat absorption and chylomicron synthesis (4), modulates the activation of LPL by apolipoprotein C-II (21), and activates lecithin-cholesterol acyltransferase (22). Previous information on altered postprandial lipemia depending on APOA4 SNPs has been published. The carriers of the minor allele of the Gln360His variant have an increased postprandial lipemia when exposed to a saturated fatty acid-rich diet, as they have increased peaks of small TRL-cholesterol (CHOL), small TRL-TG, and large TRL-TG particles (23), but, interestingly, in a case control study (offspring of fathers who suffered a myocardial infarction before the age of 55 years vs. controls), the subgroup of controls who were carriers of the minor allele of this SNP presented lower body mass indices (BMIs), fasting cholesterol, and TG concentrations. Furthermore, and after consumption of an oral fat load, obese participants of this study (but not the rest of the population) who were carriers of the minor allele had significantly reduced postprandial lipemia (24). Carriers of the rare allele of Thr347Ser show a lower postprandial response in the TG levels of TRL remnants (25). APOA4 N147S has been linked to fasting triglyceride levels but its importance to postprandial lipemia remains unknown (26). APO4T29T is a synonymous SNP located in the coding region of APOA4 that has not been previously tested as a functional SNP. Both because of its location and its being a synonymous SNP, we selected it for the present study.
Apolipoprotein V (APOA5) regulates TG metabolism by mechanisms that include hepatic VLDL and TRL catabolism (27). Some variations in this gene (T-1131C and Ser19Trp) have been linked to altered fasting and postprandial TG (28, 29). Haplotype analysis based on five polymorphisms (1131T>C, c.-3A>G, c.56C>G, IVS3+476G>A, and c.1259T>C) in the APOA5 gene define three common haplotypes (APOA5*1, APOA5*2, and APOA5*3) (30). APOA5*2 and APOA5*3 carriers have a higher postprandial response, higher area under the curve of total plasma TG, large TRL-TG, small TRL-TG, small TRL-CHOL, and large TRL-CHOL than subjects with the APOA5*1 haplotype (31).
In the intergenic region between APOA4 and APOA5 resides A4A5_inter (rs1263177) SNP. Although its location suggests it might be a nonfunctional variant, an association of this SNP with altered TG levels has been reported. Talmud et al. (32), found a lower level of fasting TG in homozygotes for the minor allele compared with common allele homozygotes. Furthermore, the same SNP was further studied by the same group, but interestingly, this time the results were distinct; although the SNP did not affect the postprandial lipids after an oral fat tolerance test, homozygosity for the rare allele was associated with higher waist-to-hip ratio, systolic blood pressure, and area under the curve (AUC) and peak of insulin after an oral glucose tolerance test (33). These contradicting reports and the repetitive positive phenotype-genotype results made this SNP a good candidate for eventual study in lipid metabolism.
The precise role of the regulatory elements present throughout the APOA1/C3/A4/A5 cluster is complex and incomplete, with numerous interactions with different transcription factors described in the immediate upstream regions of each gene (8). Both the study of the regulatory elements in the cluster region as well as the significance of the tight linkage disequilibrium (LD) between the SNPs in this region (highly influenced by the population under study) remain an active field of research (34–37).
The specific purpose of this study was to further characterize SNPs in the APOA1/C3/A4/A5 cluster, describing their relationship with postprandial lipemia. With this aim, we genotyped three SNPs at APOA1 (all in the promoter region: APOA1 -2630, APOA1 -2803, APOA1 -3012), three at APOC3 (APOC3 -2886, APOC3 -640, APOC3 G34G), two in the APOA4 region (APOA4 N147S, APOA4 T29T), and one SNP in the intergenic region of APOA4/APOA5 (A4A5_inter), in order to investigate possible associations to measures of blood lipids during the time of postprandial lipid metabolism.
MATERIALS AND METHODS
Subjects
Eighty-eight healthy men aged 18 to 33 years were enrolled in two studies conducted by the Lipids and Atherosclerosis Research Unit at Reina Sofia University Hospital. The first study included 50 participants and the second study included 38 patients. All tests were performed using the same methodology as described below. We included only young normolipemic APOE E3/E3 males in order to avoid possible effects of different APOE isoforms or gender. Other results of these studies have been published elsewhere (10, 23, 25, 28, 31, 38–45). No participants had diabetes or liver, renal, or thyroid disease, nor were they taking any medication. Anthropometric measures (weight, height, and BMI) and blood pressure were assessed and all subjects were encouraged to maintain regular lifestyle and levels of physical activity. All volunteers had plasma cholesterol and TG concentrations below 200 mg/dl. Baseline characteristics of the participants are summarized in Table 1
TABLE 1.
Baseline characteristics of the participants
| Variable | APOA1 -2630 | APOA1 -2803 | APOA1 -3012 | APOC3 -2886 | APOC3 -640 | APOC3 G34G | APOA4 N147S | APOA4 T29T | APOA4A5_inter |
|---|---|---|---|---|---|---|---|---|---|
| Age(years) | GG 22.1 ± 2.1 | GG 22.2 ± 2.1 | AA 22.6 ± 4.3 | AA 22.7 ± 3.9 | CC 21.7 ± 1.5 | CC 21.9 ± 2.1 | GG 22.5 ± 3.3 | AA 22.5 ± 3.4 | TT 22.3 ± 3.7 |
| GA 22.5 ± 3.8 | GA 22.4 ± 3.6 | AG 22.5 ± 1.9 | AC 22 ± 2.1 | CA 22.4 ± 3.4 | CT 22.6 ± 3.7 | GA 21.9 ± 2.6 | AG 21.6 ± 2.5 | TC 22.3 ± 2.5 | |
| AA 20.8 ± 2.1 | AA 21 ± 1.6 | GG 21.4 ± 2.9 | CC 22.2 ± 3.5 | AA 22.6 ± 3.5 | TT 21 ± 2 | AA 22.3 ± 3.1 | GG 21.3 ± 3.1 | CC 21.7 ± 2.7 | |
| p 0.47 | p 0.5 | p 0.35 | p 0.58 | p 0.52 | p 0.48 | p 0.38 | p 0.25 | p 0.74 | |
| BMI(kg/m2) | GG 25.7 ± 2.8 | GG 25.6 ± 2.9 | AA 25 ± 2.7 | AA 25.3 ± 2.6 | CC 25.5 ± 2.8 | CC 25.6 ± 2.9 | GG 25.4 ± 2.7 | AA 25.4 ± 2.8 | TT 25.6 ± 2.6 |
| GA 24.4 ± 2.7 | GA 24.7 ± 2.8 | AG 25.1 ± 2.6 | AC 25 ± 2.9 | CA 24.8 ± 2.8 | CT 24.3 ± 2.7 | GA 24.3 ± 2.9 | AG 24.1 ± 2.9 | TC 24.9 ± 3.1 | |
| AA 23.2 ± 2.5 | AA 23.1 ± 2.2 | GG 24.5 ± 3.4 | CC 24.3 ± 3 | AA 24.1 ± 2.9 | TT 24.2 ± 0.8 | AA 23.5 ± 2.8 | GG 24 ± 6.8 | CC 24.1 ± 2 | |
| p 0.12 | p 0.11 | p 0.77 | p 0.64 | p 0.37 | p 0.09 | p 0.2 | p 0.06 | p 0.22 | |
| CHOL(mg/dl) | GG 151.7 ± 18.8 | GG 152 ± 18.9 | AA 139.6 ± 38.6 | AA 140.1 ± 35.6 | CC 138 ± 35.9 | CC 150.4 ± 18.6 | GG 143.6 ± 34.6 | AA 146.3 ± 26.5 | TT 141 ± 43 |
| GA 139.7 ± 37.5 | GA 142.6 ± 36.1 | AG 147.8 ± 30 | AC 148.1 ± 26.9 | CA 148.1 ± 27 | CT 139.8 ± 37.5 | GA 150.4 ± 21.5 | AG 151.4 ± 21.1 | TC 149.5 ± 20.6 | |
| AA 141.5 ± 23.3 | AA 139.6 ± 18.1 | GG 147.5 ± 13.9 | CC 150.3 ± 18.9 | AA 151.8 ± 18.4 | TT 159.5 ± 20.6 | AA 146 ± 30.6 | GG 146 ± 30.6 | CC 142.1 ± 15.9 | |
| p 0.17 | p 0.32 | p 0.53 | p 0.44 | p 0.26 | p 0.19 | p 0.34 | p 0.38 | p 0.43 | |
| TG(mg/dl) | GG 75.4 ± 35.4 | GG 73.3 ± 31.8 | AA 80.6 ± 35.8 | AA 81.5 ± 31.8 | CC 81.3 ± 32.4 | CC 74.5 ± 35.8 | GG 81.1 ± 36.2 | AA 79.9 ± 38 | TT 92.8 ± 41.2 |
| GA 85.8 ± 34 | GA 88 ± 36.3 | AG 81.8 ± 35.7 | AC 78.4 ± 32.8 | CA 78.7 ± 32.4 | CT 86.9 ± 33 | GA 82.1 ± 34 | AG 82.3 ± 33.7 | TC 71.1 ± 22.8 | |
| AA 67.4 ± 21.4 | AA 60.4 ± 18.7 | GG 72.4 ± 24.1 | CC 86.2 ± 53.1 | AA 84.9 ± 50.3 | TT 58.9 ± 9.1 | AA 81.5 ± 35.3 | GG 81.5 ± 45.3 | CC 81.1 ± 40.5 | |
| p 0.29 | p 0.05 | p 0.54 | p 0.8 | p 0.85 | p 0.14 | p 0.9 | p 0.78 | p 0.03 | |
| HDL(mg/dl) | GG 47 ± 10.5 | GG 47.1 ± 9.1 | AA 44.8 ± 12.2 | AA 45.9 ± 9.8 | CC 46.2 ± 9.3 | CC 46.8 ± 10.8 | GG 46.6 ± 10.8 | AA 46.6 ± 10.6 | TT 45.3 ± 10.6 |
| GA 45.8 ± 10.3 | GA 45.6 ± 11 | AG 47.2 ± 10.1 | AC 45.8 ± 9.2 | CA 45.8 ± 9.4 | CT 45.8 ± 10 | GA 45.1 ± 9.5 | AG 45.1 ± 9.8 | TC 46 ± 9.4 | |
| AA 46.2 ± 7.5 | AA 47.5 ± 10.5 | GG 47.2 ± 8.3 | CC 50.5 ± 16.5 | AA 49.3 ± 16 | TT 48.8 ± 5.4 | AA 46.1 ± 10.3 | GG 44.1 ± 20.3 | CC 49 ± 11.8 | |
| p 0.86 | p 0.78 | p 0.62 | p 0.43 | p 0.61 | p 0.84 | p 0.52 | p 0.55 | p 0.5 | |
| LDL(mg/dl) | GG 89.5 ± 15.4 | GG 90.3 ± 16.2 | AA 93.2 ± 24.4 | AA 89.1 ± 23.9 | CC 88.1 ± 22.5 | CC 88.6 ± 15.7 | GG 91.3 ± 22.7 | AA 87.6 ± 19.3 | TT 95.5 ± 26.3 |
| GA 90.2 ± 26.5 | GA 91.1 ± 24.6 | AG 89.6 ± 23.6 | AC 90.8 ± 20.4 | CA 90.6 ± 21.3 | CT 89.7 ± 26.5 | GA 88.9 ± 20.2 | AG 89.8 ± 20.5 | TC 89.3 ± 18.4 | |
| AA 81.8 ± 23.3 | AA 80.1 ± 16.8 | GG 85.9 ± 11.9 | CC 82.1 ± 20.3 | AA 85.2 ± 21.4 | TT 98.9 ± 15.1 | AA 90.4 ± 21.8 | GG 90.4 ± 21.8 | CC 76.9 ± 14.1 | |
| p 0.76 | p 0.44 | p 0.5 | p 0.54 | p 0.73 | p 0.72 | p 0.63 | p 0.64 | p 0.02 | |
| APOA1(mg/dl) | GG 108.3 ± 19.3 | GG 109.1 ± 20.1 | AA 106 ± 22.4 | AA 109.4 ± 19.6 | CC 110.3 ± 19 | CC 108.8 ± 20.7 | GG 109.6 ± 20.3 | AA 110 ± 20.3 | TT 106.5 ± 18.2 |
| GA 103.9 ± 20.6 | GA 103.6 ± 19.3 | AG 107 ± 19.4 | AC 105.6 ± 19.3 | CA 105.3 ± 19.6 | CT 104.4 ± 18.7 | GA 99 ± 17.5 | AG 100.3 ± 17.2 | TC 105.7 ± 19.6 | |
| AA 114 ± 7.9 | AA 111.4 ± 20.1 | GG 105.6 ± 18.8 | CC 102.4 ± 22 | AA 101.6 ± 20.9 | TT 103.3 ± 19.1 | AA 105.8 ± 19.9 | GG 103.8 ± 19.9 | CC 108.4 ± 22.8 | |
| p 0.45 | p 0.37 | p 0.96 | p 0.56 | p 0.39 | p 0.56 | p 0.02 | p 0.04 | p 0.89 | |
| APOB(mg/dl) | GG 70.6 ± 13.3 | GG 70.4 ± 13.5 | AA 68.8 ± 19.3 | AA 67.5 ± 17.3 | CC 67.1 ± 16.4 | CC 69.4 ± 13.4 | GG 69.2 ± 16.1 | AA 67.8 ± 15.2 | TT 70.7 ± 19.2 |
| GA 65.2 ± 19.2 | GA 67.6 ± 18.4 | AG 69.1 ± 18.3 | AC 68.4 ± 15.5 | CA 68.3 ± 16.2 | CT 65.3 ± 19.2 | GA 65.8 ± 18.3 | AG 65.9 ± 18.3 | TC 67.5 ± 13.4 | |
| AA 59.8 ± 13.3 | AA 57.3 ± 11.3 | GG 64.2 ± 9 | CC 62.8 ± 20.4 | AA 63.2 ± 19.3 | TT 73.7 ± 13.6 | AA 68 ± 16.9 | GG 64 ± 24.9 | CC 60.8 ± 17.7 | |
| p 0.21 | p 0.16 | p 0.5 | p 0.65 | p 0.67 | p 0.43 | p 0.4 | p 0.63 | p 0.15 |
The p value in each cell corresponds to univariate ANOVA, with each genotype as independent factor and each phenotype variable as dependent factor. Within each cell, the upper genotype corresponds to homozygotes for the major allele, the intermediate to heterozygotes, and the lower to homozygotes for the minor allele. All values are mean ± SD. Grand means for the variables are: Age 22.46 ± 4.11; BMI 25.38 ± 3.62; CHOL 151.4 ± 23.07; TG 80.4 ± 35.1; HDL 46.3 ± 10.2; LDL 89.1 ± 21,5; APOA1 106.4 ± 19.5; APOB 67.4 ± 16.5.
The studies in which these participants were enrolled were approved by the Ethics Committee for Clinical Investigations of the Reina Sofía University Hospital in Cordoba, and participants previously signed an informed consent to join the study.
Study design
After an overnight 12-h fast, subjects were given a fatty meal enriched with 60,000 units of Vitamin A per m2 body surface area. The amount of fat given was 1 g of fat and 7 mg of cholesterol per kg body weight. This meal contained 60% of its energy in the form of fat (35% SAT, 19% MUFA, 6.3% PUFA), 15% as protein, and 25% as carbohydrate and was consumed within 20 min. After the meal, subjects were not allowed another energy intake for 11 h, but were permitted to drink water. Blood samples were drawn just prior to the meal and postprandially every h until 6 h then every 2 h and 30 min until 11 h. Taking samples at such late time points allowed many lipid measures to return to near fasting levels.
Biochemical determinations
SNP selection, DNA amplification, and genotyping.
SNPs within the four-gene cluster were selected for genotyping based on previous reports of associations to blood lipid measures and functional characterization of the alleles. We selected the SNPs at the APOA1/C3/A4/A5 gene cluster based on the following criteria in order of importance: i) validation status; that is, a proven polymorphism in Caucasians; ii) functional relevance and importance, namely, choosing potential functional SNPs residing within transcription factor binding sites, in exons that change amino acid sequences, or at exon–intron boundaries that alter mRNA splicing; iii) degree of heterozygosity; that is, minor allele frequencies greater than 0.05; and iv) earlier evidence of association with lipid measurements. To be genotyped, SNPs must meet at least one of the above criteria. Using a human codon frequency table based on 40,662,582 codons from 93,487 mRNAs (47), synonymous SNPs were assessed for a change in codon usage frequency. Incorporation of a rare or common codon can affect protein secondary structure (48). Reference genomic sequence of SNPs in gene control regions was analyzed with MAPPER to determine allele-specific transcription factor binding sites (49). Six of the selected SNPs map upstream of corresponding genes in regions likely to control transcription and are listed in Supplementary Table I with a “minus” designation. The SNP intergenic to APOA4 and APOA5(32) maps 28 kbp upstream of APOA5 and 706 bp downstream of the end site of APOA4 transcription. SNPs were genotyped using the Applied Biosystems TaqMan assay (45, 50, 51). Allele discrimination was performed on PCR products. Fluorescence data were collected by a 7900 Sequence Detection System (Applied Biosystems) (50).
Lipoprotein separation and lipid analysis.
Large and small TRLs were manually extracted after centrifugation in subdued light as previously described, and samples were stored at −70°C until analyzed (45). CHOL and TGs in plasma and lipoprotein fractions were assayed by enzymatic procedures (52, 53). APOA1 and APOB were determined by turbidimetry (54). HDL-C was measured by analyzing the supernatant obtained following precipitation of a plasma aliquot with dextran sulfate-Mg2+, as described by Warnick et al. (55). LDL-C levels were estimated using the Friedewald formula, based on the CHOL, TG, and HDL-C values (56).
Statistical analysis
Genotypic analysis.
LD was tested using Helix-Tree software pertinent tools (Helix-Tree, version 4.3.2. Golden Helix, Bozeman, MT). Likelihood ratio test was used for determining the existence of LD. When LD was observed, r2 was used to analyze the strength of association. Using r2 values, we classified LD as weak (<0.30), moderate (0.30–0.80) or strong (>0.80). Hardy-Weinberg equilibrium (HWE) was tested by the Fisher's exact test.
Analysis of lipid parameters.
The existence of a postprandial state after the high-fat meal was investigated by a repeated measures ANOVA test comparing the concentration of the different lipid particles at fasting versus 4 h after the meal. The influence of the SNPs on the size of the postprandial lipid fractions was analyzed by one-way ANOVA for AUC, defined as the area between the plasma concentration versus time curve, using the trapezoidal rule, with the SNPs included as independent factors and BMI and age as covariates. For any of the lipid fractions studied, a linear regression model was constructed to determine the influence of the covariates in the dependent variables. Bonferroni or Games-Howell adjustments were used, depending on Levene's test for homogeneity of variances. If the variances were homogeneous, the method for correction was Bonferroni adjustment, whereas nonhomogeneous variances called for using the Games-Howell method. Repeated-measures ANOVA was also used for both the overall gene influence (global ANOVA, p for gene influence), the kinetics of the response (p for time), and the interaction of both factors (time · gene). When statistical differences were found in the repeated-measures ANOVA, a multiple comparisons test adjusted by Bonferroni's rule was applied to identify differences among the genetic isoforms on each time point of extraction. When minor allele homozygotes were less than 5% or a dominant effect of the allele was inferred, we also studied the stratified data of carriers of the mutant allele versus the homozygotes for the common allele. As a multivariate approach to test the independence of the influences of the SNPs in the postprandial lipids when two or more SNPs showed influence on the same lipid parameter, these SNPs were subsequently entered as fixed factors in pairwise combinations into an ANOVA to determine if any nonindependent genetic effects were present (age and BMI were included as covariates). Time to maximum peak of TG and large TRL-TG was assessed as the central point at which the sum of three consecutive time-points values was maximum. A value of less than 0.05 was considered significant. All data presented in the text and tables are expressed as mean ± SE unless otherwise specified. SPSS 15.0 for Windows (SPSS Inc., Chicago, IL) was used for statistical comparisons.
RESULTS
The C allele of APOC3 G34G is part of a GGC codon for glycine and is found in 22.2 per 1,000 human codons (47). The rarer T allele at this position yields a codon of GGT, found in 10.8 per 1,000 codons or 2.1-fold less frequently. Similarly, the minor G allele for the ACG threonine codon of APOA4 T29T occurs in 6.1 per 1,000 codons, whereas the ACA codon occurs in 15.1 per 1,000 codons or 2.5-fold more frequently (47). Although synonymous, variants invoking a rare or common codon can affect protein secondary structure (48). Of the six SNPs mapping outside protein-coding regions, APOA1 -2630, APOC3 -2886, APOC3 -640, and APOA4A5_inter showed putative allele-specific transcription factor binding sites. The strongest predictions were an EBF1 (early B-cell factor 1) binding site for the C allele of APOC3 -640 and an SRF (serum response factor) binding site with the A allele of APOA1 -2630. These results are summarized in Supplementary Table II.
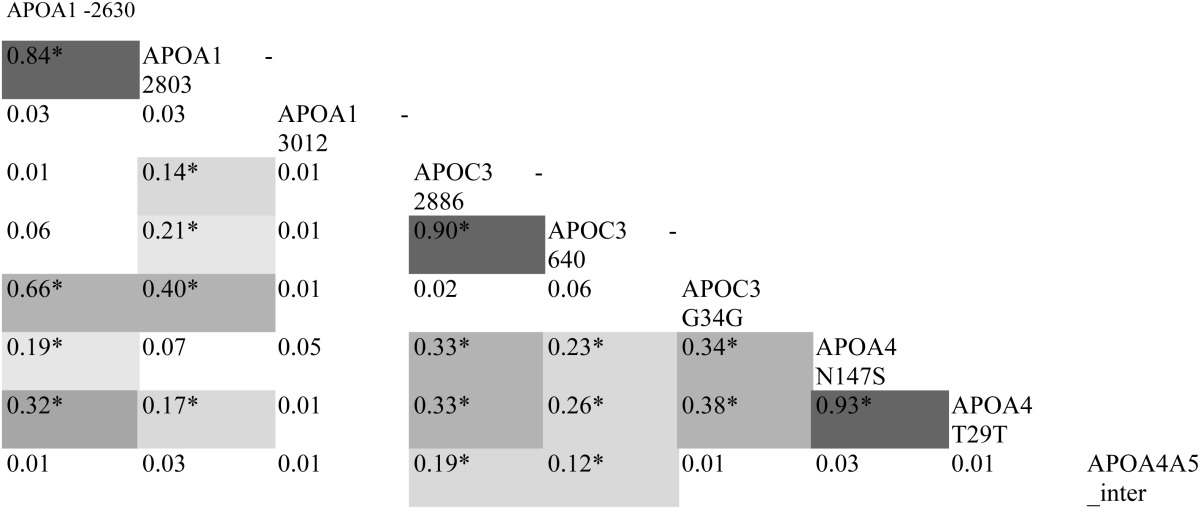
Baseline characteristics of the participants of the study are summarized in Table 1. Data on genotype distribution and HWE are shown in Table 2. The LD analysis showed the existence of LD between many of the SNPs present in this cluster. We found strong LD (r2 > 0.80) between APOA1 -2630 and -2803; between APOC3 -2886 and -640; and between APOA4 N147S and T29T. Additional LD was also found between SNPs located in different gene regions with r2 ranging from 0.14 to 0.66 as shown in Table 3. The repeated-measures ANOVA test showed that the high-fat challenge raised values of postprandial plasma TG, large TRL-TG, and small TRL-TG with respect to fasting values (all P < 0.001, data not shown). Linear regression models showed that the covariates used (BMI and age) were independent predictors of AUC of CHOL, TG, large TRL-TG, small TRL-CHOL, and APOB. Age also showed influence on large TRL-CHOL and small TRL-TG. BMI, but not age, influenced HDL-C. In our study, AUC of APOA1 and HDL-C showed a tight correlation. Pearson correlation was 0.519 (p for significance <0.001). Furthermore, in all time points the correlation was significant, all P < 0.05, with Pearson correlations ranging from 0.357 to 0.606. The influence of the SNPs on postprandial lipid levels is described below and is summarized in Table 4.
TABLE 2.
Genotype and allele frequencies of the selected SNPs, and Hardy-Weinberg equilibrium test results
| Genotype Frequency, Number of Individuals |
|||||
|---|---|---|---|---|---|
| SNP | Major Allele Homozygotes | Heterozygotes | Minor Allele Homozygotes | MAF | HWE (p) |
| APOA1 -2630(G/A)(rs613808) | 42 | 42 | 4 | 0.28 | 0.12 |
| APOA1 -2803(G/A)(rs2727784) | 47 | 32 | 9 | 0.28 | 0.43 |
| APOA1 -3012 (A/G)(rs11216158) | 25 | 39 | 24 | 0.49 | 0.39 |
| APOC3 -2886 (A/C)(rs2542051) | 31 | 47 | 10 | 0.38 | 0.26 |
| APOC3 -640 (C/A)(rs2542052) | 28 | 50 | 10 | 0.40 | 0.12 |
| APOC3 G34G (C/T)(rs4520) | 42 | 42 | 4 | 0.28 | 0.12 |
| APOA4 N147S (G/A)(rs5104) | 56 | 31 | 1 | 0.19 | 0.18 |
| APOA4 T29T (A/G)(rs5092) | 50 | 36 | 2 | 0.22 | 0.14 |
| APOA4A5_inter (T/C)(rs1263177) | 30 | 42 | 16 | 0.42 | 0.99 |
Values represent number of subjects. MAF, minor allele frequency; HWE, Hardy-Weinberg equilibrium.
TABLE 3.
Pair-wise linkage disequilibrium (LD), expressed as r2 coefficients
TABLE 4.
Area under the curve of lipid fractions in the postprandial study (mean± SE)
| APOA1 -2630 (G/A) rs613808 | APOA1 -2803(G/A) rs2727784 | APOA1 -3012 (A/G) rs11216158 | APOC3 -2886 (A/C) rs2542051 | APOC3 -640 (A/C) rs2542052 | APOC3 G34G (C/T) rs4520 | APOA4 N147S (G/A) rs5104 | APOA4 T29T (A/G) rs5092 | APOA4A5 _inter (T/C) rs1263177 | |
|---|---|---|---|---|---|---|---|---|---|
| Total TG | GG 82.6 ± 6.0 | GG 80.2 ± 6.2 | AA 92.5 ± 9.5 | AA 91.4 ± 21.4 | CC 90.2 ± 8.3 | CC 86.6 ± 17.6 | GG 89.8 ± 6.3 | AA 87.2 ± 6.8 | TT 107.2 ± 9.9d |
| GA 96.3 ± 7.3 | GA 99.3 ± 6.9 | AG 91.1 ± 7.0 | AC 90.0 ± 6.2 | CA 86.9 ± 5.7 | CT 95.4 ± 6.8 | GA 91.1 ± 7.7 | AG 89.7 ± 7.4 | TC 79.8 ± 5.3 | |
| AA 60.5 ± 4.8 | AA 55.2 ± 3.9a | GG 74.9 ± 5.5 | CC 88.6 ± 7.6 | AA 100.2 ± 21.1 | TT 80.9 ± 6.4 | AA 86.9 ± 8.7 | GG 88.5 ± 8.9 | CC 82.1 ± 10.4 | |
| CHOL | GG 98.5 ± 2.1 | GG 98.5 ± 2.4 | AA 100.6 ± 3.0 | AA 97.4 ± 4.4 | CC 96.9 ± 3.0 | CC 106.9 ± 9.5 | GG 100.4 ± 2.4 | AA 97.7 ± 2.4 | TT 103.6 ± 3.3e |
| GA 99.7 ± 3.0 | GA 100.0 ± 2.7 | AG 99.4 ± 3.0 | AC 99.3 ± 2.3 | CA 99.4 ± 2.4 | CT 99.7 ± 2.9 | GA 97.6 ± 2.7 | AG 97.9 ± 2.6 | TC 97.3 ± 2.4 | |
| AA 90.7 ± 4.4 | AA 90.2 ± 3.1 | GG 95.7 ± 2.8 | CC 98.0 ± 3.1 | AA 98.4 ± 4.0 | TT 97.1 ± 2.1 | AA 96.7 ± 2.0 | GG 96.8 ± 5.8 | CC 91.9 ± 2.6 | |
| large-TRL TG | GG 33.4 ± 3.2 | GG 34.0 ± 3.8 | AA 36.7 ± 4.8 | AA 34.8 ± 10.8 | CC 37.5 ± 4.5 | CC 44.1 ± 12.3 | GG 35.2 ± 2.9 | AA 34.6 ± 3.1 | TT 42.4 ± 4.7 |
| GA 37.4 ± 3.5 | GA 38.3 ± 3.1 | AG 37.3 ± 3.7 | AC 35.3 ± 2.9 | CA 33.8 ± 2.6 | CT 36.7 ± 3.3 | GA 37.6 ± 4.3 | AG 37.1 ± 4.1 | TC 31.2 ± 2.6 | |
| AA 23.2 ± 3.8 | AA 19.2 ± 3.0a | GG 28.1 ± 2.4 | CC 36.6 ± 4.1 | AA 40.0 ± 11.0 | TT 32.3 ± 3.7 | AA 36.5 ± 6.1 | GG 35.6 ± 6.1 | CC 35.1 ± 6.8 | |
| small-TRL TG | GG 27.3 ± 2.9 | GG 29.3 ± 3.4 | AA 25.5 ± 3.0 | AA 19.5 ± 4.5 | CC 28.7 ± 3.1 | CC 25.5 ± 3.5 | GG 30.0 ± 2.5 | AA 29.6 ± 2.8 | TT 30.7 ± 3.1 |
| GA 29.1 ± 2.2 | GA 28.5 ± 2.0 | AG 31.7 ± 3.2 | AC 30.6 ± 2.6 | CA 30.1 ± 2.4 | CT 29.6 ± 2.3 | GA 28.3 ± 2.6 | AG 29.7 ± 2.9 | TC 27.5 ± 2.7 | |
| AA 33.3 ± 11.9 | AA 24.9 ± 7.7 | GG 27.6 ± 2.9 | CC 28.5 ± 2.8 | AA 21.9 ± 4.7 | TT 27.4 ± 2.9 | AA 29.3 ± 7.9 | GG 28.6 ± 7.3 | CC 27.9 ± 3.8 | |
| large-TRL CHOL | GG 5.5 ± 0.3 | GG 5.5 ± 0.3 | AA 5.5 ± 0.4 | AA 5.7 ± 0.7 | CC 5.6 ± 3.7 | CC 5.4 ± 0.6 | GG 5.4 ± 0.3 | AA 5.4 ± 0.3 | TT 6.5 ± 0.4d |
| GA 5.3 ± 0.3 | GA 5.4 ± 2.9 | AG 5.2 ± 0.2 | AC 5.2 ± 0.3 | CA 5.2 ± 2.6 | CT 5.3 ± 0.3 | GA 5.3 ± 0.4 | AG 5.4 ± 0.4 | TC 4.8 ± 0.3 | |
| AA 4.9 ± 1.4 | AA 4.6 ± 0.7 | GG 5.4 ± 0.4 | CC 5.6 ± 0.3 | AA 5.9 ± 1.7 | TT 5.5 ± 0.3 | AA 5.6 ± 0.3 | GG 5.9 ± 0.9 | CC 5.2 ± 0.4 | |
| small-TRL CHOL | GG 7.5 ± 0.7 | GG 7.1 ± 0.6 | AA 7.1 ± 1.0 | AA 6.8 ± 2.6 | CC 7.1 ± 0.6 | CC 7.3 ± 0.2 | GG 7.5 ± 0.6 | AA 7.4 ± 0.7 | TT 8.2 ± 0.8 |
| GA 7.2 ± 0.6 | GA 7.9 ± 0.6 | AG 8.0 ± 0.7 | AC 7.5 ± 0.5 | CA 7.5 ± 0.5 | CT 7.2 ± 0.5 | GA 7.4 ± 0.6 | AG 7.7 ± 0.7 | TC 6.9 ± 0.5 | |
| AA 6.7 ± 1.5 | AA 5.2 ± 1.1 | GG 6.5 ± 0.6 | CC 7.2 ± 0.6 | AA 7.1 ± 2.3 | TT 7.4 ± 0.7 | AA 7.1 ± 3.3 | GG 7.9 ± 5.3 | CC 7.0 ± 1.1 | |
| APOA1 | GG 69.2 ± 1,8 | GG 69.6 ± 2.1 | AA 67.0 ± 2.5 | AA 65.1 ± 4.5 | CC 69.5 ± 2.1 | CC 66.6 ± 4.7 | GG 69.5 ± 1.6b | AA 69.4 ± 1.7C | TT 68.0 ± 2.0 |
| GA 66.0 ± 1,8 | GA 66.3 ± 1.5 | AG 68.1 ± 1.8 | AC 67.6 ± 1.6 | CA 67.6 ± 1.6 | CT 66.8 ± 1.6 | GA 63.7 ± 2.0 | AG 64.4 ± 1.9 | TC 67.4 ± 1.8 | |
| AA 72.6 ± 2,3 | AA 69.2 ± 4.8 | GG 68.0 ± 2.4 | CC 69.4 ± 2 | AA 64.7 ± 4.0 | TT 69.0 ± 1.9 | AA 64.0 ± 0.3 | GG 64. 0 ± 3.0 | CC 68.9 ± 2.7 | |
| APOB | GG 44.6 ± 1,4 | GG 44.6 ± 1.5 | AA 44.0 ± 2.6 | AA 39.0 ± 4.7 | CC 42.6 ± 2 | CC 47.9 ± 6.6 | GG 44.4 ± 1.6 | AA 43.4 ± 1.6 | TT 45.8 ± 2.3e |
| GA 42.2 ± 2,1 | GA 43.6 ± 1.9 | AG 43.9 ± 2 | AC 43.7 ± 1.6 | CA 44.0 ± 1.6 | CT 42.3 ± 2.1 | GA 41.5 ± 2.2 | AG 41.5 ± 2.1 | TC 43.2 ± 1.5 | |
| AA 36.8 ± 3,1 | AA 35.2 ± 2.3 a | GG 41.3 ± 1.7 | CC 43.4 ± 2.1 | AA 39.4 ± 4.2 | TT 43.7 ± 1.4 | AA 42. 0 ± 2.0 | GG 40.6 ± 1.9 | CC 37.5 ± 2.7 | |
| HDL | GG 29.9 ± 1.0 | GG 29.9 ± 1.0 | AA 29.0 ± 1.7 | AA 32.3 ± 3.7 | CC 29.0 ± 1.1 | CC 32.2 ± 3.0 | GG 29.5 ± 1.0 | AA 29.4 ± 1.0 | TT 28.5 ± 1.2 |
| GA 29.1 ± 1.1 | GA 28.9 ± 1.0 | AG 29.7 ± 1.1 | AC 29.2 ± 0.9 | CA 29.3 ± 0.9 | CT 29.4 ± 1.0 | GA 28.9 ± 1.2 | AG 28.7 ± 1.2 | TC 29.4 ± 0.9 | |
| AA 29.5 ± 2.1 | AA 30.9 ± 3.1 | GG 29.8 ± 1.0 | CC 28.9 ± 1.1 | AA 31.3 ± 3.4 | TT 29.4 ± 1.0 | AA 29.0 ± 1.0 | GG 28.3 ± 1.1 | CC 31.2 ± 2.0 |
Univariate ANOVA using BMI and age as covariates. TG, triacylglycerols; Chol, cholesterol; TRL: TG-rich lipoproteins. All values are expressed as (min·mg/dl)/103. Superscripts in cells means as follows: a=P < 0.05 APOA1 -2803 AA versus GG and AA versus GA; b=P < 0.05 APOA4 N147S GG versus GA and GG versus AA; c=P < 0.05 APOA4 T29T AA versus AG and AA versus GG; d= P < 0.05 APOA4A5_inter TT versus TC; e= P < 0.05 APOA4A5_inter TT versus CC.
APOA1
APOA1 -2803 AA (minor allele homozygotes) showed a smaller postprandial AUC of TG, large TRL-TG, and APOB than GA (P < 0.001 for TG and large TRL-TG and P = 0.013 for APOB) and GG (P = 0.005, P = 0.012, and P = 0.031, respectively) (Table 4). There was a tendency toward a smaller AUC for CHOL in AA subjects (P = 0.056 vs. GA and P = 0.086 vs. GG). The results of the repeated-measures ANOVA of the different time-point means confirmed the lower concentration of total TG and large TRL-TG for AA versus GA (P = 0.034 and P = 0.043, respectively). In the multiple comparisons analysis, the difference between AA and GA subjects for total TG was significant at baseline and at every time-point from 1 to 11 h (supplementary Fig. I). The trend for AA versus GG was similar but nonsignificant. The concentration of large TRL-TG was lower for AA versus GA at the 5-h and 6-h time points (supplementary Fig. II). APOB measures in AA subjects was lower than in GG individuals at the times of the 3-, 5-, and 11-h extractions and lower than GA at 5 and 11 h (supplementary Fig. III). We found no influence of the APOA1 -2630 or -3012 genotypes on the AUC of the different lipid fractions in the postprandial state that reached statistical significance although some trends were noted. Data are shown in Table 4.
APOC3
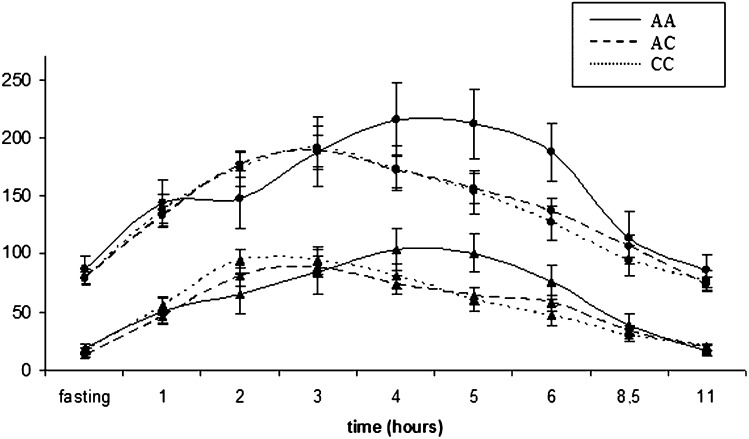
We found no influence of these APOC3 SNPs on the AUCs of the different lipid fractions. An early B-cell factor 1 (EBF1) binding site at -640 was predicted only for the C allele (supplementary Fig. IV). The EBF1 transcription factor is moderately expressed in all 75 human tissues tested but is highly expressed in human adipocytes, testis germ cells, uterus, and B cells (57). Repeated-measures ANOVA of APOC3 -640 revealed a gene*time interaction, (p for interaction = 0.004 for TG and 0.001 for large-TRL TG). The maximum peak of TG was observed at the third postprandial h (3.35 ± 0.11 h) for CC and CA and at the fourth h for AA (mean 4.03 ± 0.31 h). This difference was statistically significant in an ANOVA test between CC/CA versus AA (P = 0.047). The maximum peak for large-TRL TG was also retarded in AA homozygotes (AA 4.6 ± 0. 13 h vs. CC/CA 3.4 ± 0.40 h; P = 0.01). However, the multiple-comparisons test did not find statistically significant differences in the means of the TG concentrations at any time points (Fig. 1).
Fig. 1.
Postprandial evolution lines of total TG (upper lines, circles) and large-TRL TG (lower lines, triangles) depending on APOC3 -640 genotype.
APOA4
Heterozygotes for APOA4 N147S and T29T showed a smaller AUC of APOA1 levels than homozygotes for the common allele (APOA4 N147S GG 69531 ± 1562 vs. GA 63734 ± 2006, P = 0.040; T29T AA 69401 ± 1709 vs. AG 64408 ± 1930, P = 0.027). Repeated-measures ANOVA showed a higher concentration of APOA1 in homozygotes for the common allele versus heterozygotes for the two APOA4 SNPs (P = 0.023 for APOA4 N147S and P = 0.047 for T29T). Differences were observed at every time point from baseline to the eleventh h except at 6 h (P = 0.102) for APOA4 N147S (supplementary Fig. V), and at baseline and the first five time points for T29T (supplementary Fig. VI).
APOA4/APOA5 intergenic region
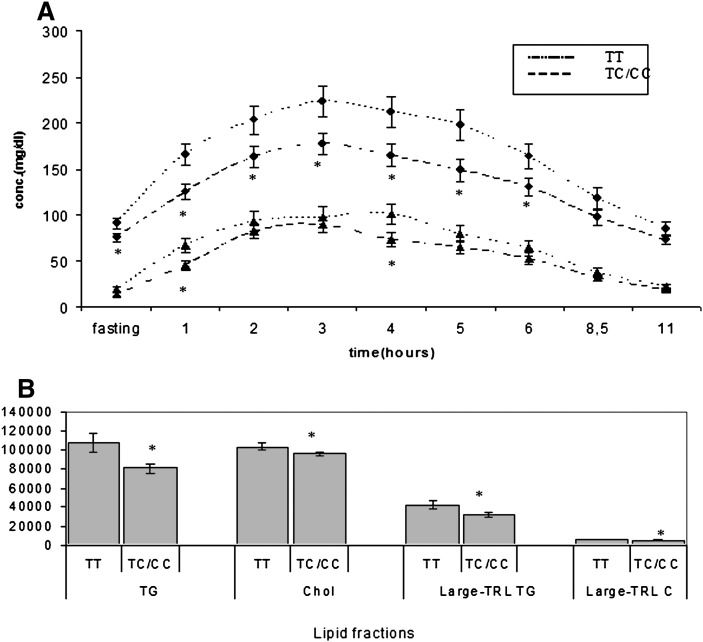
TT individuals of A4A5_inter (homozygotes for the common allele) showed a larger AUC of total TG and large TRL-CHOL than heterozygotes (P = 0.047 and P = 0.001) and a larger AUC of CHOL and APOB compared with CC group (P = 0.022 and P = 0.046 respectively) (Table 4). In view of the possibility of a dominant allele effect, we stratified the sample into carriers of the minor allele (groups TC and CC) and homozygotes for the common allele (group TT). Subsequent analyses showed a striking effect of the presence of the minor allele as it provoked lower TG (P = 0.009), CHOL (P = 0.042), large-TRL-TG (P = 0,045), and large-TRL-CHOL (P = 0.001) values (Fig. 2).
Fig. 2.
Postprandial lipid metabolism changes induced by APOA4A5_inter SNP. A: Postprandial evolution lines of Total TG (upper lines, circles) and large-TRL TG (lower lines, triangles). B: Postprandial AUC of lipid fractions. * = P < 0.05 APOA4A5_ inter TT versus TC/CC min·mg/dl.
Repeated-measures ANOVA of the means showed larger CHOL in TT compared with CC subjects (P = 0.048). These changes were observed at baseline and at every time point except at 8.5 h (supplementary Fig. VII). TG levels were higher in TT compared with TC (P = 0.019) and there was a similar but not significant trend when TT was compared with CC. The multiple-comparisons test resulted in a significantly higher mean TG at baseline and at the first six time points for TT versus TC (supplementary Fig. VIII). APOB measures showed a lower concentration in CC versus TT group (P = 0.045). The multiple-comparisons test showed differences at every time point except baseline (P = 0.054) (supplementary Fig. IX). Repeated measures for large TRL-TG global ANOVA did not show differences between genotypes (TT vs. TC, P = 0.109) but there was a gene*time interaction (P = 0.043), and the multiple-comparisons test showed a smaller large TRL-TG in TC than TT at baseline and at time points 4, 5, and 6 h (supplementary Fig. X). The evaluation of large TRL-CHOL showed a lower global mean for TC versus TT (P = 0.001) and a similar trend for CC versus TT (P = 0.076). The multiple-comparisons test showed differences at every time point for TC versus TT except at h 3 and at h 1, 2, and 8.5 for CC versus TT (supplementary Fig. XI).
Multivariate analysis
APOA1 -2803 and A4A5_inter were entered in a model to test for nonindependent gene effects in AUC of TG in a model that also included BMI and age. As expected, the model was predictive for the AUC of TG (P < 0,001; R2 = 0,358). The two SNPs remained significant in the corrected model (P = 0.001 for APOA1 -2803; P = 0.039 for A4A5_inter). The interaction of the two factors was not significant, confirming the independence of their effects on the TG. The same pair of SNPs was tested for independence of the effects shown in large TRL-TG. The subsequent model was predictive (P < 0,001; r2 = 0,324). The SNPs remained in the model (P = 0,028 for APOA1 -2803 and P = 0,049 for A4A5_inter), and the interaction factor between them was not significant.
APOA4 SNPs were also tested for independence of their effects on APOA1 concentration. In this case, when the two SNPs were entered as independent factors, the model was not predictive of the AUC of APOA1 (P = 0.266; r2 = 0.068), and significance was lost for the two SNPs, confirming that effects of these two SNPs were not independent of each other.
DISCUSSION
Although microarrays can interrogate thousands of genetic variations at once, the influence of individual or combinations of SNPs on clinical outcomes was our primary target. We selected the SNPs analyzed in the present study on the basis of their localization within the APOA1/C3/A4/A5 cluster (a region previously identified as an important regulator of lipid metabolism), previous studies on fasting lipids, and/or computational analysis of putative function. We analyzed the postprandial effects of nine SNPs in the APOA1/C3/A4/A5 cluster on various lipid fractions after administration of a meal high in saturated fatty acid content to 88 healthy young men. Homozygotes for the mutant allele for APOA1 -2803 and carriers of the minor allele of an APOA4_A5 intergenic polymorphism showed limited postprandial lipemia. The APOC3 -640 SNP showed association with altered postprandial lipemia kinetics, and APOA4 N147S and T29T SNPs were associated with changes in APOA1 concentrations during this stage. Four of the SNPs in this cluster did not display any influence on postprandial lipid metabolism in our study (APOA1 -2630, -3012; APOC3 -2886, G34G).
To the best of our knowledge, the influence of APOA1 -2803 in the postprandial lipid metabolism has never been studied. We observed that homozygotes for the minor allele displayed a lesser degree of postprandial lipemia as compared with other groups. APOA1 protein is mainly present in HDL-cholesterol. However, APOA1 is also present on the surface of nascent chylomicrons and it is in this capacity that the influence of the APOA1 -2803 variant on postprandial lipemia may be mediated by altered levels of APOA1 present on the surface of large TRL. This is supported by the fact that, despite the major influence that we have identified on TG and large TRL-TG, we found no effect of this SNP on postprandial metabolism of molecules, such as small TRL, whose surfaces do not contain APOA1. On the other hand, it appears that the modifications induced by this SNP might be due to an altered APOA1 function rather than on APOA1 transcription rate, because the presence of the minor allele did not influence APOA1 or HDL-C concentrations. The suggestion that the -2803 A/G polymorphism is in strong LD with an untested functional variant is strengthened by the lack of evidence for transcription factor binding at this position.
The APOC3 polymorphism at -640 has previously been studied in the Ashkenazi people (19), in whom homozygosity for the minor allele was over-represented in a cohort of older persons. Novelli et al. (18) eventually tested the relationship between longevity and the presence of this SNP (among other candidate SNPs) in a cohort of elderly American Caucasians but, in this case, this APOC3 SNP was not over-represented. Our study did not find any difference in the AUC of the various lipid fractions during lipemia, but a different kinetics, as the times to maximum peak of TG and large TRL-TG were delayed in subjects homozygous for the minor allele (AA). Prolonged lipemia has been linked to accelerated atherosclerosis (1), although this may not be the case, as the AUC was not affected by the presence of the minor allele. However, the different kinetics and the possible link to an increased presence of the homozygote genotype in centenarians is an interesting combination, which should be investigated further.
In our study, APOA4 N147S and T29T SNPs had an important association with APOA1 concentrations throughout postprandial lipemia. Although the clear significance of APOA4 is not stated, in vitro studies suggest that it makes an important contribution in regulating LCAT, cholesteryl ester transfer protein (CETP), and LPL activity (21, 22, 58). Current knowledge suggests an anti-atherogenic role for APOA4 based on its antioxidant and reverse cholesterol transport-enhancing properties. Data suggest a close relationship between APOA1 and APOA4 apolipoproteins, both of which are present in nascent chylomicron particles and HDL. Furthermore, the two particles share a protein functional domain with apolipoprotein E (59). Moreover, transcription of APOA1 and APOA4 are regulated synchronously in a manner that also includes APOC3 (by elements -790 to -590 bp 5′ proximal to the APOC3 mRNA start), further supporting a close relationship between APOA1 and APOA4 (60). The association of APOA4 SNPs with postprandial APOA1 concentrations may be due to a deregulation of chylomicron TG transfer to HDL or to a lower rate of APOA1 transcription following triacylglycerol stimulation. In our study, concentrations of APOA1 were highly correlated with those of HDL as previously known (61), which suggests that APOA4 SNPs somehow influence the availability of HDL in the postprandial state. Interestingly, we observed in the multivariate approach of the statistical study that the effects of the two SNPs were not independent of each other, but the results of statistical analysis do not allow quantification of the partial influence of each SNP, probably due to their tight LD.
Regarding the polymorphism intergenic to APOA4 and APOA5, we found that carriers of the variant allele show a lower postprandial accumulation of potential atherogenic particles. In partial agreement with our results is a study by Talmud et al. (32) in which a lower level of fasting TG in homozygotes was reported for the minor allele than for common allele homozygotes. However, higher concentrations of TG were noted in heterozygotes than in homozygotes for the major allele. Interestingly, the same SNP was further investigated by these same authors after oral fat and glucose tolerance tests. Although there was no influence on lipid metabolism in this second study, homozygotes for the minor allele showed a higher peak of insulin after the oral glucose tolerance test and also associated with higher systolic blood pressure and higher waist–to-hip ratio (33). The singular localization of this SNP in an intergenic region may appear surprising, making it difficult to explain its function. It is possible that this SNP could be in high LD with a functional variant in an adjacent exon or untranslated region. However, results from the recent ENCODE Study suggest that nearly half of the elements that regulate transcription of a given gene lie beyond close proximity to the transcription start site (62). Thus, this intergenic variant may affect transcription rates of any of the members of this gene cluster.
The overarching concept that can be extracted from the present study is the tight interrelationships that comprise apolipoprotein metabolism in the postprandial state. In agreement with previous reports, we believe that a broad view must be taken when evaluating the APOA1/C3/A4/A5 region. Important intra- and inter-genic LD associations have been found in this study, which replicate previous findings (8). These LD patterns in APOA1/C3/A4/A5 are rather complex and highly specific to the population under study and indicate the functional dependencies of the encoded proteins. However, it is the population-specific differences in the LD patterns that drive the need to examine the postprandial response of individuals with different haplotypes across this cluster.
When evaluating the phenotypic effects of SNPs in the APOA1/C3/A4/A5 region, a view that focuses on only one particular apolipoprotein can draw limited, if not incorrect, conclusions regarding the effects of these SNPs in total apolipoprotein metabolism. In support of this notion, we have demonstrated a very important effect of APOA4 SNPs on APOA1 concentration. Furthermore, we have shown effects of APOA1 SNPs on triglyceride levels. Although we could not find any close relation of some variants to postprandial metabolism, we did discover certain striking effects with two SNPs (APOA1 -2803 and A4A5_inter) on the postprandial metabolism of healthy young adult males. Our sample population was selected in such a way as to minimize the possibility of interactions of age and lipid metabolism. It is possible, however, that subdued effects of the SNPs may appear in persons with partially impaired lipid metabolism. To resolve this possibility, further studies that include elderly persons and patients with altered lipid metabolism (for example, metabolic syndrome, diabetes mellitus, or hypercholesterolemia) are needed.
In conclusion, we have further characterized a large number of SNPs in the APOA1/C3/A4/A5 region and have found that two SNPs in this region (APOA1 -2803 and A4A5_inter) exert major effects on the postprandial metabolism of young persons. These effects have allowed identification of a population with diminished postprandial lipemia, and, thus, reduced risk of developing atherosclerosis.
Supplementary Material
Footnotes
Abbreviations:
- APOA1
- apolipoprotein A1
- APOC3
- apolipoprotein C-III
- APOA4
- apolipoprotein A-IV
- APOA5
- apolipoprotein V
- AUC
- area under the curve
- BMI
- body mass index
- CETP
- cholesteryl ester transfer protein
- CHOL
- cholesterol
- EBF1
- early B-cell factor 1
- HWE
- Hardy-Weinberg equilibrium
- LD
- linkage disequilibrium
- SNP
- single nucleotide polymorphism
- SRF
- serum response factor
- TG
- triacylglycerol
- TRL
- TG-rich lipoprotein
This study was supported by Consejeria de Innovación, proyectos de Investigación de Excelencia, Junta de Andalucia (AGR 05/00922 to Dr. Perez-Jimenez and P06-CTS-01425 to Dr.Lopez-Miranda); Ministerio de Educación y Ciencia AGL-2006-01979/ALI to Dr. Lopez-Miranda. CIBEROBN is an initiative of ISCIII. Government of Spain.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two tables and 11 figures.
REFERENCES
- 1.Lopez-Miranda J., Perez-Martinez P., Marin C., Moreno J. A., Gomez P., Perez-Jimenez F. 2006. Postprandial lipoprotein metabolism, genes and risk of cardiovascular disease. Curr. Opin. Lipidol. 17: 132–138. [DOI] [PubMed] [Google Scholar]
- 2.Mensink R. P., Zock P. L., Kester A. D., Katan M. B. 2003. Effects of dietary fatty acids and carbohydrates on the ratio of serum total to HDL cholesterol and on serum lipids and apolipoproteins: a meta-analysis of 60 controlled trials. Am. J. Clin. Nutr. 77: 1146–1155. [DOI] [PubMed] [Google Scholar]
- 3.Parks E. J., Skokan L. E., Timlin M. T., Dingfelder C. S. 2008. Dietary sugars stimulate fatty acid synthesis in adults. J. Nutr. 138: 1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez-Miranda J., Williams C., Lairon D. 2007. Dietary, physiological, genetic and pathological influences on postprandial lipid metabolism. Br. J. Nutr. 98: 458–473. [DOI] [PubMed] [Google Scholar]
- 5.Zheng C., Khoo C., Furtado J., Ikewaki K., Sacks F. M. 2008. Dietary monounsaturated fat activates metabolic pathways for triglyceride-rich lipoproteins that involve apolipoproteins E and C–III. Am. J. Clin. Nutr. 88: 272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva K. D., Kelly C. N., Jones A. E., Smith R. D., Wootton S. A., Miller G. J., Williams C. M. 2003. Chylomicron particle size and number, factor VII activation and dietary monounsaturated fatty acids. Atherosclerosis. 166: 73–84. [DOI] [PubMed] [Google Scholar]
- 7.Williams C. M., Moore F., Morgan L., Wright J. 1992. Effects of n-3 fatty acids on postprandial triacylglycerol and hormone concentrations in normal subjects. Br. J. Nutr. 68: 655–666. [DOI] [PubMed] [Google Scholar]
- 8.Lai C. Q., Parnell L. D., Ordovas J. M. 2005. The APOA1/C3/A4/A5 gene cluster, lipid metabolism and cardiovascular disease risk. Curr. Opin. Lipidol. 16: 153–166. [DOI] [PubMed] [Google Scholar]
- 9.Reichl D., Miller N. E. 1989. Pathophysiology of reverse cholesterol transport. Insights from inherited disorders of lipoprotein metabolism. Arteriosclerosis. 9: 785–797. [DOI] [PubMed] [Google Scholar]
- 10.Marin C., Lopez-Miranda J., Gomez P., Paz E., Perez-Martinez P., Fuentes F., Jimenez-Pereperez J. A., Ordovas J. M., Perez-Jimenez F. 2002. Effects of the human apolipoprotein A-I promoter G-A mutation on postprandial lipoprotein metabolism. Am. J. Clin. Nutr. 76: 319–325. [DOI] [PubMed] [Google Scholar]
- 11.Smith J. A., Arnett D. K., Kelly R. J., Ordovas J. M., Sun Y. V., Hopkins P. N., Hixson J. E., Straka R. J., Peacock J. M., Kardia S. L. 2008. The genetic architecture of fasting plasma triglyceride response to fenofibrate treatment. Eur. J. Hum. Genet. 16: 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ooi E. M., Barrett P. H., Chan D. C., Watts G. F. 2008. Apolipoprotein C–III: understanding an emerging cardiovascular risk factor. Clin. Sci. (Lond.). 114: 611–624. [DOI] [PubMed] [Google Scholar]
- 13.Russo G. T., Meigs J. B., Cupples L. A., Demissie S., Otvos J. D., Wilson P. W., Lahoz C., Cucinotta D., Couture P., Mallory T., et al. 2001. Association of the Sst-I polymorphism at the APOC3 gene locus with variations in lipid levels, lipoprotein subclass profiles and coronary heart disease risk: the Framingham offspring study. Atherosclerosis. 158: 173–181. [DOI] [PubMed] [Google Scholar]
- 14.Tas S., Abdella N. A. 1994. Blood pressure, coronary artery disease, and glycaemic control in type 2 diabetes mellitus: relation to apolipoprotein-CIII gene polymorphism. Lancet. 343: 1194–1195. [DOI] [PubMed] [Google Scholar]
- 15.Woo S. K., Kang H. S. 2003. The apolipoprotein CIII T2854G variants are associated with postprandial triacylglycerol concentrations in normolipidemic Korean men. J. Hum. Genet. 48: 551–555. [DOI] [PubMed] [Google Scholar]
- 16.Waterworth D. M., Hubacek J. A., Pitha J., Kovar J., Poledne R., Humphries S. E., Talmud P. J. 2000. Plasma levels of remnant particles are determined in part by variation in the APOC3 gene insulin response element and the APOCI-APOE cluster. J. Lipid Res. 41: 1103–1109. [PubMed] [Google Scholar]
- 17.Minihane A. M., Finnegan Y. E., Talmud P., Leigh-Firbank E. C., Williams C. M. 2002. Influence of the APOC3–2854T>G polymorphism on plasma lipid levels: effect of age and gender. Biochim. Biophys. Acta. 1583: 311–314. [DOI] [PubMed] [Google Scholar]
- 18.Novelli V., Viviani Anselmi C., Roncarati R., Guffanti G., Malovini A., Piluso G., Puca A. A. 2008. Lack of replication of genetic associations with human longevity. Biogerontology. 9: 85–92. [DOI] [PubMed] [Google Scholar]
- 19.Atzmon G., Rincon M., Schechter C. B., Shuldiner A. R., Lipton R. B., Bergman A., Barzilai N. 2006. Lipoprotein genotype and conserved pathway for exceptional longevity in humans. PLoS Biol. 4: e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waterworth D. M., Talmud P. J., Bujac S. R., Fisher R. M., Miller G. J., Humphries S. E. 2000. Contribution of apolipoprotein C–III gene variants to determination of triglyceride levels and interaction with smoking in middle-aged men. Arterioscler. Thromb. Vasc. Biol. 20: 2663–2669. [DOI] [PubMed] [Google Scholar]
- 21.Goldberg I. J., Scheraldi C. A., Yacoub L. K., Saxena U., Bisgaier C. L. 1990. Lipoprotein ApoC-II activation of lipoprotein lipase. Modulation by apolipoprotein A-IV. J. Biol. Chem. 265: 4266–4272. [PubMed] [Google Scholar]
- 22.Steinmetz A., Utermann G. 1985. Activation of lecithin: cholesterol acyltransferase by human apolipoprotein A-IV. J. Biol. Chem. 260: 2258–2264. [PubMed] [Google Scholar]
- 23.Ostos M. A., Lopez-Miranda J., Marin C., Castro P., Gomez P., Paz E., Jimenez Pereperez J. A., Ordovas J. M., Perez-Jimenez F. 2000. The apolipoprotein A-IV-360His polymorphism determines the dietary fat clearance in normal subjects. Atherosclerosis. 153: 209–217. [DOI] [PubMed] [Google Scholar]
- 24.Fisher R. M., Burke H., Nicaud V., Ehnholm C., Humphries S. E. 1999. Effect of variation in the apo A-IV gene on body mass index and fasting and postprandial lipids in the European Atherosclerosis Research Study II. EARS Group. J. Lipid Res. 40: 287–294. [PubMed] [Google Scholar]
- 25.Ostos M. A., Lopez-Miranda J., Ordovas J. M., Marin C., Blanco A., Castro P., Lopez-Segura F., Jimenez-Pereperez J., Perez-Jimenez F. 1998. Dietary fat clearance is modulated by genetic variation in apolipoprotein A-IV gene locus. J. Lipid Res. 39: 2493–2500. [PubMed] [Google Scholar]
- 26.Kamboh M. I., Crawford M. H., Aston C. E., Leonard W. R. 1996. Population distributions of APOE, APOH, and APOA4 polymorphisms and their relationships with quantitative plasma lipid levels among the Evenki herders of Siberia. Hum. Biol. 68: 231–243. [PubMed] [Google Scholar]
- 27.Weinberg R. B., Cook V. R., Beckstead J. A., Martin D. D., Gallagher J. W., Shelness G. S., Ryan R. O. 2003. Structure and interfacial properties of human apolipoprotein A-V. J. Biol. Chem. 278: 34438–34444. [DOI] [PubMed] [Google Scholar]
- 28.Moreno R., Perez-Jimenez F., Marin C., Moreno J. A., Gomez P., Bellido C., Perez-Martinez P., Jimenez-Gomez Y., Fuentes F. J., Lopez-Miranda J. 2005. A single nucleotide polymorphism of the apolipoprotein A-V gene -1131T>C modulates postprandial lipoprotein metabolism. Atherosclerosis. 189: 163–168. [DOI] [PubMed] [Google Scholar]
- 29.Vrablik M., Horinek A., Ceska R., Adamkova V., Poledne R., Hubacek J. A. 2003. Ser19→Trp polymorphism within the apolipoprotein AV gene in hypertriglyceridaemic people. J. Med. Genet. 40: e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olivier M., Wang X., Cole R., Gau B., Kim J., Rubin E. M., Pennacchio L. A. 2004. Haplotype analysis of the apolipoprotein gene cluster on human chromosome 11. Genomics. 83: 912–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno-Luna R., Perez-Jimenez F., Marin C., Perez-Martinez P., Gomez P., Jimenez-Gomez Y., Delgado-Lista J., Moreno J. A., Tanaka T., Ordovas J. M., et al. 2007. Two independent apolipoprotein A5 haplotypes modulate postprandial lipoprotein metabolism in a healthy Caucasian population. J. Clin. Endocrinol. Metab. 92: 2280–2285. [DOI] [PubMed] [Google Scholar]
- 32.Talmud P. J., Hawe E., Martin S., Olivier M., Miller G. J., Rubin E. M., Pennacchio L. A., Humphries S. E. 2002. Relative contribution of variation within the APOC3/A4/A5 gene cluster in determining plasma triglycerides. Hum. Mol. Genet. 11: 3039–3046. [DOI] [PubMed] [Google Scholar]
- 33.Martin S., Nicaud V., Humphries S. E., Talmud P. J. 2003. Contribution of APOA5 gene variants to plasma triglyceride determination and to the response to both fat and glucose tolerance challenges. Biochim. Biophys. Acta. 1637: 217–225. [DOI] [PubMed] [Google Scholar]
- 34.Chien K. L., Chen M. F., Hsu H. C., Su T. C., Chang W. T., Lee C. M., Lee Y. T. 2008. Genetic association study of APOA1/C3/A4/A5 gene cluster and haplotypes on triglyceride and HDL cholesterol in a community-based population. Clin. Chim. Acta. 388: 78–83. [DOI] [PubMed] [Google Scholar]
- 35.Hamon S. C., Kardia S. L., Boerwinkle E., Liu K., Klos K. L., Clark A. G., Sing C. F. 2006. Evidence for consistent intragenic and intergenic interactions between SNP effects in the APOA1/C3/A4/A5 gene cluster. Hum. Hered. 61: 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi L., Liu S., Rifai N., Hunter D., Hu F. B. 2007. Associations of the apolipoprotein A1/C3/A4/A5 gene cluster with triglyceride and HDL cholesterol levels in women with type 2 diabetes. Atherosclerosis. 192: 204–210. [DOI] [PubMed] [Google Scholar]
- 37.Suviolahti E., Lilja H. E., Pajukanta P. 2006. Unraveling the complex genetics of familial combined hyperlipidemia. Ann. Med. 38: 337–351. [DOI] [PubMed] [Google Scholar]
- 38.Perez-Martinez P., Lopez-Miranda J., Ordovas J. M., Bellido C., Marin C., Gomez P., Paniagua J. A., Moreno J. A., Fuentes F., Perez-Jimenez F. 2004. Postprandial lipemia is modified by the presence of the polymorphism present in the exon 1 variant at the SR-BI gene locus. J. Mol. Endocrinol. 32: 237–245. [DOI] [PubMed] [Google Scholar]
- 39.Moreno J. A., Perez-Jimenez F., Marin C., Perez-Martinez P., Moreno R., Gomez P., Jimenez-Gomez Y., Paniagua J. A., Lairon D., Lopez-Miranda J. 2005. The apolipoprotein E gene promoter (-219G/T) polymorphism determines insulin sensitivity in response to dietary fat in healthy young adults. J. Nutr. 135: 2535–2540. [DOI] [PubMed] [Google Scholar]
- 40.Moreno J. A., Lopez-Miranda J., Marin C., Gomez P., Perez-Martinez P., Fuentes F., Fernandez de la Puebla R. A., Paniagua J. A., Ordovas J. M., Perez-Jimenez F. 2003. The influence of the apolipoprotein E gene promoter (-219G/ T) polymorphism on postprandial lipoprotein metabolism in young normolipemic males. J. Lipid Res. 44: 2059–2064. [DOI] [PubMed] [Google Scholar]
- 41.Mata P., Lopez-Miranda J., Pocovi M., Alonso R., Lahoz C., Marin C., Garces C., Cenarro A., Perez-Jimenez F., de Oya M., et al. 1998. Human apolipoprotein A-I gene promoter mutation influences plasma low density lipoprotein cholesterol response to dietary fat saturation. Atherosclerosis. 137: 367–376. [DOI] [PubMed] [Google Scholar]
- 42.Lopez-Miranda J., Ordovas J. M., Ostos M. A., Marin C., Jansen S., Salas J., Blanco-Molina A., Jimenez-Pereperez J. A., Lopez-Segura F., Perez-Jimenez F. 1997. Dietary fat clearance in normal subjects is modulated by genetic variation at the apolipoprotein B gene locus. Arterioscler. Thromb. Vasc. Biol. 17: 1765–1773. [DOI] [PubMed] [Google Scholar]
- 43.Gomez P., Miranda J. L., Marin C., Bellido C., Moreno J. A., Moreno R., Perez-Martinez P., Perez-Jimenez F. 2004. Influence of the -514C/T polymorphism in the promoter of the hepatic lipase gene on postprandial lipoprotein metabolism. Atherosclerosis. 174: 73–79. [DOI] [PubMed] [Google Scholar]
- 44.Bellido C., Lopez-Miranda J., Blanco-Colio L. M., Perez-Martinez P., Muriana F. J., Martin-Ventura J. L., Marin C., Gomez P., Fuentes F., Egido J., et al. 2004. Butter and walnuts, but not olive oil, elicit postprandial activation of nuclear transcription factor kappaB in peripheral blood mononuclear cells from healthy men. Am. J. Clin. Nutr. 80: 1487–1491. [DOI] [PubMed] [Google Scholar]
- 45.Delgado-Lista J., Perez-Jimenez F., Tanaka T., Perez-Martinez P., Jimenez-Gomez Y., Marin C., Ruano J., Parnell L., Ordovas J. M., Lopez-Miranda J. 2007. An apolipoprotein A-II polymorphism (-265T/C, rs5082) regulates postprandial response to a saturated fat overload in healthy men. J. Nutr. 137: 2024–2028. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y., Ordovas J. M., Gao G., Province M., Straka R. J., Tsai M. Y., Lai C. Q., Zhang K., Borecki I., Hixson J. E., et al. 2009. Pharmacogenetic association of the APOA1/C3/A4/A5 gene cluster and lipid responses to fenofibrate: the genetics of lipid-lowering drugs and diet network study. Pharmacogenet. Genomics. 19: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Codon Usage Database. www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=9606. accessed June 2009.
- 48.Kahali B., Basak S., Ghosh T. C. 2007. Reinvestigating the codon and amino acid usage of S. cerevisiae genome: a new insight from protein secondary structure analysis. Biochem. Biophys. Res. Commun. 354: 693–699. [DOI] [PubMed] [Google Scholar]
- 49.Marinescu V. D., Kohane I. S., Riva A. 2005. MAPPER: a search engine for the computational identification of putative transcription factor binding sites in multiple genomes. BMC Bioinformatics. 6: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osgood-McWeeney D., Galluzzi J. R., Ordovas J. M. 2000. Allelic discrimination for single nucleotide polymorphisms in the human scavenger receptor class B type 1 gene locus using fluorescent probes. Clin. Chem. 46: 118–119. [PubMed] [Google Scholar]
- 51.Perez-Martinez P., Ordovas J. M., Lopez-Miranda J., Gomez P., Marin C., Moreno J., Fuentes F., Fernandez de la Puebla R. A., Perez-Jimenez F. 2003. Polymorphism exon 1 variant at the locus of the scavenger receptor class B type I gene: influence on plasma LDL cholesterol in healthy subjects during the consumption of diets with different fat contents. Am. J. Clin. Nutr. 77: 809–813. [DOI] [PubMed] [Google Scholar]
- 52.Allain C. C., Poon L. S., Chan C. S., Richmond W., Fu P. C. 1974. Enzymatic determination of total serum cholesterol. Clin. Chem. 20: 470–475. [PubMed] [Google Scholar]
- 53.Bucolo G., David H. 1973. Quantitative determination of serum triglycerides by the use of enzymes. Clin. Chem. 19: 476–482. [PubMed] [Google Scholar]
- 54.Riepponen P., Marniemi J., Rautaoja T. 1987. Immunoturbidimetric determination of apolipoproteins A-1 and B in serum. Scand. J. Clin. Lab. Invest. 47: 739–744. [PubMed] [Google Scholar]
- 55.Warnick G. R., Benderson J., Albers J. J. 1982. Dextran sulfate-Mg2+ precipitation procedure for quantitation of high-density-lipoprotein cholesterol. Clin. Chem. 28: 1379–1388. [PubMed] [Google Scholar]
- 56.Friedewald W. T., Levy R. I., Fredrickson D. S. 1972. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 18: 499–502. [PubMed] [Google Scholar]
- 57.Su A. I., Cooke M. P., Ching K. A., Hakak Y., Walker J. R., Wiltshire T., Orth A. P., Vega R. G., Sapinoso L. M., Moqrich A., et al. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. USA. 99: 4465–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ordovas J. M., Cassidy D. K., Civeira F., Bisgaier C. L., Schaefer E. J. 1989. Familial apolipoprotein A-I, C–III, and A-IV deficiency and premature atherosclerosis due to deletion of a gene complex on chromosome 11. J. Biol. Chem. 264: 16339–16342. [PubMed] [Google Scholar]
- 59.Marchler-Bauer A., Anderson J. B., Cherukuri P. F., DeWeese-Scott C., Geer L. Y., Gwadz M., He S., Hurwitz D. I., Jackson J. D., Ke Z., et al. 2005. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33: D192–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zannis V. I., Kan H. Y., Kritis A., Zanni E., Kardassis D. 2001. Transcriptional regulation of the human apolipoprotein genes. Front. Biosci. 6: D456–D504. [DOI] [PubMed] [Google Scholar]
- 61.Fuster V., Ross R., Topol E. J. 1996. Atherosclerosis and Coronary Disease. Illustrated edition Lippincott-Raven; Philadelphia. [Google Scholar]
- 62.Birney E., Stamatoyannopoulos J. A., Dutta A., Guigo R., Gingeras T. R., Margulies E. H., Weng Z., Snyder M., Dermitzakis E. T., Thurman R. E., et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 447: 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.