Abstract
Purpose
To develop and apply DTI based normalization methodology for the detection and quantification of traumatic brain injury (TBI) and the impact of injury along specific brain pathways in: a) individual TBI subjects, and b) a TBI group.
Materials and Methods
Normalized DTI tractography was conducted in the native space of 12 TBI and 10 age-matched control subjects using the same number of seeds in each subject, distributed at anatomically equivalent locations. Whole-brain tracts from the control group were mapped onto the head of each TBI subject. Differences in the Fractional Anisotropy (FA) maps between each TBI subject and the control group were computed in a common space using a t-test, transformed back to the individual TBI subject's head-space, and thresholded to form Regions of Interest (ROIs) that were used to sort tracts from the control group and the individual TBI subject. Tract-counts for a given ROI in each TBI subject were compared to group mean for the same ROI to quantify impact of injury along affected pathways. Same procedure was used to compare TBI group to control group in a common space.
Results
Sites of injury within individual TBI subjects and affected pathways included hippocampal/fornix, inferior fronto-occipital, inferior longitudinal fasciculus, corpus callosum (genu and splenium), cortico-spinal tracts and the uncinate fasciculus. Most of these regions were also detected in the group study.
Conclusions
The DTI normalization methodology presented here enables automatic delineation of ROIs within the heads of individual subjects (or in a group). These ROIs not only localize and quantify the extent of injury, but also quantify the impact of injury on affected pathways in an individual or a group of TBI subjects.
Keywords: Diffusion Tensor Imaging, Tractography, Normalization, Traumatic Brain Injury
1. Introduction
This paper presents a novel methodology to use DTI (Diffusion Tensor Imaging) in the detection and quantification of traumatic brain injury (TBI) and brain pathways (tracts) affected by the injury. TBI is a growing health problem in the US, commonly attributed to motor vehicular accidents and sports injuries but recently also to war-related injuries sustained near an explosion. One of the serious consequences of TBI is diffuse axonal injury (DAI), or white matter injuries, induced by sudden acceleration/deceleration and/or rotational/vibrational forces causing a shearing of nerve fibers [1-4]. In addition to diffuse injury, local shearing of axons at the gray/white interface is also possible. Diffuse and local axonal shearing both disrupt axonal connections critical to brain function, and DAI commonly refers to both types of injury. DAI has been identified as one of the key reasons for permanent disability or death. In general, DAI can be very debilitating, leading to a wide range of neurological impairments from mild memory deficits to persistent vegetative states. Though CT and MRI are routinely employed to evaluate trauma and DAI, several reports suggest that CT and commonly used T1- and T2-weighted MRI protocols are unable to detect the full extent of injury and likely to underestimate the consequences of DAI, resulting in a poor correlation between diagnosis and final outcome [5,6]. Typically these patients exhibit a high rate of morbidity without any evidence of lesions on CT or MRI. It has even been suggested that “DAI can only be definitely diagnosed postmortem” [7].
Histologically DAI is characterized by disruption of the cytoskeletal network and axonal membranes, leading to impaired axonal transport [3]. Thus a thorough evaluation of DAI requires an imaging method able to quantify the integrity of axons and the network of connections required to support normal brain function. For example, a person may look fully recovered months after surviving a nearby explosion, but cognitive function may be abnormal due to loss of connectivity among key brain regions. Some of the sites frequently damaged in TBI include the corpus callosum, fornix and cingulum [8]. Because of these locations, memory and language deficits are common as are certain prefrontal syndromes such as dysexecutive, disinhibited and apathetic behavior. Often these deficits are present without any gross lesions on the CT scan and the etiology is attributed to DAI. Moreover, approaches to identify DAI through antibodies targeting amyloid precursor protein (APP) [9] may not always succeed because some axons demonstrate cytoskeletal alteration and detachment without axonal swelling and are thus not identifiable by markers of axonal swelling such as APP [10].
DTI has been used over the past several years to detect injured regions in TBI (e.g., [3,4,7,8,11-14]). DTI is sensitive to the biological diffusion of water molecules, hindered by extracellular and restricted by intracellular components [15]. When there is no obstruction to diffusion, the diffusion tensor is isotropic. However, in the presence of axons and their myelinated sheath, diffusion becomes anisotropic and quantifiable, which reveals the direction and integrity of axons. Though myelin is not necessary, it influences several diffusion anisotropy metrics. Thus damage to the myelin sheath and/or axons is potentially detectable by DTI. Maps of several diffusion anisotropy metrics, e.g., Fractional Anisotropy (FA) -- the normalized differences among the three eigen values of the tensor; Mean Diffusivity (MD) -- the average of the three eigen values; Axial Diffusivity (DA) -- the first eigen value oriented along the long axis of the axons; Radial Diffusivity (DR) -- the average of second and third eigen values; Volume Ratio (VR) -- the normalized product of the three eigen values, and the ratio DA/DR have been evaluated in human and animal DTI studies [16-21].
The exact model of axonal damage leading to changes in the diffusivity and resulting diffusion anisotropy measures such as FA is not well-understood. There is converging opinion that DAI represents a progressive injury, beginning with local swelling of axons, followed by cytoskeletal perturbations including misalignment of fibers and eventual disconnection [3,4,14,22]. It has been hypothesized [3] that the consequences of TBI on DTI would be a decrease in FA and an increase in MD, attributed mainly to an increase in DR and a decrease in diffusivity along the principal direction (i.e., a reduction in DA). Indeed, significant FA reductions but less significant changes in MD have been reported in the internal capsule and corpus callosum during the first 24 hours of injury [3]. Changes in FA, however, were significantly less at one month, suggesting the dynamic nature of the injury. A 20-subject study of changes in the FA maps in DAI [4] with significant correlation with the Glasgow Coma Scale or GCS (r = 0.65−0.74, p < 0.001) also concluded that FA values were significantly reduced within the internal capsule and splenium. MD was not analyzed in that study but no significant changes were found in the Apparent Diffusion Coefficient (ADC), which is a less sensitive marker of diffusivity. A previous study also reported an increase in MD but unchanged FA [23], and another study found reduced FA in the corpus callosum, internal capsule, and centrum semiovale, with significant increase in MD in the corpus callosum and internal capsule [12]. Similar results have also been reported very recently [24] for the fornix body, all sub-regions of the corpus callosum and peduncular projections, with the additional observation that not only did DTI detect loss of white matter integrity at the beginning of the injury process but the DTI metrics also correlate strongly to functional outcome.
A recent study also compared mild TBI to moderate-to-severe TBI and found that the FA in moderate-to-severe TBI was reduced in the posterior corona radiata, cortico-spinal tracts, cingulum, external capsule, forceps minor and major, genu, body and splenium of the corpus callosum, inferior fronto-occipital fasciculus, superior longitudinal fasciculus and sagittal stratum. FA reduction in mild TBI, however, was detected only in the cortico-spinal tract, sagittal stratum and the superior longitudinal fasciculus [25]. This study also examined DR and DA and found that both DR and DA were increased in moderate-to-severe TBI, which could explain the decrease in FA. However only DA was found to increase in some regions in mild TBI while DR did not show any significant increase in any region. It is not clear how the FA would decrease under these circumstances as FA is likely to increase if DA increases and DR remains unchanged. This specific issue was not addressed by the authors, but the authors suggest that their results are consistent with the notion that damage to myelin in mild TBI is less common, whereas both the axons and myelin are likely to be damaged in moderate to severe injury.
Contrary to the above studies, an increase in mean diffusivity and an increase in FA in an infant with severe TBI has also been reported [26], though the authors suggest that the increase in FA is probably a transient effect due to an orderly disruption of cellular membrane in combination with cellular and vasogenic edema that could temporarily increase both the FA and diffusivity. Also, an increase in FA and a decrease in MD have been reported very recently [14] in a 6-subject study of acute mild TBI. A significant increase in FA in the posterior corpus callosum and significant decrease in MD in the left anterior internal capsule within 72 hours of injury was observed and though these results were highly correlated with post-concussive symptoms (PCS) and neurobehavioral tests, the authors [14] acknowledged that their results were not consistent with previous studies of mild TBI [3,12]. The explanation offered by the authors was that as most DTI measurements observe diffusion in water contained in the space between axons (intercellular space) rather than the axons themselves (intracellular space), the swelling of axons in the acute injury phase would constrict the intercellular space, leading to a decrease in MD and an increase in FA. This model is plausible but awaits validation with a larger number of patients. Other possible explanations include coregistration/normalization errors and the presence of multiple fibers within a voxel. If more than one fiber were present in a voxel, and one of them was selectively damaged, the diffusion anisotropy around the remaining intact fibers would be enhanced, leading to a higher FA for that voxel. Also a recent longitudinal study of severe TBI found that though the FA was reduced in all investigated regions, mainly due to a decrease in DA and an increase in DR, the FA had increased in the internal capsule and the centrum semiovale at a mean 12-month follow-up [27]. The increase in FA, which now reached normal to above normal values, primarily in patients with favorable outcome, was attributed to an increase in DA with no change in DR. The FA remained depressed in patients with unfavorable outcome.
Though the model for observed FA changes in TBI is not well-established, it is apparent that maps of anisotropy changes can be, and have been, used to detect injured regions. However these maps alone are inadequate to isolate specific brain pathways disrupted by the injury or to quantify the loss of brain connectivity along affected pathways. DTI-tractography, which creates 3D maps of axonal connections, provides a mechanism to localize and quantify pathways affected by the injury.
Though there are many computational schemes to conduct tractography, the commonly used streamline tractography procedure tracks the direction of the first eigen value of an assumed rank-2 diffusion tensor per voxel until either the FA falls below the threshold or the orientation shows an abrupt angular change exceeding a specified threshold [28-31]. A 3D interpolation of the tensor matrix is usually employed to display tracts with sub-voxel resolution. Although human DTI resolution limitations (approximately 2mm) limit tractography to mapping bundles of tightly packed axons (tracts), this is not expected to be a major weakness since DAI suffered in head trauma is likely to disrupt tracts and not merely single axons. This capability to directly visualize axonal connections and quantify connectivity among specified regions is a particular strength of DTI tractography not achievable with any other imaging modality at the present time. When combined with methods to detect voxel-based anisotropy changes, DTI including tractography provides a unique capability to localize and quantify injured regions and brain pathways affected by the injury.
Anisotropy changes are detected by first normalizing the maps of anisotropy metrics (scalars) from all subjects (subject space) to a common space (normalized space) and then performing a t-test or similar statistical analysis. There are numerous publications on voxel-based whole-brain anisotropy comparisons, particularly FA in TBI [8,24,27,32,33]. Statistical comparisons of whole-brain tractography, however, are more complex as normalization of tensors or tracts requires additional eigen vector corrections. A previous approach to normalize tractography relied on averaging tensors in normalized space [34] but this does not fully account for inherent primary eigen vector variations in individuals. The commonly used current approaches to normalize whole-brain tractography incorporate a rotational correction factor that realigns the eigen vectors (or gradients before estimating the eigen vectors) after mapping all images into a standard or normalized space, consistent with the non-linear mapping transformations for every voxel [35-37]. However there are several inherent problems with tractography when it is conducted after spatial transformations, namely: a) spatial mapping exacerbates partial volume problems due to the required averaging of neighboring voxels during spatial interpolation, b) it may not maintain tract topography when non-linear normalization is used, and c) frequently known continuous tracts are divided into non-contiguous segments. To avoid performing whole-brain tractography in a common space, an alternative approach is to first identify ROIs in normalized space by, for example a FA comparison [8], and based on the anatomical locations of these ROIs, or based on a priori hypotheses [24], subjectively draw these ROIs in each subject's head-space. These ROIs are then used to sort tracts from whole-brain tractography conducted in each subject's native space. However, manually outlining ROIs is subjective and tracts sorted by such ROIs are prone to relatively large errors as even small errors in the location of seed points propagate and accumulate along tracts spanning a large number of voxels.
In an attempt to improve tract normalization and quantification methodology for application to TBI, the primary objectives of this work were to identify injured regions in a TBI patient, quantify the injury in terms of DTI anisotropy metrics, identify brain pathways (tracts) affected by the injury and quantify the effect on impacted pathways. A preliminary study where affected pathways in four TBI subjects were identified in a standard normalized space was reported by us recently at a MRI conference [38]. However, as neurosurgical or other individualized interventions rely on the anatomy of the patient's own brain, one of the key goals was to quantify injury related tractography changes in the patient's own 3D head space, which requires mapping of whole-brain tractographies of a control group to the 3D brain coordinates of an individual patient. In addition, a prerequisite was not to require any a priori hypotheses or manual outlining of ROIs to reduce errors of subjectivity. A secondary objective was to conduct a similar study comparing a group of TBI patients to a group of control subjects in a common 3D space to detect and quantify patterns of injury and affected pathways in TBI patients, also without the need for any a priori hypotheses or manually drawn ROIs. Both objectives require novel tractography normalization and quantification methods. Details of the methodology and results from a small sample of TBI patients (n=12) and normal subjects (n=10) are presented in this paper.
2. Method
2.1. Patients and Control Subjects
Patients with mild to moderate brain injuries (loss of consciousness for less than 1 hour) primarily due to falls, assault, or traffic accidents, referred to the LAC+USC Brain Injury Clinic consented to a DTI scan as part of their medical workup and care. IRB approval was obtained to process existing DTI data from 12 such TBI subjects who had been identified by a neurosurgeon based on their clinical history and no evidence of additional central nervous system disease. The mean and standard deviation of the TBI and control subjects’ ages were 28±11.4 and 27±8.1 years respectively, and the mean time interval between TBI and the DTI scan was approximately one month. All 12 patients were ambulatory (without assistance) and all could communicate well enough to provide a history of their injury and care. Patients were interviewed in their primary language (Spanish or English) regarding symptoms of post-concussive syndrome (headache, dizziness, tinnitus, depression, irritability, sexual dysfunction, etc.) and every patient who agreed to DTI had at least one post-concussive symptom. None of the patients had hemiparesis or plegia. There were no aphasic patients, nor any patients who were blind or deaf. The only cranial nerve deficit was anosmia in 2 patients. Existing DTI data that had been acquired on the same scanner using the same protocol as the TBI patients from a group of 10 age-matched volunteers, with no known history of central nervous system disease and no evidence of neurological disease on MR imaging, were used as controls.
2.2. MRI Protocol
The available DTI data from 12 TBI and 10 normal control subjects had been acquired with a whole-brain single shot Echo Planar Imaging (EPI) DTI pulse sequence on a 1.5T GE EXCITE scanner with an 8-element phased array RF coil at TR=10.3s, field-of-view 26cm, 128×128 matrix, 28 contiguous 4mm thick slices using 25 isotropic gradient directions with b=1000s/mm2, one b=0 acquisition, and number of excitations (NEX)=2 for a total acquisition time of 7min 50s. The voxel size was 2.03×2.03×4.0mm3. A 512×512 axial Fluid-Attenuated-Inversion-Recovery (FLAIR) had also been acquired from the TBI patients using TR/TE/Inversion time of 8402/146.3/2100 ms, 20 slices, slice thickness 7mm, planar resolution 0.43×0.43mm2.
2.3. DTI Data Processing
The b=1000s/mm2 images were corrected for distortions and misregistration due to eddy currents and any movements during the DTI acquisition using the eddy current correction module of FSL (The Oxford Centre for Functional Magnetic Resonance Imaging of the Brain Software Library: http://www.fmrib.ox.ac.uk/fsl/fdt). The correction relies on a 12-parameter affine transformation. Maps of diffusion anisotropy parameters including FA, DA, DR, and MD were reconstructed using in-house developed DTI software incorporating a signal-to-noise-ratio weighted multivariate least-square fitting approach [39]. Whole-brain tractography was conducted using an in-house developed C-code version of streamline tractography.
2.4. Detection and Quantification of Anisotropy Changes
As several previous studies suggest FA is altered significantly in TBI, identification of the injured regions was based on changes in an individual's FA map with respect to a group of normals. This comparison was carried out in the standard MNI (Montreal Neurological Institute) space using SPM (Statistical Parametric Mapping, http://www.fil.ion.ucl.ac.uk/spm) based forward and inverse normalization procedures [40,41]. In forward normalization, each point ‘x’ within subject space image f(x) is mapped by a unique warping function M(·) onto its equivalent point ‘y’ of a template in MNI space to generate the normalized image, g(y). Inverse normalization does the reverse, i.e., maps from MNI space back to an individual subject's space. As each point within the template space maps to exactly one point in subject space, a unique inverse mapping from the MNI space to the subject space, M−1(·), exists between all points of both spaces. SPM normalization relying on a 12-parameter affine transformation to register voxels of the subject space to those of the template space followed by a nonlinear warping transformation to estimate the 3D deformation field at each point ‘x’ was used. SPM uses a linear combination of three dimensional discrete cosine transform (DCT) basis functions in three orthogonal directions to model the deformation. The DCT coefficients are iteratively optimized via Gauss-Newton strategy to minimize the bending energies of the deformation fields as well as the residual squared difference between f(x) and g(y). We used fourth order DCT basis functions, 16 iterations, and tri-linear interpolation in this work.
An FA template was constructed from the 10 normal control subjects using a two-step procedure where segmented white matter voxels within each normal subject's b=0 image were first mapped to the MNI white-matter template using SPM 12-parameter affine/non-linear transformation. Segmentation of the b=0 images into white-matter voxels was also done by SPM retaining only those voxels that displayed >50% probability of being classified as white matter. The forward mapping parameters obtained in the first step were applied to whole-brain b=0 images and refined in a second step by mapping the resulting images to the MNI EPI template. The resulting parameters were applied to individual FA maps of normal control subjects to create the FA template in MNI space from an average of the 10 control subjects. The FA maps of all control and TBI subjects were then individually remapped to the FA template and data were analyzed to determine the voxel based FA differences between: a) each TBI subject and the control group as described below, and b) the TBI group and control group in MNI space.
2.4.1. Individual TBI subject vs. Control Group
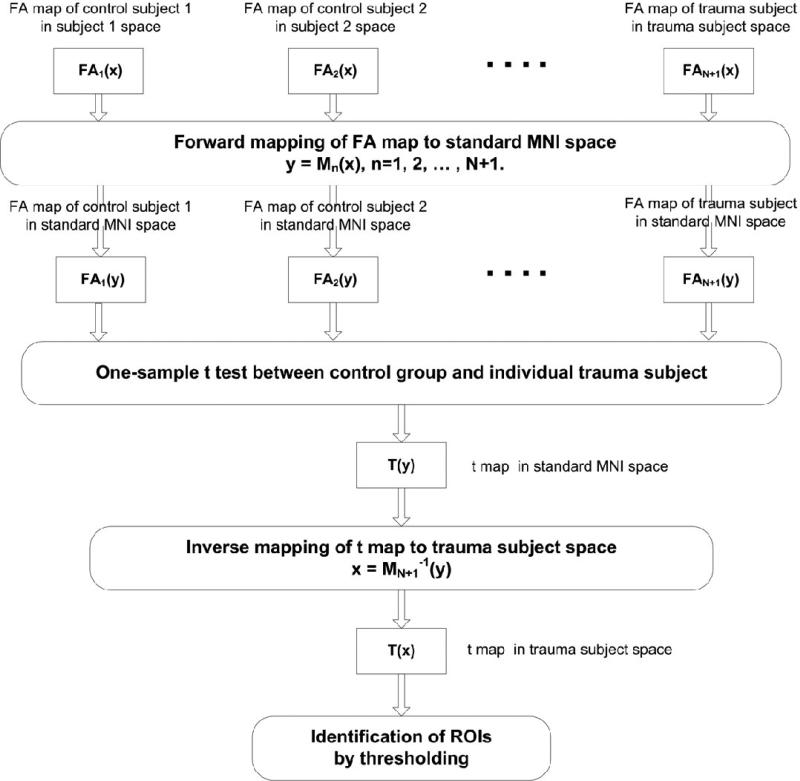
A block diagram illustrating the procedure to determine the differences between the control group and individual TBI subjects is presented in Fig. 1.
Fig. 1.
Block diagram of the procedure to detect FA changes between an individual TBI subject and a group of controls following normalization of all FA images to a FA template in MNI space. The t-map of the FA differences in MNI space was inverse mapped to the TBI subject's space and thresholded to generate regions of interest (ROIs) showing significant FA changes due to injury.
The FA map of an individual was first compared voxel-by-voxel to the FA maps of the control group in MNI space by computing a modified t-score defined as:
| (1) |
where FAi (control), FAi (individual) and σi represent mean FA of the control group, FA of an individual and the standard deviation in the FA values of the control group respectively for the i-th voxel. The t-score in Eq. (1) thus reflects the number of standard deviations by which the FA of an individual is reduced with respect to the control group FA mean, equivalent to a one-sample t-test. The t-score distribution in MNI space for an individual was then mapped back to the individual subject's head by using inverse normalization and thresholded (t ≥ 3.0, cluster size k ≥ 12) to localize the injured regions. Voxels satisfying the threshold criteria were clustered automatically into ROIs relying on contiguity of voxels along any direction. In addition to FA, other diffusion anisotropy metrics, e.g., DA, DR, MD were also computed for these ROIs. The t-score map of an individual in MNI space was also mapped onto FLAIR images using the inverse of the FLAIR to MNI T1-template normalization done by a 12-parameter affine transformation followed by non-linear warping. After mapping, the t-score distribution was thresholded as before (t ≥ 3.0, k ≥ 12) to show FA-changed regions superimposed on FLAIR images.
2.4.2. TBI Group vs. Control Group
Standard SPM group comparison procedure was used to determine FA differences between the TBI group and the control group in MNI space. A 2-sample t-test was used to detect ROIs where FA had changed significantly (pFDR ≤ 0.05 and k ≥ 12) in MNI space, where pFDR denotes the false detection rate corrected p-value.
2.5. Seed Placement and Tractography
As the number and characteristics of tracts are critically dependent on the number of seeds and how seeds are distributed in the head, it is critical to account for these variations among subjects to normalize tracts. We achieved a normalized distribution of seeds in individuals by inverse mapping seeds from the FA template onto each subject's native space, thereby distributing the same number of seeds in each subject at anatomically equivalent locations throughout the brain to account for variations in head size, shape, and white-matter distribution. The MNI space was masked to create a volume that corresponded to the common brain volume scanned in each subject's head. The center coordinates of voxels in the common brain volume were considered to be seeds for all subjects. The mm-space coordinates of these seeds were then mapped on a point-to-point basis to the native space of each subject in “floating point” using inverse mapping from MNI space to subject space, as described in Sec. 2.4., without any interpolation. The mapped MNI space seed-points were used to conduct whole-brain tractography in each subject. An in-house developed streamline tractography code based on Euler's method and a step size of 0.2mm with tensor interpolation [31,42,43] was used to generate tracts from all seeds (whole-brain tractography), where the number of seeds and their distribution were normalized in each subject's head as described above. Tracts were propagated along the direction indicated by the primary eigen vector and continued until either a FA threshold was not met or the deflection angle exceeded 45°.
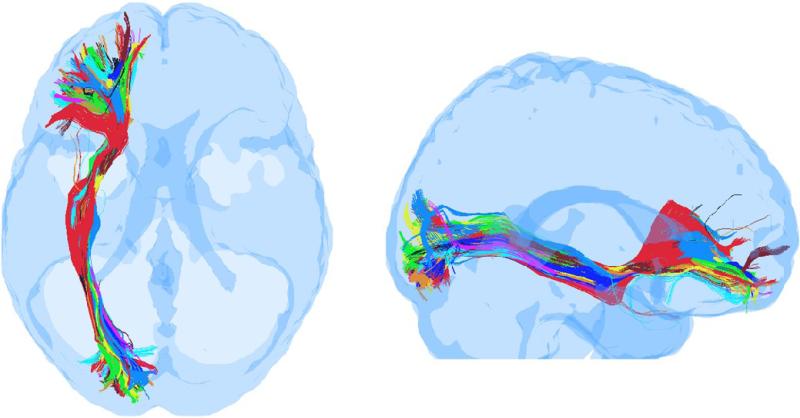
A similar procedure was first proposed by Clayden [44,45] but apparently not pursued due to registration errors in their approach. With our FA template based normalization, we have been able to achieve very good registration among the ten control subjects used in this work suggesting the viability of this approach. An example of the registration accuracy is presented in Fig. 2, where whole-brain tractography was first conducted in each control subject using an equal number of seeds obtained by inverse mapping, and then all tracts from all control subjects were mapped onto a TBI subject's head (see Sec. 2.6.1 below) and sorted in the TBI subject to extract fronto-occipital tracts using planar filters as suggested by Mori et al. [46]. It is visually apparent that the overlap among the ten subjects is very good in the middle portion of the tracts while there are inter-subject variations (to be expected) as the tracts propagate toward cortical boundaries.
Fig. 2.
Superposition of fronto-occipital tracts from 10 control subjects, obtained after mapping whole-brain tracts from all control subjects onto the head of a TBI subject and sorting in the TBI subject's head-space using a common set of frontal and occipital regions as filters. (left) Axial view, (right) Sagittal view.
2.6. Detection and Quantification of Affected Tracts
2.6.1. Individual TBI subject vs. Control Group
Whole-brain tractographs of each TBI subject were filtered by FA reduced ROIs identified in the subject's native space as described above (Sec. 2.4.1) to isolate corresponding pathways affected by each injured region. Quantification of connectivity along the affected pathways was then performed by comparing the number of tracts traversing a given ROI in the TBI subject to the mean number of tracts from the control group also traversing the same ROI in the TBI subject's space after mapping whole-brain tracts of the entire control group to the individual TBI subject's space as described below. Connectivity here is defined by the number of tracts traversing an ROI along a specified pathway. When small step sizes (such as the 0.2mm step size used in this work) are used with interpolation after each step in streamline tractography, and when only one-seed-one-eigen-vector is used per voxel, then each of the ‘n’ voxels lying on a tract emanating from the ith seed will generally also “send” a tract back to the ith voxel. Thus the number of tracts intersecting a voxel is a reflection of how many voxels are connected to it, which provides a convenient metric to quantify connectivity. In this work, the number of tracts (referred to as the tract-count) traversing voxels within a ROI along a specific pathway was used as the metric to quantify the reduction in connectivity in a TBI subject with respect to a control group along the same pathway at a given FA threshold.
A two-step procedure was implemented to map whole-brain tracts from each of the control subjects to the TBI subject's space. Tracts from all control subjects were first mapped to the MNI space and then the TBI subject specific inverse parameters were used to map all tracts from the MNI space to the TBI subject's space. Point-to-point mapping of all points lying on each tract was accomplished in the following way. SPM normalization relies on one-to-one mapping of the vertices of tetrahederal volume elements [47] from one space to another. Let us assume that y = M(x) represents the mapping, via FA template based spatial normalization, of a seed x = [x1, x2, x3], lying at the vertex of a tetrahedral in subject space, to point y = [y1, y2, y3], also lying on a vertex of a tetrahedral in standard MNI space. Then the mapped point y′ corresponding the point x′ on a tract following a small step can be obtained from the following equation,
| (2) |
where the step in any direction is represented by [∂x1 ∂x2 ∂x3], and Jx defines the Jacobian of the deformation at the point x = [x1, x2, x3]. The elements of the Jacobian were determined by the multiplication of partial derivatives of the deformation field along pairs of two orthogonal axes. Similarly the tract point y′= [y1+∂y1, y2+∂y2, y3+∂y3] in the standard MNI space can be transformed back to the point x′ = [x1′, x2′, x3′] in a specific subject space via the following equation,
| (3) |
where M−1(y) indicates a one-to-one mapping of the vertex point y in the standard MNI space to the vertex point x in the subject space. The deformation derivatives at the point y and its increments are represented by the Jacobian matrix Jy and [∂y1 ∂y2 ∂y3] respectively.
Based on the forward and inverse mapping described in Eqs. (2) and (3), all points on any tract in any subject 1 can be transformed to the standard MNI space and then inverse mapped to the subject space of any other subject 2 using the inverse mapping function of subject 2, thereby making it possible to map all tracts from a group of control subjects onto the subject space of an individual TBI subject. In conjunction with the normalized seed-tractography described in Section 2.5, it now becomes possible to compare quantitatively a specific tract identified in a TBI subject to corresponding tracts in a control group, without requiring any manual drawing of any ROI.
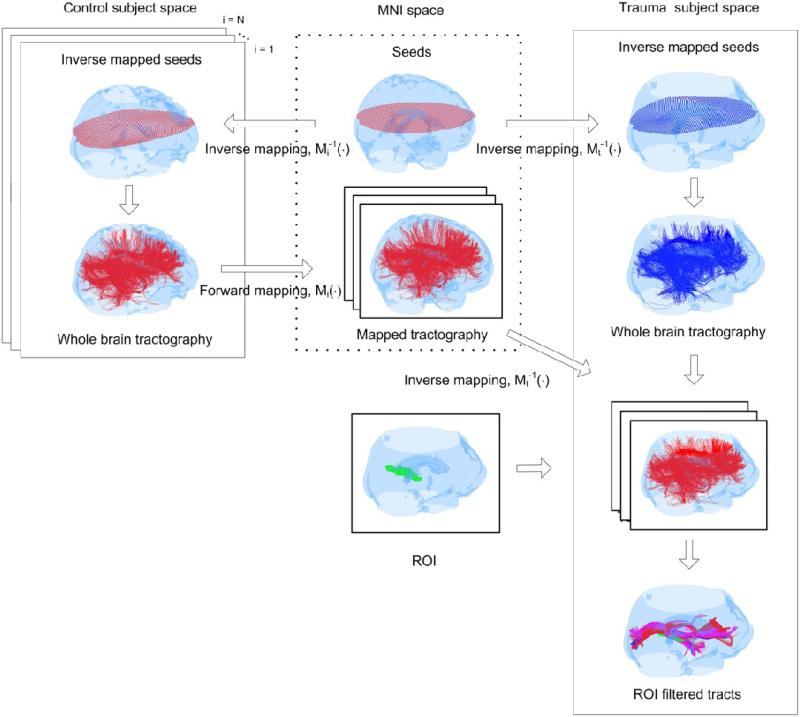
The overall approach to normalize and map tracts from control subjects onto the head of a TBI subject is conceptualized in Fig. 3. Seeds were first distributed in each subject's head using inverse mapping of MNI seeds as shown in the top row, and whole-brain streamline tractography was conducted in each subject's native space from these mapped seeds. All tracts from each of the 10 control subjects (left box in Fig 3) were first mapped onto MNI space using non-linear warping, and then mapped to the head of a TBI subject (right box) where they were sorted by ROIs obtained from the FA comparison (Sec. 2.4.1). An example of tracts sorted by a particular ROI (shown as a green cluster) in the TBI subject space is also shown in Fig. 3, where in this particular case, the ROI cluster identified the fronto-occipital and hippocampal/fornix pathways. In situations like this where more than one pathways were identified by a ROI, the ROI was subdivided into multiple sub-ROIs by identifying voxels along each pathway and grouping voxels (within the ROI) that were common to at least 50% of the control subjects along a given pathway. This procedure was used to mitigate effects of inter-subject tract variations and co-registration errors remaining after our normalization procedure.
Fig. 3.
The procedure used to normalize and sort tracts in each TBI subject's space. (top row) Seeds from the standard MNI space were first distributed within each control and TBI subject's head using inverse normalization to maintain the same number of seeds and anatomical equivalency of seed-distribution in each subject. (left to right blocks) Whole-brain tractography was conducted in each subject and all tracts from all control subjects were first mapped to MNI space and then mapped onto the head space of individual TBI subjects. ROIs were identified in the TBI subject's head using the procedure outlined in Fig. 1. Individual ROIs were then extracted one-by-one from the FA-difference map between the TBI subject and controls, and used to sort tracts. An example of a particular ROI (green) is shown at the bottom of the second column. Tracts were sorted in the TBI subject's space using this ROI from all control subjects and the TBI subject, and the mean number of tracts from the control subjects were compared to those from the TBI subject to generate the tract-count metric for this ROI. The sorted and superimposed tracts for this ROI from two normal subjects (in red and purple respectively) and the TBI subject (blue) are shown at the bottom of the third column.
The number of tracts passing through each ROI (or sub-ROI) were counted in the TBI subject and compared to the mean and standard deviation of the normalized tracts from all control subjects passing through the same ROI (or sub-ROI) in the TBI subject space to quantify the reduction along a specific pathway. When multiple pathways were identified within an ROI, the FA and other diffusion anisotropy metrics were also computed for the sub-ROIs.
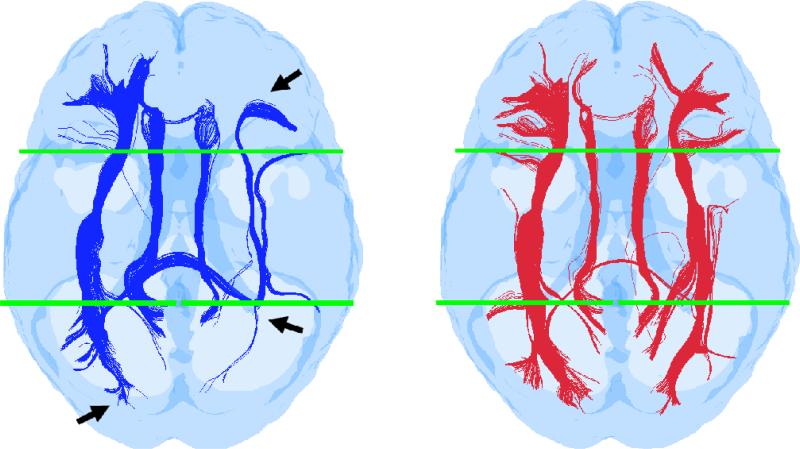
The key feature of our tract normalization approach is to conduct tractography in subject space using a unique seed distribution and then transform each tract individually, first to MNI space and then to the head-space of another subject. Contrary to previous approaches [35,36], our approach does not warp voxels but instead takes each individual tract in subject space composed of a string of points located 0.2mm apart, transforms these points to normalized space using a point-to-point mapping, and connects these mapped points to regenerate the new tract in normalized space. Mapping these individual 0.2mm spaced points and reconnecting them in normalized space is equivalent to reorienting the primary eigen vectors along each tract from subject to normalized space consistent with the non-linear transformation, thereby overcoming the problem of correcting eigen vectors used in previous normalization approaches [35,36]. To validate this key feature, a study was performed to compare tract-normalizations obtained with our approach and the commonly used previous approach [35,36] where the b =0 and all b=1000s/mm2 images are first transformed to MNI space and then the perturbations in eigen vectors are corrected to account for spatial transformations. A common set of sorting filters were employed in both cases to compare the integrity of known fronto-occipital connections after normalization. The results of this comparison are presented in Fig. 4. Though both approaches show recovery of the Inferior Fronto-Occipital (IFO) and Superior Longitudinal Fasciculus (SLF) by the filters used to sort these tracts, our method shows better continuity and branching near the ends (consistent with subject-space mapping) whereas the previous approach shows discontinuities apparently caused by the voxel-based normalization. Also the inherent smoothing in voxel-based normalization increases partial volume artifacts causing a portion of the right IFO tracts to turn around the ventricle, whereas this effect is much reduced in our approach (Fig. 4.).
Fig. 4.
Comparison of previous tensor reorientation-based normalized tractography (left, in blue) to our normalized approach where all subject space tracts are individually mapped to normalized (MNI-space) using point-to-point transformation (right, in red). Sorting of both sets of normalized tractography was conducted with identical ROIs, located in the frontal and occipital areas (shown in green) in template space. The tracts generated with the previous voxel-based normalization and reoriented tensor method tend to suffer from discontinuity toward the ends. Compared to our approach, the previous method also appears to increase the confounds of partial volume effects (arrows indicate areas of tract discontinuity and redirection around the posterior right corner of the ventricle) due to the interpolation inherent to voxel based normalization.
2.6.2. TBI Group vs. Control Group
The group analysis was done in MNI space. The same procedure as described above was used to map all tracts from all subjects including the TBI and control subjects into the common MNI space, and ROIs defined from the FA group comparison in MNI space (Sec. 2.4.2) were used to isolate pathways affected by specific ROIs in each subject. Tract counts along these pathways were compared between the two groups to quantify the reduction in connectivity.
3. Results
3.1. Individual TBI subjects
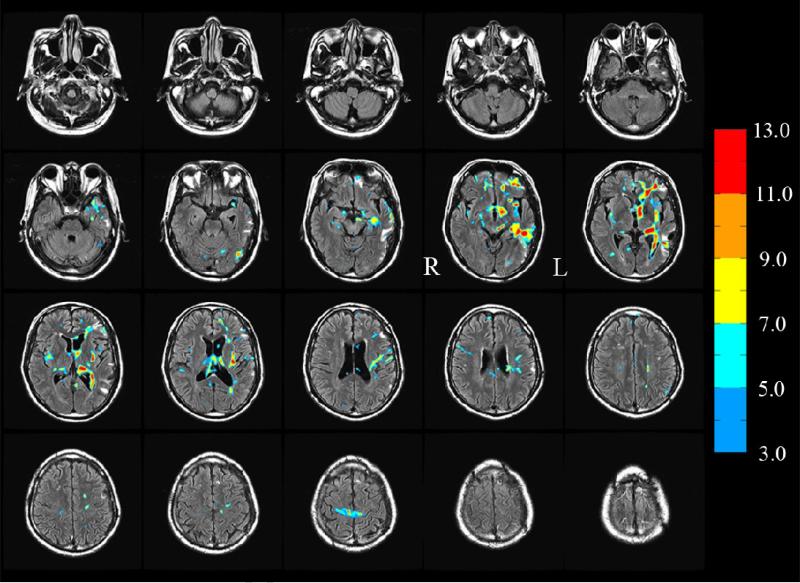
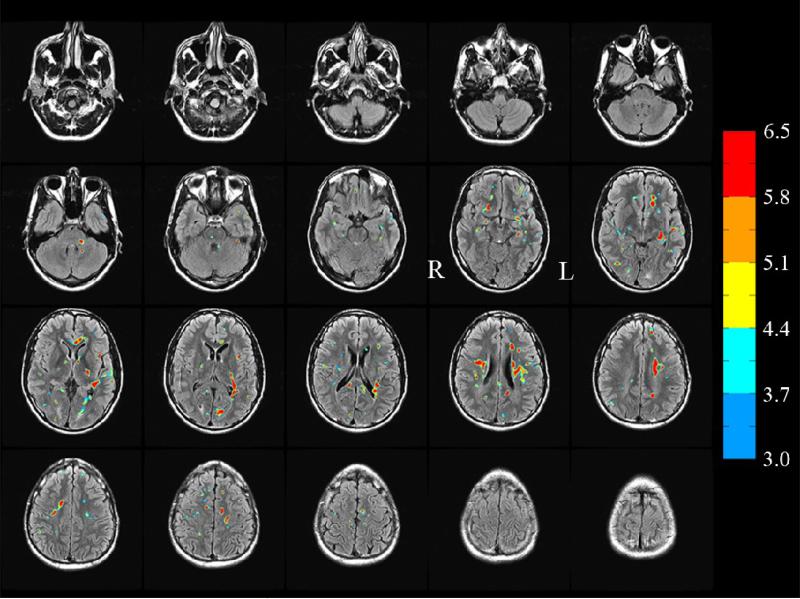
An example of t-score maps corresponding to significant reduction in FA (t ≥ 3.0, see color bar) and cluster extent k ≥ 12 between a TBI subject (subject 1) and 10 controls is presented in Fig. 5 where the FA reduced and regions have been superimposed on TBI subject's FLAIR images. It can be seen that there is partial overlap between the FLAIR spots (which mainly highlight edema resulting from cellular injury) and the FA reduced regions though there are regions highlighted in FLAIR but not observed in the FA reduced maps and vice versa, consistent with previous studies [8,24,27,33,48]. These results are also consistent with a recent study [49] where partial overlap between the FLAIR and FA-reduced regions has been explained by a model of the contrast mechanism in each modality.
Fig. 5.
FA reduced regions corresponding to t ≥ 3.0 (see color bar) subject 1. An extent threshold k ≥12 was also used to identify clusters. Some FA-reduced identify regions overlap completely, others partially and some do not overlap with the FLAIR spots. (In this montage, the left hemisphere L appears at the right in each image).
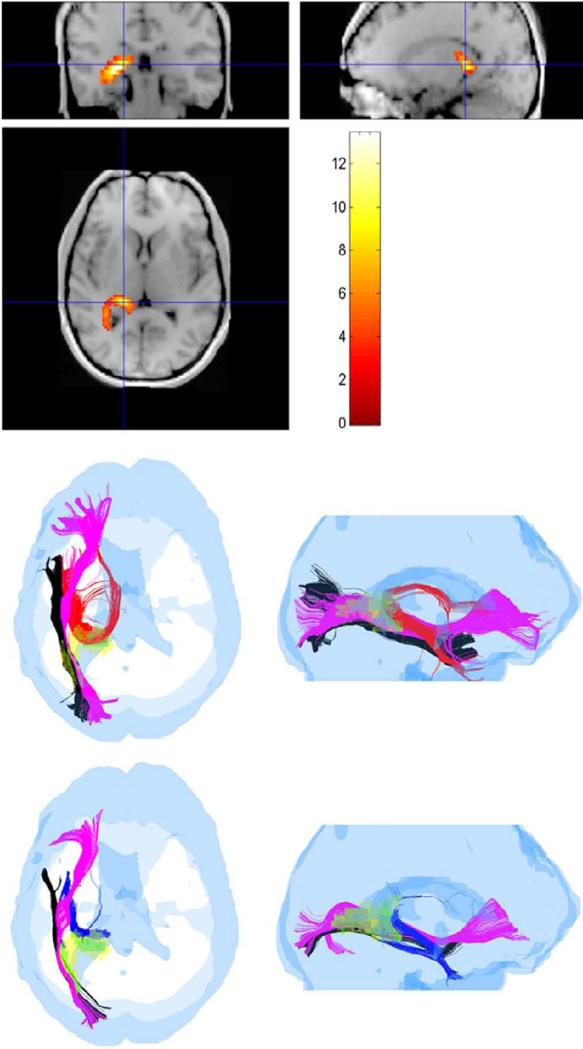
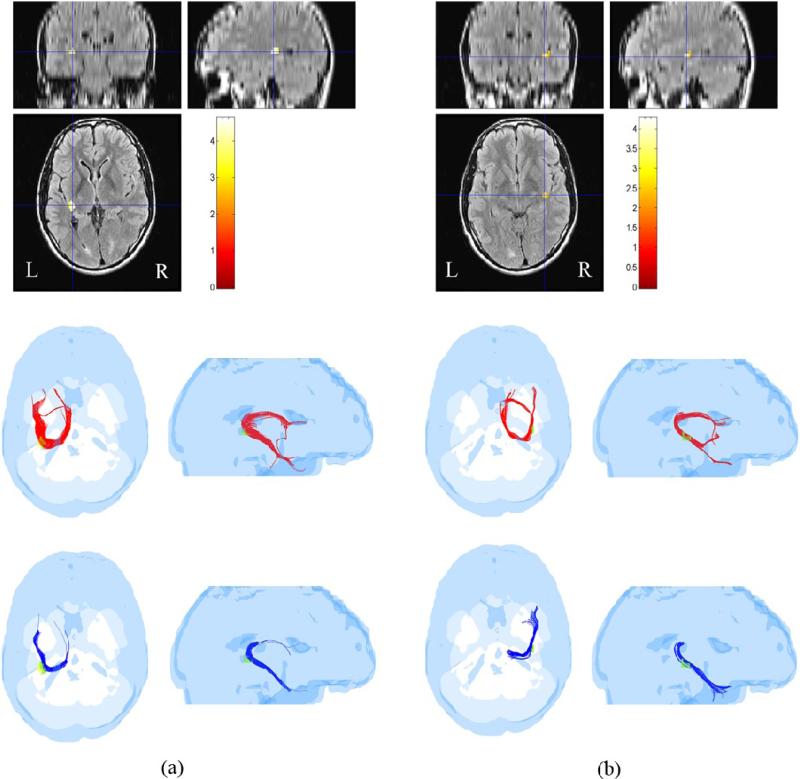
Using the FA-reduced regions of Fig. 5 as ROIs, the sorted tracts in TBI subject 1 were compared to sorted tracts obtained from each of the 10 control subjects for the same ROIs after mapping all tracts of all control subjects to the head of the TBI subject as described in Sec. 2.6.1. As an example, pathways detected by the ROI corresponding to the highest t-score in TBI subject 1 are shown in Fig. 6 at FA ≥ 0.15.
Fig. 6.
Three pathways, hippocampal/fornix (HC/FX), inferior fronto-occipital (IFO) and inferior longitudinal fasciculus (ILF) identified as crossing the voxels of the highest t-score ROI in a TBI subject (subject 1). (top panel) Coronal, sagittal and axial views of the ROI with color-coding of the t-score as indicated in the color bar. (middle row) The three pathways (HC/FX-red, IFO-magenta, ILF-black) in a normal subject shown in axial and sagittal views. (bottom row) The same three pathways (HC/FX-blue, IFO-magenta, ILF-black) in a TBI subject. (The yellow and green colors in the middle and bottom row pictures were used to identify sub-ROIs within the ROI as described in the text).
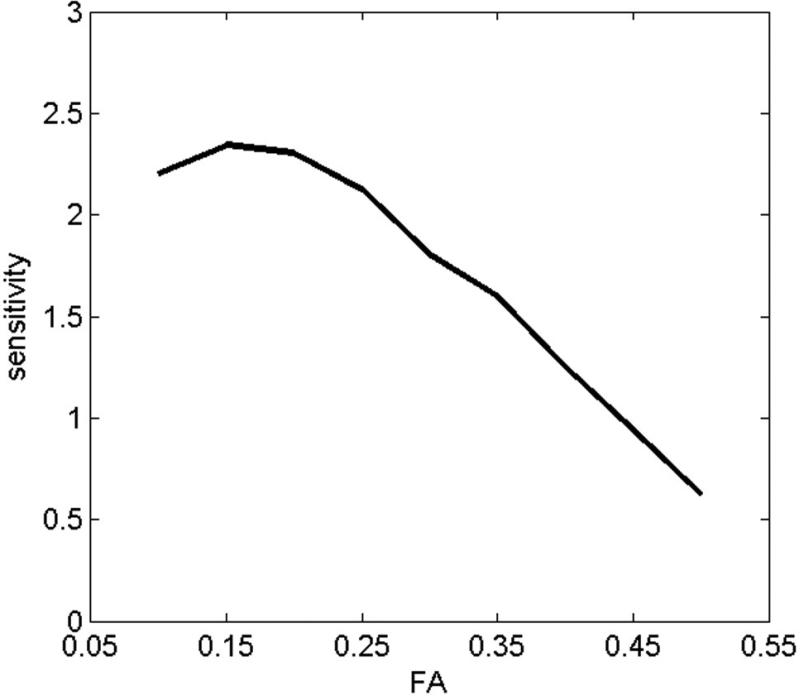
The FA threshold has a significant impact on tract-counts. FA thresholds ranging from 0.10 to 0.3 have been commonly used in streamline tractography, representing a tradeoff between noise (which increases as the FA threshold is lowered) and sensitivity (which decreases as the FA threshold is raised). A previous study where the influence of FA threshold on the accuracy of DTI tractography was evaluated by comparing histology (ground truth) to DTI tractography at FA thresholds varying from 0 to 0.60 per every 0.05 [50] reported that the best visual agreement was achieved at a FA threshold of 0.10 in that work. To determine an optimal FA for this study, we measured the sensitivity of tract-counts to differentiate between controls and individual TBI subjects as a function of FA threshold. A tract-count ‘TC’ based effect size was defined as:
where TCi (control), TCi (individual) and σi represent mean TC of the control group, TC of an individual TBI subject and the standard deviation in the TC values of the control group respectively for the i-th ROI. The effect size was computed for 28 FA-reduced ROIs identified in 2 TBI subjects at the t ≥ 3.0, k ≥ 12 mean effect size over these 28 ROIs as a function of FA is presented in Fig. 7, showing that the effect size peaks at a FA threshold of about 0.15. Thus an FA threshold of 0.15 was used in this work though the results would not change significantly for FA thresholds ranging from 0.1 to 0.2. Examples of pathways identified by two other relatively high t-score FA-reduced regions for TBI subject 1 are shown in Fig. 8 at FA ≥ 0.15.
Fig. 7.
Tract count-based sensitivity (effect size) to differentiate TBI subjects from controls as a function of FA.
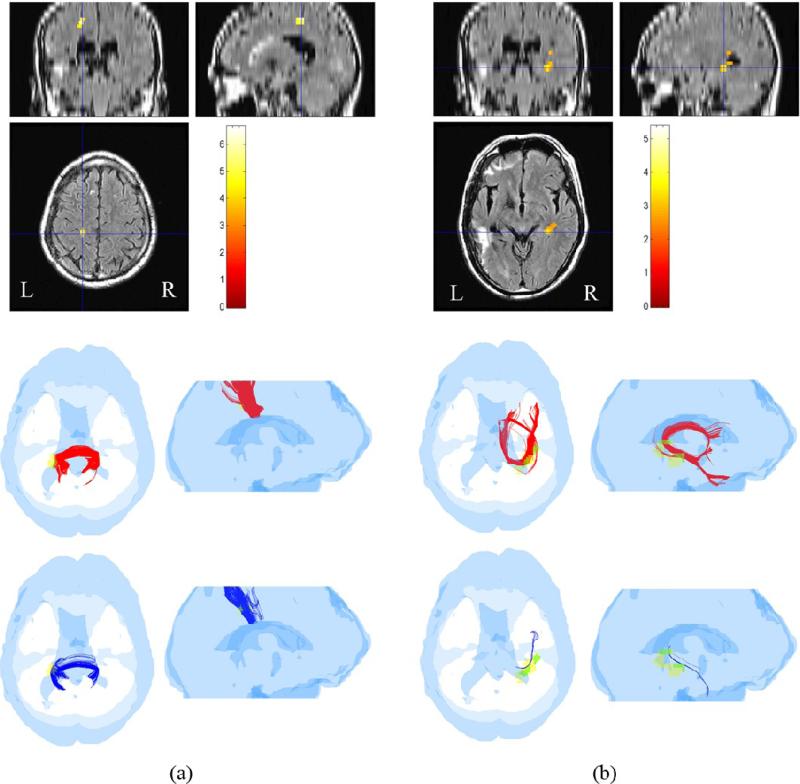
Fig. 8.
Similar to Fig. 6, pathways associated with two other ROIs within TBI subject 1 are shown in (a) and (b) respectively. Coronal, sagittal and axial views of the two ROIs with color-coding of the t-score as indicated in the color bar are shown in the top row. The ROI in (a) identified tracts through the posterior portion of the corpus callosum, whereas (b) identified the right HC/FX tracts. Like Fig. 6, tracts for a normal subject are shown in red and those in the TBI subject are shown in blue. The reduction of tract-counts in the TBI subject with respect to the control subject is obvious for these ROIs.
Though most ROIs identified only one pathway or tract, some ROIs, for example the ROI shown in Fig. 6, identified three distinct pathways going through it, namely hippocampal/fornix (HC/FX), inferior fronto-occipital (IFO) and inferior longitudinal fasciculus (ILF). In situations like this where the ROI contained multiple pathways, or the situation where some voxels within a ROI did not show any pathway as their FA values were below the FA threshold, the ROI was partitioned into multiple sub-ROIs by grouping voxels within each sub-ROI that were common to tracts along specific pathways in at least 50% of the control subjects. The diffusion anisotropy and tract-count metrics were then computed within these sub-ROIs. A summary of these metrics and the pathways identified by the ROIs/sub-ROIs is presented in Table 1. The values of FA (and other diffusion metrics) for all voxels contained within the ROIs were pooled for the ten control subjects to compute the mean and standard deviation. When a ROI indicated multiple pathways, only those voxels within the ROI that contained tracts from at least 50% of the subjects along a specific pathway were considered. In addition to FA, the anisotropy metrics DA, DR and MD were also computed as listed in Table 1.
Table 1.
List of affected pathways and diffusion metrics for 12 ROIs identified by FA-reduction in a TBI subject (subject 1) compared to a group of 10 normal controls (t ≥ 3.00, k ≥ 12). All analysis was done in the TBI subject's head-space. HC: hippocampus; FX: fornix; IFO: inferior fronto-occipital fasciculus; ILF: inferior longitudinal fasciculus; CC-s: corpus callosum splenium; CC-g: corpus callosum genu; UF: uncinate fasciculus; CST: corticospinal tract; CG: cingulum; SLF: superior longitudinal fasciculus; FA: fractional anisotropy; DA: axial diffusivity; DR: radial diffusivity; MD: mean diffusivity; TC: tract count.
| ROI | t -score | Pathway | FA | Effect Size | TC | Effect Size | ||
|---|---|---|---|---|---|---|---|---|
| Control | TBI | Control | TBI | |||||
| 1 | 5.35 (1.68) | HC/FX | 0.32 (0.10) | 0.17 (0.11) | 1.43 | 367.80 (88.58) | 135 | 2.63 |
| IFO-L | 0.47 (0.08) | 0.21 (0.12) | 2.55 | 421.50 (114.29) | 119 | 2.65 | ||
| ILF-L | 0.49 (0.08) | 0.24 (0.09) | 2.94 | 404.30 (180.02) | 121 | 1.57 | ||
| 2 | 5.29 (1.59) | IFO-L | 0.28 (0.06) | 0.11 (0.04) | 3.33 | 409.60 (150.97) | 62 | 2.30 |
| 3 | 5.00 (0.83) | CC-g | 0.47 (0.03) | 0.33 (0.01) | 6.26 | 238.00 (55.04) | 171 | 1.22 |
| UF-R | 0.42 (0.04) | 0.31 (0.02) | 3.48 | 113.80 (41.11) | 55 | 1.43 | ||
| 4 | 4.84 (0.85) | CC-s | 0.45 (0.05) | 0.28 (0.07) | 2.79 | 375.80 (116.83) | 169 | 1.77 |
| 5 | 4.80 (1.14) | FX-L | 0.29 (0.10) | 0.16 (0.09) | 1.37 | 253.50 (110.68) | 91 | 1.47 |
| IFO-L | 0.47 (0.07) | 0.28 (0.07) | 2.71 | 347.80 (108.94) | 74 | 2.51 | ||
| ILF-L | 0.48 (0.06) | 0.29 (0.07) | 2.91 | 410.10 (181.88) | 115 | 1.62 | ||
| 6 | 4.77 (0.93) | CC-g | 0.44 (0.09) | 0.27 (0.08) | 2.00 | 589.70 (184.80) | 571 | 0.10 |
| IFO-L | 0.38 (0.06) | 0.25 (0.05) | 2.35 | 526.20 (120.94) | 81 | 3.68 | ||
| 7 | 4.32 (0.53) | CG/FX | 0.25 (0.07) | 0.10 (0.04) | 2.63 | 215.40 (64.76) | 10 | 3.17 |
| 8 | 4.51 (0.64) | IFO-L | 0.35 (0.07) | 0.18 (0.04) | 2.98 | 285.80 (102.36) | 51 | 2.29 |
| 9 | 4.67 (1.22) | SLF-L | 0.43 (0.09) | 0.30 (0.10) | 1.37 | 957.10 (146.47) | 730 | 1.55 |
| 10 | 4.58 (0.76) | CST | 0.30 (0.12) | 0.16 (0.09) | 1.32 | 594.50 (205.11) | 15 | 2.83 |
| 11 | 4.18 (0.45) | UF-L | 0.22 (0.05) | 0.11 (0.03) | 2.67 | 75.80 (46.05) | 16 | 1.30 |
| 12 | 3.13 (0.61) | HC/FX-R | 0.32 (0.09) | 0.22 (0.09) | 1.11 | 232.90 (70.22) | 8 | 3.20 |
| ROI | Pathway | MD [x 10−3 mm2/s] | Effect Size | DA [x 10−3 mm2/s] | Effect Size | DR [x 10−3 mm2/s] | Effect Size | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | TBI | Control | TBI | Control | TBI | |||||
| 1 | HC/FX | 0.78 (0.20) | 1.91 (0.55) | 2.73 | 1.05 (0.23) | 2.16 (0.48) | 2.95 | 0.65 (0.20) | 1.78 (0.58) | 2.60 |
| IFO-L | 0.83 (0.20) | 1.83 (0.59) | 2.27 | 1.28 (0.20) | 2.14 (0.49) | 2.30 | 0.61 (0.21) | 1.67 (0.64) | 2.23 | |
| ILF-L | 0.74 (0.11) | 1.51 (0.44) | 2.40 | 1.18 (0.14) | 1.84 (0.39) | 2.25 | 0.53 (0.11) | 1.35 (0.47) | 2.40 | |
| 2 | IFO-L | 0.72 (0.06) | 1.21 (0.18) | 3.65 | 0.94 (0.07) | 1.34 (0.18) | 2.93 | 0.61 (0.07) | 1.15 (0.18) | 3.95 |
| 3 | CC-g | 0.74 (0.03) | 0.81 (0.01) | 3.13 | 1.17 (0.04) | 1.13 (0.01) | −1.37 | 0.52 (0.03) | 0.66 (0.01) | 6.26 |
| UF-R | 0.75 (0.03) | 0.81 (0.01) | 2.68 | 1.12 (0.06) | 1.10 (0.02) | −0.45 | 0.56 (0.04) | 0.67 (0.02) | 3.48 | |
| 4 | CC-s | 0.64 (0.03) | 0.72 (0.03) | 2.67 | 0.97 (0.07) | 0.92 (0.04) | −0.88 | 0.47 (0.03) | 0.62 (0.05) | 3.64 |
| 5 | FX-L | 0.97 (0.24) | 1.64 (0.51) | 1.68 | 1.24 (0.24) | 1.85 (0.50) | 1.56 | 0.83 (0.24) | 1.54 (0.52) | 1.75 |
| IFO-L | 0.76 (0.07) | 1.11 (0.24) | 1.98 | 1.17 (0.11) | 1.41 (0.23) | 1.33 | 0.55 (0.07) | 0.96 (0.26) | 2.15 | |
| ILF-L | 0.73 (0.07) | 1.02 (0.21) | 1.85 | 1.15 (0.11) | 1.31 (0.22) | 0.92 | 0.52 (0.07) | 0.87 (0.22) | 2.14 | |
| 6 | CC-g | 0.82 (0.20) | 1.18 (0.51) | 0.93 | 1.23 (0.25) | 1.48 (0.54) | 0.59 | 0.61 (0.19) | 1.02 (0.50) | 1.08 |
| IFO-L | 0.72 (0.06) | 0.97 (0.22) | 1.55 | 1.04 (0.09) | 1.21 (0.20) | 1.10 | 0.56 (0.07) | 0.85 (0.24) | 1.64 | |
| 7 | CG/FX | 1.22 (0.43) | 2.25 (0.42) | 2.42 | 1.54 (0.48) | 2.44 (0.42) | 2.00 | 1.07 (0.41) | 2.15 (0.42) | 2.60 |
| 8 | IFO-L | 0.72 (0.04) | 0.97 (0.06) | 4.90 | 1.01 (0.06) | 1.14 (0.06) | 2.17 | 0.58 (0.06) | 0.89 (0.07) | 4.76 |
| 9 | SLF-L | 0.62 (0.03) | 0.83 (0.17) | 1.72 | 0.94 (0.10) | 1.08 (0.16) | 1.05 | 0.46 (0.05) | 0.70 (0.19) | 1.73 |
| 10 | CST | 0.94 (0.21) | 1.81 (0.43) | 2.57 | 1.23 (0.24) | 2.04 (0.38) | 2.55 | 0.80 (0.23) | 1.69 (0.46) | 2.45 |
| 11 | UF-L | 0.76 (0.07) | 1.45 (0.14) | 6.23 | 0.94 (0.07) | 1.60 (0.12) | 6.72 | 0.67 (0.08) | 1.38 (0.16) | 5.61 |
| 12 | HC/FX-R | 0.87 (0.21) | 1.32 (0.48) | 1.21 | 1.15 (0.21) | 1.58 (0.48) | 1.16 | 0.73 (0.21) | 1.20 (0.49) | 1.25 |
The FA-reduced regions of another TBI subject (subject 2), detected at t ≥ 3.0, k ≥ 12, superimposed on the subject's FLAIR images, are presented in Fig. 9, and, as an example of quantifying pathways affected by injury, the tracts traversing two of the FA-reduced regions in this subject are shown in Fig. 10. The anisotropy metrics and the tract-counts for all ROIs at t ≥ 3.0, k ≥ 12 are summarized in Table 2 for TBI subject 2.
Fig. 9.
Similar to Fig. 5, the FA reduced regions for TBI subject 2 superimposed on the FLAIR images of the subject. (In this montage, the left hemisphere L appears at the right in each image).
Fig. 10.
Similar to Fig. 6, pathways associated with two ROIs in TBI subject 2 are shown in (a) and (b) respectively. The ROI in (a) identified the left HC/FX tracts, whereas (b) identified the right HC/FX tracts. Like Fig. 6, tracts for a normal subject are shown in red and those in the TBI subject are shown in blue. The reduction of tract-counts in the TBI subject with respect to the control subject is obvious in these ROIs.
Table 2.
List of affected pathways and diffusion metrics for 11 ROIs identified by FA-reduction in a TBI subject (subject 2) compared to a group of 10 normal controls (t ≥ 3.0, k ≥ 12). All analysis was done in the TBI subject's head-space. CC-b: corpus callosum body; CC-s: corpus callosum splenium; HC: hippocampus; FX: fornix; ILF: inferior longitudinal fasciculus; CC-g: corpus callosum genu; CST: corticospinal tract; SLF: superior longitudinal fasciculus; FA: fractional anisotropy; DA: axial diffusivity; DR: radial diffusivity; MD: mean diffusivity; TC: tract count.
| ROI | t -score | Pathway | FA | Effect Size | TC | Effect Size | ||
|---|---|---|---|---|---|---|---|---|
| Control | TBI | Control | TBI | |||||
| 1 | 4.13 (0.91) | CC-b | 0.29 (0.08) | 0.09 (0.02) | 3.43 | 225.50 (111.24) | 2 | 2.01 |
| 2 | 4.09 (0.82) | CC-s | 0.49 (0.07) | 0.33 (0.05) | 2.63 | 422.50 (136.31) | 249 | 1.27 |
| 3 | 4.03 (0.93) | CC-s | 0.37 (0.08) | 0.17 (0.08) | 2.50 | 543.80 (141.35) | 174 | 2.62 |
| 4 | 3.89 (0.53) | ILF-L | 0.46 (0.07) | 0.26 (0.09) | 2.48 | 277.10 (117.08) | 8 | 2.30 |
| 5 | 3.79 (0.54) | CC-g | 0.35 (0.04) | 0.23 (0.02) | 3.79 | 316.20 (118.45) | 317 | −0.01 |
| 6 | 3.73 (0.56) | CST | 0.48 (0.05) | 0.38 (0.05) | 2.00 | 433.00 (151.25) | 355 | 0.52 |
| 7 | 3.71 (0.62) | CC-b | 0.51 (0.03) | 0.39 (0.01) | 5.37 | 108.80 (94.95) | 30 | 0.83 |
| SLF-L | 0.31 (0.07) | 0.25 (0.07) | 0.86 | 45.00 (41.46) | 15 | 0.72 | ||
| 8 | 3.67 (0.47) | SLF-L | 0.49 (0.05) | 0.36 (0.07) | 2.14 | 536.00 (179.76) | 177 | 2.00 |
| 9 | 3.58 (0.38) | CC-s | 0.47 (0.11) | 0.16 (0.03) | 3.85 | 658.50 (109.39) | 206 | 4.14 |
| 10 | 3.54 (0.45) | SLF-L | 0.39 (0.05) | 0.27 (0.03) | 2.91 | 307.00 (146.48) | 168 | 0.95 |
| 11 | 2.67 (0.59) | HC/FX-R | 0.33 (0.06) | 0.27 (0.05) | 1.09 | 117.40 (53.08) | 78 | 0.74 |
| ROI | Pathway | MD [x 10−3 mm2/s] | Effect Size | DA [x 10−3 mm2/s] | Effect Size | DR [x 10−3 mm2/s] | Effect Size | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | TBI | Control | TBI | Control | TBI | |||||
| 1 | CC-b | 0.65 (0.05) | 0.92 (0.09) | 3.71 | 0.85 (0.07) | 1.00 (0.10) | 1.74 | 0.54 (0.07) | 0.88 (0.09) | 4.22 |
| 2 | CC-s | 0.67 (0.03) | 0.79 (0.03) | 4.00 | 1.08 (0.09) | 1.03 (0.05) | −0.69 | 0.47 (0.04) | 0.66 (0.04) | 4.75 |
| 3 | CC-s | 0.72 (0.06) | 0.85 (0.09) | 1.70 | 1.02 (0.10) | 0.99 (0.07) | −0.35 | 0.57 (0.07) | 0.78 (0.11) | 2.28 |
| 4 | ILF-L | 0.68 (0.03) | 0.83 (0.05) | 3.64 | 1.06 (0.07) | 1.05 (0.06) | −0.15 | 0.49 (0.06) | 0.72 (0.07) | 3.53 |
| 5 | CC-g | 0.68 (0.04) | 0.81 (0.02) | 4.11 | 0.95 (0.05) | 1.01 (0.04) | 1.33 | 0.55 (0.05) | 0.72 (0.03) | 4.12 |
| 6 | CST | 0.62 (0.02) | 0.69 (0.01) | 4.43 | 0.98 (0.07) | 1.00 (0.06) | 0.31 | 0.44 (0.02) | 0.54 (0.02) | 5.00 |
| 7 | CC-b | 0.63 (0.03) | 0.71 (0.02) | 3.14 | 1.03 (0.04) | 1.02 (0.02) | −0.32 | 0.43 (0.03) | 0.55 (0.02) | 4.71 |
| SLF-L | 0.62 (0.05) | 0.75 (0.14) | 1.24 | 0.82 (0.06) | 0.95 (0.11) | 1.47 | 0.52 (0.06) | 0.66 (0.15) | 1.23 | |
| 8 | SLF-L | 0.69 (0.04) | 0.82 (0.03) | 3.68 | 1.09 (0.09) | 1.15 (0.08) | 0.70 | 0.48 (0.04) | 0.66 (0.05) | 3.98 |
| 9 | CC-s | 0.89 (0.23) | 0.88 (0.10) | −0.06 | 1.38 (0.28) | 1.04 (0.12) | −1.58 | 0.64 (0.23) | 0.81 (0.10) | 0.96 |
| 10 | SLF-L | 0.73 (0.04) | 0.88 (0.03) | 4.24 | 1.07 (0.08) | 1.14 (0.05) | 1.05 | 0.56 (0.04) | 0.75 (0.04) | 4.75 |
| 11 | HC/FX-R | 0.83 (0.09) | 0.93 (0.08) | 1.17 | 1.12 (0.11) | 1.19 (0.10) | 0.67 | 0.69 (0.10) | 0.79 (0.08) | 1.10 |
3.2 Group Study: Control vs. TBI group
The results of the group comparison between 10 normal controls and 12 TBI subjects are presented in Figs. 11-12. The FA-reduced regions in the TBI group at pFDR ≤ 0.05, k ≥ 12 are shown in Fig. 11, where these FA-reduced regions have been superimposed on our FA template in MNI space to show their relative location with respect to some of the affected pathways. As an example, the pathways obtained by using two of these ROIs to sort tracts from whole-brain tracts, are shown in Fig. 12. Just like individual TBI subjects, when multiple pathways were detected within an ROI, for example as indicated in Fig. 12(a) where the HC/FX, IFO and a portion of the ILF were detected, the anisotropy and tract-count metrics were computed within sub-ROIs formed by voxels lying at the intersection of tracts from at least 50% of the subjects. Also as in single TBI subject studies, the same procedure was used to refine those ROIs where some voxels did not contain any tracts as their FA values were below the threshold.
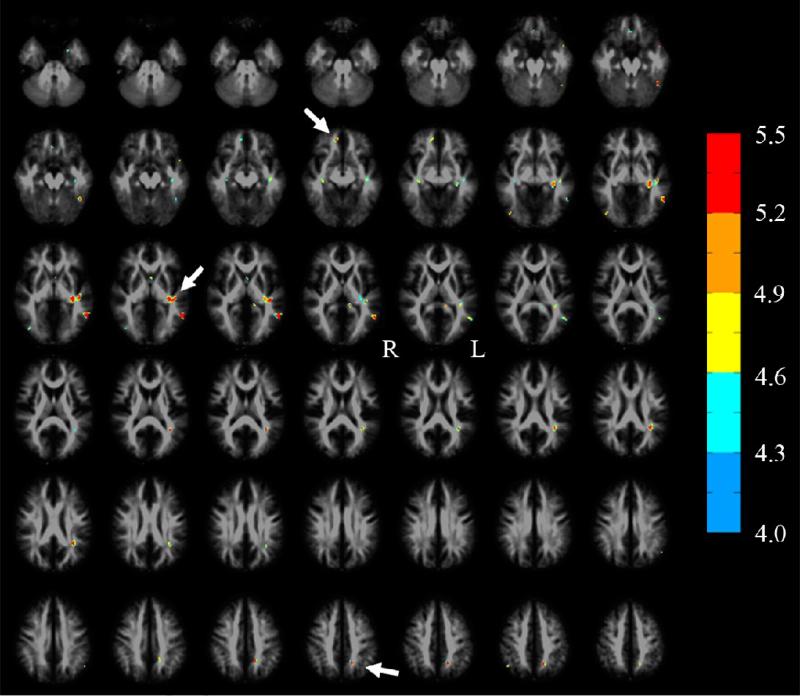
Fig. 11.
A superposition of the FA reduced regions (in color) detected by a SPM based statistical comparison of the TBI group (n=12) to the normal group (n=10) at pFDR ≤0.05, k ≥ 12 on the FA template in MNI space. Arrows point to the three ROIs (genu of the corpus callosum, hippocampal region, splenium of the corpus callosum) used in Fig. 12
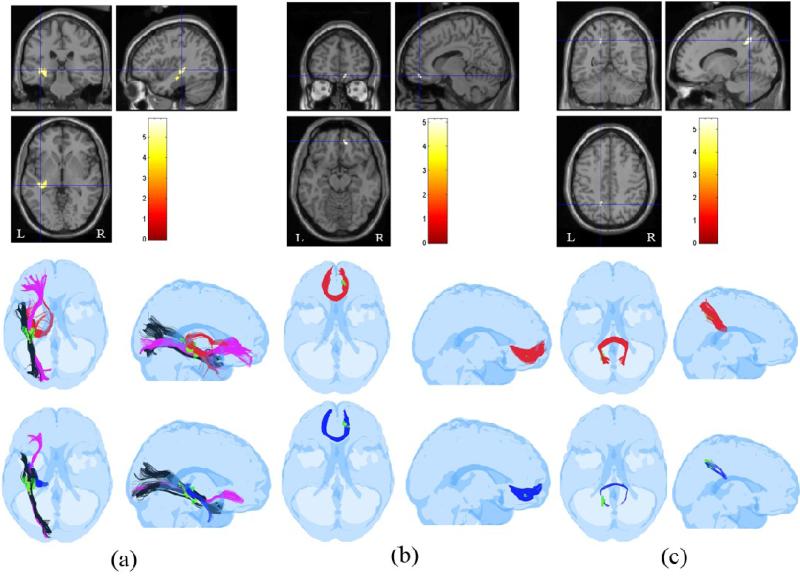
Fig. 12.
Similar to Fig. 6, pathways associated with three ROIs identified as FA-reduced regions in the group study between 10 controls and 12 TBI subjects are shown in (a) , (b), and (c) respectively. The ROI in the middle row of (a) identified the left IFO, HC/FX and a portion of the ILF for a normal control subject (IFO: purple, ILF: black, HC/FX: red), (b) identified the anterior portion of the corpus callosum (red) , and (c) identified the posterior portion of the corpus callosum (red). The bottom row shows corresponding tracts identified by the same ROIs in a TBI subject. (bottom row, a) IFO: purple, ILF: black, HC/FX: blue. (bottom row, b) anterior corpus callosum: blue. (bottom row, c) posterior corpus callosum: blue. The reduction of tract-counts in the TBI subject with respect to the control subject is obvious in these ROIs.
The anterior and inferior portions of the corpus callosum were frequently injured in this cohort of TBI subjects. An example of the ROI associated with injury to the anterior (genu) of the corpus callosum and affected tracts is shown in Fig. 12(b). All the detected ROIs with their associated pathways, and the anisotropy and tract-count metrics for these ROIs, are listed in Table 3.
Table 3.
List of affected pathways and diffusion metrics for 12 ROIs identified by SPM group analysis as FA-reduced regions in the TBI group (n=12) compared to the normal control group (n=10) (pFDR ≤ 0.05, k ≥ 12). SLF: superior longitudinal fasciculus; CC-s: corpus callosum splenium; HC: hippocampus; FX: fornix; IFO: inferior fronto-occipital fasciculus; ILF: inferior longitudinal fasciculus; CC-g: corpus callosum genu; UF: uncinate fasciculus; Cg: cingulum; FA: fractional anisotropy; DA: axial diffusivity; DR: radial diffusivity; MD: mean diffusivity; TC: tract count.
| ROI | t -score | Pathway | FA | Effect Size | TC | Effect Size | ||
|---|---|---|---|---|---|---|---|---|
| Control | TBI | Control | TBI | |||||
| 1 | 5.33 (0.15) | ILF-L | 0.12 (0.03) | 0.08 (0.02) | 1.57 | 109.60 (81.72) | 16.08 (31.53) | 1.51 |
| 2 | 4.92 (0.43) | SLF-L | 0.26 (0.09) | 0.14 (0.08) | 1.41 | 749.30 (170.43) | 284.5 (212.85) | 2.41 |
| 3 | 4.87 (0.46) | ILF-L | 0.16 (0.05) | 0.12 (0.04) | 0.88 | 76.70 (40.40) | 13.5 (13.59) | 2.10 |
| 4 | 4.75 (0.34) | CC-s-L | 0.47 (0.08) | 0.38 (0.08) | 1.13 | 343.40 (118.87) | 217.25 (115.19) | 1.08 |
| 5 | 4.7 (0.40) | ILF-L | 0.16 (0.04) | 0.13 (0.03) | 0.85 | 118.60 (43.23) | 63.17 (72.98) | 0.92 |
| UF-L | 0.16 (0.04) | 0.14 (0.03) | 0.57 | 41.40 (37.17) | 20.08 (15.04) | 0.75 | ||
| 6 | 4.66 (0.30) | HC/FX-L | 0.36 (0.11) | 0.28 (0.11) | 0.73 | 417.40 (69.89) | 339.33 (83.81) | 1.01 |
| IFO-L | 0.43 (0.07) | 0.38 (0.06) | 0.77 | 322.30 (159.08) | 203.67 (153.30) | 0.76 | ||
| ILF-L | 0.48 (0.07) | 0.40 (0.08) | 1.06 | 365.60 (157.05) | 202.33 (122.52) | 1.16 | ||
| 7 | 4.55 (0.29) | CC-g-L | 0.36 (0.08) | 0.28 (0.08) | 1.00 | 393.50 (141.60) | 269.5 (111.55) | 0.97 |
| 8 | 4.55 (0.19) | SLF-L | 0.13 (0.02) | 0.09 (0.03) | 1.57 | 80.40 (46.18) | 22.33 (16.54) | 1.67 |
| 9 | 4.53 (0.22) | ILF-R | 0.24 (0.08) | 0.14 (0.06) | 1.41 | 292.60 (122.30) | 168.58 (97.20) | 1.12 |
| 10 | 4.51 (0.26) | UF-L | 0.16 (0.04) | 0.13 (0.04) | 0.75 | 75.50 (46.91) | 39.25 (25.54) | 0.96 |
| 11 | 4.41 (0.15) | HC-R | 0.29 (0.08) | 0.26 (0.08) | 0.38 | 214.10 (69.60) | 168.08 (59.74) | 0.71 |
| IFO-R | 0.41 (0.02) | 0.37 (0.06) | 0.89 | 518.00 (88.66) | 250.42 (54.99) | 3.63 | ||
| 12 | 4.38 (0.11) | FX | 0.19 (0.04) | 0.15 (0.03) | 1.13 | 52.30 (21.47) | 12.5 (12.24) | 2.28 |
| ROI | Pathway | MD [x 10−3 mm2/s] | Effect Size | DA [x 10−3 mm2/s] | Effect Size | DR [x 10−3 mm2/s] | Effect Size | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | TBI | Control | TBI | Control | TBI | |||||
| 1 | ILF-L | 0.77 (0.05) | 0.92 (0.22) | 0.94 | 0.86 (0.05) | 0.99 (0.22) | 0.81 | 0.73 (0.06) | 0.88 (0.23) | 0.89 |
| 2 | SLF-L | 0.70 (0.06) | 0.93 (0.27) | 1.18 | 0.88 (0.07) | 1.05 (0.27) | 0.86 | 0.61 (0.08) | 0.87 (0.28) | 1.26 |
| 3 | ILF-L | 0.73 (0.04) | 0.85 (0.12) | 1.34 | 0.84 (0.05) | 0.94 (0.12) | 1.09 | 0.67 (0.05) | 0.81 (0.12) | 1.52 |
| 4 | CC-s-L | 0.68 (0.04) | 0.76 (0.05) | 1.77 | 1.07 (0.10) | 1.09 (0.09) | 0.21 | 0.49 (0.05) | 0.60 (0.07) | 1.81 |
| 5 | ILF-L | 0.81 (0.07) | 0.94 (0.12) | 1.32 | 0.94 (0.08) | 1.06 (0.12) | 1.18 | 0.74 (0.08) | 0.88 (0.12) | 1.37 |
| UF-L | 0.80 (0.08) | 0.95 (0.11) | 1.56 | 0.93 (0.08) | 1.08 (0.12) | 1.47 | 0.74 (0.08) | 0.88 (0.11) | 1.46 | |
| 6 | HC/FX-L | 0.81 (0.15) | 1.04 (0.37) | 0.81 | 1.12 (0.17) | 1.32 (0.36) | 0.71 | 0.65 (0.17) | 0.90 (0.38) | 0.85 |
| IFO-L | 0.73 (0.07) | 0.84 (0.11) | 1.19 | 1.09 (0.10) | 1.19 (0.13) | 0.86 | 0.55 (0.08) | 0.67 (0.11) | 1.25 | |
| ILF-L | 0.71 (0.05) | 0.81 (0.07) | 1.64 | 1.12 (0.09) | 1.18 (0.11) | 0.60 | 0.51 (0.06) | 0.62 (0.09) | 1.44 | |
| 7 | CC-g-L | 0.71 (0.05) | 0.82 (0.07) | 1.81 | 1.01 (0.11) | 1.07 (0.09) | 0.60 | 0.56 (0.05) | 0.70 (0.09) | 1.92 |
| 8 | SLF-L | 0.75 (0.05) | 0.92 (0.15) | 1.52 | 0.86 (0.05) | 1.00 (0.15) | 1.25 | 0.70 (0.05) | 0.88 (0.16) | 1.52 |
| 9 | ILF-R | 0.69 (0.05) | 0.81 (0.10) | 1.52 | 0.86 (0.05) | 0.92 (0.10) | 0.76 | 0.60 (0.07) | 0.75 (0.11) | 1.63 |
| 10 | UF-L | 0.83 (0.09) | 0.96 (0.13) | 1.16 | 0.96 (0.09) | 1.08 (0.13) | 1.07 | 0.76 (0.09) | 0.90 (0.14) | 1.19 |
| 11 | HC-R | 0.86 (0.13) | 0.98 (0.15) | 0.85 | 1.11 (0.14) | 1.23 (0.14) | 0.86 | 0.73 (0.14) | 0.86 (0.16) | 0.86 |
| IFO-R | 0.72 (0.06) | 0.83 (0.09) | 1.44 | 1.03 (0.09) | 1.15 (0.11) | 1.19 | 0.57 (0.04) | 0.67 (0.09) | 1.44 | |
| 12 | FX | 1.19 (0.17) | 1.50 (0.33) | 1.18 | 1.41 (0.17) | 1.70 (0.34) | 1.08 | 1.09 (0.18) | 1.40 (0.33) | 1.17 |
4. DISCUSSION
This paper presents a methodology not only to identifying and quantify the location and extent of brain regions injured in TBI, but more importantly, also to quantify the impact of injury on specific pathways in an individual. This information should be vital to diagnose and monitor objectively the progression of the injury in TBI as well as to assess the outcome of treatment.
Identification of injured regions was based on detecting voxel-based changes in FA values between an individual TBI subject and a group of age-matched control subjects after normalization of all FA maps onto a common FA template in MNI space. The FA-reduced regions were subsequently mapped back to each subject's head- space and used as objective ROIs to sort tracts in each subject's head. A unique seed-placement procedure where the number of seeds and their corresponding anatomical locations were equalized in all subjects was used to attain tractography normalization for quantitative comparisons. A unique procedure was also devised to map tracts from any subject onto the head of any other subject to perform quantitative comparisons within the head-space of any individual subject. Thus it now becomes possible to visualize, quantify and compare any specific tract or pathway in an individual patient to corresponding tracts or pathways in a group of normal subjects, all normalized and co-registered to the individual patient's brain anatomy, which should be of particular significance to an individual based interventional procedure such as neurosurgery.
After normalization and mapping of tracts from a control group onto the head-space of an individual, a given voxel in the individual should ideally show the same number of tracts and the same pathways for any control subject. However co-registration is never perfect and one would also expect some level of inter-subject variability in brain connections. Indeed, in this work we found that tracts sorted by a given ROI in the TBI subject's head-space did not overlap completely among the ten control subjects used in this work. To mitigate inter-subject variability, we refined the ROIs by subdividing them into smaller ROIs representing regions where specific tracts from at least 50% of the control subjects overlapped completely. All anisotropy and tract-count metrics were subsequently computed within these sub-ROIs.
As specific brain pathways are correlated to specific brain function and behavior, identification and quantification of damage to specific pathways due to TBI provides a metric to correlate DTI findings to neurocognitive and other behavior/clinical test scores. For example, consistent with previous studies [13,24], we observed that the hippocampal/fornix pathway (HC/FX) was frequently affected by TBI injury in the subjects reported here. This observation in individuals was validated by the group study where the HC/FX pathway was identified by one of the high t-score FA-reduced ROIs (ROI 2 in Table 3) as a commonly injured regions in this cohort of TBI subjects. Consistent with injury to the HC/FX pathways, many of the TBI subjects also subjectively reported short-term memory loss during their clinical interviews. Future work should investigate the correlation between neuro-psychological tests designed to probe specific cognitive function, such as memory, and the DTI metrics including anisotropy metrics and tract-counts along specific pathways, such as HC/FX, to validate the DTI findings. If validated, the DTI metrics would provide a unique and powerful approach to objectively monitor TBI progression.
The ROIs used in this work derive from a significant reduction in FA between an individual TBI subject and a group of age-matched controls. MD was found to increase significantly in these regions. We also investigated whether there were regions in the TBI subjects where FA had increased significantly with respect to the controls but were unable to identify any such regions in our TBI cohort. As described in Sec. 1., though many other investigators have reported a decrease in FA and an increase in MD in injured regions, an increase in FA and a decrease in MD in acute TBI have also been reported with plausible explanations for how FA could increase initially and decrease subsequently following injury. However the exact model of how FA or other anisotropy metrics would change in TBI is not known and future studies are expected to shed more light on the dynamics of FA changes in TBI.
In addition to FA, we investigated the difference maps for MD, DR and DA and found that except for FA, the other measures showed relatively diffuse and non-specific changes, distributed across the entire brain and appeared to affect almost all brain pathways. This observation is consistent with a recent report [51]. As our key objective in this work was to localize injured regions and specific pathways affected by the injury, and FA changes were much better localized than the other diffusion measures, we chose to identify injured regions from their FA changes and then compute the MD, DR and DA changes over these regions. A detailed study of all of these diffusion measures would be desirable in the future.
It is interesting to note that though DR was significantly higher in TBI compared to controls (which is expected as there would be more free space for water molecules to diffuse radially in the absence of barriers posed by intact axons and their myelin sheath), DA was also consistently higher in TBI than normals in our study. The ratio DA/DR, however, was significantly lower in TBI than control subjects. The increase in DA in TBI is puzzling at first glance but is consistent with the observation that intact axons and their myelin sheath would present some restriction to free diffusion along the axial direction, which would be lifted following axonal degeneration in TBI, leading to higher DA. The significantly higher values of the ratio DA/DR in controls compared to TBI clearly imply that diffusion along the radial direction is much more restricted when intact axons are present.
It is important to note that the number of tracts is a function of the FA and deflection thresholds in streamline tractography and hence the tract-count metric computed here is valid only for the FA threshold of 0.15 and deflection angle <45° used in this work. The FA threshold was determined objectively to maximize the tract-count difference related effect size between the TBI and controls.
Previous reports have suggested [52] that the distribution of FA (and other diffusion parameters) is not strictly Gaussian. Thus the precise statistical significance of the single subject as well as the group study are not established in this work. A non-parametric comparison such as the permutation-based approaches [53] would lead to more accurate statistical comparisons in future work.
Finally the accuracy of DTI metrics including tract-counts is ultimately limited by the signal to noise ratio of the raw data, partial volume problems due to the limited spatial resolution of the EPI images, susceptibility artifacts at highfields, micro-movement and motion artifacts, corrections for eddy currents, co-registration between the EPI and high-resolution anatomical images to identify anatomical regions, and the accuracy of spatial normalization procedures. In addition, multiple-fibers within a voxel present challenges to quantification. It is very likely that several voxels contain multiple crossing fibers, resulting in erroneous estimation of the DTI tensor's eigen values and eigen vectors that are based on a single rank-2 tensor. Errors in the eigen values would affect the anisotropy metrics and errors in the primary eigen vector would propagate tracts incorrectly leading to early termination or misinterpretation. Work is in progress in our lab and other institutions to reduce these errors.
Acknowledgment
This research was supported by Contract No. W81XWH-07-1-0015 awarded by USA Medical Research and Material Command, TATRC, and partially supported by grant NIA-NIH P50 AG05142. The authors would like to thanks Ms. Amrita Rajagopalan and Mr. Sinchai Tsao for their assistance with the tract normalization code.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Strich SJ. Shearing of nerve fibers as a cause of brain damage due to head injury: a pathological study of twenty cases. Lancet. 1961;2:443–8. [Google Scholar]
- 2.Adams JH, Graham DI, Murray LS, Scott G. Diffuse axonal injury due to nonmissile head injury in humans: an analysis of 45 cases. Ann Neurol. 1982;12:557–63. doi: 10.1002/ana.410120610. [DOI] [PubMed] [Google Scholar]
- 3.Arfanakis K, Haughton V, Carew JD, Rogers BP, Dempsey RJ, Meyerand ME. Diffusion Tensor MR Imaging in diffuse axonal Injury. AJNR Am J Neuroradiol. 2002;23:794–802. [PMC free article] [PubMed] [Google Scholar]
- 4.Huisman TA, Schwamm LH, Shaefer PW, Koroshetz WJ, Neeta SA, Ozsunar Y, Wu O, Sorensen AG. Diffusion tensor imaging as potential biomarker of white matter injury in diffuse axonal injury. Am J Neuroradiol. 2004;25:370–6. [PMC free article] [PubMed] [Google Scholar]
- 5.Gentry LR, Godersky JC, Thompson B. MR imaging of head trauma: review of the distribution and radiopathologic features of traumatic lesions. Am J Roentgenol. 1988;9:101–10. doi: 10.2214/ajr.150.3.663. [DOI] [PubMed] [Google Scholar]
- 6.Kelly AB, Zimmerman RD, Snow RB, Gandy SE, Deck MDF. Head trauma: comparison of MR and CT-experience in first 100 patients. Am J Neuroradiol. 1988;9:699–708. [PMC free article] [PubMed] [Google Scholar]
- 7.Hurley RA, MsGowan JC, Arfanakis K, Taber KH. Traumatic axonal injury: novel insights into evolution and identification, Windows to the Brain. J Neuropsychiatry Clin Neurosci. 2004;16:1–7. doi: 10.1176/jnp.16.1.1. [DOI] [PubMed] [Google Scholar]
- 8.Rutgers DR, Toulgoat F, Cazejust J, Fillard P, Lasjaunias P, Ducreux D. White matter abnormalities in mild traumatic brain injury: a diffusion tensor imaging study. Am J Neuroradiol. 2008;29:514–9. doi: 10.3174/ajnr.A0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahuquillo J, Poca MA. Diffuse axonal injury after head trauma: A review. Adv Tech Stand Neurosurg. 2002;27:23–86. doi: 10.1007/978-3-7091-6174-6_2. [DOI] [PubMed] [Google Scholar]
- 10.Stone JR, Singleton RH, Povlishock JT. Intra-axonal neurofilament compaction does not evoke local axonal swelling in all traumatically injured axons. Exp Neurol. 2001;172:320–31. doi: 10.1006/exnr.2001.7818. [DOI] [PubMed] [Google Scholar]
- 11.Ptak T, Sheridan RL, Rhea JT, Gervasini L, Novelline RA. Cerebral fractional anisotropy score in trauma patients: a new indicator of white matter injury after trauma. Am J Roentgenol. 2003;181:1401–7. doi: 10.2214/ajr.181.5.1811401. [DOI] [PubMed] [Google Scholar]
- 12.Inglese M, Markani S, Johnson G, Cohen BA, Silver JA, Gonen O, Grossman RI. Diffuse axonal injury in mild traumatic brain injury: a diffusion tensor imaging study. J Neurosurg. 2005;103:298–303. doi: 10.3171/jns.2005.103.2.0298. [DOI] [PubMed] [Google Scholar]
- 13.Salmond CH, Menon DK, Chatfield DA, Williams GB, Pena A, Sahakian BJ, Pickard JD. Diffusion tensor imaging in chronic head injury survivors: correlations with learning and memory indices. Neuroimage. 2006;29:117–24. doi: 10.1016/j.neuroimage.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 14.Bazarian JJ, Zhong J, Blythe B, Zhu T, Kavcic V, Peterson D. Diffusion tensor imaging detects clinically important axonal damage after mild traumatic brain injury: a pilot study. J Neurotrauma. 2007;24:1447–59. doi: 10.1089/neu.2007.0241. [DOI] [PubMed] [Google Scholar]
- 15.Assaf Y, Freidlin RZ, Rohde GK, Bassar PJ. New modeling and experimental framework to characterize hindered and restricted water diffusion in brain white matter. Magn Reson Med. 2004;52:965–78. doi: 10.1002/mrm.20274. [DOI] [PubMed] [Google Scholar]
- 16.Mori S, Itoh R, Zhang J, Kaufman WE, van Zijl PC, Solaiyappan M, Yarowsky P. Diffusion tensor imaging of the developing mouse brain. Magn Reson Med. 2001;46:18–23. doi: 10.1002/mrm.1155. [DOI] [PubMed] [Google Scholar]
- 17.Neil J, Miller J, Mukherjee P, Hüppi PS. Diffusion tensor imaging of normal and injured developing human brain—A technical review. NMR Biomed. 2002;15:543–52. doi: 10.1002/nbm.784. [DOI] [PubMed] [Google Scholar]
- 18.Song SK, Sun SW, Ramsbottom MJ, Chang C, Russell J, Cross AH. Dysmyelination revealed through MRI as increased radial (but unchanged axial) diffusion of water. NeuroImage. 2002;17:1429–36. doi: 10.1006/nimg.2002.1267. [DOI] [PubMed] [Google Scholar]
- 19.Zhang RL, Zhang L, Zhang ZG, Morris D, Jiang Q, Wang L, Zhang LJ, Chopp M. Migration and differentiation of adult rat subventricular zone progenitor cells transplanted into the adult rat striatum. Neuroscience. 2003;116:373–82. doi: 10.1016/s0306-4522(02)00696-6. [DOI] [PubMed] [Google Scholar]
- 20.Nair G, Tanahashi Y, Low HP, Billings-Gagliardi S, Schwartz WJ, Duong TQ. Myelination and long diffusion times alter diffusion-tensor-imaging contrast in myelin-deficient shiverer mice. Neuroimage. 2005;28:16574. doi: 10.1016/j.neuroimage.2005.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tyszka JM, Readhead C, Bearer EL, Pautler RG, Jacobs RE. Statistical diffusion tensor histology reveals regional dysmyelination effects in the shiverer mouse mutant. Neuroimage. 2006;29:1058–65. doi: 10.1016/j.neuroimage.2005.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buki A, Povlishock JT. All roads lead to disconnections-Traumatic axonal injury revisited. Acta Neurochir. 2006;148:181–94. doi: 10.1007/s00701-005-0674-4. [DOI] [PubMed] [Google Scholar]
- 23.Rugg-Gunn FJ, Symms MR, Baker GJ, Greenwood R, Duncan JS. Diffusion imaging shows abnormalities after blunt head trauma when conventional magnetic resonance imaging is normal. J. Neurol Neurosurg Psychiatry. 2001;70:530–3. doi: 10.1136/jnnp.70.4.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang JY, Bakhadirov K, Devous MD, Abdi H, McColl R, Moore C, Marquez de la Plata CD, Ding K, Whittemore A, Babcock E, Rickbeil T, Dobervich J, Kroll D, Dao B, Mohindra N, Madden CJ, Diaz-Arrastia R. Diffusion Tensor Tractography of Traumatic Diffuse Axonal Injury. Arch Neurol. 2008;65(5):619–626. doi: 10.1001/archneur.65.5.619. [DOI] [PubMed] [Google Scholar]
- 25.Kraus MF, Susmaras T, Caughlin BP, Walker CJ, Sweeney JA, Little DM. White matter integrity and cognition in chronic traumatic brain injury: a diffusion tensor imaging study. Brain. 2007;130(10):2508–19. doi: 10.1093/brain/awm216. [DOI] [PubMed] [Google Scholar]
- 26.Field AS, Hasan K, Jellison BJ, Arfanakis K, Alexander AL. Diffusion tensor imaging in an infant with traumatic brain swelling. Am J Neuroradiol. 2003;24:1461–4. [PMC free article] [PubMed] [Google Scholar]
- 27.Sidaros A, Engberg AW, Sidaros K, Liptrot MG, Herning M, Petersen P, Paulson OB, Jernigan TL, Rostrup E. Diffusion tensor imaging during recovery from severe traumatic brain injury and relation to clinical outcome: a longitudinal study. Brain. 2008;131:559–72. doi: 10.1093/brain/awm294. [DOI] [PubMed] [Google Scholar]
- 28.Conturo TE, Lori NF, Cull TS, Akbudak E, Snyder AZ, Shimony JS, McKinstry RC, Burton H, Zaichle ME. Tracking neuronal fiber pathways in the living human brain. Proc Natl Acad Sci USA. 1999;96:10422–7. doi: 10.1073/pnas.96.18.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones D, Horsfield M, Simmons A. Optimal strategies for measuring diffusion in anisotropic systems by magnetic resonance imaging. Magn Reson Med. 1999;42:515–25. [PubMed] [Google Scholar]
- 30.Mori S, Kaufmann W, Pearlson G, Crain B, Stieltjes B, Solaiyappan M, van Zijl P. In vivo visualization of human neural pathways by RI. Ann Neurology. 2000;47:412–414. [PubMed] [Google Scholar]
- 31.Basser P, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vitro fiber tractography using DT-MRI data. Magn Reson Med. 2000;44:625–32. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 32.Gupta RK, Saksena S, Agarwal A, Hasan KM, Husain M, Gupta V, Narayana PA. Diffusion tensor imaging in late posttraumatic epilepsy. Epilepsia. 2005;46:1465–71. doi: 10.1111/j.1528-1167.2005.01205.x. [DOI] [PubMed] [Google Scholar]
- 33.Wilde EA, McCauley SR, Hunter JV, Bigler ED, Chu Z, Wang ZJ, Hanten GR, Troyanskaya M, Yallampalli R, Li X, Chia J, Levin HS. Diffusion tensor imaging of acute mild traumatic brain injury in adolescents. Neurology. 2008;70:948–55. doi: 10.1212/01.wnl.0000305961.68029.54. [DOI] [PubMed] [Google Scholar]
- 34.Jones DK, Griffin LD, Alexander DC, Catani M, Horsfield MA, Howard R, Williams SC. Spatial normalization and averaging of diffusion tensor MRI data sets. Neuroimage. 2002;17:592–617. [PubMed] [Google Scholar]
- 35.Alexander DC, Pierpaoli C, Basser PJ, Gee JC. Spatial transformations of diffusion tensor magnetic resonance imaging. IEEE Trans Med Imaging. 2001;20:1131–9. doi: 10.1109/42.963816. [DOI] [PubMed] [Google Scholar]
- 36.Xu D, Mori S, Shen D, Zijl PCM, Davatzikos C. Spatial Normalization of Diffusion Tensor Fields. Magn Reson Med. 2003;50:175–82. doi: 10.1002/mrm.10489. [DOI] [PubMed] [Google Scholar]
- 37.Müller HP, Unrath A, Ludolph AC, Kassubek J. Preservation of diffusion tensor properties during spatial normalization by use of tensor imaging and fibre tracking on a normal brain database. Phys Med Biol. 2007;52:N99–N109. doi: 10.1088/0031-9155/52/6/N01. [DOI] [PubMed] [Google Scholar]
- 38.Singh M, Jeong JW. DTI-Tractography to detect and quantify brain pathways affected in traumatic brain injury. Proc Intl Soc Mag Reson. 2008;16:2269. [Google Scholar]
- 39.Basser PJ, Mattielo J, Le Bihan D. MR diffusion tensor spectroscopy and imaging. Biophys J. 1994;66:259–7. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashburner J, Friston KJ. Nonlinear spatial normalization using basis functions. Hum Brain Mapp. 1999;7:254–66. doi: 10.1002/(SICI)1097-0193(1999)7:4<254::AID-HBM4>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friston KJ, Ashburner J, Kiebel S, Nichols T, Penny W. Statistical Parametric Mapping. Academic press; 2007. [Google Scholar]
- 42.Mori S, van Zijl PC. Fiber tracking: principles and strategies-a technical review. NMR Biomed. 2002;15:468–80. doi: 10.1002/nbm.781. [DOI] [PubMed] [Google Scholar]
- 43.Singh M, Hwang D, Sungkarat W, Veera K. Evaluation of MRI DTI-tractography by tract-length histogram. Proc of SPIE medical imaging: physiology, function, and structure from medical images. 2005;5746(1):138–147. [Google Scholar]
- 44.Clayden J, Marjoram D, Bastin ME, Johnston E, Lawrie S. Towards an automated method for white matter integrity comparison between populations.. Proc ESMRME 22nd Annu Meeting.2005. p. 508. [Google Scholar]
- 45.Clayden J, Bastin M, Storkey A. Improved segmentation reproducibility in group tractography using a quantitative tract similarity measure. Neuroimage. 2006;33:482–92. doi: 10.1016/j.neuroimage.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 46.Mori S, Kaufmann W, Davatzikos C, Stieltjes B, Amodei L, Fredericksen K, Pearlson G, Melhem E, Solaiyappan M, Raymond G, Moser H, van Zijl P. Imaging cortical association tracts in the human brain using diffusion-tensor-based axonal tracking. Magn Reson Med. 2002;47:215–223. doi: 10.1002/mrm.10074. [DOI] [PubMed] [Google Scholar]
- 47.Ashburner J, Andersson JL, Friston KJ. Image registration using a symmetric prior - in three-dimensions. Hum Brain Mapp. 2000;9:212–25. doi: 10.1002/(SICI)1097-0193(200004)9:4<212::AID-HBM3>3.0.CO;2-#. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ducreux D, Huynh I, Fillard P, Renoux J, Petit-Lacour MC, Marsot-Dupuch K, Lasjaunias P. Brain MR diffusion tensor imaging and fibre tracking to differentiate between two diffuse axonal injuries. Neuroradiology. 2005;47:604–8. doi: 10.1007/s00234-005-1389-1. [DOI] [PubMed] [Google Scholar]
- 49.Zhan W, Zhang P, Lorenzen P, Mueller SG, Schuff N, Weiner MW. Correlations between DTI and FLAIR images reveal the relationship of microscopic and macroscopic white matter degeneration in elderly subjects. Proc of SPIE medical imaging: physiology, function, and structure from medical images, SPIE. 2008;691609:1–9. [Google Scholar]
- 50.Dauguet J, Peled S, Berezovskii V, Delzescaux T, Warfield SK, Born R, Westin CF. Comparison of fiber tracts derived from in-vivo DTI tractography with 3D histological neural tract tracer reconstruction on a macaque brain. Neuroimage. 2007;37:530–38. doi: 10.1016/j.neuroimage.2007.04.067. [DOI] [PubMed] [Google Scholar]
- 51.Bendlin BB, Ries ML, Lazar M, Alexander AL, Dempsey RJ, Rowley HA, Sherman JE, Johnson SC. Longitudinal changes in patients with traumatic brain injury assessed with diffusion-tensor and volumetric imaging. NeuroImage. 2008;42:503–14. doi: 10.1016/j.neuroimage.2008.04.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones DK, Symms MR, Cercignani M, Howard RJ. The effect of filter size on VBM analyses of DT-MRI data. NeuroImage. 2005;26:546–54. doi: 10.1016/j.neuroimage.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 53.Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Human Brain Mapping. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]