Abstract
MUTYH-associated polyposis (MAP) is the only inherited colorectal cancer syndrome that is associated with inherited biallelic mutations in a base excision repair gene. The MUTYH glycosylase plays an important role in preventing mutations associated with 8-oxoguanine (OG) by removing adenine residues that have been misincorporated opposite OG. MAP-associated mutations are present throughout MUTYH, with a large number coding for missense variations. To date the available information on the functional properties of MUTYH variants is conflicting. In this study, a kinetic analysis of the adenine glycosylase activity of MUTYH and several variants was undertaken using a correction for active fraction to control for differences due to overexpression and purification. Using these methods, the rate constants for steps involved in the adenine removal process were determined for the MAP variants Y165C, G382D, P391L and Q324R MUTYH. Under single-turnover conditions, the rate of adenine removal for these four variants was found to be 30–40% of WT MUTYH. In addition, the ability of MUTYH and the variants to suppress mutations and complement for the absence of MutY in E. coli was assessed using rifampicin resistance assays. The presence of WT and Q324R MUTYH resulted in complete suppression of the mutation frequency, while G382D MUTYH showed reduced ability to suppress the mutation frequency. In contrast, the mutation frequency observed upon expression of P391L and Y165C MUTYH were similar to the controls, suggesting no activity toward preventing DNA mutations. Notably, though all variations studied herein resulted in similar reductions in adenine glycosylase activity, the effects in the bacterial complementation are quite different. This suggests that the consequences of a specific amino acid variation on overall repair in a cellular context may be magnified.
Keywords: 8-oxoguanine, MAP, MUTYH, P391L MUTYH, Q324R MUTYH, adenine glycosylase, oxidative stress
1. Introduction
An unavoidable consequence of life in an aerobic environment is the presence of reactive oxygen species (ROS) such as hydrogen peroxide, superoxide, and hydroxyl radicals [1,2]. DNA modifications due to reactions with ROS have been linked to the initiation and progression of cancer [1–3]. A common product of oxidative damage to 2′-deoxyguanosine is 7,8-dihydro-8-oxo-2′-deoxyguanosine (OG) [4,5]; in fact, the amount of OG is often used as an indicator of the level of oxidative stress in cells [2]. In bacterial and mammalian cells, in the absence of repair, OG has been shown to be highly mutagenic, producing G → T transversion mutations [4]. This is consistent with the miscoding properties of OG that lead to misinsertion of A during DNA replication to form a stable OG:A mismatch [3,4]. In humans, three enzymes work in concert to prevent mutations that arise from OG in DNA [3]. The human MutY homologue (MUTYH) is responsible for removing adenine bases from OG:A mismatches, while the human OG glycosylase (hOGG1) removes damaged OG bases when paired with C. These two enzymes initiate base excision repair (BER), which is then completed by other enzymes in this pathway that act sequentially at the base-less site to restore the proper nucleotides and preserve the correct sequence of base pairs [6]. A third enzyme, the human MutT homologue (hMTH1) is responsible for hydrolysis of d(OG)TP to d(OG)MP, thereby preventing incorporation of OG-containing nucleotides into DNA by polymerases.
The direct link between a familial form of colorectal cancer (CRC), familial adenomatous polyposis (FAP), and inherited variations in the DNA glycosylase MUTYH has further underscored the crucial nature of repair mechanisms for mitigating the mutagenic potential of OG [3]. The MUTYH-CRC link was first discovered in a British family in which three siblings possessed clinical symptoms characteristic of FAP, yet lacked the characteristic hallmark of FAP of inherited mutations in the adenomatous polyposis coli (APC) gene [7]. Instead, afflicted family members harbored compound heterozygous biallelic MUTYH mutations coding for two missense variants, Y165C and G382D MUTYH. Functional defects in the MUTYH variant enzymes are proposed to be responsible for the observation of inactivating G → T transversion mutations in APC leading to the polyposis phenotype. There has been considerable work that has firmly established the relationship between MUTYH germline mutations, inactivating somatic mutations in APC and colorectal polyposis leading to the designation of a distinct cancer pre-disposition mechanism: MUTYH-associated polyposis (MAP) [8]. In addition to the variants described above, approximately eighty new germline mutations in MUTYH have been identified in patients with colorectal adenomas and carcinomas [8]. These include missense and truncating mutations, in-frame deletions, frame shifts, and splice-site mutations. Notably, several novel germline MUTYH mutations have been observed in specific populations [8,9].
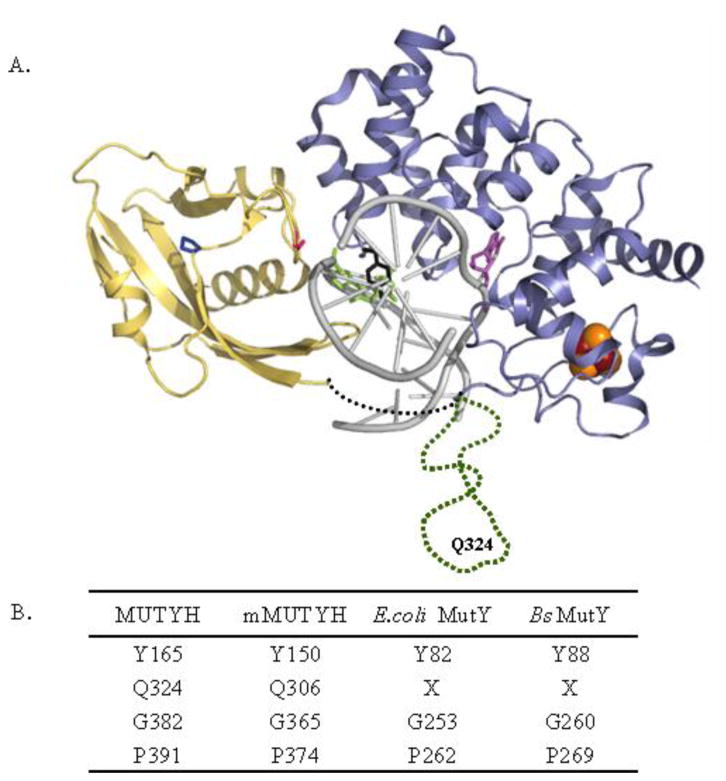
An important component of the initial discovery of MAP was the analysis of the enzymatic activity of the highly homologous E. coli enzyme, which has proven to be an excellent model system for the study of mammalian MutY homologues [7]. The E. coli MutY mutants (Y82C and G253D MutY) corresponding to those in MAP showed reduced adenine removal activity in vitro with 30-bp duplex substrates containing a G:A mismatch or when analyzed with the corresponding OG:A mismatch substrate at low temperature (2 °C) [7]; however, only the Y82C MutY enzyme showed reduced activity under conditions of physiological temperature and salt [10,11]. Both mutated enzymes were shown to have a reduced affinity for duplexes containing OG opposite a noncleavable analog of A, 2′-deoxy-2′-fluoroadenosine (FA), indicating a compromised ability to recognize OG [11]. The intimate role these two amino acids play in recognizing OG was further illuminated in the X-ray crystal structure of an inactive mutant of Bacillus stearothermophilus MutY (D144N Bs MutY) bound to an OG:A- containing duplex (Figure 1)[12].
Figure 1.
MUTYH variations associated with MAP. (A) Bacillus stearothermophilus MutY-DNA cocrystal structure (1RRQ) with residues corresponding to variations in MUTYH involved in MAP highlighted: Y88, black; G260, pink; P269, dark blue; FeS cluster; brown and orange; adenine, violet; OG, green; DNA, grey; N-terminal domain, light blue; C-terminal domain, yellow, linker region between domains, black dotted line. The sequence present only in MUTYH shown in green dotted line harbors position of Q324. This region is not based on the structure, and was added into a structure generated from coordinates (1RRQ) from the reported structures. (B) Summary of conserved residues across different species. X denotes no matching residue found on sequence alignment using Clustal W.
Several studies have examined the enzymatic activity of MUTYH variant proteins associated with MAP; however, these studies have not provided a consistent and clear picture of the consequences of these variations on the adenine glycosylase activity. For example, while studies of the murine equivalents to Y165C and G382D MUTYH indicated reduced activity and binding affinity [13] analogous to the results with the bacterial enzyme, the first report on Y165C and G382D MUTYH suggested that both variants are completely devoid of activity [14]. More recently, Ali et al. [15] reported that the Y165C variant was severely defective in both its glycosylase activity and binding affinity, while G382D was partially active in its DNA binding and glycosylase activity. Controversies regarding the intrinsic enzyme activity of MutY enzymes are not new [16,17]. In fact, previous work from our laboratory was instrumental in sorting out the origin of controversial data and establishing protocols for quantitative analysis of the activity of the bacterial enzyme that has allowed for reproducible and comparable results between different laboratories [16,18–20]. Activity analyses of the human enzyme may also be complicated by varying amounts of folded and active protein isolated after purification from protein overexpression in bacteria. In fact, even wild-type (WT) E. coli MutY exhibits variability in active enzyme fraction among different preparations [11,16,21]. In addition, the stability of a particular variant may be different than the WT over the time course of the reaction. Effects on intrinsic catalytic activity and stability are two pieces of information needed to understand the role of a particular variant MUTYH protein in CRC predisposition. Resolving some of the basic issues associated with the activity of WT MUTYH and specific variants will be important for teasing out the relevant information related to the pathogenicity of a given variant, and predicting the potential impact of new variants on OG-mediated mutagenesis.
In this study, in addition to a thorough examination of WT MUTYH and the two most commonly occurring MAP variants, Y165C and G382D MUTYH, two other novel variants, P391L and Q324R MUTYH, have been characterized. We also characterized D222N MUTYH as a control that would be expected to be inactive based on the corresponding mutated bacterial enzymes [12,22]. The Q324R MUTYH variant is a novel variant, though other mutations at this codon have been observed. Most notably, Q324H MUTYH is thought to be a common polymorphism [8,23], though this variant has also been suggested to be associated with colorectal cancer in specific populations [15,24]. In addition, a truncated enzyme (Q324X MUTYH) has been observed in some patients due to a mutation at the Gln codon that converts it into a stop codon [8]. The Q324R MUTYH variant was reported by Myriad Genetics in the screening of a cohort of patients found to be negative for mutations in the genes coding for mismatch repair proteins MLH1 and MSH2 associated with hereditary nonpolyposis colon cancer (HNPCC) [25].
Three different groups have reported the mutations encoding P391L MUTYH independently and in patients with different ethnicity. Kanter–Smoler et al.[26] reported this mutation in Swedish patients where the mode of inheritance appeared to be dominant rather than recessive. In one patient, the mutation was found on one allele, with the other allele harboring the mutation encoding Y165C MUTYH, while an unrelated patient was homozygous for the P391L mutation. In a different study, Aretz et al.[27] reported the presence of P391L MUTYH along with G382D MUTYH in German patients who displayed an attenuated polyposis phenotype. Middeldorp et al.[28] observed the P391L variation frequently in Dutch patients in both homozygous contexts or in combination with other known MUTYH variants.
Surprisingly, in the kinetic analysis herein, all of the MUTYH variants studied, Y165C, G382D, P391L, and Q324R, exhibited similarly compromised adenine glycosylase activity that was 30–40% of the activity of the WT enzyme. Notably, the assays were performed using correction for active fraction to provide the intrinsic reduction in the enzyme activity caused by the amino acid changes. In the preparations for this study, the amount of active enzyme relative to total protein after purification was typically less than with WT, suggesting that these mutations also affect proper folding and stability during the purification process. The ability of WT MUTYH and the variants to suppress mutations and complement for the absence of the bacterial MutY enzyme in E. coli was assessed using rifampicin resistance assays. It was observed that the presence of WT and Q324R MUTYH completely suppressed the mutation frequency and fully complement for the absence of the bacterial MutY enzyme. In contrast, the presence of Y165C and P391L MUTYH provided for no significant reduction in the mutation frequency. The results with G382D MUTYH indicate a modest reduction of mutation frequency. Taken together, these results show that a reduced glycosylase activity as measured in vitro is only one aspect that suggests a reduced ability to suppress DNA mutations and is highly dependent on the type and position of the amino acid variation. Other factors, such as protein stability and folding may also contribute significantly to the ability to prevent mutations in vivo and therefore, consequences of specific amino acid variations on activity may be magnified in a cellular context.
2. Materials and methods
2.1 General methods, reagents and bacterial strains
All routine chemicals used to make buffers and other reagents were purchased from Fisher Scientific, VWR or Sigma. E. coli strain BL21(DE3) was used for protein overexpression and GT100 mutY:mini-Tn10 mutM for the bacterial complementation assays. Standard protocols were followed for DNA manipulation [29]. UV-visible spectroscopy was performed on a HP8452A diode array spectrophotometer. Storage phosphor autoradiography was performed on a Typhoon Trio phosphorimager system (GE Healthcare). Data analysis was performed using ImageQuaNT (Version 5.2a) and the rate constants were determined from fitting of the data with GraFit 5.0. PCR was performed on a GeneAmp PCR system 2400 from Perkin Elmer. DNA sequencing and synthesis was provided by core facilities at the University of Utah Medical School and University of California, Davis. 7,8-Dihydro-8-oxo-2′-deoxyguanosine phosphoramidite was purchased from Glen Research. Oligonucleotides used for mutagenesis via PCR were purified using oligonucleotide purification cartridges (OPC). All other oligonucleotides were purified via HPLC on a Beckman Gold Nouveau system with a C18 RCM column from Waters. [γ-32P]-ATP and T4-polynucleotide kinase (T4-PNK) used for end-labeling were purchased from Perkin-Elmer and New England Biolabs, respectively. ProbeQuantG-50 spin columns (GE Healthcare Life Sciences) were used for purifying the labeled oligonucleotides following the manufacturer’s protocol.
2.2 Gene cloning and site-directed mutagenesis of MUTYH
The pQEMUTYH plasmid containing the MUTYH gene in which the first seven arginine codons had been modified to those more commonly found in bacteria was generously shared by M.M. Slupska and J.H. Miller (UCLA). This modified gene was subcloned into the BamHI site of the pMal-c2x vector (NEB). An additional stop codon (TAA) was engineered into the gene after the original stop codon. DNA sequencing was performed to confirm the presence and orientation of the MUTYH gene. The resulting plasmid was named pMalMUTYH WT. Mutations in the MUTYH coding region for Y165C, G382D, P391L, D222N, and Q324R MUTYH were made by site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene) and the following primers where the underlined sequence defines codon change:
Y165C-1: 5′-GGGCTGGCCTGGGCTGCTATTCTCGTGG-3′
Y165C-2: 5′-CCACGAGAATAGCAGCCCAGGCCAGCCC-3′
G382D-1: 5′-CAGTCCTGCCAGCAGATCTGAGTTGGGCC-3′
G382D-2: 5′-GGCCCAACTCAGATCTGCTGGCAGGACTG-3′
P391L-1: 5′-GGACTGTGGGAGTTCCTGTCCGTGACCTGGG-3′
P391L-2: 5′-CCCAGGTCACGGACAGGAACTCCCACAGTCC-3′
Q324R-1: 5′-CCCAACACTGGACGGTGCCACCTGTGC-3′
Q324R-2: 5′-GCACAGGTGGCACCGTCCAGTGTTGGG-3′
D222N-1:5′-ACCGGTGTGGTGAATGGCAACGTAGC-3′
D222N-2:5′-GCTACGTTGCCATTCACCACACCGG-3
Plasmid DNA was isolated from XL-1 Blue E. coli cells using a Wizard Plus DNA purification kit (Promega) according to manufacturer’s protocol. Presence of the mutations of interest were confirmed by DNA sequencing. These plasmids were named pMalMUTYH Y165C, pMalMUTYH G382D, pMalMUTYH P391L, pMalMUTYH D222N and pMalMUTYH Q324R respectively.
2.3 Enzyme Expression and Purification
BL21 (DE3) E. coli were made chemically competent by calcium chloride using standard protocols [29] and then transformed with the expression vectors pMalMUTYH WT, Y165C, G382D, P391L, D222N or Q324R using standard heat-shock procedures. Colonies were selected using 100 μg/mL ampicillin by growth on LB agar plates. For protein expression, a single colony was chosen to inoculate an overnight starter culture of LB media containing 100 μg/mL ampicillin and supplemented with 2 mg/mL glucose. The starter culture was then used to inoculate 1 L of LB growth containing ampicillin and glucose at the same concentrations. The cells were grown at 37 °C to an OD600 of 0.8 and protein expression was induced by addition of a final concentration of 300 μM IPTG and then incubated at 30 °C for 3 hours. The cells were then collected by centrifugation, resuspended in 20 mL of column buffer A (20 mM Tris-HCl pH 7.5, 200 mM NaCl, 1 mM EDTA, 1 mM DTT), supplemented with 1 mM PMSF, and stored at −80 °C. All subsequent protein purification steps were performed at 4 °C unless otherwise noted. Cells were thawed slowly at 37 °C and an additional 1 mM PMSF was added once completely thawed. Sonication was performed with six series of 30-second pulses using a Branson sonifier 250 at 70% power. After centrifugation to remove cellular debris, the supernatant was batch bound with 1 mL of amylose resin for one hour. The resin-supernatant mix was then loaded onto a column and washed with ten column volumes of column buffer A and the protein was eluted in five column volumes of column buffer B (same as column buffer A with the addition of 10 mM maltose). The protein was concentrated approximately 10-fold using an Amicon stirred cell concentrator (YM 10 membrane), aliquoted into single-use fractions, and stored at −80 °C. The purity of the protein sample was assessed by Sypro orange (Invitrogen)-stained SDS-PAGE and the presence of the MBP-MUTYH construct confirmed by Western blot analysis using an antibody to the MBP tag (NEB).
2.4 General features of glycosylase assay and substrate DNA preparation
The following oligonucleotide duplex was used for these experiments:
d(5′-CGATCATGGAGCCACXAGCTCCCGTTACAG-3′)
d(3′-GCTAGTACCTCGGTGATCGAGGGCAATGTC-5′)
Where X = 7,8-dihydro-8-oxo-2′-deoxyguanosine or 2′-deoxyguanosine and A = 2′-deoxyadenosine. The A-containing strand (2.5 pmoles) was radiolabeled on the 5′ end with [γ-32P]-ATP using T4-PNK. For glycosylase assays under multiple-turnover conditions additional unlabeled A-containing oligonucleotide strand was added such that the final concentration would contain approximately 5% [γ-32P]-phosphate labeled DNA. The complementary X-containing strand was then added in 10 % excess to the A-containing strand in annealing buffer (20 mM Tris-HCl pH 7.6, 10 mM EDTA, 150 mM NaCl). The DNA duplex was then annealed by heating to 90 °C for five minutes followed by slow cooling to room temperature overnight. For glycosylase assays under single-turnover conditions and binding experiments, 10 % excess of the X-containing strand was added to 100 % labeled A-containing DNA in the annealing buffer.
Adenine glycosylase activity assays of WT MUTYH and variant enzymes Y165C, G382D, P391L, Q324R and D222N MUTYH were performed similarly as reported previously for E. coli MutY and murine Mutyh by our laboratory [16,30]. In general, the enzyme activity is monitored by assessing the amount of strand scission at the abasic site produced by MUTYH at the A opposite OG in the duplex. The standard assay buffer consists of 40 mM Tris-HCl pH 7.5, 10 mM EDTA, 0.1 mg/mL BSA, and 150 mM NaCl. Enzyme and DNA substrate were incubated at 37 °C and aliquots were removed at various times (20 sec to 60 min). These aliquots were quenched with 0.2 M NaOH and heated to 90 °C for 5 min. An equal volume of formamide loading dye (0.025 % bromophenol blue, 0.025% xylene cyanol, and 80% formamide in 1X TBE) was added and the samples were heated at 90 °C for an additional 5 min. The samples were resolved on a 15 % (19:1) denaturing polyacrylamide gel in 1X TBE at 1500 V for 1.5 hours and visualized by storage phosphor autoradiography.
2.5 Glycosylase assays under multiple-turnover conditions: Active site titrations, stability assays, and determination of k3
For active site titration (AST) and multiple-turnover experiments, the removal of adenine from 5–10 nM DNA substrate is monitored with the amount of MUTYH enzyme adjusted to give a burst amplitude in a detectable range, typically 20% of the DNA concentration. The analysis of the data is analogous to that previously reported for E. coli MutY and murine Mutyh [16,30]. The data from these experiments were fitted with equation 1 to determine the amplitude of the burst (A0), kobs (rate constant for the burst phase) and kss (rate constant for the linear phase). Under conditions where [E] > Kd, A0 = [active MUTYH]. All concentrations listed are active site concentrations, corrected on the basis of the active site titrations.
| (1) |
The stability of MUTYH and the four variants was assessed by monitoring the amplitude of the burst phase of the reactions after pre-incubation of the enzyme at 37 °C in the glycosylase buffer for time ranging from 0–10 minutes, prior to addition of the labeled substrate. The rate constant k3 = kss/[A]0 as described previously for MutY [16].
2.6 Glycosylase Assays under single-turnover conditions: Determination of rate constant k2 and dissociation constant for MUTYH with an OG:A-containing substrate
Single-turnover ([MUTYH] > [DNA]) kinetic assays were performed in general as described above using a DNA concentration of 100 pM and an active MUTYH concentration that was as high as possible (1–2 nM). Data from single-turnover experiments were used to determine the observed rate of product formation (kobs) from fitting of the data using equation 2. Under conditions of single-turnover where [E] > Kd, kobs = k2 (Scheme 1). That appropriate single-turnover conditions were present under these conditions is consistent with no observed change in maximal kobs if the DNA (0.5 nM) or enzyme (2.0 nM) concentration was increased. Unfortunately, enzyme solubility issues precluded obtaining active [MBP-MUTYH] concentrations significantly above 2 nM. Lower DNA concentrations (0.01 nM) in Kd measurements also gave similar maximal kobs values.
Scheme 1.
Minimal kinetic scheme used in the analysis of the adenine glycosylase activity of MUTYH [16].
| (2) |
For kinetic determination of the dissociation constant (Kd) the DNA concentration was reduced to 10 pM and enzyme concentrations were varied between 100 pM and 2 nM. Values for Kd were determined by fitting the observed rate of adenine removal (kobs) versus log[enzyme] to the one-site ligand binding equation using the GraFit 5.0 program.
2.7 Complementation Assays
Complementation experiments performed herein were followed as outlined previously [31]. The WT MUTYH gene or gene expressing Y165C, G382D, P391L and Q324R MUTYH were used in the pMAL-c2x expression constructs described above. The relevant plasmids, as well as the parent pMAL-c2x plasmid, were individually transformed into the E. coli strain GT100 mutY:mini-Tn10 mutM. A series of overnight cultures (at least ten) were grown in LB media containing 100 μg/mL ampicillin and 15 μg/mL tetracycline antibiotics from the transformed colonies. Cells from the same culture were plated on LB agar media containing the appropriate antibiotics supplemented with 100 μg/mL rifampicin. To determine the number of viable cells, a 107-fold dilution from each independent culture was plated on LB agar plates containing the same antibiotics, incubated at 37 °C overnight, and the resulting colonies were counted. The mean number of colonies from the rifampicin containing plates was divided by the average value for the viable cells to calculate the mutation frequency (f) as described previously [32].
3. Results
3.1 Overexpression and Purification
An expression vector containing the gene encoding MUTYH with an N-terminal His6-tag (pQEMUTYHWT) was used that had been modified from the previously published construct [33] to convert seven rare arginine codons present in the region coding for the N-terminal domain into those more commonly found in bacteria (AGG → CGC, AGA → CGC). Initial overexpression studies using this construct provided low amounts of MUTYH protein which was unstable to purification and contained significant amounts of degraded MUTYH fragments. Consequently, the MUTYH gene was sub-cloned into a pMAL-c2x vector and expressed as a fusion protein with an N-terminal maltose binding protein (MBP) tag in protease deficient BL21 (DE3) cells. The addition of an MBP tag often aids in overexpression, solubility and stability of proteins [34,35]. We have previously observed 20-fold greater levels of overexpression of MBP-MutY relative to untagged versions in E. coli [36]. The recombinant MBP-MUTYH protein was purified in a one-step affinity chromatography procedure using amylose resin. The purified protein contained mainly MBP-MUTYH, with Western blot analysis using an MBP antibody compared to total protein staining with Sypro orange showing that the major contaminants are fragments that contain MBP, either due to premature translation termination after MBP, or due to proteolytic cleavage in the linker region between MUTYH and the MBP-tag (Supplementary Figure 1).
The variant enzymes associated with MAP, namely Y165C, G382D, Q324R and P391L, were overexpressed and purified in an analogous manner, and the spectrum of protein products is similar to that observed with WT MUTYH (Supplementary Figure 1). The overall yield of total protein is similar for the variants and WT MUTYH, and is comparable to the GST-fusion constructs reported previously[15]. Notably, we detected intrinsic glycosylase activity for all variants, which is discussed in more detail below. In order to establish that the activity observed for the overexpressed variants was not due to contaminating E. coli MutY, an inactive version of MUTYH (D222N MUTYH) was also prepared using the identical approach. The resulting D222N MUTYH exhibited no detectable glycosylase activity, establishing that the purification strategy had removed all contaminating E. coli MutY. In addition, the amount of the D222N MUTYH protein present was determined by Western blot analysis with an anti-MBP antibody to establish that the expression levels of this mutated enzyme were similar to the WT (data not shown).
3.2 General features of MUTYH activity: Active site titration and stability
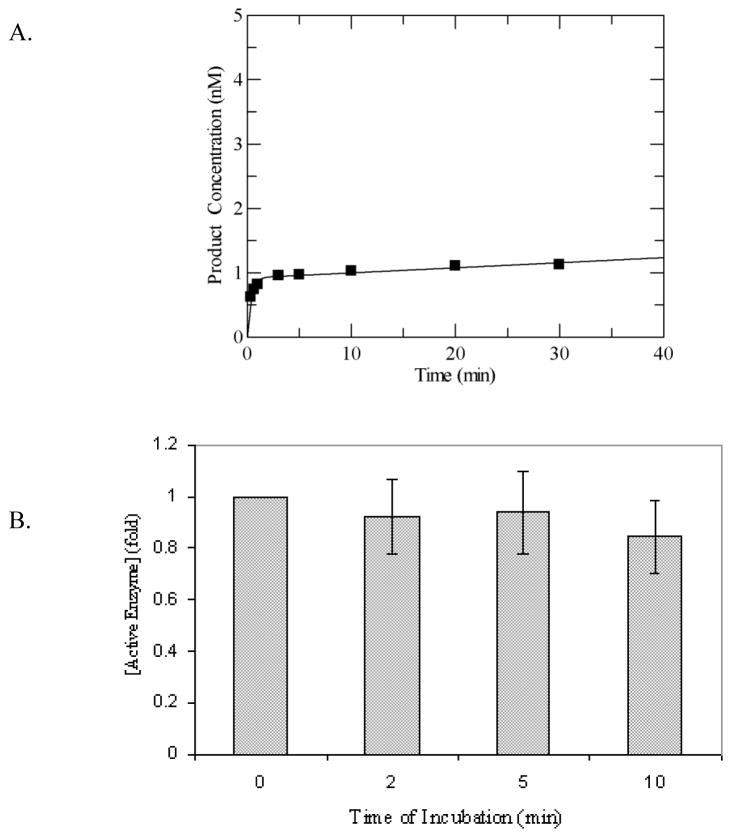
Although it has been reported previously that MUTYH is able to act upon OG:A mismatches [33], the activity has not been evaluated using a quantitative kinetic approach. Our laboratory has previously shown with several glycosylases that a detailed kinetic approach can unveil intricacies of the enzyme action that explain apparent discrepancies [16,37–39]. For this reason, we have used a similar approach to analyze the activity of MUTYH. Under multiple-turnover conditions with a 30-bp duplex containing a centrally located OG:A mismatch, in which [MUTYH] < [DNA], WT MUTYH displayed an initial exponential burst of product formation followed by a slower linear steady-state phase (Figure 2A). This type of “burst” kinetics is often seen in enzyme-substrate reactions where a slow step occurs after the initial chemistry step and may be exploited to determine the amount of active enzyme since this is related to the amplitude of the burst phase [40].
Figure 2.
Adenine glycosylase assays of WT MUTYH under multiple-turnover conditions (A) Representative plot of adenine removal activity by WT MUTYH under multiple-turnover conditions at 37 °C and 150 mM buffer salt concentration with an OG:A-containing duplex DNA substrate (10 nM). Line represents fit to equation 1 to determine A0 and k3, which are = 0.92 and 0.008 min−1, respectively, for this particular data set. (B) Stability assays of WT MUTYH based on A0 values determined after incubation of WT MUTYH in assay buffer show no significant reduction in enzyme activity as a function of time. The activity of MUTYH without prior incubation was normalized to 1. Aliquots were removed at intervals of 2, 5 and 10 minutes for adenine glycosylase assays under multiple-turnover conditions, and active enzyme concentrations were determined from the burst amplitudes of the progress curves. The adenine glycosylase activity at each time point was measured in at least three separate experiments. The grey bars represent the average on the basis of the initial amplitude and the error bars represent the standard deviation from the average (as a percentage of the normalized value).
In order to establish that the biphasic behavior is due to a slow step (i.e. product release) following chemistry, we determined the stability of MUTYH during the time-course of the reaction since loss of activity or inactivation with time may also provide a biphasic progress curve. The enzyme stability was assessed by analyzing the amplitude of the burst phase after incubation of the enzyme under the reaction conditions at 37°C for several time periods. Aliquots removed at different time intervals were assayed for the adenine removal activity under multiple-turnover conditions to determine the amplitude of the burst phase, which was then compared to that obtained with no pre-incubation of the enzyme. These experiments were performed at both 100 mM and 150 mM NaCl. The results, illustrated in Figure 2B for the reactions at 150 mM NaCl, show that the amplitude of burst phase remains nearly constant for the first ten minutes of incubation at 37 °C in the assay buffer indicating that the protein is stable during the reaction time course. Notably, the burst phase is complete within the first minute, well before the time that the enzyme loses significant activity establishing that the burst amplitude accurately reflects the active enzyme fraction in the overexpressed protein sample. Thus, all enzyme concentrations listed in this work are based on the active site concentration determined by this method. These experiments also indicated similar stability at 100 and 150 mM NaCl; consequently, the higher salt concentration was used in all subsequent experiments.
3.3 Active site titration and stability of MUTYH variants
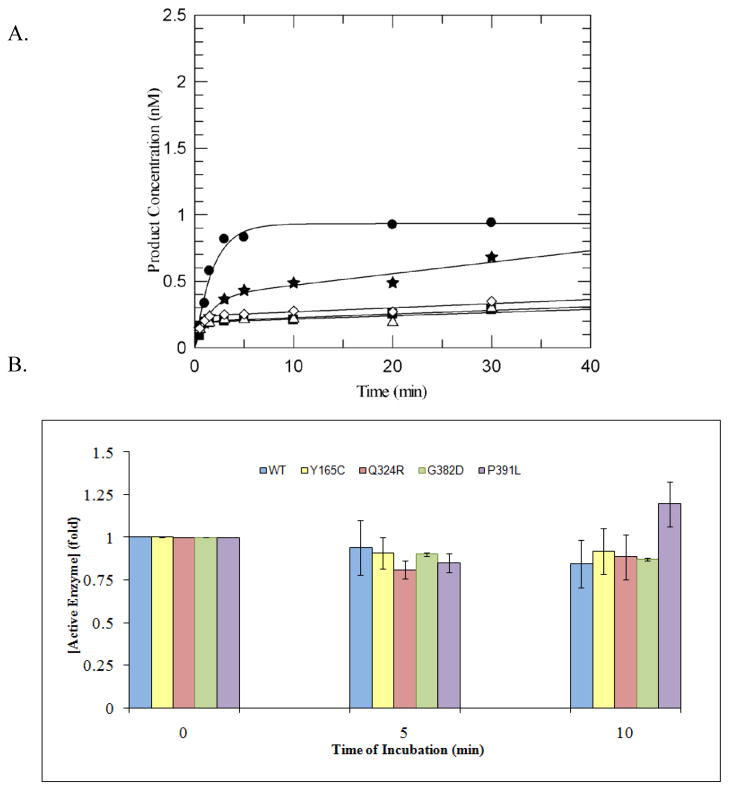
All of the variants displayed biphasic kinetic profiles similar to that of WT MUTYH enzyme under multiple-turnover conditions (Figure 3A). In this experiment, though the same amount of total protein for each variant and WT was used, significant differences in burst amplitudes are observed indicating differences in active fraction among the variants; the WT enzyme at the same total protein concentration consistently has the largest burst amplitude. In addition, the variants were assayed by monitoring the burst amplitude in a manner similar to WT MUTYH to determine the relative stability under the assay conditions. These experiments showed that the loss of enzyme activity for the variants is similar to the WT protein, and also confirms the appropriateness of using the burst amplitude to reflect upon active concentration (Figure 3B). Of note, the burst amplitude for P391L MUTYH increases slightly by the end of the incubation period. Using the amplitude of the burst phase of the reaction with the OG:A substrate from variant enzymes, the active enzyme concentration from enzyme isolated from several different protein preparations was determined (Table 1). Generally, the variants Y165C, G382D, and P391L MUTYH had an active yield that was lower than the wild type protein. Q324R MUTYH preparations generally exhibited active fractions that were the most similar to those of the WT enzyme. These values are reasonably reflective of several different preparations; however, just as the active fraction of WT MUTYH and E. coli MutY vary from different preparations, so does the active fraction of the variants. This underscores the importance of evaluating the active site concentration of a given preparation, since this ensures that the glycosylase reactions are performed under the desired conditions of enzyme in defect or excess (i.e. multiple-turnover or single-turnover, respectively) and allows for reliable and reproducible determination of the relevant kinetic rate constants. The fact that many variants generally provide for lower yields of active fraction is highly suggestive that proper folding of the enzymes into the active conformation may be influenced by a specific amino acid variation.
Figure 3.
Comparison of burst amplitudes of MUTYH variants to the WT enzyme.
(A) Representative plot of adenine removal activity by WT MUTYH and variants under multiple-turnover conditions at 37 °C and 150 mM buffer salt concentration with an OG:A containing duplex DNA substrate (10 nM). WT MUTYH (closed circles), Y165C (closed squares), Q324R (closed stars), G382D (open triangles), P391L (open rhombus) were added to the reaction mix to provide equal total protein concentrations (30 ug as measured by Bradford assay). Lines represent fit to equation 1, with values of A0 obtained for the various enzymes of WT = 0.93; Y165C = 0.19; Q324R= 0.38; G382D = 0.19; P391L = 0.24.
(B) Stability assays of WT MUTYH and variants in assay buffer as measured by glycosylase assays show no significant reduction in enzyme activity as a function of time. Aliquots were removed at intervals of 0, 5 and 10 minutes for adenine glycosylase assays under multiple-turnover conditions and fit to equation 1 to determine the burst amplitudes, A0. The activity of MUTYH without prior incubation was normalized to 1. The bars at 5 and 10 min represent the average loss of activity on incubation. The adenine glycosylase activity at each time point was measured in at least three separate experiments and the error bars represent one standard deviation from the average (as a percentage of the normalized value).
Table 1.
Active yield and rate constants determined for WT MUTYH and variants with an OG:A-containing duplex DNA substrate at 37°C in 150 mM NaCl-containing assay buffer. Reported rate constants have been averaged over at least three separate experiments and the error reported is one standard deviation from the average. Rate constants k3 were determined under conditions of multiple-turnover ([DNA] > [Enzyme]) while rate constants k2 were determined under single-turnover conditions.([DNA] < [Enzyme]).
| Enzyme | Active Yield (ng) per one liter culture | k3 (min−1) | k2 (min−1) | % of WT k2 |
|---|---|---|---|---|
| WT MBP-MUTYH | 143, 195, 850a | 0.011 ± 0.004 | 1.6 ± 0.2 | 100 |
| Y165C MBP-MUTYH | 19, 29, 122 | 0.006 ± 0.001 | 0.5 ± 0.1 | 31 |
| Q324R MBP-MUTYH | 159, 163 | 0.012 ± 0.005 | 0.6 ± 0.1 | 37 |
| G382D MBP-MUTYH | 17, 45 | 0.006 ± 0.002 | 0.5 ± 0.1 | 34 |
| P391L MBP-MUTYH | 20, 135 | 0.002 ± 0.001 | 0.5 ± 0.1 | 29 |
| WT His6-MUTYHb | Not measured | 0.013 ± 0.005 | 1.2 ± 0.5 | NA |
| WT His6-mMutyh | Not measured | 0.002 ± 0.004c | 1.0 ± 0.2 | NA |
Amount extrapolated from a 200 mL growth culture for WT MBP-MUTYH
Rate constants measured at 100 mM buffer NaCl concentration
Rate constant previously published [30].
3.4 Determination of product release rate, k3, from glycosylase activity of WT MUTYH and variants
Since WT MUTYH and all of the variants displayed a similar kinetic behavior to that observed previously with E. coli MutY[16,18], we have used the same minimal kinetic scheme (Scheme 1) and approach in our analyses of the glycosylase activity. Using this simplified scheme, reactions were performed under single- and multiple-turnover conditions to determine the relative rate constants, k2 and k3 [16].
The rate constant k3 can be derived from the slope of the linear portion of the production curves under multiple-turnover conditions and likely reflects the rate of release of MUTYH from the DNA product. However, if the enzyme loses activity over the time course of the reaction, equating the rate of turnover to k3 is not appropriate as the rate of enzyme activity decay has not been accounted for. The active site titration assays performed above (Section 3.2) establish that MUTYH loses minimal activity during the reaction period and therefore the rate determined from the linear phase of the curve may be related to the rate constant k3. The values of k3 determined from at least three separate reactions are listed in Table 1. The rate constant k3 was calculated to be 0.011 ± 0.006 min−1 for WT MUTYH suggesting the OG:AP product is released at a very slow rate. This k3 value is very similar to that observed for E. coli MutY at 150 mM NaCl with the same duplex substrate [41].
Based on the fitting of the linear phase from the biphasic plots, the rate constant k3 (0.012 ± 0.005 min−1) for Q324R MUTYH is similar to the WT enzyme. The values of k3 for G382D and Y165C (0.006 ± 0.002 and 0.006 ± 0.001 min−1, respectively) were twofold decreased compared to the WT protein. Of note, the most altered k3 compared to the other enzymes was for P391L MUTYH (0.002 ± 0.001 min−1).
3.5 Single-turnover reactions of glycosylase activity of WT MUTYH and variants: k2 and Kd measurements
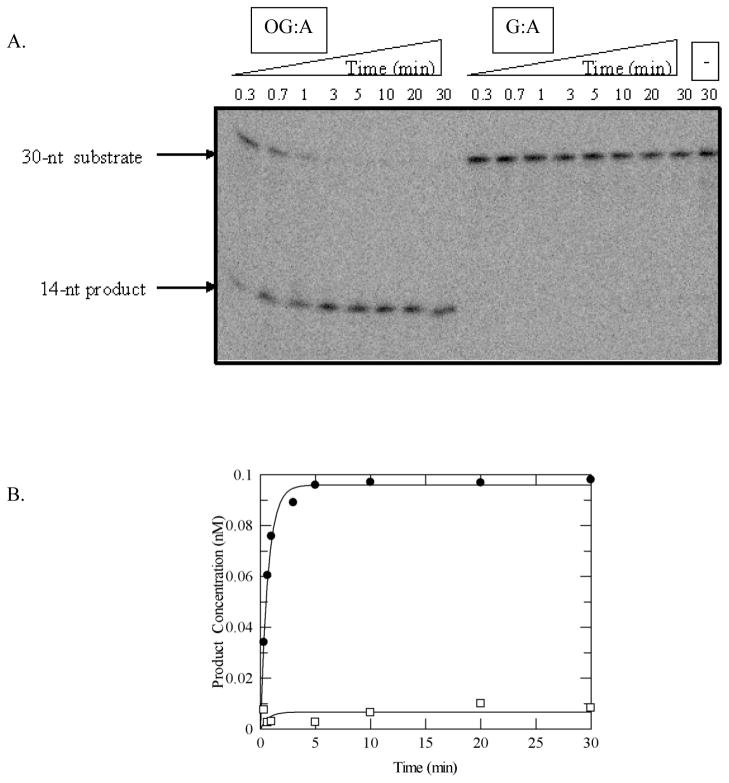
In order to determine the rate constant k2, reactions were performed under single-turnover conditions where the concentration of the enzyme is in excess of the DNA substrate [16]. The observed rate constant of the glycosylase reaction, kobs, is related to the rate constant k2 and depends on the enzyme concentration and the dissociation constant Kd under conditions of rapid equilibrium between the enzyme and substrate DNA. If the enzyme concentration is above the Kd, then it is valid to assume that the observed rate constant is the true rate constant for the base removal step(s) (k2 ≅ kobs). The value for the rate constant k2 determined for the adenine excision by MBP-MUTYH (1.6 ± 0.02 min−1) is quite similar to that obtained for reactions with His6-murine Mutyh [30] and His6-MUTYH with the same OG:A-containing 30-bp duplex (Table 1). Surprisingly, however, the adenine excision of MBP-MUTYH with the G:A containing DNA substrate under the same conditions (Figure 4) resulted in a minimal reaction (kobs < 0.002 min−1).
Figure 4.
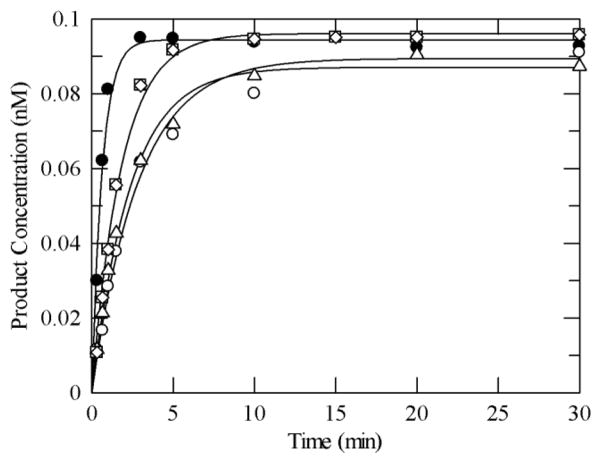
Adenine glycosylase assays of MUTYH under single-turnover conditions with OG:A and G:A substrates. (A) Representative storage phosphor autoradiogram of denaturing PAGE experiment. Bands derived from substrate and product are shown. The minus (−) lane represents the G:A-containing DNA with no enzyme added as a control. (B) Plot of adenine removal activity by WT MUTYH with OG:A- (close circles) and G:A- (open squares) containing substrates at 37 °C. Lines represent fits of the data to a single exponential (equation 2). For this particular experiment, k2 = 1.5 min−1 and <0.002 min−1, for OG:A and G:A, respectively. Reaction conditions: 0.1 nM DNA, 1.5 nM active enzyme in 150 mM NaCl-containing assay buffer.
Representative plots of the reaction of the variant MUTYH enzymes with the OG:A substrate under single-turnover conditions, compared to the WT enzyme, are shown in Figure 5 and the values for k2 determined from at least four separate experiments are listed in Table 1. Single-turnover experiments were also performed at the highest concentrations of the variant enzymes that could be attained without enzyme precipitation, as well as a ten-fold increase in DNA; no change in kobs for the reaction for either condition was observed indicating that the kobs ≈ k2 assumption is reasonable. Under these conditions, all of the variants examined exhibited a k2 value with the same OG:A substrate that was three-fold reduced compared to the wild type protein under similar reaction conditions (Table 1).
Figure 5.
Representative plot of adenine removal activity by WT MUTYH and variants under single-turnover conditions with an OG:A-containing duplex DNA substrate at 37 °C in 150 mM buffer salt concentration. Reaction conditions include 0.1 nM DNA and 1.5 nM active enzyme. WT MUTYH (closed circle), Y165C (open circle), Q324R (open triangle), G382D (open square), P391L (open rhombus). The lines represent fits to a single exponential curve (equation 2) to obtain kobs (= k2). The values for the rate constants determined from at least three measurements for each enzyme (from different enzyme preparations) were averaged and are listed in Table 1.
Under single-turnover conditions the dissociation constant Kd can be determined since this is related to the rate constant k2 through equation 3, where [E0] is the initial concentration of the enzyme. In this case, the DNA concentration was lowered to 0.01 nM to make sure this was below the expected Kd values.
| (3) |
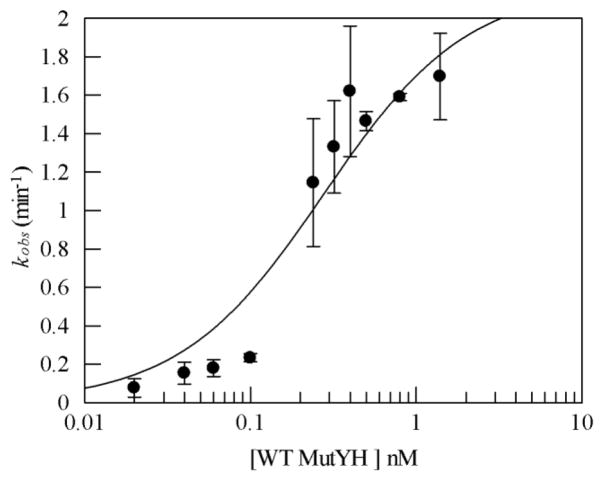
Kd is equal to the enzyme concentration when the kobs equals half of the k2. Therefore, it is possible to evaluate kobs as a function of enzyme concentration under single-turnover conditions and determine the dissociation constant with the substrate. The DNA concentration was kept constant at 0.01 nM while WT MUTYH concentration was varied between 0.025 nM to 1.5 nM. The kobs for the wild type enzyme increased with increasing enzyme concentration up to a MUTYH concentration of 1 nM. However, increasing the enzyme concentration to 1.5 nM did not produce any significant change in the kobs for the WT protein. A plot of kobs as a function of enzyme concentration is shown in Figure 6. WT MUTYH binds to an OG:A-containing DNA substrate tightly with the dissociation constant Kd of 0.3 ± 0.1 nM. This value is similar in magnitude to that observed with murine Mutyh binding to the same OG:A mismatch substrate (0.4 ± 0.1 nM) [30]. Similar titrations were performed with the variant enzymes (Supplementary Figure 2) indicating a similar affinity for the OG: A substrate.
Figure 6.
Representative binding data of WT MUTYH. Rate of adenine removal (kobs) was determined at 37°C and 150 mM buffer NaCl concentration. Reaction conditions included and OG:A mismatch-containing duplex DNA substrate (0.01 nM) and enzyme concentrations between 0.02 and 1.5 nM. The kobs value at each concentration was determined at least three times to provide an average value and the error bars represent the standard deviation from the average. The line represents the fit of the data to a single binding site isotherm and provided a Kd of 0.3 ± 0.1 nM.
3.6 Mutation frequency complementation assays
The ability of MUTYH and MutY to prevent mutations can be evaluated in E. coli using a rifampicin resistance assay [10,31,33,42]. This assay involves observing spontaneous mutations in the rifampicin binding site of E. coli RNA polymerases. If DNA repair activity is low, the accumulation of mutations in an RNA polymerase will render rifampicin less effective as a block to transcription, allowing cell growth in the presence of the drug. The mutation frequency (f) can then be related to the number of rifampicin resistant colonies relative to the control plates for viable titer [32].
The rifampicin complementation assays were performed in an E. coli strain (GT100 y-m-) that lacks functional MutY and the bacterial OG glycosylase MutM [10,43] to allow for differences in mutation frequency between cells expressing WT MUTYH and variants to be readily detected (Table 2). Expression of WT MBP-MUTYH in the GT100 y-m- strain reduced the number of colonies on rifampicin-containing plates providing for a low value for the mutation frequency (f = 1.9). This represents a reduction in mutation frequency of 6-fold and 12-fold with respect to the vector only control and the GT100 y-m- cells. This result indicates that recombinant MUTYH expressed in bacteria can complement for the absence of MutY in E. coli. Sequencing of the rpoB gene from colonies observed on rifampicin-containing plates revealed only G→T transversion mutations, which is consistent with the defect in the GT100 y-m- strain (data not shown).
Table 2.
Rifampicin resistance assay data for MUTYH and variants in GT100 mutY mutM E. coli.a
| Plasmid | f (10−8)b | Increase (fold) over WT |
|---|---|---|
| none | 22 (17–41) | 12 |
| pMAL-c2x | 12 (9.0–23) | 6 |
| pMalMUTYH WT | 1.9 (1.3–2.1) | 1 |
| pMalMUTYH Y165C | 23 (22–27) | 12 |
| pMalMUTYH Q324R | 1.9 (1.4–2.7) | 1 |
| pMalMUTYH G382D | 6.6 (5.4–9.5) | 3.5 |
| pMalMUTYH P391L | 17 (16–32) | 9 |
The rpoB mutation frequency (f) per cell was calculated by dividing the median number of mutants by the average number of cells in a series of cultures.
95% confidence limits based on the mean value are listed in parentheses.
The mutation frequencies with the four variants Y165C, G382D, Q324R, and P391L MUTYH were determined in a similar fashion. The presence of Q324R MUTYH provided a mutation frequency nearly identical to WT MUTYH. In contrast, the mutation frequency observed with Y165C and P391L MUTYH variants (f = 23 and 17, respectively) was similar to that observed in the GT100 y-m- strain. This represents an approximate 10-fold increase in mutation frequency with these variants compared to the presence of WT MUTYH. The mutation frequency observed with G382D MUTYH (f = 6.6) was between that observed with WT MUTYH and the other variants corresponding to a 4-fold increased mutation frequency compared to WT MUTYH. Differences in mutation suppression could not be related to differing amounts of protein since expression levels determined in total cell lysates using Western blot analysis were found to be similar (data not shown). In the presence of all of the variants, sequencing the rpoB gene in the rifampicin resistant colonies indicated that the predominant mutations were G→T transversions (data not shown).
4. Discussion
MUTYH is a unique glycosylase that catalyzes the removal of an undamaged adenine that has been misincorporated opposite the lesion OG. In 2002, MUTYH was thrust into the limelight when inherited mutations in the gene were correlated with colorectal cancer [7]. MUTYH-associated polyposis (MAP) is the only documented form of cancer that originates from inherited biallelic mutations in a base excision repair gene [3]. The initial mutations in MUTYH that were uncovered encoded the variants Y165C and G382D MUTYH [7]. Subsequent work has uncovered a plethora of other missense mutations, as well as truncating and splice site mutations associated with MAP; however, these two original mutations presently represent 70–80% of the known mutations in MUTYH [8]. Though the clinical data related to MAP has been accumulating rapidly, the amount of functional data on MUTYH and variants is limited. Moreover, the reported results from enzyme assays of MUTYH variants are conflicting and provide a murky picture of the relationship between the glycosylase activity and pathogenicity of a given MUTYH variant [14,15,44–46]. We believe the conflicting results are a consequence of the variable amounts of active MUTYH obtained from bacterial overexpression that complicate the analysis of the enzymatic activity. The aim of this study was to provide an approach for characterizing the glycosylase activity of MUTYH and variants associated with MAP. We have analyzed the kinetics of adenine removal from an OG:A-containing substrate with several common MAP variants, namely P391L, Q324R, Y165C, and G382D MUTYH. These results, in combination with the ability of the variant MUTYH enzymes to suppress DNA mutations in rifampicin resistance assays, provide new insight into the factors influencing the ability of MUTYH and variants to prevent mutations associated with OG.
In order to obtain enough high quality protein to analyze the adenine glycosylase activity of MUTYH in vitro, we have overexpressed MUTYH as a fusion-protein with the maltose-binding protein at the N-terminus in E. coli. The expression as an MBP-fusion coupled with the modified MUTYH gene that contains optimal Arg codons resulted in enough active MUTYH protein to use the same approach used previously to analyze the adenine removal activity of E. coli MutY. A critical aspect of this approach is measurement of the burst amplitude from pre-steady state reactions of MUTYH with an OG:A substrate to determine the amount of active enzyme. The challenges associated with expression and isolation of MUTYH from bacteria results in significant fractions of the overexpressed protein that may be misfolded and therefore inactive. Indeed, we observe considerable variations in active fraction with different preparations of WT MUTYH. However, using the active site concentration allows for correction between preparations and reproducible determination of the kinetic rate constants k2 and k3 that correspond to steps involved in the base excision process and product release, respectively. The rate constant k3 is similar for MUTYH to E. coli MutY under the same conditions indicating a similarly high affinity for the DNA product [41]. Interestingly, the rate constant k2 for the intrinsic base excision step measured for WT MUTYH with an OG:A substrate is similar to the corresponding rate constant with the murine Mutyh measured previously [30]. However, WT MUTYH showed minimal activity toward the G:A-containing substrate under the assay conditions approximating physiological salt (150 mM NaCl). This is consistent with results obtained with E. coli MutY, where there is a 70-fold lower k2 rate constant for the adenine glycosylase activity for the G:A containing duplex substrate compared to the OG:A counterpart at higher salt concentrations [41]. In the same study, when the same sequence context for G:A was evaluated in a cellular repair assay, nominal MutY-mediated repair at the G:A bp was observed, while the OG:A mismatch was completely repaired. Such results are consistent with the supposition that OG:A mismatches are the most physiologically relevant substrates for both the bacterial and mammalian forms of this BER glycosylase [33,47].
Using the same kinetic approach, the adenine glycosylase activity of Y165C, G382D, Q324R, and P391L MUTYH variants was examined. Single-turnover experiments were used to determine the rate constant k2 for these variants (Table 1). Our results show a 3-fold reduction in rate for each variant compared to WT MUTYH. This is quite different from previous reported results for Y165C and G382D MUTYH [14,15,44]. For example, the results of Wooden et al. with cell extracts containing Y165C and G382D MUTYH expressed as an N-terminal GST fusion in a CC104 mutY- strain indicated that the variant enzymes were completely devoid of adenine glycosylase activity [14]. A more recent study reported measurements of the rate constant k2 with purified GST-fusion constructs of Y165C and G382D MUTYH expressed in a bacterial strain harboring the tRNAs for the rare Arg codons [15]. The authors concluded that the glycosylase activity of Y165C MUTYH was severely compromised while that of G382D MUTYH was only slightly compromised compared to the WT MUTYH enzyme [15]. We attribute the differences in the previous results to those reported herein to the lack of consideration and correction for active enzyme fraction. We have observed different active enzyme fractions from different preparations of the same enzyme form, including WT MUTYH. In fact, in preliminary work using an expression vector without the Arg codon changes encoding His6-tagged WT MUTYH, some enzyme preparations exhibited barely detectable adenine removal activity. Correction for active site concentration is critical, since differences in active site fraction among different preparations of the same enzyme can be as large as those between different variant enzymes. In the study of Wooden et al., the reaction of WT MUTYH with the OG:A substrate was not complete within the 30 minute incubation-time analyzed indicating a low amount of active enzyme in the reaction [14]. In the later study by Ali et al. [15], the reaction of the OG:A substrate with WT GST-MUTYH was shown to go to completion indicating a higher active enzyme concentration by expression in E. coli harboring the rare Arg tRNAs; however, in the determinations of the rate constant k2, the reactions were performed using the same concentration of total enzyme rather than active enzyme. This type of analysis is misleading since different amounts of active protein may lead to differences in the observed rates. In such cases, attributing these differences to the mutation of a specific amino acid would misrepresent the pathogenicity of a given variant. Moreover, ensuring that the observed rate approximates the rate constant k2 requires that the active enzyme concentration be above Kd and the DNA concentration. Appropriate analysis of the kinetic parameters allows for accurate assessment of the extent with which a modification in an amino acid affects a specific aspect of the mismatch recognition or base excision process. Indeed, such knowledge may also allow for more accurate prediction of the potential outcomes of amino acid changes at other positions in MUTYH. It should also be noted that reproducibly lower amounts of active enzyme fraction for a given variant compared to the WT form may be due to the amino acid change that alters the proper folded conformation required for catalysis; therefore, this is an additional piece of information on the consequences of an amino acid variation in MUTYH.
The kinetic results from adenine glycosylase assays of the Y165C and G382D MUTYH variants in this study are consistent with previous work from our laboratory on the bacterial [10,11] and murine Mutyh [13] enzymes, consistent with the high degree of sequence conservation between MutY enzymes from bacteria to humans. The major difference is that previous functional work on E. coli MutY showed a more dramatic decrease in adenine glycosylase activity caused by the Tyr to Cys change [11]. In addition, the affects on the activity and binding are consistent with the location of the corresponding Tyr and Gly residues in the Bs MutY-DNA structure [12] that place both residues in close proximity to OG. Thus, the structural and functional data are consistent with an increased frequency of G to T transversions in a variety of genes, most conspicuously APC, leading to MAP.
P391L MUTYH is a variant that has been reported in several MAP cases [8,26–28]. Pro 391 in MUTYH corresponds to Pro 269 in Bs MutY and is highly conserved across species (Figure 1). The P391L variant is particularly interesting since the Pro residue is located quite remotely from the OG and adenine binding sites as well as the DNA backbone (Figure 1). This residue is located in the C-terminal domain, which makes extensive contacts with the OG-containing strand and interfaces with the catalytic domain of Bs MutY [12]. Indeed, functional studies have established that this domain plays an important role in the recognition of OG within OG:A mismatches [48,49]. Recently studies using a cell-based repair assay showed that identification of OG rather than the adenine base has a greater influence on location and repair of the mismatch by MutY [41]. Though seemingly far away from the target OG:A bp in the Bs MutY-DNA structure, Pro 269 resides near several other hydrophobic residues and together these residues form a hydrophobic core within the C-terminal domain [12]. Moreover, Pro 269 is involved in a turn region between two β-sheets that are strongly packed with other portions of the C-terminal domain. Disruption of the intricate contacts within this domain by introduction of the large leucine residue might result in destabilization or altered flexibility of this domain that ultimately hamper the enzyme’s ability to recognize OG [26].
The region of MUTYH corresponding to Gln 324 is located in a region not found in the bacterial enzymes that represents an extension of the N-terminal domain before the flexible linker between the two domains (Figure 1). This location makes it difficult to make structural prediction on the possible consequences to DNA mismatch binding and catalysis caused by mutations at this position. Interestingly, the codon for Gln 324 is frequently mutated. The commonly observed polymorph Q324H MUTYH [8] was shown by Ali et al. to be slightly less active than the wild type protein [15], while Shinmura et al. reported that the activity of this enzyme form is identical to the WT protein [50]. In the case of the mutation of Gln to Arg in Q324R MUTYH, we observe that the rate constant k2 for adenine removal from OG:A substrates is reduced three-fold compared to the WT protein. Western blot analysis with whole cell lysates also shows that the bacterial expression levels are comparable to the WT protein (data not shown). Q324R MUTYH is also able to suppress the mutation frequency to WT levels in the rifampicin assays in the absence of endogenous MutY. Thus, in this case, though this variant showed a reduced rate for adenine excision in the glycosylase assays, the enzyme activity was sufficient to mediate repair and prevent mutations in a cellular context. This suggests that enough active enzyme with a sufficient rate of adenine excision was present, providing full complementation for the bacterial enzyme. The idea that some reduction in adenine removal activity may be tolerated is consistent with recent work in our laboratory that showed that the modified substrate base pair, OG:Z3 (where Z3 = 3-deaza-2′-deoxyadenosine), was fully repaired to G:C in E. coli despite having a 100-fold reduction in the measured k2 for base removal in vitro with MutY [41]. However, it was noted that MutY exhibits high affinity for OG:Z3 base pairs suggesting that a reduction in catalytic adenine removal may be more readily tolerated in terms of overall repair if the mismatch is effectively recognized and intercepted prior to replication.
Despite having similar reductions in the rate of adenine removal as Q324R MUTYH compared to WT MUTYH (Table 1), the Y165C, G382D, and P391L MUTYH variants are all less able to suppress mutations as determined in the rifampicin resistance assays. In the presence of G382D MUTYH, the mutation frequency is increased 4-fold over the WT enzyme and is only 2-fold reduced compared to the expression vector control lacking MUTYH. The presence of Y165C or P391L MUTYH provided mutation frequency values that are greater than the vector control indicating an absence of any ability to prevent DNA mutations. The compromised ability to prevent mutations of these three variants could be a result of the reduced stability and the active fraction of these variants in E. coli. Western blot analyses indicated that total amounts of the variant proteins were similar to the WT MUTYH enzyme; this is an advantage of using the MBP tag since its presence equalizes protein expression compared to other tag constructs. However, the concentration of active protein for the Y165C, P391L and G382D MUTYH proteins measured via the glycosylase reaction are typically less than the WT protein. Reduced amounts of active protein coupled with slower adenine removal capacity would effectively further reduce OG:A repair and increase the mutation frequency. It is also possible that the intrinsic activity and active fraction of these variants may be further compromised in the cellular environment. Indeed, we previously observed that the rate constant k2 from adenine glycosylase assays of both the Y150C and G365D murine Mutyh variants was reduced by the presence of the AP endonuclease while the corresponding k2 measurements with WT murine Mutyh enzyme was not affected [13]. This suggested that other proteins may be able to compete with the variants, but not the WT enzyme, for the DNA substrate, thus magnifying any defects in activity in a cellular setting. Interestingly, our previous work showed that Y82C MutY was more severely compromised in terms of catalysis of adenine removal compared to G253D MutY [10]. In addition, the rifampicin assays herein show that the mutation frequency was reduced, albeit modestly (only 2-fold), in the presence of G382D MUTYH, while no mutation suppression was observed with Y165C MUTYH. These results suggest that some specific amino acid substitutions may be more sensitive and consequences due to the presence of the variant MUTYH may be more readily revealed in cellular assays than in vitro. Notably, a recent clinical study showed that the presence of the Y165C MUTYH variant is associated with an earlier onset and a more severe CRC phenotype than G382D MUTYH [51].
P391L MUTYH resulted in mutation frequency results similar to Y165C MUTYH. With this variant the results are not as easily rationalized as those for Y165C MUTYH. For example, the analogous E. coli enzyme (P262L MutY) was found to be identical to the WT MutY in a variety of aspects, including kinetics, mismatch affinity, stability, and ability to suppress DNA mutations [52]. A potential explanation of the seemingly conflicting results with P391L MUTYH may be a result of magnification of defects in locating the mismatch in a large DNA substrate in a cellular context that are not apparent in the bacterial enzyme or the in vitro experiments with a short defined duplex containing a single lesion. In addition, the results can not simply be explained by instability of the enzyme leading to its degradation and absence, since Western blot analyses showed that the level of total protein in cell lysates was similar for P391L and WT MUTYH. However, active fraction on average is reduced for P391L MUTYH and may be magnified in the cellular context. A distinct feature of the kinetics of P391L MUTYH is the considerably reduced rate constant k3 indicating a product release rate that is even slower than WT MUTYH. In a cellular context, MUTYH not only initiates BER but also makes sure that the DNA intermediate is passed on to the next protein in the pathway (APE1) [53,54]. Though this is a bacterial assay, the fact that MUTYH is able to complement for MutY suggests that initiation of BER and coordination with downstream BER enzymes is still maintained. The magnitude of the rate of product release may be quite important to ensure a “safe transition” between the glycosylase and the AP endonuclease. If the release is too fast, this may release the toxic and mutagenic OG:AP site product intermediate prior to full BER. Failure to release the product at some point may also be problematic since this may also thwart full repair of the mismatched site to a G:C base pair.
Clearly many unanswered questions remain as to the extent that defects in the catalytic rates of adenine glycosylase activity translate to reduction in repair of OG:A mismatches and increases in mutation frequency in APC. In the case of Q324R MUTYH, the fact that this enzyme was able to complement for E. coli MutY in the rifampicin assay suggests that this enzyme is functional to reduce mutations despite its reduced glycosylase activity. Together these results suggest that this variant may be less deleterious compared to the other variants that show defects in both types of assays. However, caution should be taken when translating these results to a clinical outcome since other factors may also modulate the activity of the MUTYH protein in colonic cells. For example, post-translational modifications should be considered with eukaryotic enzymes. Indeed, previous work has suggested that MUTYH is phosphorylated [44,55]. In addition, MUTYH has been shown to interact with several proteins associated with the repair and replication machinery [56]. In considering these issues, it is possible that the activity of variants such as Q324R and G382D MUTYH may be more reduced in human cells. Deficiency in the glycosylase activity of MUTYH may only be one of the pieces in the context of a larger repair puzzle. Hence, in order gain a better understanding of the potential pathogenic properties of MUTYH variants, several different types of analyses should be undertaken. For example, determining the effects of the mutations on stability, protein expression levels, recognition and binding of the OG:A mismatch, rate of base removal, interaction with other BER and replication machinery proteins, and cellular mutation frequency, would be particularly illuminating. These results can be correlated with clinical data on the phenotypic presentation associated with a given variant. One might expect that variants that exhibit defects in a variety of assays will be the most strongly correlated with detrimental effects to human health. This is an exceptional example of a cancer mechanism where functional data from biochemical and cellular experiments coupled with structural insight from X-ray crystallography can be correlated with clinical data on patients to provide a comprehensive picture of the disease mechanism.
Supplementary Material
Acknowledgments
We are extremely grateful to M. M. Slupska and Jeff Miller for providing the pQEMUTYHWT vector that contained the modified codons used in this work. We also thank Alan Raetz for help in sequencing the rpoB gene. This work is supported by a grant from the National Cancer Institute of the National Institutes of Health (CA67985).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ames BN, Gold LS. Endogenous mutagens and the causes of aging and cancer. Mutat Res. 1991;250:121. doi: 10.1016/0027-5107(91)90157-j. [DOI] [PubMed] [Google Scholar]
- 2.Klaunig JE, Kamendulis LM. The Role of Oxidative Stress in Carcinogenesis. Annu Rev Pharmacol Toxicol. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 3.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neeley WL, Essigmann JM. Mechanisms of Formation, Genotoxicity, and Mutation of Guanine Oxidation Products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 5.Burrows CM, Muller J. Oxidative Nucleobase Modifications Leading to Stand Scission. Chem Reviews. 1998;98:1109–1151. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 6.Sung JS, Demple B. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. FEBS J. 2006;273:1620–1629. doi: 10.1111/j.1742-4658.2006.05192.x. [DOI] [PubMed] [Google Scholar]
- 7.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Rhodri-Davies D, David SS, Sampson JR, Cheadle JP. Inherited variants of MYH associated with somatic G:C to T:A mutations in colorectal tumors. Nature Gen. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 8.Cheadle JP, Sampson JR. MUTYH-associated polyposis-From defect in base excision repair to clinical genetic testing. DNA Repair. 2007;6:274–279. doi: 10.1016/j.dnarep.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Yanaru-Fujisawa R, Matsumoto T, Ushijima Y, Esaki M, Hirahashi M, Gushima MY, Nakabeppu TY, Iida M. Genomic and functional analyses of MUTYH in Japanes patients with adenomatous polyposis. Clin Genet. 2008;73:545–553. doi: 10.1111/j.1399-0004.2008.00998.x. [DOI] [PubMed] [Google Scholar]
- 10.Chmiel NH, Livingston AL, David SS. Insight into the Functional Consequences of Inherited Variants of the hMYH Adenine Glycosylase Associated with Colorectal Cancer: Complementation Assays with hMYH Variants and Pre-steady-state kinetics of the Corresponding Mutated E. coli Enzymes. J Mol Biol. 2003;327:431–443. doi: 10.1016/s0022-2836(03)00124-4. [DOI] [PubMed] [Google Scholar]
- 11.Livingston AL, Kundu S, Henderson-Pozzi M, Anderson DW, David SS. Insight into the Roles of Tyrosine 82 and Glycine 253 in the Escherichia coli Adenine Glycosylase MutY. Biochemistry. 2005;44:14179–14190. doi: 10.1021/bi050976u. [DOI] [PubMed] [Google Scholar]
- 12.Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- 13.Pope MA, Chmiel NH, David SS. Insight into the Functional Consequences of hMYH variants associated with colorectal cancer: distinct differences in the adenine glycosylase activity and the response to AP endonuclease of Y150C and G365D murine MYH. DNA Repair. 2005;4:315–325. doi: 10.1016/j.dnarep.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Wooden SH, Bassett HM, Wood TG, McCullough AK. Identification of critical residues required for the mutation avoidance function of human MutY (hMYH) and implications in colorectal cancer. Cancer Lett. 2004;205:89–95. doi: 10.1016/j.canlet.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Ali M, Kim H, Cleary S, Cupples C, Gallinger S, Bristow R. Charcterization of mutant MUTYH proteins associated with familial colorectal cancer. Gastroenterology. 2008;135:499–507. doi: 10.1053/j.gastro.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porello SL, Leyes AE, David SS. Single-turnover and Pre-Steady-State Kinetics of the Reaction of the Adenine Glycosylase MutY with Mismatch-Containing DNA substrates. Biochemistry. 1998;37:14756–14764. doi: 10.1021/bi981594+. [DOI] [PubMed] [Google Scholar]
- 17.David SS, Williams SD. Chemistry of Glycosylases and Endonucleases Involved In Base-Excision Repair. Chem Rev. 1998;98:1221–1261. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 18.McCann JAB, Berti Adenine PJ. Release is Fast in MutY-catalyzed Hydrolysis of G:A and 8-oxo-G:A DNA Mismatches. J Biol Chem. 2003;278:29587–29592. doi: 10.1074/jbc.M212474200. [DOI] [PubMed] [Google Scholar]
- 19.Bernards AS, Miller JK, Bao KK, Wong Flipping I. Duplex DNA Inside Out: A double base-flipping reaction mechanism by Escherichia coli MutY Adenine Glycosylase. J Biol Chem. 2002;277:20960–20964. doi: 10.1074/jbc.C200181200. [DOI] [PubMed] [Google Scholar]
- 20.Wiederholdt CJ, Delaney MO, Pope MA, David SS, Greenberg MM. Repair of DNA Containing FapydG and its C-Nucleoside Analogue by Formamidopyrimidine DNA Glycosylase and MutY. Biochemistry. 2003;42:9755–9760. doi: 10.1021/bi034844h. [DOI] [PubMed] [Google Scholar]
- 21.Francis AW, Helquist SA, Kool ET, David SS. Probing the Requirements for Recognition and Catalysis in Fpg and MutY with Nonpolar Adenine Isosteres. J Am Chem Soc. 2003;125:16235–16242. doi: 10.1021/ja0374426. [DOI] [PubMed] [Google Scholar]
- 22.Guan Y, Manuel RC, Arvai AS, Parikh SS, Mol CD, Miller JH, Lloyd RS, Tainer JA. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nature Struct Biol. 1998;5:1058–1064. doi: 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- 23.Sampson JR, Dolwani S, Jones S, Eccles D, Ellis A, Evan DG, Frayling I, Pigatto F, Jordan S, Mak T, Maher ER, Maynard J, Shaw J, Cheadle JP. MYH Polyposis: a new autosomal recessive form of familial adenomatous polyposis demanding reappraisal of genetic risk and family management. Lancet. 2003;362:39–41. doi: 10.1016/S0140-6736(03)13805-6. [DOI] [PubMed] [Google Scholar]
- 24.Kasahara M, Osawa K, Yoshida K, Miyaishi A, Osawa Y, Inoue N, Tsutou A, Tabuchi Y, Tanaka K, Yamamoto M, Shimada E, Takahashi J. Association of MUTYH Gln324His and APEX1 Asp148Glu with colorectal cancer and smoking in a Japanese population. J Exp Clin Canc Res. 2008;27 doi: 10.1186/1756-9966-27-49. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eliason K, Judkins T, Hendrickson BC, Lyon E, Norton M, Thompson V, Gresko S, Leclair B, Barrus J, David S, Livingson A, Reid J, Ward BE, Noll WW, Scholl T. Indentification of novel mutations in MYH in North American patients demonstrates a requirement for whole-gene mutation screening [abstract 476]. Presented at the annual meeting of the American Society of Human Genetics; 2004. [Google Scholar]
- 26.Kanter-Smoler G, Bjork J, Fritzell K, Engwall Y, Hallberg B, Karlsson G, Gronberg H, Karlsson P, Wallgren A, Wahlstrom J, Hultcrantz R, Nordling M. Novel findings in Swedish patients with MYH-associated polyposis: mutation detection and clinical characterization. Clin Gastroenterol Hepatol. 2006;4:499–506. doi: 10.1016/j.cgh.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Aretz S, Uhlhaas S, Georgens H, Siberg K, Vogel M, Pagenstecher C, Mangold E, Caspari R, Propping P, Friedl W. MUTYH-associated polyposis: 70 of 71 patients with biallelic mutations present with an attenuated or atypical phenotype. Int J Cancer. 2006;119:807–814. doi: 10.1002/ijc.21905. [DOI] [PubMed] [Google Scholar]
- 28.Middledorp A, van Pujenbroek M, Nielsen M, Corver WE, Jordanova ES, ter Haar N, Tops C, Vasen H, Lips E, van Eijk R, Hes FJ, Oosting J, Wijnen J, van Wezel T, Morreau H. High frequency of copy-neutral LOH in MUTYH-associated polyposis carcinomas. J Pathol. 2008;216:25–31. doi: 10.1002/path.2375. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch EF, Maniatis T. A laboratory manual. 2. Cold Spring Harbor Laboratory Press; 1989. Molecular cloning. [Google Scholar]
- 30.Pope MA, David SS. DNA damage recognition and repair by the murine MutY homologue. DNA Repair. 2005;4:91–102. doi: 10.1016/j.dnarep.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Golinelli MP, Chmiel NH, David SS. Site-Directed Mutagenesis of the Cysteine Ligands to the [4Fe-4S] Cluster of Escherichia coli MutY. Biochemistry. 1999;38:6997–7007. doi: 10.1021/bi982300n. [DOI] [PubMed] [Google Scholar]
- 32.Wolff E, Kim M, Hu K, Yang H, Miller JH. Polymerases Leave Fingerprints: Analysis of the Mutational Spectrum in Escherichia coli rpoB to Assess the Role of Polymerase IV in Spontaneous Mutation. J Bacteriol. 2004;186:2900–2905. doi: 10.1128/JB.186.9.2900-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slupska MM, Luther WM, Chiang JH, Yang H, Miller JH. Functional Expression of hMYH, a Human Homolog of the Escherichia coli MutY Protein. J Bacteriol. 1999;181:6210–6213. doi: 10.1128/jb.181.19.6210-6213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Planson AG, Guijarro JI, Goldberg ME, Chaffotte AF. Assistance of Maltose Binding Protein to the in vivo Folding of the Disulfide-Rich C-terminal Fragment from Plasmodium falciparum Merozite Surface Protein 1 Expressed in Escherichia coli. Biochemistry. 2003;42:13202–13211. doi: 10.1021/bi035321c. [DOI] [PubMed] [Google Scholar]
- 35.Watkins HA, Baker EN. Cloning, expression, purification and preliminary crystallographic analysis of teh RNase HI domain of the Mycobacterium tuberculosis protein Rv2228C as a maltose-binding protein fusion. Acta Crystallographica Section F Structural Biology and Crystallography Communication. 2008;64:746–749. doi: 10.1107/S1744309108021118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boon EM, Livingston AL, Chmiel NH, David SS, Barton JK. DNA-mediated charge transport for DNA repair. Proc Natl Acad Sci USA. 2003;100:12543–12547. doi: 10.1073/pnas.2035257100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams SD, David SS. Evidence that MutY is a monofunctional glycosylase capable of forming a covalent Schiff base intermediate with substrate DNA. Nuc Acids Res. 1998;26:5123–5133. doi: 10.1093/nar/26.22.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams SD, David SS. A Single Engineered Point Mutation in the Adenine Glycosylase MutY Confers Bifunctional Glycosylase/AP Lyase Activity. Biochemistry. 2000;39:10098–10109. doi: 10.1021/bi0004652. [DOI] [PubMed] [Google Scholar]
- 39.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior Removal of Hydantoin Lesions Relative to Other Oxidized Based by the Human DNA Glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fersht A. Enzyme Structure and Mechanism. W. H. Freeman; New York: 1985. [Google Scholar]
- 41.Livingston AL, O’Shea VL, Kim T, Kool ET, David SS. Unnatural substrates reveal the importance of 8-oxoguanine for in vivo mismatch recognition and repair by the MutY glycosylase. Nat Chem Biol. 2008;4:51–58. doi: 10.1038/nchembio.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai H, Lu AL. Physical and Functional Interactions between Escherichia coli MutY Glycosylase and Mismatch Repair Protein MutS. J Bacteriol. 2007;189:902–909. doi: 10.1128/JB.01513-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michaels ML, Cruz C, Grollman AP, Miller JH. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker AR, Sieber OM, Shi C, Hua L, Takao M, Tomlinson I, Eschleman JR. Cells with pathogenic biallelic mutations in the human MUTYH genes are defective in DNA damage binding and repair. Carcinogenesis. 2005;26:2010–2018. doi: 10.1093/carcin/bgi166. [DOI] [PubMed] [Google Scholar]
- 45.Bai H, Grist S, Gardner J, Suthers G, Wilson TM, Lu AL. Functional characterization of human MutY homolog (hMYH) missense mutation (R231L) that is linked with hMYH-associated polyposis. Cancer Lett. 2007;250:74–81. doi: 10.1016/j.canlet.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai H, Jones S, Guan X, Wilson TM, Sampson JR, Cheadle JP, Lu AL. Functional characterization of two human MutY homolog (hMYH) missense mutations (R227W and V232F) that lie within the putative hMSH6 binding domain and are associated with hMYH polyposis. Nucleic Acids Res. 2005;33:597–604. doi: 10.1093/nar/gki209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michaels ML, Tchou J, Grollman AP, Miller JH. A Repair System for 8-oxo-7,8-dihydrodeoxyguanosine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- 48.Noll DM, Gogos A, Granek JA, Clarke ND. The C-terminal Domain of the Adenine-DNA Glycosylase MutY Confers Specificity of 8-Oxoguanine-Adenine Mispairs and May have evolved from MutT, an 8-oxo-dGTPase. Biochemistry. 1999;38:6374–6379. doi: 10.1021/bi990335x. [DOI] [PubMed] [Google Scholar]
- 49.Chmiel NH, Golinelli MP, Francis AW, David SS. Efficient Recognition of substrates and substrate analogs by the adenine glycosylase MutY requires the C-terminal domain. Nucleic Acids Res. 2001;29:553–564. doi: 10.1093/nar/29.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shinmura K, Yamaguchi S, Saitoh T, Takeuchi-Sasaki M, Kim SR, Nohmi T, Yokota J. Adenine excisional repair function of MYH protein on the adenine:8-hydroxyguanine base-pair in double-stranded DNA. Nucleic Acid Res. 2000;28:4912–4918. doi: 10.1093/nar/28.24.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nielsen MJ, Joernik-Van De Bel MC, Jones N, Vogt S, Tops CM, Vasen HFA, Sampson JR, Aretz S, Hes FJ. Analysis of MUTYH Genotypes and Colorectal Phenotypes in Patients with MUTYH-Associated Polyposis. Gastroenterology. 2008 doi: 10.1053/j.gastro.2008.10.056. in press. [DOI] [PubMed] [Google Scholar]
- 52.Livingston AL, Kundu S. unpublished results. [Google Scholar]
- 53.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: Implications for DNA damage recognition, removal and repair. DNA Repair. 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 54.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nature Struct Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 55.Gu Y, Parker A, Wilson TM, Bai H, Chang DY, Lu AL. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J Biol Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- 56.Parker AR, Eschleman JR. Human MutY: gene structure, protein functions and interactions, and role in carcinogenesis. Cell Mol Life Sci. 2003;60:2064–2083. doi: 10.1007/s00018-003-3053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.