Abstract
Recognition of stop codons by class I release factors is a fundamental step in the termination phase of protein synthesis. Since premature termination is costly to the cell, release factors have to efficiently discriminate between stop and sense codons. In order to understand the mechanism of discrimination between stop and sense codons, we developed a new, pre-steady state kinetic assay to monitor the interaction of RF1 with the ribosome. Our results show that RF1 associates with similar association rate constants to ribosomes programmed with a stop or sense codons. However, dissociation of RF1 from sense codons is as much as three orders of magnitude faster than from stop codons. Interestingly, the affinity of RF1 for ribosomes programmed with different sense codons does not correlate with the defects in peptide release. Thus, discrimination against sense codons is achieved, both, by increasing the dissociation rates and by decreasing the rate of peptide release. These results suggest that sense codons inhibit conformational changes necessary for RF1 to stably bind to the ribosome and catalyze peptide release.
Keywords: Termination, ribosome, release factor, decoding, translation
Termination of protein synthesis is triggered when the nearly universal stop codons UAA, UAG, or UGA enter the decoding center of the small ribosomal subunit (1). Recognition of a stop codon by class I release factors (RF) leads to peptidyl-tRNA hydrolysis and the release of the newly synthesized protein from the ribosome (2). In bacteria, the stop codons in the mRNA sequence are recognized by two release factors: RF1 and RF2. RF1 recognizes UAA and UAG, while RF2 recognizes UAA and UGA (3). In eukaryotes, a single release factor (eRF1) recognizes all three stop codons (4). Stop codons are recognized by RFs with remarkably high accuracy (error frequency of 1 × 10-3 to 1 × 10-6), even without a proofreading mechanism, indicating that the RFs have a sophisticated mechanism for distinguishing the three stop codons from the sixty-one sense codons (5) (6).
RF1 and RF2 consist of 4 domains (7) (8). Genetic and biochemical studies identified a ‘tripeptide anticodon’ motif in domain 2 of RF1 and RF2 [P(A/V)T in RF1 and SPF in RF2] that is important for stop codon recognition (9). Additionally, a universally conserved GGQ motif located in domain 3 of RF1 and RF2 is important for peptide-tRNA hydrolysis suggesting that RF1 and RF2 spans the ≈75 Å distance between the decoding and the peptidyl transferase centers (10). This was confirmed by hydroxyl-radical probing experiments (11) (12), cryoelectron microscopy (cryoEM) (13) (14) (15) and crystal structures of RF1 or RF2 bound to the ribosome (16). In contrast, crystal structures of unbound RF1 and 2 show the factors in a closed conformation with the tripeptide anticodon and GGQ motif only 25 Å apart (7) (8). This has led to the suggestion that RFs bind to the ribosome in a closed conformation and extend into the peptidyl transferase center after binding (14). Solution X-ray scattering (SAXS) experiments show that RF 1 and 2 exist in an ensemble of open and closed forms in solution (17) (18). What form RFs are in when binding to the ribosome and what conformational changes they undergo remain open questions.
Recent x-ray crystal structures of RF1 or RF2 bound to the ribosome have revealed in exquisite detail the structural basis for stop codon recognition (19) (20, 21). The ‘anticodon tripeptide’ motif in RF1 and RF2 interact precisely with the stop codons in the decoding center (Figure 1). Interestingly, the structures showed that other residues in RF1 and RF2, in addition to the tripeptide motif, are also important for stop codon recognition. The first position of the stop codon (U1) interacts with a conserved glycine in domain 2 of RF1 or RF2. Additionally, specific hydrogen bonds are formed by U1 with conserved residues in the tripeptide motifs of RF1 and RF2. These interactions strongly discriminate against a purine and also explain the preference for a uridine at the first position. The second position of the stop codon (A2 or G2) stacks against conserved residues in the RFs and forms hydrogen bonds with the threonine or serine in the tripeptide motifs of RF1 or RF2, respectively. However, it is not clear how the RFs discriminate against pyrimidines at the second position, other than the loss of packing interactions. Finally, the third position of the stop codon (A3 or G3) is unstacked from the second position of the codon by a histidine from RF inserted between the two bases and stacks instead on G530 of the 16S rRNA. The third position forms several hydrogen bonds with specific residues in RF1 or RF2, which explains the preference for an adenine or a guanine by RF1 and the preference for an adenine by RF2.
Figure 1. Interaction of RF1 with the ribosome.
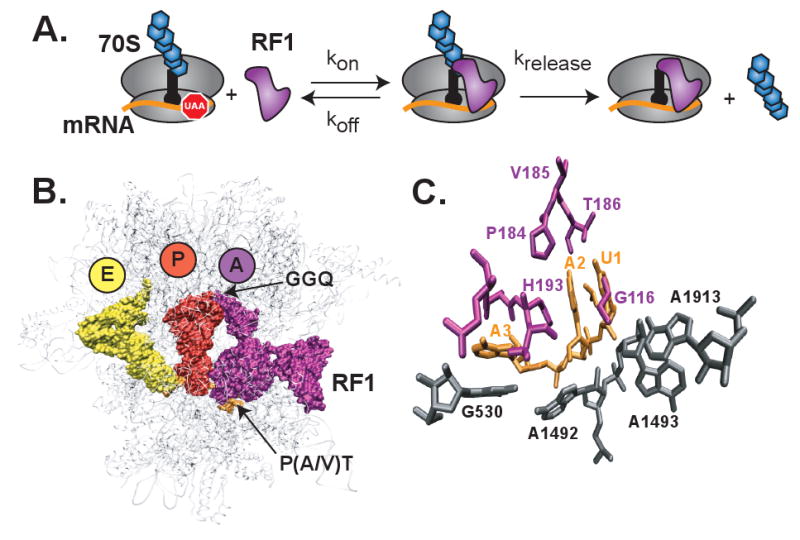
(A) A kinetic model for RF1 binding to the ribosome followed by hydrolysis of the newly synthesized protein attached to the P site tRNA. Ribosome (grey), mRNA (orange), P site tRNA (black) with attached protein (blue hexagons), and RF1 (purple). (B) Structure of RF1 bound to the ribosome. Ribosome (grey), E site tRNA (yellow), P site tRNA (red), and RF1 (purple). (C) Recognition of the stop codon in the decoding center by RF1. Stop codon U1A2A3 (orange), RF1 residues (purple), and bases in 16S and 23S rRNAs (grey).
While the x-ray crystal structures provide a rationale for specific recognition of stop codons by RF1 and RF2, it is not known how the dynamics of RF binding is influenced by stop and sense codons in the ribosome. This is an important problem because the kinetics of RF association and dissociation must be finely tuned so that stop codons are efficiently recognized without inhibiting the elongation phase of protein synthesis by competing with aminoacyl-tRNAs for binding to the A site. A landmark study with a variety of sense codons that differed by a single nucleotide from the stop codon showed that the catalytic rate constant (kcat) of peptide release was reduced by 2 -to 180-fold, while the KM of RF1 increased by 400- to 3,000-fold (6). However, this study did not monitor RF binding directly but relied on KM measurements to distinguish between defects in binding from catalysis. Recent studies suggest that the conformational changes induced by RF1 with stop codons versus sense codons are different leading to differences in the kcat of peptide release (22). This makes it difficult to interpret the molecular basis for the observed changes in KM and kcat with ribosomes having sense codons in the A site.
In order to directly monitor the interaction of RF1 with the ribosome, we have developed a fluorescence based, pre-steady state kinetic assay for RF binding. Our kinetic studies show that the association rate constant of RF1 is not significantly affected with a stop or sense codon in the decoding center. In contrast, the dissociation rate constant of RF1 differs by as much as a 4,000-fold depending on whether a stop or sense codon is present in the decoding center. Interestingly, the binding kinetics of RF1 does not always correlate with the rate of peptide hydrolysis suggesting that conformational changes, following stop codon recognition, are important for preventing premature termination on sense codons.
Experimental Procedures
Buffers, Ribosomes, tRNA, mRNA, and RF1
All experiments were performed in 20 mM Hepes-KOH (pH 7.6), 6 mM MgCl2, 150 mM NH4Cl, 4 mM β-mercaptoethanol, 0.05 mM spermine, 2 mM spermidine (23). Tight coupled 70S ribosomes were isolated from E. coli MRE600 cells, essentially as described (24). Native tRNAfMet, was purchased from Sigma. mRNAs were purchased from Dharmacon and labeled with pyrene succinimide as previously described (25). His-tagged RF1 was purified essentially as described in the QIAexpressionist manual (Qiagen). Fractions containing RF1 were pooled and concentrated in an Amicon 10 kDa cutoff filter. Buffer exchange was performed in the Amicon filter to greater than 3,000 fold dilution of the unretained buffer. RF1 was then quantitated by the Bradford assay, flash frozen in liquid nitrogen, and stored at -80°C.
Fluorescence Measurements of RF1 Binding
Release complexes were formed by heat activating 0.25 μM tight-coupled 70S ribosomes at 42 °C for 10 min. Ribosomes were then cooled to 37 °C for 10 min. 0.33 μM pyrene labeled mRNA was added and incubated for 10 min. at 37 °C. 0.5 μM tRNAfMet was then added and incubated at 37°C for 30 min.
Fluorescence emission scans were performed with a Fluoromax-P (J. Y. Horiba, Inc. USA) using an excitation and emission bandpass of 1 nm. Samples were excited at 343 nm and emission scans from 360 to 420 nm were taken before and after the addition of 0.5 μM RF1 to termination complexes.
Equilibrium KD titrations were performed by mixing the indicated amounts of RF1 with 0.25 μM release complexes in a 1 mL fluorescence cuvette at 25 °C. The samples were excited with 343 nm wavelength light and the emission at 376 nm was read 5 min after mixing. In parallel, the fluorescence emission of RF1 added to buffer was measured and subtracted from the data to account for light scattering at high protein concentrations. Experiments were repeated at least three times. Data were fit to the equilibrium KD equation below using Graphpad Prism as described (26).
Stopped-flow measurements were performed at 25 °C on a μSFM-20, BioLogic stopped-flow instrument. The samples were excited at 343 nm (band pass 10 nm) and the fluorescence emission was measured at 376 nm after passing a longpass filter 361 AELP (Omega Optical, VT, USA) installed in front of the detector. 0.25 μM (final concentration) termination complexes were mixed with varying amounts of RF1. Individual time courses were fit to the first-order rate equation (Y = b + C*exp(-k*x)).
Peptide Release Assay
Release complexes for peptide release assays were formed by heat activating 1.0 μM 70S at 42 °C for 10 min. Samples were then cooled to 37 °C for 10 min. 1.3 μM mRNA was added and incubated at 37 °C for 10 min. In parallel, tRNAfMet was aminoacylated by mixing 2 μM tRNAfMet, 3 μM [35S] methionine, 0.4 mM N10-formyltetrahydrofolic acid, 3 mM ATP, 10% (v/v) MetRS, and 10% (v/v) MTF. MetRS and MTF were purified as previously described (27). The aminoacylation reaction was then mixed with the 70S/mRNA complex and the incubation was continued for 30 min at 37 °C. Excess [35S] Met was washed away by filtration in a Microcon YM-100 centrifugal filter to a dilution of greater than 1,000 fold and the volume was adjusted to obtain pre-termination complexes with a final concentration of 0.5 μM.
Peptide release time courses were performed by mixing 0.25 μM (final concentration) release complex with varying amounts of RF1. Reactions were quenched with 25% formic acid, spotted on a TLC, separated and analyzed as described previously (28). All experiments were repeated at least three times.
Results
Fluorescence based method to measure RF1 binding to the ribosome
Rapid kinetic methods have been very valuable for understanding the mechanism of translation initiation, tRNA selection, translocation, and ribosome recycling (29) (30) (25) (31). However, no rapid kinetic methods have been developed to examine the intermediates in the mechanism of stop codon recognition by RF1 or RF2 (32). To determine the pre-steady state kinetics of RF1 binding to the ribosome, we developed a fluorescence-based method. We attached the fluorescent probe, pyrene, to the 3′ end of a short mRNA (Figure 2A). The pyrene dye is located 3 bases away from the A-site codon where the release factor binds. Based upon crystal structures, the probe would be located in the mRNA channel between the head and shoulder of the 30S subunit, approximately 25 Å away from the middle position of the codon in the A-site (33).
Figure 2. Fluorescence assay to monitor RF1 binding.
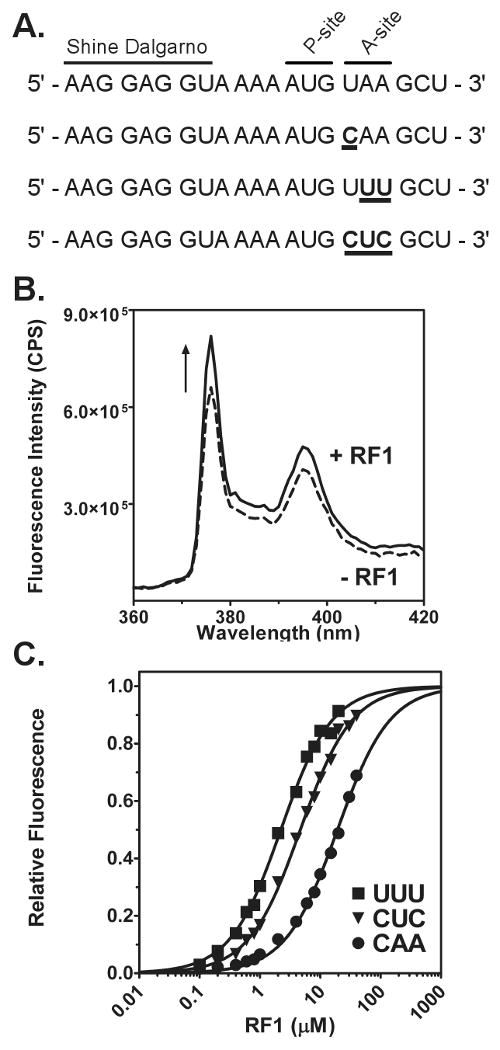
(A) Sequence of four model mRNAs used to measure RF1 binding to stop or sense codons. The Shine-Dalgarno sequence, P-site, and A-site codons are labeled. Changes from the UAA stop codon are shown in bold and underlined. (B) Increase in fluorescence intensity due to RF1 binding. Fluorescence emission scans before (dotted line) and after addition (black line) of RF1 to release complex with a UAA stop codon in the A site. (C) Examples of fluorescence titrations to determine the KD of RF1 for sense codons: UUU (square), CUC (triangle), and CAA (circle). Data were analyzed by fitting to a quadratic equation (black line) and normalized from zero to one based upon the best-fit line.
To monitor the binding of RF1 to the ribosome, we formed release complexes (RC) by sequentially adding pyrene-labeled mRNA and tRNAfMet to 70S ribosomes. The mRNA has a start codon (AUG) at the first position and a stop codon (UAA) at the second position. Binding of tRNAfMet to the P site will position the stop codon in the A site. Upon addition of RF1 to release complexes, an increase in the fluorescence intensity of the pyrene probe was observed (Figure 2B). A likely explanation for the increase in fluorescence intensity is the exclusion of solvent from the ribosomal A-site upon binding of RF1. Direct interactions between the probe and RF1 appear unlikely based on the X-ray crystal structures (19) (20, 21).
Affinities of RF1 for stop and sense codons
Four model mRNAs were synthesized in order to investigate the mechanism of RF1 discrimination of stop and sense codons (Figure 2A). The sense codons were selected based upon interactions observed in the crystal structure of RF1 bound to a UAA stop codon (19), previous biochemical data on release factor discrimination (6), and work done on tRNA selection (34) in order to facilitate comparison between these processes. The UAA stop codon was chosen in order to characterize the correct stop codon recognition pathway. The second mRNA has a CAA codon in the A site. The single incorrect C in the first position should not create any obvious steric clashes with the release factor but will result in loss of hydrogen bonding with the release factor in this position. A third mRNA with a UUU A-site codon was chosen because UUU is by far the most commonly used codon for in vitro ribosome experiments and other than loss of packing interactions with the release factor, it is unclear from crystallography how RF1 discriminates against pyrimidines in the second and third positions of the A-site codon (19). The last mRNA tested had a CUC codon in the A-site, which is different from a stop codon at all three positions while still avoiding obvious steric clashes with the ribosome or release factor.
Previous work addressing discrimination of sense codons from stop codons by RF1 at the binding step was primarily done by measuring the KM of peptide release (6). As KM measurements are dependent on catalytic activity, discrimination of stop and sense codons by RF1 at the binding step have never been addressed independent of catalysis (32). In order to determine the affinity of RF1 for stop and sense codons, we performed fluorescence titrations of RF1 with ribosomes programmed with each of the above mRNAs (Figure 2C). Increasing amounts of RF1 were added to a fixed concentration of RC programmed with each of the mRNAs. The fluorescence emission was measured after each addition of release factor. In parallel, a blank titration was performed to account for increase in fluorescence due to light scattering under conditions of high protein concentration. The affinity of RF1 for a UAA stop codon programmed A-site could not be accurately determined due to the extremely tight binding affinity of the release factor for this codon. Titrations at the lowest measurable concentration of labeled termination complexes showed that the KD of RF1 for UAA programmed ribosomes was less than 3.5 nM (data not shown). Fluorescence titrations performed on each of the sense codons showed at least 1000-fold increase in the KD of RF1 for the A-site. Among sense codons, RF1 was found to bind most tightly to the UUU codon with a KD of 1.6 μM. Release factor bound less stably to the CUC codon with a KD of 6.5 μM. Surprisingly, RF1 bound least stably to the CAA codon, with a KD of 15.4 μM, even though it varies by only one nucleotide from a cognate stop codon.
Kinetics of RF1 binding to stop and sense codons
To investigate the kinetics of RF1 binding to stop and sense codons we determined the time course of fluorescence change with a stopped-flow fluorimeter. Release complexes were mixed with varying amounts of RF1 and the increase in fluorescence intensity was monitored over time. Time courses of the fluorescence change were described well by single exponential fits, which were used to determine the observed rate of RF1 binding to the ribosome. (Figure 3A). Plotting the observed rate of fluorescence change versus release factor concentration showed a linear relationship (Figure 3B). The linear concentration dependence indicates a second order reaction with the slope of the line equal to the association rate constant and the y-intercept equal to the dissociation rate constant (35). RF1 bound to UAA programmed ribosomes with a rate constant of 34.4 μM-1s-1. This association rate constant was reduced by two-fold or less in the cases of sense codons CAA, UUU or CUC, indicating that the association of the factor to the A-site is not a significant source of stop versus sense codon discrimination. Due to the small value of the y-intercept of concentration dependence curves of RF1 binding to the UAA stop codon and errors magnified by extrapolation of the data back to the y-axis, the values of the dissociation rate constants were calculated from directly measured values of KD and kon (Table 1). Dissociation rate constants were found to vary over a nearly 15-fold range among sense codons and overall to be at least 250-fold faster than dissociation from the UAA stop codon. Trends in dissociation rate constants were the same when comparing y-intercepts and calculations from KD measurements
Figure 3. Kinetics of RF1 binding to stop and sense codons.
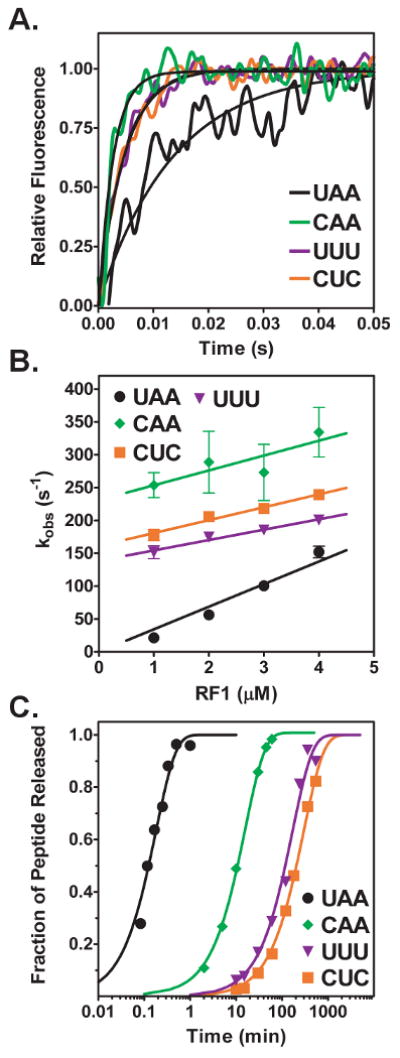
(A) Stopped-flow time course of RF1 (2 μM) binding to release complexes (0.25 μM) with UAA (black trace), CAA (green trace), UUU (purple trace), or CUC (orange trace) codon in the A site. The time courses were fit to a single exponential equation (black line) to determine the observed rate of RF 1 binding (kobs). (B) Concentration-dependence of RF1 binding. Observed rates are plotted versus RF1 concentration for UAA (circle), CAA (diamond), UUU (triangle), and CUC (square). The standard errors from at least three independent experiments are shown. Plots were fit to a linear equation to determine the association (kon) and dissociation (koff) rate constants. In the case of UAA, the y-intercept was constrained to be a positive value. (C) Examples of peptide release time course at saturating RF1 concentrations. Peptide release from release complexes with UAA (circle), CAA (diamond), UUU (triangle), or CUC (square) codons in the A site are indicated. The concentration of RF1 was 9 μM for the stop codon and 200 μM for the sense codons. Data were normalized and fit to a single exponential equation (black line) to determine the rate of peptide release (krelease).
Table 1. Kinetic and thermodynamic parameters for RF1 binding.
| Codon | KD (μM) | kon (μM-1 s-1) | koff (s-1)a | krelease (s-1) |
|---|---|---|---|---|
| UAA | < 0.0035 | 34.4 | < 0.1 | 9.0 × 10-2 ± 0.1 × 10-2 |
| CAA | 15.4 ± 4.6 | 22.6 | 348 | 9.9 × 10-4 ± 0.9 × 10-4 |
| UUU | 1.6 ± 0.3 | 15.8 | 25.3 | 9.4 × 10-5 ± 0.4 × 10-5 |
| CUC | 6.5 ± 3.1 | 19.6 | 127.4 | 6.6 × 10-5 ± 1.4 × 10-5 |
, koff was calculated from KD and kon.
Kinetics of peptide release on stop and sense codons
Peptide release time courses were performed by forming release complexes containing each mRNA to be tested and [S35]fMet-tRNAfMet in the P-site, then mixing release complexes with saturating amounts of RF1. RF1-catalyzed release of [S35]fMet was analyzed by separating the released [S35]fMet by electrophoretic thin layer chromatogrphy (eTLC) and quantitated on a phosphorimager. In order to verify that saturation had been reached, time courses were also performed with half the concentration of RF1 with identical kinetics obtained. As expected, peptide release was significantly slower on sense codons than on the stop codon; however; the kinetics of peptide release surprisingly did not follow the same trends as binding (compare Figure 3C and Table 1). Peptide release was catalyzed with a rate of 9.0 × 10-2 s-1 on the cognate UAA stop codon, which is similar to previously determined rates of release (22). Among sense codons, RF1 was able to most efficiently catalyze peptide release on the CAA sense codon to which it bound least stably (krelease = 9.9 × 10-4 s-1). Peptide release on the CAA sense codon was 10-15 times faster than on UUU or CUC sense codons (krelease = 9.4 × 10-5 s-1 and 6.6 × 10-5 s-1 for UUU and CUC, respectively).
Discussion
The simplest kinetic model for peptide release involves two steps: (1) Binding of RF1 or RF2 to ribosomes with a stop codon in the A site and (2) Release of the newly synthesized protein by the catalytic activity of RF1 or RF2 (Figure 1A). Much work has been done investigating the mechanism of peptide release but this work has focused exclusively on the catalytic step (32). In order to differentiate the contribution of binding and catalysis to correct stop codon selection by a release factor, we have developed a fluorescence-based method to monitor RF1 binding to the ribosome. Fluorescence titrations on four model mRNAs designed to disrupt interactions with RF1 in a variety of ways were performed to look at the affinity of release factor for sense codons. As expected, RF1 bound best to the correct UAA stop codon but, surprisingly, had the lowest affinity for the CAA codon, which only has a single base change from a cognate stop codon, compared to UUU and CUC which have 2 and 3 base changes, respectively. The binding kinetics of RF1 to the A-site was also determined. Association rate constant of release factor to the A-site was only slightly affected by the codon. In contrast, dissociation rate constants were found to increase by at least 250-fold when a stop codon was replaced with a sense codon (Table 1). Furthermore, dissociation from sense codons varied over a 15-fold range. In agreement with the equilibrium KD measurements, the rate of dissociation of RF1 was greater from ribosomes with the CAA than CUC codon in the A site.
It is unclear why RF1 would bind less stably to CAA than to CUC. Both have the first, and most sensitive, position of the codon changed from U to C but the two remaining nucleotides are correct for a stop codon in CAA and incorrect for a stop codon in CUC. The crystal structure of RF1 bound to a UAA stop codon shows that the second and third nucleotides in the codon are unstacked with a histidine residue from the release factor inserted between them (Figure 1C) (19). It appears from the structural data that this unfavorable event could at least be partially compensated for by stacking of A in the second position of the codon with H193 of RF1 and stacking of the third position A with G530 of the 16S rRNA (Figure 1C). The difference in stability of RF1 binding to a CAA or CUC codon however suggests that unstacking of the AA residues is not entirely compensated for by these alternative interactions. Although, thermodynamically unfavorable, unstacking of the AA residues could be evolutionarily conserved if it plays an important role in conformational changes associated with the catalytic step of peptide release. This does appear to be the case as peptide release is catalyzed more efficiently on a CAA codon than CUC despite the less favorable binding interactions.
Consistent with our results, a previous study on release factor fidelity showed that RF1 is most sensitive to changes in the first position of the stop codon (CAA or CAG). A CAA codon in the A-site increased the KM of peptide release by 2,000-fold and decreased the kcat by 180-fold (6). We observed a greater than 4,000-fold defect in KD and a 90-fold defect in peptide release. The difference in the effect of CAA on the KD and KM is likely due to the fact that the active state of peptide release is at least somewhat induced on the CAA codon, resulting in less RF1 required to complete hydrolysis. This agrees with our observation that a lower concentration RF1, relative to the KD, was required to reach saturation on a CAA codon than the other sense codons tested (data not shown).
Measurement of peptide release rates showed that the efficiency of catalysis of peptide release on sense codons is not directly related to the affinity of release factor to the A-site. Among sense codons tested, peptide release was most efficiently catalyzed on a CAA sense codon, which had the lowest affinity for RF1 (Table 1). Especially interesting is the 10-fold higher rate of peptide release on CAA compared to CUC codon. A potential explanation for why this occurs comes from X-ray crystallography (19). Stacking of the third residue in the codon with G530 of 16S rRNA is more favorable with a purine than a pyrimidine. By unstacking the third residue from the rest of the codon, the backbone of the mRNA is distorted. A1492 comes out of its helix and packs against this distorted mRNA backbone. Movement of A1492 is important for release factor function as it opens up space for A1913 of the 23S rRNA to stack with A1493 of the 16S rRNA, allowing RF1 to undergo its putative extension into the peptidyl transferase center after binding or to bind in the fully extended form (19). Without this conformational change, A1913 would block binding of RF1. A purine in the third codon position could be important to precipitate the conformational changes seen in the decoding center, explaining why peptide release is better on a CAA codon than CUC despite the greater binding defects.
It has often been suggested that release factors act as molecular mimics of tRNAs (32). However, there are clear differences in the pathway of codon selection by release factors and tRNAs. First, release factors are able to discriminate stop from sense codons without a high energy intermediate, which is required for tRNA selection by the kinetic proofreading mechanism (6) (36). Second, there appears to be no nonspecific binding intermediate limiting the association or dissociation of RF1 from the ribosome as has been suggested in the mechanism of tRNA selection (30, 37). This is important as the uniformity in RF1 association rates indicates that RF1 is able to scan each codon position and only remains stably bound when a stop codon is encountered. If dissociation rates of RF1 on sense codons were not sufficiently fast, an overall inhibition of protein synthesis would be observed. Third, the affinity of RF1 for all codons is not as predictable as Watson-Crick base pairing between tRNAs and mRNA. We have seen that single base changes from a cognate stop codon can result in a 10-fold lower affinity than when two bases are changed even in the limited set of sense codons tested.
Decoding by release factors and tRNA selection do appear to share an induced fit mechanism, however. In tRNA selection, Watson-Crick base pairing between the anticodon of the tRNA and codon of the mRNA in the A-site of the ribosome induces an acceleration of the forward rates of GTP hydrolysis by EF-Tu and peptidyl transfer (38, 39). Modulation of these rates could be achieved through acceleration of the conformational changes leading to catalysis or improvement in the transition state stabilization. Similarly, for codon selection by RF1, an increase in the rate of peptide hydrolysis is seen independently from defects in the binding step. It seems reasonable that an active site such as the peptidyl transferase center would be dependent upon conformational changes caused by substrate binding because it must be able to accept a variety of very distinct substrates and catalyze two distinct reactions depending upon interactions 75 Å away in the decoding center, while protecting the currently bound substrate from spontaneous hydrolysis by solvent (40).
We have shown that steps after the simple recognition of the codon by release factor are important in selection of the correct stop codon. Changes in the measured value of the krelease step could encompass conformational changes proposed to occur after release factor binding such as full opening of RF after binding, decoding center rearrangement, changes in the peptidyl transferase center to accommodate the RF and attacking water molecule, or misalignment of catalytic residues in the peptityl transferase center leading to poor transition state stabilization. Determination of relevant reaction intermediates and their role in correct stop codon selection is essential to understand the early steps of termination of protein synthesis.
Acknowledgments
We thank Oliver Scholz (University of Hanover, Germany) for performing some of the early experiments during a summer internship and Ulrich Muller for comments on the manuscript.
Abbreviations
- RF1
release factor 1
- rRNA
ribosomal RNA
- mRNA
messenger RNA
- tRNA
transfer RNA
Footnotes
This work was supported by a NIH Molecular Biophysics Training grant (GM08326 to B.H.) and a NIH grant (GM 065265 to S.J.).
References
- 1.Brenner S, Stretton AO, Kaplan S. Genetic code: the ‘nonsense’ triplets for chain termination and their suppression. Nature. 1965;206:994–998. doi: 10.1038/206994a0. [DOI] [PubMed] [Google Scholar]
- 2.Capecchi MR. Polypeptide chain termination in vitro: isolation of a release factor. Proc Natl Acad Sci U S A. 1967;58:1144–1151. doi: 10.1073/pnas.58.3.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scolnick E, Tompkins R, Caskey T, Nirenberg M. Release factors differing in specificity for terminator codons. Proc Natl Acad Sci U S A. 1968;61:768–774. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konecki DS, Aune KC, Tate W, Caskey CT. Characterization of reticulocyte release factor. J Biol Chem. 1977;252:4514–4520. [PubMed] [Google Scholar]
- 5.Jorgensen F, Adamski FM, Tate WP, Kurland CG. Release factor-dependent false stops are infrequent in Escherichia coli. J Mol Biol. 1993;230:41–50. doi: 10.1006/jmbi.1993.1124. [DOI] [PubMed] [Google Scholar]
- 6.Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M. The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci U S A. 2000;97:2046–2051. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vestergaard B, Van LB, Andersen GR, Nyborg J, Buckingham RH, Kjeldgaard M. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol Cell. 2001;8:1375–1382. doi: 10.1016/s1097-2765(01)00415-4. [DOI] [PubMed] [Google Scholar]
- 8.Shin DH, Brandsen J, Jancarik J, Yokota H, Kim R, Kim SH. Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome. J Mol Biol. 2004;341:227–239. doi: 10.1016/j.jmb.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 9.Ito K, Uno M, Nakamura Y. A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature. 2000;403:680–684. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- 10.Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson KS, Ito K, Noller HF, Nakamura Y. Functional sites of interaction between release factor RF1 and the ribosome. Nat Struct Biol. 2000;7:866–870. doi: 10.1038/82818. [DOI] [PubMed] [Google Scholar]
- 12.Scarlett DJ, McCaughan KK, Wilson DN, Tate WP. Mapping functionally important motifs SPF and GGQ of the decoding release factor RF2 to the Escherichia coli ribosome by hydroxyl radical footprinting. Implications for macromolecular mimicry and structural changes in RF2. J Biol Chem. 2003;278:15095–15104. doi: 10.1074/jbc.M211024200. [DOI] [PubMed] [Google Scholar]
- 13.Klaholz BP, Pape T, Zavialov AV, Myasnikov AG, Orlova EV, Vestergaard B, Ehrenberg M, van Heel M. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature. 2003;421:90–94. doi: 10.1038/nature01225. [DOI] [PubMed] [Google Scholar]
- 14.Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- 15.Rawat U, Gao H, Zavialov A, Gursky R, Ehrenberg M, Frank J. Interactions of the release factor RF1 with the ribosome as revealed by cryo-EM. J Mol Biol. 2006;357:1144–1153. doi: 10.1016/j.jmb.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 16.Petry S, Brodersen DE, Murphy FVt, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 17.Vestergaard B, Sanyal S, Roessle M, Mora L, Buckingham RH, Kastrup JS, Gajhede M, Svergun DI, Ehrenberg M. The SAXS solution structure of RF1 differs from its crystal structure and is similar to its ribosome bound cryo-EM structure. Mol Cell. 2005;20:929–938. doi: 10.1016/j.molcel.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 18.Zoldak G, Redecke L, Svergun DI, Konarev PV, Voertler CS, Dobbek H, Sedlak E, Sprinzl M. Release factors 2 from Escherichia coli and Thermus thermophilus: structural, spectroscopic and microcalorimetric studies. Nucleic Acids Res. 2007;35:1343–1353. doi: 10.1093/nar/gkl696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 20.Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci U S A. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Youngman EM, He SL, Nikstad LJ, Green R. Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol Cell. 2007;28:533–543. doi: 10.1016/j.molcel.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Bartetzko A, Nierhaus KH. Mg2+/NH4+/polyamine system for polyuridine-dependent polyphenylalanine synthesis with near in vivo characteristics. Methods Enzymol. 1988;164:650–658. doi: 10.1016/s0076-6879(88)64075-4. [DOI] [PubMed] [Google Scholar]
- 24.Powers T, Noller HF. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10:2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 26.Studer SM, Joseph S. Binding of mRNA to the bacterial translation initiation complex. Methods Enzymol. 2007;430:31–44. doi: 10.1016/S0076-6879(07)30002-5. [DOI] [PubMed] [Google Scholar]
- 27.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 28.Feinberg JS, Joseph S. A conserved base-pair between tRNA and 23 S rRNA in the peptidyl transferase center is important for peptide release. J Mol Biol. 2006;364:1010–1020. doi: 10.1016/j.jmb.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 29.Studer SM, Joseph S. Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol Cell. 2006;22:105–115. doi: 10.1016/j.molcel.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Rodnina MV, Fricke R, Wintermeyer W. Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry. 1994;33:12267–12275. doi: 10.1021/bi00206a033. [DOI] [PubMed] [Google Scholar]
- 31.Peske F, Rodnina MV, Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Youngman EM, McDonald ME, Green R. Peptide release on the ribosome: mechanism and implications for translational control. Annu Rev Microbiol. 2008;62:353–373. doi: 10.1146/annurev.micro.61.080706.093323. [DOI] [PubMed] [Google Scholar]
- 33.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 34.Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- 35.Johnson KA. Rapid kinetic analysis of mechanochemical adenosinetriphosphatases. Methods Enzymol. 1986;134:677–705. doi: 10.1016/0076-6879(86)34129-6. [DOI] [PubMed] [Google Scholar]
- 36.Rodnina MV, Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu Rev Biochem. 2001;70:415–435. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- 37.Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. Embo J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daviter T, Gromadski KB, Rodnina MV. The ribosome's response to codon-anticodon mismatches. Biochimie. 2006;88:1001–1011. doi: 10.1016/j.biochi.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 40.Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]


