Abstract
Genome-wide association studies result in inflated false positive results when unrecognized cryptic relatedness exists. A number of methods have been proposed for testing association between markers and disease with a correction for known pedigree-based relationships. However, in most case-control studies, relationships are generally unknown, yet the design is predicated on the assumption of at least ancestral relatedness among cases. Here, we focus on adjusting cryptic relatedness when the genealogy of the sample is unknown, particularly in the context of samples from isolated populations where cryptic relatedness may be problematic. We estimate cryptic relatedness using maximum-likelihood methods and use a corrected chi-square test with estimated kinship coefficients for testing in the context of unknown cryptic relatedness. Estimated kinship coefficients characterize precisely the relatedness between truly related people, but are biased for unrelated pairs. The proposed test substantially reduces spurious positive results, producing a uniform null distribution of p-values. Especially with missing pedigree information, estimated kinship coefficients can still be used to correct non-independence among individuals. The corrected test was applied to real data sets from genetic isolates and created a distribution of p-value that was close to uniform. Thus the proposed test corrects the non-uniform distribution of p-values obtained with the uncorrected test and illustrates the advantage of the approach on real data.
Keywords: Cryptic Relatedness, Kinship Coefficient, Corrected χ2 test, Type I error, Genome scan
INTRODUCTION
Genome-wide association studies have been proposed as an efficient and powerful method of uncovering genetic variants that contribute to complex disease [Hirschhorn and Daly, 2005; The Wellcome Trust Case Control Consortium, 2007]. Especially when genes have modest effects on disease risk and have common risk allele frequencies, association studies are believed to be more powerful than linkage studies, which are widely used for detecting genes of a major effect [Risch and Merikangas, 1996; Cardon and Bell, 2001; Carlson et al., 2004]. However association testing can lead to spurious positive results when unrecognized population structure exists [Hirschhorn and Daly, 2005; Cardon and Bell, 2001]. This has motivated the development and use of robust association methods to correct for effects of population structure caused by stratification, including the Transmission Disequilibrium Test (TDT) [Spielman et al., 1993; Ewans and Spielman, 1995], and generalizations implemented in the Family Based Association Test (FBAT) [Laid et al., 2000], even though such family-based designs diminish statistical efficiency. In population-based association studies, several statistical methods have also been developed to detect population stratification and to account for its effect on testing [Pritchard and Rosenberg, 1999; Devlin and Roeder, 1999; Bacanu et al., 2000; Shmulewitz et al., 2004; Price et al., 2006; Zang et al., 2007].
Another way to avoid effects of population stratification is to use population isolates. With a small number of founders and long isolation, population isolates have long been exceptional resources for genetic studies of simple genetic diseases. Researchers have also argued that population isolates provide advantages for mapping complex traits [Wright et al., 1999; Peltonen et al., 2000; Peltonen, 2000; Shifman and Darvasi, 2001; Escamilla, 2001; Venken and Del-Favero, 2007]. In addition to reducing genetic heterogeneity, such isolates have more homogeneous environmental backgrounds than are typical in larger heterogeneous populations [Peltonen, 2000; Shifman and Darvasi, 2001]. With less population stratification, Hardy-Weinberg Equilibrium (HWE) is more likely to hold in population isolates. [Peltonen et al., 2000]. These factors increase the validity of testing for differences in allele frequencies between cases and controls. However, a challenge to the use of population isolates is that classical statistical methods for ideal population data, which consist of independent individuals, may not be applicable: an important feature of population isolates is that the relatedness of two random individuals may be non-negligible [Bourgain and Genin, 2005]. Such cryptic relatedness may also be found in outbred populations as a source of population structure, and cryptic relatedness among affected individuals might be another serious source of spurious positive results [Devlin and Roeder, 1999; Bacanu et al., 2000] because cases share a genetic disorder [Bourgain and Genin, 2005; Voight and Pritchard, 2005].
There are a number of statistical methods that are designed to overcome this problem. When the entire genealogy of the sample is known, one suggested approach is to use the Armitage trend test with a correction factor that is computed conditional on the marker genotypes and pedigree structure [Slager and Schaid, 2001]. With the same idea, the standard χ2 test for comparing allele frequencies between cases and controls can be modified with a correction factor to account for relatedness among individuals (corrected χ2 test) [Bourgain et al., 2003]. This correction factor depends only on kinship and inbreeding coefficients derived directly from the pedigree information. While a modified Armitage trend test is not applicable to complicated pedigree structures, the corrected χ2 test can be used for any population structure for which kinship coefficients among pairs of individuals and inbreeding coefficients of every individual are known [Bourgain et al., 2003]. Furthermore, the χ2 test can be easily performed for multiallelic data, including the presence of rare alleles. To improve power, a quasi-likelihood score (QLS) test has also been proposed [Bourgain et al., 2003]. The QLS test is derived from the quasi-likelihood framework and also accounts for the correlations among individuals. However, the QLS test can lead to negative values for estimated allele frequencies, particularly when alleles are rare, thus making it less applicable to general situations than the corrected χ2 test. Finally, all such proposed corrections for relatedness to date assume knowledge of the relationships in the sample.
Since relationships will be unknown in most case-control studies, it is useful to consider approaches that do not depend on the existence of known relationships. Genetic relatedness among individuals can be estimated if genetic markers are available. Under the assumption that the loci are unlinked, several methods for estimating relatedness have been developed including traditional maximum-likelihood estimation [Thompson, 1975] and other estimators based on method-of moments approaches [Ritland, 1996; Lynch and Ritland, 1999; Wang, 2002]. Compared to other estimators, maximum-likelihood estimators of relatedness exhibit the desirable features of having consistently lower standard errors and being adaptable to many sampling situations [Milligan, 2003; Thompson, 1975].
In this paper, our goals are (1) to characterize the effect of cryptic relatedness on testing in case-control samples, particularly from an isolated population, (2) to estimate cryptic relatedness with such samples, and (3) to develop an approach for testing for association with a correction for cryptic relatedness. To test for association between markers and disease and to simultaneously account for unknown relatedness among individuals, we propose to modify the corrected χ2 test by using estimated, rather than assigned, coefficients of relatedness. We focus on the corrected χ2 test because of its desirable statistical behavior, ease of computation, and applicability to a variety of marker and data types. We perform simulations to evaluate the statistical properties of the corrected χ2 test for use in the context of case-control data, and we illustrate the advantage of the proposed method by application to two samples from genetic isolates.
METHODS
RELATIONHIP ESTIMATION
The simplest summary of the degree of a pairwise relationship is the kinship coefficient, ϕ, which is the probability that a pair of homologous alleles chosen from two individuals are identical by descent (IBD). In this section, we briefly describe how to estimate ϕ by estimating the set of three k-coefficients in a non-inbred population [Crow and Kimura, 1970].
The three k-coefficients, k0, k1 and k2, are defined as the probabilities that a pair of individuals share neither, one or both, respectively, of their two alleles at a locus IBD.
| (1) |
where ki ≥ 0, k0 + k1 + k2 = 1 and X is the number of alleles shared IBD. The kinship coefficient can be computed as ϕ = 0.5k2 + 0.25k1.
Assume that individuals are from a non-inbred population in Hardy-Weinberg Equilibrium (HWE) and that each marker locus is segregating independently. For given IBD modes, the conditional probabilities of the seven possible identity by state (IBS) modes Si are shown in Table I. For a single locus of observed IBS Si, the likelihood of the k-coefficients is [Thompson, 1975] :
Table I.
Probability for seven identity-by-state modes
| Si | Genotypes | Pr(Si | k) | |
|---|---|---|---|
| S 1 | AiAi, AiAi |
|
|
| S 2 | AiAi, AjAj |
|
|
| S 3 | AiAi, AiAj |
|
|
| S 4 | AiAi, AjAm |
|
|
| S 5 | AiAj, AiAj |
|
|
| S 6 | AiAj, AiAm |
|
|
| S 7 | AiAj, AmAl | 4k0pipjpmpl |
| (2) |
Assuming independent unlinked loci, the likelihood for the overall genome is then simply the product of the single locus likelihoods. We used the EM algorithm [Dempster et al., 1977] to find maximum-likelihood estimators for the k-coefficients [McPeek and Sun, 2000] and then obtained an estimate of the kinship coefficient ϕ̂ = 0.5k̂2 + 0.25k̂1. The EM algorithm provides more efficient computation than the simplex method, a hill-climbing optimization technique, which has been used previously [Milligan, 2003].
Relationship estimation can be extended to inbred populations by estimating probabilities of Jacquard’s nine IBD modes instead of three k-coefficients [Jacquard, 1972; Anderson and Weir, 2007; Weir et al., 2006]. The inbreeding coefficient, f, which is the probability that a person carries two alleles IBD at a locus, can also be estimated by maximizing the likelihood using the EM algorithm [Dempster et al., 1977; Thompson, 2000]. In human genetic isolated populations, it is rare to have inbreeding within an individual even though there are higher kinships among individuals [Agarwala et al., 2001].
THE CORRECTION OF TYPE 1 ERROR FOR ALLELIC ASSOCIATION TESTING
A corrected χ2 test has previously been proposed to correct for inflated false positive rates for association tests in the context of known relationships [Bourgain et al., 2003]. We begin by outlining this test for the diallelic case. Suppose we have Nc sampled individuals in a case group and Nt sampled individuals in a control group. Let Y = (Y1, …Yi, …YN)T where Yi = 0.5 × (the number of alleles of type 1 in individual i) and N = Nc + Nt. Let p be the frequency of allele 1, 0 < p < 1. Under the null hypothesis of no association between a given marker and disease and HWE for the given marker, E0(Y) = p1N and , where
| (3) |
where fi is the inbreeding coefficient of individual i, and ϕij is the kinship coefficient between two individuals i and j, 1 ≤ i, j ≤ N. Here fi = 0 if we assume a non-inbred population.
To test for an association between the marker alleles and the disease, we consider the model: E(Y) = μ = (μ1, ⋯, μN)T with μi = p + r if i is a case and μi = p if i is a control. The null hypothesis is H0 : r = 0. The proposed corrected χ2 test is one that extends the χ2 test by taking into account the correlation structure among individuals. The test statistic of the corrected χ2 test is [Bourgain et al., 2003] :
| (4) |
where , and 1c is a vector of length N with ith entry 1 if individual i is a case and 0 if i is a control. The statistic follows a χ2 distribution with 1 degree of freedom asymptotically under the null hypothesis [Thornton and McPeek, 2007]. When all kinship and inbreeding coefficients are zero, the test statistic in (5) is the χ2 test statistic,
| (5) |
The corrected χ2 test can be easily extended to the multiallelic case and under the null distribution, the test statistic asymptotically follows a χ2 distribution with (a − 1) degrees of freedom, where a is the number of alleles, as described previously [Bourgain et al., 2003].
The corrected χ2 test requires known kinship coefficients and inbreeding coefficients. We propose replacing these coefficients with their MLEs. We call this statistic a corrected χ2 test with estimated coefficients, and this is same as except that we use Φ̂ rather than the assigned Φ from a known pedigree structure. With estimates of kinship and inbreeding coefficients, unknown background relatedness or cryptic relatedness caused by common population history can be accommodated. An estimate of kinship would also be useful in the case that a sample includes a mixture of known and unknown relationships or any missing/misspecified pedigree relationships.
EVALUATION OF THE PROPOSED METHOD
To investigate the performance of kinship estimation, we estimated kinship coefficients based on both simulated data and CEPH family data. The simulated data was used to evaluate performance for use with multiallelic marker and with varying available relationship information, and the CEPH data was used to evaluate performance for use with denser SNP markers. We also performed a simulation study to explore the null distribution of p-values and to compare the power of the corrected χ2 tests using different types of kinship coefficients with the classical χ2 test. Four kinship coefficients were considered: actual (ϕact), pedigree-based (ϕped), estimated (ϕest), and posterior (ϕpost) kinship coefficients. The actual kinship coefficient, ϕact, is computed based on the underlying truth in the simulation and the pedigree-based kinship coefficient, ϕped, is assigned from a known pedigree structure. The estimated kinship coefficient, ϕest, is the maximum-likelihood estimate computed based on genome data only, without pedigree information. The posterior kinship coefficient, ϕpost, is also an estimated coefficient, but based on both genome data and pedigree information. In all cases, we used the average of posterior kinship coefficients over the whole genome panel of markers. Merlin 1.1.2 was used to compute the posterior IBD and kinship coefficients [Abecasis et al., 2002].
We focused only on individuals and families from non-inbred populations, which means that inbreeding coefficients were assumed zero. The program genedrop in Morgan 2.8.2 [Thompson, 1995; Thompson, 2000b; Thompson, 2005] was used for generating marker genotypes for the simulated data sets. For each individual, we simulated 400 microsatellite markers based on Rutgers map positions and allele frequencies from the version 10 CEPH data at ftp://ftp.cephb.fr/ceph_genotype_db/. These markers were chosen randomly, subject to the condition that they were separated by an average of 10cM. Except where otherwise noted, we used all 400 markers for further analyses.
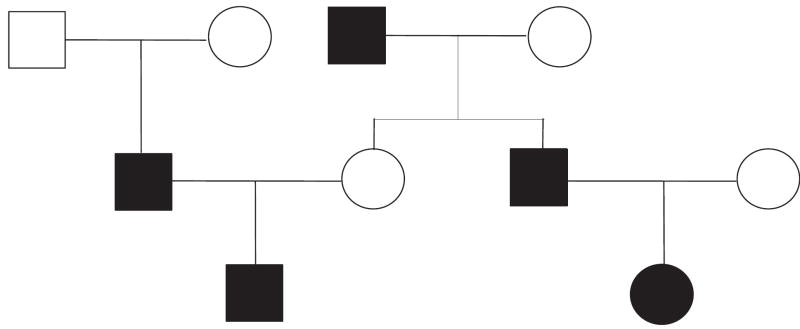
The simulation studies were performed based on three relationship scenarios. (1) In Scenario I, the case group consisted of 250 individuals from 50 pedigrees as seen in Figure 1 with each pedigree having five cases. The entire pedigree information was assumed known for the case group. The control group simply included 250 independent and unrelated individuals. (2) Scenario II was the same as Scenario I except the cases were chosen from a large pedigree of 13 generations and 4070 individuals to simulate the background correlation among individuals. Each set of parents in the 13 generations had one or two offspring, and 1124 individuals were unrelated founders. We assumed that the genealogy of only the last two generations was available. (3) In Scenario III, samples were simulated for 500 related and 500 independent unrelated people. The related individuals were in the final generations of a large pedigree of 13 generations as in Scenario II, but the pedigree information was assumed known for the last three generations. Controls in each of these three scenarios were unrelated, which is the most ideal situation. Cases of Scenario I were from an entirely known pedigree without any background correlation. Scenario II and III simulated background correlation among cases with limited pedigree information, which commonly occurs in genetic studies. Scenario II had less pedigree information than Scenario III, so we could explore more efficiently the effect of lack of pedigree information on ϕped, ϕpost and ϕest.
Figure 1.
Pedigree for simulation study: black is affected and white is unaffected.
PERFORMANCE OF KINSHIP ESTIMATION
To evaluate the performance of kinship estimation, we carried out a simulation study based on Scenario I. Multiallelic marker simulation was done without reference to the fixed disease status, thus representing genotypes generated under H0. The relationships in this sample are 150 parent-offspring (PO), 50 avuncular (AV), 100 grand parent-grand child (GG) and 50 first cousin (CO) pairs. Also, 124,400 unrelated pairs (UN) were included in the sample. For each individual, we simulated 400 microsatellite markers as described above and among these simulated markers, subsets of 50, 100, 200 and 400 markers were used to calculate MLEs of kinship coefficients using the EM algorithm. Allele frequencies were assumed unknown and sample allele frequencies were used for such estimation.
We also investigated the performance of kinship estimation based on diallelic markers by estimating kinship coefficients of CEPH families based on the real data, single-nucleotide polymorphism (SNP) markers from the version 10 CEPH data at ftp://ftp.cephb.fr/ceph_genotype_db/. We selected 91 individuals of 12 pedigrees with more than 80% genotyped markers. To investigate the effect of use of different numbers of markers, three marker panels were used: 500 SNP markers, 5,000 SNP markers, and all of the 16,977 SNP markers of the whole genome. In the first two panels, the markers were randomly chosen from the 16,977 SNP markers. Kinship coefficients were estimated for all possible pairs of 91 individuals based on these two marker panels. The entire pedigree information was known and estimated kinship coefficients were compared with pedigree-based kinship coefficients.
NULL DISTRIBUTION OF THE P-VALUES OF THE CORRECTED χ2 TEST
In Scenario I, we evaluated whether resulting p-values were distributed uniformly as would be expected under the null distribution. The χ2 test and the corrected χ2 test with ϕact, ϕped, ϕest, and ϕpost were performed for each multiallelic marker and Q – Q plots were used to examine the uniformity of the p-value distributions. Note that the same markers are used for both the estimation of kinship coefficients in the previous section and for the test of association.
ASSESSMENT OF THE EFFECT OF MISSING PEDIGREE INFORMATION
We further investigated the null distribution of p-values for Scenario II where only part of the genealogy was known. We assumed only part of the genealogy known, and ϕped and ϕpost were computed based on the pedigree information and the pedigree plus multiallelic marker information of the last two generations. Under the null distribution, we performed the classical χ2 test and the corrected χ2 test with ϕact, ϕped, ϕest, and ϕpost for each simulated marker. Q – Q plots were used to present the null distribution of p-values.
To assess the performance of various kinship coefficients based on limited pedigree information, ϕped, and ϕest were compared with ϕact in Scenario III. The genealogy of only the last three generations was used for the computation of ϕped and ϕpost to mimic a typical situation in which more distant relationships are not known.
POWER OF THE χ2 AND CORRECTED χ2 TESTS
For comparing the power of the classical χ2 test and the corrected χ2 test with various kinship coefficients, we performed simulations based on Scenario III. The genotype of the trait locus was simulated under a disease allele frequency of 0.5, with affected status assigned based on the simulated genotypes at the trait locus and the penetrance probabilities shown in Table III. This yielded a variable number of cases and controls over replicates. The mean sizes of the case and control samples were reported with their standard deviations in Table III. The power of the χ2 test and the corrected χ2 test were estimated from 10,000 replicates.
Table III.
Penetrance probabilities and mean (sd) case-control sample sizes for power analysis
| Model | Pr(aff | aa) | Pr(aff | Aa) | Pr(aff | AA) | Control | Case |
|---|---|---|---|---|---|
| Additive | 0.25 | 0.4 | 0.55 | 299.97 (11.60) | 200.93 (11.63) |
| Dominant | 0.25 | 0.55 | 0.55 | 263.34 (11.81) | 236.69 (11.84) |
| Recessive | 0.25 | 0.25 | 0.55 | 336.77 (11.12) | 163.23 (11.21) |
APPLICATION TO REAL DATA
The method was applied to two real data sets, one from Guam and the other from Kosrae. Both studies were approved by University of Washington Institutional Review board (IRB). The data set from Guam consisted of a genome scan for association in a case-control sample collected to complement a family-based linkage study. The disorder of cases has characteristics of both amyotrophic lateral sclerosis and parkinsonism combined with dementia and is prevalent in the Chamorros, the indigenous people of Guam. A sample of 140 cases and 88 age-matched Chamorro controls were available with no known relationships among these subjects at the time they were sampled. The markers consisted of a standard genome-scan panel of 402 multiallelic markers. For each pair of individuals, the kinship and inbreeding coefficients were estimated using the methods described above. For each marker, allele frequencies of cases and controls were compared using the standard and the corrected χ2 tests.
The data set from Kosrae consisted of a genome scan for schizophrenia [Wijsman et al., 2003]. A sample of 36 cases and 76 unrelated controls from the island was available with the genealogy known for the case group. Even though the pedigree information of the cases was available, we suspected there might be additional relatedness among cases and cryptic relatedness among controls since the data were collected from a relatively small population. The genome scan consisted of a standard panel of 379 microsatellite markers. As in the analysis of the Guam data, estimation of relatedness and association tests were performed. Also, using the known genealogy, the corrected χ2 test with pedigree-based assigned kinship coefficients was performed.
RESULTS
EVALUATION OF THE PROPOSED METHOD
PERFORMANCE OF KINSHIP ESTIMATION
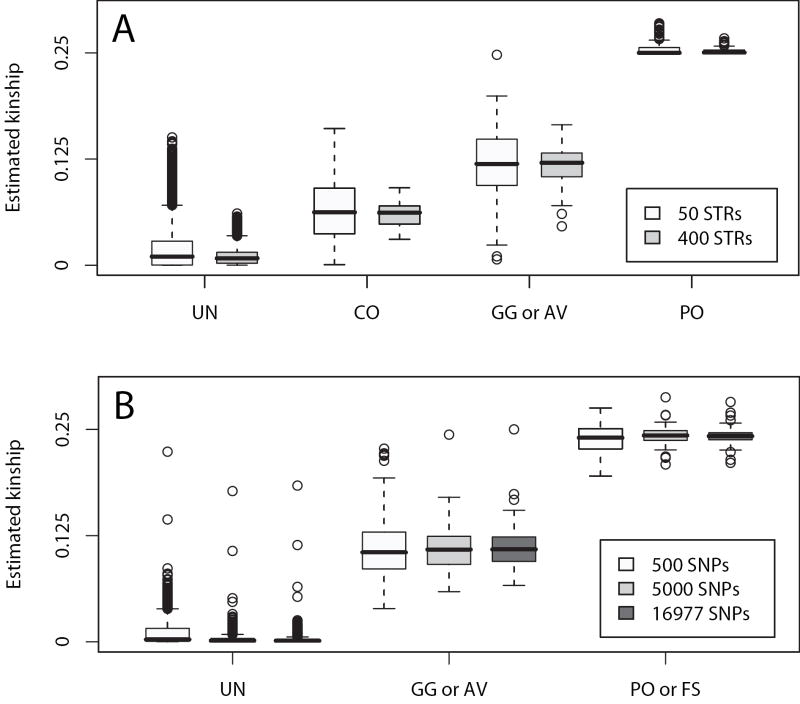
With increasing number of markers used for estimation, ϕest was closer to ϕped, with also less variation in the estimates from multiallelic markers. The values of the pedigree-based k-coefficients and kinship coefficients for non-inbred individuals are shown in Table II. Figure 2 shows the relation between ϕest and ϕped using 50 and 400 multiallelic markers (Panel A). For most relationships, ϕest was close to the pedigree-based expectation, ϕped, with the accuracy of the estimate increasing with the number of markers. For example, in the case of PO pairs, the mean of ϕest was 0.255 (s.d. 0.009), 0.254 (0.006), 0.253 (0.005) and 0.252 (0.003) when 50, 100, 200 and 400 multiallelic markers were used, respectively. Estimation of other relationships showed a similar trend (not shown).
Table II.
Pedigree-based assigned kinship coefficients for non-inbred relatives
| Relationship | k 0 | k 1 | k2 | ϕ |
|---|---|---|---|---|
| Unrelated | 1.00 | 0 | 0 | 0 |
| Parent-offspring | 0 | 1.00 | 0 | 0.25 |
| Grandparent-grandchild | 0.50 | 0.50 | 0 | 0.125 |
| Avuncular | 0.50 | 0.50 | 0 | 0.125 |
| First cousin | 0.75 | 0.25 | 0 | 0.0625 |
Figure 2.
Box plots of estimated kinship coefficients. A: the simulated sample of Scenario I using 50 and 400 microsatellite markers. B: CEPH families using 500, 5,000 and 16,977 SNP markers.
Similarly, in the case of diallelic markers, ϕest was closer to ϕped when more markers were used. Figure 2 shows the the relation between ϕest and ϕped in the diallelic case of CEPH families with 500, 5,000 and 16,977 SNP markers (Panel B). Panel B also shows the similar results of kinship estimation from 5,000 and 16,977 markers, which indicates that the number of SNPs does not need to be too large. The estimate of the kinship coefficient for parent-offspring was more accurate than that for full-siblings in the CEPH data. In the case of the 5,000 diallelic marker panel, the means of the estimated kinship coefficients were 0.243 (0.026, N=8) for full-sibling and 0.243 (0.008, N=87) for parent-offspring. The means of the parent-offspring and full-sibling relationships were similar, but that for the parent-offspring relationship had a smaller standard deviation even with the larger sample size. However, kinship coefficients were overestimated for unrelated pairs for both the simulated and CEPH data.
The EM algorithm was used for estimating relatedness with computation time proportioned to MN2, where M is the number of markers and N is the number of individuals. We implemented the method by using R-2.5.0. In the case of the CEPH data (91 individuals), computation times on a linux AMD Opteron 1.8GHz computer were 804.41 sec for 500 markers, 1617.68 sec for 1,000 markers and 7712.80 sec (2.14 hrs) for 5,000 markers.
NULL DISTRIBUTION OF THE P-VALUES OF THE CORRECTED χ2 TEST
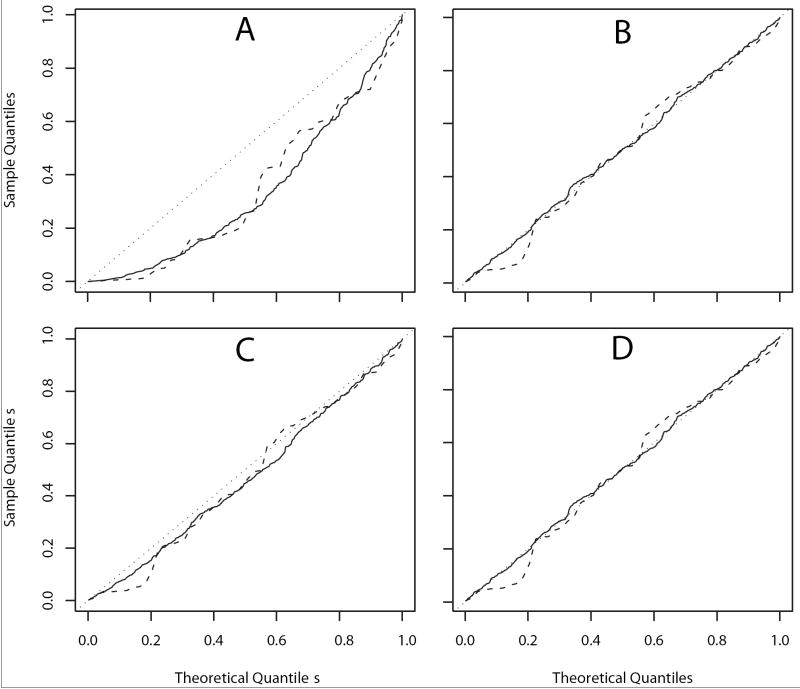
The corrected χ2 test using kinship coefficients reduced the false positive rate substantially. The Q – Q plots of p-values of the four association tests for 50 and 400 markers are shown in Figure 3. Results for 100 and 200 multiallelic markers were consistent with these (not shown). Since genotypes of all markers were simulated under the null hypothesis of no association, p-values are expected to be uniformly distributed. Figure 3A suggests a high false positive rate when the classical χ2 test was used. Panels B, C and D of Figure 3 demonstrate that the corrected χ2 test reduces the false positive rate, producing a distribution of p-values that is close to uniform. Since the entire genealogy was known, ϕped was close to ϕact and the p-values were uniformly distributed (Figure 3D). The ϕpost had the same results as the pedigree-based results (not shown). When ϕest was used, the Q – Q plots of p-values were slightly different from the uniform distribution (Figure 3C). Overestimated kinship coefficients for UN pairs, especially when one individual is from the case group and the other is from the control group, may lead to slightly inflated false positive rates.
Figure 3.
Quantile-quantile plots of χ2 test and corrected χ2 tests for 50 markers (dashed line) and 400 markers (solid line) in the simulated sample of Scenario I. A: Classical χ2 test. B: Corrected χ2 test with actual kinships. C: Corrected χ2 test using estimated kinships. D: Corrected χ2 test using pedigree-based kinships.
ASSESSMENT OF THE EFFECT OF MISSING PEDIGREE INFORMATION
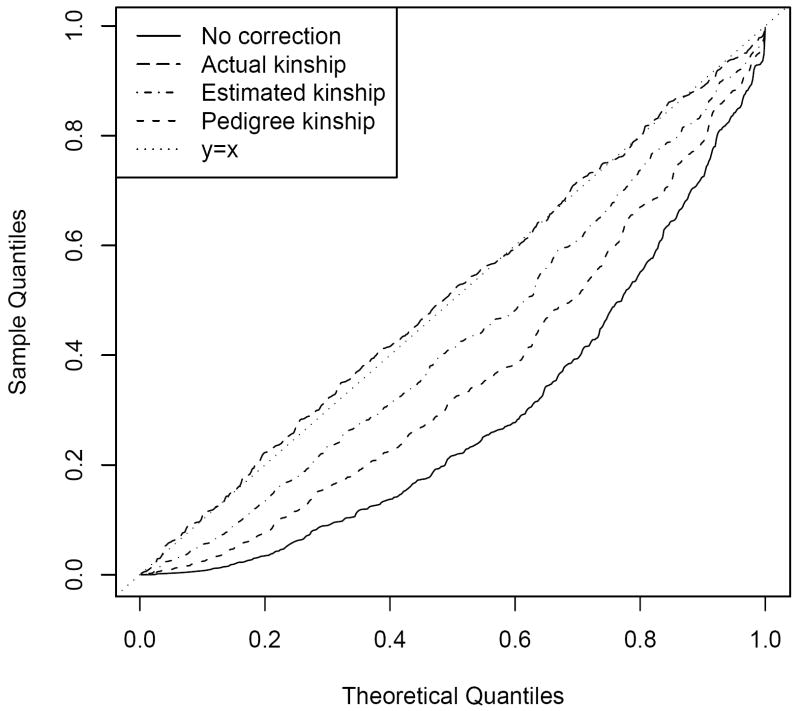
With missing pedigree information, the corrected χ2 test using ϕest reduces the rate of spurious positive results more than when using ϕped or ϕpost. Figure 4 presents the Q – Q plots of p-values of the χ2 and the corrected χ2 tests when only part of the genealogy is known. As with the previous simulation, the χ2 test had a high false positive rate, and the p-values of the corrected χ2 test with ϕact were uniformly distributed (Figure 4). However, missing pedigree information resulted in undercorrection of false positives when ϕped was used. In the case of ϕest, the Q – Q plot was consistent with the previous simulation (with the entire genealogy) since the performance of kinship estimation was not affected by missing pedigree information.
Figure 4.
Quantile-quantile plots of χ2 test and corrected χ2 tests in the simulated sample of Scenario II.
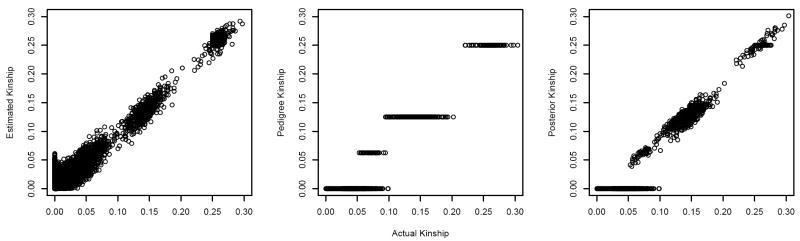
With the incomplete genealogy, ϕest gave more precise estimates for related pairs, but ϕped and ϕpost characterized unrelated pairs better because of the intrinsic overestimation of kinship coefficients in this case (Figure 5). Because MLEs of the kinship coefficient of UN pairs were overestimated, this approach may not distinguish unrelated people from people sharing a common genetic background. On the other hand, based on only the part of the pedigree information used in the analysis, ϕped and ϕpost were 0 for some of the pairs that shared their unknown genetic background. This leads to an under-correction of relatedness and an inflated false positive rate. For related pairs, the mean squared error (MSE) of ϕest, ϕped and ϕpost was 1.1 ×10−4, 3.3 × 10−4 and 3.2 × 10−4 respectively. For unrelated pairs, the MSE of ϕest was 1.36 × 10−4 and the MSEs of ϕped and ϕpost were 0.
Figure 5.
Comparison of kinship coefficients: actual kinships vs. estimated, pedigree-based and posterior kinships in the simulated sample of Scenario III.
POWER OF THE CORRECTED χ2 TEST USING ESTIMATED AND ACTUAL KINSHIPS
Overall, the χ2 test is most powerful at the nominal significance level, as expected, given the inflated false positive rate. Table IV shows the estimated power for the χ2 test and the corrected χ2 tests with ϕact, ϕped, ϕest and ϕpost. At the nominal significance level, the corrected χ2 test has lower power than the χ2 test, and in particular, the corrected χ2 test with ϕact has the least power. The corrected χ2 test with ϕest is more powerful than the corrected χ2 test with ϕped and ϕpost. This result shows that the correction for kinship may cause apparent loss of power given a nominal significance level. However the corrected tests give the proper null distribution of p-values as we showed previously. As a result, the test has the correct type I error in contrast to the χ2 test, which is too liberal. Because power in our simulation was estimated under the nominal significance level, but the tests have different type I errors, it is not meaningful to compare actual power in this situation.
Table IV.
Estimated Power (sd) of the classical χ2 test and the corrected χ2 test with actual and several estimated kinship coefficients with significance level 0.05
| Additive | Dominant | Recessive | |
|---|---|---|---|
| No correction | .788 (0.004) | .699 (0.005) | .865 (0.003) |
| Φact | .657 (0.005) | .532 (0.005) | .779 (0.004) |
| Φest | .719 (0.004) | .607 (0.005) | .819 (0.004) |
| Φped | .701 (0.005) | .586 (0.005) | .808 (0.004) |
| Φpost | .701 (0.005) | .586 (0.005) | .809 (0.004) |
APPLICATION TO REAL DATA
ESTIMATION OF CRYPTIC RELATIONSHIPS
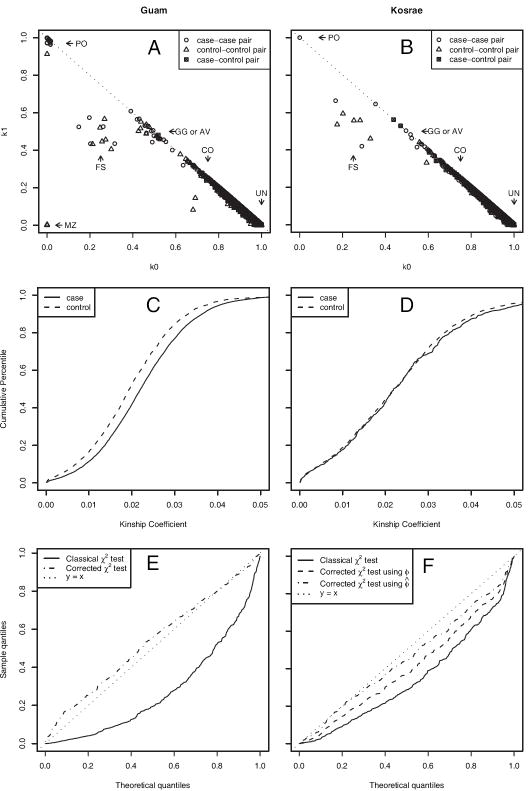
Estimated kinship coefficients suggested that there were unknown relationships in the Guam and Kosrae data. Even though some pedigree information in the Kosrae data was known, more relationships were found by estimating kinship coefficients. Panel A and B of Figure 6 presents the estimated k-coefficients and Panel C and D of Figure 6 shows the cumulative distribution of the estimated kinship coefficients in the two samples. Estimation of k-coefficients allows us to quantify not only well-defined and possibly verifiable relatives such as parent-offspring, full sibling or cousin pairs, but also continuous degrees of relatedness. In the Guam data, the means and standard deviations of the estimated kinship coefficients are 0.023 (0.016) for the case group and 0.021 (0.020) for the control group with the case group having slightly higher average estimated kinship coefficients. This is consistent with the expectation that affected individuals may be more closely related because they share an ancestral mutation leading to the genetic disorder [Voight and Pritchard, 2005]. In the Kosrae data, the mean estimated kinship coefficients in the case group is 0.026 (0.025), which is also slightly higher than 0.024 (0.017) in the control group.
Figure 6.
Estimated k-coefficients of pairs in (A) Guam and (B) Kosrae samples. Cumulative distribution of estimated kinship coefficients of cases and controls in the (C) Guam and (D) Kosrae data sets. Quantile-quantile plot of p-values of χ2 test and corrected χ2 test in (E) Guam and (F) Kosrae samples. In the Kosrae study, ϕ is the pedigree-based kinship coefficient and ϕ̂ is the estimated kinship coefficient.
TESTING ASSOCIATIONS
The corrected χ2 test based on ϕest successfully reduced spurious associations between markers and disease. Q – Q plots of the p-values are shown in Panel E and F of Figure 6. In the Guam data analysis, p-values of the uncorrected χ2 tests are not uniformly distributed, but p-values of the corrected χ2 tests based on ϕest are close to uniformly distributed. These results show that the false positive rate was reduced by correcting for relatedness. If the distribution of the p-values were exactly uniform, the expected 1% and 5% quantiles would be 0.01 and 0.05, but the 1% and 5% quantiles of the χ2 test are 0.0007 and 0.007, which are considerably smaller than the nominal quantiles. For the corrected χ2 test, the 1% and 5 % quantiles are less extreme at 0.02 and 0.098, and are actually conservative. When the uncorrected χ2 test was used, 95 markers were significantly associated with the disease at significance level 0.05, but only 10 markers remained at this significance level with the corrected χ2 test.
Similar to the Guam data analysis, a Q – Q plot of p-values in the Kosrae data analysis reveals that the χ2 test results in a high false positive rate, but these false associations diminished in two corrected χ2 tests (Figure 6F). Since, in the Kosrae study, the pedigree information of the case group was available, the corrected χ2 tests were performed with ϕped. However, since the known genealogy does not include complete information of relatedness among individuals, the corrected χ2 tests work better with ϕest than with ϕped. The 1% and 5% quantiles of the χ2 test are 0.0014 and 0.0095, and the 1% and 5 % quantiles of the corrected χ2 test with ϕest are 0.0095 and 0.036, much closer to their nominal levels. We had 50 significant markers at significance level 0.05 with the χ2 test, but 22 of these markers were not significant after correcting relatedness by using ϕest.
After correction of relatedness among individuals, the p-values increased, which implies less significant associations between disease and markers. The ten markers with the smallest p-values are reported in Table V for the Guam study and Table VI for the Kosrae study. For the Guam study, the correction for cryptic relatedness gave a p-value that was 1-3 orders of magnitude higher than that from the uncorrected test, while for the Kosrae study, the p-values for all these extreme markers increased by one order of magnitude.
Table V.
Guam Data results
| Chr | Marker | Position | χ2 test | Corrected χ2 test with ϕ̂ |
|---|---|---|---|---|
| 4 | D4S403 | 27.250 | 2.30 × 10−5 | 2.80 × 10−3 |
| 6 | D6S434 | 109.241 | 7.33 × 10−5 | 1.30 × 10−2 |
| 14 | D14S258 | 65.035 | 2.52 × 10−4 | 1.40 × 10−2 |
| 4 | D4S1572 | 112.691 | 7.46 × 10−4 | 2.32 × 10−2 |
| 21 | D21S263 | 32.143 | 2.20 × 10−4 | 2.58 × 10−2 |
| 21 | D21S266 | 54.988 | 9.24 × 10−4 | 3.79 × 10−2 |
| 3 | D3S1304 | 20.465 | 1.40 × 10−3 | 3.84 × 10−2 |
| 4 | D4S1575 | 136.798 | 5.81 × 10−3 | 3.85 × 10−2 |
| 2 | D2S142 | 168.287 | 2.87 × 10−3 | 4.45 × 10−2 |
| 5 | D5S428 | 102.903 | 1.79 × 10−3 | 4.46 × 10−2 |
Table VI.
Kosrae Data results
| Chr | Marker | Position | χ2 test | Corrected χ2 test with ϕ̂ |
|---|---|---|---|---|
| 2 | D2S305 | 40.761 | 4.73 × 10−5 | 6.12 × 10−4 |
| 15 | D15S127 | 96.023 | 3.66 × 10−5 | 9.83 × 10−4 |
| 4 | D4S2935 | 13.531 | 2.56 × 10−4 | 1.81 × 10−3 |
| 7 | D7S550 | 184.839 | 1.30 × 10−3 | 8.47 × 10−3 |
| 3 | D3S1267 | 134.568 | 1.42 × 10−3 | 9.80 × 10−3 |
| 1 | D1S252 | 152.755 | 1.85 × 10−3 | 1.11 × 10−2 |
| 5 | D5S408 | 209.550 | 2.45 × 10−3 | 1.16 × 10−2 |
| 6 | D6S441 | 163.648 | 1.92 × 10−3 | 1.40 × 10−2 |
| 10 | D10S1693 | 140.353 | 4.76 × 10−3 | 1.93 × 10−2 |
| 16 | D16S3136 | 63.737 | 5.74 × 10−3 | 2.04 × 10−2 |
DISCUSSION
We have presented here an evaluation of the usefulness of estimated kinship coefficients to account for cryptic relatedness in a population-based association study. Such cryptic relatedness may occur even when there is no population structure and when pedigree information is available. For example, the pedigrees may not fully describe the actual relatedness among individuals, such as when individuals share a long common genetic background history, or in other situations such as the presence of non-paternity or unreported adoption. We showed that the use of whole-genome scan markers to estimate relationships and to determine an appropriate variance correction reduces the false positive rate that otherwise results from use of the data without such a correction.
While part of our investigation involved simulation, simulation cannot evaluate every possible situation. In particular, in isolated populations, there may be variable levels of inbreeding, common genetic background from the shared complex and deep genealogies, or differences between social kinship and biological kinship. Such isolated populations motivated us to explore Scenario II and III which simulated the relatively simple situation of the availability of only partial information about relatedness among samples. Real situations probably involve much deeper genealogies, but even these simple situations demonstrated the impact of the missing relationship information.
To properly reduce spurious positive evidence of association, it is important to get accurate estimates of relationships. Our results indicate that estimated kinship coefficients can be more accurate than are pedigree-based or posterior kinship coefficients. The reasons for this improved accuracy are that pedigree-based kinship coefficients allow no variability within a class of relative pair, and posterior kinship coefficients do not allow for sources of relationship other than the pedigree. The one exception to this is the case of unrelated pairs, for which MLEs can be biased because only non-negative values are allowed for estimated kinship coefficients, thereby resulting in overcorrection of the test statistic. An ad hoc method might be considered to avoid overcorrecting for such related pairs, such as setting kinship coefficients to 0 for pairs with estimated kinship coefficients smaller than a specific threshold. This might improve the over correction of inflated false positive rates. Another concern in estimating relatedness is potential violation of the HWE assumption which might affect the resulting MLEs if there is Hardy-Weinberg Disequilibrium (HWD) in the sample. We have not formally evaluated the effect of HWD on the resulting estimates. Since substantial HWD is rare in unstructured populations and inbreeding (leading to HWD) levels are small even in genetic isolates that have been evaluated [Agarwala et al., 2001], it is likely that the impact of HWD is minimal.
We estimated relationships based on diallelic markers with comparable accuracy to those based on multiallelic markers. As we showed, the 5,000 and 16,977 marker panels had similar variances for kinship estimates, indicating that all markers in the dense SNP panel are not necessarily needed. The subset of markers that were spaced suffciently far apart provides enough information for estimating kinship coefficients and removes the need to account for linkage-disequilibrium (LD) among SNP dense markers. We can therefore consider SNP markers as the simplest case of multiallelic markers and the analysis performed on multiallelic markers in this study can be applicable and valid to SNP markers as well. Computation time for SNPs was also not exorbitant, compared to smaller number of multiallelic markers, even for our R code used to carry out these analyses. Computation time could be improved by carrying out some computations with C or C++.
In the proposed method, the idea of correcting the variance using genomic data is similar to the idea behind Genomic Control (GC) [Devlin and Roeder, 1999]. Both approaches use a constant correction for tests based on all genome markers. However, GC corrects the test statistics after inflated test statistics are observed, while the proposed method uses the genome data to adjust the variance so that the test statistic is not inflated in the first place. This also explains why the proposed test is straightforward for more complicated markers such as multiallelic markers, while GC needs separate correction factors for each type of marker, defined by the number of alleles. GC suffers when used for multiallelic markers because a separate correction for each type of marker is needed, and because a large number of markers of each type of marker are required to achieve an adequate correction [Marchini et al., 2004]. In general, multiallelic markers or haplotypes based on multiple SNPs can be considerably more informative than diallelic markers for association testing [Ott and Rabinowitz, 1997; Chapman and Wijsman, 1998]. For use with multiallelic markers or haplotype data, the easy extension and computation of the corrected χ2 test to the multiallelic situation is an advantage over GC.
A previous study recommended use of pedigree-based (prior) kinship coefficients because of the theoretical justification for the test statistic [Thornton and McPeek, 2007]. While we have not studied the asymptotic properties for the corrected χ2 test based on estimated kinships, as shown in the simulation study, p-values are uniformly distributed under the null hypothesis, so that the corrected χ2 test based on estimated kinships is useful in practice and is also applicable to testing associations. We believe the corrected χ2 test with estimated kinships would be more appropriate for reducing the false positive rate if there is background correlation among individuals beyond what is reflected in the known pedigree structures. Especially among cases, individuals are more likely to share their ancestral background and to be genetically similar to one another than are controls [Voight and Pritchard, 2005]. Estimated kinship coefficients would also be more appropriate for adjusting an unbalance of relatedness between the case and control group. We believe that the proposed approach will provide a practical tool for association studies to reduce false positive results caused by the cryptic relatedness beyond a known or unknown genealogy.
Acknowledgments
This study was supported by National Institutes of Health grants GM075091, AG11762, AG05136, and AG14382.
APPENDIX
EM ALGORITHM FOR MLE OF K-COEFFICIENTS
Start with arbitrary initial values of . In the E-step, for given and the observed IBS Si,m of genotypes at marker m, find the probability of IBD state Xm = j, j = 0, 1, 2.
| (1) |
The probability of genotypes of IBS Si,m for given Xm is shown in Table I.
In the M-step, for given Pr(Xm = j|Si,m), m = 1…M, update k0, k1 and k2, which maximize the complete likelihood,
| (2) |
The updated estimate of k-coefficients is . Then, repeat the E-step and M-step until k̂0, k̂1 and k̂2 converge.
WEB RESOURCES
The R and Perl code for the proposed method will be available at http://faculty.washington.edu/wijsman/software.shtml
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Agarwala R, Schaffer AA, Tomlin JF. Towards a complete north American Anabaptist genealogy II: analysis of inbreeding. Hum Biol. 2001;73(4):533–545. doi: 10.1353/hub.2001.0045. [DOI] [PubMed] [Google Scholar]
- Anderson AD, Weir BS. A maximum-likelihood method for the estimation of pairwise relatedness in structured populations. Genetics. 2007;176:421–440. doi: 10.1534/genetics.106.063149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacanu SA, Devlin B, Roeder K. The power of genomic control. Am J Hum Genet. 2000;66:1933–1944. doi: 10.1086/302929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgain C, Genin E. Complex trait mapping in isolated populations: Are specific statistical methods required? Eur J Hum Genet. 2005;13:698–706. doi: 10.1038/sj.ejhg.5201400. [DOI] [PubMed] [Google Scholar]
- Bourgain C, Hoffjan S, Nicolae R, Newman D, Steiner L, Walker K, Reynoldes R, Ober C, McPeek MS. Novel case-control test in a founder population identifies P-Selection as an atopy-susceptibility locus. Am J Hum Genet. 2003;72:612–626. doi: 10.1086/378208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91–99. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Kruglyak L, Nickerson DA. Mapping complex disease loci in whole-genome association studies. Nature. 2004;429:446–452. doi: 10.1038/nature02623. [DOI] [PubMed] [Google Scholar]
- Chapman NH, Wijsman EM. Genome screens using linkage disequilibrium tests: optimal marker characteristics and feasibility. Am J Hum Genet. 1998;63:1872–1885. doi: 10.1086/302139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF, Kimura M. An introduction to population genetics theory. Harper & Row; New York, Evanston, and London: 1970. [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Stat Soc B. 1977;39:1–38. [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Escamilla MA. Population isolates: their special value for locating genes for bipolar disorder. Bipolar Disord. 2001;3:299–317. doi: 10.1034/j.1399-5618.2001.30605.x. [DOI] [PubMed] [Google Scholar]
- Ewens WJ, Spielman RS. The transmission/disequilibrium test: history, subdivision and admixture. Am J Hum Genet. 1995;57:455–464. [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Jacquard A. Genetic information given by a relative. Biometrics. 1972;28:1101–1114. [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family based tests of association. Genet Epidemiol. 2000;19(Suppl 1):S36–42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Lynch M, Ritland K. Estimation of pairwise relatedness with molecular markers. Genetics. 1999;152:1753–1766. doi: 10.1093/genetics/152.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan BG. Maximum-likelihood estimation of relatedness. Genetics. 2003;163:1153–1167. doi: 10.1093/genetics/163.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J, Rabinowitz D. The effect of marker heterozygosity on the power to detect linkage disequilibrium. Genetics. 1997;147:927–930. doi: 10.1093/genetics/147.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen L. Positional cloning of disease genes: advantages of genetic isolates. Hum Hered. 2000;50:66–75. doi: 10.1159/000022892. [DOI] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1:182–190. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Ritland K. Estimators for pairwise relatedness and inbreeding coefficients. Genet Res. 1996;67:175–186. [Google Scholar]
- Shifman S, Darvasi A. The value of isolated populations. Nat Genet. 2001;28:309–310. doi: 10.1038/91060. [DOI] [PubMed] [Google Scholar]
- Shmulewitz D, Zhang J, Greenberg DA. Case-control association studies in mixed populations: correcting using genomic control. Hum Hered. 2004;58:145–153. doi: 10.1159/000083541. [DOI] [PubMed] [Google Scholar]
- Slager SL, Schaid DJ. Evaluation of candidate genes in case-control studies: a statistical method to account for related subjects. Am J Hum Genet. 2001;68:1457–1462. doi: 10.1086/320608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–683. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EA. The estimation of pairwise relationships. Ann Hum Genet. 1975;39:173–188. doi: 10.1111/j.1469-1809.1975.tb00120.x. [DOI] [PubMed] [Google Scholar]
- Thompson EA. Monte Carlo in Genetic Analysis Technical report No 294. Department of Statistics, University of Washington; 1995. [Google Scholar]
- Thompson EA. Statistical inference from genetic data on pedigrees. Vol. 6. IMS/ASA; 2000. [Google Scholar]
- Thompson EA. Statistical Inferences from Genetic Data on Pedigrees NSF-CBMS Regional Conference Series in Probability and Statistics. Vol. 6. IMS; Beachwood, OH: 2000. [Google Scholar]
- Thompson EA. MCMC in the Analysis of Genetic Data on Pedigrees. In: Liang F, Wang J-S, Kendall W, editors. Markov Chain Monte Carlo: Innovations and Applications. Lecture Note Series of the IMS National University of Singapore. World Scientific Co Pte Ltd; Singapore: 2005. pp. 183–216. [Google Scholar]
- Thornton T, McPeek MS. Case-control association testing with related individuals: a more powerful quasi-likelihood score test. Am J Hum Genet. 2007;81:321–337. doi: 10.1086/519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken T, Del-Favero J. Chasing genes for mood disorders and schizophrenia in genetically isolated populations. Hum Mutat. 2007;28:1156–1170. doi: 10.1002/humu.20582. [DOI] [PubMed] [Google Scholar]
- Voight BJ, Pritchard JK. Confounding from cryptic relatedness in case-control association studies. PLoS Genetics. 2005;uc1(3):e32. doi: 10.1371/journal.pgen.0010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. An estimator for pairwise relatedness using molecular markers. Genetics. 2002;160:1203–1215. doi: 10.1093/genetics/160.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Anderson AD, Hepler AB. Genetic relatedness analysis: modern data and new challenges. Nat Rev Genet. 2006;7:771–780. doi: 10.1038/nrg1960. [DOI] [PubMed] [Google Scholar]
- Wijsman EM, Rosenthal EA, Hall D, Blundell ML, Sobin C, Heath SC, Williams R, Brownstein MJ, Gogos JA, Karayiorgou M. Genome-wide scan in a large complex pedigree with predominantly male schizophrenics from the island of Kosrae: evidence for linkage to chromosome 2q. Mol Psychiatry. 2003;8:695–705. doi: 10.1038/sj.mp.4001356. [DOI] [PubMed] [Google Scholar]
- Wright AF, Carothers AD, Pirastu M. Population choice in mapping genes for complex diseases. Nat Genet. 1999;23:397–404. doi: 10.1038/70501. [DOI] [PubMed] [Google Scholar]
- Zang Y, Zhang H, Yang Y, Zheng G. Robust genomic control and robust delta centralization tests for case-control association studies. Hum Hered. 2007;63:187–195. doi: 10.1159/000099831. [DOI] [PubMed] [Google Scholar]