Abstract
The expression of the mouse Cr2 gene has been shown to be restricted to mature B cells, follicular dendritic cells and, in some reports, to a minor population of activated T cells. In this report, we demonstrate that the expression of antigen(s) recognized by the anti-CR2 antibody on the surface of T cells is coincident with T cell apoptotic death. Two distinct regions of the Cr2 gene have been implicated as critical for specific expression, the promoter region at the transcription start site and a control region within the first intron of the gene, approximately 1,500bp from the transcription start site. We have created a mouse that is lacking this intronic control sequence which, in the wild type (WT) mouse, contains multiple known binding sites for RBP-jκ, Oct, NFAT and YY1 proteins. The analysis of this mouse named Cr2iΔ (Cr2 intron deletion) demonstrated normal tissue specific expression of the Cr2 gene including a lack of expression in mouse T cells. B cell expression of the Cr2 gene products, CR1 and CR2, is normal compared to WT, and the FDC of these mice continue to express Cr2 gene products. Therefore the intronic control region of the Cr2 gene, defined in transfection-based reporter gene assays as instrumental in controlling the cell specific expression profile of Cr2, does not influence the expression of the Cr2 gene in vivo nor alter the relative production of the CR1 and CR2 proteins via alternative slicing of Cr2 gene products
Keywords: complement receptor, transcription, T cells, activation
Introduction
The human CR2 (CD21) gene encodes a single protein, CR2, that is expressed on mature B cells (Tedder et al., 1984), follicular dendritic cells (FDC) (Reynes et al., 1985) and some T cells. The mouse Cr2 (CD21) gene encodes two proteins (termed CR1 and CR2) that are expressed by mature B cells, FDC and, reportedly, memory or activated T cells (Jacobson and Weis, 2008; Kaya et al., 2001; Kaya et al., 2005; Kurtz et al., 1990; Qin et al., 1998). Expression of the mouse CD21 proteins coincides with the T1 to T2 transition of splenic B cells and these proteins continue to be expressed by marginal zone (MZ) and follicular (Fo) B cells. There are a number of reports of CD21 expression by T cells (Fischer et al., 1991; Fischer et al., 1999; Tsoukas and Lambris, 1988) including T cell lines and activated primary T cells. The human CR2 protein has also been shown to be a co-factor for HIV infection in that complement opsonized HIV is concentrated on the surface of CR2-expressing T cells allowing for efficient infection (Delibrias et al., 1993). Previous publications have shown the mouse CD21 proteins to be expressed by a subset of memory T cells (CD44hi, CD62Llo, CD3+ cells) suggesting that CD21 proteins play a specific role in regulating the adaptive immune response (Kaya et al., 2001). Subsequently, it was suggested that the CD21 gene products expressed by T cells helped regulate those cells responses to LPS (Kaya et al., 2005). In addition, two reports have suggested that allo-antigen primed mouse T cells can also express the CD21 proteins (Pratt et al., 2002; Qian et al., 2005).
The mouse and human CD21 genes appear to possess similar transcriptional control pathways (Rayhel et al., 1991; Tolnay et al., 1997; Zabel and Weis, 2001). Both mouse and human CD21 gene promoters are active in transfection analyses in B and T cell lines even though only B cells express the endogenous gene (Hu et al., 1997; Makar et al., 1998; Makar et al., 2001). Previously, we showed that introduction of the first intron of the mouse CD21 gene into Cr2 reporter constructs silenced CD21 gene expression in T cell transfections (Hu et al., 1997). This silencing is CD21 promoter specific since the use of a different promoter was not affected by the CD21 intron sequence (Zabel et al., 2000). The silencing site was mapped to a repetitive series of binding sites for NFAT family members, octamer sites, and YY1 binding sites (Zabel et al., 2002). Similar data was obtained from analyses of the human CD21 intronic element that mapped the silencing sequence to a single RBP-Jκ site (Makar et al., 2001). The sequences within the first intron of the mouse and human genes have been conserved between species, thus, this area was termed the mouse human homology box (MH box).
In this report we have analyzed the expression of the Cr2 gene products in native and activated T cells and do not demonstrate binding with anti-Cr2 antibodies unless the cells are undergoing apoptosis. In addition, we have created a mouse lacking the key regulatory sites previously implicated in the tissue specific control of Cr2 gene expression and find the expression of the gene to be indistinguishable from wild type. The possibility that the cluster of transcription factor binding sites is involved in controlling the expression of other genes is proposed.
Materials and Methods
Mice
BALB/c mice were acquired from Charles River Laboratories (Wilmington, MA) while A/J, C57BL/6, and B6.MRL-Faslpr mice were obtained from The Jackson Laboratory (Bar Harbor, ME). DO11.10 transgenic mice (Murphy et al., 1990), with a T cell receptor specific for ovalbumin (OVA), were a generous gift from Dr. Raymond Daynes (University of Utah). All mice were kept under pathogen free conditions at the Comparative Medicine Facility at the University of Utah and used under the regulations of the Institutional Animal Care and Use Committee.
Generation of intron deletion mouse
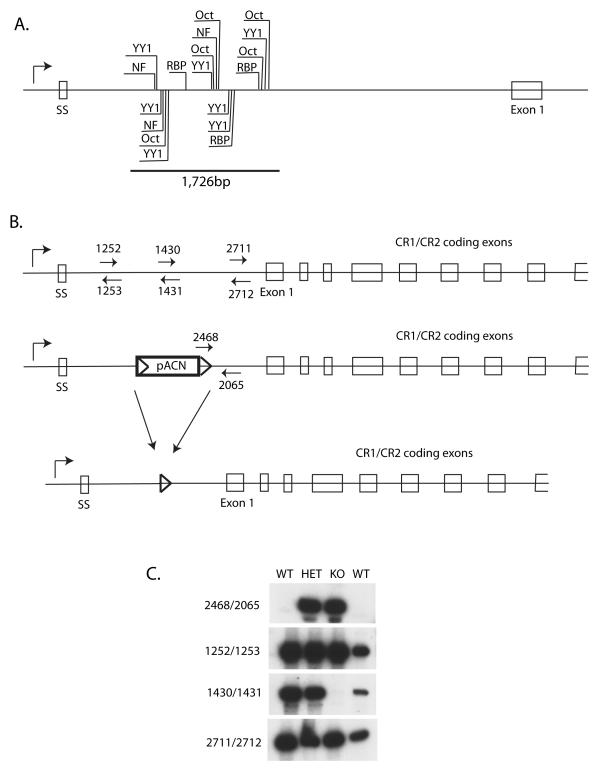
A homologous recombination construct was prepared from 129/Sv genomic DNA. The construct contained the first 1035bp of the first intron, the pACN vector, followed by a Cr2 genomic fragment that started at 2,761bp in the first intron and extended 3′, creating a construct that would, upon homologous recombination, remove 1,726bp of the Cr2 first intron containing the MH box sequences. A single ES clone was identified as possessing the correct recombined fragments and was used to generate the Cr2iΔ line of mice. Knockout mice were genotyped via a PCR assay that detected remnants of the introduced DNA construct (oligo numbers 2468/2065), the absence of intronic sequence (oligo’s 1430/1431) and confirms the presence of flanking intronic sequence (oligo’s 1252/1253 and 2711/2712). Sequences for the PCR oligonucleotides are as follows: #1252 5′ –TTCTGGTAGTGAAGGGATCAGG- 3′, #1253 5′ –AACTGAGGGTCAGGGCTCTAAC- 3′, #2468 5′ – AGCTTGATATCGAATTCCTGC- 3′, #2065 5′ – GCTCACAGTCATCTATTGGATG- 3′, #2711 5′ –CCTCCATTGAGGGCTCTG- 3′, and #2712 5′ –CCTCAACCTCTCTAGTACTC- 3′. DNA sequence for oligonucleotides #1430 and #1431 has been previously published (Zabel et al., 2002). Genomic tail DNA was amplified using the air thermocycler (Idaho Technologies) and visualized on a 6% acrylamide sequencing gel via radioactive detection (Zabel et al., 1999). Mice containing the deletion were backcrossed with C57BL/6 mice for five generations. The amount of remaining 129/Sv DNA on Chromosome 1 in knockout mice was detected by PCR using D1Mit303 (MGI:101463), D1Mit217 (MGI:91597), D1 Mit286 (MGI:91673), D1Mit102 (MGI:91471), D1Mit36 (MGI:91688) and D1Mit17 polymorphism markers and visualized via radioactive detection. DNA from 129/Sv, C57BL/6 and Cr2-deficient C57BL/6 mice were used for comparison.
Immunization strategy
In experiments using hen egg lysozyme (HEL) (Sigma) as an antigen, mice were immunized subcutaneously with 50 μg HEL in PBS or emulsified in Complete Freund’s Adjuvant (CFA) (Sigma). DO11.10 mice were immunized subcutaneously with 10 μg ovalbumin (Sigma) resuspended in alum or PBS. Unimmunized or mice immunized with an equal volume of PBS were used as controls in all experiments.
Activation strategy
Splenocytes were isolated from mice and cultured in RPMI medium (supplemented with 10% FCS and 1% Pen-strep) in the presence or absence of 1 μM phorbol 12-myristate 13-acetate (PMA) (Sigma) plus 25 ng/ml ionomycin (Calbiochem) for 24 hrs.
Flow cytometry analysis
Single cell suspensions from the spleen and thymus were obtained as previously published (Roundy et al., 1999) Superficial inguinal lymph nodes were removed from the animal and cells were obtained by similar treatment as the other tissues. Staining of cells was also completed as previously published (Roundy et al., 1999). The following antibodies were used for analysis of cell populations: CD3ε (Clone 145-2C11), CD8 (53-6.7), CD21 (7G6), CD35 (8C12) from BDBiosciences and B220 (RA3-6B2), CD4 (GK1.5), CD1d (1B1), CD5 (53-7.3), CD19 (eBio1D3), CD23 (B3B4) from eBioscience (San Diego, CA). Memory T cells were identified with CD44 (IM7) and CD62L (MEL-14). Activated cells were identified with CD25 (PC61) (BD Biosciences) and LPS activation. Apoptotic cells were analyzed by staining with Annexin V (BD Biosciences) and 7-AAD (BD Biosciences) in a flow cytometry assay. Viability assays used 7-AAD or TO-PRO3 (Molecular Probes) to eliminate dead cells. Data was collected using the BD FACScan and analyzed with CellQuest software.
Isolation of follicular dendritic cells
FDC were isolated from the spleens of age-matched female C57BL/6 and Cr2iΔ mice using a modified version of previously published protocols (Kosco et al., 1992; Sukumar et al., 2006). Basically, spleens were minced into smaller pieces and enzymatically digested in 2 ml RPMI (Invitrogen) containing 2500U DNase I (Sigma, D-4527) and 0.28U/ml Liberase Blendzyme 2 (Roche). Spleens were then incubated at 37°C with agitation for 30 min. After 30 min, the culture media was collected, placed on ice, and replaced with fresh enzymatic cocktail. The remaining tissue was subjected to an additional 30 min incubation, after which the media was collected and pooled with the first collected volume. The collected supernatant was centrifuged for 5 min at 1000 rpm to pellet the released cells. Red blood cells were lysed with ACK lysis buffer. Remaining cells were incubated with FDC-M1 antibody (BD Biosciences) on ice for 45 min. Cells were then rinsed and incubated with biotinylated mouse anti-rat Ig (MRK-1, BD Biosciences) for 30 min. on ice. FDC-M1+ cells were isolated from total splenocytes by using streptavidin magnetic beads according to the manufacturer’s protocol (Miltenyi Biotec). RNA was isolated from FDC-M1+ cells using the Illustra RNA isolation kit (GE Healthcare) and reverse-transcribed into cDNA to be used in quantitative PCR.
Quantitative RT-PCR
Transcript levels for Cr1, Cr2, Mfge8, Cxcl13 and Clu were quantified using the Roche LightCyler (Indianapolis, IN) according to previously published protocols (Morrison et al., 1999; Zabel et al., 1999). Oligonucleotides used for quantitative PCR included the following: Cr1-F 5′ –GGTTCGCTCTGGGTTTTCTTCAC- 3′ and Cr1-R 5′ –CTGACCACTTGGAGGTTTCTAAGC- 3′; Mfge8-F 5′ –ATATGGGTTTCATGGGCTTG- 3′ and Mfge8-R 5′ –GAGGCTGTAAGCCACCTTGA- 3′; Clu-F 5′ –GAGAAGGTGAAGATGACCGCAC- 3′ and Clu-R 5′ –GGGCAGGATTGTTGGTTGAAC- 3′. Oligonucleotide sequences for Cr2, Cxcl13, and β-actin were previously published (Debnath et al., 2007; Hojgaard et al., 2006). Gene expression levels are displayed relative to β-actin expression levels.
Results
Analysis of CD21 expression by BALB/c, MRL-Faslpr and A/J T cells
In previous assays using reporter constructs, we demonstrated that a minimal Cr2 gene promoter could drive transcription in mouse T and B cell lines. The addition of intron sequences from the MH box, however, suppressed the expression of the reporter construct in T cells but allowed transcription in B cells. Thus we had two primary goals in the research described in this report: to define the parameters (if any) for allowing Cr2 expression by murine T cells, and to determine the effect on Cr2 transcription in various cell types by removing the MH box sequences from the endogenous gene.
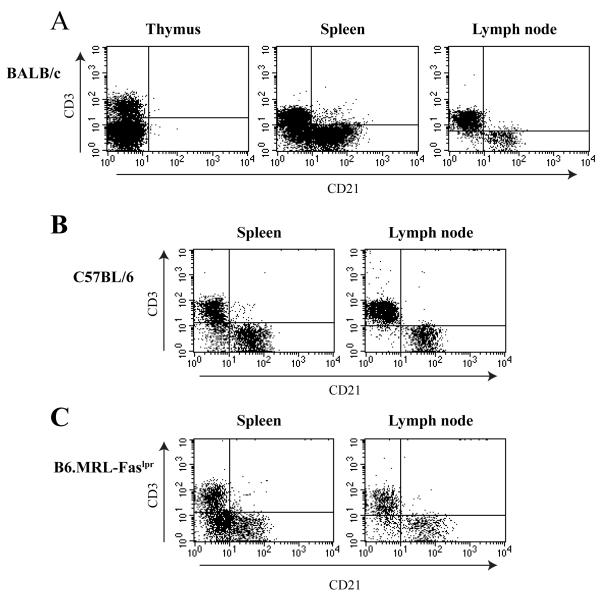
To examine the expression of Cr2 gene products by T cells, we analyzed naïve and activated cells from a variety of strains that, in the literature, have been proposed to express CD21. Initially, we first examined T cells from BALB/c animals for CD21 expression. Total naïve cell populations obtained from the spleen, thymus and lymph node were analyzed by flow cytometry using the CD3 T cell marker and the anti-CD21 antibody that recognizes both the CR1 and CR2 proteins. As shown in Figure 1A, there is a small fraction (upper right quadrant) of cells in the spleen that appear to possess CD3 and the CD21 proteins. Such double positive cells are largely absent in the samples taken from the thymus and the lymph node. Similar data are obtained from cells from C57BL/6 mice (Figure 1B).
Figure 1. Analysis of potential CD21+ T cell populations in BALB/c naïve animals.
A. Spleen, thymus, and superficial inguinal lymph nodes were removed from mature BALB/c female mice. Cells were analyzed for expression of CD21 in correlation with T cell markers. Spleen and thymus data were collected from 10,000 live events while lymph node plots are based on 5,000 total events. Results shown are from one animal but representative of three independent experiments. B. Cells from naïve spleen and lymph nodes isolated from mature C57BL/6 female mice were analyzed as described above. C. Cells from naïve spleen and lymph nodes isolated from mature B6.MRL-Faslpr female mice were analyzed as described above.
The B6.MRL-Faslpr mouse possesses a defect in the Fas receptor such that hyper proliferation of T cells is evident in aged animals, and the development of autoimmune responses is progressive (Rieux-Laucat et al., 2003). The MRL/lpr mutation has been bred upon a strain of mouse lacking the CD21 proteins; such mice demonstrate an accelerated autoimmune response (Boackle et al., 2004). We hypothesized that inappropriate expression of the CD21 proteins (CR1 and CR2) by lpr T cells could concentrate complement-bound immune complexes onto the surface of such cells potentially leading to the inappropriate activation of antigen-specific B cell. Naïve splenocytes and lymph node cells were obtained from aged lpr mice and analyzed for T cells (CD3+) expressing CD21 (Figure 1C). Only a very small subset of cells in the tissues from such mice could be interpreted as possessing both the CD21 proteins and the T cell marker, CD3. There was no significant difference in the percentages of CD21+/CD3+ cells in the tissues of the B6.MRL-Faslpr mouse compared to WT C57BL/6 animals.
One previous documentation of CD21 expression by activated T cells utilized the A/J stain of mice. Accordingly, we immunized A/J mice with HEL (or mock immunized with PBS) and analyzed draining lymph node cells and splenocytes for co-incident expression of T cell markers and CD21. Similar to that seen for the BALB analyses, only a few CD4+/CD21+ cells could be detected in the naïve and immunized samples (data not shown). We repeated this immunization strategy with a more potent adjuvant, complete Freunds adjuvant (CFA), to determine if we could enrich for the expression of CD21 expressing T cells. Cell recruitment into the draining lymph node (A) but not the spleen (B) is evident following immunization, and there is a slight increase in the population of CD21+/CD4+ T cells (denoted by the oval on the FACS plot) in the immunized lymph node samples but less so for the spleen (Supplemental Figure 1). It should be noted that the percentage of T cells within the lymph node sample declined from about 55% to 39% (upper left quadrant) following immunization, due to the increase in B cells, but the overall percentage of cells within the CD4+/CD21+ quadrant (upper right) in those samples remained at about 1-2% of the total cells for the naïve and immunized samples. These data suggest that the addition of CFA to the immunization protocol did slightly increase the percentage of CD21+/CD4+ cells in the draining lymph nodes.
One difficulty in analyzing antigen-specific T cells following an immunization protocol is that such cells represent only a small subset of total cells. This concern can be overcome by using a mouse line possessing a transgenic T cell receptor (TCR) complex where every T cell develops with the specificity of that single antigen, and the expression of coordinated proteins can be observed from the majority of the activated T cells. The D011.10 mouse expresses a TCR with specificity for OVA such that all CD4+ T cells in the animal express that TCR and will respond equivalently. Accordingly, we obtained and immunized the D011.10 mouse with ovalbumin (OVA) (Murphy et al., 1990). Examination of draining lymph nodes of immunized mice (Supplemental Figure 2A) demonstrated CD4+ cells with increased CD44 staining (and decreased CD62l staining) indicative of the generation of memory T cells but little appreciable increase of CD4+/CD21+ cells. By comparison, analysis of CD4+/CD21+ cells in the spleen (Supplemental Figure 2B) did not generate a significant memory response in splenic CD4+ T cells and did not result in an increase in CD4+/CD21+ cells.
CD21 expression by murine T cells is limited to dead and dying cells
The previous experiments were designed to determine if the generation of in vivo activated and memory T cells would enrich for T cells expressing the CD21 gene products. One concern with such protocols is that they may not provide the T cells with the appropriate signal to induce transcription and expression of the CD21 gene products. PMA and ionomycin are known to be very potent T cell activators, acting by dramatically increasing the activity of protein kinase C and releasing intracellular calcium. Therefore we utilized these T cell activators to determine if in vitro treated T cells would now preferentially express the CD21 gene products.
Naïve C57BL/6 splenocytes (and C57BL/6 Cr2-/- deficient splenocytes) (Haas et al., 2002) were treated with PMA and ionomycin for 6 and 24 hours, and analyzed for the coincident expression of CD4 and CD21 (Supplemental Figure 3). Cells for both time points are clearly activated as determined by the increased expression of CD25 compared to the control cells incubated in media alone (data not shown). The 6-hour and 24hr PMA/ionomycin incubations showed a slight increase in the numbers of T cells expressing CD21. Cells obtained from the CD21 deficient line demonstrated CD21 staining on CD4+ (Supplemental Figure 3) or CD8+ cells (data not shown) similar to that of the WT samples. Similar analyses were performed on splenocytes from the lpr animal; there was no significant increase in CD4+/CD21+ of cells following PMA/ionomycin activation compared to the control animal cells (data not shown).
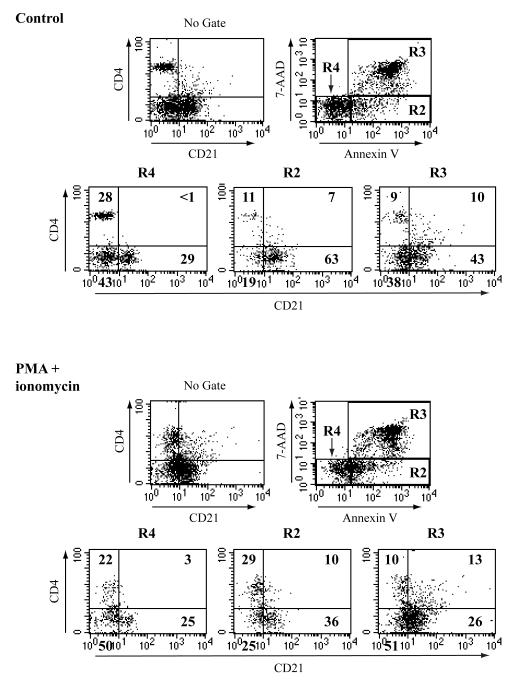
The previous figure suggested a slight increase in T cells staining with the anti-CD21 antibody after treatment with PMA and ionomycin. We analyzed this response further using counterstaining with the viability marker dye 7-AAD which is taken up by dead and dying cells and Annexin V that binds to the phosphatidyl serine residues exposed in the surface of cells undergoing apoptosis. At the same time, the cells were labeled with the anti-CD4 and anti-CD21 antibodies. The first plot “no gate” simply shows total cells for CD4 and CD21 staining. Total cells were then gated into the R2, R3 and R4 populations, based upon 7-AAD and Annexin V staining, and then re-assessed for CD4 and CD21 surface staining. Cells in the R4 gate are viable (double negatives for 7AAD and Annexin-V), those in R2 are dying and those in R3 are dead cells. The same experiment was carried out for either 6 hours (Supplemental Figure 4) or 24 hours (Figure 2). Live cells (R4 gate) in the control and activated cell samples clearly stain with CD21 or CD4; there are no double positive cells. Dying cells (R2 gate) do contain CD4+/CD21+ cells, especially after 24hrs of culture. Dead cells in the R3 gate in both the control and treated samples contain a significant number of CD4+/CD21+ cells. These data suggest that dead (or dying) T cells induce the expression of antigens that are recognized by the anti-CD21 antibody.
Figure 2. CD21 expression by murine T cells is coincidental with cell death.
Total splenocytes from mature C57BL/6 mice were collected and cultured in the presence or absence (control) of PMA+ ionomycin for 24 hours and then analyzed for apoptosis via Annexin V staining. Based on 7-AAD and Annexin V staining, gated cells were then analyzed for CD4 and CD21 staining. The results from a single set of experiments are shown but are representative of multiple similar analyses.
Creation and analysis of a mouse line lacking the putative Cr2 intron 1 control elements
In previous studies on the mouse and human Cr2/CR2 genes, sequences within the first intron of the genes dictated cell specific expression using reporter gene constructs and transient and stable transfection into cell lines. In addition, this region was shown in transgenic mouse lines to confer cell specific expression. As show in Figure 3A, the first intron MH box region (within the 1,726bp region) contains a number of transcription factor binding sites including the NFAT proteins, YY1, Octamer proteins, and RBPjκ. A knockout construct was prepared using mouse 129/Sv DNA that removed 1,726bp of intronic sequence that included all of these putative binding sites (Figure 3B). The intron sequences were replaced with pACN, which is flanked by lox sites, and contains a Neo gene for positive selection and a testes-specific Cre expression cassette that leads to the loss of the entire insert, except for a single lox site, after male transmission of the transgene. One ES cell demonstrated appropriate homologous recombination and it was used to create a line of mice bearing this germline deletion (Cr2iΔ) (Figure 3C). The homozygous KO mouse lacks sequences from this region of Cr2 intron 1 but is normal for flanking sequences. This construct did not influence the viability of the animal.
Figure 3. Generation of a mouse line lacking the MH box sequences.
A. Line drawing showing location of the MH box sequences, transcription factor binding sites and the position of the 1,726bp deletion created within the construct. SS encodes the signal sequence, and exon 1 encodes the first SCR domain of the CR1 protein. B. Line drawing showing the location of oligonucleotide probes for PCR analysis of WT and Cr2iΔ animals. After passage through sperm, the pACN cassette is removed leaving only a single Lox site within the Cr2 gene. All figures not drawn to scale. C. PCR analysis of genomic DNA from wildtype (WT/C57BL/6), heterozygote (HET) and knockout (KO) mice. DNA fragments were resolved on a 6% polyacrylamide gel. Locations for the oligonucleotides used in this assay are shown in Panel B.
The reporter construct experiments using regions of the MH box suggested that lacking such sequences should allow for robust Cr2 gene expression in T cells. Accordingly, the expression of transcripts encoding CR1 and CR2 were evaluated using spleen, marrow and thymus samples. As shown in Supplemental Figure 5, the expression of the Cr2 gene for the CR1 and CR2 gene products was not altered by the removal of the intron 1 sequences. These data demonstrate that the expression of the Cr2 gene was not, as expected, enhanced in T cells and that the alternative splicing seen in the Cr2 gene to produce the CR1 and CR2-specific transcripts was not lost with the intronic deletion.
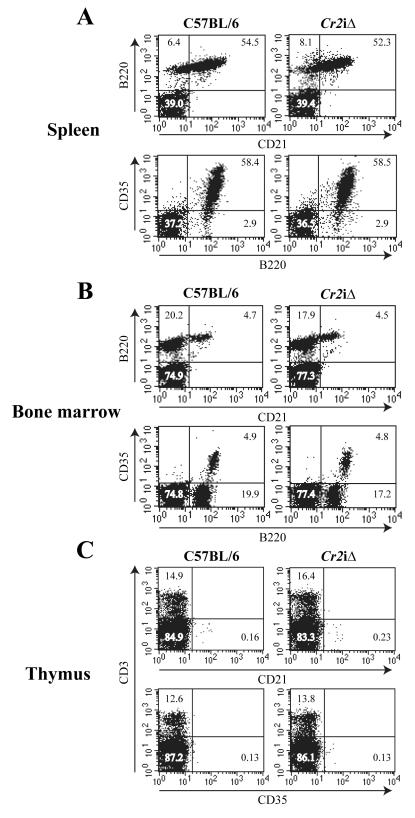
To analyze the expression of the Cr2 gene products on the single cell level, flow cytometry experiments were performed on tissues obtained from the WT and Cr2iΔ mice. As shown in Figure 4, the surface expression of CR1 (CD35) and CR2 (CD21) was identical between the WT and Cr2iΔ tissues. There was no enhanced expression of the Cr2 gene products on non-B220 cells in the spleen or bone marrow, nor was there any expression of the CR1 or CR2 proteins on the surface of CD3+ cells of the thymus. Thus, the removal of the intron 1 MH box control region did not alter the expression of the Cr2 gene in naïve cell types of the mouse.
Figure 4. Flow cytometry analysis of CD21 and CD35 expression in the Cr2iΔ animals.
Total cells from WT (C57BL/6) and Cr2iΔ spleen (A), Bone marrow (B) and thymus (C) were analyzed for CD21 (CR2) or CD35 (CR1) expression. Antibodies for B220 (B cells) and CD3 (T cells) were used to counterstain. Percentages of cells in each quadrant are marked.
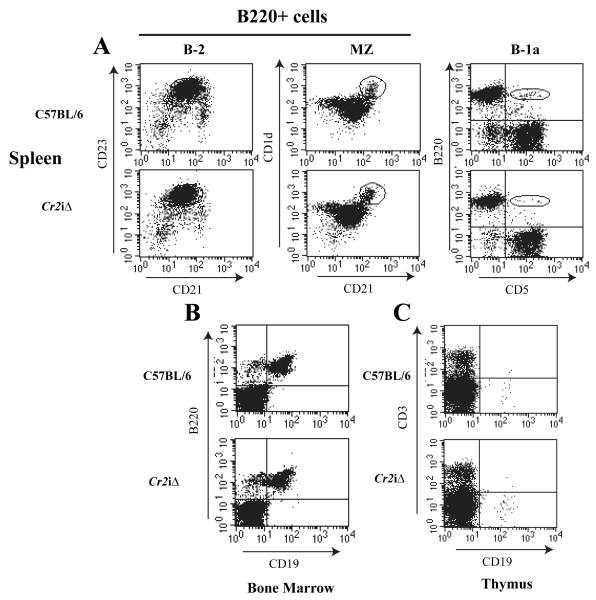
Loss of MH box sequences does not alter B cell types or FDC expression of Cr2
Cr2 is expressed in multiple lineages of B cells and at various times during B cell development. The expression of the Cr2 gene in B-2 B cells becomes evident at the T1 to T2 transition stage, and continues in mature MZ and FM cells. Plasma cells lose CD21 expression due to the loss of Pax5, which is a positive transcriptional regulator of Cr2. Cr2 is also expressed by B-1 B cells. Since the MH box sequences did not appear to be instrumental in the specific expression of Cr2 (B cells versus T cells) perhaps these sequences are involved in regulating the stage specific expression of Cr2 in maturing B-1 and B-2 B cells. In Figure 5A, the expression of CR2 (CD21) is analyzed in splenic B220+ WT and mice by flow cytometry using CD23 and CD1d as secondary markers. These data indicate that there is no discernable difference between WT and Cr2iΔ animals in CD21 expression on B-2 cells or in the percentages of B cell types. Additionally, there is no significant difference in the number of splenic B-1a B cells in the spleen of the animal compared to WT. The same analyses were performed on total bone marrow cells (Fig 5B) and total thymocytes (Fig 5C) and demonstrated no difference in the percentages of B cells in the marrow or thymus. Supplemental Table I delineates the data from a number of different experiments and shows that there is no alteration in the numbers of cells within B cell subsets comparing the WT animals to the animals.
Figure 5. Flow cytometry analysis of cell populations in the Cr2iΔ animals.
A. B220+ spleen cells from WT (C57BL/6) and Cr2iΔ animals were analyzed for B-2 cell populations using CD21/CD23 and marginal zone B cells (MZ) using CD1d/CD21 staining (circled gate). B-1a B cells were identified by co-staining with antibodies specific for the B220 and CD5 antigens (oval area). B. Bone marrow populations analyzed with anti-B220 and anti-CD19. C. Thymus populations analyzed with anti-CD3 and anti-CD19.
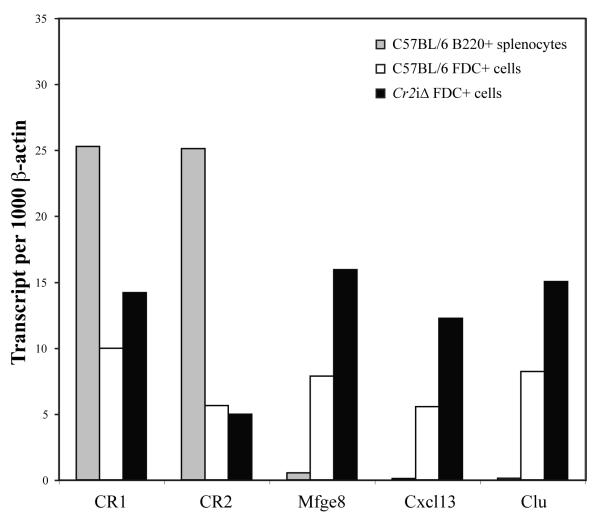
The second major cell type of the animal that expresses Cr2 is the FDC, which also produces both the CR1 and CR2 protein products. The sequences of the Cr2 gene that define FDC-specific expression are not known, therefore, we examined FDC-specific Cr2 expression from FDC obtained from WT and mice for alterations in Cr2 expression. FDC were enriched by magnetic bead isolation using FDC-M1 antibody that recognizes a unique epitope on FDC. FDC-enriched samples were obtained from mature WT and Cr2iΔ mice, total RNA was isolated, and RT-PCR carried out using CR1 and CR2 specific oligonucleotides as well as those for milk fat globule-EGF factor 8 protein (Mfge8) (Kranich et al., 2008), chemokine (C-X-C motif) ligand 13 (Cxcl13) (Allen and Cyster, 2008) and clusterin (Clu) (Huber et al., 2005) which are known FDC specific/enriched products. As shown in Figure 6, the expression of the CR1 and CR2 gene products is the same for FDC samples obtained from the WT and mice indicating that the deletion of the intronic sequences had no effect upon FDC expression of the Cr2 gene, either in total or in CR1 or CR2 specific gene products. The expression of the Mfge8, Cxcl13 and Clu genes was as expected for purified FDC. Therefore, in the absence of the intronic control region, Cr2 gene expression is apparently unaffected in B cell and FDC lineages
Figure 6. CR1 and CR2 gene products are produced by FDC from Cr2iΔ animals.
Quantitative RT-PCR analysis of transcripts encoding CR1, CR2, Mfge8, Cxcl13 and Clu using RNA prepared from total C57BL/6 B220+ splenocytes (gray bar), FDC cells from C57BL/6 splenocytes (open bar) and FDC enriched from Cr2iΔ splenocytes (black bar).
Discussion
In our previous studies using reporter gene constructs and transient transfection analyses, we determined that sequences within the MH box of the Cr2 intron 1 were instrumental in controlling the cell specific expression of that gene (Hu et al., 1997; Zabel et al., 2000; Zabel et al., 2002). Similar results were also obtained from studies with the human CR2 gene (Makar et al., 1998; Makar et al., 2001) although those studies focused upon a single functional RBP-jκ binding site while our mouse analyses included sequences for RBPjκ, YY1, and Oct and NFAT family members. Therefore we expected that the removal of these intronic control sequences would allow for Cr2 gene expression in other lymphoid cells and could alter the B cell stage specific expression profile of the gene. However, while reporter gene assays can be useful in defining critical factor binding sites, they are artificial in that large quantities of plasmid DNA is transiently transfected into permissive, immortalized cell lines and the processes of transcription/translation are monitored using an artificial assay, i.e. luciferase. In the past decade, our understanding of how chromosomal alterations can influence gene expression has raised serious questions as to the validity of reporter assays and if the data derived from such assays can be artifactual. With these concerns in mind, we chose to stringently test the requirement of the MH box sequences in the control of the Cr2 gene by creating a mouse that lacks them.
The data presented in this manuscript focuses upon the expression of the Cr2 gene products by naïve or activated T cells, and whether the removal of the MH box sequences would now allow promiscuous expression of CR1/CR2 on these cells as suggested by the reporter assays. We show that the detection of Cr2 gene products using anti-CD21 antibody on the surface of naïve or activated T cells is coincident with the death of such cells. It is likely that a neo-antigen is expressed by dying cells that is recognized by the 7G6 antibody. We also demonstrate that removal of the MH box sequences from the Cr2 gene did not allow for expression of this gene in T cell lineages using both transcript and flow cytometry analyses. In addition, B cell subsets are normal in the Cr2iΔ mouse and FDC expression of Cr2 is indistinguishable from WT. Thus this region of the mouse Cr2 gene, although dense with transcription factor binding confirmed in vivo by ChIP analysis, is not required for the normal expression of the gene.
If the MH box is not essential for the expression of the Cr2 gene, then could it be required for expression of neighboring genes? Chromatin conformation capture experiments (3C) have demonstrated in a number of mammalian systems that transcriptional control regions can act upon genes distal on the same or different chromosomes (de Laat et al., 2008; Dekker et al., 2002; Liu and Garrard, 2005; Murrell et al., 2004; Spilianakis et al., 2005). Thus the MH box could act as a positive or negative control element regulating genes other than Cr2. Preliminary gene grid array analysis of splenocytes from the WT and Cr2iΔ animal suggests altered expression of a number of genes on mouse chromosome 1 and elsewhere in the genome. However, the altered expression of such genes may be due to either the loss of the MH box sequences, or due to 129/Sv allelic differences between the Cr2iΔ gene and the rest of mouse chromosome 1: (after 5 backcrosses of the founder 129/Sv Cr2iΔ animal to C57BL/6 animals, the genome of the Cr2iΔ mouse is all C57BL/6 except for the final 48Mbp of mouse chromosome 1)(data not shown). Therefore to determine that the MH box sequences are indeed controlling the expression of genes other than Cr2, additional backcrosses to the C57BL/6 line must be performed as well as 3C experiments (Dekker, 2006) in order to trap chromosomal interactions between the MH box sequences and those of affected genes.
Supplementary Material
Table 1.
Altered gene expression in the Cr2iΔ mouse compared to wild type. Shown are B cell specific or other selected transcripts that are not localized to mouse chromosome 1. FC (fold change) positive numbers denotes higher in WT compared to Cr2iΔ and negative numbers denote higher in Cr2iΔ compard to WT.
| Probe Set ID | Gene Title | Gene Symbol | FC | Genome |
|---|---|---|---|---|
| 1428358_at | RIKEN cDNA 1810010M01 gene | 1810010M01Rik | 4.83 | chr7 |
| 1439475_at | zinc finger protein 429 | Zfp429 | 4.39 | chr13 |
| 1434171_at | zinc finger protein 847 (RIKEN C330011K17) | Zfp847 | 3.39 | chr13 |
| 1434441_at | RIKEN cDNA 1110018J18 gene | 1110018J18Rik | 3.27 | chr13 |
| 1455886_at | Casitas B-lineage lymphoma | Cbl | −2.92 | chr9 |
| 1418600_at | Kruppel-like factor 1 (erythroid) | Klf1 | −3.21 | chr8 |
| 1419477_at | C-type lectin domain family 2, member d | Clec2d | −4.41 | chr6 |
| 1425385_a_at | Immunoglobulin heavy chain complex | Igh-6 | −5.72 | chr12 |
| 1442544_at | Immunoglobulin heavy chain complex | Igh-6 | −5.77 | chr12 |
| 1427756_x_at | Immunoglobulin heavy chain complex | Igh-6 | −5.96 | chr12 |
| 1424631_a_at | Immunoglobulin heavy chain (gamma) | Ighg | −6.28 | chr12 |
| 1426174_s_at | immunoglobulin heavy chain 3 (serum IgG2b) | Igh-3 | −6.62 | chr12 |
| 1425247_a_at | immunoglobulin heavy constant gamma 1 | Ighg1 | −8.05 | chr12 |
| 1427870_x_at | Immunoglobulin heavy chain complex | Igh-6 | −8.33 | chr12 |
| 1425324_x_at | Immunoglobulin heavy chain complex | Igh-6 | −9.25 | chr12 |
Acknowledgements
The authors would like to thank Elena Enioutina and Raymond Daynes for providing the D011.10 mouse strain and advice for the use of the animals. We thank Ann Sweeny for excellent technical assistance. We would also like to thank all the members of the Weis laboratories for critical analysis of the work. This work was supported by NIH RO1 AI-24158 (to JHW) and NIH grants AI-32223 (to JJW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Allen CD, Cyster JG. Follicular dendritic cell networks of primary follicles and germinal centers: phenotype and function. Semin Immunol. 2008;20:14–25. doi: 10.1016/j.smim.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boackle SA, Culhane KK, Brown JM, Haas M, Bao L, Quigg RJ, Holers VM. CR1/CR2 deficiency alters IgG3 autoantibody production and IgA glomerular deposition in the MRL/lpr model of SLE. Autoimmunity. 2004;37:111–23. doi: 10.1080/08916930410001685063. [DOI] [PubMed] [Google Scholar]
- de Laat W, Klous P, Kooren J, Noordermeer D, Palstra RJ, Simonis M, Splinter E, Grosveld F. Three-dimensional organization of gene expression in erythroid cells. Curr Top Dev Biol. 2008;82:117–39. doi: 10.1016/S0070-2153(07)00005-1. [DOI] [PubMed] [Google Scholar]
- Debnath I, Roundy KM, Weis JJ, Weis JH. Defining in vivo transcription factor complexes of the murine CD21 and CD23 genes. J Immunol. 2007;178:7139–50. doi: 10.4049/jimmunol.178.11.7139. [DOI] [PubMed] [Google Scholar]
- Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Delibrias CC, Kazatchkine MD, Fischer E. Evidence for the role of CR1 (CD35), in addition to CR2 (CD21), in facilitating infection of human T cells with opsonized HIV. Scand J Immunol. 1993;38:183–9. doi: 10.1111/j.1365-3083.1993.tb01711.x. [DOI] [PubMed] [Google Scholar]
- Fischer E, Delibrias C, Kazatchkine MD. Expression of CR2 (the C3dg/EBV receptor, CD21) on normal human peripheral blood T lymphocytes. J Immunol. 1991;146:865–9. [PubMed] [Google Scholar]
- Fischer EM, Mouhoub A, Maillet F, Fremeaux-Bacchi V, Krief C, Gould H, Berrih-Aknin S, Kazatchkine MD. Expression of CD21 is developmentally regulated during thymic maturation of human T lymphocytes. Int Immunol. 1999;11:1841–9. doi: 10.1093/intimm/11.11.1841. [DOI] [PubMed] [Google Scholar]
- Haas KM, Hasegawa M, Steeber DA, Poe JC, Zabel MD, Bock CB, Karp DR, Briles DE, Weis JH, Tedder TF. Complement receptors CD21/35 link innate and protective immunity during Streptococcus pneumoniae infection by regulating IgG3 antibody responses. Immunity. 2002;17:713–23. doi: 10.1016/s1074-7613(02)00483-1. [DOI] [PubMed] [Google Scholar]
- Hojgaard A, Close R, Dunn DM, Weiss RB, Weis JJ, Weis JH. Altered localization of CXCL13 expressing cells in mice deficient in Pactolus following an inflammatory stimulus. Immunology. 2006;119:212–23. doi: 10.1111/j.1365-2567.2006.02426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Martin BK, Weis JJ, Weis JH. Expression of the murine CD21 gene is regulated by promoter and intronic sequences. Journal of Immunology. 1997;158:4758–68. [PubMed] [Google Scholar]
- Huber C, Thielen C, Seeger H, Schwarz P, Montrasio F, Wilson MR, Heinen E, Fu YX, Miele G, Aguzzi A. Lymphotoxin-beta receptor-dependent genes in lymph node and follicular dendritic cell transcriptomes. J Immunol. 2005;174:5526–36. doi: 10.4049/jimmunol.174.9.5526. [DOI] [PubMed] [Google Scholar]
- Jacobson AC, Weis JH. Comparative functional evolution of human and mouse CR1 and CR2. J Immunol. 2008;181:2953–9. doi: 10.4049/jimmunol.181.5.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya Z, Afanasyeva M, Wang Y, Dohmen KM, Schlichting J, Tretter T, Fairweather D, Holers VM, Rose NR. Contribution of the innate immune system to autoimmune myocarditis: a role for complement. Nat Immunol. 2001;2:739–45. doi: 10.1038/90686. [DOI] [PubMed] [Google Scholar]
- Kaya Z, Tretter T, Schlichting J, Leuschner F, Afanasyeva M, Katus HA, Rose NR. Complement receptors regulate lipopolysaccharide-induced T-cell stimulation. Immunology. 2005;114:493–8. doi: 10.1111/j.1365-2567.2004.02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosco MH, Pflugfelder E, Gray D. Follicular dendritic cell-dependent adhesion and proliferation of B cells in vitro. J Immunol. 1992;148:2331–9. [PubMed] [Google Scholar]
- Kranich J, Krautler NJ, Heinen E, Polymenidou M, Bridel C, Schildknecht A, Huber C, Kosco-Vilbois MH, Zinkernagel R, Miele G, Aguzzi A. Follicular dendritic cells control engulfment of apoptotic bodies by secreting Mfge8. J Exp Med. 2008;205:1293–302. doi: 10.1084/jem.20071019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz CB, O’Toole E, Christensen SM, Weis JH. The murine complement receptor gene family. IV. Alternative splicing of Cr2 gene transcripts predicts two distinct gene products that share homologous domains with both human CR2 and CR1. J Immunol. 1990;144:3581–91. [PubMed] [Google Scholar]
- Liu Z, Garrard WT. Long-range interactions between three transcriptional enhancers, active Vkappa gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol Cell Biol. 2005;25:3220–31. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makar KW, Pham CT, Dehoff MH, O’Connor SM, Jacobi SM, Holers VM. An intronic silencer regulates B lymphocyte cell- and stage-specific expression of the human complement receptor type 2 (CR2, CD21) gene. Journal of Immunology. 1998;160:1268–78. [PubMed] [Google Scholar]
- Makar KW, Ulgiati D, Hagman J, Holers VM. A site in the complement receptor 2 (CR2/CD21) silencer is necessary for lineage specific transcriptional regulation. Int Immunol. 2001;13:657–64. doi: 10.1093/intimm/13.5.657. [DOI] [PubMed] [Google Scholar]
- Morrison TB, Ma Y, Weis JH, Weis JJ. Rapid and sensitive quantification of Borrelia burgdorferi-infected mouse tissues by continuous fluorescent monitoring of PCR. J Clin Microbiol. 1999;37:987–92. doi: 10.1128/jcm.37.4.987-992.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KM, Heimberger AB, Loh DY. Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo. Science. 1990;250:1720–3. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–93. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Pratt JR, Basheer SA, Sacks SH. Local Synthesis of Complement Component C3 Regulates Acute Renal Transplant Rejection. Nature Medicine. 2002;8:582–587. doi: 10.1038/nm0602-582. [DOI] [PubMed] [Google Scholar]
- Qian Z, Bieler JG, Baldwin WM, Wasowska BA. Expression of CR1/2 Receptor on Alloantigen-Stimulated Mouse T Cells. Transplantation Proceedings. 2005;37:32–34. doi: 10.1016/j.transproceed.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Qin D, Wu J, Carroll MC, Burton GF, Szakal AK, Tew JG. Evidence for an important interaction between a complement-derived CD21 ligand on follicular dendritic cells and CD21 on B cells in the initiation of IgG responses. Journal of Immunology. 1998;161:4549–54. [PubMed] [Google Scholar]
- Rayhel EJ, Dehoff MH, Holers VM. Characterization of the human complement receptor 2 (CR2, CD21) promoter reveals sequences shared with regulatory regions of other developmentally restricted B cell proteins. Journal of Immunology. 1991;146:2021–6. [PubMed] [Google Scholar]
- Reynes M, Aubert JP, Cohen JH, Audouin J, Tricottet V, Diebold J, Kazatchkine MD. Human follicular dendritic cells express CR1, CR2, and CR3 complement receptor antigens. J Immunol. 1985;135:2687–94. [PubMed] [Google Scholar]
- Rieux-Laucat F, Le Deist F, Fischer A. Autoimmune lymphoproliferative syndromes: genetic defects of apoptosis pathways. Cell Death Differ. 2003;10:124–33. doi: 10.1038/sj.cdd.4401190. [DOI] [PubMed] [Google Scholar]
- Roundy K, Kollhoff A, Eichwald EJ, Weis JJ, Weis JH. Microphthalmic mice display a B cell deficiency similar to that seen for mast and NK cells. Journal of Immunology. 1999;163:6671–8. [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–45. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Sukumar S, Conrad DH, Szakal AK, Tew JG. Differential T cell-mediated regulation of CD23 (Fc epsilonRII) in B cells and follicular dendritic cells. J Immunol. 2006;176:4811–7. doi: 10.4049/jimmunol.176.8.4811. [DOI] [PubMed] [Google Scholar]
- Tedder TF, Clement LT, Cooper MD. Expression of C3d receptors during human B cell differentiation: immunofluorescence analysis with the HB-5 monoclonal antibody. J Immunol. 1984;133:678–83. [PubMed] [Google Scholar]
- Tolnay M, Lambris JD, Tsokos GC. Transcriptional regulation of the complement receptor 2 gene: role of a heterogeneous nuclear ribonucleoprotein. Journal of Immunology. 1997;159:5492–501. [PubMed] [Google Scholar]
- Tsoukas CD, Lambris JD. Expression of CR2/EBV receptors on human thymocytes detected by monoclonal antibodies. Eur J Immunol. 1988;18:1299–302. doi: 10.1002/eji.1830180823. [DOI] [PubMed] [Google Scholar]
- Zabel MD, Byrne BL, Weis JJ, Weis JH. Cell-specific expression of the murine CD21 gene depends on accessibility of promoter and intronic elements. J Immunol. 2000;165:4437–45. doi: 10.4049/jimmunol.165.8.4437. [DOI] [PubMed] [Google Scholar]
- Zabel MD, Weis JH. Cell-specific regulation of the CD21 gene. Int Immunopharmacol. 2001;1:483–93. doi: 10.1016/s1567-5769(00)00046-1. [DOI] [PubMed] [Google Scholar]
- Zabel MD, Weis JJ, Weis JH. Lymphoid transcription of the murine CD21 gene is positively regulated by histone acetylation. Journal of Immunology. 1999;163:2697–703. [PubMed] [Google Scholar]
- Zabel MD, Wheeler W, Weis JJ, Weis JH. Yin Yang 1, Oct1, and NFAT-4 form repeating, cyclosporin-sensitive regulatory modules within the murine CD21 intronic control region. J Immunol. 2002;168:3341–50. doi: 10.4049/jimmunol.168.7.3341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.