Abstract
Two orders of magnitude more protein sequences can be modeled by comparative modeling than have been determined by X-ray crystallography and NMR spectroscopy. Investigators have nevertheless been cautious about using comparative models for ligand discovery because of concerns about model errors. We suggest how to exploit comparative models for molecular screens, based on docking against a wide range of crystallographic structures and comparative models with known ligands. To account for the variation in the ligand-binding pocket as it binds different ligands, we calculate “consensus” enrichment by ranking each library compound by its best docking score against all available comparative models and/or modeling templates. For the majority of the targets, the consensus enrichment for multiple models was better or comparable to that of the holo and apo X-ray structures. Even for single models, the models are significantly more enriching than the template structure if the template is paralogous and shares more than 25% sequence identity with the target.
Keywords: comparative modeling, docking screens, consensus enrichment
Introduction
Structure-based methods have been widely used in ligand design and discovery.1-4 In particular, ligands can be identified among a large library of small molecules by virtual screening.1, 5-11 Each library molecule is docked into a binding site, then scored and ranked by its complementarity to the protein. High-ranking docked molecules are subsequently tested experimentally. The two critical parts of this process are the docking to search through plausible binding modes for candidate compounds and the scoring to distinguish ligands from nonbinding “decoys”.
Docking a ligand to a protein structure is most successful when the shape of the binding site is similar to that found in the protein-ligand complex. Therefore, the protein structure for docking is best determined in complex with a ligand that is similar to the ligand being docked, by X-ray crystallography or NMR spectroscopy. Induced fit and differences between conformations of the protein bound to different ligands limit the utility of the unbound structure (apo structure) and even complex structures (holo structures) obtained for dissimilar ligands. The problem of the protein conformational heterogeneity is especially difficult to surmount in virtual screening, which involves docking of many different ligands, each one of which may in principle bind to a different protein conformation.
An even greater challenge is that many interesting targets have no experimentally determined structures at all, especially in the early phases of ligand discovery. During the last five years, the number of experimentally determined structures deposited in the Protein Data Bank (PDB) increased from 23,096 to 52,821 (September 2008).12 However, over the same period, the number of sequences in the Universal Protein Resource (UniProt) increased from 1.2 million to 6.4 million.13, 14 This rapidly growing gap between the sequence and structure databases can be bridged by protein structure prediction15, including comparative modeling, threading, and de novo methods. Despite progress in de novo prediction,16, 17 comparative modeling remains the most reliable method that can sometimes predict the structure of a protein with an accuracy comparable to a low-resolution, experimentally determined structure.18 Comparative modeling benefits from structural genomics.19 In particular, the Protein Structure Initiative (PSI) aims to determine representative atomic structures of most major protein families by X-ray crystallography or NMR spectroscopy, so that most of the remaining protein sequences can be characterized by comparative modeling (http://www.nigms.nih.gov/Initiatives/PSI/).20, 21 Currently, the fraction of sequences in a genome for whose domains comparative models can be obtained varies from approximately 20% to 75%, increasing the number of structurally characterized protein sequences by two orders of magnitude relative to the entries in the PDB.14 Therefore, comparative models in principle greatly extend the applicability of virtual screening, compared to using only the experimentally determined structures.22
Comparative models have in fact been used in virtual screening to detect novel ligands for many protein targets,22 including the G-protein coupled receptors (GPCR),23-35 protein kinases,36-39 nuclear hormone receptors, and several different enzymes.40-53 Nevertheless, the relative utility of comparative models versus experimentally determined structures has only been sparsely assessed.23, 36, 37, 54-56 In a study of ten enzyme targets, the X-ray structure of a ligand-bound target often provided the best enrichment for known binders.56 The comparative models yielded better enrichment than random selection and performed comparably to the holo X-ray structure in two cases. The relationship between the sequence identity on which a model is based and the screening accuracy was addressed by virtual screening against eight and four comparative models for factor VIIa and cyclin-dependent kinase 2 (CDK2), respectively.37 It was shown that a 5-fold enrichment over random selection was obtained using comparative models based on templates with sequence identities greater than 50%. In an intriguing study, Gilson and co-workers observed that whereas docking against comparative models could lead to substantial enrichment,55 there was little correlation between target-template sequence identity and the success of the docking screen. Furthermore the templates on which the modeled structures were based (i.e., proteins with putatively incorrect sequences) could perform just as well in the docking screens, as the comparative models built form them with the correct sequence.
The lack of consensus in these observations, and arguably the absence of a large-scale benchmarking study, inspired us to investigate the following questions. How does docking against comparative models compare to random selection and to docking against the template structures, over a large number of proteins? If multiple models are calculated on the basis of different templates, can any of them outperform apo and even holo X-ray structures of the target? If so, can one reliably identify which model will do so, or even perform optimally among a set of modeled structures — are there sequence and/or structural attributes (i.e., the overall target-template sequence identity, the binding site target-template sequence identity, and the predicted accuracy of a model) that reliably predict the accuracy of ligand docking? Can the docking screens be improved by employing multiple models, or will using multiple models merely increase the noise from decoys?
Here, we answered these questions with the aid of 38 protein targets selected from “directory of useful decoys” (DUD).57 For each target, DUD contains known ligands as well as decoys with similar physical properties but dissimilar chemical structures. Our analysis proceeded in three steps, performed independently for each of the 38 targets. First, comparative models were calculated based on different template structures. Second, all compounds in DUD were docked against the holo and apo (if available) X-ray structure of the target, the comparative models, and the template X-ray structures. Third, the docking was evaluated based on the enrichment for known ligands with respect to the whole DUD. The enrichments achieved with modeled structures were compared to those achieved with X-ray structures of the corresponding target and templates. We correlate the success of docking with a variety of target/template attributes. In addition, we evaluated a consensus enrichment that combines docking scores from independent virtual screens against different comparative models of the same target.
We begin by describing the target set, the automated modeling and docking pipeline, methods to evaluate the model accuracy, to evaluate the accuracy of virtual ligand screening, and to compare ligand enrichments yielded by different structures (Methods). We then describe the relative utilities of comparative models and X-ray structures for virtual screening, as well as the correlation between the screening performance and various template/model/target similarity measures (Results). Finally we discuss the implications of the current approach and answer the questions we asked above, given our modeling, docking and benchmark (Discussion and Conclusions).
Methods
Target set
DUD contains 2,950 annotated ligands and 95,316 corresponding decoys for 40 protein targets.57 Here, we used the 38 targets of DUD for which holo X-ray structures are available in the PDB (Table 1). These targets are organized into two classes: enzymes (six of which are kinases), and nuclear hormone receptors. For each target, the holo X-ray structure in the original study of DUD was used, except for androgen receptor for which a more recently deposited structure was selected (PDBID 2ao6). An apo structure with the highest sequence identity to the holo structure was also used; no apo structure was found for 4 hormone receptors. For each target sequence, templates for comparative modeling were obtained from the PDB. We aimed to include one holo and one apo template structure for each of the eight 10% sequence identity ranges from 20% to 100%. Structural alignments were performed with the MAMMOTH program58 via the DBALI server.59
Table 1. Protein targets for virtual ligand screening.
| Protein target |
PDB code (Holo) |
PDB code (Apo) |
Number of ligands |
Sequence identity of templates [%] |
Number of templates (Holo) |
Number of templates (Apo) |
|---|---|---|---|---|---|---|
| ACE | 1o86 | 2iul | 49 | 42 – 55 | 3 | 1 |
| ADA | 1ndw | 1vfl | 23 | 26 – 85 | 1 | 1 |
| COMT | 1h1d | 2avd | 12 | 20 | 1 | 0 |
| PDE5 | 1xp0 | 2h40 | 51 | 24 – 96 | 2 | 2 |
| DHFR | 3dfr | 1zdr | 201 | 20 – 37 | 5 | 3 |
| GART | 1c2t | 3gar | 21 | 40 – 42 | 1 | 1 |
| FXa | 1f0r | 1c5m | 142 | 24 – 64 | 3 | 4 |
| Thrombin | 1ba8 | 2afq | 65 | 23 – 40 | 3 | 3 |
| Trypsin | 1bju | 2tga | 44 | 21 – 79 | 5 | 5 |
| AChE | 1eve | 1qih | 105 | 28 – 93 | 4 | 4 |
| ALR2 | 1ah3 | 1xgd | 26 | 20 – 87 | 5 | 6 |
| AmpC | 1xgj | 1l0d | 21 | 20 – 77 | 3 | 4 |
| COX-1 | 1q4g | 1prh | 25 | 62 – 63 | 1 | 1 |
| COX-2 | 1cx2 | 5cox | 349 | 64 – 65 | 1 | 1 |
| GPB | 1a8i | 1gpb | 52 | 42 – 81 | 2 | 0 |
| HIVPR | 1hpx | 3phv | 53 | 23 – 97 | 7 | 5 |
| HIVRT | 1rt1 | 1rtj | 40 | 98 – 99 | 1 | 1 |
| HMGR | 1hw8 | 1r7i | 35 | 100 | 2 | 0 |
| InhA | 1p44 | 2ied | 85 | 20 – 29 | 2 | 2 |
| NA | 1a4g | 1ns b | 49 | 29 – 34 | 2 | 1 |
| PARP | 1efy | 2paw | 33 | 46 – 87 | 1 | 1 |
| PNP | 1b8o | 1pbn | 25 | 20 – 93 | 5 | 3 |
| SAHH | 1a7a | 1ky4 | 33 | 49 – 85 | 2 | 0 |
| CDK2 | 1ckp | 1hcl | 50 | 21 – 95 | 6 | 6 |
| EGFr | 1m17 | 1m14 | 444 | 20 – 38 | 3 | 3 |
| FGFr1 | 1agw | 1fgk | 118 | 20 – 51 | 4 | 4 |
| HSP90 | 1uy6 | 1uyl | 24 | 20 – 70 | 5 | 3 |
| P38 MAP | 1kv2 | 1p38 | 256 | 20 – 62 | 5 | 4 |
| SRC | 2src | 1fmk | 162 | 20 – 82 | 7 | 4 |
| TK | 1kim | 1e2h | 22 | 21 – 40 | 3 | 0 |
| AR | 2ao6 | — | 74 | 20 – 84 | 5 | 1 |
| ERagonist | 1l2i | 2b23 | 67 | 20 – 93 | 7 | 2 |
| ERantagonist | 3ert | 2b23 | 39 | 21 – 93 | 6 | 3 |
| GR | 1m2z | — | 78 | 22 – 56 | 6 | 1 |
| MR | 2aa2 | — | 15 | 20 – 57 | 3 | 1 |
| PPARg | 1fm9 | 1prg | 81 | 20 – 65 | 3 | 2 |
| PR | 1sr7 | — | 27 | 20 – 54 | 4 | 1 |
| RXRa | 1mvc | 1g1u | 20 | 20 – 89 | 5 | 4 |
Protein target: ACE, angiotensin-converting enzyme; ADA, Adenosine deaminase; COMT, catechol O-methyltransferase; PDE5, Phosphodiesterase 5; DHFR, dihydrofolate reductase; GART, glycinamide ribonucleotide transformylase; FXa, factor Xa; AChE, acetylcholinesterase; ALR2, aldose reductase; AmpC, AmpC beta-lactamase; COX-1, cyclooxygenase-1; COX-2, cyclooxygenase-2; GPB, glycogen phosphorylase beta; HIVPR, HIV protease; HIVRT, HIV reverse transcriptase; HMGR, hydroxymethylglutaryl-CoA reductase; InhA, enoyl ACP reductase; NA, Neuraminidase; PARP, poly(ADP-ribose) polymerase; PNP, purine nucleoside phosphorylase; SAHH, S-adenosyl-homocysteine hydrolase; CDK2, cyclin-dependent kinase 2; EGFr, epidermal growth factor receptor kinase domain; FGFr1, fibroblast growth factor receptor kinase; HSP90, human heat shock protein 90; P38 MAP, P38 mitogen activated protein; SRC, tyrosine kinase SRC ; TK, thymidine kinase; AR, androgen receptor; ER, estrogen receptor; GR, glucocorticoid receptor; MR, mineralocorticoid receptor; PPARg, peroxisom e proliferator activated receptor gamma; PR, progesterone receptor; RXRa, retinoic X receptor alpha. PDB code (Holo), the X-ray structure of the protein target with a bound ligand. Number of ligands, the number of annotated binders of the protein target in the database. Sequence identity of templates, the range of sequence identity from all templates with respect to the holo X-ray structure of the protein target. Number of templates (Holo), the number of template structures cocrystallized with a ligand.
Comparative modeling
For each target-template pair, a comparative model was generated using MODELLER-9v2.60 The target-template alignment was calculated by profile-profile alignment as implemented in the “profile.scan” routine of MODELLER.61 For each alignment, 50 models were calculated with the standard “automodel” class. Next, the binding site loops were defined as those binding site residues that were not aligned to the template structure; the binding site residues, in turn, were defined as the residues with more than one non-hydrogen atom within 10 Å of any ligand atom in the target structure. The model with the best MODELLER objective function value was then subjected to a refinement of binding site loops with the standard “loopmodel” class; all the binding site loops were optimized simultaneously. 2500 target conformations were generated and the single refined model with the best objective function value was selected for virtual screening. Ligands, ions, and cofactors in the templates were copied to the target models and treated as rigid during both initial model building and binding site refinement, using the “BLK” functionality of MODELLER. In total, 222 models were generated based on 222 templates for the 38 test proteins (Table S3). 172 of the 222 templates are determined by X-ray crystallography at resolution better than 2.5 Å.
Evaluation of comparative models
The comparative models were assessed by the root-mean-square deviation of the non-hydrogen atoms in binding site residues between a model and the holo X-ray structure (RMSDbs), after superposition of the binding site atoms by MODELLER’s “superpose” routine. The holo X-ray structures and comparative models were also assessed by an atomic distance-dependent statistical potential called Discrete Optimized Protein Energy (DOPE).62 The normalized score (N-DOPE) derived from the raw DOPE score was used.63 The N-DOPE score can often discriminate between near-native (native overlap “NO3.5Å” > 0.8; N-DOPE < −1.0) and inaccurate models (NO3.5Å < 0.3; N-DOPE > 1.0).
Molecular docking
DUD ligands and decoys (98,266 molecules) were screened against the holo X-ray structure, the apo X-ray structure, the comparative models, and the template X-ray structures of each target. The apo X-ray structure, the comparative models, and the template structures were superposed on the corresponding holo X-ray structure using the binding site residues only (for template structures, the residues aligned to the binding site residues were used). Virtual screening against the 38 holo X-ray structures, 34 apo X-ray structures, 222 comparative models, and 222 template structures was performed using a semi-automated docking pipeline.57 Briefly, the protocol includes binding site preparation, sphere (hot spot) generation, scoring grids construction, and docking of DUD. The solvent-accessible molecular surface was calculated for the binding site residues using the program DMS.64 For the holo X-ray structure, 35 matching spheres serving to orient DUD compounds in the site were generated by augmenting the ligand-derived spheres with the receptor-derived spheres. The ligand-derived spheres were represented by the positions of non-hydrogen atoms of the ligand in the holo X-ray structure. The receptor-derived spheres were calculated using the program SPHGEN65 from the molecular surface of the binding site. The matching spheres generated in the holo X-ray structures were employed for the corresponding apo X-ray structures, the comparative models, and the template structures. The program DOCK 3.5.54 was used for docking.66-68 Presampled conformations of each compound were docked to the binding site and ranked by an energy function consisting of protein-ligand van der Waals interactions, protein-ligand electrostatic interactions, and a correction for ligand desolvation.
Evaluation of virtual screening results
The accuracy of virtual screening was evaluated by the enrichment for the known ligands among the top scoring DUD compounds. The conformation with the best docking energy of each compound was ranked and the enrichment factor was defined as
| (1) |
where ligandtotal is the number of known ligands in a database containing Ntotal compounds and ligandselected is the number of ligands found in a given subset of Nsubset compounds. EFsubset reflects the ability of virtual screening to find true positives among the decoys in the database compared to a random selection. An enrichment curve is obtained by plotting the percentage of actual ligands found (y-axis) within the top ranked subset of all database compounds (x-axis on logarithmic scale). To measure the enrichment independently of the arbitrary value of Nsubset, we also calculated the area under the curve (logAUC) of the enrichment plot:
| (2) |
where Δx is 0.1 in this study. A random selection (ligandselected / ligandtotal = Nsubset / Ntotal) of compounds from the mixture of actual ligands and decoys yields a logAUC of 14.5. A mediocre selection that picks twice as many ligands at any Nsubset as a random selection has logAUC of 24.5 (ligandselected / ligandtotal = 2 * Nsubset / Ntotal; Nsubset / Ntotal ≤ 0.5). A highly accurate enrichment that produces ten times as many ligands than the random selection has logAUC of 47.7 (ligandselected / ligandtotal = 10 * Nsubset / Ntotal; Nsubset / Ntotal ≤ 0.1).
It was suggested that the area under the curve for ROC plots (AUC) is used for reporting the enrichment results.69 In addition, to characterize the “early” enrichment, enrichment percentages at 0.5, 1, 2 and 5% of the docking database can be used. Here, we use a recently described measure that captures both aspects in one number, by using the logarithmic scale for the x-axis.70
When multiple structures are available (either models or templates), a consensus enrichment was calculated by combining the docking results of multiple structures. For each docked compound, the best docking score across all structures was used for ranking the compound. Thus, the ranking relied on optimizing the protein conformation as well as protein-ligand complementarity.
Comparison of ligand enrichment
A difference in ligand enrichment values for two individual structures or models was defined to be significant if larger than 3 in the logAUC units; otherwise, the enrichment values were considered to be comparable.
Statistical significance of the difference between the accuracies of two docking protocols was calculated by the following procedure. For each protocol, an enrichment value was calculated for each target in the benchmark set. The distribution of the protocol difference in enrichment values for each target was compared with the normal distribution using the Shapiro-Wilk test.71 If the difference was distributed normally at the confidence level of 95%, the dependent t-test for paired samples was performed to determine whether or not the differences between the two samples were statistically significant at this confidence level.72 Otherwise, the Wilcoxon signed-rank test was used.73 These analyses were preformed using the statistical package R (http://www.r-project.org).
Results
We begin by evaluating the comparative models in terms of their N-DOPE62, 63 score and RMSDbs from the holo X-ray structure of the target. For each of the 38 targets in the benchmark, the ability to identify known ligands is then compared for the holo X-ray structure, the apo X-ray structure, the individual comparative models, and their templates. Next, a consensus enrichment is calculated for each of the 38 targets by combining the docking scores against a variety of models (each based on a different template) and evaluated against enrichment values derived for single structures and models; a detailed analysis is performed for 8 targets. Finally, we describe 6 examples of docking against a comparative model that reproduced the known binding geometry of the ligand.
Comparative model evaluation
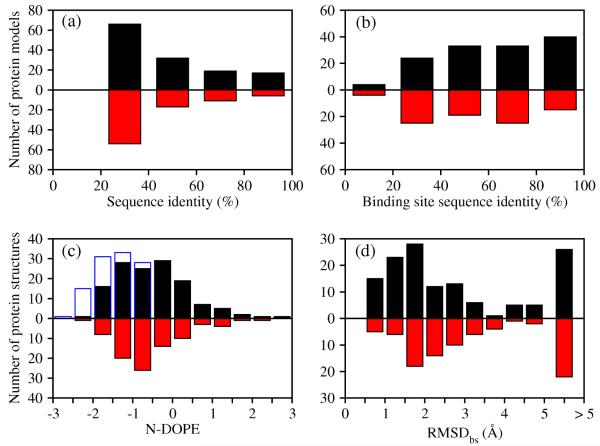
Among the 222 templates used for model generation, 134 templates are holo X-ray structures and 88 templates are apo X-ray structures. For both holo and apo templates, the number of structures decreases with an increase in the target-template sequence identity (Figure 1). In contrast, the sequence identity of the binding sites is distributed evenly, reflecting the relative conservation of the functional regions in evolution. The holo X-ray structures and comparative models of the 38 targets were assessed by the normalized DOPE score (N-DOPE). 34% of the models based on holo templates received N-DOPE scores below −1.0, 74% of the models received scores below 0.0, and 93% of the models received scores below 1.0. Nearly the same distribution of N-DOPE scores was found for models based on apo templates (33% below −1.0, 78% below 0.0, and 93% below 1.0). 49% of the models based on holo X-ray templates had RMSDbs to their holo X-ray structure of less than 2.0 Å. A smaller fraction (33%) of the models based on apo templates showed the same deviation. Thus, we expect that the comparative models used here are representative of the whole range of comparative models likely to be used for ligand docking in real applications.
Figure 1. Composition of the benchmark.
(a) Distribution of the target-template sequence identity for holo templates (black bars) and apo templates (red bars). (b) Distribution of the target-template binding site sequence identity. (c) Distribution of N-DOPE statistical potential score for comparative models (black and red bars for models based on holo and apo templates, respectively) and target X-ray structures (empty blue bars). (d) Distribution of root-mean-square deviation of the non-hydrogen atoms in binding site residues (RMSDbs) between a comparative model and a target X-ray structure.
Ligand enrichment for an individual model
The performance of comparative models in virtual ligand screening was evaluated by the percentage of area under the enrichment curve (logAUC) and compared to that from both the holo and apo X-ray structures (Table 2). For 14 targets, the holo X-ray structure outperformed all comparative models; for 9 targets, the enrichment of the holo X-ray structure was comparable to that of the most enriching model; for the remaining 15 targets, the most enriching model outperformed the holo X-ray structure. Averaged over all 38 targets, the average logAUC for the holo X-ray structures is 30.6, which is only slightly higher than the average logAUC for the most enriching models (28.7). The difference between a holo X-ray structure and a most enriching model is not statistically significant. Considering the 34 targets for which both holo and apo X-ray structures were available, the average logAUC is 29.8 for the holo X-ray structures, 20.2 for the apo X-ray structures, and 28.7 for the most enriching models. For either the 38-target sample or the 34-target sample, the most enriching model tends to be significantly more enriching than the apo X-ray structure. We calculated binding site volumes for all models of the 38 targets.74 There is no significant correlation between the enrichment and model binding site volume: The most enriching model has the largest / smallest volume of all target models in 7 / 6 out of the 38 cases.75
Table 2. Ligand enrichment using X-ray structures and multiple comparative models.
| Protein target |
Most enriching model | Best model by sequence identity |
Consensus Enrichment |
Holo X-ray Enrichment |
Apo X-ray Enrichment |
||
|---|---|---|---|---|---|---|---|
| Sequence Identity [%] |
Enrichment | Sequence Identity [%] |
Enrichment | ||||
| ACE | 44.1 | 50.2 | 54.8 | 37.9 | 48.9 | 40.6 | 44.1 |
| ADA | 26.5 | 40.3 | 84.8 | 38.8 | 41.1 | 22.7 | 46.3 |
| COMT | 20.1 | 0.0 | 20.1 | 0.0 | 0.0 | 27.6 | 1.9 |
| PDE5 | 30.4 | 22.1 | 95.5 | 20.5 | 18.6 | 12.1 | 26.3 |
| DHFR | 27.9 | 34.6 | 37.3 | 13.1 | 20.3 | 18.9 | 67.2 |
| GART | 40.4 | 27.5 | 41.5 | 0.6 | 27.4 | 35.3 | 3.7 |
| FXa | 30.4 | 52.1 | 63.8 | 27.2 | 49.6 | 41.8 | 19.1 |
| Thrombin | 30.0 | 43.4 | 39.5 | 36.9 | 42.4 | 29.4 | 56.2 |
| Trypsin | 78.8 | 38.0 | 78.8 | 38.0 | 32.3 | 29.3 | 9.5 |
| AChE | 38.0 | 35.7 | 92.7 | 17.3 | 29.1 | 38.5 | 15.5 |
| ALR2 | 68.7 | 41.9 | 86.6 | 39.5 | 37.1 | 39.7 | 24.6 |
| AmpC | 76.6 | 46.6 | 76.6 | 37.2 | 40.3 | 47.4 | 1.3 |
| COX-1 | 63.4 | 26.2 | 63.4 | 26.2 | 25.3 | 28.3 | 24.1 |
| COX-2 | 64.4 | 13.3 | 64.7 | 8.2 | 12.4 | 40.8 | 39.3 |
| GPB | 41.8 | 8.9 | 80.7 | 6.5 | 6.8 | 17.1 | 23.9 |
| HIVPR | 49.0 | 30.5 | 96.9 | 4.8 | 23.7 | 11.9 | 31.3 |
| HIVRT | 97.7 | 12.9 | 98.9 | 0.0 | 12.9 | 25.8 | 5.2 |
| HMGR | 100.0 | 43.2 | 100.0 | 43.2 | 41.5 | 40.9 | 44.7 |
| InhA | 29.0 | 14.6 | 29.0 | 14.6 | 11.8 | 8.2 | 10.1 |
| NA | 28.8 | 51.2 | 34.2 | 20.4 | 42.6 | 47.6 | 33.9 |
| PARP | 46.4 | 9.2 | 86.8 | 6.4 | 6.3 | 8.2 | 7.3 |
| PNP | 36.9 | 33.5 | 93.1 | 19.3 | 23.9 | 49.1 | 18.7 |
| SAHH | 84.7 | 82.4 | 84.7 | 82.4 | 78.4 | 82.8 | 20.3 |
| CDK2 | 27.6 | 13.5 | 94.9 | 10.1 | 11.7 | 11.3 | 10.4 |
| EGFr | 37.7 | 13.2 | 38.1 | 12.3 | 10.7 | 21.5 | 16.0 |
| FGFr1 | 41.3 | 12.3 | 51.0 | 6.2 | 11.9 | 6.7 | 4.9 |
| HSP90 | 62.4 | 18.7 | 70.4 | 14.5 | 11.5 | 24.6 | 4.1 |
| P38 MAP | 29.4 | 31.4 | 62.2 | 21.2 | 15.6 | 12.5 | 10.4 |
| SRC | 68.8 | 13.8 | 82.0 | 13.4 | 12.3 | 9.5 | 10.3 |
| TK | 20.7 | 13.4 | 40.4 | 3.0 | 2.9 | 63.5 | 3.9 |
| AR | 56.0 | 31.7 | 84.3 | 31.5 | 14.5 | 48.2 | — |
| ERagonist | 93.1 | 41.0 | 93.1 | 41.0 | 19.6 | 55.4 | 40.3* |
| ERantagonist | 21.0 | 19.2 | 93.0 | 9.0 | 14.7 | 23.2 | 2.6 |
| GR | 27.4 | 18.8 | 56.6 | 15.2 | 16.7 | 20.5 | — |
| MR | 56.8 | 30.5 | 56.8 | 30.5 | 29.5 | 57.0 | — |
| PPARg | 20.2 | 13.2 | 65.0 | 9.2 | 8.8 | 4.4 | 11.3 |
| PR | 53.5 | 35.9 | 53.5 | 35.9 | 11.8 | 23.2 | — |
| RXRa | 87.0 | 26.9 | 88.8 | 2.0 | 11.2 | 37.9 | 0.0 |
Sequence identity, the sequence identity of the complete template and target sequences. Enrichment, the ligand enrichment is represented by logAUC. Consensus Enrichment, logAUC calculated from docking scores against all target models. Holo X-ray Enrichment, the enrichment for the holo X-ray structure of the target. The enrichment for the models is in bold when it is larger by 3 units than that for the holo X-ray structure.
the ER apo structure used for both the ER agonist and antagonist contains no ligands but adopts the conformation close to the ER agonist due to the binding of a glucocorticoid receptor-interacting protein 1 NR box II peptide.
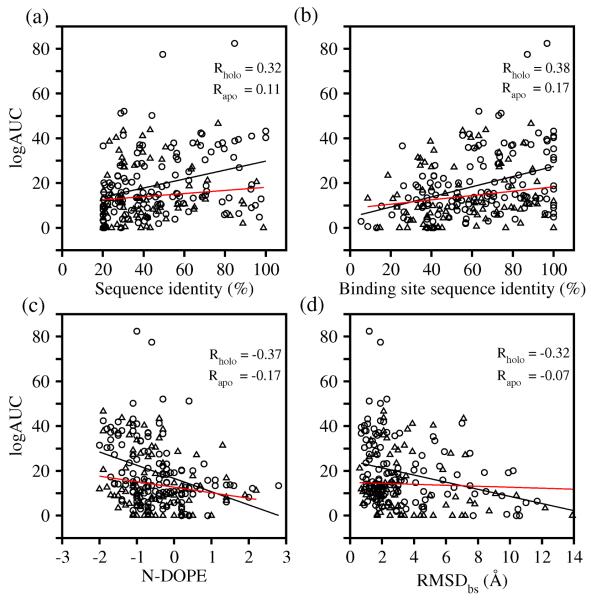
These results suggest that the ability to identify the most enriching model of the target in the absence of knowing the actual ligands would be invaluable. Such an identification might be achieved by one or more predictors of model accuracy. Therefore, we asked whether there were model properties that might predict the most enriching model among multiple models based on different template structures. In fact, we found no good criterion for such a prediction. There is essentially no correlation between the ligand enrichment and template resolution ( R = -0.16 ). The ligand enrichment is correlated only weakly with the accuracy of the binding site as measured by RMSDbs ( R = -0.32 ) (Figure 2), which is in any case unknown in the absence of the defined holo structure. We also calculated correlation coefficients between the ligand enrichment (logAUC) and three different properties of the model (Figure 2). There were only weak correlations of enrichment with the binding-site sequence identity ( R = 0.38 ), the overall sequence identity ( R = 0.32 ), and the N-DOPE score ( R = -0.37 ). For 38 targets, the average logAUC of the most enriching models was significantly higher than that of the models predicted to have the best enrichment using any of three criteria (average logAUC were 21.0, 20.9, and 18.6 for the best-assessed models selected by binding-site sequence identity, the overall sequence identity, and the N-DOPE score, respectively). These criteria were thus unreliable predictors of docking success judged by ligand enrichment.
Figure 2. Model features do not predict docking success.
Circles and triangles correspond to models built from holo- and apo-templates, respectively.
Ligand enrichment for multiple models
Because the tested sequence and/or structural attributes were unreliable predictors of ligand enrichment, we turned to calculating enrichment using a “consensus” compound rank by combining multiple models, each based on a different template (Figure 3). For 11 and 7 of the 38 targets, the consensus enrichment for multiple models is respectively better and comparable to that for the holo X-ray structure. This compares to 15 and 9 for the most enriching model decided post hoc. For 16 and 8 of the 34 targets with an apo structure, the consensus enrichment is respectively better and comparable to that for apo structures; the average consensus enrichment is 23.6. The consensus enrichment is significantly better than the enrichment for the best-assessed model identified by the sequence identity. The difference between the consensus enrichment and the enrichment for the apo X-ray structure is not distinguishable at the 95% confidence level, but is statistically significant at the 85% confidence level. Consensus enrichment benefits from a de facto treatment of receptor flexibility through consideration of multiple target conformations, each one corresponding to a different comparative model based on a different template structure.
Figure 3. Enrichment plots for the 38 protein targets.
Enrichment curves for the holo X-ray structure (black line), the apo X-ray structure (blue line), the consensus enrichment for multiple models (red line), and random selection (dotted line).
Two protocols further exploring the conformational space of the target were compared by their corresponding enrichments. In the first protocol, four new templates with sequence identities spanned by the original set of templates were added for each target; thus the number of models used for consensus enrichment calculations varied from 5 to 13 per target. In the second protocol, the original set of templates was used, but for each template the five models with the best MODELLER objective function values were used, not only the best scoring model; thus, the number of models used for consensus enrichment calculations was five times the number of templates, varying from 5 to 45.
These two protocols were tested on 10 of the original 38 targets (including six enzymes, two of which are kinases, and four hormone receptors) (Table 3). Using the first protocol, 4 targets yielded improved enrichment and 6 targets yielded enrichment comparable to that for the initial set of models; in no case did enrichment decrease. Using the second protocol, no clear trends were observed: 1 target yielded improved enrichment, 2 targets yielded worse enrichment, and 7 targets yielded enrichment comparable to that for the initial set of models. Thus, it appears that ligand enrichment is better optimized by adding additional templates than by adding models based on the same template and with suboptimal values of the MODELLER objective function.
Table 3. Consensus enrichment using additional models.
| Protein target | Initial | Addition of templates | Addition of suboptimal models |
|---|---|---|---|
| COMT | 0.0 | 10.2 | 0.0 |
| GART | 27.4 | 27.5 | 20.6 |
| Thrombin | 42.4 | 55.8 | 47.1 |
| GPB | 6.8 | 6.9 | 6.2 |
| EGFr | 10.7 | 9.9 | 10.2 |
| TK | 2.9 | 2.5 | 3.0 |
| AR | 14.5 | 14.7 | 11.6 |
| ERantagonist | 14.7 | 14.1 | 16.9 |
| MR | 29.5 | 43.9 | 21.0 |
| PPARg | 8.8 | 31.0 | 7.4 |
Initial, the consensus enrichment achieved by combining docking score for a set of models, each one of which is based on a different template (Table 2). Addition of templates, 4 templates were added to the initial set of templates. Addition of suboptimal models, the models with top five MODELLER scores (not only the top model) were selected for each of the original templates.
Docking to templates instead of comparative models
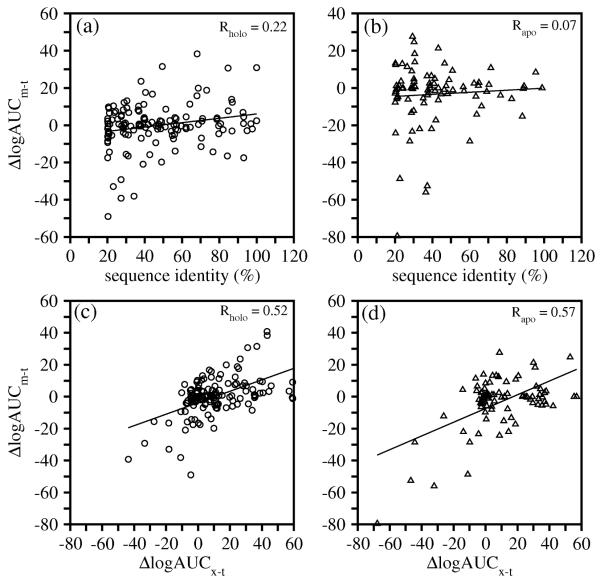
Comparative models rather than template structures are often used as targets for ligand screening. The comparative models and the corresponding template structures determined by X-ray crystallography were compared by their ability to identify the known ligands (Table S1). To our surprise, no statistically distinguishable difference in ligand enrichment was found between the modeled structures and the templates, using either the most enriching model/template, the model/template with the highest sequence identity, or the consensus enrichment combining all models/templates. We examined the difference between the enrichments for each model-template pair ( Δ log AUCm-t = log AUCmodel - log AUCtemplate ). Among the 222 model-template pairs, models were better, comparable and worse than their templates for 30%, 39% and 31% of the pairs, respectively.
Δ log AUCm-t is correlated weakly with the target-template sequence identity ( Rholo = 0.22, Rapo = 0.07 ) (Figure 4a, 4b). Several models based on 20 – 40% sequence identity to their templates yielded much worse enrichments than their templates. A much stronger correlation was found between Δ log AUCm-t and the enrichment difference between the holo X-ray structure of the target and the template ( Δ log AUCx-t ) (Figure 4c, 4d). For both holo and apo model-template pairs, the correlation coefficients are above 0.5. In other words, the enrichment for a comparative model relative to its template is correlated with the enrichment for the target relative to its template. For instance, the target structure (PDBID 3dfr) of dihydrofolate reductase (DHFR) from Lactobacillus casei yielded a ligand enrichment of 18.9. One template (PDBID 1rc4), which is also a DHFR structure but from Escherichia coli, yielded an enrichment of 62.5. The comparative model based on this template showed an enrichment of 23.3, higher than that of the target structure. This result suggests that a template orthologous to the target may be more enriching than the target itself, whether the target structure is determined by X-ray crystallography or predicted by comparative modeling.
Figure 4. Comparing ligand enrichments for comparative models and their templates.
Circles and triangles correspond to models based on holo- and apo-templates, respectively. (a, b) Scatter plots of the difference between the enrichments for a comparative model and the corresponding template ( Δ log AUCm-t = log AUCmodel - log AUCtemplate ) versus the target-template sequence identity. (c, d) Scatter plots of Δ log AUCm-t versus the difference between the enrichments for the target holo X-ray structure and the template ( Δ log AUCx-t = log AUCx-ray - log AUCtemplate ).
We hypothesized that comparative models achieve a better enrichment relative to the template structures when the target and template binding profiles are dissimilar. We obtained the corresponding subset of target-template pairs by eliminating orthologous pairs72 as well as nuclear hormone receptor pairs involving AR, GR and MR that bind very similar ligands. For the remaining 124 paralagous target-template pairs, models were better, comparable and worse than their templates for 34%, 40%, and 26% of the pairs, respectively, in agreement with our hypothesis. Moreover, we also hypothesized that comparative models achieve a better enrichment relative to the template structures when comparative models are relatively accurate (i.e., the targets are not too different from the templates). Thus, we also removed from our benchmark the 37 target-template pairs with less than 25% sequence identity, leaving 39% of the original 222 target-template pairs. As a result, the models were better, comparable and worse than their templates for 42%, 41%, and 17% of the pairs, respectively, again in agreement with our hypothesis.
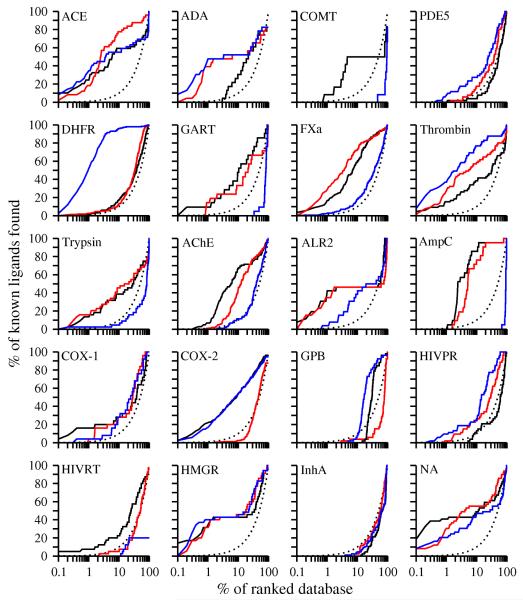
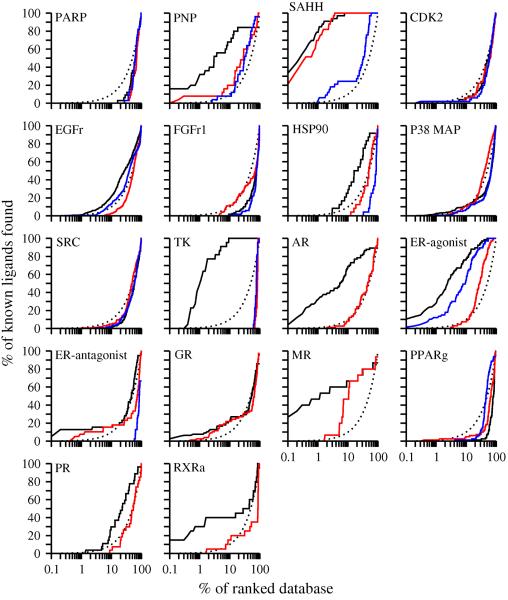
Detailed analysis of ligand enrichment for 8 targets
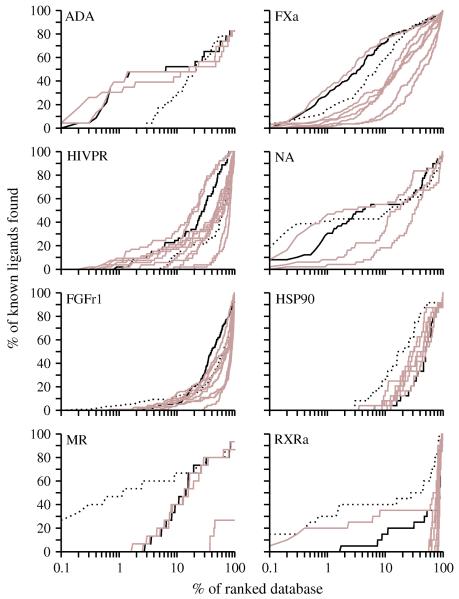
The enrichment for 8 targets, including 6 enzymes and 2 hormone receptors, was examined in detail (Figure 5).
Figure 5. Sample enrichment curves.
For 8 targets (6 enzymes, one of which is a kinase, and 2 hormone receptors), enrichment curves are plotted for the holo X-ray structure (dotted line), the consensus based on multiple models (black line), and each single model (brown lines).
Adenosine deaminase (ADA) is a metalloenzyme, having a large binding pocket with a catalytic zinc ion coordinated by three histidine residues.76, 77 Electrostatic interactions play an important role during ligand binding to this enzyme. The holo X-ray structure of the target is bound to a non-nucleoside inhibitor. Two other X-ray structures of adenosine deaminase were used as templates: one is an apo structure with 26% sequence identity to ADA; the other binds a purine nucleoside analog and has 85% sequence identity to ADA. The enrichments (logAUC) for the two models were 40.3 and 38.8, much better than the logAUC of 22.7 for the holo X-ray structure (Table 2). Combining the docking results for both models, the consensus enrichment increased to 41.1. The poorer enrichment using the holo X-ray structure is not surprising in this case. Most known ligands of ADA are nucleoside analogs that form the major part of the ADA-like ligands in DUD. The target structure, binding to a relatively hydrophobic non-nucleoside inhibitor, undergoes a conformational change that exposes hydrophobic patches. These patches may not be exposed when the target binds less hydrophobic ligands in DUD, rationalizing why the holo X-ray structure with a non-nucleoside inhibitor is less enriching than comparative models based on a more representative holo X-ray structure. In fact, the two templates also yielded better enrichment than the target structure (28.9 and 31.5, respectively). This example illustrates the benefits of virtual screening with multiple models based on different templates.
Factor Xa (FXa) converts prothrombin to thrombin, which is the last enzyme in the coagulation cascade and is responsible for fibrin formation, platelet activation, and other physiological events.78, 79 The binding site of FXa can accommodate structurally diverse inhibitors. In the holo X-ray structure, the inhibitor binds in an extended conformation through an ionic S1 pocket and a hydrophobic S4 pocket.80 In the holo X-ray structure, the S2 pocket is blocked by Tyr99. Seven templates were used, with sequence identity to the target ranging from 24% to 64%. The model based on a holo template with 30% sequence identity yielded the highest enrichment 52.1, while the model based on an apo template with 24.0% sequence identity yielded the lowest enrichment 15.3. The consensus enrichment was 49.6, close to that achieved by the most-enriching model. The binding site in the most enriching model was similar to that in the holo X-ray structure. The S1 subsite mimicked the X-ray conformation and formed hydrogen bonds with most ligands through the anchor residues Asp189 and Gly219. The S4 subsite is lined by the aromatic rings of Phe174 and Tyr99 that are almost parallel with each other and perpendicular to the Trp215 ring. In the holo X-ray structure, these residues close down on the ligand and make the nearby subsite S2 inaccessible. In the most enriching model, the sidechain of Tyr99 moved toward the outside of the S4 pocket and changed its χ1 by 90°. The modeled S4 pocket thus became 14% larger in volume than that in the holo X-ray structure and presumably capable of binding a larger variety of ligands.75 This hypothesis is supported by the improved enrichment relative to that for the holo X-ray structure. Thus, FXa provides another example of why multiple comparative models may be more useful for virtual screening than a single holo X-ray structure.
HIV protease (HIVPR) is a key enzyme for the production of infectious virus particles and is an important target for antiviral AIDS drugs. Numerous classes of substrate- and structure-based inhibitors have been designed, tested, and co-crystallized with the enzyme. The active site contains two extended β-hairpins (i.e., flaps) that sequester it from water and two catalytic aspartyl residues (Asp25 and Asp25′) at the bottom of the ligand binding cavity.81 In the holo X-ray structure of the target, the protein binds a pentapeptide mimic in an extended conformation.82 This X-ray structure didn’t yield a high enrichment (11.9) because many other known ligands do not fit into this particular conformation. 12 templates were found for HIVPR with a wide range of sequence identity from 23% to 97%. The highest enrichment, 30.5, was obtained from a model based on an apo template with 49% sequence identity. The improved enrichment for this model benefited from both the opening of the flaps that allowed most ligands to fit into the cavity, and the conservation of sidechain conformations at the bottom of the cavity, including the two catalytic Asp residues. The consensus enrichment was 23.7, lower than that for the most enriching model but better or comparable to the enrichments for all other models. The docking energies used to calculate the consensus enrichment were gathered from 11 models, among which 6 models contributed more than 10% of the scores of the known ligands each. This case again illustrates the advantages of using comparative models in different conformational states over a single holo X-ray structure.
Neuroaminidase (NA) is one of two glycoproteins expressed on the surface of the influenza virus and is responsible for the release of viruses from infected cells; the enzyme is the target for the anti-influenza drugs such as oseltamivir (Tamiflu) and zanamivir (Relenza), the latter of which was developed by structured-based design.83 The holo X-ray structure84 showed a better enrichment for a small fraction of the ranked database than two models (Figure 5), with a logAUC of 47.6. One model based on a holo template with 29% sequence identity yielded the highest enrichment, 51.2. This modeled binding site was similar to that of the holo X-ray structure. The consensus enrichment was 42.6, again close to that for the most enriching model.
Fibroblast growth factor receptor kinase (FGFr1) is a difficult target for virtual screening due to the receptor flexibility and the exposed binding site. Neither the holo X-ray structure in the DFG-in conformation nor the eight comparative models yielded enrichments above random. The highest enrichment was 12.3 from the model based on an apo template with 41% sequence identity. The consensus enrichment was 11.9, close to the highest enrichment observed for this target.
Heat shock protein 90 (HSP90) is an ATP binding protein responsible for stabilizing partially folded forms of many proteins. The holo X-ray structure yielded the enrichment for 24.6. Among the eight models built for this target, the model based on an apo template with 62% sequence identity achieved the highest enrichment for 18.7. However, two models, one based on a holo template with 20% sequence identity and the other one on an apo template with 43% sequence identity failed because of the distortion of the binding site. The consensus enrichment was 11.5, worse than the enrichments for the other six models.
Mineralocorticoid receptor (MR) is a hormone receptor that integrates hormonal signaling and activates the expression of aldosterone target genes, which control several physiological processes including disorders of the nervous system, hypertension, and cardiac failure. The holo X-ray structure of the target binds aldosterone in a fully enclosed pocket. The A-ring ketone of aldosterone makes hydrogen bonds with the sidechains of Gln776 and Arg817. On the other side of the cavity, the hydroxyl and ketone groups in the C-ring and D-ring make hydrogen bonds with the sidechains of Asn770 and Thr94585. The X-ray structure ligand is further pinned down by hydrophobic interactions with the binding site. The enrichment obtained using the holo X-ray structure is 57.0. Four models were generated based on templates with sequence identities from 20% to 57%. Among them, the model based on the holo X-ray structure of progesterone receptor (PR) with 57% sequence identity yielded the best enrichment, 30.5. The consensus enrichment for multiple models is 29.5, close to the highest enrichment obtained. The PR structure used as the template for the most enriching model binds a nonsteroidal ligand tanaproget (TNPR).86 In comparison to the aldosterone binding in the holo X-ray structure of the actual target, the 1-methyl-1H-pyrrole-2-carbonitrile ring of TNPR lies approximately between the A and B rings of aldosterone, while the 1,4-dihydro-3,1-benzoxazine-2-thione moiety lies close to the C and D rings of aldosterone. Like the A-ring ketone in aldosterone, the nitrile group of TNPR also hydrogen bonds with the mainchain amido groups of the Gln and Arg residues. The benzoxazine amide group at the distal end of TNPR forms hydrogen bonds with the Asn sidechain. TNPR binds similarly as the aldosterone, forming hydrophobic contacts with the PR cavity. The geometry of hydrogen bonds and hydrophobic interactions in the most enriching model was similar to those in the holo X-ray structure with aldosterone. However, two differences were observed. First, the Arg817 sidechain moved out of the modeled binding site because of the nitrile group in TNPR. Arg817 thus became less capable of forming hydrogen bonds with the A-ring ketone. Second, the Thr945 sidechain in the target structure formed hydrogen bonds with the aldosterone, while the corresponding Thr in the PR template formed no hydrogen bonds with TNPR. Thus, Thr945 in the modeled binding site based on the PR template was incapable of hydrogen bonding with MR ligands. These two changes explain the decrease in the ligand enrichment obtained using the most enriching model relative to the holo X-ray structure.
Retinoic X receptor α (RXRa) is also a hormone receptor, mediating the biological effects of retinoids. RXRa uniquely forms heterodimers with other nuclear receptors, including peroxisome proliferator-activated receptor (PPAR). Furthermore, RXRa can heterodimerize in response to its ligands. Heterodimerization of RXRa can mediate diverse endocrine signaling pathways.87 Hydrophobic interactions play an important role in the binding of the synthetic ligand BMS649 to the X-ray structure of the target. The carboxylate group of BMS649 also forms ionic interactions with the basic residue Arg316 and a hydrogen bond with the backbone amide of Ala327. Nine templates with a range of sequence identity from 20% to 89% were used to build comparative models. The enrichment obtained using the holo X-ray structure is 37.9. Among the 9 models, only the model based on a holo template with 87% sequence identity yielded an enrichment above random (26.9). The consensus enrichment was 11.2, which is comparable to random selection. The decrease of ligand enrichment using the most enriching model relative to the holo X-ray structure is rationalized by a distortion of the anchor residue Arg316. In the holo X-ray structure of the target, both amines contributed to the ionic interactions with BMS649. However, in the modeled binding site, the Arg316 sidechain is rotated, so that only one guanidinium nitrogen points into the pocket. This rationalization is supported by the docking poses and energies of the known ligands, in comparison with BMS649. For instance, the known ligand Bexarotene docked into the modeled binding site similarly as in the holo X-ray structure, but received a less favorable docking energy due to a decrease in the electrostatic interaction energy.
Accuracy of the docking geometry in modeled binding sites
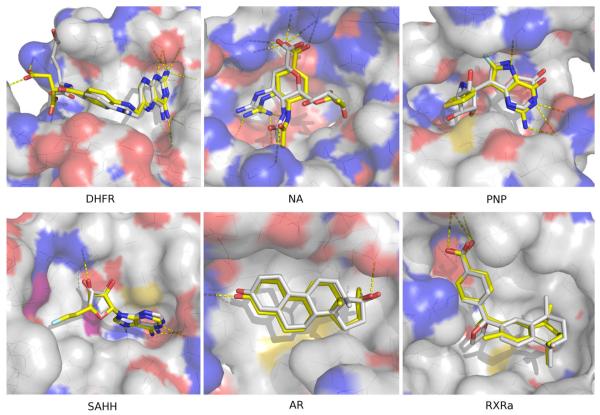
Modeling and docking were evaluated primarily by their ability to identify known ligands among top-ranking docked molecules. We have not focused on predicting the conformation of each target in complex with its known ligands. Nevertheless, the predicted poses of a number of ligands turned out to be modeled relatively accurately, as illustrated by four enzymes and two hormone receptors (Figure 6).
Figure 6a. Binding poses of 6 protein targets.
For each target, the docking pose of one known binder with one comparative model of the target was selected. The ligand in complex with the crystallographic structure of the target protein was always located in the background. The targets include two hormone receptors AR and RXRa, four enzymes DHFR, NA, PNP and SAHH.
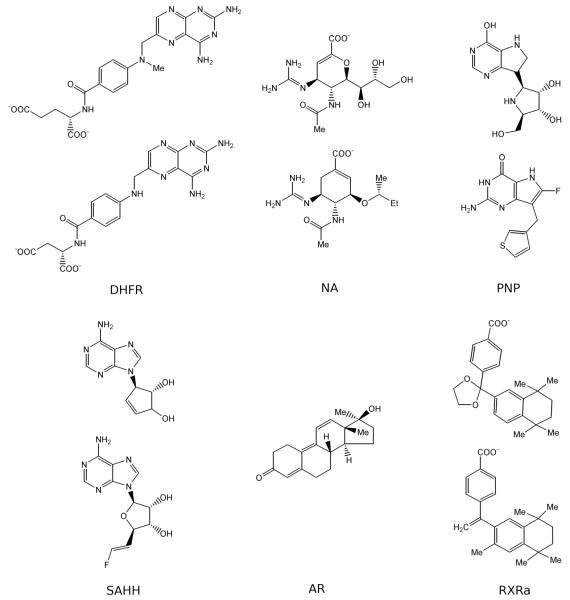
In the target structure of DHFR, methotrexate binds to the large half-buried binding site with a neighbouring cofactor NADP. A comparative model was generated using another DHFR structure as a template that shares 27% sequence identity to the target but only binds methotrexate without NADP. Aminoanfol, which contains the same diamino-pteridine and benzoic acid groups as methotrexate, was ranked 72 among DUD molecules (top 0.1%). The docked pose of aminoanfol accurately reproduced that of the co-crystallized methotrexate, resulting in an overlap of the equivalent diamino-pteridine and the benzoic acid groups. In the target structure of NA, zanamivir binds to the shallow and solvent-exposed binding site with essential hydrogen bonding interactions. A comparative model was calculated based on another neuraminidase structure with 29% sequence identity to the target and a bound sialic acid. In the modeled binding site, a carboxylic acid analog, ranked 102 among DUD molecules, overlapped with the co-crystallized zanamivir on the guanidine, acetamido and carboxylic acid groups (Figure 6b), and reproduced the hydrogen bonding interactions in the target structure. The target structure of purine nucleoside phosphorylase (PNP) binds simultaneously immucillin H and an inorganic phosphate. A comparative model is generated using another PNP structure as a template that shares 37% sequence identity to the target and binds both an immucillin H analogue and a phosphate. The purine group of a ligand, ranked 9 among DUD molecules, overlapped with the purine group of the immucillin H in the target. The target structure of s-adenosyl-homocysteine hydrolase (SAHH) binds an adenosine derivative and a neighboring cofactor NADP. A comparative model was calculated based on another SAHH structure with 49% sequence identity to the target, a bound adenosine and a neighboring cofactor NADP. In the modeled binding site, an adenosine derivative, ranked 15 among DUD molecules, overlapped with the adenosine in the target structure on both the adenine and the ribose rings. In the target structure of androgen receptor (AR), metribolone binds to the deeply buried binding site through hydrophobic interactions as well as hydrogen bonds using its ketone and hydroxyl groups. A comparative model was generated using another AR structure as a template that shares 70% sequence identity to the target and binds tetrahydrogestrinone. In the modeled binding site, metribolone, ranked 1167, accurately reproduced its binding geometry in the target. In the target structure of retinoic X receptor α (RXRa), the ligand BMS649 binds to the L-shaped and elongated binding pocket through hydrophobic interactions87 as well as a hydrogen bond using its carboxylate group. A comparative model was calculated based on another RXR structure with 87% sequence identity to the target and a bound retinoic acid. Using this model, bexarotene, ranked 26 among DUD molecules, overlapping with the co-crystallized BMS649 on both the benzoic acid group and the tetramethylnaphthalene group.
Figure 6b. 2D images of ligands in Figure 5a.
The ligands used in Figure 5a are presented. For AR, the ligands crystallized in the structures of target proteins are the same as the docked ligands. For the other 5 targets, the X-ray structure ligands are shown on top of the docked ligands.
Receptor-based matching spheres
In this study, the docking protocol depends on the protein and ligand “matching spheres” to position small molecules in the binding site. Ideally, to reduce the size of the search space, the protein matching spheres should cover only the binding site surface and not other regions of the protein; similarly, the ligand matching spheres should cover only the space occupied by the ligands. For a holo X-ray structure, the matching spheres can be calculated in a straightforward manner from the knowledge of the protein-ligand complex coordinates (Methods), although even in that case they may under- or over-estimate the available protein and ligand volumes for other ligands. For the apo X-ray structures and comparative models, exactly the same matching spheres were used in our benchmark. This computation may have introduced a bias in our results; in realistic applications, the holo X-ray structure will often not be available. In principle, this bias could be either positive or negative. For example, using the “holo X-ray” matching spheres can productively restrict the sampling space compared to a looser definition of the binding site and lead to a reasonable enrichment (e.g., the most enriching model of HIVPR); alternatively, restricting the sampling too much based on a small ligand in the holo X-ray structure may hurt identifying larger ligands (e.g., using the receptor-based spheres, the most enriching thrombin model yielded enrichment better than that using the “holo X-ray” matching spheres).
We investigated a potential benchmarking bias resulting from using “holo X-ray” matching spheres for docking against apo structures and models. For each of the 38 targets, 35 “binding site” matching spheres were generated independently for each structure (the holo X-ray structure, the apo X-ray structure, and each of the comparative models), based on the binding site residues identified in the holo X-ray structure (in many real applications, this information would have to be obtained from some other source); the ligand matching spheres were not used to restrict the sampling of the ligand position and conformation. On average, the best enrichment and consensus enrichment decrease by 10% (3 units) and 16% (4 units) respectively, when using “binding site” matching spheres instead of “holo X-ray” matching spheres (Table S2), presumably due to omitting ligand matching spheres. However, the order of enrichment for different types of structures and models remains unchanged, thus supporting our benchmarking.
Discussion
Overview
Three key results emerge from this study. First, if multiple models based on different templates are used, one can frequently find at least one model that outperforms even the holo X-ray structure of the target. This encouraging result is mitigated by our inability to predict the optimal model for ligand enrichment in the absence of knowing the actual ligands — the enrichment shows little relationship to standard metrics of model attributes such as sequence identity or protein-structure-based scores.
Second, we find that over the DUD set the template structures typically perform as well as the comparative models that are built from them. This result is dispiriting at a first glance, suggesting that models having the correct sequence are not better targets than the template experimental structures from which they are built. However, it is 2.5 times as likely that a model is more enriching than the corresponding template, when the target and template binding profiles are dissimilar (i.e., the target and template are not orthologs) and when comparative models are relatively accurate (i.e., they are based on more than 25% sequence identity).
These first two observations lead to a final point: If models built based on different templates are considered, the corresponding consensus enrichment frequently outperforms the enrichment yielded by the apo X-ray structure of the same target. Whereas these consensus enrichments still tend to be outperformed by the enrichments yielded by the holo X-ray structures, they are often competitive with them. We consider each of these points in turn.
Models versus target structure
We assessed the utility of comparative models in virtual ligand screening, through comparison with the holo and apo X-ray structures. The results showed that docking against comparative models frequently substantially enriches known ligands (Table 2). For 15 targets, the most enriching model is better for virtual screening than the holo X-ray structure; for 9 targets, the most enriching model is as good as the holo X-ray structure. 27 most enriching models and 29 holo X-ray structures yielded better ligand enrichment than random selection. This result suggests that the conformational space spanned by multiple comparative models, each based on a different template, will overlap to some degree with an ensemble of conformations for the receptor in complex with different ligands. However, we could not find any features of comparative models or templates that reliably predict the most enriching model (Figure 2). This result is consistent with two earlier studies39, 55 and can be rationalized as follows. The docking library contains multiple ligands for proteins in the target family. These ligands may have different affinities for different receptor conformations. One comparative model may yield a better enrichment than another model based on a more similar template sequence but lesser binding affinity for the target ligands.
Model versus template structure
Our results are consistent with the observation on a smaller benchmark that docking to a comparative model is on average as successful as docking to its template.55 In fact, for our benchmark as a whole, we found that it is approximately equally likely a model will be more enriching, comparable, or less enriching than a template (Table S1, Figure 4). There are two explanations for this observation.
First, many target-template pairs are orthologous (i.e., belonging to the same family and thus having very similar ligand binding profiles). In such cases, there is no reason to expect templates would on average result in a lower enrichment for the target ligands than a target model or even the target X-ray structure (Figure 4c, 4d). For example, in the case of dihydrofolate reductase (DHFR), models were built based on 8 templates that are orthologous DHFRs from different species. 6 of these 8 templates yielded better ligand enrichments than the target structure, and 4 out of the 6 templates resulted in comparative models that yielded an enrichment better or comparable to that of the holo X-ray structure. As expected, when the orthologous template-target pairs are excluded, comparative models enrich the ligands slightly better than the templates.
Second, models and templates also result in comparable (low) enrichment when both the template and the model are an inaccurate representation of the target holo structure. Such a situation tends to arise when the target-template sequence similarity is low (i.e., less than 25% sequence identity). As a special case, errors in the modeling of induced fit can explain why the holo models are more likely than the apo models to be more enriching than their templates (Figure 4a, 4b). When the 37 target-template pairs with less than 25% sequence identity are removed from the 124 paralogous target-template pairs (out of the 222 pairs in our benchmark), the enrichment obtained for models was better, comparable and worse than that of their templates for 42%, 41%, and 17% of the pairs, respectively.
Consensus enrichment
Most proteins are flexible, adopting different conformations when binding to different ligands. Thus, methods that can handle this variability are needed.10, 11, 88-92 The receptor variability has been considered during docking by using a “soft” representation of the receptor,93-95 minimizing the scoring function with respect to sidechain orientations,96-102 and allowing protein flexibility via molecular dynamics (MD) and Monte Carlo (MC) simulations.103-116 The receptor variability has also been partly addressed by docking to an ensemble of static receptor conformations. This ensemble can be derived experimentally by X-ray crystallography or NMR spectroscopy;117-123 alternatively, the ensemble can be derived computationally by MD/MC simulations,124-130 normal mode analysis,131 or protein structure prediction.132, 133 In this study, multiple comparative models were built for the target sequence, each based on a different template structure. We now turn to calculating a “consensus” enrichment based on multiple models and/or templates, motivated by the observations that the most enriching of the multiple models frequently yields a better enrichment than the holo X-ray structure of the target and that no model/template feature can accurately predict the most enriching model.
Overall, the consensus enrichment outperformed the enrichments for both the apo X-ray structures and the best-assessed models (by sequence identity or the N-DOPE score). For 11/7 and 16/8 targets, the consensus enrichment is better or comparable to the enrichment obtained using the holo and the apo X-ray structures, respectively (Figure 3). For 23 targets, the consensus enrichment was better than random selection. Consensus enrichment benefits from maximizing the number of binding modes presented by different template structures (Table 3). This interpretation is supported by examples, such as phosphodiesterase 5 (PDE5), DHFR, and thrombin. For these targets, the consensus scores of top-ranked ligands were contributed by docking screens against different comparative models, although we didn’t find a clear difference in the chemotypes of the top-ranked ligands preferred by different models. The combination of individual docking screens against different models can frequently rescue ligands that are missed in a docking screen to a single inaccurate model.
Consensus enrichment was also calculated for each target-template pair, combining the docking results of a single model and the corresponding template (data not shown). For the 222 target-template pairs, the consensus enrichment was better, comparable and worse than the enrichment for 23% (23%), 72% (67%) and 5% (10%) of the models (templates), respectively. When orthologous target-template pairs,72 hormone receptor pairs involving AR, GR and MR, and pairs with less than 25% sequence identity were removed, further improvement of the consensus enrichment over the template-based enrichment was observed. For the 87 target-template pairs, the consensus enrichment was better and worse than the template enrichment in 33% and 3% of the cases, respectively. Thus, it is 11 times as likely that a model-template pair is more enriching than the template alone, when the target and the template are not orthologs and the model is based on a template with more than 25% sequence identity to the target.
Consensus enrichment of multiple templates was also shown to be better than that of a single, best-assessed template (Table S1), suggesting that the consensus enrichment can be used in docking screens where multiple experimentally determined structures are available. Furthermore, when the docking results of all models and templates were combined, the resulting consensus enrichment is frequently better (29%) or comparable (47%) to the higher of the values for the consensus enrichment of all models and the consensus enrichment of all templates.
In summary, applications, such as protein function prediction or ligand discovery, could benefit from the consensus enrichment that combines docking screens against multiple experimentally determined structures of the protein target, multiple comparative models, and the templates on which these models are based.
Several “consensus” scoring methods have been suggested.134 In distinction to our approach, however, these methods use multiple scoring functions, not multiple comparative models based on different templates. In principle, both types of consensus scoring could be combined with each other.
Conclusions
We conclude by returning to the questions we asked in Introduction, for the modeling, docking, and benchmark. As for all quantifications of enrichment differences, a difference in logAUC of 3 units or more is considered significant.
How does docking against comparative models compare to random selection?
Comparative models typically outperform random selection significantly, doing so for 27 out of the 38 targets (Table 2).
How does docking against comparative models compare to docking against the template structures?
For the entire benchmark, comparative models are on average no more enriching than the corresponding templates. This measurement, however, is confounded by the likelihood of orthologous templates genuinely recognizing the ligands for the modeled target. Conversely, a modeled structure based on a paralogous template with at least 25% sequence identity to the target is 2.5 times more likely to be significantly more enriching than the template (“Docking to templates instead of comparative models” section in Results).
If multiple models are calculated for a target, each one based on a different template, can any of them outperform apo and even holo X-ray structures of the target?
Typically, the holo x-ray structure returns the best enrichments, but the modeled structures are often competitive. For 15 of the 38 targets, the most enriching model is better for virtual screening than the holo X-ray structure; for 9 targets, the most enriching model is as good as the holo X-ray structure (Table 2). Compared to apo X-ray structures, the model performance is better still.
Can one reliably identify which model will be most enriching?
No, none of the tested sequence or structural attributes (i.e., the overall target-template sequence identity, the binding site target-template sequence identity, and the predicted accuracy of a model) can reliably predict the accuracy of ligand docking (Table 2, Figure 2).
Can the docking screens be improved by employing multiple models instead of a single model?
Yes. For the 38 targets, the enrichment of the model based on the highest sequence identity is better or comparable to the enrichment for the apo and holo X-ray structures in 65% and 45% cases, respectively (Table 2); in contrast, the consensus enrichment for multiple models (and templates) is better or comparable to the enrichment for the apo and holo X-ray structures in 70% (79%) and 47% (50%) cases, respectively (Table 2, Table S1). For the 222 target-template pairs, the consensus enrichment is better and worse than the template enrichment in 23% and 10% of the cases, respectively. For the 87 paralogous target-template pairs related at more than 25% sequence identity, the consensus enrichment is better and worse than the template enrichment in 33% and 3% of the cases, respectively (“Consensus enrichment” section in Discussion).
In conclusion, these results suggest techniques to best exploit comparative models in molecular docking screens: Whether one or multiple templates are available, comparative models are best used via consensus enrichment calculations that include multiple models as well as templates. For a single template, however, the corresponding comparative model tends to be more enriching than the template only if the template is paralogous and shares more than 25% sequence identity with the target.
Supplementary Material
Acknowledgement
We thank Dr. Niu Huang for discussions about the DUD database and the automated docking pipeline. We thank Dr. Peter Kolb for discussion about the manuscript. We acknowledge funds from Sandler Family Supporting Foundation and National Institutes of Health (P01 GM71790 and R01 GM54762 to Dr. Andrej Sali and R01 GM59957 to Dr. Brian Shoichet). We are also grateful to Ron Conway, Mike Homer, Hewlett-Packard, IBM, NetApp, and Intel for hardware gifts.
References
- 1.Kuntz ID. Structure-Based Strategies for Drug Design and Discovery. Science. 1992;257(5073):1078–1082. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- 2.Klebe G. Recent developments in structure-based drug design. J. Mol. Med. 2000;78(5):269–281. doi: 10.1007/s001090000084. [DOI] [PubMed] [Google Scholar]
- 3.Dailey MM, Hait C, Holt PA, Maguire JM, Meier JB, Miller MC, Petraccone L, Trent JO. Structure-based drug design: From nucleic acid to membrane protein targets. Exp. Mol. Pathol. 2009;86(3):141–150. doi: 10.1016/j.yexmp.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ealick SE, Armstrong SR. Pharmacologically relevant proteins. Curr. Opin. Struct. Biol. 1993;3(6):861–867. [Google Scholar]
- 5.Gschwend DA, Good AC, Kuntz ID. Molecular docking towards drug discovery. J. Mol. Recognit. 1996;9(2):175–186. doi: 10.1002/(sici)1099-1352(199603)9:2<175::aid-jmr260>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann D, Kramer B, Washio T, Steinmetzer T, Rarey M, Lengauer T. Two-stage method for protein-ligand docking. J. Med. Chem. 1999;42(21):4422–4433. doi: 10.1021/jm991090p. [DOI] [PubMed] [Google Scholar]
- 7.Stahl M, Rarey M. Detailed analysis of scoring functions for virtual screening. J. Med. Chem. 2001;44(7):1035–1042. doi: 10.1021/jm0003992. [DOI] [PubMed] [Google Scholar]
- 8.Charifson PS, Corkery JJ, Murcko MA, Walters WP. Consensus scoring: A method for obtaining improved hit rates from docking databases of three-dimensional structures into proteins. J. Med. Chem. 1999;42(25):5100–5109. doi: 10.1021/jm990352k. [DOI] [PubMed] [Google Scholar]
- 9.Abagyan R, Totrov M. High-throughput docking for lead generation. Curr. Opin. Chem. Biol. 2001;5(4):375–382. doi: 10.1016/s1367-5931(00)00217-9. [DOI] [PubMed] [Google Scholar]
- 10.Klebe G. Virtual ligand screening: strategies, perspectives and limitations. Drug Discov. Today. 2006;11(1314):580–594. doi: 10.1016/j.drudis.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sperandio O, Miteva MA, Delfaud F, Villoutreix BO. Receptor-based computational screening of compound databases: The main docking-scoring engines. Curr. Protein Peptide Sci. 2006;7(5):369–393. doi: 10.2174/138920306778559377. [DOI] [PubMed] [Google Scholar]
- 12.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bairoch A, Bougueleret L, Altairac S, Amendolia V, Auchincloss A, Puy GA, Axelsen K, Baratin D, Blatter MC, Boeckmann B, Bollondi L, Boutet E, Quintaje SB, Breuza L, Bridge A, Saux VBL, deCastro E, Ciampina L, Coral D, Coudert E, Cusin I, David F, Delbard G, Dornevil D, Duek-Roggli P, Duvaud S, Estreicher A, Famiglietti L, Farriol-Mathis N, Ferro S, Feuermann M, Gasteiger E, Gateau A, Gehant S, Gerritsen V, Gos A, Gruaz-Gumowski N, Hinz U, Hulo C, Hulo N, Innocenti A, James J, Jain E, Jimenez S, Jungo F, Junker V, Keller G, Lachaize C, Lane-Guermonprez L, Langendijk-Genevaux P, Lara V, Le Mercier P, Lieberherr D, Lima TD, Mangold V, Martin X, Michoud K, Moinat M, Morgat A, Nicolas M, Paesano S, Pedruzzi I, Perret D, Phan I, Pilbout S, Pillet V, Poux S, Pozzato M, Redaschi N, Reynaud S, Rivoire C, Roechert B, Sapsezian C, Schneider M, Sigrist C, Sonesson K, Staehli S, Stutz A, Sundaram S, Tognolli M, Verbregue L, Veuthey AL, Vitorello C, Yip L, Zuletta LF, Apweiler R, Alam-Faruque Y, Barrell D, Bower L, Browne P, Chan WM, Daugherty L, Donate ES, Eberhardt R, Fedotov A, Foulger R, Frigerio G, Garavelli J, Golin R, Horne A, Jacobsen J, Kleen M, Kersey P, Laiho K, Legge D, Magrane M, Martin MJ, Monteiro P, O’Donovan C, Orchard S, O’Rourke J, Patient S, Pruess M, Sitnov A, Whitefield E, Wieser D, Lin Q, Rynbeek M, di Martino G, Donnelly M, van Rensburg P, Wu C, Arighi C, Arminski L, Barker W, Chen YX, Crooks D, Hu ZZ, Hua HK, Huang HZ, Kahsay R, Mazumder R, McGarvey P, Natale D, Nikolskaya AN, Petrova N, Suzek B, Vasudevan S, Vinayaka CR, Yeh LS, Zhang J, Consortium, U. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pieper U, Eswar N, Webb B, Eramian E, Kelly L, Barkan DT, Carter H, Mankoo P, Karchin R, Marti-Renom MA, Davis FP, Sali A, Sanchez R. MODBASE, a database of annotated comparative protein structure models, and ssociated resources. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkn791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294(5540):93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 16.Baker D. A surprising simplicity to protein folding. Nature. 2000;405(6782):39–42. doi: 10.1038/35011000. [DOI] [PubMed] [Google Scholar]
- 17.Bonneau R, Baker D. Ab initio protein structure prediction: Progress and prospects. Annu. Rev. Biophys. Biomol. Struct. 2001;30:173–189. doi: 10.1146/annurev.biophys.30.1.173. [DOI] [PubMed] [Google Scholar]
- 18.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 19.Sali A. 100,000 protein structures for the biologist. Nat. Struct. Biol. 1998;5:1029–1032. doi: 10.1038/4136. [DOI] [PubMed] [Google Scholar]
- 20.Chandonia JM, Brenner SE. The impact of structural genomics: Expectations and outcomes. Science. 2006;311(5759):347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 21.Liu JF, Montelione GT, Rost B. Novel leverage of structural genomics. Nat. Biotechnol. 2007;25(8):850–853. doi: 10.1038/nbt0807-849. [DOI] [PubMed] [Google Scholar]
- 22.Jacobson M, Sali A. Comparative protein structure modeling and its applications to drug discovery. Annu. Rep. Med. Chem. 2004;39:259–276. 39. [Google Scholar]
- 23.Bissantz C, Bernard P, Hibert M, Rognan D. Protein-based virtual screening of chemical databases. II. Are homology models of G-protein coupled receptors suitable targets? Proteins: Struct. Funct. Genet. 2003;50(1):5–25. doi: 10.1002/prot.10237. [DOI] [PubMed] [Google Scholar]
- 24.Cavasotto CN, Orry AJW, Abagyan RA. Structure-based identification of binding sites, native ligands and potential inhibitors for G-protein coupled receptors. Proteins: Struct. Funct. Genet. 2003;51(3):423–433. doi: 10.1002/prot.10362. [DOI] [PubMed] [Google Scholar]
- 25.Evers A, Klebe G. Ligand-supported homology modeling of G-protein-coupled receptor sites: Models sufficient for successful virtual screening. Angewandte Chemie-International Edition. 2004;43(2):248–251. doi: 10.1002/anie.200352776. [DOI] [PubMed] [Google Scholar]
- 26.Evers A, Klebe G. Successful virtual screening for a submicromolar antagonist of the neurokinin-1 receptor based on a ligand-supported homology model. J. Med. Chem. 2004;47(22):5381–5392. doi: 10.1021/jm0311487. [DOI] [PubMed] [Google Scholar]
- 27.Evers A, Klabunde T. Structure-based drug discovery using GPCR homology modeling: Successful virtual screening for antagonists of the Alpha1A adrenergic receptor. J. Med. Chem. 2005;48(4):1088–1097. doi: 10.1021/jm0491804. [DOI] [PubMed] [Google Scholar]
- 28.Moro S, Deflorian F, Bacilieri M, Spalluto G. Novel strategies for the design of new potent and selective human A(3) receptor antagonists: An update. Curr. Med. Chem. 2006;13(6):639–645. doi: 10.2174/092986706776055670. [DOI] [PubMed] [Google Scholar]
- 29.Nowak M, Kolaczkowski M, Pawlowski M, Bojarski AJ. Homology modeling of the serotonin 5-HT1A receptor using automated docking of bioactive compounds with defined geometry. J. Med. Chem. 2006;49(1):205–214. doi: 10.1021/jm050826h. [DOI] [PubMed] [Google Scholar]
- 30.Chen JZ, Wang JM, Xie XQ. GPCR structure-based virtual screening approach for CB2 antagonist search. J. Chem. Inf. Model. 2007;47(4):1626–1637. doi: 10.1021/ci7000814. [DOI] [PubMed] [Google Scholar]
- 31.Zylberg J, Ecke D, Fischer B, Reiser G. Structure and ligand-binding site characteristics of the human P2Y(11) nucleotide receptor deduced from computational modelling and mutational analysis. Biochem. J. 2007;405:277–286. doi: 10.1042/BJ20061728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radestock S, Weil T, Renner S. Homology model-based virtual screening for GPCR ligands using docking and target-biased scoring. J. Chem. Inf. Model. 2008;48(5):1104–1117. doi: 10.1021/ci8000265. [DOI] [PubMed] [Google Scholar]
- 33.Singh N, Cheve G, Ferguson DM, McCurdy CR. A combined ligand-based and target-based drug design approach for G-protein coupled receptors: application to salvinorin A, a selective kappa opioid receptor agonist. J. Comput.-Aided Mol. Des. 2006;20(78):471–493. doi: 10.1007/s10822-006-9067-x. [DOI] [PubMed] [Google Scholar]
- 34.Kiss R, Kiss B, Konczol A, Szalai F, Jelinek I, Laszlo V, Noszal B, Falus A, Keseru GM. Discovery of novel human histamine H4 receptor ligands by large-scale structure-based virtual screening. J. Med. Chem. 2008;51(11):3145–3153. doi: 10.1021/jm7014777. [DOI] [PubMed] [Google Scholar]
- 35.de Graaf C, Foata N, Engkvist O, Rognan D. Molecular modeling of the second extracellular loop of G-protein coupled receptors and its implication on structure-based virtual screening. Proteins: Struct. Funct. Bioinform. 2008;71(2):599–620. doi: 10.1002/prot.21724. [DOI] [PubMed] [Google Scholar]
- 36.Diller DJ, Li RX. Kinases, homology models, and high throughput docking. J. Med. Chem. 2003;46(22):4638–4647. doi: 10.1021/jm020503a. [DOI] [PubMed] [Google Scholar]
- 37.Oshiro C, Bradley EK, Eksterowicz J, Evensen E, Lamb ML, Lanctot JK, Putta S, Stanton R, Grootenhuis PDJ. Performance of 3D-database molecular docking studies into homology models. J. Med. Chem. 2004;47(3):764–767. doi: 10.1021/jm0300781. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen TL, Gussio R, Smith JA, Lannigan DA, Hecht SM, Scudiero DA, Shoemaker RH, Zaharevitz DW. Homology model of RSK2 N-terminal kinase domain, structure-based identification of novel RSK2 inhibitors, and preliminary common pharmacophore. Bioorg. Med. Chem. 2006;14(17):6097–6105. doi: 10.1016/j.bmc.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Rockey WM, Elcock AH. Structure selection for protein kinase docking and virtual screening: Homology models or crystal structures? Curr. Protein Peptide Sci. 2006;7(5):437–457. doi: 10.2174/138920306778559368. [DOI] [PubMed] [Google Scholar]
- 40.Schapira M, Abagyan R, Totrov M. Nuclear hormone receptor targeted virtual screening. J. Med. Chem. 2003;46(14):3045–3059. doi: 10.1021/jm0300173. [DOI] [PubMed] [Google Scholar]
- 41.Marhefka CA, Moore BM, Bishop TC, Kirkovsky L, Mukherjee A, Dalton JT, Miller DD. Homology modeling using multiple molecular dynamics simulations and docking studies of the human androgen receptor ligand binding domain bound to testosterone and nonsteroidal ligands. J. Med. Chem. 2001;44(11):1729–1740. doi: 10.1021/jm0005353. [DOI] [PubMed] [Google Scholar]
- 42.Kasuya A, Sawada Y, Tsukamoto Y, Tanaka K, Toya T, Yanagi M. Binding mode of ecdysone agonists to the receptor: comparative modeling and docking studies. J. Mol. Model. 2003;9(1):58–65. doi: 10.1007/s00894-002-0113-x. [DOI] [PubMed] [Google Scholar]
- 43.Li RS, Chen XW, Gong BQ, Selzer PM, Li Z, Davidson E, Kurzban G, Miller RE, Nuzum EO, McKerrow JH, Fletterick RJ, Gillmor SA, Craik CS, Kuntz ID, Cohen FE, Kenyon GL. Structure-based design of parasitic protease inhibitors. Bioorg. Med. Chem. 1996;4(9):1421–1427. doi: 10.1016/0968-0896(96)00136-8. [DOI] [PubMed] [Google Scholar]
- 44.Selzer PM, Chen XW, Chan VJ, Cheng MS, Kenyon GL, Kuntz ID, Sakanari JA, Cohen FE, McKerrow JH. Leishmania major: Molecular modeling of cysteine proteases and prediction of new nonpeptide inhibitors. Exp. Parasitol. 1997;87(3):212–221. doi: 10.1006/expr.1997.4220. [DOI] [PubMed] [Google Scholar]
- 45.Enyedy IJ, Ling Y, Nacro K, Tomita Y, Wu XH, Cao YY, Guo RB, Li BH, Zhu XF, Huang Y, Long YQ, Roller PP, Yang DJ, Wang SM. Discovery of small-molecule inhibitors of bcl-2 through structure-based computer screening. J. Med. Chem. 2001;44(25):4313–4324. doi: 10.1021/jm010016f. [DOI] [PubMed] [Google Scholar]
- 46.de Graaf C, Oostenbrink C, Keizers PHJ, van der Wijst T, Jongejan A, Vemleulen NPE. Catalytic site prediction and virtual screening of cytochrome P450 2D6 substrates by consideration of water and rescoring in automated docking. J. Med. Chem. 2006;49(8):2417–2430. doi: 10.1021/jm0508538. [DOI] [PubMed] [Google Scholar]
- 47.Katritch V, Byrd CM, Tseitin V, Dai DC, Raush E, Totrov M, Abagyan R, Jordan R, Hruby DE. Discovery of small molecule inhibitors of ubiquitin-like poxvirus proteinase I7L using homology modeling and covalent docking approaches. J. Comput.-Aided Mol. Des. 2007;21(1011):549–558. doi: 10.1007/s10822-007-9138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukherjee P, Desai PV, Srivastava A, Tekwani BL, Avery MA. Probing the structures of leishmanial farnesyl pyrophosphate synthases: Homology modeling and docking studies. J. Chem. Inf. Model. 2008;48(5):1026–1040. doi: 10.1021/ci700355z. [DOI] [PubMed] [Google Scholar]
- 49.Song L, Kalyanaraman C, Fedorov AA, Fedorov EV, Glasner ME, Brown S, Imker HJ, Babbitt PC, Almo SC, Jacobson MP, Gerlt JA. Prediction and assignment of function for a divergent N-succinyl amino acid racemase. Nat. Chem. Biol. 2007;3(8):486–491. doi: 10.1038/nchembio.2007.11. [DOI] [PubMed] [Google Scholar]
- 50.Kalyanaraman C, Imker HJ, Federov AA, Federov EV, Glasner ME, Babbitt PC, Almo SC, Gerlt JA, Jacobson MP. Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening. Structure. 2008;16:1668–1677. doi: 10.1016/j.str.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rotkiewicz P, Sicinska W, Kolinski A, DeLuca HF. Model of three-dimensional structure of vitamin D receptor and its binding mechanism with 1 alpha,25-dihydroxyvitamin D-3. Proteins: Struct. Funct. Genet. 2001;44(3):188–199. doi: 10.1002/prot.1084. [DOI] [PubMed] [Google Scholar]
- 52.Que XC, Brinen LS, Perkins P, Herdman S, Hirata K, Torian BE, Rubin H, McKerrow JH, Reed SL. Cysteine proteinases from distinct cellular compartments are recruited to phagocytic vesicles by Entamoeba histolytica. Mol. Biochem. Parasitol. 2002;119(1):23–32. doi: 10.1016/s0166-6851(01)00387-5. [DOI] [PubMed] [Google Scholar]
- 53.Parrill AL, Echols U, Nguyen T, Pham TCT, Hoeglund A, Baker DL. Virtual screening approaches for the identification of non-lipid autotaxin inhibitors. Bioorg. Med. Chem. 2008;16(4):1784–1795. doi: 10.1016/j.bmc.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 54.Fernandes MX, Kairys V, Gilson MK. Comparing ligand interactions with multiple receptors via serial docking. J. Chem. Inf. Comput. Sci. 2004;44(6):1961–1970. doi: 10.1021/ci049803m. [DOI] [PubMed] [Google Scholar]
- 55.Kairys V, Fernandes MX, Gilson MK. Screening drug-like compounds by docking to homology models: A systematic study. J. Chem. Inf. Model. 2006;46(1):365–379. doi: 10.1021/ci050238c. [DOI] [PubMed] [Google Scholar]
- 56.McGovern SL, Shoichet BK. Information decay in molecular docking screens against holo, apo, and modeled conformations of enzymes. J. Med. Chem. 2003;46(14):2895–2907. doi: 10.1021/jm0300330. [DOI] [PubMed] [Google Scholar]
- 57.Huang N, Shoichet BK, Irwin JJ. Benchmarking sets for molecular docking. J. Med. Chem. 2006;49(23):6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ortiz AR, Strauss CEM, Olmea O. MAMMOTH (Matching molecular models obtained from theory): An automated method for model comparison. Protein Sci. 2002;11(11):2606–2621. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marti-Renom MA, Pieper U, Madhusudhan MS, Rossi A, Eswar N, Davis FP, Al-Shahrour F, Dopazo J, Sali A. DBAli tools: mining the protein structure space. Nucleic Acids Res. 2007;35:W393–W397. doi: 10.1093/nar/gkm236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sali A, Blundell TL. Comparative Protein Modeling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 61.Marti-Renom MA, Madhusudhan MS, Sali A. Alignment of protein sequences by their profiles. Protein Sci. 2004;13(4):1071–1087. doi: 10.1110/ps.03379804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15(11):2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eramian D, Eswar N, Shen MY, Sali A. Can the accuracy of comparative protein structure models be predicted? Protein Sci. 2008 doi: 10.1110/ps.036061.108. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ferrin TE, Huang CC, Jarvis LE, Langridge R. The Midas Display System. J. Mol. Graphics. 1988;6(1):13. &. [Google Scholar]
- 65.Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. A Geometric Approach to Macromolecule-Ligand Interactions. J. Mol. Biol. 1982;161(2):269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 66.Lorber DM, Shoichet BK. Hierarchical docking of databases of multiple ligand conformations. Curr. Top. Med. Chem. 2005;5(8):739–749. doi: 10.2174/1568026054637683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meng EC, Shoichet BK, Kuntz ID. Automated Docking with Grid-Based Energy Evaluation. J. Comput. Chem. 1992;13(4):505–524. [Google Scholar]
- 68.Wei BQQ, Baase WA, Weaver LH, Matthews BW, Shoichet BK. A model binding site for testing scoring functions in molecular docking. J. Mol. Biol. 2002;322(2):339–355. doi: 10.1016/s0022-2836(02)00777-5. [DOI] [PubMed] [Google Scholar]
- 69.Jain AN, Nicholls A. Recommendations for evaluation of computational methods. J. Comput.-Aided Mol. Des. 2008;22(34):133–139. doi: 10.1007/s10822-008-9196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Irwin JJ, Shoichet BK, Mysinger MM, Huang N, Colizzi F, Wassam P, Cao Y. Automated docking screens: A feasibility study. J. Med. Chem. 2009;52(18):5712–5720. doi: 10.1021/jm9006966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shapiro SS, Wilk MB. An Analysis of Variance Test for Normality (Complete Samples) Biometrika. 1965;52:591. &. [Google Scholar]
- 72.Marti-Renom MA, Madhusudhan MS, Fiser A, Rost B, Sali A. Reliability of assessment of protein structure prediction methods. Structure. 2002;10(3):435–440. doi: 10.1016/s0969-2126(02)00731-1. [DOI] [PubMed] [Google Scholar]
- 73.Wilcoxon F. Individual comparisons by ranking methods. Biometrics Bull. 1945;1(6):80–83. [Google Scholar]
- 74.Ferrara P, Jacoby E. Evaluation of the utility of homology models in high throughput docking. J. Mol. Model. 2007;13(8):897–905. doi: 10.1007/s00894-007-0207-6. [DOI] [PubMed] [Google Scholar]
- 75.Laskowski RA. Surfnet - a Program for Visualizing Molecular-Surfaces, Cavities, and Intermolecular Interactions. J. Mol. Graphics. 1995;13(5):323–330. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 76.Terasaka T, Nakanishi I, Nakamura K, Eikyu Y, Kinoshita T, Nishio N, Sato A, Kuno M, Seki N, Sakane K. Structure-based de novo design of non-nucleoside adenosine deaminase inhibitors. Bioorg. Med. Chem. Lett. 2003;13(22):4147–4147. doi: 10.1016/s0960-894x(03)00026-x. vol 13, pg 1115, 2003. [DOI] [PubMed] [Google Scholar]
- 77.Terasaka T, Kinoshita T, Kuno M, Nakanishi I. A highly potent non-nucleoside adenosine deaminase inhibitor: Efficient drug discovery by intentional lead hybridization. J. Am. Chem. Soc. 2004;126(1):34–35. doi: 10.1021/ja038606l. [DOI] [PubMed] [Google Scholar]
- 78.Prager NA, Abendschein DR, Mckenzie CR, Eisenberg PR. Role of Thrombin Compared with Factor Xa in the Procoagulant Activity of Whole-Blood Clots. Circulation. 1995;92(4):962–967. doi: 10.1161/01.cir.92.4.962. [DOI] [PubMed] [Google Scholar]
- 79.Brandstetter H, Kuhne A, Bode W, Huber R, vonderSaal W, Wirthensohn K, Engh RA. X-ray structure of active site-inhibited clotting factor Xa - Implications for drug design and substrate recognition. J. Biol. Chem. 1996;271(47):29988–29992. doi: 10.1074/jbc.271.47.29988. [DOI] [PubMed] [Google Scholar]
- 80.Maignan S, Guilloteau JP, Pouzieux S, Choi-Sledeski YM, Becker MR, Klein SI, Ewing WR, Pauls HW, Spada AP, Mikol V. Crystal structures of human factor Xa complexed with potent inhibitors. J. Med. Chem. 2000;43(17):3226–3232. doi: 10.1021/jm000940u. [DOI] [PubMed] [Google Scholar]
- 81.Brik A, Wong CH. HIV-1 protease: mechanism and drug discovery. Org. Biomol. Chem. 2003;1(1):5–14. doi: 10.1039/b208248a. [DOI] [PubMed] [Google Scholar]
- 82.Baldwin ET, Bhat TN, Gulnik S, Liu BS, Topol IA, Kiso Y, Mimoto T, Mitsuya H, Erickson JW. Structure of Hiv-1 Protease with Kni-272, a Tight-Binding Transition-State Analog Containing Allophenylnorstatine. Structure. 1995;3(6):581–590. doi: 10.1016/s0969-2126(01)00192-7. [DOI] [PubMed] [Google Scholar]
- 83.Vonitzstein M, Wu WY, Kok GB, Pegg MS, Dyason JC, Jin B, Phan TV, Smythe ML, White HF, Oliver SW, Colman PM, Varghese JN, Ryan DM, Woods JM, Bethell RC, Hotham VJ, Cameron JM, Penn CR. Rational Design of Potent Sialidase-Based Inhibitors of Influenza-Virus Replication. Nature. 1993;363(6428):418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 84.Abu Hammad AM, Afifi FU, Taha MO. Combining docking, scoring and molecular field analyses to probe influenza neuraminidase-ligand interactions. J. Mol. Graphics Model. 2007;26(2):443–456. doi: 10.1016/j.jmgm.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 85.Bledsoe RK, Madauss KP, Holt JA, Apolito CJ, Lambert MH, Pearce KH, Stanley TB, Stewart EL, Trump RP, Willson TM, Williams SP. A ligand-mediated hydrogen bond network required for the activation of the mineralocorticoid receptor. J. Biol. Chem. 2005;280(35):31283–31293. doi: 10.1074/jbc.M504098200. [DOI] [PubMed] [Google Scholar]
- 86.Zhang ZM, Olland AM, Zhu Y, Cohen J, Berrodin T, Chippari S, Appavu C, Li S, Wilhem J, Chopra R, Fensome A, Zhang PW, Wrobel J, Unwalla RJ, Lyttle R, Winneker RC. Molecular and pharmacological properties of a potent and selective novel nonsteroidal progesterone receptor agonist tanaproget. J. Biol. Chem. 2005;280(31):28468–28475. doi: 10.1074/jbc.M504144200. [DOI] [PubMed] [Google Scholar]
- 87.Egea PF, Mitschler A, Moras D. Molecular recognition of agonist Ligands by RXRs. Mol. Endocrinol. 2002;16(5):987–997. doi: 10.1210/mend.16.5.0823. [DOI] [PubMed] [Google Scholar]
- 88.Teague SJ. Implications of protein flexibility for drug discovery. Nat. Rev. Drug Discov. 2003;2(7):527–541. doi: 10.1038/nrd1129. [DOI] [PubMed] [Google Scholar]
- 89.Rester U. Dock around the clock - Current status of small molecule docking and scoring. QSAR Comb. Sci. 2006;25(7):605–615. [Google Scholar]
- 90.Totrov M, Abagyan R. Flexible ligand docking to multiple receptor conformations: a practical alternative. Curr. Opin. Struct. Biol. 2008;18(2):178–184. doi: 10.1016/j.sbi.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cozzini P, Kellogg GE, Spyrakis F, Abraham DJ, Costantino G, Emerson A, Fanelli F, Gohlke H, Kuhn LA, Morris GM, Orozco M, Pertinhez TA, Rizzi M, Sotriffer CA. Target Flexibility: An Emerging Consideration in Drug Discovery and Design. J. Med. Chem. 2008;51(20):6237–6255. doi: 10.1021/jm800562d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Teodoro ML, Kavraki LE. Conformational flexibility models for the receptor in structure based drug design. Curr. Pharm. Des. 2003;9(20):1635–1648. doi: 10.2174/1381612033454595. [DOI] [PubMed] [Google Scholar]
- 93.Jiang F, Kim SH. Soft Docking - Matching of Molecular-Surface Cubes. J. Mol. Biol. 1991;219(1):79–102. doi: 10.1016/0022-2836(91)90859-5. [DOI] [PubMed] [Google Scholar]
- 94.Schnecke V, Swanson CA, Getzoff ED, Tainer JA, Kuhn LA. Screening a peptidyl database for potential ligands to proteins with side-chain flexibility. Proteins: Struct. Funct. Genet. 1998;33(1):74–87. [PubMed] [Google Scholar]
- 95.Apostolakis J, Pluckthun A, Caflisch A. Docking small ligands in flexible binding sites. J. Comput. Chem. 1998;19(1):21–37. [Google Scholar]
- 96.Leach AR. Ligand Docking to Proteins with Discrete Side-Chain Flexibility. J. Mol. Biol. 1994;235(1):345–356. doi: 10.1016/s0022-2836(05)80038-5. [DOI] [PubMed] [Google Scholar]
- 97.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267(3):727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 98.Schaffer L, Verkhivker GM. Predicting structural effects in HIV-1 protease mutant complexes with flexible ligand docking and protein side-chain optimization. Proteins: Struct. Funct. Genet. 1998;33(2):295–310. doi: 10.1002/(sici)1097-0134(19981101)33:2<295::aid-prot12>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 99.Anderson AC, O’Neil RH, Surti TS, Stroud RM. Approaches to solving the rigid receptor problem by identifying a minimal set of flexible residues during ligand docking. Chem. Biol. 2001;8(5):445–457. doi: 10.1016/s1074-5521(01)00023-0. [DOI] [PubMed] [Google Scholar]
- 100.Althaus E, Kohlbacher O, Lenhof HP, Muller P. A combinatorial approach to protein docking with flexible side chains. J. Comput. Biol. 2002;9(4):597–612. doi: 10.1089/106652702760277336. [DOI] [PubMed] [Google Scholar]
- 101.Kairys V, Gilson MK. Enhanced docking with the mining minima optimizer: Acceleration and side-chain flexibility. J. Comput. Chem. 2002;23(16):1656–1670. doi: 10.1002/jcc.10168. [DOI] [PubMed] [Google Scholar]
- 102.Zavodszky MI, Kuhn LA. Side-chain flexibility in protein-ligand binding: The minimal rotation hypothesis. Protein Sci. 2005;14(4):1104–1114. doi: 10.1110/ps.041153605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dinola A, Roccatano D, Berendsen HJC. Molecular-Dynamics Simulation of the Docking of Substrates to Proteins. Proteins: Struct. Funct. Genet. 1994;19(3):174–182. doi: 10.1002/prot.340190303. [DOI] [PubMed] [Google Scholar]
- 104.Luty BA, Wasserman ZR, Stouten PFW, Hodge CN, Zacharias M, Mccammon JA. A Molecular Mechanics Grid Method for Evaluation of Ligand-Receptor Interactions. J. Comput. Chem. 1995;16(4):454–464. [Google Scholar]
- 105.Wasserman ZR, Hodge CN. Fitting an inhibitor into the active site of thermolysin: A molecular dynamics case study. Proteins: Struct. Funct. Genet. 1996;24(2):227–237. doi: 10.1002/(SICI)1097-0134(199602)24:2<227::AID-PROT9>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 106.Nakajima N, Higo J, Kidera A, Nakamura H. Flexible docking of a ligand peptide to a receptor protein by multicanonical molecular dynamics simulation. Chem. Phys. Lett. 1997;278(46):297–301. [Google Scholar]
- 107.Pak YS, Wang SM. Application of a molecular dynamics simulation method with a generalized effective potential to the flexible molecular docking problems. J. Phys. Chem. B. 2000;104(2):354–359. [Google Scholar]
- 108.Kua J, Zhang YK, McCammon JA. Studying enzyme binding specificity in acetylcholinesterase using a combined molecular dynamics and multiple docking approach. J. Am. Chem. Soc. 2002;124(28):8260–8267. doi: 10.1021/ja020429l. [DOI] [PubMed] [Google Scholar]
- 109.Wu GS, Robertson DH, Brooks CL, Vieth M. Detailed analysis of grid-based molecular docking: A case study of CDOCKER - A CHARMm-based MD docking algorithm. J. Comput. Chem. 2003;24(13):1549–1562. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
- 110.Camacho CJ. Modeling side-chains using molecular dynamics improve recognition of binding region in CAPRI targets. Proteins: Struct. Funct. Bioinform. 2005;60(2):245–251. doi: 10.1002/prot.20565. [DOI] [PubMed] [Google Scholar]
- 111.Sivanesan D, Rajnarayanan RV, Doherty J, Pattabiraman N. In-silico screening using flexible ligand binding pockets: a molecular dynamics-based approach. J. Comput.-Aided Mol. Des. 2005;19(4):213–228. doi: 10.1007/s10822-005-4788-9. [DOI] [PubMed] [Google Scholar]
- 112.Zhu J, Fan H, Liu HY, Shi YY. Structure-based ligand design for flexible proteins: Application of new F-DycoBlock. J. Comput.-Aided Mol. Des. 2001;15(11):979–996. doi: 10.1023/a:1014817911249. [DOI] [PubMed] [Google Scholar]
- 113.Krol M, Tournier AL, Bates PA. Flexible relaxation of rigid-body docking solutions. Proteins: Struct. Funct. Bioinform. 2007;68(1):159–169. doi: 10.1002/prot.21391. [DOI] [PubMed] [Google Scholar]
- 114.Caflisch A, Fischer S, Karplus M. Docking by Monte Carlo minimization with a solvation correction: Application to an FKBP-substrate complex. J. Comput. Chem. 1997;18(6):723–743. [Google Scholar]
- 115.Trosset JY, Scheraga HA. Flexible docking simulations: Scaled collective variable Monte Carlo minimization approach using Bezier splines, and comparison with a standard Monte Carlo algorithm. J. Comput. Chem. 1999;20(2):244–252. [Google Scholar]
- 116.Verkhivker GM, Rejto PA, Bouzida D, Arthurs S, Colson AB, Freer ST, Gehlhaar DK, Larson V, Luty BA, Marrone T, Rose PW. Parallel simulated tempering dynamics of ligand-protein binding with ensembles of protein conformations. Chem. Phys. Lett. 2001;337(13):181–189. [Google Scholar]
- 117.Claussen H, Buning C, Rarey M, Lengauer T. FlexE: Efficient molecular docking considering protein structure variations. J. Mol. Biol. 2001;308(2):377–395. doi: 10.1006/jmbi.2001.4551. [DOI] [PubMed] [Google Scholar]
- 118.Osterberg F, Morris GM, Sanner MF, Olson AJ, Goodsell DS. Automated docking to multiple target structures: Incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins: Struct. Funct. Bioinform. 2002;46(1):34–40. doi: 10.1002/prot.10028. [DOI] [PubMed] [Google Scholar]
- 119.Ferrari AM, Wei BQQ, Costantino L, Shoichet BK. Soft docking and multiple receptor conformations in virtual screening. J. Med. Chem. 2004;47(21):5076–5084. doi: 10.1021/jm049756p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wei BQ, Weaver LH, Ferrari AM, Matthews BW, Shoichet BK. Testing a flexible-receptor docking algorithm in a model binding site. J. Mol. Biol. 2004;337(5):1161–1182. doi: 10.1016/j.jmb.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 121.Cavasotto CN, Abagyan RA. Protein flexibility in ligand docking and virtual screening to protein kinases. J. Mol. Biol. 2004;337(1):209–225. doi: 10.1016/j.jmb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 122.Knegtel RMA, Kuntz ID, Oshiro CM. Molecular docking to ensembles of protein structures. J. Mol. Biol. 1997;266(2):424–440. doi: 10.1006/jmbi.1996.0776. [DOI] [PubMed] [Google Scholar]
- 123.Damm KL, Carlson HA. Exploring experimental sources of multiple protein conformations in structure-based drug design. J. Am. Chem. Soc. 2007;129(26):8225–8235. doi: 10.1021/ja0709728. [DOI] [PubMed] [Google Scholar]
- 124.Lin JH, Perryman AL, Schames JR, McCammon JA. Computational drug design accommodating receptor flexibility: The relaxed complex scheme. J. Am. Chem. Soc. 2002;124(20):5632–5633. doi: 10.1021/ja0260162. [DOI] [PubMed] [Google Scholar]
- 125.Lin JH, Perryman AL, Schames JR, McCammon JA. The relaxed complex method: Accommodating receptor flexibility for drug design with an improved scoring scheme. Biopolymers. 2003;68(1):47–62. doi: 10.1002/bip.10218. [DOI] [PubMed] [Google Scholar]
- 126.McCammon JA. Target flexibility in molecular recognition. Biochim. Biophys. Acta-Proteins Proteomics. 2005;1754(12):221–224. doi: 10.1016/j.bbapap.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 127.Wong CF, Kua J, Zhang YK, Straatsma TP, McCammon JA. Molecular docking of balanol to dynamics snapshots of protein kinase A. Proteins: Struct. Funct. Bioinform. 2005;61(4):850–858. doi: 10.1002/prot.20688. [DOI] [PubMed] [Google Scholar]
- 128.Pang YP, Kozikowski AP. Prediction of the Binding-Sites of Huperzine-a in Acetylcholinesterase by Docking Studies. J. Comput.-Aided Mol. Des. 1994;8(6):669–681. doi: 10.1007/BF00124014. [DOI] [PubMed] [Google Scholar]
- 129.Gorfe AA, Caflisch A. Functional plasticity in the substrate binding site of beta-secretase. Structure. 2005;13(10):1487–1498. doi: 10.1016/j.str.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 130.Broughton HB. A method for including protein flexibility in protein-ligand docking: Improving tools for database mining and virtual screening. J. Mol. Graphics Model. 2000;18(3):247. doi: 10.1016/s1093-3263(00)00036-x. + [DOI] [PubMed] [Google Scholar]
- 131.Cavasotto CN, Kovacs JA, Abagyan RA. Representing receptor flexibility in ligand docking through relevant normal modes. J. Am. Chem. Soc. 2005;127(26):9632–9640. doi: 10.1021/ja042260c. [DOI] [PubMed] [Google Scholar]
- 132.Meiler J, Baker D. ROSETTALIGAND: Protein-small molecule docking with full side-chain flexibility. Proteins: Struct. Funct. Bioinform. 2006;65(3):538–548. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- 133.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006;49(2):534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 134.Feher M. Consensus scoring for protein-ligand interactions. Drug Discovery Today. 2006;11(910):421–428. doi: 10.1016/j.drudis.2006.03.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.