Abstract
Silk proteins belong to a class of unique, high molecular weight, block copolymer-like proteins that have found widespread use in biomaterials and regenerative medicine. The useful features of these proteins, including self-assembly, robust mechanical properties, biocompatibility and biodegradability can be enhanced through a variety of chemical modifications. These modifications provide chemical handles for the attachment of growth factors, cell binding domains and other polymers to silk, expanding the range of cell and tissue engineering applications attainable. This review focuses on the chemical reactions that have been used to modify the amino acids in silk proteins, and describes their utility in biomedical applications.
1. Introduction
Bombyx mori silkworms produce cocoons made of a fascinating protein-based material called silk fibroin (SF). SF is one of the strongest natural fibers, and this strength can be attributed to the chemical structure of the protein itself. The amino acid sequence of SF contains repetitive glycine-alanine-glycine-alanine-glycine-serine (GAGAGS) repeats1 which self-assemble into an anti-parallel β-sheet structure1,2 (Figure 1). These β-sheets are highly crystalline and essentially crosslink the protein through strong intra- and inter-molecular hydrogen bonds, as well as strong van der Waals interactions between stacked β-sheets, giving the material robust mechanical properties. Advantages of using SF in biomedical applications include the excellent mechanical properties,2 slow degradation profile,3 and aqueous processibility.4–13 The extent of β-sheet structure can be controlled through physical9,14 or chemical methods,15–17 leading to materials with controlled crystallinity and degradation rate. The crystalline, hydrophobic β-sheet domains prevent the penetration of water and proteases resulting in slow biodegradation of silk in vivo.18
Fig. 1.
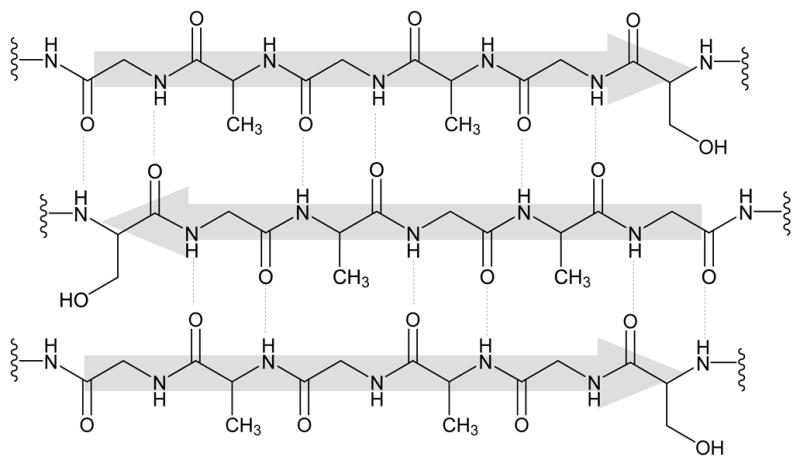
Silk is primarily composed of (Gly-Ala-Gly-Ala-Gly-Ser)6 amino acid repeat units that self-assemble into an anti-parallel β-sheet structure.
Raw and regenerated versions of SF have been used extensively in biomedical applications such as sutures, coatings for cell culture,19 drug delivery matrices,4,5,20–22 and 3D scaffolds for ligament,23,24 bone,25–28 cartilage,29–31 fat,32 and vasculature33–35 engineering. Cocoons from B. Mori contain SF and a glue-like protein called sericin, which is typically extracted (degummed) prior to use, as sericin has been found to illicit inflammatory responses in vivo.24 Extensive testing of degummed SF has revealed that only very mild inflammatory responses to SF occur in vivo,18 and these responses are much less than what is observed with commonly used biomaterials such as collagen or poly(lactic acid).18,36,37
Degummed SF fibers can be used directly for applications such as sutures or ligament engineering, but SF is more commonly used in the regenerated form. SF fibers can be dissolved at elevated temperatures (60–80 °C) in several aqueous solutions including concentrated lithium bromide, lithium thioisocyanate, or a mixture of CaCl2:water:ethanol (in a 1:8:2 molar ratio) then dialyzed into water to giving aqueous silk solutions that can be processed into films, fibers, gels or 3D porous structures.24 Additionally, SF fibers can be dissolved in the organic solvent hexafluoroisopropanol and then processed.38,39
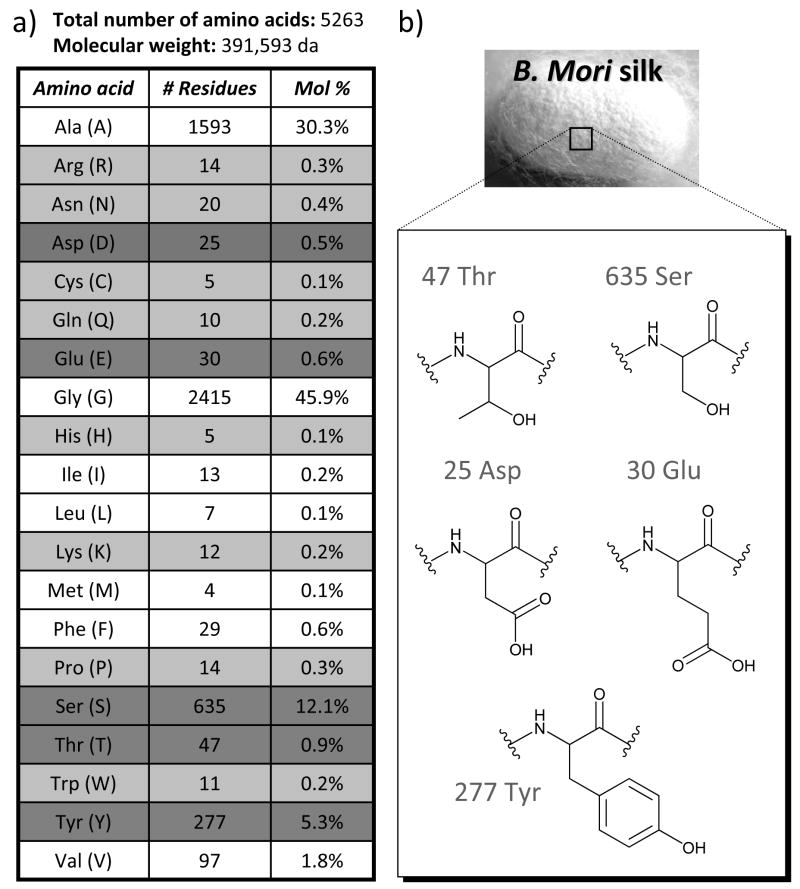
The ability to further tune the surface chemistry of silk materials is desirable to control the interaction between silk and living systems. Biomaterial surface chemistry is known to influence a variety of cell responses ranging from changes in surface adhesion to activation of biochemical pathways regulating cellular proliferation, differentiation, and survival.38,40–42 The presence of several reactive amino acids in SF allow chemical modification strategies to be utilized to tailor the protein for a desired application. As shown in Figure 2, SF is a large protein containing over 5000 amino acids, where the majority of the protein is composed of the non-reactive amino acids glycine and alanine. However, SF does contain significant quantities of serine, threonine, aspartic and glutamic acid, and tyrosine that can all be modified with known chemistries. This review will highlight methods used to chemically modify these residues, and will discuss how the resulting materials have been used in different biomedical applications.
Fig. 2.
a) Amino acid composition of the heavy chain of silk fibroin (accession number P05790). Reactive amino acids present in less than 0.5% are highlighted in light gray, while reactive amino acids present in more than 0.5% are highlighted in dark gray. b) Chemical structures of the most abundant reactive amino acids in silk fibroin.
2. Chemical modification of silk fibroin
2.1 Coupling reactions
Cyanuric chloride-activated coupling
Several reports on the use of cyanuric chloride-activated coupling of molecules to the tyrosine residues in silk have been made. Molecules bearing nucleophilic hydroxyl or amino functional groups were first conjugated to cyanuric chloride, then reacted with the tyrosine residues in SF under basic aqueous conditions to form modified silks such as that shown in Scheme 1. This reaction method is highly compatible with silk, as SF is stable at the basic pH needed for the reaction and there are many tyrosine residues available for reaction (5 mol%, ~277 residues).1 Lysine residues in SF can also participate in this reaction, but are only present in very low quantities (0.3 mol %, ~12 residues).1
Scheme 1.
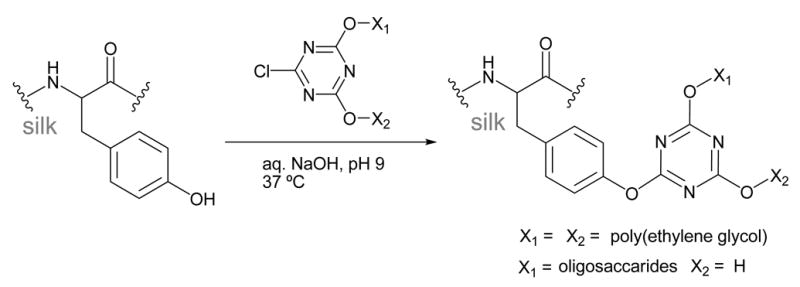
Cyanuric chloride-activated coupling
Gotoh and coworkers were able to attach approximately 210 poly(ethylene glycol) (PEG) molecules (MW ~104 Da) to each silk molecule using this method.43,44 This corresponds to modification of ~75% of the tyrosine residues present in SF. The addition of PEG to SF was found to promote β-sheet formation, and increase the hydrophilicity, reducing the contact angle of SF from 47° to 33°.
Lactose derivatives45,46 and N-acetyl-chito-oligosaccharides47 were also attached to SF using this method. 1H-NMR was used to confirm the attachment of these oligosaccharides to tyrosine and it was determined that ~75% of the tyrosines were modified, similar to the PEG reactions described above. Further characterization of the lactose-SF derivatives demonstrated that the sugars were still accessible after conjugation to SF, as lectin-induced aggregation was observed.45 The tensile strength after modification with lactose was 20–40% lower than native SF.46,48
Carbodiimide coupling
Carbodiimide coupling is a standard method used to react primary amines with carboxylic acids resulting in the formation of an amide bond. This reaction has broad use in protein chemistry as many proteins contain these functional groups in their amino acid side chains or at the N- and C-termini. SF contains a fair number of aspartic (0.5 mol%, ~25 residues)1 and glutamic acids (0.6 mol%, ~30 residues),1 as well as a small number of lysine residues (0.2 mol%, ~12 residues)1 that can participate in this reaction.
The acidic side chains in SF are more commonly targeted for carbodiimide modification, as there are significantly more aspartic and glutamic acids in SF than lysine. A number of researchers have conjugated molecules such as bone morphogenic protein-2 (BMP-2),12 the adhesion peptide RGD27,28,40,48 and horseradish peroxidase49 to SF through lysine residues or the N-terminus of these protein-based molecules (Scheme 2). Coupling reactions can be carried out on the soluble protein in aqueous solutions,27,28 or on the surface of solid silk fibers,50 films,40,51 and scaffolds.49 Reported reaction conditions typically activate the carboxylic acid residues in SF with a mixture of the water soluble 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) and N-hydroxysuccinimide (NHS) in 2-(N-morpholino) ethanesulfonic acid (MES) or phosphate buffer at pH 6.0–6.5, followed by reaction with the desired amine-bearing biomolecule.
Scheme 2.
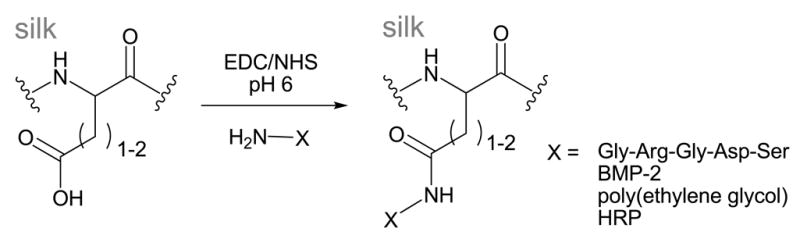
Carbodiimide coupling
Characterization of these peptide-protein conjugation reactions are difficult, as the large background signal from the silk protein backbone makes it hard to distinguish the addition of a small amount of protein or peptide. One reported chemical characterization method measured iodine content with XPS of iodine-labeled GRYDS peptides (as a control for GRGDS) coupled to the SF using the carbodiimide coupling conditions described above.28,40 For solid phase reactions on the surface of silk films, it was found that approximately one peptide was attached to each silk molecule giving a surface peptide density of 24 ± 3 pmol/cm2.40,48 To our knowledge, solution phase yields using this method have not been reported. Due to the difficulty in chemical characterization, activity assays or cell interactions with the resulting SF conjugates are more commonly used as a measure of reaction rather than direct characterization, and will be discussed below.
One report has also been made of coupling poly(D, L-lactic acid) (PLA) to SF through the small number of lysine residues.52 In this case, silk was partially hydrolyzed by treatment with NaOH, then coupled to the surface of PLA activated with EDC. Surface coverage was estimated to be ~65% using elemental analysis.52 The addition of SF fragments to the surface of PLA increased the hydrophilicity, dropping the contact angle from 69±3 to 54±4 degrees.52
Glutaraldehyde
Glutaraldehyde is commonly used as a fixing agent for biological tissues, as it contains two aldehyde groups that can crosslink biopolymers containing amines. However, this reagent can also be used as a non-specific coupling agent for amine-containing molecules and polymers. In this manner, glutaraldehyde has been used to conjugate insulin53 and L-asparaginase9 to SF to increase the stability of these drugs. In both cases, the SF protein was partially hydrolyzed prior to reaction to obtain small silk fragments ranging from 40–120 kDa. The level of insulin conjugated to SF was determined using an ELISA assay specific for insulin,53 while L-asparaginase incorporation was characterized using activity assays for the enzyme.9
2.2 Amino acid modification
Arginine masking
The small amount of positively charged arginine residues in regenerated SF (0.3 mol%, ~14 residues) can be reacted with 1,2-cyclohexandione under basic aqueous conditions to form an uncharged imidazolidinone product,54,55 as shown in Scheme 3. The reaction was estimated to go to completion using amino acid analysis of the hydrolyzed protein. No significant change was observed in the protein secondary structure by FTIR or CD, likely due to the low level of arginine originally present in SF. The basicity of SF was estimated to decrease following modification, but was not directly measured.
Scheme 3.
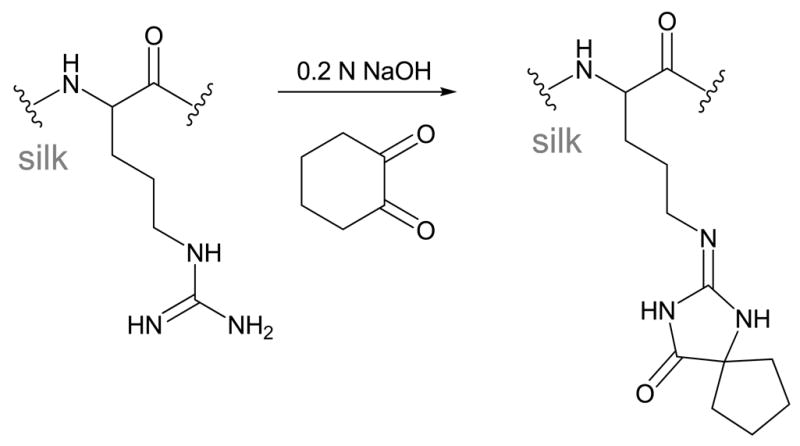
Arginine Masking
Sulfation of tyrosine
The tyrosine and serine residues in SF can be sulfated by treating degummed fibers with chlorosulfonic acid in pyridine at 70° C, as shown in Scheme 4.56,57 These reaction conditions cause hydrolysis of the silk protein backbone, resulting in fragments with molecular weight of ~20 kDa.56 Sulfate incorporation was confirmed with FTIR and 1H NMR, and the maximum sulfate incorporation was estimated by titration to be ~1.0 mmol/g after reaction for 10 h at 80°. However, severe hydrolysis and protein loss during purification resulted in total protein yields of less than 10%.56
Scheme 4.
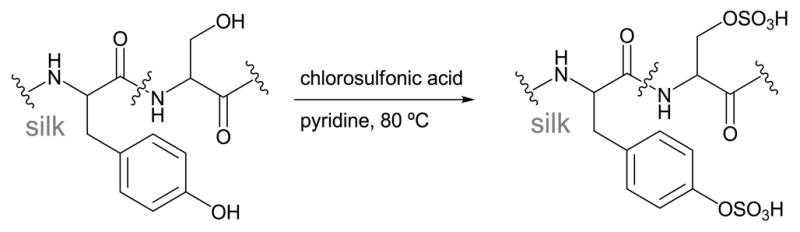
Reaction with chlorosulfonic acid
Another group also reported using sulfuric acid for the same purpose, although it seems from their data that the sulfuric acid completely hydrolyzes the protein as little or no amide bonds could be observed by FTIR or high resolution XPS after treatment.58
Azo-modified tyrosine
Methods to functionalize SF using diazonium coupling chemistry have recently been developed.59 Diazonium reactions with silk involve an electrophilic aromatic substitution reaction between the tyrosine side chains and a diazonium salt resulting in an azobenzene derivative, as shown in Scheme 5. SF is very stable in the basic pH conditions needed for this reaction, so this is a suitable reaction to modify the relatively large number of tyrosines (5.3 mol%, ~277 residues). In addition, the reaction is rapid, all the necessary reagents are commercially available, and the mild conditions allow >90% of the modified protein to be recovered after the reaction. This method can be used to install small molecules with various non-natural functional groups into SF including sulfonic acids, carboxylic acids, ketones and alkanes, resulting in hydrophobic and hydrophilic SF derivatives.59
Scheme 5.
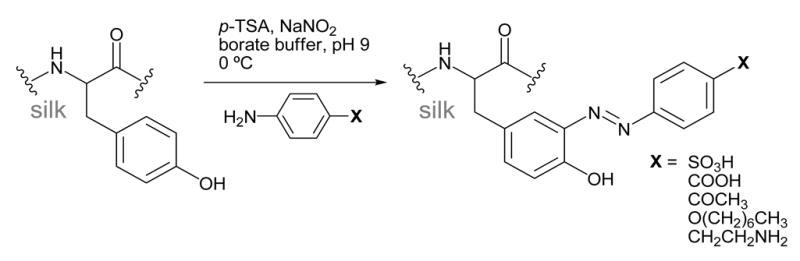
Diazonium coupling
This reaction can be carried out by treating dissolved SF in borate buffer (pH 9.0) with the diazonium salt of the desired aniline derivative, followed by purification using gel filtration columns. The molar ratio of diazonium salt to tyrosine can be tailored to produce the desired level of modification.59 Alternatively, solid silk films and scaffolds can also be modified by first soaking in borate buffer, followed by treatment with the diazonium salt (unpublished).
The extent of the diazonium coupling with these aniline derivatives can be followed with 1H-NMR and UV/vis, as the newly formed azobenzene group has a unique absorbance around 325 nm. When the reaction is carried out in solution with electron-withdrawing anilines, approximately ~70% diazonium salt added results in azo formation. The extent of modification depends on the silk concentration. In reactions using 3% (wt/v) silk solutions, the tyrosine modification level is limited to ~40%, as the diazonium salts have limited solubility in water.59 Higher modification levels can be obtained by diluting the silk concentration, or when the reactions are carried out on solid silk substrates.
Chemical modification using diazonium coupling was also found to influence the fibroin protein structure, where incorporation of sulfonic acid groups was found to inhibit β-sheet self-assembly, while addition of low levels of hydrophobic groups catalyzed β-sheet assembly resulting in rapid hydrogel formation.59
2.3 Grafting reactions
Tyrosinase-catalyzed grafting
Enzyme-catalyzed grafting of biopolymers to SF can be achieved using tyrosinase enzymes.60,61 In the presence of oxygen, tyrosinase is known to hydroxylate and oxidize tyrosine amino acids to form o-quinone derivatives. These quinones can either condense with each other to form crosslinks, or react with molecules containing nucleophiles such as amines or thiols, as shown in Scheme 6.
Scheme 6.
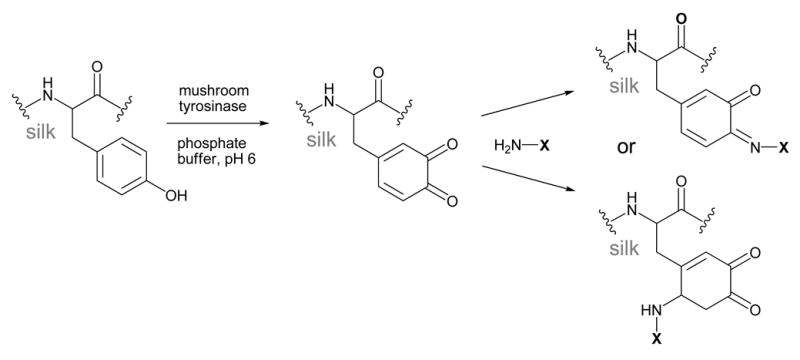
Tyrosinase-catalyzed grafting
The ability of mushroom tyrosinase to oxidize the tyrosine residues in SF by was confirmed using UV absorbance, amino acid analysis, and FTIR.60 The reactive o-quinones formed were further exploited to graft chitosan polysaccharides to SF.60,61 Chitosan is a deacetylated version of the natural polysaccharide chitin, and is composed of β-1,4-linked glucosamine units. The primary amines found in glucosamine can react with the o-quinones formed in SF resulting in either a Schiff-base or Michael addition product, as shown in Scheme 6.
When this grafting reaction was carried out on aqueous solutions of SF in phosphate buffer, it was found that ~30% of the tyrosine residues could be oxidized by tyrosinase and undergo subsequent reaction with chitosan.60 When the same reaction was done on solid SF samples, only ~10% of the tyrosine residues were accessible for reaction in silk gels, and little or no reaction was detected on SF powders and fibers.61 Successful grafting of chitosan to oxidized SF was confirmed using FTIR, DSC and amino acid analysis.60,61
The authors hypothesized that low conversion to the oxidized species in solution (~30%) may be because the majority of the tyrosine residues are buried in hydrophobic domains making them inaccessible for reaction with the enzyme.60 In our hands we have also observed this phenomenon, and found that buffers such as borate buffer are more effective at unfolding the SF protein than phosphate buffer, allowing for higher levels of reaction (unpublished results). Further studies of this reaction in different buffers may allow for higher conversion to the oxidized species.
Poly(methacrylate) grafting
Attachment of acrylate monomers to silk followed by radical polymerization is another polymer grafting approach used to modify the surface of SF fibers. As shown in Scheme 7, the nucleophilic amino acids in degummed SF fabric such as serine (12.1 mol%, ~635 residues) and lysine (0.2 mol%, ~12 residues) were first reacted with 2-methacryloyloxyethyl isocyanate to introduce a double bond capable of radical polymerization.62 The extent of reaction was evaluated with FTIR, and by measuring weight gain. Modified SF samples where approximately 3.5 mol% of the amino acids were modified with 2-methacryloyloxyethyl isocyanate were used in subsequent polymerization reactions. Graft polymerization of 2-methacryloyloxyethyl phosphorylcholine (Scheme 7) onto the isocyanate modified SF was conducted using 2,2′-azobis[2-(2-imidazolin-2-yl)propane dihydrochloride] as an azo polymerization initiator. The weight gain during polymerization reached a plateau after ~2 hours at a level of 26 wt%.62 The resulting polymer-grafted SF was analyzed with FTIR, where peaks corresponding to the added phosphate and quaternary amine functional groups could be distinguished.62
Scheme 7.
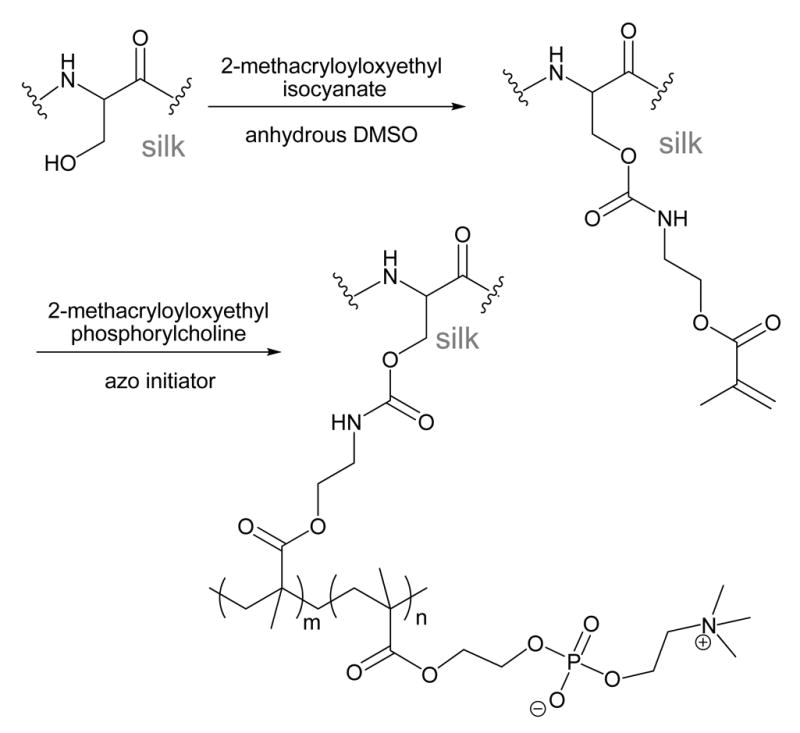
Poly(methacrylate) grafting
3. Biomedical applications of chemically-modified silks
3.1 Coatings for 2D-cell culture
Several modified SF derivatives have been used as coatings on 2D-cell culture substrates to investigate how the surface chemistry or conjugation of biomolecules to the surface of SF can effect cell attachment, growth and differentiation.
Modified surface chemistry
Conjugation of SF to poly(D, L-lactic acid) was found to increase the hydrophilicity of SF, decreasing the contact angle from 69° to 52°. The increased hydrophilicity resulted in increased osteoblast cell attachment and proliferation.52,63 Modification of SF with poly(ethylene glycol) chains was also found to dramatically increase the hydrophilicity, resulting in SF derivatives with a contact angle of 33°. However, these SF-PEG derivatives showed decreased cell attachment of fibroblast cells (L-929).44 Masking the small number of positively charged arginine residues resulted in slightly different levels of initial cell attachment, but no significant differences in cell growth.64
Incorporation of azo groups containing different functional groups that either increased or decreased the hydrophilicity also influenced growth and proliferation of human mesenchymal stem cells (hMSCs).59 hMSCs proliferated more extensively on hydrophilic SF derivatives with contact angles ranging from 43–55°, yet hMSCs were unable to form confluent monolayers on these substrates. In contrast, hMSCs formed confluent monolayers on the hydrophobic (contact angle > 70°) azosilk derivatives.
Direct comparison of these results is difficult, as each study has looked at adhesion and growth of different cell types, which each may have different optimal growth conditions.65,66 However, these results are consistent with literature reports that mammalian cells prefer to adhere to and proliferate on surfaces with a moderate hydrophilicity (water contact angle = 50–70°).67,68 Therefore, increasing the hydrophilicity silk substrates can aid in cell adhesion, but very hydrophilic SF derivatives can negatively effect cell binding. Rather than directly influencing cell behavior, it is likely that the surface hydrophilicity mediates absorption of serum proteins onto the surface, indirectly resulting in increased cell attachment and growth.42,69 The ability to tune the surface chemistry of SF through the modification reactions described above make it a good candidate for more in depth studies of the effects of surface chemistry on cell behavior.
Biomolecule conjugates
Cell attachment to SF can also be enhanced by conjugating small molecules such as lactose to the surface. Sugars immobilized in polymers such as polyacrylamide have been found to increase hepatocyte attachment, presumably through interactions with cell surface glycoproteins.70 Likewise, modification of SF with lactose increased the ability of hepatocytes to adhere to SF, but cells were not able to spread.45 However, fibroblasts cultured on lactose-modified SF showed increased cell attachment, proliferation and survival over cells cultured on unmodified SF.46 In addition, it was found that myofibroblast differentiation was suppressed in fibroblastic cells cultured on both SF and lactose-SF films. This result suggests that these SF materials would be useful for wound healing applications since suppression of fibroblast differentiation into myofibroblasts can reduce hypertrophic scar tissue formation.
3.2 Tissue engineering
One of the major applications for chemically-modified SF is in the field of tissue engineering, which seeks new methods and materials to create synthetic tissue mimics that can be implanted in vivo to spur regeneration of injured or diseased tissue. A typical engineered tissue consists of cells grown within a biodegradable 3D scaffold that is designed to mimic the structural, mechanical and biochemical properties of a native tissue. SF is a promising material for tissue engineering due to its robust mechanical properties, biocompatibility and biodegradability. 3D scaffolds of native and regenerated SF have demonstrated use in ligament,23,24 bone,25–28 cartilage,29–31 fat,32 and vasculature33–35 engineering. However, further optimization of these engineered tissues requires the ability to fine tune the interface between the SF scaffolds and the cells grown within.
Several groups using SF matrices for tissue engineering have explored the effects of covalent attachment of RGD peptides to SF via carbodiimide coupling. The RGD peptide is known to engage integrins located on the surface of many cell types, and facilitate cellular adhesion to substrates displaying the RGD sequence.71 Several studies suggest that integrin binding can also initiate several internal cell signaling pathways, such as those involved in differentiation.72–74 As expected, attachment of RGD peptides to the surface of silk films40,48 and 3D scaffolds results in marked improvement in cell attachment and differentiation in engineered ligament,50 bone,28 and dental51,75 tissue constructs.
Covalent attachment of protein-based growth factors to SF has also been explored as a means to enhance differentiation of cells cultured within SF scaffolds. One commonly explored growth factor for bone tissue engineering is bone morphogenic protein (BMP-2), which plays an important role in bone formation and regeneration. BMP-2 can be directly immobilized on SF films,51 scaffolds,7,76 and electrospun mats6 via carbodiimide coupling. In all cases, hMSCs cultured on SF substrates containing immobilized BMP-2 exhibited significantly higher expression of osteogenic markers when subjected to osteogenic stimulants.
3.3 Blood-contacting materials
Several approaches have been taken to chemically-modify SF to improve its blood compatibility. Many approaches have been modeled after the structure of the highly sulfated polysaccharide heparin, which is a commonly used anticoagulant. In order to mimic the heparin structure, strategies have been developed to incorporate sulfate groups into SF. SF derivatives produced through the reaction with chlorosulfonic acid,56 and sulfonated silk blends58 were shown to be effective anticoagulants, suggesting that this type of chemical modification of SF would be useful for applications where these materials will be in contact with blood. Anti-HIV activity was also demonstrated with sulfated SF derivatives.57
A second approach to improving the blood compatibility of SF involved grafting of 2-methacryloyloxyethyl phosphorylcholine (MPC) onto silk fabric.62 Grafting MPC onto polymers such as cellulose had been previously shown to reduce protein and cell adhesion to membranes used in blood dialysis. Likewise, incorporation of PC into silk was found to significantly reduce platelet adhesion, which may reduce inflammatory reactions to these materials in vivo.62
3.4 Drug delivery
Utilization of SF in drug delivery applications has also been investigated due to the biocompatibility and relatively slow degradation of the silk protein in vivo.22 Due to slow degradation, the primary mechanism for drug release from silk matrices is diffusion. Therefore, the release rate is highly dependant on both the conditions used to process the silk,17,22 and the chemical properties of the drug.22,77 Small molecules and proteins can be non-specifically encapsulated within SF films5,20,22 microspheres,4,78 or tablets,79,80 and released by passive diffusion.
However, drugs can also be covalently bound to silk to limit their diffusion from the SF matrix or to increase their stability. For example, the biological stability of insulin53 and L-asparaginase10 was increased when these drugs were covalently attached to SF via glutaraldehye crosslinking. In addition, SF-asparaginase conjugates showed higher enzyme activity, and reduced immunogenicity than the enzyme alone.10 These results suggest that covalent attachment of drugs to SF is a promising approach to increase stability, circulation time, and effectiveness of many protein based drugs and enzymes.
4. Conclusion
A variety of silk protein modification chemistries have been reported. These approaches exploit the amino acid composition and block copolymer nature of the protein, expanding the utility of this protein family in a range of biomaterial needs. These chemical modifications have been shown to alter important silk material features such as hydrophobicity, β-sheet content, solution behavior and materials morphology. Additional chemical modifications to alter cell interactions and functions on silk-based biomaterials have been explored and demonstrate the versatility and utility of these approaches to optimize silk-based biomaterials for a range of needs in the field of regenerative medicine.
Supplementary Material
Acknowledgments
A.M. would like to thank the Training in Education and Critical Research Skills Program at Tufts University and the Ruth L. Kirschstein National Research Service Award Postdoctoral Fellowship supported by NIH/NIAMS for funding. We also thank the NIH via the Tissue Engineering Resource Center (P41 EB002520) for additional support.
Biographies

Amanda Murphy obtained her Ph.D. in Chemistry at the University of California at Berkeley in 2006, working with Professor Jean Fréchet. Currently she is a postdoctoral fellow in the laboratory of Professor David Kaplan in the Biomedical Engineering Department at Tufts University.

David Kaplan holds the Stern Family Professor of Engineering Endowed Chair at Tufts University. He is Professor & Chair of the Department of Biomedical Engineering. His research focus is on biopolymer engineering to understand structure-function relationships, with emphasis on studies related to self-assembly, biomaterials engineering and functional tissue engineering.
References
- 1.Zhou CZ, Confalonieri F, Jacquet M, Perasso R, Li ZG, Janin J. Proteins. 2001;44:119–122. doi: 10.1002/prot.1078. [DOI] [PubMed] [Google Scholar]
- 2.Heslot H. Biochimie. 1998;80:19–31. doi: 10.1016/s0300-9084(98)80053-9. [DOI] [PubMed] [Google Scholar]
- 3.Horan RL, Antle K, Collette AL, Wang Y, Huang J, Moreau JE, Volloch V, Kaplan DL, Altman GH. Biomaterials. 2005;26:3385–3393. doi: 10.1016/j.biomaterials.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Wenk E, Matsumoto A, Meinel L, Li C, Kaplan DL. J Control Release. 2007;117:360–370. doi: 10.1016/j.jconrel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Hu X, Daley A, Rabotyagova O, Cebe P, Kaplan DL. J Control Release. 2007;121:190–199. doi: 10.1016/j.jconrel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li C, Vepari C, Jin HJ, Kim HJ, Kaplan DL. Biomaterials. 2006;27:3115–3124. doi: 10.1016/j.biomaterials.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 7.Karageorgiou V, Tomkins M, Fajardo R, Meinel L, Snyder B, Wade K, Chen J, Vunjak-Novakovic G, Kaplan DL. J Biomed Mater Res A. 2006;78:324–334. doi: 10.1002/jbm.a.30728. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Kim HJ, Xu P, Matsumoto A, Kaplan DL. Langmuir. 2005;21:11335–11341. doi: 10.1021/la051862m. [DOI] [PubMed] [Google Scholar]
- 9.Kim UJ, Park J, Kim HJ, Wada M, Kaplan DL. Biomaterials. 2005;26:2775–2785. doi: 10.1016/j.biomaterials.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 10.Nazarov R, Jin HJ, Kaplan DL. Biomacromolecules. 2004;5:718–726. doi: 10.1021/bm034327e. [DOI] [PubMed] [Google Scholar]
- 11.Kim UJ, Park J, Li C, Jin HJ, Valluzzi R, Kaplan DL. Biomacromolecules. 2004;5:786–792. doi: 10.1021/bm0345460. [DOI] [PubMed] [Google Scholar]
- 12.Jin HJ, Chen J, Karageorgiou V, Altman GH, Kaplan DL. Biomaterials. 2004;25:1039–1047. doi: 10.1016/s0142-9612(03)00609-4. [DOI] [PubMed] [Google Scholar]
- 13.Jin HJ, Fridrikh SV, Rutledge GC, Kaplan DL. Biomacromolecules. 2002;3:1233–1239. doi: 10.1021/bm025581u. [DOI] [PubMed] [Google Scholar]
- 14.Valluzzi R, Winkler S, Wilson D, Kaplan DL. Philos Trans R Soc Lond B Biol Sci. 2002;357:165–167. doi: 10.1098/rstb.2001.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winkler S, Wilson D, Kaplan DL. Biochemistry. 2000;39:12739–12746. doi: 10.1021/bi001335w. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto A, Chen J, Collette AL, Kim UJ, Altman GH, Cebe P, Kaplan DL. J Phys Chem B. 2006;110:21630–21638. doi: 10.1021/jp056350v. [DOI] [PubMed] [Google Scholar]
- 17.Jin HJJP, Karageorgiou V, Kim UJ, Valluzzi R, Cebe P, Kaplan DL. Advanced Functional Materials. 2005;15:1241–1247. [Google Scholar]
- 18.Wang Y, Rudym DD, Walsh A, Abrahamsen L, Kim HJ, Kim HS, Kirker-Head C, Kaplan DL. Biomaterials. 2008;29:3415–3428. doi: 10.1016/j.biomaterials.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inouye K, Kurokawa M, Nishikawa S, Tsukada M. J Biochem Biophys Methods. 1998;37:159–164. doi: 10.1016/s0165-022x(98)00024-4. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Zhang X, Castellot J, Herman I, Iafrati M, Kaplan DL. Biomaterials. 2008;29:894–903. doi: 10.1016/j.biomaterials.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Wenk E, Hu X, Castro GR, Meinel L, Li C, Merkle H, Kaplan DL. Biomaterials. 2007;28:4161–4169. doi: 10.1016/j.biomaterials.2007.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofmann S, Foo CT, Rossetti F, Textor M, Vunjak-Novakovic G, Kaplan DL, Merkle HP, Meinel L. J Control Release. 2006;111:219–227. doi: 10.1016/j.jconrel.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Altman GH, Horan RL, Lu HH, Moreau J, Martin I, Richmond JC, Kaplan DL. Biomaterials. 2002;23:4131–4141. doi: 10.1016/s0142-9612(02)00156-4. [DOI] [PubMed] [Google Scholar]
- 24.Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen J, Lu H, Richmond J, Kaplan DL. Biomaterials. 2003;24:401–416. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- 25.Kim HJ, Kim UJ, Leisk GG, Bayan C, Georgakoudi I, Kaplan DL. Macromol Biosci. 2007;7:643–655. doi: 10.1002/mabi.200700030. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann S, Hagenmuller H, Koch AM, Muller R, Vunjak-Novakovic G, Kaplan DL, Merkle HP, Meinel L. Biomaterials. 2007;28:1152–1162. doi: 10.1016/j.biomaterials.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 27.Meinel L, Betz O, Fajardo R, Hofmann S, Nazarian A, Cory E, Hilbe M, McCool J, Langer R, Vunjak-Novakovic G, Merkle HP, Rechenberg B, Kaplan DL, Kirker-Head C. Bone. 2006;39:922–931. doi: 10.1016/j.bone.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 28.Meinel L, Karageorgiou V, Hofmann S, Fajardo R, Snyder B, Li C, Zichner L, Langer R, Vunjak-Novakovic G, Kaplan DL. J Biomed Mater Res A. 2004;71:25–34. doi: 10.1002/jbm.a.30117. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Blasioli DJ, Kim HJ, Kim HS, Kaplan DL. Biomaterials. 2006;27:4434–4442. doi: 10.1016/j.biomaterials.2006.03.050. [DOI] [PubMed] [Google Scholar]
- 30.Hofmann S, Knecht S, Langer R, Kaplan DL, Vunjak-Novakovic G, Merkle HP, Meinel L. Tissue Eng. 2006;12:2729–2738. doi: 10.1089/ten.2006.12.2729. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Kim UJ, Blasioli DJ, Kim HJ, Kaplan DL. Biomaterials. 2005;26:7082–7094. doi: 10.1016/j.biomaterials.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 32.Mauney JR, Nguyen T, Gillen K, Kirker-Head C, Gimble JM, Kaplan DL. Biomaterials. 2007;28:5280–5290. doi: 10.1016/j.biomaterials.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Baughman CB, Kaplan DL. Biomaterials. 2008;29:2217–2227. doi: 10.1016/j.biomaterials.2008.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soffer L, Wang X, Zhang X, Kluge J, Dorfmann L, Kaplan DL, Leisk G. J Biomater Sci Polym Ed. 2008;19:653–664. doi: 10.1163/156856208784089607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lovett M, Cannizzaro C, Daheron L, Messmer B, Vunjak-Novakovic G, Kaplan DL. Biomaterials. 2007;28:5271–5279. doi: 10.1016/j.biomaterials.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meinel L, Hofmann S, Karageorgiou V, Kirker-Head C, McCool J, Gronowicz G, Zichner L, Langer R, Vunjak-Novakovic G, Kaplan DL. Biomaterials. 2005;26:147–155. doi: 10.1016/j.biomaterials.2004.02.047. [DOI] [PubMed] [Google Scholar]
- 37.Acharya C, Ghosh SK, Kundu SC. J Mater Sci Mater Med. 2008;19:2827–2836. doi: 10.1007/s10856-008-3408-3. [DOI] [PubMed] [Google Scholar]
- 38.Kim HJ, Kim UJ, Vunjak-Novakovic G, Min BH, Kaplan DL. Biomaterials. 2005;26:4442–4452. doi: 10.1016/j.biomaterials.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 39.Zhao C, Yao J, Masuda H, Kishore R, Asakura T. Biopolymers. 2003;69:253–259. doi: 10.1002/bip.10350. [DOI] [PubMed] [Google Scholar]
- 40.Sofia S, McCarthy MB, Gronowicz G, Kaplan DL. J Biomed Mater Res. 2001;54:139–148. doi: 10.1002/1097-4636(200101)54:1<139::aid-jbm17>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 41.Uebersax L, Hagenmuller H, Hofmann S, Gruenblatt E, Muller R, Vunjak-Novakovic G, Kaplan DL, Merkle HP, Meinel L. Tissue Eng. 2006;12:3417–3429. doi: 10.1089/ten.2006.12.3417. [DOI] [PubMed] [Google Scholar]
- 42.Keselowsky BG, Collard DM, Garcia AJ. Proc Natl Acad Sci U S A. 2005;102:5953–5957. doi: 10.1073/pnas.0407356102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gotoh Y, Tsukada M, Minoura N. Bioconjug Chem. 1993;4:554–559. doi: 10.1021/bc00024a020. [DOI] [PubMed] [Google Scholar]
- 44.Gotoh Y, Tsukada M, Minoura N, Imai Y. Biomaterials. 1997;18:267–271. doi: 10.1016/s0142-9612(96)00137-8. [DOI] [PubMed] [Google Scholar]
- 45.Gotoh Y, Niimi S, Hayakawa T, Miyashita T. Biomaterials. 2004;25:1131–1140. doi: 10.1016/s0142-9612(03)00633-1. [DOI] [PubMed] [Google Scholar]
- 46.Acharya C, Hinz B, Kundu SC. Biomaterials. 2008;29:4665–4675. doi: 10.1016/j.biomaterials.2008.08.033. [DOI] [PubMed] [Google Scholar]
- 47.Gotoh Y, Tsukada M, Aiba S, Minoura N. Int J Biol Macromol. 1996;18:19–26. doi: 10.1016/0141-8130(95)01039-4. [DOI] [PubMed] [Google Scholar]
- 48.Kardestuncer T, McCarthy MB, Karageorgiou V, Kaplan D, Gronowicz G. Clin Orthop Relat Res. 2006;448:234–239. doi: 10.1097/01.blo.0000205879.50834.fe. [DOI] [PubMed] [Google Scholar]
- 49.Vepari CP, Kaplan DL. Biotechnol Bioeng. 2006;93:1130–1137. doi: 10.1002/bit.20833. [DOI] [PubMed] [Google Scholar]
- 50.Chen J, Altman GH, Karageorgiou V, Horan R, Collette A, Volloch V, Colabro T, Kaplan DL. J Biomed Mater Res A. 2003;67:559–570. doi: 10.1002/jbm.a.10120. [DOI] [PubMed] [Google Scholar]
- 51.Karageorgiou V, Meinel L, Hofmann S, Malhotra A, Volloch V, Kaplan D. J Biomed Mater Res A. 2004;71:528–537. doi: 10.1002/jbm.a.30186. [DOI] [PubMed] [Google Scholar]
- 52.Cai K, Yao K, Lin S, Yang Z, Li X, Xie H, Qing T, Gao L. Biomaterials. 2002;23:1153–1160. doi: 10.1016/s0142-9612(01)00230-7. [DOI] [PubMed] [Google Scholar]
- 53.Zhang YQ, Ma Y, Xia YY, Shen WD, Mao JP, Zha XM, Shirai K, Kiguchi K. J Biomed Mater Res B Appl Biomater. 2006;79:275–283. doi: 10.1002/jbm.b.30539. [DOI] [PubMed] [Google Scholar]
- 54.Gotoh Y, Tsukada M, Minoura N. Int J Biol Macromol. 1996;19:41–44. doi: 10.1016/0141-8130(96)01098-7. [DOI] [PubMed] [Google Scholar]
- 55.Gotoh Y, Tsukada M, Minoura N. Int J Biol Macromol. 1992;14:198–200. doi: 10.1016/s0141-8130(05)80027-3. [DOI] [PubMed] [Google Scholar]
- 56.Tamada Y. Biomaterials. 2004;25:377–383. doi: 10.1016/s0142-9612(03)00533-7. [DOI] [PubMed] [Google Scholar]
- 57.Gotoh K, Izumi H, Kanamoto T, Tamada Y, Nakashima H. Biosci Biotechnol Biochem. 2000;64:1664–1670. doi: 10.1271/bbb.64.1664. [DOI] [PubMed] [Google Scholar]
- 58.Ma X, Cao C, Zhu H. J Biomed Mater Res B Appl Biomater. 2006;78:89–96. doi: 10.1002/jbm.b.30466. [DOI] [PubMed] [Google Scholar]
- 59.Murphy AR, St John P, Kaplan DL. Biomaterials. 2008;29:2829–2838. doi: 10.1016/j.biomaterials.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sampaio S, Taddei P, Monti P, Buchert J, Freddi G. J Biotechnol. 2005;116:21–33. doi: 10.1016/j.jbiotec.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 61.Freddi G, Anghileri A, Sampaio S, Buchert J, Monti P, Taddei P. J Biotechnol. 2006;125:281–294. doi: 10.1016/j.jbiotec.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 62.Furuzono T, Ishihara K, Nakabayashi N, Tamada Y. Biomaterials. 2000;21:327–333. doi: 10.1016/s0142-9612(99)00177-5. [DOI] [PubMed] [Google Scholar]
- 63.Cai K, Yao K, Cui Y, Yang Z, Li X, Xie H, Qing T, Gao L. Biomaterials. 2002;23:1603–1611. doi: 10.1016/s0142-9612(01)00287-3. [DOI] [PubMed] [Google Scholar]
- 64.Gotoh Y, Tsukada M, Minoura N. J Biomed Mater Res. 1998;39:351–357. doi: 10.1002/(sici)1097-4636(19980305)39:3<351::aid-jbm2>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 65.Rappaport C. In Vitro Cell Dev Biol Anim. 2003;39:187–192. doi: 10.1290/1543-706X(2003)039<0187:RICAPO>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 66.Jacobson BS, Ryan US. Tissue Cell. 1982;14:69–83. doi: 10.1016/0040-8166(82)90008-8. [DOI] [PubMed] [Google Scholar]
- 67.Kim MS, Shin YN, Cho MH, Kim SH, Kim SK, Cho YH, Khang G, Lee IW, Lee HB. Tissue Eng. 2007;13:2095–2103. doi: 10.1089/ten.2006.0062. [DOI] [PubMed] [Google Scholar]
- 68.Grinnell F. Int Rev Cytol. 1978;53:65–144. doi: 10.1016/s0074-7696(08)62241-x. [DOI] [PubMed] [Google Scholar]
- 69.Garcia AJ, Vega MD, Boettiger D. Mol Biol Cell. 1999;10:785–798. doi: 10.1091/mbc.10.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weigel PH, Schmell E, Lee YC, Roseman S. J Biol Chem. 1978;253:330–333. [PubMed] [Google Scholar]
- 71.Hersel U, Dahmen C, Kessler H. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 72.Takeuchi Y, Suzawa M, Kikuchi T, Nishida E, Fujita T, Matsumoto T. J Biol Chem. 1997;272:29309–29316. doi: 10.1074/jbc.272.46.29309. [DOI] [PubMed] [Google Scholar]
- 73.Xiao G, Wang D, Benson MD, Karsenty G, Franceschi RT. J Biol Chem. 1998;273:32988–32994. doi: 10.1074/jbc.273.49.32988. [DOI] [PubMed] [Google Scholar]
- 74.Keselowsky BG, Wang L, Schwartz Z, Garcia AJ, Boyan BD. J Biomed Mater Res A. 2007;80:700–710. doi: 10.1002/jbm.a.30898. [DOI] [PubMed] [Google Scholar]
- 75.Xu WP, Zhang W, Asrican R, Kim HJ, Kaplan DL, Yelick PC. Tissue Eng Part A. 2008;14:549–557. doi: 10.1089/tea.2007.0227. [DOI] [PubMed] [Google Scholar]
- 76.Kirker-Head C, Karageorgiou V, Hofmann S, Fajardo R, Betz O, Merkle HP, Hilbe M, von Rechenberg B, McCool J, Abrahamsen L, Nazarian A, Cory E, Curtis M, Kaplan D, Meinel L. Bone. 2007;41:247–255. doi: 10.1016/j.bone.2007.04.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fang JY, Chen JP, Leu YL, Wang HY. Chem Pharm Bull (Tokyo) 2006;54:156–162. doi: 10.1248/cpb.54.156. [DOI] [PubMed] [Google Scholar]
- 78.Wilz A, Pritchard EM, Li T, Lan JQ, Kaplan DL, Boison D. Biomaterials. 2008;29:3609–3616. doi: 10.1016/j.biomaterials.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Katayama H, Issiki M, Yoshitomi H. Biol Pharm Bull. 2000;23:1229–1234. doi: 10.1248/bpb.23.1229. [DOI] [PubMed] [Google Scholar]
- 80.Bayraktar O, Malay O, Ozgarip Y, Batigun A. Eur J Pharm Biopharm. 2005;60:373–381. doi: 10.1016/j.ejpb.2005.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.