This paper reports a strategy to perform functional assays of membrane proteins that are immobilized to biochips. The work is significant because it combines the use of nanodiscs to compartmentalize membrane proteins within a lipid bilayer environment and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) to detect binding interactions with other protein partners in a label-independent manner. Proteins that span the lipid bilayer membrane—including cell adhesion molecules, growth factor and cytokine receptors, and transporters—mediate a broad range of interactions of the cell with its environment and are prominent targets in many drug discovery programs. The difficulties associated with traditional cell-based and micelle-solubilized assays have motivated significant work in the past decade to immobilize proteins within lipid bilayers supported on flat surfaces,[1–7] and has made possible the reconstitution of membrane proteins and assays of their activities by fluorescence and evanescent wave methods.
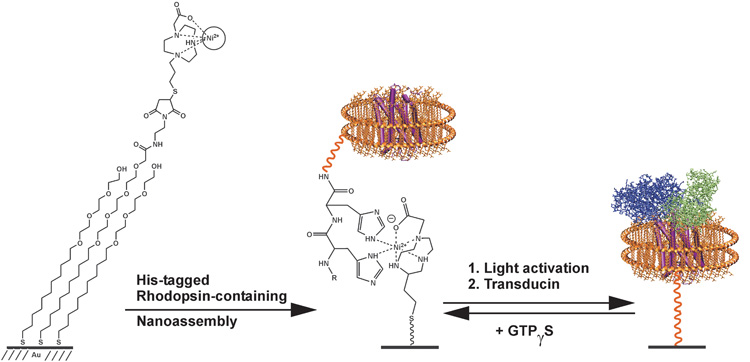
Our objective in the current work was to use mass spectrometry (MS) to perform functional assays of membrane resident proteins. MS methods have the benefits that they do not require labels—and, therefore, simplify and increase the scope of assays and provide molecular information about the analyte. Our design is presented in Figure 1 and begins with solubilization of the seven transmembrane photoreceptor rhodopsin in a nanodisc. The nanodisc strategy is based on the self-assembly of lipid molecules within a membrane scaffold protein, giving a circular patch of soluble lipid bilayer that can additionally contain a transmembrane protein.[8–10] We expressed a membrane scaffold protein (MSP) having a hexa-his tag at its N-terminus and prepared nanodiscs containing the rhodopsin protein and the lipid 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) as described in the supplementary material.[11–16] A MALDI-TOF mass spectrum of the nanodiscs showed a clear peak at m/z = 25.6 kDa for the membrane scaffold protein, and at 760.1 for the lipid molecule.
Figure 1.
The seven-transmembrane protein rhodopsin is immobilized to a self-assembled monolayer by way of a his-tagged membrane scaffold protein. Upon activation of rhodopsin with light, the receptor binds the transducin complex, and can be detected using SAMDI-TOF mass spectrometry.
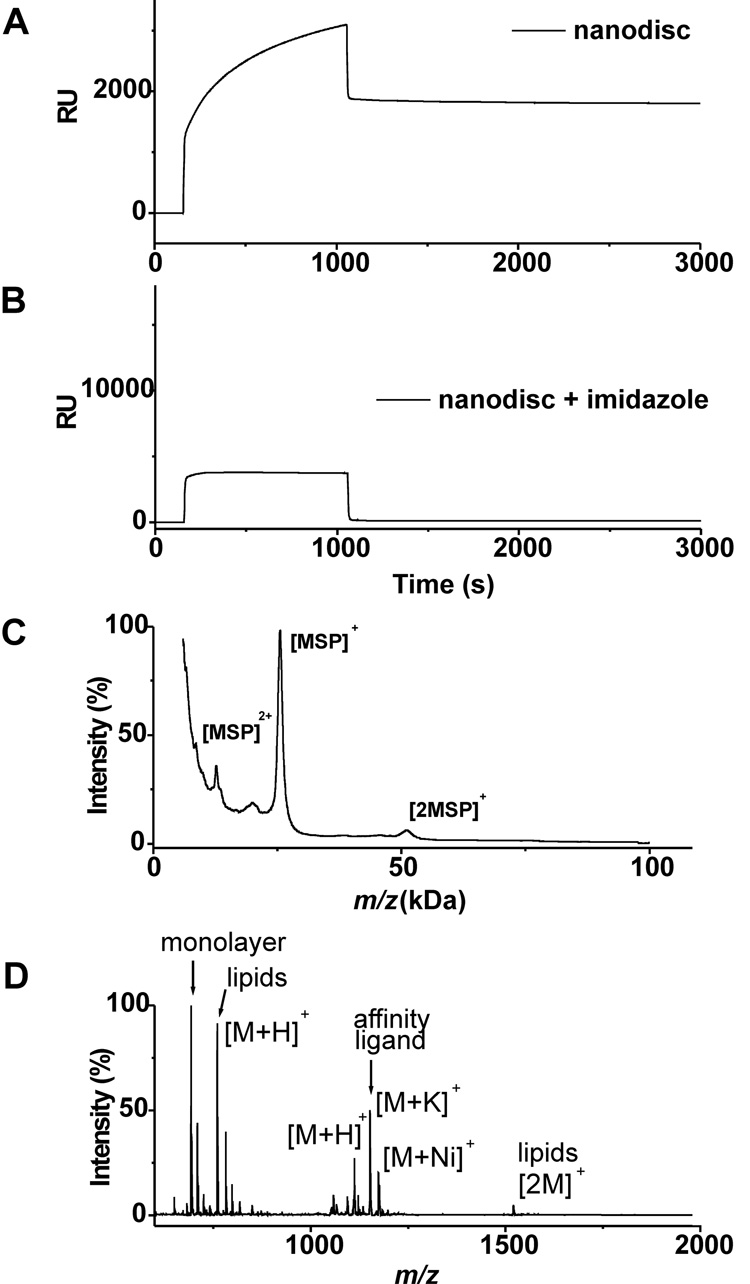
We first characterized the immobilization of the lipid-loaded nanodiscs to a self-assembled monolayer. To prepare this surface, we immobilized a thiol-terminated triazacyclononane-derived ligand (Actacn), which is functionally analogous to the common NTA ligand for immobilization of his-tagged proteins,[17, 18] to a monolayer presenting maleimide and tri(ethylene glycol) groups (see supplementary materials).[19] The former provides a defined density of attachment sites for the nanodiscs and the latter are effective at preventing non-specific interactions of proteins with the surface. We prepared nanodisc assemblies containing only the membrane scaffold protein and POPC and monitored the immobilization of this construct using surface plasmon resonance spectroscopy (SPR) (Figure 2A). The nanodisc efficiently bound to the Actacn-Ni(II) complex on the monolayer and remained bound during a wash with buffer. The addition of imidazole, which complexes the Ni(II) ion, prevented binding of the his-tagged nanodisc and demonstrates the effectiveness of the oligo(ethylene glycol) groups to prevent unwanted protein adsorption. Following immobilization of the nanodisc, we used MALDI-TOF mass spectrometry to characterize the surface. This technique, known as SAMDI,[20–23] shows a peak at [M]+ m/z = 25.6 kDa for the his-tagged membrane scaffold protein (Figure 2C). Another spectrum acquired at lower mass range also revealed peaks at [M]+ m/z = 760.1 and [2M]+ at m/z = 1520.2 that correspond to the lipid molecules of the nanodisc (Figure 2D). Hence, the nanodiscs could be efficiently immobilized to self-assembled monolayers and observed by both surface plasmon resonance spectroscopy and SAMDI mass spectrometry.
Figure 2.
Characterization of his-tagged nanoassemblies containing lipids immobilized onto Actacn-presenting monolayers. SPR binding curve of nanodiscs in the absence (A) and presence of imidazole (150 mM). SAMDI-TOF MS spectra of duplicate samples prepared as in panel A. Sinapinic acid was used as a matrix to observe protein (C) and THAP was used to observe small molecules (D).
Nanodiscs containing the protein rhodopsin were subsequently self assembled at a mole ratio of 700:10:1 POPC:MSP:rhodopsin. Again, SPR and SAMDI MS confirmed the immobilization of the protein-loaded nanodiscs to monolayers. The SAMDI spectrum revealed peaks at [M]+ m/z = 25.6 KDa and [2M]+ at m/z = 12.8 KDa for the MSP and a peak at [M]+ m/z = 43.6 KDa for rhodopsin (Figure 3A).
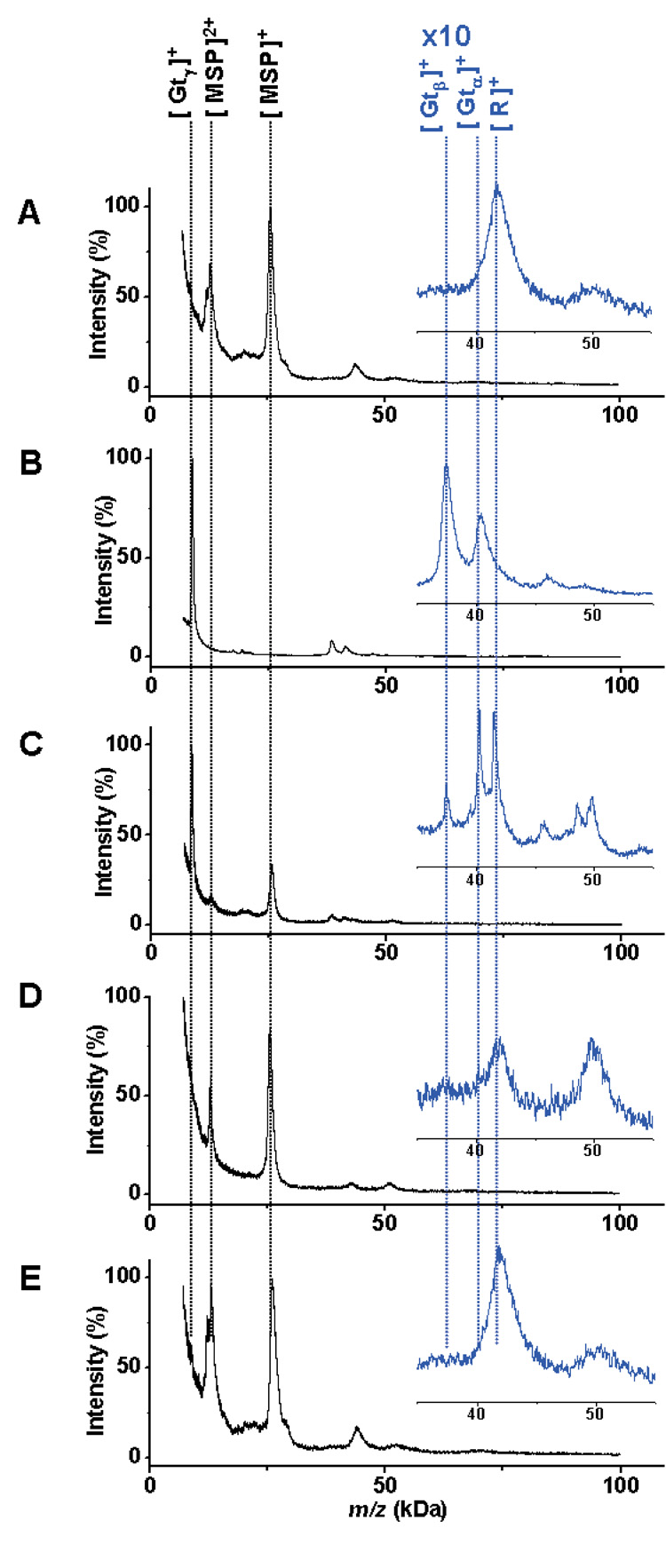
Figure 3.
A) SAMDI-TOF MS spectrum of rhodopsin-loaded nanoassemblies immobilized to a monolayer. B) MALDI-TOF MS spectrum of transducin. C) Immobilized rhodopsin exposed to light binds to the transducin protein complex. Peaks for the transducin α (Gtα), β (Gtβ) γ (Gtγ), rhodopsin (R) and the membrane scaffold protein (MSP) are indicated at the top. D) A control experiment performed in the dark prevented binding of transducin to immobilized rhodopsin, as did inclusion of the nucleotide inhibitor GTPγS (E).
We then performed a functional assay based on the photoactivation of rhodopsin, whereby photoisomerization of 11-cis retinal, a lysine-bound chromophore, leads to a conformational change that allows for binding of the transducin protein complex.[24] We incubated the Actacn-presenting monolayers with a mixture of the rhodopsin-loaded nanodiscs and transducin (10 µM) for 30 minutes at room temperature and in the field of ambient light. A SAMDI spectrum revealed peaks corresponding to both MSP and transducin (Figure 3C). The peak for the γ-subunit of transducin has a greater intensity than those for the α and β subunits, or for rhodopsin. We frequently find that lower molecular weight proteins yield more intense peaks. Hence, a comparison of peaks for γ-transducin and MSP provides the clearest evidence for activity of the rhodopsin receptor. A control experiment performed in the dark, which prevented the activation of the rhodopsin receptor, was analyzed by SAMDI and revealed peaks corresponding only to the MSP and rhodopsin (Figure 3D). This protein-protein interaction is also regulated by small molecules, including the nonhydrolysable nucleotide GTPγS which binds to the α subunit of transducin (Gtα) and blocks its interaction with rhodopsin.[25] We treated a monolayer presenting the rhodopsin-transducin complex with an excess of GTPγS. Analysis of this substrate by SAMDI showed that the inhibitor led to dissociation of transducin from the nanodisc-bound rhodopsin (Figure 3E).
This example establishes a route to performing functional assays of membrane bound proteins that are immobilized to biochip surfaces. Several aspects of this approach are significant. Nanodiscs provide a means to control the size of the bilayer,[11] the lipid composition[26] and the stoichiometry of the incorporated membrane protein,[27] permitting these macromolecules to be treated as soluble proteins. The self-assembled monolayers provide well-defined substrates for protein immobilization. The density of the immobilized proteins can be controlled by adjusting the density of the Actacn ligand and therefore a more uniform activity of protein. We note that a range of other immobilization chemistries are available, including the cutinase affinity tag that gives a covalent immobilization of protein.[28–30]
Finally, the monolayers enable the use of mass spectrometry to directly observe the proteins immobilized to the monolayer and any interacting partners. This strategy obviates the need for fluorescent or radioisotopic labels, provide mass information on the species bound to biochip and, therefore, can be effective at identifying (and ignoring) background species, at identifying post-translational modifications of immobilized proteins, and at performing multi-analyte assays. These attributes bring to membrane-bound proteins the full range of techniques that have been important for assaying the activities of soluble proteins.
Supplementary Material
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Acknowledgments
This work was supported primarily by the Nanoscale Science and Engineering Initiative of the National Science Foundation under NSF Award Number EEC-0647560.
Footnotes
Publisher's Disclaimer: This PDF receipt will only be used as the basis for generating PubMed Central (PMC) documents. PMC documents will be made available for review after conversion (approx. 2–3 weeks time). Any corrections that need to be made will be done at that time. No materials will be released to PMC without the approval of an author. Only the PMC documents will appear on PubMed Central -- this PDF Receipt will not appear on PubMed Central.
Contributor Information
Dr. Violeta L. Marin, Department of Chemistry and Howard Hughes Medical Institute, University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA), Fax: (+1) 773-702-1677
Dr. Timothy H. Bayburt, Department of Biochemistry, University of Illinois, Urbana, IL 61801 (USA)
Prof. Stephen G. Sligar, Departments of Chemistry and Biochemistry, University of Illinois, Urbana, IL 61801 (USA), E-mail: s-sligar@uiuc.edu
Prof. Milan Mrksich, Department of Chemistry and Howard Hughes Medical Institute, University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA), Fax: (+1) 773-702-1677, E-mail: mmrksich@uchicago.edu
References
- 1.Fang Y, Frutos AG, Lahiri J. Chem.BioChem. 2002;3:987. doi: 10.1002/1439-7633(20021004)3:10<987::AID-CBIC987>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 2.Fang Y, Frutos AG, Lahiri J. J. Am. Chem. Soc. 2002;124:2394. doi: 10.1021/ja017346+. [DOI] [PubMed] [Google Scholar]
- 3.Hong Y, Webb BL, Pai S, Ferrie A, Peng J, Lai F, Lahiri J, Biddlecome G, Rasnow B, Johnson M, Min H, Fang Y, Salon J. J. Biomol. Screen. 2006;11:435. doi: 10.1177/1087057106287139. [DOI] [PubMed] [Google Scholar]
- 4.Kraft ML, Fishel SF, Marxer CG, Weber PK, Hutcheon ID, Boxer SG. Appl. Surf. Sci. 2006;252:6950. [Google Scholar]
- 5.Lahiri J, Kalal P, Frutos AG, Jonas SJ, Schaeffler R. Langmuir. 2000;16:7805. [Google Scholar]
- 6.Rigler P, Ulrich W-P, Vogel H. Langmuir. 2004;20:7901. doi: 10.1021/la049002d. [DOI] [PubMed] [Google Scholar]
- 7.Yoshina-Ishii C, Boxer SG. J. Am. Chem. Soc. 2003;125:3696. doi: 10.1021/ja029783+. [DOI] [PubMed] [Google Scholar]
- 8.Bayburt TH, Grinkova YV, Sligar SG. Nano Lett. 2002;2:853. [Google Scholar]
- 9.Bayburt TH, Sligar SG. Protein Sci. 2003;12:2476. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11509. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. J. Am. Chem. Soc. 2004;126:3477. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 12.Ernst OP, Bieri C, Vogel H, Hofmann KP. Vertebrate Phototransduction and the Visual Cycle, Part A. 2000;Vol. 315:471. [Google Scholar]
- 13.Fahmy K, Sakmar TP. Biochemistry. 1993;32:7229. doi: 10.1021/bi00079a020. [DOI] [PubMed] [Google Scholar]
- 14.Litman BJ, Aton B, Hartley JB. Vision Res. 1982;22:1439. doi: 10.1016/0042-6989(82)90206-1. [DOI] [PubMed] [Google Scholar]
- 15.Papermaster DS. Meth. Enzymol. 1982;81:48. doi: 10.1016/s0076-6879(82)81010-0. [DOI] [PubMed] [Google Scholar]
- 16.Wessling Resnick M, Johnson GL. J. Biol. Chem. 1987;262:12444. [PubMed] [Google Scholar]
- 17.Johnson DL, Martin LL. J. Am. Chem. Soc. 2005;127:2018. doi: 10.1021/ja045084g. [DOI] [PubMed] [Google Scholar]
- 18.Warden A, Graham B, Hearn MTW, Spiccia L. Org. Lett. 2001;3:2855. doi: 10.1021/ol016291d. [DOI] [PubMed] [Google Scholar]
- 19.Houseman BT, Gawalt ES, Mrksich M. Langmuir. 2003;19:1522. [Google Scholar]
- 20.Min D-H, Su J, Mrksich M. Angew. Chem., Int. Ed. 2004;43:5973. doi: 10.1002/anie.200461061. [DOI] [PubMed] [Google Scholar]
- 21.Min D-H, Tang W-J, Mrksich M. Nat. Biotechnol. 2004;22:717. doi: 10.1038/nbt973. [DOI] [PubMed] [Google Scholar]
- 22.Su J, Mrksich M. Angew. Chem., Int. Ed. 2002;41:4715. doi: 10.1002/anie.200290026. [DOI] [PubMed] [Google Scholar]
- 23.Su J, Mrksich M. Langmuir. 2003;19:4867. [Google Scholar]
- 24.Pepe IM. J. Photochem. Photobiol., B. 1999;48:1. doi: 10.1016/S1011-1344(99)00200-6. [DOI] [PubMed] [Google Scholar]
- 25.Noel JP, Hamm HE, Sigler PB. Nature. 1993;366:654. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- 26.Shaw AW, Pureza VS, Sligar SG, Morrissey JH. J. Biol. Chem. 2007;282:6556. doi: 10.1074/jbc.M607973200. [DOI] [PubMed] [Google Scholar]
- 27.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. J. Biol. Chem. 2007;282:14875. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 28.Kwon Y, Han ZZ, Karatan E, Mrksich M, Kay BK. Anal. Chem. 2004;76:5713. doi: 10.1021/ac049731y. [DOI] [PubMed] [Google Scholar]
- 29.Hodneland CD, Lee YS, Min DH, Mrksich M. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5048. doi: 10.1073/pnas.072685299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeo W-S, Min D-H, Hsieh RW, Greene GL, Mrksich M. Angew. Chem., Int. Ed. 2005;44:5480. doi: 10.1002/anie.200501363. [DOI] [PubMed] [Google Scholar]
- 31.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. J. Am. Chem. Soc. 2004;126:3477. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 32.Fahmy K, Sakmar TP. Biochemistry. 1993;32:7229. doi: 10.1021/bi00079a020. [DOI] [PubMed] [Google Scholar]
- 33.Ernst OP, Bieri C, Vogel H, Hofmann KP. Meth. Enzymol. 2000;315:471. doi: 10.1016/s0076-6879(00)15862-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.