Abstract
The granzymes are cell death–inducing enzymes, stored in the cytotoxic granules of cytotoxic T lymphocytes and natural killer cells, that are released during granule exocytosis when a specific virus-infected or transformed target cell is marked for elimination. Recent work suggests that this homologous family of serine esterases can activate at least three distinct pathways of cell death. This redundancy likely evolved to provide protection against pathogens and tumors with diverse strategies for evading cell death. This review discusses what is known about granzyme-mediated pathways of cell death as well as recent studies that implicate granzymes in immune regulation and extracellular proteolytic functions in inflammation.
Keywords: cytotoxic T lymphocyte, cytotoxic granule, natural killer cell, serine protease, granule exocytosis, serpin
INTRODUCTION
The granzymes (granule enzymes) are a family of highly homologous serine proteases contained in cytotoxic granules of innate and adaptive immune killer cells. Their major job is to induce cell death to eliminate viruses and tumor cells. The granzymes may also play a role in immune regulation by controlling the survival of activated lymphocytes and may also regulate inflammation by acting on extracellular substrates. There are five human granzymes and ten mouse granzymes, expressed from three gene clusters. Granzyme A and granzyme B (GzmA, GzmB) are the most abundant granzymes. GzmB, which cleaves after aspartic acid residues in many of the same substrates as the caspases, has been the most extensively studied, but recent studies have begun to elucidate the roles of and cell death pathways activated by GzmA and the other (so-called orphan) granzymes. Killer cells, including natural killer (NK) cells, cytotoxic CD4 and CD8 T cells, and even some regulatory T cells (Tregs), express highly variable and tightly regulated patterns of granzymes that depend on both cell type and mode of activation, but investigators are only beginning to study what controls the expression of each of the granzymes. The cytotoxic granules also contain perforin, needed to deliver the granzymes into the target cell. When cytotoxic T lymphocytes (CTLs) and NK cells form an immune synapse with a specifically recognized target cell destined for elimination, cytotoxic granules move to the immune synapse where the cytotoxic granule membrane fuses with the killer cell membrane, releasing the granule contents into the synaptic cleft. The granzymes are then delivered into the target cell (but not the killer cell), where they initiate at least three distinct pathways of programmed cell death. The killer cell is a serial killer that escapes this encounter unharmed and can then seek and destroy another target cell.
This review focuses on what is known about the biochemistry, gene regulation, cell biology, functions, and inhibitors of the granzyme family, with special attention to what has been learned in the past five years since the last comprehensive review in this series by Russell & Ley (1).
GENE EXPRESSION
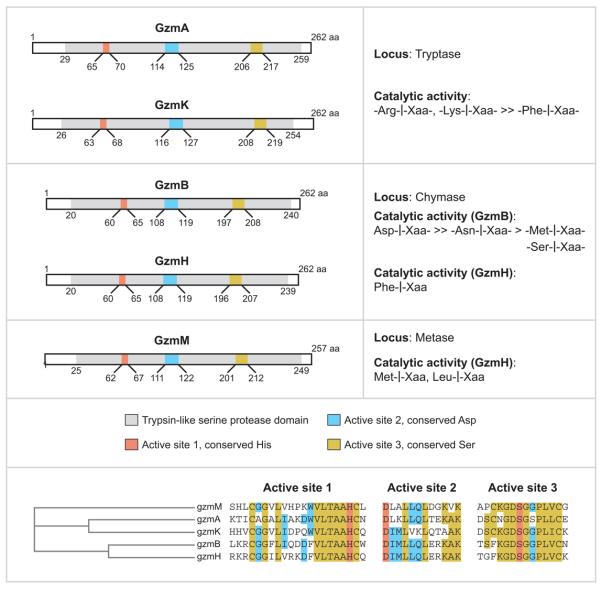
The granzyme serine proteases are encoded in three distinct clusters in humans and mice: GzmA and GzmK, both tryptases, on chromosome 5 (human) and 13 (mouse); GzmB and GzmH on chromosome 14 (human) and their mouse counterparts (GzmB and GzmC) also on chromosome 14; and GzmM, which cleaves after Met or Leu, on chromosome 19 (human) and chromosome 10 (mouse) (Figure 1). GzmB cleaves after aspartic acid, whereas GzmC and GzmH have the specificity of chymotrypsin, cleaving after aromatic amino acids. The GzmB cluster also encodes for cathepsin G and mast cell chymase. The mouse GzmB cluster is uniquely expanded by multiple gene duplications to encode, in addition, GzmD, E, F, G, L, and N. Nothing is known about these mouse-specific enzymes, but investigators have hypothesized that they may have evolved to defend against specific common mouse pathogens (1).
Figure 1.
The family of human granzymes are encoded in three clusters.
Expression in Noncytolytic Lymphoid Cells
Granzymes in the past were thought to be expressed uniquely only by NK cells and CTLs, which could be either CD8+ or cytolytic CD4+ cells, usually of the Th1 lineage (1). However, granzyme transcripts can be amplified by RT-PCR from mouse progenitor T cells (2). GzmB transcripts are found in prothymocytes in fetal liver, whereas mRNA for GzmA, B, and C are found in immature double-negative thymocytes. Although GzmA transcripts are found in thymocytes with the potential to develop into CD8+ cells, GzmA activity is detected only in the most mature CD4−CD8+ thymocytes. These results suggest post-transcriptional regulation of granzyme protein (see below for additional examples). Whether there might be any function for granzyme transcripts in early progenitor T cells is unknown. In the past few years, it has become clear that granzymes are expressed in a broader array of cells and have additional noncytolytic functions in regulating lymphocyte survival and immune tolerance, as well as promoting inflammation and potentially enhancing lymphocyte migration by proteolyzing extracellular proteins or cell surface receptors. Some of these additional functions do not require perforin and may be activated by cells that express granzymes, but not perforin. GzmB, but not GzmA, is expressed in Tregs and plays an important perforin-dependent role in Treg function in mice (3, 4). B-chronic lymphocytic leukemia (B-CLL) cells treated with interleukin-21 (IL-21) produce low levels of GzmB, which can be enhanced by adding either CpG oligodeoxynucleotide or anti–B cell receptor (anti-BCR) antibody (5). In this setting, GzmB induces apoptosis of both the GzmB-expressing cells and untreated bystander B-CLL cells. Similarly, the combination of IL-21 and anti-BCR induces GzmB in benign human B cells, Epstein-Barr virus–transformed lymphoblasts, and many lymphoma cell lines. These results suggest that inducing GzmB expression could open new approaches to the therapy of B-CLL and other B cell malignancies.
Expression of Granzymes in Nonlymphoid Cells
GzmB is expressed in many different types of myeloid cells, generally without perforin. Within the immune system, GzmB is expressed in human plasmacytoid dendritic cells (pDCs) (6). There are comparable levels of GzmB transcripts in resting and activated pDCs, but significantly higher levels of GzmB protein in activated cells, suggesting post-transcriptional regulation of expression. GzmB is also expressed in both normal and neoplastic human mast cells in vitro and in vivo (7). GzmB localizes to specific granules in human mast cells and is secreted upon activation as it is in CTL. Low levels of perforin mRNA and protein can be detected in one human mast cell line, HMC-1, but not in another (LAD-2) or in primary mast cells derived from cord blood and skin. In mice, only skin-associated mast cells and bone marrow–derived in vitro–differentiated mast cells make GzmB protein, but lung mast cells do not (8). Neither GzmA nor perforin are detected in mouse mast cells. Because the GzmB gene is encoded within a few hundred kilobases of mast cell proteases, the GzmB genomic region may be open and active in mast cells, allowing coordinated GzmB expression with mast cell chymase and tryptase. In human basophils, which are developmentally related to mast cells, IL-3-mediated GzmB induction in the absence of GzmA and perforin expression has also been reported (9). Expression of GzmB in mast cells and basophils suggests a role of GzmB in mediating allergic disease. In fact, GzmB has been found in bronchoalveolar lavage fluid after allergen exposure. Several studies have also provided evidence for the expression of GzmB and perforin in human neutrophils, but this is controversial (10-13).
GzmB is also expressed in the absence of perforin in the human reproductive system. It is detected by immunohistochemistry in developing spermatocytes and in placental trophoblasts (14). It is also produced in response to follicle-stimulating hormone by granulosa cells of the human ovary (15). In addition, GzmB protein has been detected in a subset of primary human breast carcinomas and in chondrocytes of articular cartilage (16). The specific expression of GzmB, but not perforin, in these different cell types suggests a noncytotoxic role for GzmB in these cells. The other granzymes expressed in nonlymphoid cells are mouse GzmK and GzmM. The GzmM transcript is expressed at low levels in the photoreceptor cells of the retina in the mouse (17). Interestingly, an alternatively spliced form (aGM) is exclusively expressed in these cells at much higher levels. Like GzmM, GzmK has an alternatively spliced form exclusively expressed in the brain (18). The physiological significance of the alternative transcripts of GzmM and GzmK is yet to be determined.
Extracellular Signals Regulating Granzyme Expression
Early studies with T cell populations showed that naive T cells do not express granzymes, and most activated CD8 T cells coexpress granzymes and perforin. However, the kinetics and expression level of the individual granzymes and perforin vary in different clonal populations in vitro and in vivo and depend on how they are activated (19-21). Most circulating CD8+ T lymphocytes that express any granzyme express both GzmA and GzmB, but some cells are positive for only one granzyme. Single-cell expression profiles of granzymes, perforin, and IFN-γ have been investigated in in vitro– and in vivo–activated CD8+ T cells using RT-PCR in mice (22) and intracellular staining and flow cytometry in humans (4). Individual T cells show unexpected diversity in the expression of these genes. Although some pairs of genes (perforin and IFN-γ) are expressed more frequently than others, no specific combination of genes is consistently coexpressed. During in vitro activation of mouse naive lymphocytes with antibodies to CD3, CD8, CD11a, and IL-2, the expression of GzmA and GzmC is consistently delayed compared with cytolytic activity and expression of perforin and GzmB (22). When mouse CTLs are activated in vivo by influenza virus infection, cytotoxicity is evident in the lung but not in the mesenteric lymph nodes. Most antigen-specific tetramer+ CD8 T cells in the lung one week after infection express both GzmA and GzmB, and about one-third of them also express perforin. Moreover, there is no difference in the kinetics of induction in vivo of GzmA, GzmB, or perforin. In addition, GzmC is not induced by influenza infection in vivo. The diversity of expression of individual granzyme and perforin genes suggests that each gene is regulated independently, although these genes will likely share some common transcription factor recognition sites and epigenetic changes. It is likely that differences in TCR avidity, costimulatory and inhibitory receptor engagement, cytokine milieu, type and state of activation of the antigen-presenting cell, and presence of helper or regulatory CD4 T cells will influence the induction of the granzyme and perforin genes. Moreover, the cell's prior history of activation will affect cytolytic gene expression during subsequent encounters with antigen. Surprisingly little is known about this subject.
The perforin and granzyme genes are induced during T cell activation. However, the only signal shown to upregulate GzmA and B and perforin consistently is IL-2 (23). Previously, the pleiotropic properties of IL-2 made dissociating its effects on T cell survival and proliferation from its effects on gene expression difficult (24). However, a recent study in mice showed that IL-2 regulates perforin and granzyme expression directly and independently of its effect on CD8+ T cell survival and proliferation (25). Mice genetically deficient in IL-2 retain the ability to elicit a CTL response against many viruses, tumors, and allografts (26, 27), although there are deficiencies in cytotoxicity under certain conditions (28). The other γc-dependent cytokines (IL-4, IL-7, IL-9, IL-15, and IL-21) are the most likely candidates for substituting for IL-2 in its absence. IL-15 is particularly important because along with the γ-chain it also shares the β-chain with the IL-2 receptor. IL-15 induces the expression of perforin, GzmA, GzmB, IFN-γ, and Fas ligand in primary mouse lymphocytes (29). IL-21 works synergistically with IL-15 to upregulate GzmA and GzmB expression in mouse CD8 T cells (30). In vivo in mice, IL-21 exhibits potent antitumor function by enhancing NK and CD8 T cell cytotoxicity (31). Similarly, in human peripheral blood CD8 T cells, IL-15 and IL-21 both activate GzmB and perforin expression, but interestingly IL-21 does so without inducing CD8 T cell proliferation (32).
Other cytokines implicated in regulating granzymes and perforin are the IL-6/IL-12 family. IL-12 induces the cytotoxic activity of NK cells and enhances their expression of perforin (33). IL-27 treatment of activated CD8+ T cells significantly augments GzmB and, to a lesser extent, perforin expression (34).
Transcriptional Regulation of Granzymes
Induction of the expression of granzyme transcripts requires at least two independent stimuli—activation of the TCR and costimulation via cytokines of the γc family. The signals from several distinct signal transduction pathways are integrated in the nucleus in the form of transcription factors that bind to granzyme gene regulatory elements and activate transcription. Early studies identified a 243-bp fragment upstream of the mouse GzmB transcription start site that potentially regulates GzmB transcription (35). This region contains binding sites for two ubiquitous transcription factors, activating transcription factor/cyclic AMP-responsive element binding protein (ATF/CREB) and activator protein-1 (AP-1), and two lymphoidspecific factors, Ikaros and core-binding factor (CBF/PEBP2) (36). Several of these transcription factor–binding sites are evolutionarily conserved between the human and mouse GzmB promoters (37, 38). Analysis of reporter assays using promoters that had been systematically mutated at these sites in primary cells and cell lines revealed subtle differences in the importance of some transcription factors in primary cells versus cell lines. For example, AP-1, CREB, and CBF were not as important for transcription in primary cells as they appeared to be in cell lines (37, 39). These studies suggested that combinations of transcription factors (particularly AP-1 and CBF) are required to activate GzmB expression in primary cells. The most compelling difference between the mouse and human GzmB gene promoter is the importance of the Ikaros site in human GzmB expression. Mutations introduced into the Ikaros-binding site of the human, but not mouse, GzmB promoter abrogate expression in primary T cells (37, 39).
Two recent studies have shown a role for signal transducer and activator of transcription 1 (STAT1) in the transcriptional regulation of GzmB. Mouse splenocytes stimulated with IFN-α typically express GzmB. This IFN-α effect is abrogated in STAT1-deficient mice (40). IL-27-mediated enhancement of GzmB and perforin expression is also impeded in STAT1-deficient mice (34). IL-27-induced augmentation of GzmB and perforin expression is also dependent on the T-box family transcription factor T-bet (34). Another Tbox family member, Eomesodermin, was recently identified in activated CD8+ T cells in mice and shown to drive perforin, GzmB, and IFN-γ expression when ectopically expressed in mouse Th2 CD4+ cells (41). In the converse experiment, expression of a dominant-negative Eomesodermin impaired GzmB expression and cytolytic activity in CD8+ T cells. Thus, these two T-box family members, T-bet and Eomesodermin, appear to regulate GzmB expression cooperatively.
Little is known regarding the transcriptional regulation of GzmA. Two groups looking to identify transcriptional targets of the glucocorticoid receptor found that GzmA expression is significantly upregulated in a glucocorticoid-sensitive human pre-B acute lymphocytic leukemia line (42–44). Interestingly, GzmA expression in these cells induces apoptosis, which can be prevented by treatment with a synthetic GzmA inhibitor (43). Further study of the GzmA promoter did not find a glucocorticoid responsive element (GRE) upstream of the transcriptional start site, but did find a GRE in the first intron (42). In fact, a novel 5′ variant GzmA transcript starts 290 bp downstream of this intronic GRE (45). These alternative promoters generate two GzmA transcripts with different first exons (exon1a and 1b). GzmA transcripts isolated from CTL contain exon1a, which encodes an N-terminal leader peptide to direct the nascent protein into the lumen of the endoplasmic reticulum (ER). Glucocorticoid treatment induces the exon1b-containing transcript and also suppresses the exon1a transcript (45). The exon1b encodes a shorter leader peptide, which may cause aberrant subcellular localization of mature GzmA. Glucocorticoid-induced apoptosis of leukemia cells might then be due to the GzmA promoter switch and production of GzmA that is atypically released into the cytosol.
Post-Transcriptional Regulation of Granzymes
An intriguing recent study suggests that GzmB and perforin gene expression may be post-transcriptionally regulated (46). Mouse NK cells in a resting state from pathogen-free animals have abundant GzmB and perforin mRNA but no corresponding protein and no substantial cytotoxicity. Upon activation, these cells show a dramatic increase in GzmB and perforin protein, but a minimal change in mRNA levels. Both resting and activated mouse NK cells have high levels of GzmA. This report appears to contradict previous studies showing that resting mouse and human NK cells do not need to be activated to develop cytotoxicity (47-49). The reason for the discrepancy is unclear but could result from low levels of asymptomatic infection in mice used in earlier studies, leading to prior activation of NK cell precursors. Unlike sterile laboratory mice, human subjects are chronically exposed to commensal organisms that activate the innate immune system as well as to a variety of pathogens. Therefore, human NK cells constitutively express perforin and GzmA and GzmM (but not GzmB). However, to our knowledge, no one has looked at granzyme expression in human NK cells in the immediate postnatal setting.
In addition to the difference in granzyme mRNA and protein in T cell progenitors mentioned above, there are other examples that suggest post-transcriptional gene regulation. There are comparable levels of GzmB transcripts in resting and activated human pDC, but significantly higher levels of GzmB protein in activated cells (6). Human mast cells upon activation express higher levels of GzmB mRNA than are found in CTL, but the corresponding protein levels are considerably lower in mast cells (7). Mouse memory CTLs express abundant GzmB mRNA but no protein (50). All these results point toward a general mechanism of prearming cytotoxic lymphocytes with effector mRNAs, allowing these cells to respond rapidly to external stimuli. This type of gene regulation is well known to regulate cytokine expression, presumably for the same purpose.
GRANZYME STRUCTURE
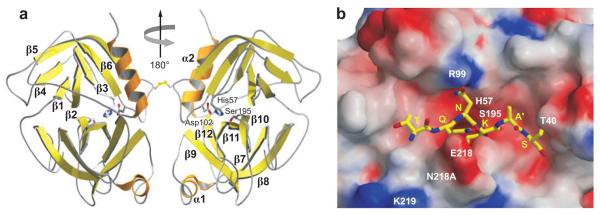
The granzymes are homologous to trypsin, and their overall structure is similar to trypsin and other related serine proteases. Human GzmA, human and rat GzmB, and human pro-GzmK have all now been crystalized to yield high resolution structures (51-54a) (Figure 2). The active granzymes are produced by cleavage of a dipeptide from the N terminus of the proenzyme. Activation is likely accompanied by a radical conformational change because a monoclonal antibody to GzmA does not recognize pro-GzmA. Pro-GzmK has a more rigid structure lacking an open active site, compared with the solved structures for the active granzymes. Detailed information about the conformation surrounding the active site of GzmA and GzmB has provided the structural basis for understanding how subtle differences in the active site conformation lead to substantial differences in substrate specificity. As a consequence, mouse GzmB is preferentially able to cleave mouse procaspase-3, whereas human GzmB is better able to cleave the human homolog. GzmA differs from the other granzymes in forming a covalent homodimer; the other granzymes are monomeric. Dimerization creates an extended site for substrate binding that is believed to confer a high degree of specificity to GzmA for its substrates. In particular, because of the extended exosite for substrate binding, GzmA substrates do not share a common short peptide sequence around the cleavage site. The crystal structures for the granzymes should provide useful tools for identifying small molecule inhibitors.
Figure 2.
Crystal structure of the GzmA homodimer. (a) GzmA is a disulfide-linked dimer in which the two active sites, indicated on the right (His57-Asp102-Ser195), face in opposite directions. The surface of the molecule contains concentrations of basic amino acids, which may explain the preference for acidic protein substrates through binding outside the active site through an extended exosite. (b) The SET protein is an important target of GzmA, whose cleavage triggers its unique pathway of DNA damage. Model of how the SET peptide surrounding the GzmA cleavage site fits into the GzmA active site. [Figures based on the structure obtained by Hink-Shauer and colleagues (54a), reprinted with permission.]
GRANZYME BIOSYNTHESIS AND STORAGE IN TARGET CELLS
The granzymes are expressed with a signal sequence that directs them to the ER. Cleavage of the signal peptide produces an inactive proenzyme that contains an N-terminal dipeptide that needs to be cleaved to produce an active protease. In the Golgi, a mannose-6-phosphate tag is added, a sorting signal for transporting the proenzyme to lysosomes. Cytotoxic granules are specialized secretory lysosomes, maintained at an acidic pH with a distinctive electron dense core on electron micrographs. Within the cytotoxic granule, the N-terminal dipeptide is removed by cathepsin C (dipeptidyl peptidase I) (55). However, both mice and humans genetically deficient in cathepsin C have only partially reduced granzyme activity and cytolytic function and modestly reduced immune defense against viral infection (56, 57). This suggests that there are alternate ways to activate the proenzyme. In fact, IL-2 treatment stimulates cathepsin C–independent dipeptide cleavage in NK cells from patients with PapillonLefevre syndrome who have loss of function of cathepsin C (58). Granzymes, which are highly basic, are bound along with perforin to the acidic serglycin proteoglycan within the granule, which probably keeps them inactive. Serglycin is responsible for the electron dense core and may help to enhance the granzyme storage capability of the granules (59). Granzyme proteolytic activity is also negligible at acidic pH. Therefore, during protein synthesis and processing and when stored within the granule, several mechanisms are at work to make sure the granzymes are not proteolytically active.
GRANZYME RELEASE, UPTAKE, AND TRAFFICKING IN TARGET CELLS
When a CD8 T cell or NK cell is activated by its antigen receptor, the lytic granules move to cluster around the microtubule organizing center and then align along the immunological synapse (reviewed in 60). The granule membrane fuses with the killer cell plasma membrane, releasing its contents, including perforin and granzymes, into the synapse. In CTL, granule fusion appears to localize to a distinct (secretory) region of the central cluster (c-SMAC) that is separate from the signaling domain containing the T cell receptor and associated kinases (61). However, the organization of granule fusion at the synapse may differ among types of killer cells, and cytotoxicity and granule fusion may occur even in the absence of a stable synapse (62). Granzymes likely dissociate from serglycin before they enter target cells (63). Granzymes bind to the target cell membrane by electrostatic interactions (granzymes are very positively charged with pIs ~9–11, and the cell surface is negatively charged) (64-66) and also by specific receptors, such as the cation-independent mannose-6-phosphate receptor (67). However, specific receptors are not required for binding and cytotoxicity (64, 68, 69).
Entry of granzymes into the target cell cytosol is generally mediated by perforin, but how perforin accomplishes this is still unclear (reviewed in 70). The original model of granzyme entry through plasma membrane pores formed by perforin (perforin is a pore-forming protein with homology to complement) is generally no longer considered valid. A revised model posits that perforin makes microscopic holes in the plasma membrane that cause a calcium influx, which triggers a cellular plasma membrane response and rapid endocytosis of granzymes and anything else bound to the cell surface (71). In fact, entry is dynamin-dependent (72) and results in the formation of giant endosomes containing both granzymes and perforin (71; D. Keefe, J. Thiery, and J. Lieberman, manuscript in preparation). Within minutes, the granzymes escape (through perforin pores in the endo-some?) and find their way into the cytosol. Although some key granzyme targets are cytosolic [i.e., BH3 interacting domain death agonist (bid) and inhibitor of caspase activated DNase (ICAD) for GzmB], other important targets are in other membrane-bound cellular compartments, including the nucleus and mitochondrion. GzmA and GzmB rapidly translocate to the nucleus (73, 74), where proteolytic cleavage of key substrates is important to induce programmed cell death by both GzmA (SET, Ape1, lamins, histones, Ku70, PARP1) and GzmB (lamin B, PARP1). Nuclear translocation of the granzymes may be mediated by importin-α (75). We have recently found that GzmA also traffics into the mitochondrial matrix, which is necessary for it to initiate mitochondrial damage (D. Martinvalet, D.M. Dykxhoorn, R. Ferrini, J. Lieberman, manuscript submitted). Genetic mutations that affect perforin function or granule exocytosis are associated with profound immunodeficiency and the familial hemophagocytic lymphohistocytosis syndrome (77-81).
The killer cell on the other side of the synapse is not injured by its own granules. One important protective mechanism against autodestruction is provided by the expression in the killer cell cytoplasm of irreversible granzyme inhibitors called serpins (see next section). However, although serpins that inactivate GzmB have been shown in killer cells, no serpins are known that can inactivate GzmA. Another protective mechanism occurs via externalization of a cytotoxic granule membrane protein (cathepsin B), capable of proteolytically inactivating perforin, to the killer cell plasma membrane during granule fusion (82). Cathepsin B is thought to protect the killer cell membrane from any perforin that is redirected to the CTL side of the synapse. However, killer cells from cathepsin B–knockout mice survive encounters with target cells unharmed (83). This suggests that other mechanisms likely exist to protect killer cells from their own agents of destruction.
Although perforin is the major molecule responsible for granzyme delivery, under some circumstances other molecules may serve that function. For example, bacterial and viral endosomolysins can substitute for perforin in vitro [and are widely used as laboratory reagents for intracellular delivery (84)] and may potentially play a similar role in vivo. The heat shock protein hsp70, which is known to chaperone some peptides across cell membranes, can also carry GzmB (and presumably other granyzmes) into cells (85). Hsp70 is found on the surface of some stressed cells or tumor cells and may help to remove these cells from the body.
GRANZYME INHIBITORS—ENDOGENOUS, VIRAL, AND SYNTHETIC
The regulation of proteolytic enzymes in tissues by endogenous inhibitors is critical to maintaining homeostasis and preventing undesirable damage. Although the trafficking of granzymes within CTLs is designed to minimize leakage of active enzyme out of granules, any stray molecules in the cytoplasm could cause cell death (86). During granule exocytosis, some granzymes might inadvertently reenter the effector cells. Because CTLs typically kill several targets in succession without harming themselves, an important question is how CTLs protect themselves from their own cytotoxic molecules. One way is by expressing granzyme-specific inhibitors, members of the serpin (serine proteinase inhibitor) superfamily (87). Serpins are the largest and most broadly distributed superfamily of protease inhibitors, with more than 1500 family members (88, 89). Serpins inactivate their targets either by covalently and irreversibly binding to the active site of the enzyme or by forming noncovalent complexes that are tight enough to resist the denaturing conditions of SDS-PAGE gel electrophoresis (89, 90).
GzmB Endogenous Inhibitors
The only intracellular inhibitor of human GzmB is the nucleocytoplasmic serpin, proteinase inhibitor-9 (PI-9). PI-9 is expressed by lymphocytes (87, 91), dendritic cells (DCs) (92), cells at immune privileged sites (testis and placenta) (14, 93, 94), endothelial and mesothelial cells (95), and finally mast cells (96). This in vivo distribution pattern supports the idea that PI-9 protects effector, accessory, and bystander cells from ectopic GzmB during an immune response. PI-9 gene expression is induced by modulators of inflammation like lipopolysaccharide, IFN-γ, and IL-1β (97, 98). PI-9 expression is enhanced by estrogen and hypoxia because of estrogen responsive elements and hypoxia inducible factor 2 (HIF-2)-binding sites, respectively, in the PI-9 promoter (99, 100). In particular, PI-9 is induced by hypoxia in neuroblastomas (101).
The mouse counterpart of PI-9 is serine proteinase inhibitor-6 (SPI-6) (102). SPI-6 is expressed in CTL and NK cells (103) and is upregulated during DC maturation (104). Overexpression of SPI-6 in target cells protects them from CTL killing (105). In SPI-6 transgenic mice, increased numbers of CTLs persist long after viral clearance, suggesting that SPI-6 protects CTLs from self-destruction (103). However, somewhat paradoxically, GzmB-deficient mice do not have increased numbers of CTL after viral infection, raising questions about the interpretation of the SPI-6 transgenic study (103). However, CTL from mice genetically deficient in SPI-6 have increased cytosolic GzmB and reduced viability (106). One surprising finding is a breakdown of the integrity of the cytotoxic granules in SPI-6-deficient CTLs (106). The exact mechanism behind this collapse of cytotoxic granules is unclear.
Early studies with solid tumors and lymphomas in humans and mice suggested that overexpression of PI-9 or SPI-6 may be a mechanism by which tumors evade the GzmB/perforin pathway (105, 107). In these studies there is no comparison of serpin expression in tumor cells relative to corresponding normal tissues, thus making the results difficult to interpret. Also, only a small subset of human lymphoma cell lines express PI-9 (107). These issues have been addressed in recent studies. In cultured human hepatoma cells, induction of endogenous PI-9 by IFN-γ or estrogen partially blocks CTLand NK-induced apoptosis (98, 99). Similarly, induction of increasing amounts of endogenous PI-9 by estrogen in a human breast cancer line (MCF-7) progressively increases its resistance to NK-mediated cytolysis (108). PI-9 expression in pediatric acute lymphoblastic leukemias also correlates with resistance to cytolysis in vitro (109). Importantly, PI-9 expression is an important determinant of disease-free survival time of melanoma patients following immunotherapy (110). However, endogenous PI-9 and Bcl-2 expression by some human lymphomas do not confer any resistance to cytolysis by in vitro–activated CTLs or NK cells (111). This study assumes that the cytotoxicity of an in vitro–activated cytotoxic lymphocyte is comparable to the in vivo scenario. Measuring cytotoxicity using highly activated cytotoxic lymphocytes in vitro may exaggerate the effectiveness of these cells (112) and thus underestimate the protective capacity of antiapoptotic molecules. The ability of serpins to make tumors resistant to immune cell destruction will likely depend on the level of expression of the serpin and of other antiapoptotic molecules, such as bcl-2 family members and survivin.
GzmA Endogenous Inhibitors
No intracellular inhibitors of GzmA have yet been identified. However, some trypsin inhibitors also inhibit GzmA. GzmA is bound and irreversibly inhibited in the circulation by two trypsin inhibitors, α-2 macroglobulin and antithrombin III (113). Extracellular GzmA complexed to proteoglycans is resistant to these two protease inhibitors (114). A recent study identified another GzmA inhibitor, pancreatic secretory trypsin inhibitor (PSTI), from pancreatic secretions (115). PSTI is found in the blood, particularly in patients with severe inflammation and tissue destruction (116, 117). Extrapancreatic or blood PSTI might regulate extracellular activity of GzmA. Unlike the other two GzmA inhibitors, PSTI inhibits GzmA complexed to proteoglycans (115). It is still not clear whether any of these GzmA inhibitors are expressed in cytotoxic lymphocytes.
Viral Granzyme Inhibitors
The pox virus–encoded cytokine response modifier A gene (CrmA) is the first viral inhibitor that was found to inhibit GzmB (118). CrmA directly binds and inhibits GzmB both in vitro and in vivo. Overexpression of CrmA in target cells inhibits CTL-mediated cell death. However, CrmA also strongly binds and inhibits caspases-1 and -8 and weakly inhibits other caspases like caspase-3; therefore, it is difficult to pinpoint the importance of GzmB inhibition in these studies (119). Parainfluenza virus type 3 specifically inhibits GzmB by degrading GzmB mRNA in infected T cells (120). Importantly, GzmA transcripts are not affected by this virus. The mechanism of virus-mediated GzmB mRNA decay is not known.
Human GzmB is inhibited by the adenoviral assembly protein (Ad5-100K) by a unique unserpin-like mechanism (121). In adenovirus-infected cells, Ad5-100K rapidly complexes with GzmB and gets cleaved very slowly at specific sites. GzmB that enters the infected target cell upon CTL attack is saturated by the abundant Ad5-100K protein. Importantly, the slow kinetics of the cleavage reaction ensures that there is always a molar excess of Ad5-100K protein relative to GzmB. Unlike CrmA, which is just an antiapoptotic factor, Ad5-100K is also necessary for virus assembly (121). It impedes human GzmB but does not inhibit caspases or other apoptotic pathways (122). Interestingly, the inhibitory activity of Ad5-100K is specifically directed against human GzmB and not its mouse or rat homolog.
A recent study shows how CTLs have gotten around adenovirus resistance. In adenovirus-infected cells, the Ad5-100K-mediated GzmB inhibition is released by the action of an orphan granzyme, GzmH. GzmH cleaves Ad5-100K to rescue GzmB activity (123). Both GzmB and GzmH target the same adenoviral proteins, DNA-binding protein (DBP) and Ad5-100K. The direct cleavage of essential viral proteins by granzymes is a novel mechanism by which cytotoxic cells rapidly and directly block viral replication (123). Additionally, the different specificities of the granzymes allow distinct substrate processing, leading to synergistic antiviral activity. Viruses have evolved pathways to evade or inhibit granzymes and block apoptosis. This is the first example of how the unique catalytic specificities of granzymes combine to counter a viral challenge.
Synthetic Inhibitors
Synthetic inhibitors of granzymes are powerful tools both for research (identification of peptide substrate specificity and determination of granzyme function) and potentially for therapeutic applications (immune suppression during autoimmune diseases and organ transplantation). There are several classes of granzyme inhibitors, including isocoumarin derivatives, peptide chloromethyl ketones, and peptide phosphonates, but the major limitation has been a lack of specificity (113). Modifications that increase specificity generally diminish efficiency. Thornberry and colleagues (124) recently reported the identification of a novel class of human GzmB inhibitors. The key feature of these compounds is a 1,2,3-triazole moiety that is crucial for their selectivity and cellular efficacy. Future work with these inhibitors will determine their importance in studying the biology of granzymes.
PROGRAMMED CELL DEATH PATHWAYS INITIATED BY THE GRANZYME ALPHABET SOUP
We are just beginning to understand how granzymes, other than GzmB, activate cell death, as laboratories have begun to express active recombinant forms of many of these enzymes. Now all five of the human enzymes have been expressed. Granzymes likely activate at least three (and probably more) distinct pathways of cell death (Figure 3, Table 1). We know most about cell death by GzmB, which activates the caspase apoptotic pathway by cleaving caspase-3 and also proteolyzes many of the important caspase substrates directly. However, there is good evidence that GzmB can also activate other pathways of cell death (particularly in the mitochondrion) that remain to be worked out. GzmA activates cell death that has all the morphological features of apoptosis but is completely caspase-independent and involves novel mitochondrial and DNA damage pathways. GzmC (in mouse) and GzmH (in humans) also activate caspase-independent cell death with a pronounced mitochondrial phenotype. There is some evidence that GzmM may activate autophagy. Understanding in detail the workings of these multiple roads to cell death will help us understand the many strategies for protection against intracellular pathogens and cellular transformation and how viruses and tumors evade immune destruction. Inevitably, research in this area will also help us comprehend important mechanisms in normal cellular metabolism and some of the strategies cells have at their disposal to deal with outside stress. Most of the focus has been on unveiling mitochondrial and DNA damage pathways. The molecular mechanisms behind other aspects of cell death, such as how plasma membrane integrity is disrupted (a critical feature of cell death), are still not well understood.
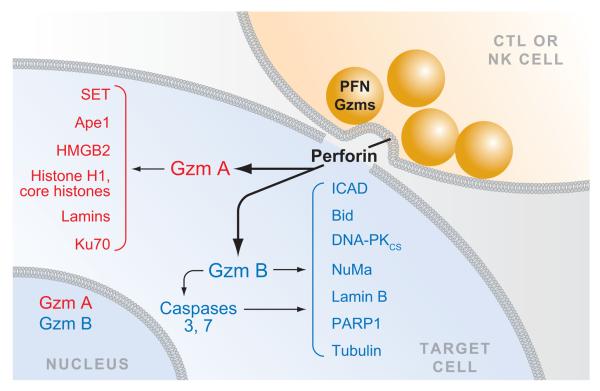
Figure 3.
Granule exocytosis-mediated cell death. When a CTL or NK cell recognizes a target cell, cytolytic granules containing perforin (PFN) and granzymes move to the immune synapse, and the granule membranes fuse with the killer cell plasma membrane, releasing PFN and granzymes into the synapse. PFN facilitates the entry of granzymes into the cytosol of the target cell. The most abundant granzymes are GzmA and GzmB. GzmA activates cell death independently of the caspases, whereas GzmB activates the caspase pathway both directly by cleaving the caspases and indirectly by cleaving key caspase substrates. Some of the key substrates of human GzmA and GzmB are shown. Both GzmA and GzmB traffic to the nucleus by an unknown pathway, where many of the nuclear substrates are cleaved.
Table 1.
Features of the distinct cell death pathways induced by the granzymes
| Granzyme | A | B | C/H | K | M |
|---|---|---|---|---|---|
| Expression | |||||
| Cytolytic CD8 T cells | ++ | ++ | + | + | |
| Cytolytic CD4 T cells | + | + | |||
| CD4 Tregs | − | + | |||
| NK cells | + | +/− | + | ||
| Myeloid cells | − | + | |||
| Common features | |||||
| Rapid loss of membrane integrity | + | + | + | + | ? |
| Annexin V staining | + | + | + | + | ? |
| Chromatin condensation | + | + | + | + | ? |
| DNA damage | + | + | + | + | ? |
| Mitochondrial depolarization | + | + | + | + | ? |
| Caspase activation | − | + | − | − | ? |
| Type of DNA damage | |||||
| Oligonucleosomal DNA fragmentation | − | + | − | − | ? |
| Single-stranded DNA nicks | + | − | + | + | − |
| TdT labeling | + | + | + | + | ? |
| Klenow labeling | + | + | + | + | −? |
| Type of mitochondrial damage | |||||
| Inhibition by bcl-2 overexpression | − | + | ? | ? | ? |
| Cytochrome c release | − | + | +? | ? | ? |
| Mitochondrial swelling | + | + | ++ | + | + |
| Autophagy | − | − | − | − | +? |
GRANZYME A
GzmA is a tryptase that induces caspase-independent cell death, which is morphologically indistinguishable from apoptosis (125, 126). Tryptases with homology to GzmA have been found in cytotoxic cells as far back in evolution as bony fish (127). Although GzmA was the first granzyme to be described and is the most widely expressed, much less is known about it than about GzmB. Cells treated with GzmA and perforin die rapidly—within minutes they undergo membrane blebbing, lose plasma membrane integrity (take up propidium iodide), and have evidence of mitochondrial dysfunction [increased reactive oxygen species (ROS), loss of mitochondrial transmembrane potential (Δψm), disruption of mitochondrial morphology] (128; D. Martinvalet, D.M. Dykxhoorn, R. Ferrini, J. Lieberman, manuscript submitted). Within an hour or two, the slower-onset hallmarks of apoptosis are apparent: externalization of phosphatidyl serine (measured by annexin V staining) and DNA damage. DNA is damaged by single-stranded cuts into megabase fragments that are much larger than the oligonucleosomal fragments generated during caspase- or GzmB-activated cell death (129). Because the DNA fragments are too large to be released from the nucleus, assays that measure DNA release into culture supernatants are typically negative until many hours later. Until recently, this was incorrectly interpreted as meaning that GzmA induces a slow, nonapoptotic death. Indeed, the caspases are not activated, and cell death proceeds unabated in the presence of pancaspase inhibition or in cells overexpressing bcl-2 family members or other inhibitors of caspase-mediated apoptosis (125). Moreover, mitochondria are damaged without mitochondrial outer membrane permeabilization (MOMP) or release of proapoptotic mediators, such as cytochrome c, from the mitochondrial intermembrane space (128).
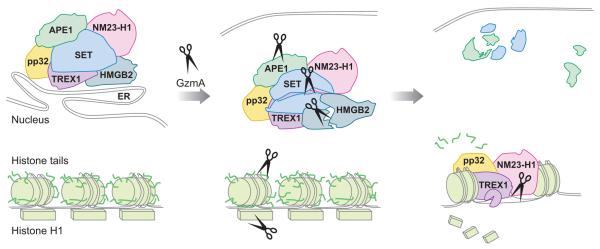
The molecular basis for the parallel caspase-independent programmed cell death pathway activated by GzmA has begun to be elucidated. Triggering mitochondrial damage is key to cell death induction because treating target cells with superoxide scavengers completely blocks cell death (and also blocks cell death by CTLs expressing all granzymes) (128). From the cytosol, GzmA is transported by an unknown mechanism [that may involve its ability to bind to the mitochondrial chaperone heat shock proteins (130; D. Martinvalet, D.M. Dykxhoorn, R. Ferrini, J. Lieberman, manuscript submitted)] into the mitochondrial matrix, where it cleaves a component of the electron transport chain complex I to interfere with mitochondrial redox function, ATP generation, and maintenance of Δψm and to generate superoxide anion (128, 131; D. Martinvalet, D.M. Dykxhoorn, R. Ferrini, J. Lieberman, manuscript submitted). The su-peroxide generated by damaged mitochondria drives an ER-associated oxidative stress response complex, called the SET complex, which plays a critical role in GzmA-induced nuclear damage, into the nucleus (128, 129). The SET complex contains three nucleases [the base excision repair (BER) endonuclease Ape1, an endonuclease NM23-H1, and a 5′-3′ exonuclease Trex1]; the chromatin-modifying proteins SET and pp32, which are also inhibitors of the tumor suppressor protein phosphatase 2A; and a DNA-binding protein that recognizes distorted DNA, HMGB2 (132-136). One of the functions of the complex is to repair abasic sites in DNA generated by oxidative damage. GzmA, which traffics to the nucleus by an unknown mechanism, converts this DNA repair complex into an engine for DNA destruction by cleaving SET, an inhibitor of the endonuclease NM23-H1 (134) (Figure 2). This allows NM23-H1 to nick DNA; the exonuclease Trex1 then extends the break (133) (Figures 3 and 4). At the same time, GzmA cleaves and inactivates HMGB2 and Ape1 to interfere with BER (135, 136). In addition to disabling BER, GzmA also interferes with DNA repair more generally by interfering with the recognition of damaged DNA by cleaving and inactivating Ku70 (137) and PARP-1 (P. Zhu, D. Martinvalet, D. Zhang, A. Schlesinger, D. Chowdhury, J. Lieberman, manuscript in preparation). Within the nucleus, GzmA also opens up chromatin by cleaving the linker his-tone H1 and removing the tails from the core histones, making DNA more accessible to any nuclease, and disrupts the nuclear envelope by cleaving lamins (139, 140). It is noteworthy that all the GzmA substrates mentioned above are cleaved both in vitro and in vivo. When a mutant form of the key substrates that lacks the GzmA cleavage site is expressed in target cells, the target cell is less susceptible to GzmA-mediated cell death.
Figure 4.
The GzmA pathway of DNA damage. ROS generated by GzmA in mitochondria drives the ER-associated SET complex into the nucleus. GzmA also enters the nucleus by an unknown pathway. In the nucleus, GzmA cleaves three components of the SET complex (SET, HMGB2, and APE1) to activate two nucleases in the complex to make single-stranded DNA lesions—NM23-H1 makes a nick, which is extended by the exonuclease TREX1. GzmA also degrades the linker histone H1 and removes the tails from the core histones, opening up chromatin and making it more accessible to these nucleases.
GRANZYME B
GzmB is unique among serine proteases because it cleaves after aspartic acid residues, like the caspases (reviewed in 141, 142). It induces target cell apoptosis by activating the caspases, particularly the key executioner caspase, caspase-3 (143, 144) (Figure 3). Human GzmB, but not the mouse enzyme, also activates cell death by directly cleaving the key caspase substrates, bid and ICAD, to activate the same mitochondrial and DNA damage pathways, respectively, as the caspases (53, 145-151) (Figure 3). As a consequence, caspase inhibitors have little effect on human GzmB-mediated cell death and DNA fragmentation, whereas the same inhibitors significantly block the action of the mouse enzyme. Therefore, human CTLs and NK cells may be more effective than mouse killer cells at eradicating virus-infected cells or tumors that have developed methods for evading the caspases. Both human and mouse enzymes cleave many of the same substrates as the caspases (including PARP-1, lamin B, NuMa, DNA-PKcs, tubulin) and have substrate specificity close to that of caspases-6, −8, and −9 (152). However, human GzmB cleaves optimally after the tetrapeptide IEPD, whereas mouse GzmB has somewhat different peptide specificity, preferring to cleave after IEFD (53, 145). Moreover, other regions, including the P′ region (C-terminal to the cleavage site) and more distal regions, contribute to substrate specificity. As a consequence of subtle differences in sequence, the human and mouse GzmB can differ in important ways with respect to substrates and the efficiency with which they are cleaved.
The GzmB (and caspase) mitochondrial pathway leads to ROS generation, dissipation of Δψm and MOMP, with release of cytochrome c and other proapoptotic molecules from the mitochondrial intermembrane space. Human GzmB activates this pathway directly by cleaving bid, whereas mouse GzmB activates it indirectly. However, GzmB targets the mitochondria in other ways that remain to be worked out. Loss of Δψm, but not cytochrome c release, occurs in the presence of pan-caspase inhibitors (even using mouse GzmB) and in mice genetically deficient for bid, bax, and bak (the latter two bcl-2 family members are required for bid-induced mitochondrial damage) (153-156). Mitochondrial damage is key to cell death induction because treatment of target cells with superoxide scavengers that neutralize ROS completely blocks cell death by CTLs expressing all granzymes (128). DNA damage by GzmB is mediated primarily by the activation of the caspase-activated DNase (CAD) following proteolytic cleavage of its inhibitor ICAD either directly by human GzmB or indirectly by executioner caspases, such as caspase-3.
In humans, there is a common polymorphism of GzmB in which three amino acids (Q48, P88, Y245) are mutated to R48A88H24 (157). This polymorphism does not seem to affect cytotoxicity and probably does not have any clinical significance (158).
GRANZYMES C AND H
Mouse GzmC and human GzmH, homologous granzymes that lie downstream from GzmB, are predicted to have chymotryptic activity and cleave substrates after aromatic residues (159, 160). GzmH is thought to have arisen during primate evolution, independently of GzmC, in an intergenic recombination event between GzmB and a mast cell chymase (161). Both induce caspase-independent death with hallmarks of programmed cell death—ROS generation, dissipation of Δψm, chromatin condensation, and nuclear fragmentation (159, 160). DNA destruction by GzmC (and probably GzmH as well) is via single-stranded nicks and does not involve CAD. Rapid mitochondrial swelling and disruption of mitochondrial ultrastructure are particularly striking in cells treated with GzmC. The mitochondrial pathways activated by GzmC and GzmH may be different; GzmC has been reported to trigger cytochrome c release, a sign of MOMP, whereas GzmH does not seem to cause cytochrome c release (159, 160).
Although no normal cellular substrates have yet been identified for GzmC or GzmH, GzmH cleaves two adenoviral proteins— DBP (also a GzmB substrate) and the adenovirus 100K assembly protein, a previously described inhibitor of GzmB (121, 123). Cleavage of DBP interferes with viral DNA replication, whereas cleavage of 100K restores GzmB function in adenovirus-infected cells. Therefore, GzmH may play a special role in adenoviral immune defense. Because GzmH is expressed in NK cells, it may be used to eliminate adenovirus early in infection, before adaptive immunity has had a chance to develop.
GRANZYME K
GzmK (also known as Gzm 3) is another tryptase found in mice, rats, and humans, encoded downstream close to GzmA on human 5q11–12 (or the syntenic region of mouse chromosome 13). It is expressed much less than GzmA and, unlike GzmA, is a monomer, not a dimer. Mice genetically deficient in GzmA express GzmK, which may explain the lack of a significant phenotype of GzmA−/− mice, except when challenged with some viruses (162, 163). Purified rat and recombinant human GzmK has been available for some time (164, 165), but little was known about its cell death activation until recently. Like GzmA, purified rat GzmK efficiently induces caspase-independent cell death, characterized by mitochondrial dysfunction without MOMP (ROS and loss of Δψm, but without cytochrome c release) (154). However, unlike GzmA, rat GzmK–induced cell death was originally reported to be inhibited in cells overexpressing bcl-2 (154). This finding was surprising because bcl-2 inhibits MOMP, which leads to cytochrome c release, which was not detected in GzmK-treated cells. In fact, a more recent study found that cell death by recombinant human GzmK did not activate caspase-3 and was unaffected by caspase inhibitors or bcl-xL overexpression (166). GzmK mimics GzmA DNA damage (166)—it causes caspase-independent nuclear fragmentation and nuclear condensation and single-stranded DNA breaks by targeting the SET complex. Like GzmA, GzmK causes SET complex nuclear translocation and hydrolyzes and inactivates SET, Ape1, and HMGB2 in the SET complex (166). Presumably, cleavage of SET, the inhibitor of NM23-H1, triggers DNA damage by the GzmA-activated DNases, NM23-H1 and Trex1, in the SET complex (133, 134). The same group recently reported that GzmK causes mitochondrial damage that includes not only ROS generation and dissipation of Δψm, but also bid cleavage (to a fragment that appears to be the same size as is generated by GzmB) and MOMP with release of cytochrome c and endoG (167). This finding needs to be verified because rat GzmK does not cause cytochrome c release (154), and this same group showed that caspases are not activated by GzmK, nor does overexpression of bcl-xL interfere with human GzmK–induced cell death (166), as would be expected if MOMP is triggered. Although GzmK appears to duplicate GzmA's nuclear damage pathway, further studies are needed to determine whether the mitochondrial GzmK pathway resembles that activated by GzmA (no MOMP) or GzmB (bid cleavage, MOMP) or is a hybrid of both.
GRANZYME M
GzmM is the most distinctive of the granzymes. It likely arose from a gene duplication of a neutrophil protease because it is encoded near a cluster of other neutrophil proteases in human chromosome 19p13.3 (or a synteic region of mouse chromosome 10) and is slightly more homologous to one of them (complement factor D) than to the other granzymes (168). Unlike the other granzymes, GzmM cuts after Met or Leu (169, 170). None of the serine protease inhibitors that block the other granzymes, including the pangranzyme inhibitor 3,4-dichloroisocoumarin, effectively inhibit GzmM (171). Moreover, GzmM appears to function primarily in innate immunity, as it is expressed mostly in NK cells and γδ T cells and only in the subset of CD56+ T cells (112). Until recently it was not clear whether GzmM induces cell death (172). GzmM−/− mice have unimpaired NK and T cell development and NK cell-mediated cytotoxicity but are less able to defend against mouse cytomegalovirus infection (173).
The literature does not agree about the type of cell death activated by GzmM. On the one hand, Kelly et al. (172), using recombinant human GzmM expressed from baculovirus in insect cells, found that GzmM induced rapid, caspase-independent cell death that looked like autophagic death and did not find evidence for DNA fragmentation, mitochondrial depolarization, phosphatidyl serine externalization, or caspase activation. On the other hand, using human GzmM expressed in yeast, the Fan laboratory (174, 175) argued that GzmM activated caspase-dependent cell death with phosphatidyl serine externalization, caspase activation, ICAD cleavage, and CAD activation with oligonucleosomal DNA laddering, PARP cleavage, and mitochondrial disruption with MOMP (mitochondrial swelling, dissipation of Δψm, ROS generation, cytochrome c release). This group also provides evidence that another GzmM substrate may be TRAP75, a heat shock protein that inhibits GzmM-induced ROS generation (175). However, one aspect of this study that may not be completely consistent with what is known about GzmM is that the Fan paper (174, 175) claims that GzmM cleaves ICAD after a Ser residue, whereas peptides containing Ser at the P1 site are not substrates of GzmM expressed in yeast. Therefore, further work is needed to determine whether GzmM activates GzmB-like caspase-dependent cell death or a completely novel pathway distinct from any of the other granzymes. Examining cell death induced by native purified GzmM may be necessary to determine what type of cell death is induced by this enzyme. One other intriguing activity of GzmM may be to cleave and inactivate the GzmB serpin inhibitor PI-9, which it does in vitro (170). If this proves to be a physiologically relevant substrate in cells, then one function of GzmM may be to potentiate the activity of GzmB.
EXTRACELLULAR ROLES OF GRANZYMES
Although most research has focused on the cell death–inducing properties of granzymes, there is an older and growing literature to suggest that granzymes may have extracellular functions in promoting inflammation and degrading extracellular matrix, potentially to allow cytotoxic cells access to target cells within tissue or to induce death by anoikis of anchorage-dependent cells. It is unclear whether, during killer cell degranulation, the immune synapse forms a perfectly tight gasket that completely prevents granzymes from leaking out into the extracellular space. Moreover, increasing evidence suggests that granzymes may be expressed without perforin and secreted by other types of white blood cells during inflammation, including mast cells, neutrophils, activated macrophages, as well as potentially some nonhematopoietic cells, such as UV-damaged keratinocytes (7, 10, 11, 176-178). GzmB is even expressed by developing germ cells in the testes and by syncytial trophoblasts in the placenta (14).
Low concentrations of GzmA, GzmB, and GzmK have been detected in the serum of healthy donors (179). During inflammation and infection, elevated levels of granzymes exist in both serum and other bodily fluids. Examples in which extracellular granzymes have been detected include the serum of patients undergoing acute cytomegalovirus infection or chronic HIV infection, the joints of rheumatoid arthritis patients, and the bronchoalveolar lavage fluid of allergen-challenged patients with asthma and patients with chronic obstructive pulmonary disease (9, 114, 179-183). Elevated granzyme levels also occur in the serum of patients with endotoxemia and bacteremia, supporting the idea that granzymes are expressed and secreted by activated leukocytes, not just by lymphocytes (184). In fact, in sepsis patients, not only is serum GzmK elevated, but its natural inhibitor (inter-α protein) is depleted, so the free active form of the enzyme is circulating and may cause damage (185). GzmB has also been detected in macrophages of atheromatous lesions and rheumatoid joints (176). Proteolysis by extracellular granzymes is inhibited to some extent by serum and extracellular protease inhibitors, such as the trypsin inhibitors antithrombin III and alpha-2-macroglobulin (114). Some conditions that induce extracellular granzymes may also increase the release of intracellular serpins (186).
Although these extracellular granzymes may not be able to get into the cytoplasm of cells to induce cell death without a high local concentration of perforin, they could proteolyze cell surface receptors or extracellular proteins. The extracellular functions of the granzymes that have been reported are summarized below, but these proteases, despite their high degree of substrate specificity, likely have multiple, as yet unappreciated, destructive effects, particularly if present at high concentrations at inflamed sites in the absence of natural inhibitors.
The known extracellular activities of GzmA suggest a proinflammatory effect. GzmA can activate the proinflammatory cytokine IL-1β directly by cleaving its propep-tide (187). Other reports suggest that GzmA may proteolytically activate macrophages to secrete cytokines (188). It can also cause neurite retraction on astrocytes and inhibit thrombin-induced platelet aggregation by cleaving the thrombin receptor (189, 190). One study suggests another anticoagulant effect via activating prourokinase to activate plasminogen (191). Other papers suggest possible roles in degrading extracellular matrix proteins, including heparin sulfate proteogly-cans, collagen type IV, and fibronectin (192-194). Moreover, binding of GzmA to basement membrane proteoglycans may protect it from extracellular tryptase inhibitors and serve to release growth factors, such as bFGF, that are stored bound to the extracellular matrix (194).
GzmB also remodels the extracellular matrix by direct cleavage of vitronectin, fibronectin, and laminin (195). In fact, GzmB can cleave after the RGD integrin-binding domain of vitronectin. Proteolysis of the extracellular matrix may cause anchorage-independent cell death, restrict tumor cell invasion, facilitate lymphocyte migration to sites of infection or inflammation, or cause tissue destruction at sites of inflammation (195, 196). In addition, GzmB can degrade cartilage proteoglycans potentially to exacerbate autoimmune or inflammatory arthritis (197, 198). In the central nervous system, GzmB cleaves a glutamate receptor (GluR3), potentially contributing to immunoneurotoxicity, excitation, and autoimmunity in the brain (199, 200). GzmB on its own causes death of neurons in a pertussis toxin-sensitive manner, suggesting possible cleavage or involvement of G protein-coupled receptors (201). Other potential GzmB receptor targets are Notch1 and FGFR1, which may inhibit growth signals to developing or malignant cells (202).
LESSONS FROM KNOCKOUT MICE
GzmA-, GzmB-, and perforin-deficient mice were generated a decade ago, and early studies have been extensively reviewed (1). Although perforin-deficient mice are severely immunodeficient and compromised in their ability to defend against viruses and tumors, mice deficient in any one of the ten granzymes, or even of the GzmB cluster, have only subtle differences compared with wild-type animals. These experiments highlight the functional redundancy of the granzymes. Although only one molecule (perforin) effectively delivers the granzymes into target cells, each of the granzymes can trigger cell death. However, target cells may be selectively resistant to one or another of the granzymes, i.e., by bcl-2 overexpression or by expression of viral ser-pins, and granzyme expression also varies in different types of immune responders. Specific requirements for a single granzyme have been shown in some cases by specific immune challenges. For example, GzmA-deficient mice are particularly susceptible to the pox virus ectromelia (203), and GzmB-deficient mice have a markedly attenuated incidence of graft-versus-host disease (204). In constructing genetically deficient mice, genetic alterations of one gene can affect the expression of nearby granzyme genes. In the original GzmB-knockout mice, the presence of a phosphoglycerate kinase promoter-neomycin resistance gene cassette in the GzmB locus impedes the expression of the GzmB-proximal genes (GzmC, D, and F). The GzmB gene has also been deleted while keeping the expression of GzmC, D, and F intact (205). CTL from the GzmB-specific deletion mouse are significantly more effective at inducing apoptosis than the earlier GzmB cluster–knockout animal, underlining the importance of the other GzmB cluster granzymes.
Because GzmA and GzmB are the most abundantly expressed granzymes in T cells, GzmA/B doubly deficient mice are more immunodeficient than the single knockouts (206-208). CTLs from GzmA/B-deficient mice, although somewhat impaired in cytotoxicity relative to wild-type cells, nonetheless largely retain the ability to kill target cells (131, 209, 210). However, the timing of key molecular events during apoptosis, such as membrane ruffling and externalization of phosphatidylserine (annexin V staining), is markedly different during cell death induced by wild-type CTLs versus GzmA/Bdeficient CTLs (210). CTLs lacking GzmA and GzmB induce a modified form of cell death that is morphologically distinct from either perforin-mediated necrosis or wild-type CTL-mediated apoptosis (210). These differences could potentially be physiologically significant. In contrast to perforindeficient mice, GzmA/B-deficient animals develop normally, do not develop spontaneous tumors, and clear many viruses normally. The most economic explanation of these results is that the other death-inducing, perforin-dependent granzymes (particularly C, K, and M) (154, 159, 172) substitute for GzmA and GzmB. GzmM, abundantly expressed in NK cells, may be particularly important in early innate defense to contain the spread of infection or recognize transformed cells.
SUMMARY: WHY SO MANY GRANZYMES?
Recent studies have begun to define multiple death pathways activated by individual granzymes and potentially important extracellular roles of these enzymes. The granzymes can trigger at least three distinct cell death pathways, which are just being elucidated with the recent availability of recombinant active forms of many of the granzymes. The immune system needs to contend with a wide variety of tumors and infections, which have elaborated multiple strategies to evade apoptosis and immune destruction. Moreover, the granzyme-perforin system may also play a significant role in regulating immune cell numbers and function and disarming specific intracellular pathogens. The redundancy of granzymes makes sense, given the variety of tasks they need to accomplish. The example of the interplay between GzmB and GzmH and adenovirus illustrates why multiple granzymes may have evolved to eliminate important pathogens (121-123). Although both enzymes can cleave and inactivate at least two adenoviral proteins, the virus has also developed a way of inactivating GzmB. It appears that GzmH can potentiate the effect of GzmB by destroying the GzmB inhibitor. In the future, careful in vitro and in vivo studies of immune protection from important pathogens will likely define the specific role of each of the granzymes and help us understand their evolution.
ACKNOWLEDGMENTS
We thank members of the Lieberman laboratory and Mark Smyth for useful discussions and Rohit Panchakshari for help with Figure 1. This work was supported by NIH grants AI45587 and AI63430 (J.L.) and a Leukemia and Lymphoma Society fellowship (D.C.).
LITERATURE CITED
- 1.Russell JH, Ley TJ. Lymphocyte-mediated cytotoxicity. Annu. Rev. Immunol. 2002;20:323–70. doi: 10.1146/annurev.immunol.20.100201.131730. [DOI] [PubMed] [Google Scholar]
- 2.Ebnet K, Levelt CN, Tran TT, Eichmann K, Simon MM. Transcription of granzyme A and B genes is differentially regulated during lymphoid ontogeny. J. Exp. Med. 1995;181:755–63. doi: 10.1084/jem.181.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gondek DC, Lu LF, Quezada SA, Sakaguchi S, Noelle RJ. Cutting edge: contact-mediated suppression by CD4+CD25+ regulatory cells involves a granzyme B-dependent, perforin-independent mechanism. J. Immunol. 2005;174:1783–86. doi: 10.4049/jimmunol.174.4.1783. [DOI] [PubMed] [Google Scholar]
- 4.Grossman WJ, Verbsky JW, Tollefsen BL, Kemper C, Atkinson JP, Ley TJ. Differential expression of granzymes A and B in human cytotoxic lymphocyte subsets and T regulatory cells. Blood. 2004;104:2840–48. doi: 10.1182/blood-2004-03-0859. [DOI] [PubMed] [Google Scholar]
- 5.Jahrsdorfer B, Blackwell SE, Wooldridge JE, Huang J, Andreski MW, et al. B-chronic lymphocytic leukemia cells and other B cells can produce granzyme B and gain cytotoxic potential after interleukin-21-based activation. Blood. 2006;108:2712–19. doi: 10.1182/blood-2006-03-014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rissoan MC, Duhen T, Bridon JM, Bendriss-Vermare N, Peronne C, et al. Subtractive hybridization reveals the expression of immunoglobulin-like transcript 7, EphB1, granzyme B, and 3 novel transcripts in human plasmacytoid dendritic cells. Blood. 2002;100:3295–303. doi: 10.1182/blood-2002-02-0638. [DOI] [PubMed] [Google Scholar]
- 7.Strik MC, de Koning PJ, Kleijmeer MJ, Bladergroen BA, Wolbink AM, et al. Human mast cells produce and release the cytotoxic lymphocyte associated protease granzyme B upon activation. Mol. Immunol. 2007;44:3462–72. doi: 10.1016/j.molimm.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 8.Pardo J, Wallich R, Ebnet K, Iden S, Zentgraf H, et al. Granzyme B is expressed in mouse mast cells in vivo and in vitro and causes delayed cell death independent of perforin. Cell Death Differ. 2007;14:1768–79. doi: 10.1038/sj.cdd.4402183. [DOI] [PubMed] [Google Scholar]
- 9.Tschopp CM, Spiegl N, Didichenko S, Lutmann W, Julius P, et al. Granzyme B, a novel mediator of allergic inflammation: its induction and release in blood basophils and human asthma. Blood. 2006;108:2290–99. doi: 10.1182/blood-2006-03-010348. [DOI] [PubMed] [Google Scholar]
- 10.Hochegger K, Eller P, Huber JM, Bernhard D, Mayer G, et al. Expression of granzyme A in human polymorphonuclear neutrophils. Immunology. 2007;121:166–73. doi: 10.1111/j.1365-2567.2006.02551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner C, Iking-Konert C, Denefleh B, Stegmaier S, Hug F, Hansch GM. Granzyme B and perforin: constitutive expression in human polymorphonuclear neutrophils. Blood. 2004;103:1099–104. doi: 10.1182/blood-2003-04-1069. [DOI] [PubMed] [Google Scholar]
- 12.Martin P, Wallich R, Pardo J, Mullbacher A, Munder M, et al. Quiescent and activated mouse granulocytes do not express granzyme A and B or perforin: similarities or differences with human polymorphonuclear leukocytes? Blood. 2005;106:2871–78. doi: 10.1182/blood-2005-04-1522. [DOI] [PubMed] [Google Scholar]
- 13.Metkar SS, Froelich CJ. Human neutrophils lack granzyme A, granzyme B, and perforin. Blood. 2004;104:905–6. doi: 10.1182/blood-2004-03-0888. author reply 907–8. [DOI] [PubMed] [Google Scholar]
- 14.Hirst CE, Buzza MS, Sutton VR, Trapani JA, Loveland KL, Bird PI. Perforin-independent expression of granzyme B and proteinase inhibitor 9 in human testis and placenta suggests a role for granzyme B-mediated proteolysis in reproduction. Mol. Hum. Reprod. 2001;7:1133–42. doi: 10.1093/molehr/7.12.1133. [DOI] [PubMed] [Google Scholar]
- 15.Sasson R, Dantes A, Tajima K, Amsterdam A. Novel genes modulated by FSH in normal and immortalized FSH-responsive cells: new insights into the mechanism of FSH action. FASEB J. 2003;17:1256–66. doi: 10.1096/fj.02-0740com. [DOI] [PubMed] [Google Scholar]
- 16.Horiuchi K, Saito S, Sasaki R, Tomatsu T, Toyama Y. Expression of granzyme B in human articular chondrocytes. J. Rheumatol. 2003;30:1799–810. [PubMed] [Google Scholar]
- 17.Taniguchi M, Tani N, Suemoto T, Ishimoto I, Shiosaka S, Yoshida S. High expression of alternative transcript of granzyme M in the mouse retina. Neurosci. Res. 1999;34:115–23. doi: 10.1016/s0168-0102(99)00036-x. [DOI] [PubMed] [Google Scholar]
- 18.Suemoto T, Taniguchi M, Shiosaka S, Yoshida S. cDNA cloning and expression of a novel serine protease in the mouse brain. Brain Res. Mol. Brain Res. 1999;70:273–81. doi: 10.1016/s0169-328x(99)00166-7. [DOI] [PubMed] [Google Scholar]
- 19.Prendergast JA, Helgason CD, Bleackley RC. Quantitative polymerase chain reaction analysis of cytotoxic cell proteinase gene transcripts in T cells. Pattern of expression is dependent on the nature of the stimulus. J. Biol. Chem. 1992;267:5090–95. [PubMed] [Google Scholar]
- 20.Garcia-Sanz JA, MacDonald HR, Jenne DE, Tschopp J, Nabholz M. Cell specificity of granzyme gene expression. J. Immunol. 1990;145:3111–18. [PubMed] [Google Scholar]
- 21.Ebnet K, Chluba-de Tapia J, Hurtenbach U, Kramer MD, Simon MM. In vivo primed mouse T cells selectively express T cell-specific serine proteinase-1 and the proteinase-like molecules granzyme B and C. Int. Immunol. 1991;3:9–19. doi: 10.1093/intimm/3.1.9. [DOI] [PubMed] [Google Scholar]
- 22.Kelso A, Costelloe EO, Johnson BJ, Groves P, Buttigieg K, Fitzpatrick DR. The genes for perforin, granzymes A-C and IFN-γ are differentially expressed in single CD8+ T cells during primary activation. Int. Immunol. 2002;14:605–13. doi: 10.1093/intimm/dxf028. [DOI] [PubMed] [Google Scholar]
- 23.Liu CC, Rafii S, Granelli-Piperno A, Trapani JA, Young JD. Perforin and serine esterase gene expression in stimulated human T cells. Kinetics, mitogen requirements, and effects of cyclosporin A. J. Exp. Med. 1989;170:2105–18. doi: 10.1084/jem.170.6.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaffen SL. Signaling domains of the interleukin 2 receptor. Cytokine. 2001;14:63–77. doi: 10.1006/cyto.2001.0862. [DOI] [PubMed] [Google Scholar]
- 25.Janas ML, Groves P, Kienzle N, Kelso A. IL-2 regulates perforin and granzyme gene expression in CD8+ T cells independently of its effects on survival and proliferation. J. Immunol. 2005;175:8003–10. doi: 10.4049/jimmunol.175.12.8003. [DOI] [PubMed] [Google Scholar]
- 26.Kundig TM, Schorle H, Bachmann MF, Hengartner H, Zinkernagel RM, Horak I. Immune responses in interleukin-2-deficient mice. Science. 1993;262:1059–61. doi: 10.1126/science.8235625. [DOI] [PubMed] [Google Scholar]
- 27.Steiger J, Nickerson PW, Steurer W, Moscovitch-Lopatin M, Strom TB. IL-2 knockout recipient mice reject islet cell allografts. J. Immunol. 1995;155:489–98. [PubMed] [Google Scholar]
- 28.Kramer S, Mamalaki C, Horak I, Schimpl A, Kioussis D, Hung T. Thymic selection and peptide-induced activation of T cell receptor-transgenic CD8 T cells in interleukin-2-deficient mice. Eur. J. Immunol. 1994;24:2317–22. doi: 10.1002/eji.1830241009. [DOI] [PubMed] [Google Scholar]
- 29.Ye W, Young JD, Liu CC. Interleukin-15 induces the expression of mRNAs of cytolytic mediators and augments cytotoxic activities in primary murine lymphocytes. Cell. Immunol. 1996;174:54–62. doi: 10.1006/cimm.1996.0293. [DOI] [PubMed] [Google Scholar]
- 30.Zeng R, Spolski R, Finkelstein SE, Oh S, Kovanen PE, et al. Synergy of IL-21 and IL-15 in regulating CD8+ T cell expansion and function. J. Exp. Med. 2005;201:139–48. doi: 10.1084/jem.20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leonard WJ, Spolski R. Interleukin-21: a modulator of lymphoid proliferation, apoptosis and differentiation. Nat. Rev. Immunol. 2005;5:688–98. doi: 10.1038/nri1688. [DOI] [PubMed] [Google Scholar]
- 32.White L, Krishnan S, Strbo N, Liu H, Kolber MA, et al. Differential effects of IL-21 and IL-15 on perforin expression, lysosomal degranulation, and proliferation in CD8 T cells of patients with human immunodeficiency virus-1 (HIV) Blood. 2007;109:3873–80. doi: 10.1182/blood-2006-09-045278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto K, Shibata F, Miyasaka N, Miura O. The human perforin gene is a direct target of STAT4 activated by IL-12 in NK cells. Biochem. Biophys. Res. Commun. 2002;297:1245–52. doi: 10.1016/s0006-291x(02)02378-1. [DOI] [PubMed] [Google Scholar]
- 34.Morishima N, Owaki T, Asakawa M, Kamiya S, Mizuguchi J, Yoshimoto T. Augmentation of effector CD8+ T cell generation with enhanced granzyme B expression by IL-27. J. Immunol. 2005;175:1686–93. doi: 10.4049/jimmunol.175.3.1686. [DOI] [PubMed] [Google Scholar]
- 35.Fregeau CJ, Bleackley RC. Transcription of two cytotoxic cell protease genes is under the control of different regulatory elements. Nucleic Acids Res. 1991;19:5583–90. doi: 10.1093/nar/19.20.5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Babichuk CK, Duggan BL, Bleackley RC. In vivo regulation of murine granzyme B gene transcription in activated primary T cells. J. Biol. Chem. 1996;271:16485–93. doi: 10.1074/jbc.271.28.16485. [DOI] [PubMed] [Google Scholar]
- 37.Wargnier A, Legros-Maida S, Bosselut R, Bourge JF, Lafaurie C, et al. Identification of human granzyme B promoter regulatory elements interacting with activated T-cell-specific proteins: implication of Ikaros and CBF binding sites in promoter activation. Proc. Natl. Acad. Sci. USA. 1995;92:6930–34. doi: 10.1073/pnas.92.15.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haddad P, Wargnier A, Bourge JF, Sasportes M, Paul P. A promoter element of the human serine esterase granzyme B gene controls specific transcription in activated T cells. Eur. J. Immunol. 1993;23:625–29. doi: 10.1002/eji.1830230307. [DOI] [PubMed] [Google Scholar]
- 39.Babichuk CK, Bleackley RC. Mutational analysis of the murine granzyme B gene promoter in primary T cells and a T cell clone. J. Biol. Chem. 1997;272:18564–71. doi: 10.1074/jbc.272.30.18564. [DOI] [PubMed] [Google Scholar]
- 40.Zimmerer JM, Lesinski GB, Radmacher MD, Ruppert A, Carson WE., III STAT1-dependent and STAT1-independent gene expression in murine immune cells following stimulation with interferon-α. Cancer Immunol. Immunother. 2007;56:1845–52. doi: 10.1007/s00262-007-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–43. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 42.U M, Shen L, Oshida T, Miyauchi J, Yamada M, Miyashita T. Identification of novel direct transcriptional targets of glucocorticoid receptor. Leukemia. 2004;18:1850–56. doi: 10.1038/sj.leu.2403516. [DOI] [PubMed] [Google Scholar]
- 43.Yamada M, Hirasawa A, Shiojima S, Tsujimoto G. Granzyme A mediates glucocorticoid-induced apoptosis in leukemia cells. FASEB J. 2003;17:1712–14. doi: 10.1096/fj.02-1116fje. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida NL, Miyashita T, U M, Yamada M, Reed JC, et al. Analysis of gene expression patterns during glucocorticoid-induced apoptosis using oligonucleotide arrays. Biochem. Biophys. Res. Commun. 2002;293:1254–61. doi: 10.1016/S0006-291X(02)00361-3. [DOI] [PubMed] [Google Scholar]
- 45.Ruike Y, Katsuma S, Hirasawa A, Tsujimoto G. Glucocorticoid-induced alternative promoter usage for a novel 5′ variant of granzyme A. J. Hum. Genet. 2007;52:172–78. doi: 10.1007/s10038-006-0099-9. [DOI] [PubMed] [Google Scholar]
- 46.Fehniger TA, Cai SF, Cao X, Bredemeyer AJ, Presti RM, et al. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26:798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 47.Vosshenrich CA, Ranson T, Samson SI, Corcuff E, Colucci F, et al. Roles for common cytokine receptor γ-chain-dependent cytokines in the generation, differentiation, and maturation of NK cell precursors and peripheral NK cells in vivo. J. Immunol. 2005;174:1213–21. doi: 10.4049/jimmunol.174.3.1213. [DOI] [PubMed] [Google Scholar]
- 48.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–40. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 49.Bratke K, Kuepper M, Bade B, Virchow JC, Jr, Luttmann W. Differential expression of human granzymes A, B, and K in natural killer cells and during CD8+ T cell differentiation in peripheral blood. Eur. J. Immunol. 2005;35:2608–16. doi: 10.1002/eji.200526122. [DOI] [PubMed] [Google Scholar]
- 50.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–51. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 51.Bell JK, Goetz DH, Mahrus S, Harris JL, Fletterick RJ, Craik CS. The oligomeric structure of human granzyme A is a determinant of its extended substrate specificity. Nat. Struct. Biol. 2003;10:527–34. doi: 10.1038/nsb944. [DOI] [PubMed] [Google Scholar]
- 52.Hink-Schauer C, Estebanez-Perpina E, Wilharm E, Fuentes-Prior P, Klinkert W, et al. The 2.2-Å crystal structure of human progranzyme K reveals a rigid zymogen with unusual features. J. Biol. Chem. 2002;277:50923–33. doi: 10.1074/jbc.M207962200. [DOI] [PubMed] [Google Scholar]
- 53.Casciola-Rosen L, Garcia-Calvo M, Bull HG, Becker JW, Hines T, et al. Mouse and human granzyme B have distinct tetrapeptide specificities and abilities to recruit the bid pathway. J. Biol. Chem. 2007;282:4545–52. doi: 10.1074/jbc.M606564200. [DOI] [PubMed] [Google Scholar]
- 54.Estebanez-Perpina E, Fuentes-Prior P, Belorgey D, Braun M, Kiefersauer R, et al. Crystal structure of the caspase activator human granzyme B, a proteinase highly specific for an Asp-P1 residue. Biol. Chem. 2000;381:1203–14. doi: 10.1515/BC.2000.148. [DOI] [PubMed] [Google Scholar]
- 54a.Hink-Shauer C, Estebanez-Perpina E, Kurschus FC, Bode W, Jenne DE. Crystal structure of the apoptosis-inducing granzyme A dimer. Nature Struct. Biol. 2003;10:535–40. doi: 10.1038/nsb945. [DOI] [PubMed] [Google Scholar]
- 55.Kummer JA, Kamp AM, Citarella F, Horrevoets AJ, Hack CE. Expression of human recombinant granzyme A zymogen and its activation by the cysteine proteinase cathepsin C. J. Biol. Chem. 1996;271:9281–86. doi: 10.1074/jbc.271.16.9281. [DOI] [PubMed] [Google Scholar]
- 56.Pham CT, Ivanovich JL, Raptis SZ, Zehnbauer B, Ley TJ. Papillon-Lefevre syndrome: correlating the molecular, cellular, and clinical consequences of cathepsin C/dipeptidyl peptidase I deficiency in humans. J. Immunol. 2004;173:7277–81. doi: 10.4049/jimmunol.173.12.7277. [DOI] [PubMed] [Google Scholar]
- 57.Sutton VR, Waterhouse NJ, Browne KA, Sedelies K, Ciccone A, et al. Residual active granzyme B in cathepsin C-null lymphocytes is sufficient for perforin-dependent target cell apoptosis. J. Cell Biol. 2007;176:425–33. doi: 10.1083/jcb.200609077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meade JL, de Wynter EA, Brett P, Sharif SM, Woods CG, et al. A family with Papillon-Lefevre syndrome reveals a requirement for cathepsin C in granzyme B activation and NK cell cytolytic activity. Blood. 2006;107:3665–68. doi: 10.1182/blood-2005-03-1140. [DOI] [PubMed] [Google Scholar]
- 59.Grujic M, Braga T, Lukinius A, Eloranta ML, Knight SD, et al. Serglycin-deficient cytotoxic T lymphocytes display defective secretory granule maturation and granzyme B storage. J. Biol. Chem. 2005;280:33411–18. doi: 10.1074/jbc.M501708200. [DOI] [PubMed] [Google Scholar]
- 60.Bossi G, Griffiths GM. CTL secretory lysosomes: biogenesis and secretion of a harmful organelle. Semin. Immunol. 2005;17:87–94. doi: 10.1016/j.smim.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 61.Stinchcombe JC, Bossi G, Booth S, Griffiths GM. The immunological synapse of CTL contains a secretory domain and membrane bridges. Immunity. 2001;15:751–61. doi: 10.1016/s1074-7613(01)00234-5. [DOI] [PubMed] [Google Scholar]
- 62.Purbhoo MA, Irvine DJ, Huppa JB, Davis MM. T cell killing does not require the formation of a stable mature immunological synapse. Nat. Immunol. 2004;5:524–30. doi: 10.1038/ni1058. [DOI] [PubMed] [Google Scholar]
- 63.Raja SM, Metkar SS, Honing S, Wang B, Russin WA, et al. A novel mechanism for protein delivery: granzyme B undergoes electrostatic exchange from serglycin to target cells. J. Biol. Chem. 2005;280:20752–61. doi: 10.1074/jbc.M501181200. [DOI] [PubMed] [Google Scholar]
- 64.Kurschus FC, Bruno R, Fellows E, Falk CS, Jenne DE. Membrane receptors are not required to deliver granzyme B during killer cell attack. Blood. 2005;105:2049–58. doi: 10.1182/blood-2004-06-2180. [DOI] [PubMed] [Google Scholar]
- 65.Bird CH, Sun J, Ung K, Karambalis D, Whisstock JC, et al. Cationic sites on granzyme B contribute to cytotoxicity by promoting its uptake into target cells. Mol. Cell. Biol. 2005;25:7854–67. doi: 10.1128/MCB.25.17.7854-7867.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi L, Keefe D, Durand E, Feng H, Zhang D, Lieberman J. Granzyme B binds to target cells mostly by charge and must be added at the same time as perforin to trigger apoptosis. J. Immunol. 2005;174:5456–61. doi: 10.4049/jimmunol.174.9.5456. [DOI] [PubMed] [Google Scholar]
- 67.Motyka B, Korbutt G, Pinkoski MJ, Heibein JA, Caputo A, et al. Mannose 6-phosphate/insulin-like growth factor II receptor is a death receptor for granzyme B during cytotoxic T cell-induced apoptosis. Cell. 2000;103:491–500. doi: 10.1016/s0092-8674(00)00140-9. [DOI] [PubMed] [Google Scholar]
- 68.Dressel R, Raja SM, Honing S, Seidler T, Froelich CJ, et al. Granzyme-mediated cytotoxicity does not involve the mannose 6-phosphate receptors on target cells. J. Biol. Chem. 2004;279:20200–10. doi: 10.1074/jbc.M313108200. [DOI] [PubMed] [Google Scholar]
- 69.Trapani JA, Sutton VR, Thia KY, Li YQ, Froelich CJ, et al. A clathrin/dynaminand mannose-6-phosphate receptor-independent pathway for granzyme B-induced cell death. J. Cell Biol. 2003;160:223–33. doi: 10.1083/jcb.200210150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pipkin ME, Lieberman J. Delivering the kiss of death: progress on understanding how perforin works. Curr. Opin. Immunol. 2007;19:301–8. doi: 10.1016/j.coi.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Keefe D, Shi L, Feske S, Massol R, Navarro F, et al. Perforin triggers a plasma membrane-repair response that facilitates CTL induction of apoptosis. Immunity. 2005;23:249–62. doi: 10.1016/j.immuni.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 72.Veugelers K, Motyka B, Frantz C, Shostak I, Sawchuk T, Bleackley RC. The granzyme B-serglycin complex from cytotoxic granules requires dynamin for endocytosis. Blood. 2004;103:3845–53. doi: 10.1182/blood-2003-06-2156. [DOI] [PubMed] [Google Scholar]
- 73.Jans DA, Briggs LJ, Jans P, Froelich CJ, Parasivam G, et al. Nuclear targeting of the serine protease granzyme A (fragmentin-1) J. Cell Sci. 1998;111(Pt 17):2645–54. doi: 10.1242/jcs.111.17.2645. [DOI] [PubMed] [Google Scholar]
- 74.Jans DA, Jans P, Briggs LJ, Sutton V, Trapani JA. Nuclear transport of granzyme B (fragmentin-2). Dependence of perforin in vivo and cytosolic factors in vitro. J. Biol. Chem. 1996;271:30781–89. doi: 10.1074/jbc.271.48.30781. [DOI] [PubMed] [Google Scholar]
- 75.Blink EJ, Jiansheng Z, Hu W, Calanni ST, Trapani JA, et al. Interaction of the nuclear localizing cytolytic granule serine protease granzyme B with importin α or β: modulation by the serpin inhibitor PI-9. J. Cell. Biochem. 2005;95:598–610. doi: 10.1002/jcb.20415. [DOI] [PubMed] [Google Scholar]
- 76. Deleted in proof.
- 77.Kagi D, Ledermann B, Burki K, Seiler P, Odermatt B, et al. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature. 1994;369:31–37. doi: 10.1038/369031a0. [DOI] [PubMed] [Google Scholar]
- 78.Stepp SE, Dufourcq-Lagelouse R, Le Deist F, Bhawan S, Certain S, et al. Perforin gene defects in familial hemophagocytic lymphohistiocytosis. Science. 1999;286:1957–59. doi: 10.1126/science.286.5446.1957. [DOI] [PubMed] [Google Scholar]
- 79.Menasche G, Feldmann J, Fischer A, de Saint Basile G. Primary hemophagocytic syndromes point to a direct link between lymphocyte cytotoxicity and homeostasis. Immunol. Rev. 2005;203:165–79. doi: 10.1111/j.0105-2896.2005.00224.x. [DOI] [PubMed] [Google Scholar]
- 80.Feldmann J, Menasche G, Callebaut I, Minard-Colin V, Bader-Meunier B, et al. Severe and progressive encephalitis as a presenting manifestation of a novel missense perforin mutation and impaired cytolytic activity. Blood. 2005;105:2658–63. doi: 10.1182/blood-2004-09-3590. [DOI] [PubMed] [Google Scholar]
- 81.Dufourcq-Lagelouse R, Pastural E, Barrat FJ, Feldmann J, Le Deist F, et al. Genetic basis of hemophagocytic lymphohistiocytosis syndrome (review) Int. J. Mol. Med. 1999;4:127–33. doi: 10.3892/ijmm.4.2.127. [DOI] [PubMed] [Google Scholar]
- 82.Balaji KN, Schaschke N, Machleidt W, Catalfamo M, Henkart PA. Surface cathepsin B protects cytotoxic lymphocytes from self-destruction after degranulation. J. Exp. Med. 2002;196:493–503. doi: 10.1084/jem.20011836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baran K, Ciccone A, Peters C, Yagita H, Bird PI, et al. Cytotoxic T lymphocytes from cathepsin B-deficient mice survive normally in vitro and in vivo after encountering and killing target cells. J. Biol. Chem. 2006;281:30485–91. doi: 10.1074/jbc.M602007200. [DOI] [PubMed] [Google Scholar]
- 84.Browne KA, Blink E, Sutton VR, Froelich CJ, Jans DA, Trapani JA. Cytosolic delivery of granzyme B by bacterial toxins: evidence that endosomal disruption, in addition to transmembrane pore formation, is an important function of perforin. Mol. Cell. Biol. 1999;19:8604–15. doi: 10.1128/mcb.19.12.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gross C, Koelch W, DeMaio A, Arispe N, Multhoff G. Cell surface-bound heat shock protein 70 (Hsp70) mediates perforin-independent apoptosis by specific binding and uptake of granzyme B. J. Biol. Chem. 2003;278:41173–81. doi: 10.1074/jbc.M302644200. [DOI] [PubMed] [Google Scholar]
- 86.Ida H, Nakashima T, Kedersha NL, Yamasaki S, Huang M, et al. Granzyme B leakage-induced cell death: a new type of activation-induced natural killer cell death. Eur. J. Immunol. 2003;33:3284–92. doi: 10.1002/eji.200324376. [DOI] [PubMed] [Google Scholar]
- 87.Sun J, Bird CH, Sutton V, McDonald L, Coughlin PB, et al. A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes. J. Biol. Chem. 1996;271:27802–9. doi: 10.1074/jbc.271.44.27802. [DOI] [PubMed] [Google Scholar]
- 88.Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, et al. An overview of the serpin superfamily. Genome Biol. 2006;7:216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 2001;276:33293–96. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 90.Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors: structure, function, and regulation. J. Biol. Chem. 1994;269:15957–60. [PubMed] [Google Scholar]
- 91.Bird CH, Blink EJ, Hirst CE, Buzza MS, Steele PM, et al. Nucleocytoplasmic distribution of the ovalbumin serpin PI-9 requires a nonconventional nuclear import pathway and the export factor Crm1. Mol. Cell. Biol. 2001;21:5396–407. doi: 10.1128/MCB.21.16.5396-5407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hirst CE, Buzza MS, Bird CH, Warren HS, Cameron PU, et al. The intracellular granzyme B inhibitor, proteinase inhibitor 9, is up-regulated during accessory cell maturation and effector cell degranulation, and its overexpression enhances CTL potency. J. Immunol. 2003;170:805–15. doi: 10.4049/jimmunol.170.2.805. [DOI] [PubMed] [Google Scholar]
- 93.Buzza MS, Hosking P, Bird PI. The granzyme B inhibitor, PI-9, is differentially expressed during placental development and up-regulated in hydatidiform moles. Placenta. 2006;27:62–69. doi: 10.1016/j.placenta.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 94.Bladergroen BA, Strik MC, Bovenschen N, van Berkum O, Scheffer GL, et al. The granzyme B inhibitor, protease inhibitor 9, is mainly expressed by dendritic cells and at immune-privileged sites. J. Immunol. 2001;166:3218–25. doi: 10.4049/jimmunol.166.5.3218. [DOI] [PubMed] [Google Scholar]
- 95.Buzza MS, Hirst CE, Bird CH, Hosking P, McKendrick J, Bird PI. The granzyme B inhibitor, PI-9, is present in endothelial and mesothelial cells, suggesting that it protects bystander cells during immune responses. Cell. Immunol. 2001;210:21–29. doi: 10.1006/cimm.2001.1806. [DOI] [PubMed] [Google Scholar]
- 96.Bladergroen BA, Strik MC, Wolbink AM, Wouters D, Broekhuizen R, et al. The granzyme B inhibitor proteinase inhibitor 9 (PI9) is expressed by human mast cells. Eur. J. Immunol. 2005;35:1175–83. doi: 10.1002/eji.200425949. [DOI] [PubMed] [Google Scholar]
- 97.Kannan-Thulasiraman P, Shapiro DJ. Modulators of inflammation use nuclear factor-κB and activator protein-1 sites to induce the caspase-1 and granzyme B inhibitor, proteinase inhibitor 9. J. Biol. Chem. 2002;277:41230–39. doi: 10.1074/jbc.M200379200. [DOI] [PubMed] [Google Scholar]
- 98.Barrie MB, Stout HW, Abougergi MS, Miller BC, Thiele DL. Antiviral cytokines induce hepatic expression of the granzyme B inhibitors, proteinase inhibitor 9 and serine proteinase inhibitor 6. J. Immunol. 2004;172:6453–59. doi: 10.4049/jimmunol.172.10.6453. [DOI] [PubMed] [Google Scholar]
- 99.Jiang X, Orr BA, Kranz DM, Shapiro DJ. Estrogen induction of the granzyme B inhibitor, proteinase inhibitor 9, protects cells against apoptosis mediated by cytotoxic T lymphocytes and natural killer cells. Endocrinology. 2006;147:1419–26. doi: 10.1210/en.2005-0996. [DOI] [PubMed] [Google Scholar]
- 100.Kanamori H, Krieg S, Mao C, Di Pippo VA, Wang S, et al. Proteinase inhibitor 9, an inhibitor of granzyme B-mediated apoptosis, is a primary estrogen-inducible gene in human liver cells. J. Biol. Chem. 2000;275:5867–73. doi: 10.1074/jbc.275.8.5867. [DOI] [PubMed] [Google Scholar]
- 101.Holmquist-Mengelbier L, Fredlund E, Lofstedt T, Noguera R, Navarro S, et al. Recruitment of HIF-1α and HIF-2α to common target genes is differentially regulated in neuroblastoma: HIF-2α promotes an aggressive phenotype. Cancer Cell. 2006;10:413–23. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 102.Sun J, Ooms L, Bird CH, Sutton VR, Trapani JA, Bird PI. A new family of 10 murine ovalbumin serpins includes two homologs of proteinase inhibitor 8 and two homologs of the granzyme B inhibitor (proteinase inhibitor 9) J. Biol. Chem. 1997;272:15434–41. doi: 10.1074/jbc.272.24.15434. [DOI] [PubMed] [Google Scholar]
- 103.Phillips T, Opferman JT, Shah R, Liu N, Froelich CJ, Ashton-Rickardt PG. A role for the granzyme B inhibitor serine protease inhibitor 6 in CD8+ memory cell homeostasis. J. Immunol. 2004;173:3801–9. doi: 10.4049/jimmunol.173.6.3801. [DOI] [PubMed] [Google Scholar]
- 104.Medema JP, Schuurhuis DH, Rea D, van Tongeren J, de Jong J, et al. Expression of the serpin serine protease inhibitor 6 protects dendritic cells from cytotoxic T lymphocyte-induced apoptosis: differential modulation by T helper type 1 and type 2 cells. J. Exp. Med. 2001;194:657–67. doi: 10.1084/jem.194.5.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Medema JP, de Jong J, Peltenburg LT, Verdegaal EM, Gorter A, et al. Blockade of the granzyme B/perforin pathway through overexpression of the serine protease inhibitor PI-9/SPI-6 constitutes a mechanism for immune escape by tumors. Proc. Natl. Acad. Sci. USA. 2001;98:11515–20. doi: 10.1073/pnas.201398198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang M, Park SM, Wang Y, Shah R, Liu N, et al. Serine protease inhibitor 6 protects cytotoxic T cells from self-inflicted injury by ensuring the integrity of cytotoxic granules. Immunity. 2006;24:451–61. doi: 10.1016/j.immuni.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 107.Bladergroen BA, Meijer CJ, ten Berge RL, Hack CE, Muris JJ, et al. Expression of the granzyme B inhibitor, protease inhibitor 9, by tumor cells in patients with non-Hodgkin and Hodgkin lymphoma: a novel protective mechanism for tumor cells to circumvent the immune system? Blood. 2002;99:232–37. doi: 10.1182/blood.v99.1.232. [DOI] [PubMed] [Google Scholar]
- 108.Bird CH, Sutton VR, Sun J, Hirst CE, Novak A, et al. Selective regulation of apoptosis: the cytotoxic lymphocyte serpin proteinase inhibitor 9 protects against granzyme B-mediated apoptosis without perturbing the Fas cell death pathway. Mol. Cell. Biol. 1998;18:6387–98. doi: 10.1128/mcb.18.11.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Classen CF, Ushmorov A, Bird P, Debatin KM. The granzyme B inhibitor PI-9 is differentially expressed in all main subtypes of pediatric acute lymphoblastic leukemias. Haematologica. 2004;89:1314–21. [PubMed] [Google Scholar]
- 110.van Houdt IS, Oudejans JJ, van den Eertwegh AJ, Baars A, Vos W, et al. Expression of the apoptosis inhibitor protease inhibitor 9 predicts clinical outcome in vaccinated patients with stage III and IV melanoma. Clin. Cancer Res. 2005;11:6400–7. doi: 10.1158/1078-0432.CCR-05-0306. [DOI] [PubMed] [Google Scholar]
- 111.Godal R, Keilholz U, Uharek L, Letsch A, Asemissen AM, et al. Lymphomas are sensitive to perforin-dependent cytotoxic pathways despite expression of PI-9 and overexpression of bcl-2. Blood. 2006;107:3205–11. doi: 10.1182/blood-2005-07-2880. [DOI] [PubMed] [Google Scholar]
- 112.Sayers TJ, Brooks AD, Ward JM, Hoshino T, Bere WE, et al. The restricted expression of granzyme M in human lymphocytes. J. Immunol. 2001;166:765–71. doi: 10.4049/jimmunol.166.2.765. [DOI] [PubMed] [Google Scholar]
- 113.Kam CM, Hudig D, Powers JC. Granzymes (lymphocyte serine proteases): characterization with natural and synthetic substrates and inhibitors. Biochim. Biophys. Acta. 2000;1477:307–23. doi: 10.1016/s0167-4838(99)00282-4. [DOI] [PubMed] [Google Scholar]
- 114.Spaeny-Dekking EH, Kamp AM, Froelich CJ, Hack CE. Extracellular granzyme A, complexed to proteoglycans, is protected against inactivation by protease inhibitors. Blood. 2000;95:1465–72. [PubMed] [Google Scholar]
- 115.Tsuzuki S, Kokado Y, Satomi S, Yamasaki Y, Hirayasu H, et al. Purification and identification of a binding protein for pancreatic secretory trypsin inhibitor: a novel role of the inhibitor as an antigranzyme A. Biochem. J. 2003;372:227–33. doi: 10.1042/BJ20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kitahara T, Takatsuka Y, Fujimoto KI, Tanaka S, Ogawa M, Kosaki G. Radioimmunoassay for human pancreatic secretory trypsin inhibitor: measurement of serum pancreatic secretory trypsin inhibitor in normal subjects and subjects with pancreatic diseases. Clin. Chim. Acta. 1980;103:135–43. doi: 10.1016/0009-8981(80)90205-3. [DOI] [PubMed] [Google Scholar]
- 117.Matsuda K, Ogawa M, Shibata T, Nishibe S, Miyauchi K, et al. Postoperative elevation of serum pancreatic secretory trypsin inhibitor. Am. J. Gastroenterol. 1985;80:694–98. [PubMed] [Google Scholar]
- 118.Quan LT, Caputo A, Bleackley RC, Pickup DJ, Salvesen GS. Granzyme B is inhibited by the cowpox virus serpin cytokine response modifier A. J. Biol. Chem. 1995;270:10377–79. doi: 10.1074/jbc.270.18.10377. [DOI] [PubMed] [Google Scholar]
- 119.Cassens U, Lewinski G, Samraj AK, von Bernuth H, Baust H, et al. Viral modulation of cell death by inhibition of caspases. Arch. Immunol. Ther. Exp. 2003;51:19–27. [PubMed] [Google Scholar]
- 120.Sieg S, Xia L, Huang Y, Kaplan D. Specific inhibition of granzyme B by parainfluenza virus type 3. J. Virol. 1995;69:3538–41. doi: 10.1128/jvi.69.6.3538-3541.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Andrade F, Bull HG, Thornberry NA, Ketner GW, Casciola-Rosen LA, Rosen A. Adenovirus L4–100K assembly protein is a granzyme B substrate that potently inhibits granzyme B-mediated cell death. Immunity. 2001;14:751–61. doi: 10.1016/s1074-7613(01)00149-2. [DOI] [PubMed] [Google Scholar]
- 122.Andrade F, Casciola-Rosen LA, Rosen A. A novel domain in adenovirus L4–100K is required for stable binding and efficient inhibition of human granzyme B: possible interaction with a species-specific exosite. Mol. Cell. Biol. 2003;23:6315–26. doi: 10.1128/MCB.23.17.6315-6326.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Andrade F, Fellows E, Jenne DE, Rosen A, Young CS. Granzyme H destroys the function of critical adenoviral proteins required for viral DNA replication and granzyme B inhibition. EMBO J. 2007;26:2148–57. doi: 10.1038/sj.emboj.7601650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Willoughby CA, Bull HG, Garcia-Calvo M, Jiang J, Chapman KT, Thornberry NA. Discovery of potent, selective human granzyme B inhibitors that inhibit CTL mediated apoptosis. Bioorg. Med. Chem. Lett. 2002;12:2197–200. doi: 10.1016/s0960-894x(02)00363-3. [DOI] [PubMed] [Google Scholar]
- 125.Beresford PJ, Xia Z, Greenberg AH, Lieberman J. Granzyme A loading induces rapid cytolysis and a novel form of DNA damage independently of caspase activation. Immunity. 1999;10:585–94. doi: 10.1016/s1074-7613(00)80058-8. [DOI] [PubMed] [Google Scholar]
- 126.Shresta S, Graubert TA, Thomas DA, Raptis SZ, Ley TJ. Granzyme A initiates an alternative pathway for granule-mediated apoptosis. Immunity. 1999;10:595–605. doi: 10.1016/s1074-7613(00)80059-x. [DOI] [PubMed] [Google Scholar]
- 127.Praveen K, Evans DL, Jaso-Friedmann L. Evidence for the existence of granzyme-like serine proteases in teleost cytotoxic cells. J. Mol. Evol. 2004;58:449–59. doi: 10.1007/s00239-003-2566-7. [DOI] [PubMed] [Google Scholar]
- 128.Martinvalet D, Zhu P, Lieberman J. Granzyme A induces caspase-independent mitochondrial damage, a required first step for apoptosis. Immunity. 2005;22:355–70. doi: 10.1016/j.immuni.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 129.Beresford PJ, Zhang D, Oh DY, Fan Z, Greer EL, et al. Granzyme A activates an endoplasmic reticulum-associated caspase-independent nuclease to induce single-stranded DNA nicks. J. Biol. Chem. 2001;276:43285–93. doi: 10.1074/jbc.M108137200. [DOI] [PubMed] [Google Scholar]
- 130.Beresford PJ, Jaju M, Friedman RS, Yoon MJ, Lieberman J. A role for heat shock protein 27 in CTL-mediated cell death. J. Immunol. 1998;161:161–67. [PubMed] [Google Scholar]
- 131.Pardo J, Bosque A, Brehm R, Wallich R, Naval J, et al. Apoptotic pathways are selectively activated by granzyme A and/or granzyme B in CTL-mediated target cell lysis. J. Cell Biol. 2004;167:457–68. doi: 10.1083/jcb.200406115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Beresford PJ, Kam CM, Powers JC, Lieberman J. Recombinant human granzyme A binds to two putative HLA-associated proteins and cleaves one of them. Proc. Natl. Acad. Sci. USA. 1997;94:9285–90. doi: 10.1073/pnas.94.17.9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, et al. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol. Cell. 2006;23:133–42. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 134.Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112:659–72. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 135.Fan Z, Beresford PJ, Zhang D, Lieberman J. HMG2 interacts with the nucleosome assembly protein SET and is a target of the cytotoxic T-lymphocyte protease granzyme A. Mol. Cell. Biol. 2002;22:2810–20. doi: 10.1128/MCB.22.8.2810-2820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fan Z, Beresford PJ, Zhang D, Xu Z, Novina CD, et al. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat. Immunol. 2003;4:145–53. doi: 10.1038/ni885. [DOI] [PubMed] [Google Scholar]
- 137.Zhu P, Zhang D, Chowdhury D, Martinvalet D, Keefe D, et al. Granzyme A, which causes single-stranded DNA damage, targets the double-strand break repair protein Ku 70. EMBO Rep. 2006;7:431–37. doi: 10.1038/sj.embor.7400622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Deleted in proof.
- 139.Zhang D, Beresford PJ, Greenberg AH, Lieberman J. Granzymes A and B directly cleave lamins and disrupt the nuclear lamina during granule-mediated cytolysis. Proc. Natl. Acad. Sci. USA. 2001;98:5746–51. doi: 10.1073/pnas.101329598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang D, Pasternack MS, Beresford PJ, Wagner L, Greenberg AH, Lieberman J. Induction of rapid histone degradation by the cytotoxic T lymphocyte protease granzyme A. J. Biol. Chem. 2001;276:3683–90. doi: 10.1074/jbc.M005390200. [DOI] [PubMed] [Google Scholar]
- 141.Lord SJ, Rajotte RV, Korbutt GS, Bleackley RC. Granzyme B: a natural born killer. Immunol. Rev. 2003;193:31–38. doi: 10.1034/j.1600-065x.2003.00044.x. [DOI] [PubMed] [Google Scholar]
- 142.Trapani JA, Sutton VR. Granzyme B: proapoptotic, antiviral and antitumor functions. Curr. Opin. Immunol. 2003;15:533–43. doi: 10.1016/s0952-7915(03)00107-9. [DOI] [PubMed] [Google Scholar]
- 143.Darmon AJ, Nicholson DW, Bleackley RC. Activation of the apoptotic protease CPP32 by cytotoxic T-cell-derived granzyme B. Nature. 1995;377:446–48. doi: 10.1038/377446a0. [DOI] [PubMed] [Google Scholar]
- 144.Adrain C, Murphy BM, Martin SJ. Molecular ordering of the caspase activation cascade initiated by the cytotoxic T lymphocyte/natural killer (CTL/NK) protease granzyme B. J. Biol. Chem. 2005;280:4663–73. doi: 10.1074/jbc.M410915200. [DOI] [PubMed] [Google Scholar]
- 145.Cullen SP, Adrain C, Luthi AU, Duriez PJ, Martin SJ. Human and murine granzyme B exhibit divergent substrate preferences. J. Cell Biol. 2007;176:435–44. doi: 10.1083/jcb.200612025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Barry M, Heibein JA, Pinkoski MJ, Lee SF, Moyer RW, et al. Granzyme B short-circuits the need for caspase 8 activity during granule-mediated cytotoxic T-lymphocyte killing by directly cleaving Bid. Mol. Cell. Biol. 2000;20:3781–94. doi: 10.1128/mcb.20.11.3781-3794.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Heibein JA, Goping IS, Barry M, Pinkoski MJ, Shore GC, et al. Granzyme B-mediated cytochrome c release is regulated by the bcl-2 family members bid and Bax. J. Exp. Med. 2000;192:1391–402. doi: 10.1084/jem.192.10.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sutton VR, Davis JE, Cancilla M, Johnstone RW, Ruefli AA, et al. Initiation of apoptosis by granzyme B requires direct cleavage of bid, but not direct granzyme B-mediated caspase activation. J. Exp. Med. 2000;192:1403–14. doi: 10.1084/jem.192.10.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Alimonti JB, Shi L, Baijal PK, Greenberg AH. Granzyme B induces BID-mediated cytochrome c release and mitochondrial permeability transition. J. Biol. Chem. 2001;276:6974–82. doi: 10.1074/jbc.M008444200. [DOI] [PubMed] [Google Scholar]
- 150.Thomas DA, Du C, Xu M, Wang X, Ley TJ. DFF45/ICAD can be directly processed by granzyme B during the induction of apoptosis. Immunity. 2000;12:621–32. doi: 10.1016/s1074-7613(00)80213-7. [DOI] [PubMed] [Google Scholar]
- 151.Sharif-Askari E, Alam A, Rheaume E, Beresford PJ, Scotto C, et al. Direct cleavage of the human DNA fragmentation factor-45 by granzyme B induces caspase-activated DNase release and DNA fragmentation. EMBO J. 2001;20:3101–13. doi: 10.1093/emboj/20.12.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 1997;272:17907–11. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 153.Heibein JA, Barry M, Motyka B, Bleackley RC. Granzyme B-induced loss of mitochondrial inner membrane potential (Δψm) and cytochrome c release are caspase independent. J. Immunol. 1999;163:4683–93. [PubMed] [Google Scholar]
- 154.MacDonald G, Shi L, Vande Velde C, Lieberman J, Greenberg AH. Mitochondria-dependent and -independent regulation of granzyme B-induced apoptosis. J. Exp. Med. 1999;189:131–44. doi: 10.1084/jem.189.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Thomas DA, Scorrano L, Putcha GV, Korsmeyer SJ, Ley TJ. Granzyme B can cause mitochondrial depolarization and cell death in the absence of BID, BAX, and BAK. Proc. Natl. Acad. Sci. USA. 2001;98:14985–90. doi: 10.1073/pnas.261581498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang GQ, Wieckowski E, Goldstein LA, Gastman BR, Rabinovitz A, et al. Resistance to granzyme B-mediated cytochrome c release in Bak-deficient cells. J. Exp. Med. 2001;194:1325–37. doi: 10.1084/jem.194.9.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.McIlroy D, Cartron PF, Tuffery P, Dudoit Y, Samri A, et al. A triple-mutated allele of granzyme B incapable of inducing apoptosis. Proc. Natl. Acad. Sci. USA. 2003;100:2562–67. doi: 10.1073/pnas.0437935100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sun J, Bird CH, Thia KY, Matthews AY, Trapani JA, Bird PI. Granzyme B encoded by the commonly occurring human RAH allele retains proapoptotic activity. J. Biol. Chem. 2004;279:16907–11. doi: 10.1074/jbc.M400563200. [DOI] [PubMed] [Google Scholar]
- 159.Fellows E, Gil-Parrado S, Jenne DE, Kurschus FC. Natural killer cell-derived human granzyme H induces an alternative, caspase-independent cell-death program. Blood. 2007;110:544–52. doi: 10.1182/blood-2006-10-051649. [DOI] [PubMed] [Google Scholar]
- 160.Johnson H, Scorrano L, Korsmeyer SJ, Ley TJ. Cell death induced by granzyme C. Blood. 2003;101:3093–101. doi: 10.1182/blood-2002-08-2485. [DOI] [PubMed] [Google Scholar]
- 161.Haddad P. The CTLA-1 gene: a member of the granzyme multigene family in human and murine cytotoxic T cells. Oxf. Surv. Eukaryot. Genes. 1991;7:143–60. [PubMed] [Google Scholar]
- 162.Shresta S, Goda P, Wesselschmidt R, Ley TJ. Residual cytotoxicity and granzyme K expression in granzyme A-deficient cytotoxic lymphocytes. J. Biol. Chem. 1997;272:20236–44. doi: 10.1074/jbc.272.32.20236. [DOI] [PubMed] [Google Scholar]
- 163.Ebnet K, Hausmann M, Lehmann-Grube F, Mullbacher A, Kopf M, et al. Granzyme A-deficient mice retain potent cell-mediated cytotoxicity. EMBO J. 1995;14:4230–39. doi: 10.1002/j.1460-2075.1995.tb00097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wilharm E, Parry MA, Friebel R, Tschesche H, Matschiner G, et al. Generation of catalytically active granzyme K from Escherichia coli inclusion bodies and identification of efficient granzyme K inhibitors in human plasma. J. Biol. Chem. 1999;274:27331–37. doi: 10.1074/jbc.274.38.27331. [DOI] [PubMed] [Google Scholar]
- 165.Shi L, Kam CM, Powers JC, Aebersold R, Greenberg AH. Purification of three cytotoxic lymphocyte granule serine proteases that induce apoptosis through distinct substrate and target cell interactions. J. Exp. Med. 1992;176:1521–29. doi: 10.1084/jem.176.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhao T, Zhang H, Guo Y, Zhang Q, Hua G, et al. Granzyme K cleaves the nucleosome assembly protein SET to induce single-stranded DNA nicks of target cells. Cell Death Differ. 2007;14:489–99. doi: 10.1038/sj.cdd.4402040. [DOI] [PubMed] [Google Scholar]
- 167.Zhao T, Zhang H, Guo Y, Fan Z. Granzyme K directly processes bid to release cytochrome c and endonuclease G leading to mitochondria-dependent cell death. J. Biol. Chem. 2007;282:12104–11. doi: 10.1074/jbc.M611006200. [DOI] [PubMed] [Google Scholar]
- 168.Smyth MJ, Sayers TJ, Wiltrout T, Powers JC, Trapani JA. Met-ase: cloning and distinct chromosomal location of a serine protease preferentially expressed in human natural killer cells. J. Immunol. 1993;151:6195–205. [PubMed] [Google Scholar]
- 169.Smyth MJ, O'Connor MD, Trapani JA, Kershaw MH, Brinkworth RI. A novel substrate-binding pocket interaction restricts the specificity of the human NK cell-specific serine protease, Met-ase-1. J. Immunol. 1996;156:4174–81. [PubMed] [Google Scholar]
- 170.Mahrus S, Kisiel W, Craik CS. Granzyme M is a regulatory protease that inactivates proteinase inhibitor 9, an endogenous inhibitor of granzyme B. J. Biol. Chem. 2004;279:54275–82. doi: 10.1074/jbc.M411482200. [DOI] [PubMed] [Google Scholar]
- 171.Rukamp BJ, Kam CM, Natarajan S, Bolton BW, Smyth MJ, et al. Subsite specificities of granzyme M: a study of inhibitors and newly synthesized thiobenzyl ester substrates. Arch. Biochem. Biophys. 2004;422:9–22. doi: 10.1016/j.abb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 172.Kelly JM, Waterhouse NJ, Cretney E, Browne KA, Ellis S, et al. Granzyme M mediates a novel form of perforin-dependent cell death. J. Biol. Chem. 2004;279:22236–42. doi: 10.1074/jbc.M401670200. [DOI] [PubMed] [Google Scholar]
- 173.Pao LI, Sumaria N, Kelly JM, van Dommelen S, Cretney E, et al. Functional analysis of granzyme M and its role in immunity to infection. J. Immunol. 2005;175:3235–43. doi: 10.4049/jimmunol.175.5.3235. [DOI] [PubMed] [Google Scholar]
- 174.Lu H, Hou Q, Zhao T, Zhang H, Zhang Q, et al. Granzyme M directly cleaves inhibitor of caspase-activated DNase (CAD) to unleash CAD leading to DNA fragmentation. J. Immunol. 2006;177:1171–78. doi: 10.4049/jimmunol.177.2.1171. [DOI] [PubMed] [Google Scholar]
- 175.Hua G, Zhang Q, Fan Z. Heat shock protein 75 (TRAP1) antagonizes reactive oxygen species generation and protects cells from granzyme M-mediated apoptosis. J. Biol. Chem. 2007;282:20553–60. doi: 10.1074/jbc.M703196200. [DOI] [PubMed] [Google Scholar]
- 176.Kim WJ, Kim H, Suk K, Lee WH. Macrophages express granzyme B in the lesion areas of atherosclerosis and rheumatoid arthritis. Immunol. Lett. 2007;111:57–65. doi: 10.1016/j.imlet.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 177.Hernandez-Pigeon H, Jean C, Charruyer A, Haure MJ, Baudouin C, et al. UVA induces granzyme B in human keratinocytes through MIF: implication in extracellular matrix remodeling. J. Biol. Chem. 2007;282:8157–64. doi: 10.1074/jbc.M607436200. [DOI] [PubMed] [Google Scholar]
- 178.Hernandez-Pigeon H, Jean C, Charruyer A, Haure MJ, Titeux M, et al. Human keratinocytes acquire cellular cytotoxicity under UV-B irradiation. Implication of granzyme B and perforin. J. Biol. Chem. 2006;281:13525–32. doi: 10.1074/jbc.M512694200. [DOI] [PubMed] [Google Scholar]
- 179.Bade B, Lohrmann J, ten Brinke A, Wolbink AM, Wolbink GJ, et al. Detection of soluble human granzyme K in vitro and in vivo. Eur. J. Immunol. 2005;35:2940–48. doi: 10.1002/eji.200526249. [DOI] [PubMed] [Google Scholar]
- 180.Spaeny-Dekking EH, Hanna WL, Wolbink AM, Wever PC, Kummer AJ, et al. Extracellular granzymes A and B in humans: detection of native species during CTL responses in vitro and in vivo. J. Immunol. 1998;160:3610–16. [PubMed] [Google Scholar]
- 181.Tak PP, Spaeny-Dekking L, Kraan MC, Breedveld FC, Froelich CJ, Hack CE. The levels of soluble granzyme A and B are elevated in plasma and synovial fluid of patients with rheumatoid arthritis (RA) Clin. Exp. Immunol. 1999;116:366–70. doi: 10.1046/j.1365-2249.1999.00881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bratke K, Bottcher B, Leeder K, Schmidt S, Kupper M, et al. Increase in granzyme B+ lymphocytes and soluble granzyme B in bronchoalveolar lavage of allergen challenged patients with atopic asthma. Clin. Exp. Immunol. 2004;136:542–48. doi: 10.1111/j.1365-2249.2004.02468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hodge S, Hodge G, Nairn J, Holmes M, Reynolds PN. Increased airway granzyme B and perforin in current and ex-smoking COPD subjects. COPD. 2006;3:179–87. doi: 10.1080/15412550600976868. [DOI] [PubMed] [Google Scholar]
- 184.Lauw FN, Simpson AJ, Hack CE, Prins JM, Wolbink AM, et al. Soluble granzymes are released during human endotoxemia and in patients with severe infection due to gram-negative bacteria. J. Infect. Dis. 2000;182:206–13. doi: 10.1086/315642. [DOI] [PubMed] [Google Scholar]
- 185.Rucevic M, Fast LD, Jay GD, Trespalcios FM, Sucov A, et al. Altered levels and molecular forms of granzyme K in plasma from septic patients. Shock. 2007;27:488–93. doi: 10.1097/01.shk.0000246905.24895.e5. [DOI] [PubMed] [Google Scholar]
- 186.Rowshani AT, Strik MC, Molenaar R, Yong SL, Wolbink AM, et al. The granzyme B inhibitor SERPINB9 (protease inhibitor 9) circulates in blood and increases on primary cytomegalovirus infection after renal transplantation. J. Infect. Dis. 2005;192:1908–11. doi: 10.1086/497606. [DOI] [PubMed] [Google Scholar]
- 187.Irmler M, Hertig S, MacDonald HR, Sadoul R, Becherer JD, et al. Granzyme A is an interleukin 1β-converting enzyme. J. Exp. Med. 1995;181:1917–22. doi: 10.1084/jem.181.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sower LE, Froelich CJ, Allegretto N, Rose PM, Hanna WD, Klimpel GR. Extra-cellular activities of human granzyme A. Monocyte activation by granzyme A vs alpha-thrombin. J. Immunol. 1996;156:2585–90. [PubMed] [Google Scholar]
- 189.Suidan HS, Bouvier J, Schaerer E, Stone SR, Monard D, Tschopp J. Granzyme A released upon stimulation of cytotoxic T lymphocytes activates the thrombin receptor on neuronal cells and astrocytes. Proc. Natl. Acad. Sci. USA. 1994;91:8112–16. doi: 10.1073/pnas.91.17.8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Suidan HS, Clemetson KJ, Brown-Luedi M, Niclou SP, Clemetson JM, et al. The serine protease granzyme A does not induce platelet aggregation but inhibits responses triggered by thrombin. Biochem. J. 1996;315:939–45. doi: 10.1042/bj3150939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Brunner G, Simon MM, Kramer MD. Activation of prourokinase by the human T cell-associated serine proteinase HuTSP-1. FEBS Lett. 1990;260:141–44. doi: 10.1016/0014-5793(90)80087-y. [DOI] [PubMed] [Google Scholar]
- 192.Simon MM, Kramer MD, Prester M, Gay S. Mouse T-cell associated serine proteinase 1 degrades collagen type IV: a structural basis for the migration of lymphocytes through vascular basement membranes. Immunology. 1991;73:117–19. [PMC free article] [PubMed] [Google Scholar]
- 193.Simon MM, Simon HG, Fruth U, Epplen J, Muller-Hermelink HK, Kramer MD. Cloned cytolytic T-effector cells and their malignant variants produce an extracellular matrix degrading trypsin-like serine proteinase. Immunology. 1987;60:219–30. [PMC free article] [PubMed] [Google Scholar]
- 194.Vettel U, Brunner G, Bar-Shavit R, Vlodavsky I, Kramer MD. Charge-dependent binding of granzyme A (MTSP-1) to basement membranes. Eur. J. Immunol. 1993;23:279–82. doi: 10.1002/eji.1830230144. [DOI] [PubMed] [Google Scholar]
- 195.Buzza MS, Zamurs L, Sun J, Bird CH, Smith AI, et al. Extracellular matrix remodeling by human granzyme B via cleavage of vitronectin, fibronectin, and laminin. J. Biol. Chem. 2005;280:23549–58. doi: 10.1074/jbc.M412001200. [DOI] [PubMed] [Google Scholar]
- 196.Choy JC, Hung VH, Hunter AL, Cheung PK, Motyka B, et al. Granzyme B induces smooth muscle cell apoptosis in the absence of perforin: involvement of extracellular matrix degradation. Arterioscler. Thromb. Vasc. Biol. 2004;24:2245–50. doi: 10.1161/01.ATV.0000147162.51930.b7. [DOI] [PubMed] [Google Scholar]
- 197.Froelich CJ, Zhang X, Turbov J, Hudig D, Winkler U, Hanna WL. Human granzyme B degrades aggrecan proteoglycan in matrix synthesized by chondrocytes. J. Immunol. 1993;151:7161–71. [PubMed] [Google Scholar]
- 198.Ronday HK, van der Laan WH, Tak PP, de Roos JA, Bank RA, et al. Human granzyme B mediates cartilage proteoglycan degradation and is expressed at the invasive front of the synovium in rheumatoid arthritis. Rheumatology (Oxford) 2001;40:55–61. doi: 10.1093/rheumatology/40.1.55. [DOI] [PubMed] [Google Scholar]
- 199.Gahring L, Carlson NG, Meyer EL, Rogers SW. Granzyme B proteolysis of a neuronal glutamate receptor generates an autoantigen and is modulated by glycosylation. J. Immunol. 2001;166:1433–38. doi: 10.4049/jimmunol.166.3.1433. [DOI] [PubMed] [Google Scholar]
- 200.Ganor Y, Teichberg VI, Levite M. TCR activation eliminates glutamate receptor GluR3 from the cell surface of normal human T cells, via an autocrine/paracrine granzyme B-mediated proteolytic cleavage. J. Immunol. 2007;178:683–92. doi: 10.4049/jimmunol.178.2.683. [DOI] [PubMed] [Google Scholar]
- 201.Wang T, Allie R, Conant K, Haughey N, Turchan-Chelowo J, et al. Granzyme B mediates neurotoxicity through a G-protein-coupled receptor. FASEB J. 2006;20:1209–11. doi: 10.1096/fj.05-5022fje. [DOI] [PubMed] [Google Scholar]
- 202.Loeb CR, Harris JL, Craik CS. Granzyme B proteolyzes receptors important to proliferation and survival, tipping the balance toward apoptosis. J. Biol. Chem. 2006;281:28326–35. doi: 10.1074/jbc.M604544200. [DOI] [PubMed] [Google Scholar]
- 203.Mullbacher A, Ebnet K, Blanden RV, Hla RT, Stehle T, et al. Granzyme A is critical for recovery of mice from infection with the natural cytopathic viral pathogen, ectromelia. Proc. Natl. Acad. Sci. USA. 1996;93:5783–87. doi: 10.1073/pnas.93.12.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Graubert TA, Russell JH, Ley TJ. The role of granzyme B in murine models of acute graft-vs-host disease and graft rejection. Blood. 1996;87:1232–37. [PubMed] [Google Scholar]
- 205.Revell PA, Grossman WJ, Thomas DA, Cao X, Behl R, et al. Granzyme B and the downstream granzymes C and/or F are important for cytotoxic lymphocyte functions. J. Immunol. 2005;174:2124–31. doi: 10.4049/jimmunol.174.4.2124. [DOI] [PubMed] [Google Scholar]
- 206.Davis JE, Smyth MJ, Trapani JA. Granzyme A and B-deficient killer lymphocytes are defective in eliciting DNA fragmentation but retain potent in vivo antitumor capacity. Eur. J. Immunol. 2001;31:39–47. doi: 10.1002/1521-4141(200101)31:1<39::aid-immu39>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 207.Pardo J, Balkow S, Anel A, Simon MM. Granzymes are essential for natural killer cell-mediated and perf-facilitated tumor control. Eur. J. Immunol. 2002;32:2881–87. doi: 10.1002/1521-4141(2002010)32:10<2881::AID-IMMU2881>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 208.Simon MM, Hausmann M, Tran T, Ebnet K, Tschopp J, et al. In vitro- and ex vivo-derived cytolytic leukocytes from granzyme A × B double knockout mice are defective in granule-mediated apoptosis but not lysis of target cells. J. Exp. Med. 1997;186:1781–86. doi: 10.1084/jem.186.10.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Krenacs L, Smyth MJ, Bagdi E, Krenacs T, Kopper L, et al. The serine protease granzyme M is preferentially expressed in NK-cell, γδ T-cell, and intestinal T-cell lymphomas: evidence of origin from lymphocytes involved in innate immunity. Blood. 2003;101:3590–93. doi: 10.1182/blood-2002-09-2908. [DOI] [PubMed] [Google Scholar]
- 210.Waterhouse NJ, Sutton VR, Sedelies KA, Ciccone A, Jenkins M, et al. Cytotoxic T lymphocyte-induced killing in the absence of granzymes A and B is unique and distinct from both apoptosis and perforin-dependent lysis. J. Cell Biol. 2006;173:133–44. doi: 10.1083/jcb.200510072. [DOI] [PMC free article] [PubMed] [Google Scholar]