Abstract
Radiation-induced free radical damage in solid-state DNA arises from direct-type damage mechanisms. In the present study, free radical yields in film and lyophilized Na DNA model systems are compared. The free radical yields in lyophilized samples are significantly greater than those in films. Since DNA conformation cannot account for the differences in free radical yields between different DNA preparations, it is proposed that the intermolecular spacing of DNA is a critical variable. The differences in the hydration dependence of free radical yields between the film and lyophilized DNA model systems are consistent with this thesis. The relatively large free radical yields observed in lyophilized DNA emphasize the fact that DNA is an extremely effective electron and hole scavenger, more so than previously thought.
INTRODUCTION
Direct-type damage in DNA exposed to low-LET radiation involves electron addition and abstraction reactions on the DNA molecule. DNA and DNA constituents, frozen in the solid state, provide excellent model systems in which to investigate direct-type processes (for reviews, see refs. 1–5). Several variables influence the extent of direct-type damage experienced by DNA, including DNA hydration (6–14), conformation (6, 8, 9, 11, 14) and packing (10–12, 15, 16). The latter, DNA packing, reflects the aggregation of DNA molecules with each other, as well as the molecular order of the surrounding DNA hydration waters. DNA hydration waters act as proton donors and proton acceptors. Proton transfer reactions are thought to stabilize free radical damage on the DNA molecule (10, 15–23). Hence the hydrogen-bonding structure of the hydration waters may influence the extent of free radical trapping on DNA.
The physical separation of DNA molecules relative to one another is also thought to influence radical stability (16). Closely aggregated DNA promotes the scavenging of electrons and holes onto separate DNA molecules, while sparsely associated DNA tends to trap electrons and holes onto isolated DNA molecules. In the former, combination is relatively impeded, resulting in more extensive free radical trapping.
In a previous study of DNA films (16), the scatter in free radical yields in variably hydrated Li DNA films was attributed to variability in DNA packing. In contrast, Na DNA afforded reproducible measurements of free radical yields, allowing hydration-dependent trends in free radical yields to be discerned. The hydration-dependent changes in Na DNA were attributed primarily to changes in DNA packing. However, the influence of conformational changes at lower DNA hydrations (less than 15 waters per nucleotide) could not be ruled out.
In the present study, the relative differences in free radical yields between different physical forms of DNA are examined. Large differences in free radical yields between different DNA preparations, hydrated to the same degree, are observed. This disparity is ascribed to differences in the DNA packing order.
The maximal observed free radical yields are significantly greater (1.5 to 2 times) than those reported previously (7, 11, 12, 24). These large free radical yields suggest that DNA, particularly well-ordered DNA, is a significantly better free radical trapping medium than previously thought.
MATERIALS AND METHODS
Sample Preparation
The Na DNA samples were prepared from calf thymus Na DNA (Sigma Chemical Co.). All samples were placed into Suprasil quartz tubes; the tubes had an outer diameter of 1.5 mm and 0.1-mm-thick walls.
DNA films were prepared as described previously (16). The DNA films were translucent but became opaque if stretched. They were not easily compressed; when pressed into a pellet, dry films (Γ < 10) were not highly cohesive. Upon hydration, however, the pellet became more opaque and remained intact. These physical properties suggest that dry films possess a regular DNA packing order, constituting a somewhat rigid matrix. Upon hydration of the DNA, this packing order becomes less defined.
With the exception of the vacuum-dried samples (~2.5 waters per nucleotide), all DNA film samples were dehydrated by equilibration against a saturated solution of NaOH (5% relative humidity, corresponding to ~5 waters per nucleotide) and were subsequently brought to a steady-state hydration under various relative humidities. Some of the film samples were brought to a high hydration (>120 waters per nucleotide) state prior to dehydration in the relative humidity chambers.
In the hydration regime of 11–13 waters per nucleotide, DNA films were equilibrated at three different temperatures: 4, 20 and 37°C. As suggested below, we believe that the temperature at which the films are equilibrated has an impact on the DNA packing order.
Solid-state DNA was also prepared by lyophilizing a flash-frozen DNA solution. This form is termed “lyophilized” DNA in the remainder of this paper; this technique of lyophilization is, however, unique by virtue of the method used for rapid freezing.
To prepare the lyophilized DNA, a Na DNA solution (~150 ml solution with A260 ~20 and 0.6 M NaCl) was ethanol-precipitated by contact with an ice bath and subsequently dialyzed against an 80:20 (v/v) ethanol: water, 0.15 M NaCl solution to control the excess salt content (25). The DNA precipitate was redissolved in ~250 ml of purified water and filtered. The DNA solution (A260 > 15 OD units) was then sprayed, as an atomized mist, into a liquid nitrogen (77 K) bath. The slurry of microscopic beads of frozen DNA and liquid nitrogen was then transferred from a Dewar to an 800-ml beaker, inside of which the aggregation of the microscopic DNA beads formed a frozen cylindrical “cake”. This DNA sample was vacuum-dried while held at −10 to −5°C. Although no attempt was made to measure the molecular weight distribution, we presume that the DNA is sheared into smaller sizes by this process.
The lyophilized DNA appeared as a fine meshwork of intermingled DNA fibers. The texture of the outer edges of the sample was crusty, possibly as a result of the physical compaction of the frozen cake as it sublimed; these areas were not used. Film-like regions were also apparent (also near the edges); these areas were also avoided. Some microscopic film-like regions may have been present, but undetected.
The lyophilized DNA was fluffy and compressible, akin to lint. When packed into quartz tubes, the DNA appeared as opaque clumps, with a somewhat glassy appearance. These properties, along with the sample opacity, suggest that the lyophilized DNA is less rigidly ordered than the film DNA. The process of atomization and rapid freezing effectively partitions the DNA molecules, preventing the formation of an extended matrix.
The lyophilized DNA samples were equilibrated against a saturated solution of NaOH and subsequently hydrated to a steady state under various relative humidities. The low-hydration lyophilized DNA samples (~2.5 waters per nucleotide) were not equilibrated against NaOH.
The degree of DNA hydration was calculated (12, 16) from sample weights measured on a Cahn C-30 Microbalance. The extent of hydration is expressed in terms of moles of water per mole of nucleotide and is represented by Γ. In the calculation of Γ, in both films and lyophilized samples, two assumptions were made: (1) 2.5 mol of water per nucleotide could not be removed by vacuum drying (8, 9), and (2) roughly 6% of the dry weight of DNA was attributable to excess salt (25).
Sample Cooling and Irradiation
Samples were cooled to 4 K in a Janis cryostat (26) and X-irradiated using a Varian/Eimac OEG-76H X-ray tube as described earlier (16). The X-ray tube was run at a voltage of 70 keV and a current of 20 mA unless otherwise noted. Under these conditions, the samples are irradiated and observed under a vacuum.
The calculation of the dose rate was also described earlier (16). The dose rate of the X-ray tube, for a sample loaded into a Suprasil quartz tube and situated in the Janis Dewar, is ~24.0 kGy/h when the tube is run at 70 keV and 20 mA. When the tube is run at 50 keV and 20 mA, the dose rate is ~14.5 kGy/h. The self-attenuation of the X-ray beam by the quartz tube is incorporated into these dose rates, since both the alanine dosimeter and the DNA samples were examined in quartz. Self-attenuation within DNA is greater than that within alanine, particularly at lower (below 50 keV) energies where the phosphate group of DNA results in slightly higher attenuation. This small difference was ignored. Theoretically, the greater attenuation of DNA relative to that of alanine could cause the doses administered to DNA to be lower, resulting in greater free radical yields than measured. However, the small breadth of the samples (≤1.3 mm) and the filtering of low-energy X rays by aluminum and beryllium windows (as well as quartz) make self-attenuation insignificant.
EPR Measurements
EPR measurements were taken with a Varian Q-band spectrometer, which has the waveguide and sample cavity incorporated into a Janis cryostat (26). The calculation of free radical yields was as described previously (16). In the calculation of free radical yield, the absorbing mass is considered to be the total sample mass (DNA plus all water).
RESULTS AND DISCUSSION
DNA Hydration
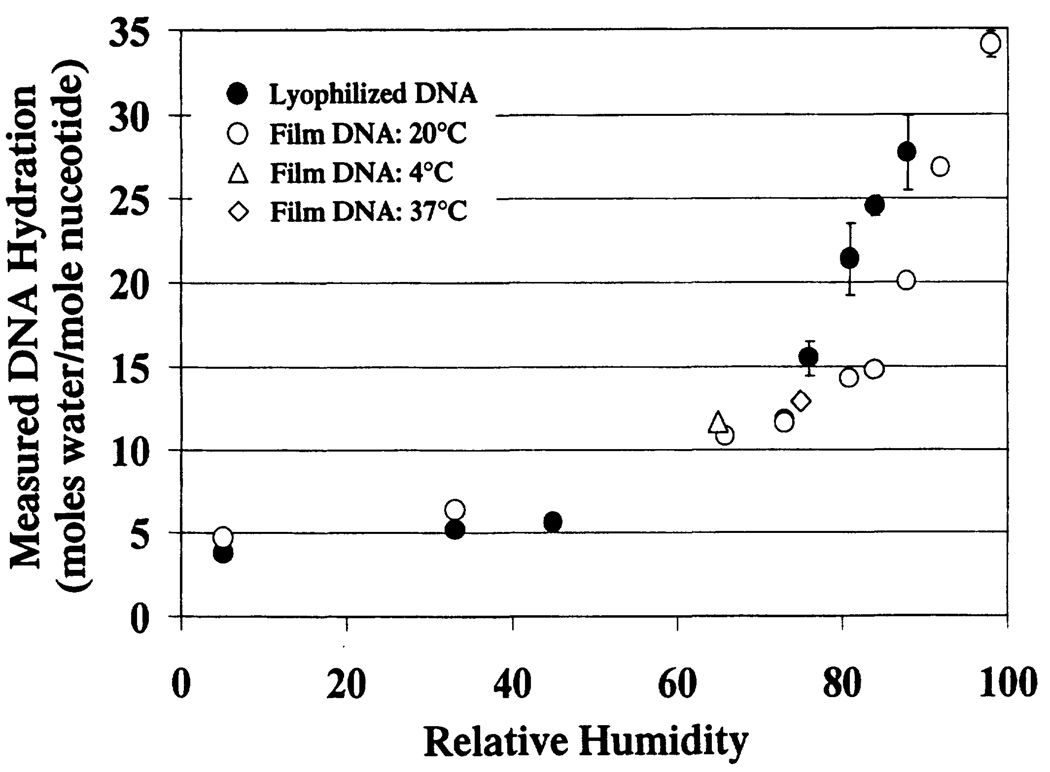
Figure 1 shows the observed relationship between relative humidity and DNA hydration in the film and lyophilized DNA samples. The dependence of DNA hydration on relative humidity is similar in both systems for 0% < relative humidity < 73%. At 73% relative humidity, the DNA hydration in both the film and lyophilized DNA systems is 12 to 13 waters per nucleotide. Above 73% relative humidity they differ, with the lyophilized DNA being more readily hydrated than the film DNA. Proposed differences in the packing order between these two model systems (see the Materials and Methods and below) can account for the observed differences in DNA hydration. The less-constrained packing order of the lyophilized DNA is considered more accessible to DNA hydration waters and is therefore more readily hydrated.
FIG. 1.
DNA hydration as a function of relative humidity for various DNA model systems. The error bars represent ± 1 SD from the average of a set of samples. In those points without error bars, errors were negligible (less than the breadth of the data point).
The scatter in DNA hydration at higher relative humidities (>75%) is greater in the lyophilized DNA than in the film DNA. This scatter may reflect differences in the amount of microscopic film-like DNA present in the lyophilized samples.
Free Radical Yields in DNA
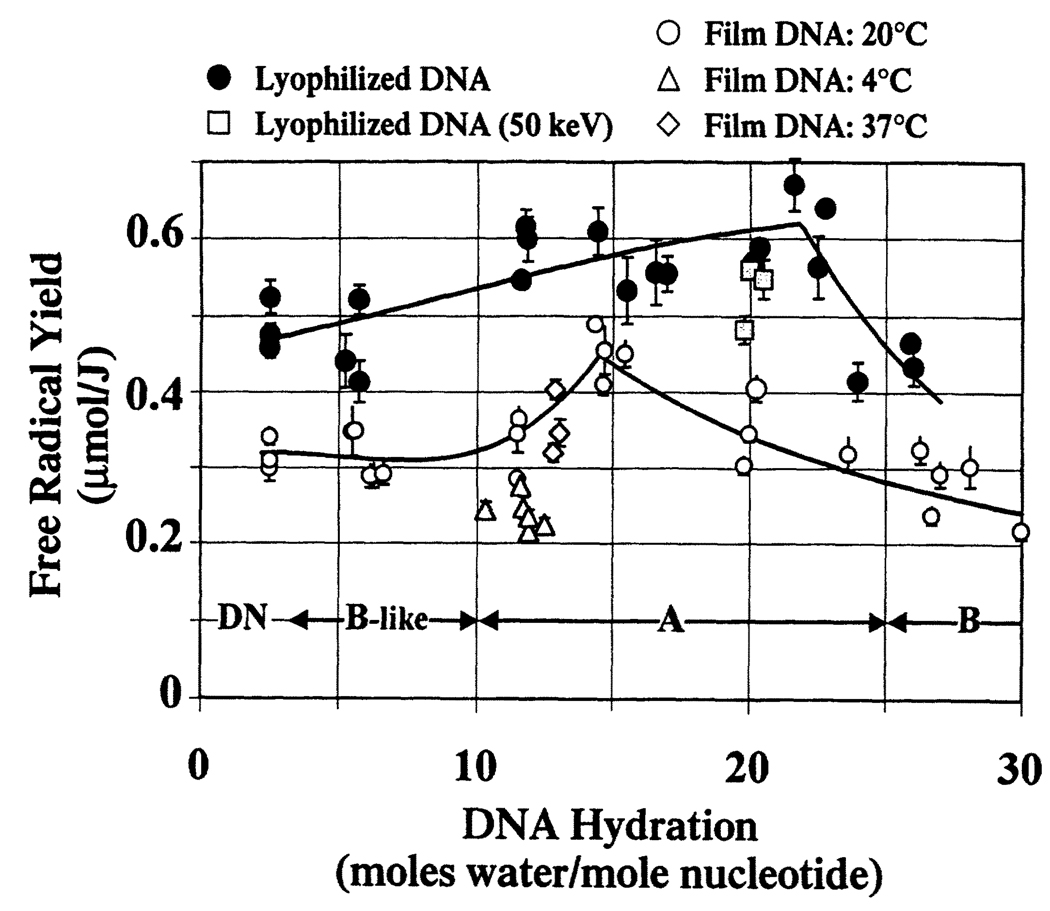
Figure 2 compares the hydration dependence of free radical yields in lyophilized DNA with that in DNA films. Many of the data points for DNA equilibrated at room temperature (~20°C) were published earlier (16) and are shown here for comparison.
FIG. 2.
Free radical yields as a function of DNA hydration for various DNA model systems. The solid lines reflect least-squares fits of the DNA film free radical yields (data for 20°C only) and lyophilized DNA free radical yields (not including 50 keV irradiated DNA free radical yields). For both sets of data, a power regression is used to model the data at and above the peak free radical yield. A third-order polynomial fit is used to model the data at and below the peak free radical yield. These two fits converge at the maximal free radical yield. Each data point represents one dose-response curve determined by four or five data points. The error bars are ±1 SD of the slope of the dose-response curve.
Effects of DNA Conformation
DNA conformation is known to vary with changes in DNA hydration (27–29). Vacuum-dried DNA [~2.5 waters per nucleotide (8, 9)] is denatured; slightly wetter DNA (~3 < Γ < ~10) is in a B-like conformation; more fully hydrated DNA (~10 < Γ < ~25) tends to be in the A conformation; at greater hydrations, DNA is in the B conformation. The hydration ranges in which the different DNA conformational forms are stable are delineated by the double-headed arrows at the bottom of Fig. 2.
For either the film or the lyophilized DNA, one might attribute the hydration dependence of free radical yields, in the range of 2 > Γ > 30, to changes in DNA conformation; conceivably, these changes could be gradual, giving rise to various blends of conformational states. This explanation cannot, however, readily account for the two different curve shapes for the two DNA sample types. Furthermore, at a given DNA hydration, where the conformation of the film DNA is expected to be the same as that of the lyophilized DNA, the free radical yields are quite different for the two DNA forms. Although DNA packing and conformation are not strictly independent variables (29), it is implausible that packing differences in our sample preparations significantly affect conformation. It is therefore necessary to consider how DNA packing could influence free radical yields.
Effects of Packing
It is evident from Fig. 2 that sample preparation influences free radical yields. While the free radical yields in DNA films equilibrated at 37°C conform to the hydration-dependent trend of free radical yields in DNA films equilibrated at 20°C, the free radical yields in films equilibrated at 4°C are significantly (on average 30%) lower. We propose that this discrepancy is primarily due to differences in packing. Upon dehydration of DNA, the volume of the sample contracts, decreasing the average interhelical spacing. At lower temperatures, we suggest that contraction of DNA is likely to be less pronounced. Consequently, the free radical yields of DNA films equilibrated at 4°C are expected to be lower by virtue of the greater interhelical spacing (16).
Comparing the data on free radical yield in film DNA to that in lyophilized DNA (Fig. 2), it is apparent that, throughout the 2.5 < Γ < 30 regime, both the hydration dependence and the magnitudes of the free radical yields differ. These differences are attributed to packing. The slow evaporation of a concentrated DNA solution into a film is believed to result in an extended, interconnecting network of DNA strands. Formation of the extended network is viewed as being promoted by the substrate (Teflon surface) on which the film is grown. The substrate, coupled with slow evaporation, reduces the extent of DNA self-aggregation. In contrast, rapid freezing promotes self-aggregation, effectively freezing out the DNA in a dilute aqueous state (which may also, to some extent, cause intermolecular aggregation). Thus rapid freezing generates relatively isolated DNA molecules. Lyophilization of this DNA will result in tight microscopic aggregates; these aggregates are themselves loosely interconnected. Thus, at a local level, the interhelical distance is constrained to larger values in the films than within the microscopic aggregates of lyophilized DNA. The physical characteristics of these two DNA forms (see the Materials and Methods) support this model. Sample history and packing have also been implicated in influencing yields of strand breaks (30).
We postulate that tight compared to loose DNA packing affects yields in two ways. One, postulated to be the dominant factor, is that smaller interhelical spacing increases free radical yields by inhibiting combination reactions (16). The other, possibly less important, is a lattice effect; that is, different hydrogen-bonding networks, in discrete DNA packing states, will result in different probabilities of proton transfer reactions (15). These proton transfer reactions return the sites of ionization to their original charge state, thereby inhibiting radical combination reactions (18, 19, 31). The degree to which the DNA’s hydration layer influences radical trapping on DNA by participating in proton transfer reactions is not known.
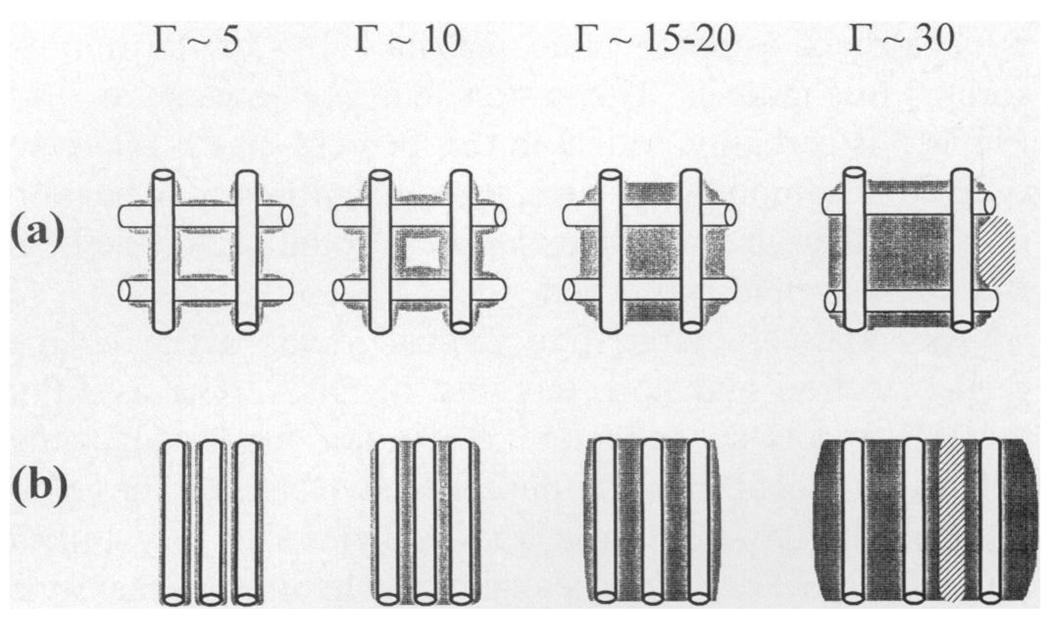
Figure 3a and b depicts, using highly simplified schematics, the proposed hydration-dependent changes in packing order in lyophilized and film DNA. At low hydrations (Γ < 6), the tight aggregates of lyophilized DNA (Fig. 3b) are loosely interconnected, while within the aggregate, strong DNA–DNA interactions enhance free radical trapping. As the DNA hydration is increased to Γ > ~20–25, the microscopic aggregates appear to coalesce, forming a gel-like aggregate. The film DNA (Fig. 3a), at low hydrations, is constrained so that the average interhelical spacing is larger than that in lyophilized DNA. The free radical yields are therefore lower due to the two mechanisms proposed above. Additional waters, up to Γ ~15, fill in the DNA solvation shell, but have little effect on interhelical spacing. The added water in this regime may enhance trapping by bridging DNA molecules with a more extensive hydrogen bond network. With further increases in hydration (15 > Γ > ~22), the film expands but remains relatively intact; the increase in interhelical spacing results in decreased free radical yields (16). Above Γ ~22, the DNA is fully solvated and the film begins to disperse; added water forms ice and the free radical yield continues to drop.
FIG. 3.
Models of packing in lyophilized DNA and film DNA. The cylinders represent DNA, the gray area is DNA hydration, the hatched area is ice, and the white area is vacuum. (a) Film DNA is proposed to consist of an extended interconnecting matrix that inhibits the formation of tightly clustered DNA. At a hydration of ~20 to 25 waters per nucleotide, the DNA film begins to expand (42–44), suggesting that the intermolecular spaces have become saturated with water and the addition of water causes the physical separation of DNA (16). (b) Depicted is the tightly packed DNA within an aggregate, formed by rapid freezing and lyophilization. In the dry state (Γ ~5), there are extensive DNA–DNA interactions. Upon hydration to Γ ~15, the water fills in the primary hydration layer. Additional hydration, Γ ~20, fills the secondary hydration layer and begins to increase the DNA–DNA separation. At greater hydrations, Γ ~30, the DNA separation increases and bulk water begins to form. Crystalline ice grows out of the bulk water phase.
As in film DNA, the incorporation of water in lyophilized DNA forms proton transfer networks, which stabilize trapped radicals. The hydration-dependent increase in free radical yields (from Γ ~5 to Γ ~15) is less pronounced in the lyophilized DNA compared to the film DNA (~45% compared to ~20%). One explanation for this difference is that, in lyophilized DNA, the efficiency of radical trapping is not as heavily dependent upon lattice waters, since the radicals are effectively shielded by the more extensive radical sequestering onto separate DNA molecules.
In both film and lyophilized DNA, damage transfer occurs from the tightly bound hydration waters to the DNA molecule (6, 7, 11, 12, 16, 32). The more extensive hydration of lyophilized DNA, at relative humidities greater than 75% (discussed above), suggests that the hydration waters are more widely (i.e. more DNA–water contacts) and more uniformly distributed throughout the DNA. At any given DNA hydration (12 > Γ > ~22), this arrangement of water would allow for more extensive damage transfer from the DNA hydration layer to DNA, and thus it may also contribute to the higher free radical yields in lyophilized DNA.
At higher DNA hydrations, a hydration-dependent drop in free radical yields is observed in both film and lyophilized DNA. At these high DNA hydrations, additional waters do not contribute to damage transfer, largely accounting for the observed drop in free radical yields. Freeze-back (7) of some hydration waters may also contribute to this drop. Freeze-back is a process whereby, once ice crystals begin to grow, the ice phase recruits water away from the DNA solvation layer. In our earlier study of film DNA (16), measurements of hydrogen-atom trapping indicated that the onset of ice formation was in the hydration region of 20 > Γ > 24. Given that the drop in free radical yield in films begins at Γ ~15, changes in packing presumably also play an important role. The present observation in lyophilized DNA, that free radical yields do not drop until Γ exceeds ~22, provides additional evidence that DNA packing influences free radical yields. With the more extensive incorporation of water in the lyophilized DNA (compared to the film DNA), the emergence of crystalline ice (and accompanying freeze-back) would not occur until higher hydrations.
The findings presented here and previously (11, 12, 16) provide evidence that packing plays a critical role in determining free radical yields in DNA. While the proposed mechanisms are consistent with the known properties of DNA films and lyophilized solutions, they are speculative since the films and powders are difficult to characterize. We have recently begun to address this difficulty by measuring the free radical yields in crystalline DNA oligomers. Preliminary results on a crystalline decamer support the hypothesis that, when DNA is tightly packed, the free radical yield is relatively large.
Single crystals of d(CCAACGTTGG) X-irradiated and measured at 4 K give a free radical yield that is of the order of 0.7 to 0.8 µmol/J (unpublished data). A more precise determination of the free radical yield is not available because the crystalline samples are exceptionally small, 10 to 100 µg. From the crystal structure, it is known that the decamers are packed so as to form a parallel array of B-DNA helices (33, 34). These decamer helices come into close contact, forming a matrix of tightly packed DNA. Because base stacking is continuous, the only feature with any significance that distinguishes the crystalline DNA from a high-molecular-weight polymer is that there is one phosphate missing for every 10 bases.
In comparing crystalline free radical yields to Na DNA free radical yields, it is informative to quote the number of radicals per nucleotide. Since the crystalline decamer lacks a 3′ terminal OPO3− group and possesses a Mg+2 counterion (one counterion per two nucleotides), the average nucleotide molecular weight (~310 g/mol) is lower than that in Na DNA (~331 g/mol). The hydration of crystalline d(CCAACGTTGG) is estimated to be roughly 12 waters per nucleotide. At this hydration, the crystalline free radical yield of ~0.36 to 0.40 radicals/nucleotide MGy−1 is ~15–25% greater than that of lyophilized Na DNA free radical yield of ~0.32 radicals/nucleotide MGy−1. The value for Na DNA is calculated from a free radical yield of 0.55 µmol/J, where the mass of excess salt is assumed to be 6% of the dry mass.
Thus we have a data point for B-DNA, with extremely well-characterized packing, at a hydration of ~12 waters per nucleotide. The free radical yield is higher than that of lyophilized DNA, which in turn is twice as high as that observed in films formed at 4°C. The greater free radical yield correlates with smaller interhelical spacing.
Absolute Values of Free Radical Yields
The irradiation conditions in the present study differ from those in previous work by Bernhard and coworkers in that the X-ray tube was run at a voltage of 70 keV rather than 50 keV. The free radical yields for samples irradiated at 50 keV (Fig. 2) showed only a minimal decrease (on average, <7 %), thus suggesting that the influence of X-ray tube voltage on free radical yields is not significant. (This slight decrease may be due to the greater self-attenuation of DNA at lower photon energies, but it is within experimental error—see the Materials and Methods.)
The errors in the absolute free radical yields are estimated to be 25–50%. These errors arise primarily from errors in measuring free radical concentration and to a much lesser extent from errors in dosimetry (16). The errors in the relative free radical yields are significantly less (estimated at 10–15%) because the absolute error of the ruby EPR standard is constant throughout.
The free radical yields reported here are 1.5 to 2 times greater than those reported previously (7, 11, 12, 24). It has been assumed that the maximal free radical yield in DNA (that obtained if all ionizations are trapped as radicals) is roughly equivalent to the initial free radical yield in water (19, 24) since the stopping power of DNA is similar to that of water (35, 36). The initial free radical yield in water is estimated to be 1.18 µmol/J (37). The data presented here suggest that DNA, at certain hydration and packing states, is capable of trapping more than 50–70% of the initially formed free radicals. The proton shuttling network of DNA (18, 19) is certainly critical in this process. In the relatively ordered DNA model systems, proton shuttling may be more effective than in amorphous DNA, accounting for the large free radical yields observed.
The relatively large free radical yields argue against facile electron and hole mobility in DNA. The assertion that DNA acts a conductor (38) is not supported by the extensive radical trapping observed in DNA. Furthermore, the relatively large free radical yield in crystalline d(CCAACGTTGG) argues against the hypothesis that well-stacked, well-ordered DNA promotes long-range electron migration (39–41). The competing proton transfer reactions in DNA would hinder any migration that might occur.
CONCLUSIONS
Free radical yields were measured in film DNA and in lyophilized DNA. Differences in both the magnitude of free radical yields and the hydration dependence of free radical yields were attributed to differences in DNA packing characteristics. It was argued that the close association of DNA molecules in the lyophilized DNA tends to promote radical trapping. The relatively large magnitude of the free radical yields indicates that DNA can act as a highly effective radical trapping medium. In summary, DNA packing is a critical variable in determining free radical yields in DNA, significantly more so than duplex conformation.
ACKNOWLEDGMENTS
We thank Kermit R. Mercer for his invaluable technical assistance and Michael G. Debije for his thoughtful discussions. The investigation was supported by PHS Grant 2-R37-CA32546, awarded by the National Cancer Institute, DHHS. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
REFERENCES
- 1.Bernhard WA. Solid state radiation chemistry of DNA: The bases. Adv. Radiat. Biol. 1981;9:199–280. [Google Scholar]
- 2.Close DM. Radical ions and their reactions in DNA constituents. ESR/ENDOR studies of radiation damage in the solid state. Radiat. Res. 1993;135:1–15. [PubMed] [Google Scholar]
- 3.Sevilla M, Becker D. Radiation damage in DNA. Electron Spin Resonance (A Specialist Report) 1994;14:130–165. [Google Scholar]
- 4.Becker D, Sevilla MD. The chemical consequences of radiation damage to DNA. Adv. Radiat. Biol. 1993;17:121–180. [Google Scholar]
- 5.O’Neill P, Fielden EM. Primary free radical processes in DNA. Adv. Radiat. Biol. 1993;17:53–120. [Google Scholar]
- 6.Wang W, Yan M, Becker D, Sevilla MD. The influence of hydration on the absolute yields of primary free radicals in gamma-irradiated DNA at 77 K. II. Individual radical yields. Radiat. Res. 1994;137:2–10. [PubMed] [Google Scholar]
- 7.Wang W, Becker D, Sevilla MD. The influence of hydration on absolute yields of primary ionic free radicals in γ-irradiated DNA at 77 K. I. Total radical yields. Radiat. Res. 1993;135:146–154. [PubMed] [Google Scholar]
- 8.Swarts SG, Becker D, Sevilla M, Wheeler KT. Radiation-induced DNA damage as a function of hydration. II. Base damage from electron loss centers. Radiat. Res. 1996;145:304–314. [PubMed] [Google Scholar]
- 9.Swarts SG, Sevilla MD, Becker D, Tokar CJ, Wheeler KT. Radiation-induced DNA damage as a function of hydration. I. Release of unaltered bases. Radiat. Res. 1992;129:333–344. [PubMed] [Google Scholar]
- 10.Mroczka NE, Mercer KR, Bernhard WA. The effects of lattice water on free radical yields in X-irradiated crystalline pyrimidines and purines: A low-temperature electron paramagnetic resonance investigation. Radiat. Res. 1997;147:560–568. [PubMed] [Google Scholar]
- 11.Mroczka N, Bernhard WA. Electron paramagnetic resonance investigation of X-irradiated poly (U), poly (A) and poly (A):poly (U). Influence of hydration, packing and conformation on radical yield at 4 K. Radiat. Res. 1995;144:251–257. [PubMed] [Google Scholar]
- 12.Mroczka N, Bernhard WA. Hydration effects on free radical yields in DNA X-irradiated at 4 K. Radiat. Res. 1993;135:155–159. [PubMed] [Google Scholar]
- 13.Hüttermann J, Röhrig M, Köhnlein W. Free radicals from irradiated lyophilized DNA: Influence of water of hydration. Int. J. Radiat. Biol. 1992;61:299–313. doi: 10.1080/09553009214550981. [DOI] [PubMed] [Google Scholar]
- 14.Gregoli S, Olast M, Bertinchamps A. Radiolytic pathways in γ-irradiated DNA: Influence of chemical and conformational factors. Radiat. Res. 1982;89:238–254. [PubMed] [Google Scholar]
- 15.Bernhard WA, Barnes J, Mercer KR, Mroczka N. The influence of packing on free radical yields in crystalline nucleic acids: The pyrimidine bases. Radiat. Res. 1994;140:199–214. [PubMed] [Google Scholar]
- 16.Milano MT, Bernhard WA. The effect of packing and conformation on free radical yields in films of variably hydrated DNA. Radiat. Res. 1998;151:39–49. [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Sevilla MD. Protonation of nucleobase anions in gamma-irradiated DNA and model systems. Which DNA base is the ultimate sink for the electron? Radiat. Res. 1994;138:9–17. [PubMed] [Google Scholar]
- 18.Steenken S. Electron transfer in DNA? Competition by ultrafast-fast proton transfer? Biol. Chem. 1997;378:1293–1297. [PubMed] [Google Scholar]
- 19.Bernhard WA, Mroczka N, Barnes J. Combination is the dominant free radical process initiated in DNA by ionizing radiation: An overview based on solid-state EPR studies. Int. J. Radiat. Biol. 1994;66:491–497. doi: 10.1080/09553009414551511. [DOI] [PubMed] [Google Scholar]
- 20.Steenken S. Electron-transfer induced acidity/basicity and reactivity changes of purine and pyrimidine bases. Consequences of redox processes for DNA base pairs. Free Radic. Res. Commun. 1992;6:349–379. doi: 10.3109/10715769209049187. [DOI] [PubMed] [Google Scholar]
- 21.Barnes J, Bernhard WA. Irreversible protonation sites of one-electron-reduced adenine: Comparisons between the C5 and the C2 or C8 protonation sites. J. Phys. Chem. 1994;98:10969–10977. [Google Scholar]
- 22.Barnes J, Bernhard WA. The protonation state of one-electron-reduced cytosine and adenine. 1. Initial protonation sites at low temperatures in glassy solids. J. Phys. Chem. 1993;97:3401–3408. [Google Scholar]
- 23.Barnes J, Bernhard WA, Mercer KR. Distribution of electron trapping in DNA: Protonation of one-electron-reduced cytosine. Radiat. Res. 1991;126:104–107. [PubMed] [Google Scholar]
- 24.Spalletta R, Bernhard WA. Free radical yields in A:T polydeoxynucleotides, oligodeoxynucleotides, and monodeoxynucleotides at 4 K. Radiat. Res. 1992;130:7–14. [PubMed] [Google Scholar]
- 25.Rupprecht A, Forslind B. Variation of electrolyte content in wetspun lithium- and sodium-DNA. Biochim. Biophys. Acta. 1970;204:304–316. doi: 10.1016/0005-2787(70)90148-6. [DOI] [PubMed] [Google Scholar]
- 26.Mercer KR, Bernhard WA. Design and operation of a variable temperature accessory for Q-band ESR. J. Magn. Reson. 1987;74:66–71. [Google Scholar]
- 27.Lindsay SM, Lee SA, Powell JW, Weidlich T, DeMarco C, Lewen GD, Tao NJ. The origin of the A to B transition in DNA fibers and films. Biopolymers. 1988;27:1015–1043. doi: 10.1002/bip.360270610. [DOI] [PubMed] [Google Scholar]
- 28.Saenger W, Hunter WH, Kennard O. DNA conformation is determined by economics in the hydration of phosphate groups. Nature. 1986;324:385–388. doi: 10.1038/324385a0. [DOI] [PubMed] [Google Scholar]
- 29.Shakked Z, Guerstein-Guzikevich G, Eisenstein M, Frolow E, Rabinovich D. The conformation of the DNA double helix in the crystal is dependent on its environment. Nature. 1989;342:456–460. doi: 10.1038/342456a0. [DOI] [PubMed] [Google Scholar]
- 30.Newton GL, Aguilera JA, Ward JF, Fahey RC. Effect of polyamine-induced compaction and aggregation of DNA on the formation of radiation-induced strand breaks: Quantitative models for cellular radiation damage. Radiat. Res. 1997;148:272–284. [PubMed] [Google Scholar]
- 31.Barnes J, Bernhard WA. The electron scavenging ability of the DNA bases in glassy matrices X-irradiated at 4 K. J. Phys. Chem. 1995;99:11248–11254. [Google Scholar]
- 32.La Vere T, Becker D, Sevilla MD. Yields of •OH in gamma-irradiated DNA as a function of DNA hydration: Hole transfer in competition with •OH formation. Radiat. Res. 1996;145:673–680. [PubMed] [Google Scholar]
- 33.Privé GG, Yanagi K, Dickerson RE. Structure of the B-DNA decamer C-C-A-A-C-G-T-T-G-G and comparison with the isomorphous decamers C-C-A-A-G-A-T-T-G-G and C-C-A-G-G-C-C-T-G-G. J. Mol. Biol. 1991;217:177–199. doi: 10.1016/0022-2836(91)90619-h. [DOI] [PubMed] [Google Scholar]
- 34.Grzeskowiak K, Yanagi K, Privé GG, Dickerson RE. The structure of B helical C-G-A-T-C-G-A-T-C-G and comparison with C-C-A-A-C-G-T-T-G-G. J. Biol. Chem. 1991;266:8861–8883. doi: 10.2210/pdb1d23/pdb. [DOI] [PubMed] [Google Scholar]
- 35.Pimblott SM, LaVerne JA, Mozumder A, Green NJB. Structure of electron tracks in water. 1. Distribution of energy deposition events. J. Phys. Chem. 1990;94:488–495. [Google Scholar]
- 36.LaVerne JA, Pimblott SM. Electron energy-loss distributions in solid, dry DNA. Radiat. Res. 1995;141:208–215. [PubMed] [Google Scholar]
- 37.Chatterjee A, Koehl P, Magee JL. Theoretical consideration of the chemical pathways for radiation-induced strand breaks. Adv. Space Res. 1986;6:97–105. doi: 10.1016/0273-1177(86)90282-6. [DOI] [PubMed] [Google Scholar]
- 38.Holmlin RE, Dandliker PJ, Barton JK. Charge transfer through the DNA base stack. Angew. Chem. Int. Ed. Engl. 1997;36:2714–2730. [Google Scholar]
- 39.Hall DB, Barton JK. Sensitivity of DNA-mediated electron transfer to the intervening π-stack: A probe for the integrity of the DNA base stack. J. Am. Chem. Soc. 1997;119:5045–5046. [Google Scholar]
- 40.Diederichsen U. Charge transfer in DNA: A controversy. Angew. Chem. Int. Ed. Engl. 1997;36:2317–2319. [Google Scholar]
- 41.Wilson EK. DNA a poor conductor new study says. Chem. Eng. News. 1998;18:6–7. [Google Scholar]
- 42.Falk M, Hartman JKA, Lord RC. Hydration of deoxyribonucleic acid I. A gravimetric study. J. Am. Chem. Soc. 1962;84:3843–3846. [Google Scholar]
- 43.Falk M, Hartman JKA, Lord RC. Hydration of deoxyribonucleic acid II. An infrared study. J. Am. Chem. Soc. 1963;85:387–391. [Google Scholar]
- 44.Tao NJ, Lindsay SM. Structure of DNA hydration shells studied by Raman spectroscopy. Biopolymers. 1989;28:1019–1030. doi: 10.1002/bip.360280509. [DOI] [PubMed] [Google Scholar]