Abstract
Mammalian class I aldehyde dehydrogenase (ALDH) plays an important role in the biosynthesis of the hormone retinoic acid (RA), which modulates gene expression and cell differentiation. RA has been shown to mediate control of human ALDH1 gene expression through modulation of the retinoic acid receptor α (RARα) and the CCAAT/enhancer binding protein β (C/EBPβ). The positive activation of these transcription factors on the ALDH1 promoter is inhibited by RA through a decrease of C/EBPβ binding to the ALDH1 CCAAT box response element. However, the mechanism of this effect remains unknown. Here we report that the RARα/retinoid X receptor β (RXRβ) complex binds to the mouse retinaldehyde dehydrogenase 1 (Raldh1) promoter at a non-consensus RA response element (RARE) with similar affinity to that of the consensus RARE. We found that C/EBPβ binds to a Raldh1 CCAAT box located at –82/–58 bp, adjacent to the RARE. Treatment with RA increases GADD153 and GADD153–C/EBPβ interaction resulting in a decreased cellular availability of C/EBPβ for binding to the Raldh1 CCAAT box. These data support a model in which high RA levels inhibit Raldh1 gene expression by sequestering C/EBPβ through its interaction to GADD153.
Keywords: Raldh1, Retinoic acid, RARα, C/EBPβ, GADD153
1. Introduction
Retinoic acid (RA) is a metabolite of vitamin A (retinol) that regulates developmental pathways in vertebrate animals. It exerts its effects through two families of nuclear receptors, RA receptors (RARs) and retinoid X receptors (RXRs). Maintenance of RA homeostasis is crucial for normal embryogenesis and development in chordate organisms.
RA synthesis involves the oxidation of retinol to retinal and the subsequent oxidation of retinal to RA by members of the aldehyde dehydrogenase family (ALDH) [1]. ALDHs are a group of NADP-dependent enzymes that catalyze the conversion of aldehydes into acid metabolites. They also participate in the metabolism of alcohols [2], biogenic amines [3], vitamins [4], steroids [5] and lipids [6,7], as well as in the biotransformation of numerous drugs and environmental agents [8].
To date, 555 ALDH genes have been described, including 172 in eukaryotes, and 17 functional genes in humans [9]. Among them, cytosolic human ALDH1, mouse retinaldehyde dehydrogenase (Raldh) 1 and mouse Raldh2 have been shown to mediate retinaldehyde oxidation and are considered to play a major role in RA synthesis [10]. A critical role for Raldh2 in mammalian development was established by the finding that embryos homozygous null for Raldh2 failed to develop due to severe abnormalities [11]. On the other hand, Raldh1 null mouse presents an insulin resistance and increased energy dissipation [12].
The mouse Raldh1 gene has similar tissue-specificity and developmental control as the human ALDH1 gene. It is highly expressed in adult mouse tissues, including liver [13,14], and plays a role in RA clearance in vivo [15]. Although the mouse Raldh1 gene has been sequenced and the promoter region has been characterized, little is known about the molecular mechanisms by which expression of this gene is regulated.
Recently, it has been shown that the aryl hydrocarbon receptor (AhR) is involved in the RA metabolism. The AhR is a ligand activated receptor [16], and a member of the basic helix–loop–helix-PAS (bHLH-Per-Arnt-Sim) transcription factor family. Upon binding ligand, the AhR translocates to the nucleus, and up-regulates the expression of a battery of genes encoding xenobiotic-metabolizing enzymes, such as cytochrome P450s (CYP1A1, CYP1A2, CYP1B1), NAD(P)H quinone oxydoreductase and UDP-glucoronosyl-transferase-6 [17]. Livers from AhR-null mice show a 3-fold increase in RA levels as well as a down regulation of Raldh1 and 2, suggesting that AhR controls RA catabolism [18]. More recently, it was determined that expression of CYP2C39, a mouse enzyme responsible for RA 4-hydroxylation, is under AhR control also [19]. These data explain the high levels of RA observed in AhR-null mouse liver. Because AhR-null mouse has a defect in RA catabolism leading to elevated RA levels and decreased Raldh1 mRNA levels, this mouse model is an important tool to study the role of RA on Raldh1 gene regulation.
We previously observed that RA mediates feedback inhibition of the human ALDH1 gene expression through decreasing C/EBPβ binding to CCAAT box response element located at the ALDH1 promoter [20]. However, the molecular mechanism of this inhibition is still unknown. In order to determine the molecular nature of this effect, we first investigated whether the mouse Raldh1 gene expression is regulated by a similar mechanism as determined for the ALDH1. As in human ALDH1, RARα and C/EBPβ bind to a novel RA response element (RARE) and to the CCAAT box, respectively, located within the Raldh1 promoter. We also show that treatment with RA increased GADD153 mRNA and GADD153–C/EBPβ interaction resulting in a sequestration of C/EBPβ and decreased C/EBPβ binding to the Raldh1 CCAAT box.
2. Methods and materials
2.1. Materials
Mouse hepatoma-derived cells (Hepa-1) were obtained from ATCC (Manassas, VA, USA). All-trans-RA was obtained from Sigma (St. Louis, MO). RARα and RXRβ antibodies were purchased from Affinity Bioreagents (Golden, CO). Raldh1 cDNA was provided by Gregg Duester (Burnham Institute, La Jolla, CA) [21] and the type II transglutaminase (TG-II) and RARα cDNA were provided by Ronald M. Evans (Howard Hughes Medical Institute, San Diego, CA) [22].
2.2. Cell culture
Hepa-1 cells were cultured in Dulbecco's modified Eagle's medium (Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA). Cells were harvested and cultured at 37 °C in a humidified atmosphere of 95% air/5% CO2.
2.3. Electrophoretic mobility shift assays (EMSA)
Double-stranded oligonucleotides complementary to the RARα-binding site (5′-CGAGTTCAGCGAGAGTTCAGC-3′) [23], the 5′ upstream mouse Raldh1 gene –82 to –58 region (5′-CTTGCCCTGAGCTGCCCATCCAATC-3’), and an unrelated double-stranded oligonucleotide (5′-CTGCAGTGACCACTGCCCCATCATTGCTGGCTC-3′) (Sigma, St. Louis, MO) were end-labeled using the Klenow fragment of DNA polymerase I and [32P]dCTP (Amersham Pharmacia Biotech, UK). Hepa-1 cell nuclear extracts (10 μg), prepared as described previously [24], were incubated for 30 min at 25 °C in a 20-μl reaction mixture (20 mM Tris–HCl, pH 8.0, 0.5 mM EDTA, 1 mM dithiothreitol, 80 mM KCl, 12% glycerol, 0.5 μg of poly(dI-dC)) (Amersham Pharmacia Biotech, UK), with 2 μl of the double-stranded oligonucleotide probe (50,000 cpm). The mixtures were subjected to electrophoresis in a 4% polyacrylamide gel (BioRad, Richmond, CA) at 4 °C. In competition assays, a large excess of unlabeled double-stranded oligonucleotide competitor was incubated together with the nuclear extract prior to adding the 32P-labeled probe. For gel mobility supershift assays, RARα and RXRβ antibodies (Affinity Bioragents, Golden, CO) were incubated with the probe-nuclear extract mixtures for additional 60 min prior to gel electrophoresis. The sequence of C/EBPβ binding motif was 5′-CTAGGGCTTGCGCAATCTATATTCG-3′ [25].
2.4. Northern blot analysis
Total RNA from mouse liver was isolated by homogenizing cells in a guanidine/phenol solution (Biotex Laboratories, Houston, TX). Total RNA (20 μg) was subjected to electrophoresis in a 1% agarose, 2.2 M formaldehyde gel and then transferred to GeneScreen Plus membranes in 20× SSC (3 M NaCl, 30 mM sodium citrate, pH 7.0) (J.T. Baker, Mexico). The RNA was fixed to the membranes by baking at 80 °C for 2 h, prehybridized in SSC/formamide solution, and hybridized at 42 °C with the 32P-labeled probes. cDNAs were labeled by random priming with DNA polymerase I Klenow fragment using [32P]dCTP (Amersham Pharmacia Biotech, UK). Labeled probes were added to the membranes at 2.0 × 106 cpm/ml. Filters were washed in 0.1× SSC and 0.5% sodium dodecyl sulfate (Invitrogen, Carlsbad, CA), and the membranes were exposed to autoradiographic film overnight at –80 °C with aid of a Phosphor screen to visualize hybridized RNA.
2.5. RT-PCR analysis
Total RNA from Hepa-1 cell cultures or mouse liver was isolated by homogenizing cells or tissue in a guanidine/phenol solution (Invitrogen, Carlsbad, CA). Reverse transcription was performed using OneStep RT-PCR kit, according to the manufacturer's instructions (Qiagen Inc., Germany). For C/EBPβ analysis the following program was used: denaturation at 94 °C for 2 min and 30 cycles of PCR consisting of denaturation at 94 °C for 2 min, annealing at 58 °C for 2 min, and extension at 72 °C for 2 min, and a final extension step at 72 °C for 5 min. For GAPDH analysis the following program was used: denaturation at 91 °C for 10 min and 30 cycles of PCR consisting of denaturation at 91 °C for 45 s, annealing at 51 °C for 20 s, and extension at 70 °C for 45 s. For GADD153 and RNAr18S analysis the following conditions were used: denaturation at 94 °C for 3 min and 30 cycles of PCR consisting of denaturation at 94 °C for 50 s, annealing at 61 °C for 40 s, and extension at 72 °C for 40 s.
Forward and reverse primers were as follows: C/EBPβ, 5′-GCGCGAGCGCAACAACATCG-3′ and 5′-CAGCACAGGCTGTTGACCATCATA-3′ [26], GADD153, 5′-CATACACCACCACACCTGAAAG-3′ and 5′-CCGTTTCCTAGTTCTTCCTTGC-3′ [27], GAPDH, 5′-CCAATATGATTCCACCCATG-3′ and 5′-AGGTCCACCACTGACACGTT-3′ [28], and 18S 5′-GGACAGGATTGACAGATTGATAG-3′ and 5′-CTCGTTCGTTATCGGAATTAAC-3′ [29]. PCR products were electrophoresed on a 2% agarose gel, and stained with ethidium bromide to visualize PCR amplification products. Relative intensity was determined using the Sigma Gel program (Jandel Scientific software, Sigma, St. Louis, MO).
2.6. Western blotting and immunoprecipitation
To determine C/EBPβ protein level and to detect C/EBPβ-associated proteins, Hepa-I cell cultures were treated with 0.1, 1.0 μM RA or 0.05% (v/v) dimethyl sulfoxide for 24 h or 72 h (Sigma, St. Louis, MO). The cells were washed with ice-cold PBS, then scraped and lysed on ice for 30 min by addition of 0.5 ml/10 cm plate of lysis buffer (25 mM Hepes, pH 7.7, 150 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.5% Tween 20, 20 mM beta-glycerophosphate and enzyme inhibitor cocktail) (Complete, Roche Diagnostics Corp., Indianapolis, IN, USA). The cell debris was removed by centrifugation at 14,000 rpm for 15 min at 4 °C. To determine C/EBPβ and actin protein levels 50 μg of cell lysates were subjected to SDS-polyacrylamide gel electrophoresis on 15% polyacrylamide gels. Proteins were transferred to nitrocellulose membranes. Polyclonal C/EBPβ and Actin antibodies (Santa Cruz Biotechnology, CA) were used at 1:200 and 1:500 dilutions, respectively. Immunoprecipitations were performed with total cell lysates (50 μg protein) using an anti-C/EBPβ rabbit polyclonal antibody (Santa Cruz Biotechnology, CA, 2 μg/ml, overnight at 4 °C), followed by adsorption to protein A/G plus-agarose (Santa Cruz Biotechnology, CA) for 2 h at room temperature. Protein A/G plus-agarose complexes were washed twice with complete washing buffer (50 mM Tris, pH 7.5, 150 mM NaC, 5 mM EDTA and 1% triton X-100) (Sigma, St. Louis, MO) and two times with washing buffer without triton X-100. The bound proteins were fractionated by 15% SDS-polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane. Membranes were blocked with 5% nonfat dry milk in Tris–buffered saline containing 0.1% Tween 20 for 1 h with gentle shaking, and incubated with a mouse monoclonal anti-GADD-153 antibody (1/1000, Santa Cruz Biotechnology, CA) overnight at 4 °C. A peroxidase-conjugated goat anti-mouse IgG antibody and the ECL-PLUS detection kit (Amersham Pharmacia Biotech, Piscataway, NJ) were used for visualization of immune complexes.
2.7. Animals
Ahr-null (targeted mutation of the aryl-hydrocarbon receptor gene) and wild-type male C57BL6N/129sv mice aged 6–8 weeks were maintained in a pathogen-free environment with a controlled temperature (22 ± 2 °C) with 40–60% humidity. The mice had 12/12 light/dark cycles and were given free access to food and water. Animal housing and care was in accordance with the Mexican Regulations of Animal Care and Maintenance (NOM-062-ZOO-1999, 2001), and with the Guide for the Care and Use of Laboratory Animals as adopted and promulgated by the U.S. National Institutes of Health.
2.8. Statistical analysis
Results are presented as the mean values ± SD. The Mann–Whitney's test was used for statistical comparisons. The criterion for significance was p < 0.05 in all cases.
3. Results
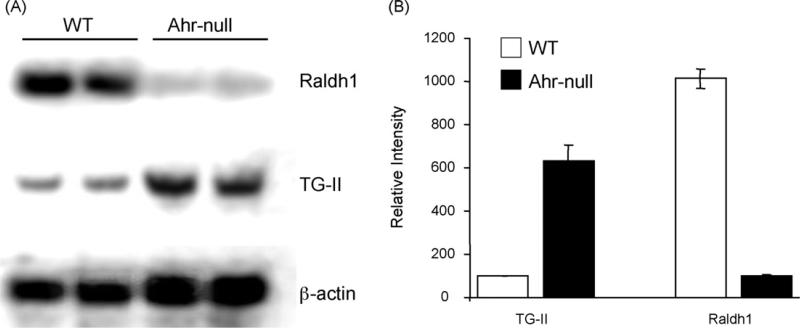
In order to confirm the alteration on RA metabolism observed in AhR-null mice, we first evaluated the expression of hepatic genes under RA control. Transglutaminase 2 (TG-II) is an inducible transamidating acyltransferase that catalizes Ca2+-dependent protein modification. Its expression is up-regulated by RA [30]. Fig. 1 shows that AhR-null mouse liver presents a 5-fold increase on TG-II mRNA levels compared with wild-type mice. These results are in agreement with previous reports describing increased TG-II activity in the Ahr-null mouse [18]. As expected, a decrease in Raldh1 mRNA levels in the Ahr-null liver relative to wild-type mouse liver was observed (Fig. 1). Once expression of Raldh1 in the AhR-null phenotype was confirmed, we proceeded to characterize the Raldh1 promoter.
Fig. 1.
Northern blot analysis of Raldh1 and transglutaminase II mRNAs. (A) Liver total RNA isolated from Ahr-null and wild-type mice. Mouse actin mRNA was used to verify integrity. (B) Hybridization signal quantification data are presented as the mean levels of expression ± SD, n = 3.
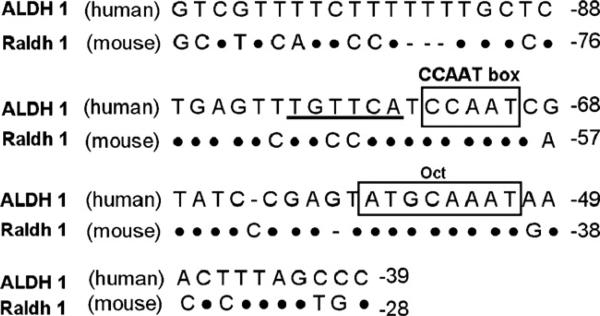
Comparison of the 5′-sequences of human ALDH1 with mouse Raldh1 genes revealed that the mouse Raldh1 sequence is similar to the human ALDH1 proximal promoter region (Fig. 2). Within the upstream 100-bp region from the transcription start site, the Raldh1 promoter included a CCAAT box and octamer binding sites. Nucleotide sequence analysis also revealed a sequence similar to the RARE human ALDH1 motif, TGCCCA, located at –65/–70 bp.
Fig. 2.
Comparison of 5′-flanking region sequences of mouse Raldh1, and human ALDH1. Identity is indicated by dots and nucleotide deletions by dashes. CAATT box and Oct regulatory elements are boxed, and human RARE-like element is underlined.
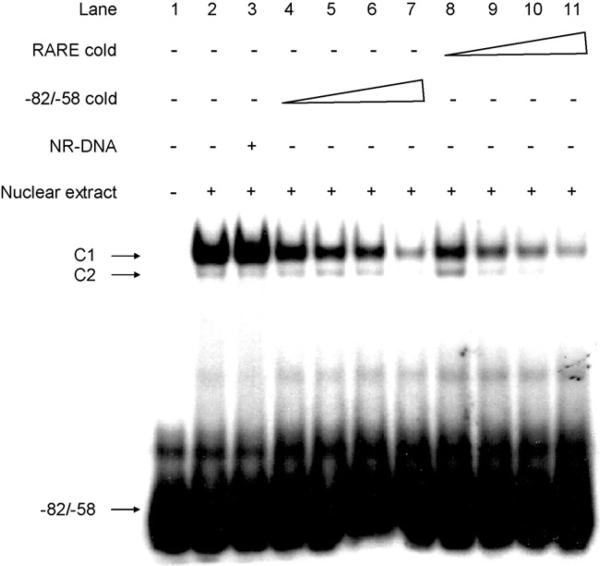
To determine whether RARα binds to the RARE-like motif present in the Raldh1 promoter, EMSA was performed (Fig. 3). Addition of Hepa-1 nuclear extracts resulted in the formation of two DNA–protein complexes. Competition of complex formation with addition of excess unlabeled –82/–58 oligonucleotide probe (lane 4–7), but not a nonspecific oligonucleotide probe (lane 3), indicated that the binding was saturated. Competition experiments using a consensus RARE oligonucleotide demonstrated decreased binding to complex 1 and 2 (lanes 8–11), suggesting that these complexes are the result of binding to the RARE.
Fig. 3.
EMSA analysis of protein binding to the mouse Raldh1 promoter region. Nuclear extracts from Hepa-1 cells were incubated with radiolabeled –82/–58 bp alone (lane 2), –82/–58 bp plus a 100-fold molar excess of a nonspecific unlabeled competitor (NR-DNA) oligonucleotide (lane 3), –82/–58 bp plus a 10, 20, 50, or 100-fold molar excess of unlabeled –82/–58 bp (lanes 4–7), or –82/–58 bp plus a 10, 20, 50, or 100-fold molar excess of unlabeled consensus RARE (lanes 8–11). Lane 1 is radiolabeled probe alone. The arrows identify the shifted bands (C1 and C2) and the unbound radiolabeled –82/–58 bp probe. A representative set of data from two independent experiments is shown.
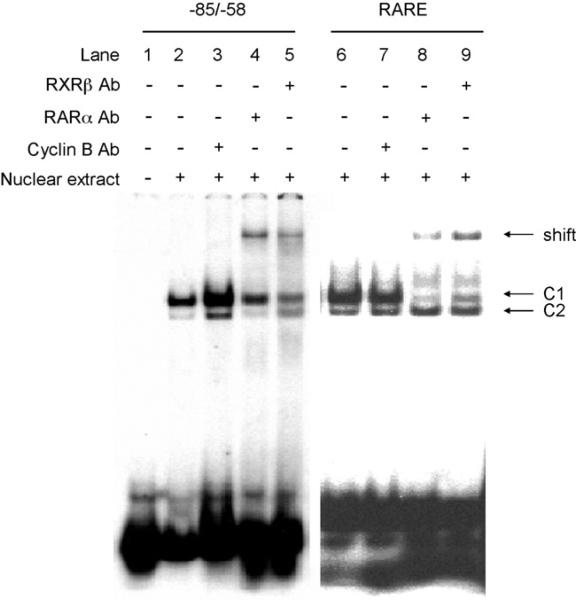
EMSA supershift analyses were used to detect endogenous RARα in nuclear extracts of Hepa-1 cells (Fig. 4). As expected, RARα and RXRβ antiserum supershifted the consensus RARE complex (lanes 8 and 9). A similar result was observed for the complex 1 bound to the mouse region –82/–58 bp (lanes 4 and 5). No supershift was observed when non-related antiserum was used (lanes 3 and 7). These data indicate that complex 1 represents the RARα/RXRβ heterodimer.
Fig. 4.
Binding of RARα to the mouse –82/–58 bp Raldh1 gene region. Nuclear extracts from Hepa-1 cells were incubated with radiolabeled –82/–58 bp (lanes 2–5), or radiolabeled consensus RARE (lanes 6–9), and then incubated with RARα antibody (Ab) (lanes 4 and 8), or RXRβ Ab (lanes 5 and 9) for 60 min. Lanes 3 and 7 were incubated with an unrelated Ab (Cyclin B). Arrows identify the shifted bands. A representative set of data from two independent experiments is shown.
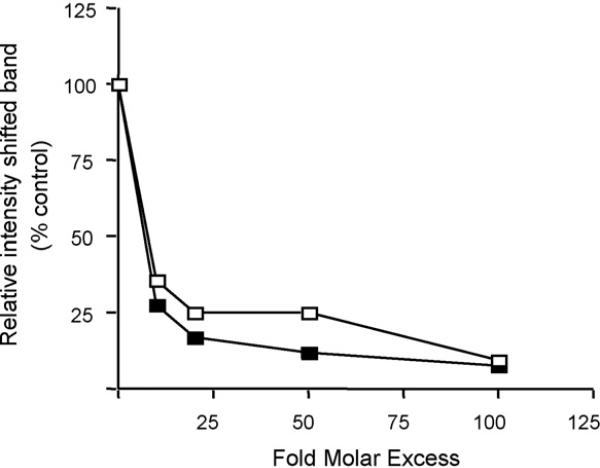
To compare the affinity of RARα for the mouse promoter region –82/–58 bp with a consensus RARE, competitive inhibition experiments were performed (Fig. 3) in which the relative intensity of complex 1 was evaluated (Fig. 5). Consensus RARE competed equivalently with the mouse –82/–58 bp oligonucleotide for binding to RARα/RXRβ, indicating that the mouse Raldh1 –82/–58 bp region is functionally similar to the consensus RARE.
Fig. 5.
Competitive binding analysis of the RARα/RXRβ complex to Raldh1 gene promoter. Nuclear extracts were incubated with radiolabeled mouse –82/–58 bp probe and varying amounts of unlabeled –82/–58 bp (open squares) and consensus RARE (closed squares) probes. Following EMSA, the intensity of shifted band, identified as a RARα/RXRβ DNA complex, was quantitated by densitometry. A representative set of data from two independent experiments is shown.
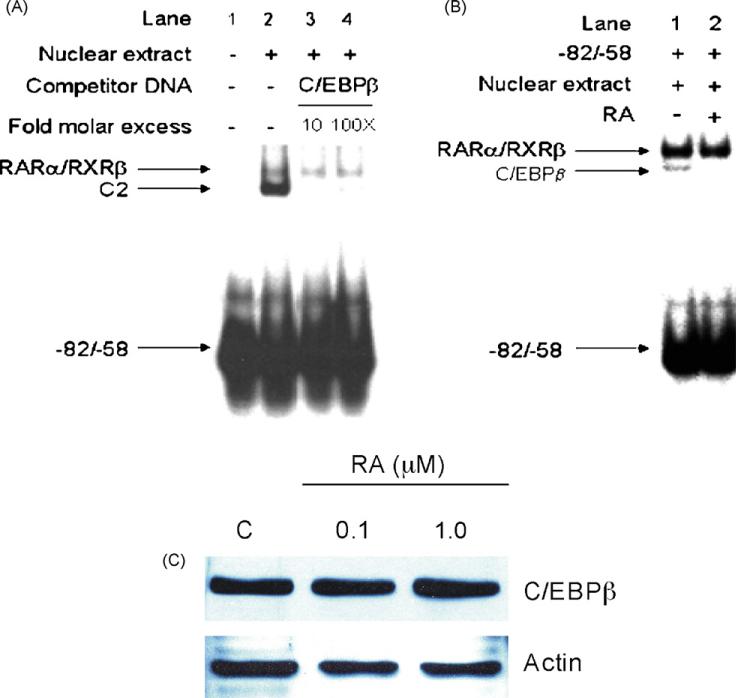
In order to determine whether the CCAAT box found in the mouse Raldh1 gene promoter region serves as a binding region for C/EBPβ, the binding affinity of C/EBPβ protein was tested in a competitive EMSA. An unlabeled oligonucleotide containing the consensus sequence of the C/EBPβ binding site was used. As Fig. 6A shows, the C/EBPβ consensus sequence specifically decreased the formation of complex 2 (Fig. 6A, lanes 3 and 4), indicating that C/EBPβ binds to the mouse –82/–58 bp region and thus may contribute to the control of Raldh1 gene transcription.
Fig. 6.
(A) Competitive EMSA to identify the CCAAT box binding protein. Unlabeled oligonucleotide containing the consensus sequence of C/EBPβ was incubated with Hepa-1 nuclear extract for 30 min at the indicated molar excess prior to addition of the –82/–58 bp labeled probe. Arrows identify the shifted bands and the unbound radiolabeled probe. (B) Effect of RA on C/EBPβ binding. Nuclear extracts from RA-treated (1.0 μM RA, 72 h) Hepa-1 cells were incubated with radiolabeled mouse –82/–58 bp probe (lane 2) and analyzed by EMSA. Arrows identify the shifted bands and the unbound radiolabeled probe. C) Effect of RA treatment on C/EBPβ protein levels. Hepa-1 cells were treated with 0.1 or 1.0 μM of RA for 72 h and cell lysates were incubated with C/EBPβ antibody. C/EBPβ protein levels were determined as described in Section 2.A representative set of data from two independent experiments is shown.
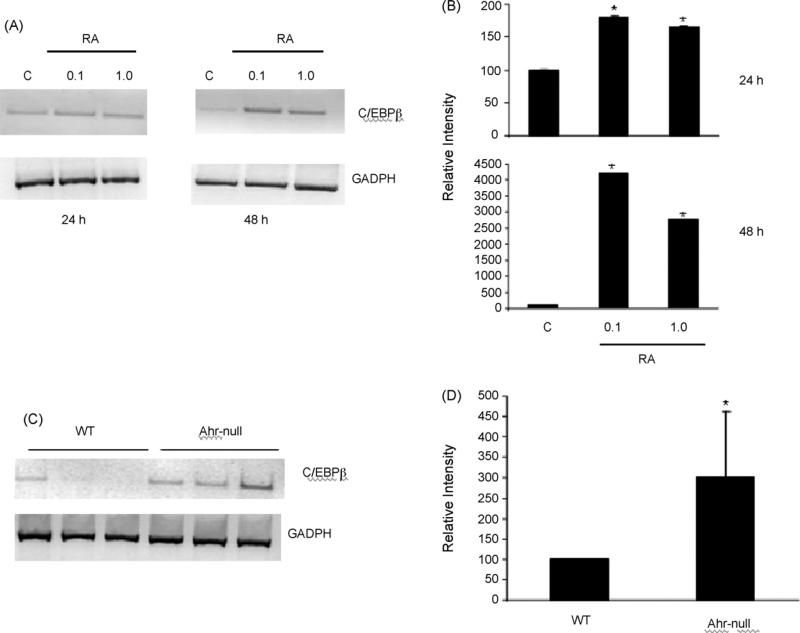
Previously, we have shown that RA decrease C/EBPβ binding to the human ALDH1 promoter [20], suggesting a possible mechanism by which RA control ALDH1 gene expression. In order to elucidate whether a similar mechanism acts on the mouse Raldh1, EMSA analysis was performed. Results revealed that nuclear extracts from Hepa-1 cell cultures treated with RA (1.0 μM) reduced C/EBPβ binding (Fig. 6B, lane 2). In order to determine whether RA modified translational or protein stability, C/EBPβ protein levels were evaluated after RA treatment. Western blotting analysis indicated that RA treatment of Hepa-1 cells did not decrease C/EBPβ protein levels (Fig. 6C). This finding confirms that RA disrupts binding of C/EBPβ to the Raldh1 gene promoter. Because a decrease in C/EBPβ transcription levels may explain the observed low binding of this transcription factor to the Raldh1 promoter, we examined the effect of RA on C/EBPβ mRNA levels. RT-PCR experiments revealed that RA treatment of Hepa-1 cultured cells increased C/EBPβ mRNA levels, in a time-dependent manner (Fig. 7A and B). In agreement with this, a 3-fold increase in the expression of C/EBPβ compared to wild-type mouse was found in Ahr-null mouse livers, which have elevated RA levels (Fig. 7C and D). These surprising results suggest that C/EBPβ gene expression is under RA control, and that RA-induced decrease of C/EBPβ binding to the CCAAT box is not mediated at transcriptional level.
Fig. 7.
Effect of RA treatment on C/EBPβ mRNA levels. (A) Hepa-1 cells were treated with 0.1 or 1.0 μM of RA for 24 or 48 h, C/EBPβ mRNA levels were determined by RT-PCR, the relative intensity of the signal was quantified (B). (C) Liver RNA, isolated from Ahr-null and wild-type mice, was subjected to RT-PCR analysis, and the relative intensity of the signal was quantified (D). Signals were normalized relative to GADPH. Data are presented as the mean levels of expression ± SD of triplicates, *p < 0.05.
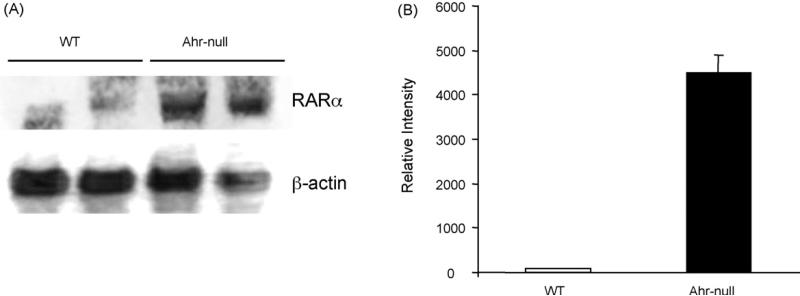
Several studies have proposed that increased RARα levels may favor protein interaction between RARα and CCAAT binding proteins [31,32]. We then investigate whether RA increase RARα mRNA levels and favor the C/EBPβ–RARα interaction. Although Ahr-null liver mouse showed a massive 36-fold increase of RARα mRNA relative to wild-type samples (Fig. 8), no interaction between C/EBPβ and RARα was observed (data not shown).
Fig. 8.
Northern blot analysis of RARα. (A) Liver total RNA, isolated from Ahr-null and wild-type mice, was subjected to Northern blot analysis. Mouse actin mRNA was used to verify integrity. (B) Following hybridization, the membrane was exposed to phosphor screen, and the signal was quantified. Data are presented as the mean levels of expression ± SD, n = 3.
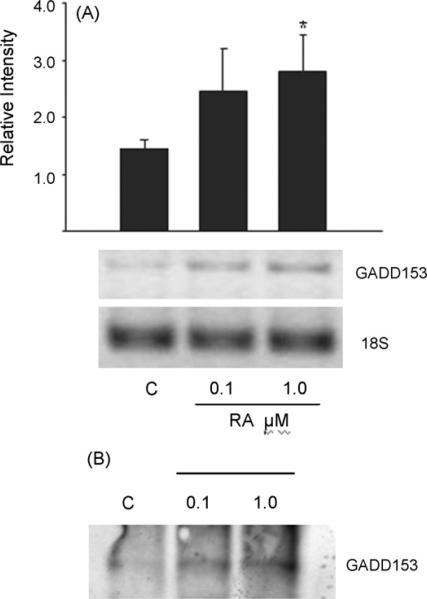
GADDD153 (also named CHOP-10) is a member of the CCAAT/enhancer binding proteins family of transcription factors that lacks a functional DNA binding domain. Its expression is ubiquitous and highly induced in response to a variety of cellular signals [33]. An interaction between GADD153 and C/EBPβ was reported during cellular stress [34]. Therefore, we investigated the effect of RA on GADD153 expression and whether this treatment favors C/EBPβ/GADD153 interaction. Fig. 9A shows that Hepa1-RA treated cultures present an increase in GADD153 mRNA levels in a dose-dependent manner. To determine whether GADD153 interacts directly with C/EBPβ in Hepa1 cells, coimmunoprecipitation experiments were carried out. Protein complexes were immunoprecipitated with C/EBPβ antibody, and this was followed by Western blot analysis with GADD153 antibody. Results indicate that GADD153 was found in complexes immunoprecipitated with C/EBPβ antibody, and that this interaction is RA-dependent, also in a dose-dependent fashion (Fig. 9B).
Fig. 9.
(A) Effect of RA treatment on GADD-153 mRNA levels. Hepa-1 cells were treated with 0.1 or 1.0 μM of RA for 24 h and GADD-153 mRNA levels were determined by RT-PCR, and the relative intensity of the signal was quantified. Data are presented as the mean levels of expression ± SD of triplicates, *p < 0.05. (B) Coimmunoprecipitation of GADD-153 and C/EBPβ. Hepa-1 cells were treated with 0.1 or 1.0 μM of RA for 24 h and cell lysates were incubated with C/EBPβ antibody. After PAGE, the membrane was incubated with GADD-153 antibody. A representative set of data from two independent experiments is shown.
4. Discussion
RA has marked effects on cell differentiation and proliferation. Since oxidation of retinaldehyde to RA is irreversible, this reaction must be tightly regulated. Therefore, understanding the molecular mechanisms controlling RA synthesis is of great importance. The present study demonstrated that RARα and C/EBPβ bind to the mouse Raldh1 gene 5′-flanking region and that this interaction resembles that reported for human ALDH1 gene regulation [20]. The mouse Raldh1 –75/–18 bp regions is highly conserved between human and mouse and it contains an octamer factor binding site, and a CCAAT box. Within this region a putative RARE was found to be located adjacent to the CCAAT box (at –82/–58 bp), and this RARE-like motif was demonstrated to bind the RARα/RXRβ heterodimer by EMSA. Although no apparent RARE consensus sequence was identified within the –82/–58 bp region, this sequence seems to belong to a RA response complex element which has few or no obvious consensus elements. The affinity to the RARα/RXRβ complex shown by this RARE-like motif was similar to that seen with the RARE/DR-5 element, which is one of the most potent inducing sequences found in RAR-responsive element promoters [35]. RXRβ antiserum super-shifted complex 1, thus indicating that this signal represents the RARα/RXRβ heterodimer. However, a complex 1 signal without shifting was still detected; suggesting that this complex may be also represented by the interaction of RARα with other RXR subtype.
The mouse Raldh2 promoter sequence contains several potential transcription factor-binding sites: CCAAT boxes, AP-1, AP-2, AP-4 and AHR/ARNT. However RARE or RXRE consensus sequences were not predicted [36]. Nevertheless, after a Raldh2 promoter analysis, we found evidence for a RARE at –2057 bp that is similar to the RARE found in the human ALDH1 and mouse Raldh1 promoters (data not shown). These data suggest that RA may generally control retinal dehydrogenase gene expression through an interaction between RARα and RA response complex elements.
In previous studies we uncovered the molecular mechanism by which RA down regulates ALDH1 gene expression [20]. This RA inhibitory effect is mediated by decreasing C/EBPβ binding to the promoter. However, the nature of this decrease was unknown. C/EBPβ also binds to the Raldh1 promoter suggesting that Raldh1 transcription may be regulated by this transcription factor. As reported previously, C/EBPβ is a positive regulator of human ALDH1, and mutation analysis showed that the CCAAT box is critical for its promoter activity. Similarly, Stewart et al. observed that disruption of the CCAAT box in mouse Raldh2 decreased transcriptional activity by 50% [25]. More recently, the rat Raldh1 promoter was characterized. The authors report a CCAAT box and octamer binding site located between –72 and –68 bp and –56 and –49 bp, respectively, from transcriptional start site. Mutations of CCAAT box abolish transactivation activity, indicating that this motif is critical for basal promoter activity [37]. These findings suggest that C/EBPβ may generally regulate the expression of retinal oxidizing ALDHs. However, no RA effects were observed on rat Raldh1 mRNA or on promoter activity. The RARE identified in human ALDH1, and the mouse, and rat Raldh1 promoters were TGTTCA, TGCCCA, and TGGCCA, respectively. These data suggest that T/G change on the putative rat RARE respect with the human is responsible for the lack of responsiveness to RA.
In the present study we found that Hepa-1 cell cultures treated with RA resulted in increased, rather than decreased, C/EBPβ mRNA levels in a time-dependent manner, ruling out that the mechanism by which RA disrupts CCAAT–C/EBPβ interaction is at transcriptional level. Since the Ahr-null mouse presents elevated hepatic RA concentrations, the finding of increased C/EBPβ mRNA levels in AhR-null liver provides further support for the idea that C/EBPβ gene expression is RA responsive. This finding suggests that RA may regulate gene expression through modulation of C/EBPβ levels. It was also reported that expression of other members of the CCAAT/enhancer binding protein family such as C/EBPε are under RA control [38], suggesting that this hormone regulates expression of this transcription factor family.
Schule et al. showed that addition of RARα reduced AP-1/DNA complex formation [39], suggesting that RARα functions as a negative regulator of AP-1 responsive genes by interfering with their binding activity. More recently, Kim et al. observed that RAR and RXR associate with the serum response factor (SRF) in a ligand-dependent manner, and inhibit SRF transactivation activity [40]. In addition, many studies have demonstrated that retinoid receptors inhibit the expression of several genes by interfering with the action of transcription factors [31,39,41,42]. Specifically, RARs and RXRs have been shown to inhibit IL-6 gene expression by antagonizing the effect of NF-IL6, a CCAAT binding factor [41,43]. The observed increase in RARα mRNA levels found in Ahr-null mice liver may favor protein interaction between RARα and C/EBPβ, which may result in a decrease of C/EBPβ/CCAAT box complex formation. However no interaction between these two proteins was found.
Alternatively, other proteins may interact with C/EBPβ. It has been demonstrated that GADD153, a member of the CCAAT enhancer binding protein family, interacts and form complexes with C/EBPβ [34]. On the other hand, GADD153 mRNA levels increase when NB4 and HL60 AML cell lines were treated with all-trans retinoic acid [44]. In the Hepa-1 cells used in the present study, RA treatment increased GADD153 mRNA levels in a dose-dependent manner. Furthermore, we demonstrate that GADD153 binds C/EBPβ, serving as a dominant negative inhibitor, decreasing DNA-binding activity of C/EBPβ to the CCAAT box in the Raldh1 promoter. Interestingly, Fawcett et al. showed that, in rat pheochromocytoma PC12 cells, C/EBPβ acts as a positive regulator of GADD153 transcription, and provided evidence supporting an autoregulatory feedback loop in which expression of GADD153 suppresses its own transcription through its interaction with C/EBPβ [34].
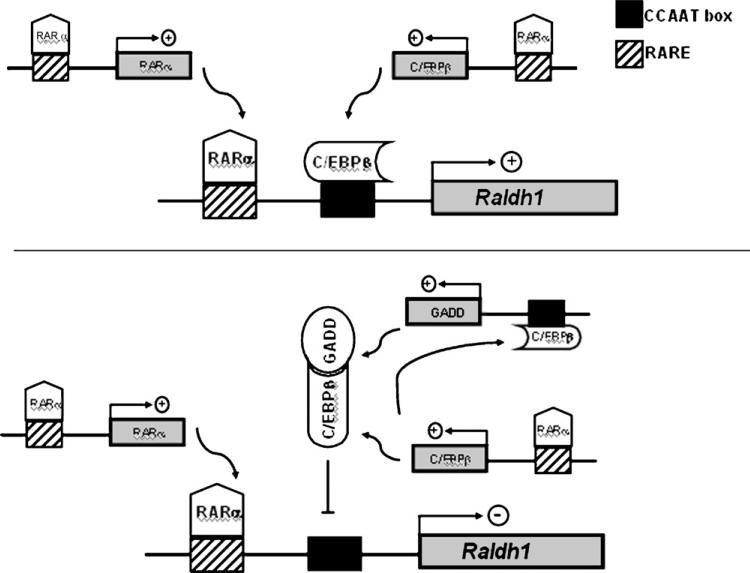
All together, these data suggest that Raldh1 transcription is under regulation of two linked negative feedback systems (Fig. 10). Raldh1, which plays a major role in RA synthesis, is under RARα and C/EBPβ transactivation control. Expression of both transcription factor genes is RA responsive, probably through RARα [23] (Fig. 10A). When RA levels increase, Raldh1 expression is inhibited. The high levels of this hormone increase C/EBPβ mRNA levels which increase GADD153 expression. GADD153 then interacts with C/EBPβ, decreasing DNA-binding activity of this transcription factor to the CCAAT box in the Raldh1 promoter (Fig. 10B).
Fig. 10.
Model of autoregulation of the Raldh1 gene promoter by heterodimerization of GADD153 and C/EBPβ. At low RA concentrations RARα and C/EBPβ transactivate the Raldh1 promoter (top panel). When RA levels increase, RARα transactivate the C/EBPβ promoter increasing the C/EBPβ abundance resulting in an increase of GADD153 amount and the formation of GADD153–C/EBPβ heterodimers (lower panel).
RA levels observed in the AhR-null mouse livers respond to a decrease on CYP2C39 transcription [19]. Therefore, AhR ligands would have the potential to alter the RA levels modifying not only Raldh1 gene expression, but also other genes under C/EBPβ-GADD145 autoregulatory feedback loop.
Finally, since C/EBPβ is a transcription factor that is critical to cellular differentiation, further studies will be necessary to elucidate the molecular mechanism by which RA induces C/EBPβ expression. The RARα RXRβ complex and C/EBPβ bind to specific sequences within the mouse Raldh1 gene promoter. These findings demonstrate that mouse Raldh1 is under similar transcriptional control as the human ALDH1 gene. We also showed that RA increases GADD153 and C/EBPβ mRNA levels, and promotes the interaction between these two proteins, resulting in the inhibition of Raldh1 expression.
REFERENCES
- 1.Duester G, Mic FA, Molotkov A. Cytosolic retinoid dehydrogenases govern ubiquitous metabolism of retinol to retinaldehyde followed by tissue-specific metabolism to retinoic acid. Chem Biol Interact. 2003;143–144:201–10. doi: 10.1016/s0009-2797(02)00204-1. [DOI] [PubMed] [Google Scholar]
- 2.Harrington MC, Henehan GT, Tipton KF. The interrelationships of alcohol dehydrogenase and the aldehyde dehydrogenases in the metabolism of ethanol in liver. Biochem Soc Trans. 1988;16(3):239–41. doi: 10.1042/bst0160239. [DOI] [PubMed] [Google Scholar]
- 3.MacKerell Jr AD, Blatter EE, Pietruszko R. Human aldehyde dehydrogenase: kinetic identification of the isozyme for which biogenic aldehydes and acetaldehyde compete. Alcohol Clin Exp Res. 1986;10(3):266–70. doi: 10.1111/j.1530-0277.1986.tb05087.x. [DOI] [PubMed] [Google Scholar]
- 4.McCaffery P, Tempst P, Lara G, Drager UC. Aldehyde dehydrogenase is a positional marker in the retina. Development. 1991;112(3):693–702. doi: 10.1242/dev.112.3.693. [DOI] [PubMed] [Google Scholar]
- 5.Han A, Marandici A, Monder C. Metabolism of corticosterone in the mouse. Identification of 11 beta, 20 alpha-dihydroxy-3-oxo-4-pregnen-21-oic acid as a major metabolite. J Biol Chem. 1983;258(22):13703–7. [PubMed] [Google Scholar]
- 6.Lindahl R, Petersen DR. Lipid aldehyde oxidation as a physiological role for class 3 aldehyde dehydrogenases. Biochem Pharmacol. 1991;41(11):1583–7. doi: 10.1016/0006-2952(91)90157-z. [DOI] [PubMed] [Google Scholar]
- 7.Pinazo-Duran MD, Verdejo C, Azorin I, Renau-Piqueras J, Iborra FJ. Colocalization of aldehyde dehydrogenases and Fe/NADPH-induced lipid peroxidation in tissue sections of rat retina. Ophthalmic Res. 2000;32(2–3):61–8. doi: 10.1159/000055591. [DOI] [PubMed] [Google Scholar]
- 8.Vasiliou V, Pappa A, Petersen DR. Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem Biol Interact. 2000;129(1–2):1–19. doi: 10.1016/s0009-2797(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 9.Sophos NA, Vasiliou V. Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem Biol Interact. 2003;143–144:5–22. doi: 10.1016/s0009-2797(02)00163-1. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida A, Hsu LC, Dave V. Retinal oxidation activity and biological role of human cytosolic aldehyde dehydrogenase. Enzyme. 1992;46(4–5):239–44. doi: 10.1159/000468794. [DOI] [PubMed] [Google Scholar]
- 11.Niederreither K, Subbarayan V, Dolle P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21(4):444–8. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- 12.Ziouzenkova O, Orasanu G, Sharlach M, Akiyama TE, Berger JP, Viereck J, et al. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat Med. 2007;13(6):695–702. doi: 10.1038/nm1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ang HL, Duester G. Retinoic acid biosynthetic enzyme ALDH1 localizes in a subset of retinoid-dependent tissues during xenopus development. Dev Dyn. 1999;215(3):264–72. doi: 10.1002/(SICI)1097-0177(199907)215:3<264::AID-AJA8>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 14.Haselbeck RJ, Hoffmann I, Duester G. Distinct functions for Aldh1 and Raldh2 in the control of ligand production for embryonic retinoid signaling pathways. Dev Genet. 1999;25(4):353–64. doi: 10.1002/(SICI)1520-6408(1999)25:4<353::AID-DVG9>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molotkov A, Duester G. Genetic evidence that retinaldehyde dehydrogenase Raldh1 (Aldh1a1) functions downstream of alcohol dehydrogenase Adh1 in metabolism of retinol to retinoic acid. J Biol Chem. 2003;278(38):36085–90. doi: 10.1074/jbc.M303709200. [DOI] [PubMed] [Google Scholar]
- 16.Whitlock Jr JP. Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol. 1999;39:103–25. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez FJ, Fernandez-Salguero P. The aryl hydrocarbon receptor: studies using the AHR-null mice. Drug Metab Dispos. 1998;26(12):1194–8. [PubMed] [Google Scholar]
- 18.Andreola F, Fernandez-Salguero PM, Chiantore MV, Petkovich MP, Gonzalez FJ, De Luca LM. Aryl hydrocarbon receptor knockout mice (AHR–/–) exhibit liver retinoid accumulation and reduced retinoic acid metabolism. Cancer Res. 1997;57(14):2835–8. [PubMed] [Google Scholar]
- 19.Andreola F, Hayhurst GP, Luo G, Ferguson SS, Gonzalez FJ, Goldstein JA, et al. Mouse liver CYP2C39 is a novel retinoic acid 4-hydroxylase. Its down-regulation offers a molecular basis for liver retinoid accumulation and fibrosis in aryl hydrocarbon receptor-null mice. J Biol Chem. 2004;279(5):3434–8. doi: 10.1074/jbc.M305832200. [DOI] [PubMed] [Google Scholar]
- 20.Elizondo G, Corchero J, Sterneck E, Gonzalez FJ. Feedback inhibition of the retinaldehyde dehydrogenase gene ALDH1 by retinoic acid through retinoic acid receptor alpha and CCAAT/enhancer-binding protein beta. J Biol Chem. 2000;275(50):39747–53. doi: 10.1074/jbc.M004987200. [DOI] [PubMed] [Google Scholar]
- 21.Hsu LC, Chang WC, Hoffmann I, Duester G. Molecular analysis of two closely related mouse aldehyde dehydrogenase genes: identification of a role for Aldh1, but not Aldh-pb, in the biosynthesis of retinoic acid. Biochem J. 1999;339(Pt 2):387–95. [PMC free article] [PubMed] [Google Scholar]
- 22.Giguere V, Ong ES, Segui P, Evans RM. Identification of a receptor for the morphogen retinoic acid. Nature. 1987;330(6149):624–9. doi: 10.1038/330624a0. [DOI] [PubMed] [Google Scholar]
- 23.Leroy P, Nakshatri H, Chambon P. Mouse retinoic acid receptor alpha 2 isoform is transcribed from a promoter that contains a retinoic acid response element. Proc Natl Acad Sci USA. 1991;88(22):10138–42. doi: 10.1073/pnas.88.22.10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19(9):2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart MJ, Dipple KM, Stewart TR, Crabb DW. The role of nuclear factor NF-Y/CP1 in the transcriptional regulation of the human aldehyde dehydrogenase 2-encoding gene. Gene. 1996;173(2):155–61. doi: 10.1016/0378-1119(96)00068-6. [DOI] [PubMed] [Google Scholar]
- 26.Sabatakos G, Davies GE, Grosse M, Cryer A, Ramji DP. Expression of the genes encoding CCAAT-enhancer binding protein isoforms in the mouse mammary gland during lactation and involution. Biochem J. 1998;334(Pt 1):205–10. doi: 10.1042/bj3340205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tajiri S, Yano S, Morioka M, Kuratsu J, Mori M, Gotoh T. CHOP is involved in neuronal apoptosis induced by neurotrophic factor deprivation. FEBS Lett. 2006;580(14):3462–8. doi: 10.1016/j.febslet.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 28.Elizondo G, Medina-Diaz IM. Induction of CYP3A4 by 1alpha, 25-dyhydroxyvitamin D3 in HepG2 cells. Life Sci. 2003;73(2):141–9. doi: 10.1016/s0024-3205(03)00262-5. [DOI] [PubMed] [Google Scholar]
- 29.Deindl E, Boengler K, van Royen N, Schaper W. Differential expression of GAPDH and beta3-actin in growing collateral arteries. Mol Cell Biochem. 2002;236(1–2):139–46. doi: 10.1023/a:1016166127465. [DOI] [PubMed] [Google Scholar]
- 30.Shimada J, Suzuki Y, Kim SJ, Wang PC, Matsumura M, Kojima S. Transactivation via RAR/RXR-Sp1 interaction: characterization of binding between Sp1 and GC box motif. Mol Endocrinol. 2001;15(10):1677–92. doi: 10.1210/mend.15.10.0707. [DOI] [PubMed] [Google Scholar]
- 31.Nagpal S, Athanikar J, Chandraratna RA. Separation of transactivation and AP1 antagonism functions of retinoic acid receptor alpha. J Biol Chem. 1995;270(2):923–7. doi: 10.1074/jbc.270.2.923. [DOI] [PubMed] [Google Scholar]
- 32.DiSepio D, Malhotra M, Chandraratna RA, Nagpal S. Retinoic acid receptor-nuclear factor-interleukin 6 antagonism. A novel mechanism of retinoid-dependent inhibition of a keratinocyte hyperproliferative differentiation marker. J Biol Chem. 1997;272(41):25555–9. doi: 10.1074/jbc.272.41.25555. [DOI] [PubMed] [Google Scholar]
- 33.Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11(4):381–9. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 34.Fawcett TW, Eastman HB, Martindale JL, Holbrook NJ. Physical and functional association between GADD153 and CCAAT/enhancer-binding protein beta during cellular stress. J Biol Chem. 1996;271(24):14285–9. doi: 10.1074/jbc.271.24.14285. [DOI] [PubMed] [Google Scholar]
- 35.de The H, Vivanco-Ruiz MM, Tiollais P, Stunnenberg H, Dejean A. Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene. Nature. 1990;343(6254):177–80. doi: 10.1038/343177a0. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Penzes P, Napoli JL. Cloning of a cDNA encoding an aldehyde dehydrogenase and its expression in Escherichia coli. Recognition of retinal as substrate. J Biol Chem. 1996;271(27):16288–93. doi: 10.1074/jbc.271.27.16288. [DOI] [PubMed] [Google Scholar]
- 37.Guimond J, Devost D, Brodeur H, Mader S, Bhat PV. Characterization of the rat RALDH1 promoter. A functional CCAAT and octamer motif are critical for basal promoter activity. Biochim Biophys Acta. 2002;1579(2–3):81–91. doi: 10.1016/s0167-4781(02)00510-9. [DOI] [PubMed] [Google Scholar]
- 38.Shiohara M, Dawson MI, Hobbs PD, Sawai N, Higuchi T, Koike K, et al. Effects of novel RAR- and RXR-selective retinoids on myeloid leukemic proliferation and differentiation in vitro. Blood. 1999;93(6):2057–66. [PubMed] [Google Scholar]
- 39.Schule R, Rangarajan P, Yang N, Kliewer S, Ransone LJ, Bolado J, et al. Retinoic acid is a negative regulator of AP-1-responsive genes. Proc Natl Acad Sci USA. 1991;88(14):6092–6. doi: 10.1073/pnas.88.14.6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim SW, Kim HJ, Jung DJ, Lee SK, Kim YS, Kim JH, et al. Retinoid-dependent antagonism of serum response factor transactivation mediated by transcriptional coactivator proteins. Oncogene. 2001;20(45):6638–42. doi: 10.1038/sj.onc.1204695. [DOI] [PubMed] [Google Scholar]
- 41.Nicholson RC, Mader S, Nagpal S, Leid M, Rochette-Egly C, Chambon P. Negative regulation of the rat stromelysin gene promoter by retinoic acid is mediated by an AP1 binding site. EMBO J. 1990;9(13):4443–54. doi: 10.1002/j.1460-2075.1990.tb07895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang-Yen HF, Zhang XK, Graupner G, Tzukerman M, Sakamoto B, Karin M, et al. Antagonism between retinoic acid receptors and AP-1: implications for tumor promotion and inflammation. New Biol. 1991;3(12):1206–19. [PubMed] [Google Scholar]
- 43.Nagpal S, Cai J, Zheng T, Patel S, Masood R, Lin GY, et al. Retinoid antagonism of NF-IL6: insight into the mechanism of antiproliferative effects of retinoids in Kaposi's sarcoma. Mol Cell Biol. 1997;17(7):4159–68. doi: 10.1128/mcb.17.7.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gery S, Park DJ, Vuong PT, Chih DY, Lemp N, Koeffler HP. Retinoic acid regulates C/EBP homologous protein expression (CHOP), which negatively regulates myeloid target genes. Blood. 2004;104(13):3911–7. doi: 10.1182/blood-2003-10-3688. [DOI] [PubMed] [Google Scholar]