Abstract
DLC1, a tumor suppressor gene that encodes a RhoGTPase-activating protein, is recurrently downregulated or silenced in various solid tumors and hematological malignancies due to epigenetic modifications or genomic deletion. Here, we identified DLC-1 promoter hypermethylation in 43 out of 44 multiple myeloma (MM) cell lines, which resulted in downregulation or silencing of DLC1 in 41 samples. High frequency of tumor-specific methylation and attenuation or silencing of DLC1 expression could serve as an independent diagnostic marker for MM. Combined treatment with demethylating and acetylating agents significantly elevated the expression of DLC1 and suppressed MM cell proliferation. Two cell lines exhibiting complete promoter methylation and absence of DLC1 expression were transduced by an adenoviral vector containing DLC1 cDNA. In both cell lines reexpression of DLC1 inhibited myeloma cell invasion and migration, reduced RhoA activity and resulted in reorganization of actin cytoskeleton. These results provide the first evidence for antiproliferative effect of DLC1 in a hematological cancer and implicate RhoA pathway in suppression of MM migration and invasion. Given the myeloma cells sensitivity to reactivation of DLC-1 function, the potential for molecular targeted therapy of DLC-1 mediated pathways as well as epigenetic therapies hold prospects.
Keywords: multiple myeloma, DLC1, tumor suppressor gene, promoter methylation, RhoGap, RhoA, migration, invasion
INTRODUCTION
Multiple myeloma (MM), a malignant B-cell tumor, is the second most frequent hematological cancer, with an estimated 16,000 new cases each year in the United States. The incidence of MM is two-fold higher in African American descendents than in Caucasians and affects men more frequently than women.1-3 The disease occurs de novo or evolves from benign monoclonal gammopathy. Clinical manifestations of MM due to accumulation of plasma cells in the bone marrow and excessive levels of monoclonal immunoglobulin can result in osteolytic lesions, anemia, myelosuppression and renal dysfunction. Ionizing radiation and certain environmental chemicals have been linked to development of MM, whereas rare familial cases are suggestive of a genetic susceptibility component. 4-6 Despite significant efforts, the therapy of MM is unsatisfactory and only a fraction of patients survive longer than 10 years after diagnosis. The poor response to therapy could be at least partially attributed to the heterogeneity of the disease that is characterized by cell populations with numerous structural and numerical chromosomal alterations .7
Rapid advancements in array technologies for genome-wide screening of changes in gene expression and DNA copy-number has raised the prospects for a better understanding the pathogenesis of MM, improvements in early detection, new prognostic classification and the identification of new therapeutic targets.2 A recent comprehensive and integrated array comparative genomic hybridization analysis illustrates the potential benefits of this approach. This analysis uncovered distinct genomic subtypes of MM and defined 87 discrete minimal regions of recurrent amplification and deletion that harbor known or still unknown oncogenes and tumor suppressor genes that may be implicated in pathogenesis of MM.6 One of recurrently deleted region in MM is 8p23.3-8p21.3 which contains deleted in liver cancer 1 (DLC1) gene encoding a RhoGTPase-activating protein (RhoGAP). 6,8 In several major types of cancer, reintroduction of DLC1 into tumor cells lacking expression of the endogenous gene suppresses tumor cell proliferation, migration and invasion, induces apoptosis in vitro and reduces or prevents the formation of tumors and metastases in nude mice, 9 and a recent study using shRNA-mediated knockdown of DLC1 expression in a mouse liver tumor model confirmed that DLC1 is a bona fide tumor suppressor gene.10
Mutations in the coding region of DLC1 are rare in human cancers while homozygous or heterozygous deletion of DLC1 were detected in several solid tumors. In liver, lung, colon, and breast tumors, the incidence of heterozygous DLC1 deletions is higher than that of the INK4/ARF, PTEN or p53 tumor suppressor genes.10 Epigenetic mechanisms are, however, predominantly responsible for down regulation and silencing of DLC1 in human cancers and particularly in hematological malignancies.9 Whereas DLC1 is not methylated in normal bone marrow and lymphocytes, DLC1 promoter hypermethylation has been detected in over 80% of patients with acute lymphoblastic leukemia and non-Hodgkin's lymphoma. 11-14 As in other hematological malignancies, DLC1 was found hypermethylated in 78% of patients with MM and in 6 out of 9 MM cell lines. 15,16 In Drosophila RhoGTPases are downstream mediators of the Wnt signaling pathway. There is conclusive evidence that the two signaling pathways are interconnected in mammalian cells, and that both are frequently activated in certain cancers.17, 18 It has been shown that Wnt-mediated migration of MM cells requires activation of RhoA GTPase and some of protein kinase C family members.19 DLC-1 is a RhoGAP specific for RhoA and may thus play a role in myeloma cell migration and invasion.
Given the well-documented antitumor effect of DLC1 in various solid neoplasms and the lack of any information regarding the mechanism of its oncosuppressive activity in hematological malignancies, we decided to examine the involvement of DLC1 in MM. Thus, we screened 44 MM cell lines for abnormal DLC1 promoter methylation and mRNA expression and found that the majority of the cell lines exhibited various degree of promoter hypermethylation that correlated with downregulation or silencing of DLC1 expression. Treatment of two cell lines lacking DLC1 expression due to full promoter hypermethylation with demethylating and acetylating agents significantly augmented the expression of DLC1 and inhibited cell proliferation. Transfer of DLC1 cDNA to these cell lines by an adenoviral vector that was previously successfully used for expressing DLC1 in prostate and hepatocellular carcinoma cells20, 21 resulted in suppression of cell migration and invasion, associated with a significant reduction of RhoA activity and in changes in the amount and distribution of actin content. These observations provide the first evidence for oncosuppressive function of DLC1 in a hematological cancer and raise the possibility of therapeutic intervention in MM.
MATERIALS AND METHODS
Methylation-specific PCR (MSP)
Genomic DNA extracted from multiple myeloma cell lines modified by sodium bisulfite using EZ DNA Methylation Kit (ZYMO Research, Orange, CA) according to the manufacturer's instructions. For detection of aberrant methylation of the DLC1 gene, modified DNA was amplified either using primers (MSP-F, MSP-R) specific for the methylated sequences22 or primers (USP-F, USP-R) specific for the unmethylated sequence of this gene. Hot-started PCR was performed in a 50μl reaction-volume using HotstarTaq DNA Polymerase (Qiagen, Valencia, CA) (condition 95°C, 94°C 30s, 58°C 30s, 72°C 30s for 35 cycles, 72°C 10 min. Eight μl of the products was run on a 2% agarose gel and visualized under UV illumination.
Plasma cell enrichment and Flow cytometry
Plasma cell enrichment was done as previously described 23and B cells were enriched from buffy coat of healthy donors by using StemSep B cell purification Ab mixtures (StemCell Technologies, Vancouver, BC Canada). Purified B cells were stained with various mAb combinations for 20 min on ice in staining buffer (1% BSA and 5% FCS in PBS). The directly conjugated mAb used were anti-CD27-PE, anti-CD19-PerCpCy5. 5, and anti-CD38-allophycocyanin (clone HB7) (BD Immunocytometry, San Jose, CA). Stained cells were washed and data were collected immediately using a four-color FACS calibur (BD Biosciences, Franklin Lakes, NJ). Data were analyzed using FlowJo software (Tree Star). B cell populations were sorted using the Dako Cytomation MoFlo (>98% pure). The plasma cell population, based on the phenotype (CD19low, CD27high, and CD38high), was isolated from the B-cell population for further analysis.
Sodium bisulfite DNA sequencing
Genomic DNA extracted from MM cell lines with either full or heavy methylation was modified by sodium bisulfite using EZ DNA Methylation Kit (ZYMO Research, Orange, CA) according to the manufacturer's instructions. The bisulfite-modified DNA was amplified by PCR using the specific primers (BIS-DLC-F, BIS-DLC-R). PCR products were then subcloned into the pCR2.1-TOPO vector using a TA cloning kit (Invitrogen Carlsbad, CA). To determine the CpG methylation status of the 5’CpG island of DLC-1 gene, 4 clones from each plate were sequenced using an ABI PRISM Dye Deoxyterminator Cycle Sequencing Kit and analyzed on an ABI 377 DNA Sequencer (Applied Biosystems, Foster City, CA).
Cell culture
Human MM cell lines U266, Delta47 and OPM2 were kindly provided by Janet Pumphrey (National Cancer Institute, Bethesda MD) Human bone marrow stromal cell line HS-5 was purchased from the American Type Culture Collection (Manassas, VA). All the cell lines were maintained in RPMI 1640 media supplemented with 10% fetal bovine serum and antibiotics (Biosource, Camarillo, CA) in 5% CO2 and 37°C.
Trichostatin A and 5-aza-2’- deoxycytidine treatment
Cell lines at a density of 2 × 106 per 25 ml flask, were treated with 1 or 5 mM 5-aza-2’-deoxycytidine (5-aza-dC), Sigma–Aldrich, St. Louis, MO) for 72 or 96 hours, 500nM trichostatin A (TSA) (Sigma–Aldrich) for 12 hours, or a combination of 5-aza-dC and TSA 5 (1 or 5 mM for 72 or 96 hours) and TSA (500nM, added during last 12 hours of experiment).
Quantitative and semiquantitative RT –PCR
Total RNA isolation and quantitative real-time RT-PCR of DLC-1 and GAPDH were described previously22. To evaluate differences in DLC1 expression between the samples, the normal bone marrow cell value was used as a calibrator, it's expression of DLC-1 was set as 1 and the difference from DLC-1 expression in myeloma cell lines was revealed by the 2-ΔΔCt method. Significant difference was considered when 2-ΔΔCt was ≥2. Semiquantitative RT-PCR was performed using PCR Supermix (Invitrogen, Carlsbad, CA) and primers DLC-1 7F and DLC-1 9R for amplification of DLC-1. Samples were considered positive when a DLC-1 PCR product was detected after 30 cycles of amplification. GAPDH was used as an internal control of amplification and PCR was carried out using primers GAPDH R and GAPDH L (Table 1).
Table 1.
Primer sequences used for RT-PCR of DLC-1 and GAPDH.
| Primer | Sequence (5' – 3') | Product size (bp) |
|---|---|---|
| DLC-1 7F | CACAGGACAACCGTTGCCTCGA | 465 |
| DLC-1 9R | CTCTTCAGGGTGTTGAGATGGA | |
| GAPDH R | AGGGGAGATTCAGTGTGGTG | 230 |
| GAPDH L | CGACCACTTTGTCAAGCTCA |
Cell proliferation assay
To determine the effect of 5-aza-dC and TSA and their combination on cell growth, 104 5-aza-dC and TSA treated cells were seeded in triplicates on a 96-well plate and incubated for 3 days. Cell proliferation was measured every day by the colorimetric MTT reduction assay (Promega, Madison, WI) using a reader (SPEKTRA Plus, PGC Scientifics, Garner, North Carolina) and filter for 490 nm.
Cell transfection
Myeloma cell lines at a density of 106 cells in 3 ml total volume (6-well plate) were transfected with an adenovirus vector pAd/CMV/V5-GW containing cDNA encoding the open reading frame of DLC-1 or LacZ, for 72 hours according to the manufacturer's protocol (Invitrogen, Carlsbad CA). Efficiency of adenovirus transfection of LacZ was checked by b-galactosidase staining kit (Invitrogen, Carlsbad, CA) according to manufacturer's protocol. Transfected cells were fixed with Fixative Solution and then stained for 4 hours in 37°C. Blue colored cells were counted under a microscope. The reexpression of DLC-1 was checked by real-time RT-PCR.
Transendothelial migration and invasion assay
Transendothelial migration of MM cell lines U266 and delta47 was performed as described previously.24 Human recombinant Insulin-like Growth Factor I (IGF) at concentration of 50ng/ml was added to the lower chamber as a decoy chemoattractant. Matrigel invasion chambers (BD Bioscience, Bedford, MA) were seeded with 5 × 104 cells/well and incubated overnight in 37°C, 50ng/ml Insulin-like Growth Factor I (IGF) was added into lower chamber as a decoy chemoattractant. At the end of incubation period inserts were cleaned from Matrigel, fixed in 75% ethanol for 5 minutes and stained by 0.05% crystal violet for 2 minutes according to manufacturer's protocol. Invading cells were counted under microscope. All migration and invasion experiments were performed in triplicates.
RhoA activation assay
The relative RhoA activity in two MM cell lines, Delta 47 and U266, was measured by using an ELISA-based RhoA activation assay kit (G-LISA, Cytoskeleton, Inc. Denver, CO), according to the manufacturer's instructions. The cells were transduced with Ad-DLC1 and Ad-LacZ for 72 hours. After serum-starvation and lysophosphatidic acid (LPA) stimulation, cells were lysed and the protein concentrations were determined according to the manufacturer's protocol. The lysates were incubated in microwells coated with the Rhotekin Rho-binding domain, and the levels of active RhoA were measured using indirect immunodetection followed by a colorimetric reaction measured by absorbance at 490 nm.
Fluorescence microscopy analysis
For detection of F-actin, MM cells were cultured as described and centrifuged onto precleaned microscopic slides using Cyto-System attachments in a Heraeus Megafuge 1R centrifuge, at 800 rpm or 1200 rpm for 5 minutes. Slides were fixed in paraformaldehyde, inspected under phase contrast microscope and stained with Rhodamine-phalloidin according to earlier published protocol.25 Multiple sequential grayscale images were recorded using Photometrics Cold Snap HQ CCD camera mounted on Zeiss Axiophot microscope equipped with motorized shutter and filter wheel and controlled by Apple Macintosh computer trough IP Lab Spectrum software.
RESULTS AND DISCUSSION
Aberrant DLC1 promoter methylation and mRNA expression
Given the high frequency of DLC1 expression downregulation or inactivation mediated by epigenetic mechanisms in various hematological malignancies, including patients with MM, we examined the DLC1 promoter methylation status and mRNA expression in 44 MM cell lines, and in samples of plasma cells derived from normal donors. As expected, we found no signs of DLC1 promoter methylation in normal plasma cells from healthy donors and in only one of the MM cell lines, H929 (Figure 1). The remaining cells lines exhibited different degrees of abnormal methylation, ranging from light to full (Table 2). The methylation patterns of the 35 CpG sites in the promoter region of the DLC1 gene were determined by bisulfite genomic DNA sequencing. To see the effect of aberrant promoter methylation on DLC1 mRNA expression, RNA from the 44 MM cell lines and from samples of plasma cells derived from normal donors were screened by real time PCR (Figure 2) and conventional RT-PCR. Only three cell lines expressed DLC1 at levels comparable with those in normal plasma cells, whereas in the remaining lines downregulation or loss of DLC1 expression was found (Table 2).
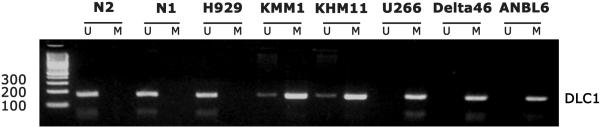
Figure 1.
Methylation-specific PCR detection of bisulfite-modified and amplified unmethylated (U) and methylated (M) sequences of DLC1 gene in two normal plasma cell samples (N1, N2) and six multiple myeloma cell lines shows different levels of DLC1 promoter methylation from none (N1, N2, H929), to moderate (KMM1, KHM11), to total methylation (U266, Delta47, ANBL6).
Table 2.
DLC1 Gene Promotor methylation status and gene's mRNA expression in 44 multiple myeloma cell lines and 2 normal control plasma cells.
| Cell Line | Promotor Methylation | DLC1 mRNA Expression | Cell Line | Promotor Methylation | DLC1 mRNA Expression |
|---|---|---|---|---|---|
| ANBL6 | Full | Undetectable | MM-M1 | Partial | Undetectable |
| ARP1 | Full | Undetectable | OCIMY1 | Partial | Undetectable |
| DELTA47 | Full | Undetectable | PE2 | Partial | Undetectable |
| FR4 | Full | Undetectable | SKMM2 | Partial | Undetectable |
| KMS12BM | Full | Undetectable | UTMC2 | Partial | Undetectable |
| KMS12PE | Full | Undetectable | XG1 | Partial | High |
| OCI-MY5 | Full | Undetectable | XG2 | Partial | Undetectable |
| U266 | Full | Undetectable | XG6 | Partial | Undetectable |
| KHM11 | Heavy | Undetectable | XG7 | Partial | Undetectable |
| KMM1 | Heavy | Undetectable | FLAM76 | Light | Undetectable |
| SKMM1 | Heavy | Undetectable | JIM3 | Light | Low |
| 8226 | Partial | Undetectable | KMS18 | Light | Undetectable |
| EJM | Partial | Undetectable | KMS26 | Light | Undetectable |
| H1112 | Partial | Undetectable | L363 | Light | Undetectable |
| INA6 | Partial | Undetectable | LP1 | Light | Low |
| JJN3 | Partial | High | MM1-144 | Light | Low |
| JK-6L | Partial | Undetectable | OCI-MY7 | Light | Undetectable |
| KARPAS 620 | Partial | Undetectable | OPM1 | Light | Undetectable |
| KHM1B | Partial | Low | OPM2 | Light | Low |
| KMS11 | Partial | Undetectable | PE1 | Light | Low |
| KMS28BE | Partial | Undetectable | H929 | None | Low |
| KMS28PE | Partial | High | NORMAL1 | None | High |
| KMS34 | Partial | Undetectable | NORMAL2 | None | High |
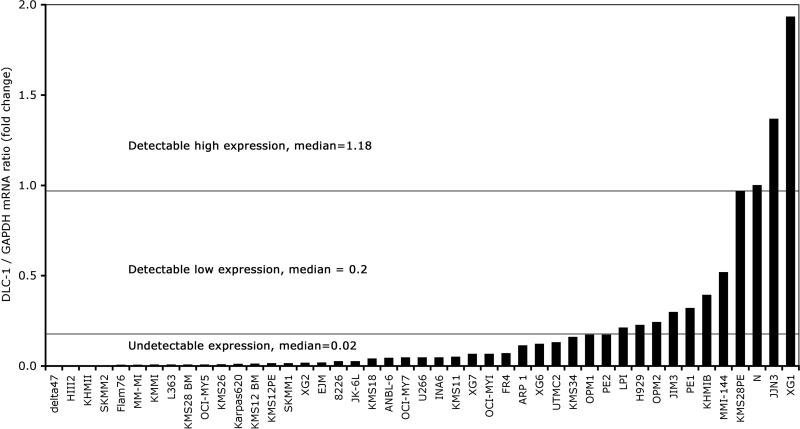
Figure 2.
Expression of DLC-1 mRNA in multiple myeloma cell lines determined by real-time RT-PCR. Gene expression was normalized to the expression of control gene GAPDH. Normal bone marrow was used as a calibrator, it's expression of DLC-1 was set at 1 and the difference from DLC-1 expression in myeloma cell lines was revealed by the 2-ΔΔCt method. Samples were divided into three subgroups according to the level of expressed DLC-1 mRNA and median of expression was determined for each subgroup.
Gene silencing by aberrant promoter methylation or histone deacetylation in various types of human cancers has been reported at least as frequently as inactivation by means of mutation or deletion.26, 27Molecular studies of MM so far have largely addressed the role of genetic abnormalities acquired through specific chromosome rearrangements, while they focused considerably less on the impact of epigenetic modifications, a major mechanism associated with loss of tumor suppressor gene function.28 The high frequency of abnormal, tumor-specific, methylation of DLC1 promoter observed in this series of MM cell lines, as well as in patients with MM,15 exceeds the rate of epigenetic silencing of several other tumor suppressor genes known to be involved in pathogenesis of this cancer.28 It, therefore, may provide a useful independent marker for diagnosis of this neoplasm. More important, however, would be to establish a correlation between DLC1 methylation pattern and clinical characteristics in a large cohort–of MM patients, as it was done in the case of p16 gene, where abnormal methylation at diagnosis was associated with unfavorable prognosis.28
Restoration of DLC-1 expression by 5-aza-2’-deoxycytidine and trichostatin A treatment
Although promoter methylation is the predominant mechanism of epigenetic DLC1 silencing, histone deacetylation is also an important component in this process, as demonstrated in experiments with DNA samples from patients and cell lines derived from MM and prostate carcinoma.15,22 Furthermore, optimal reexpression of epigenetically silenced genes in myelodysplastic syndrome or acute myeloid leukemia was achieved, both in vitro and in vivo, when application of DNA methyltransferase inhibitors was sequentially followed by histone deacetylase inhibitors.29 Given this proven approach, MM U266 and Delta47 cells, that lacked intrinsic DLC-1 expression, were treated with 5-aza-dC , TSA, or by combination of both drugs.
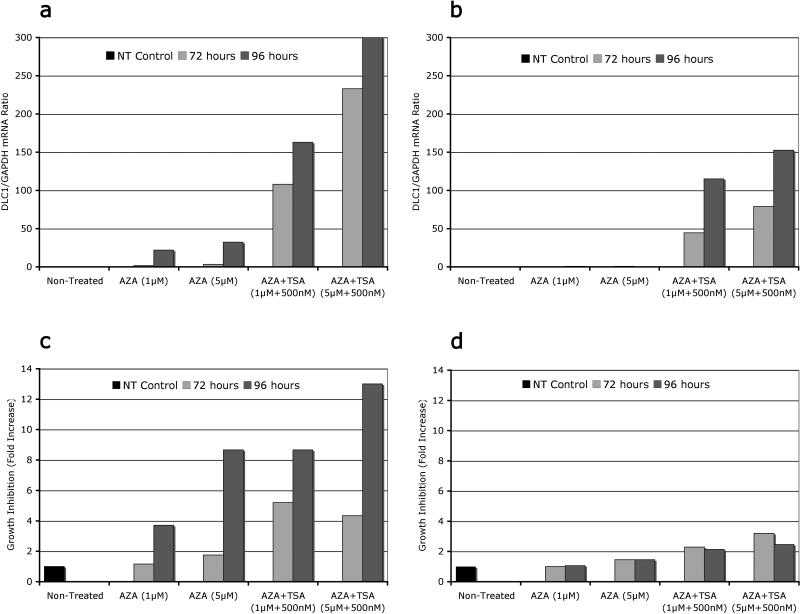
Although treatment with 5-aza-dC was able to restore DLC1 expression in Delta 47 cells, that effect in U266 was negligible. Only combined treatment with 5-aza-dC and TSA significantly restored expression of DLC1 in both cell lines (Figure 3a, b), albeit the response was again more pronounced in the case of Delta 47 cells. The response to treatment was equally dose-and time dependent. Longer treatment (96 hours) with higher dose of 5-aza-dC (5μM) invariably produced a stronger response. The 5 aza dC-and TSA-mediated induction of DLC1 mRNA was paralleled by a decrease in cell proliferation (Figure 3c, d). Again, the effect in Delta47 cells was stronger than in U266, whereas the dynamics of growth inhibition followed only partially the dose- and/or time-dependent pattern. These results have significant therapeutic implications in MM, as downregulation or silencing of tumor suppressor genes by promoter methylation and histone deacetylation makes them an attractive target for epigenetic therapy with DNA methylation inhibitors and histone deacetylase inhibitors. A powerful single or combinatorial protocol would consists of treatment with zebularine, a stable DNA cytosine DNA methylation inhibitor that preferentially targets cancer cells, and suberoylanilide hydroxamic acid (SAHA), a potent histone deacetylase inhibitor already tested in clinical trials for advanced leukemia and myelodysplastic syndromes.30, 31 This potential therapeutic approach was recently highlighted in a remarkable article outlining several therapeutic options in tumors with disabled DLC1. 32
Figure 3.
Different concentrations and durations of treatment with 5-Aza-dC (AZA) alone or 5-Aza-dC (AZA+TSA) combination, cause different levels of DLC1 reexpression (A, B) and subsequent inhibition of proliferation (C, D) in multiple myeloma cell lines Delta 47 (A, C) and U266 (B, D)
Adenovirus-mediated restoration of DLC1 expression and its effect on MM cell migration and invasion
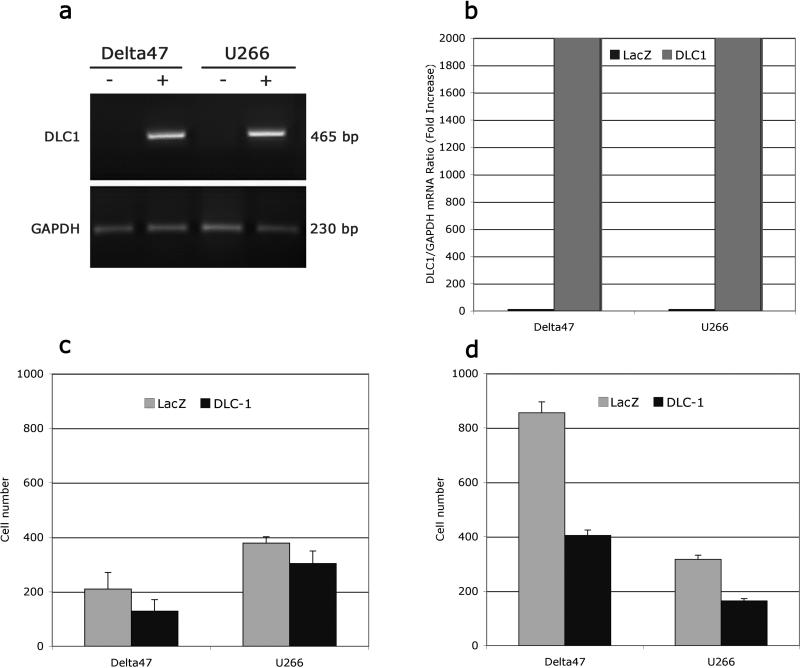
To address the role of DLC1 on the capacity of MM cells to migrate and invade, we first examined Delta 47 and U266 cells by RT-PCR and real time RT-PCR analysis after adenovirus–mediated DLC1 transduction. In both cell lines we found a robust increase in DLC1 expression in DLC1 transduced cell compared to controls (Figure 4a, b). Then we used a transwell migration assay and matrigel invasion assay to measure the ability of MM cells transfected with DLC1 or the LacZ control to penetrate the layer of stromal bone morrow cells. In both cell lines the DLC1-positive cells showed significantly a lower rate of migration (Figure 4c) than those transfected by LacZ. DLC1 expression reduced migration of Delta47 and U266 cells by 39% and 20% respectively. Experiments using Matrigel coated invasion chambers revealed that reexpression of DLC1 was considerably more effective in inhibiting MM cell invasion. AdDLC1 transduction of Delta 47 and U266 cells resulted in reduction of cell invasion of 53% and 48%, respectively, compared to LacZ control samples (Figure 4d). DLC1-mediated suppression of cell migration and invasion is of significant importance as the two processes are instrumental in the myeloma cell movement within the bone marrow and metastasis to secondary sites19. Existing evidence suggests that DLC1 meets the criteria of a metastasis suppressor gene,33 as restoration of DCL1 inhibited the dissemination of human hepatocellular carcinoma and breast cancer cells to distant organs in nude mice. 21,34
Figure 4.
Adenovirus mediated transfection of multiple myeloma Delta47 and U266 cells with the DLC1 cDNA resulted in restored expression of the gene (A) and abundance of DLC1 mRNA (B), while transfection with control LacZ vector had no effect. Only transiently DLC1-positive Delta47 and U266 cells exhibited reduced ability to migrate (C) and decreased invasive capability (D) as measured by transendothelial migration and invasion essay.
DLC1 effect on RhoA activity and cytoskeleton organization
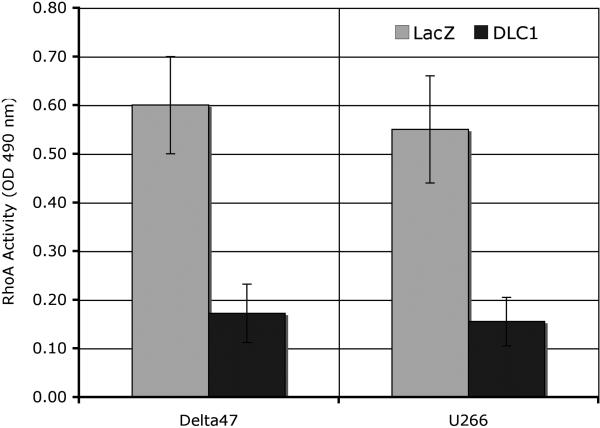
Given the findings that DLC1-mediated inhibition of liver, lung or prostate tumor cell growth is associated with suppression of RhoA activity, and evidence that the Rho pathway may play a role in malignant cell invasion in MM, we sought to see whether restoration of DLC1 function was accompanied with a reduction of RhoA activity. The levels of active, GTP-bound RhoA (Figure 5) in DLC1-transduced Delta 47 and U266 cells were 60 to 70% lower, respectively, than in LacZ-transduced cells (P=0.037). Since the identification of the small GTPase, the first human oncogene, there has been a sustained interest in whether RhoGTPase like ras contribute to cell transformation and tumor progression.35, 36 Rho-GTPase activity is frequently deregulated in human cancers and experimental evidence has implicated Rho-GTPase signaling in promoting the growth and invasiveness of tumor cells. 36-38 In the past several years it became evident that loss of DLC1 expression is one of most frequent mechanism for the aberrant activation of Rho GTPases in human oncogenesis. DLC1-mediated negative regulatory effect of cell growth and tumorigenicity is primarily due to RhoGAP ability to inactivate Rho proteins and several groups found that DLC1 is a potent GAP for Rho A. 20, 39-41
Figure 5.
Delta47 and U266 cells, in which high level of DLC1 gene expression was transiently restored by Ad-DLC1 transfection, show 60 to 70% decrease in the levels of active GTP-bound RhoA as compared to cells transfected with control vector Ad-LacZ.
While the suppressive effect of ectopic restoration of DLC1 expression on the ability of human cancer cell lines to form primary tumors and metastases was demonstrated by several independent groups, 9 the role of loss of DLC1 in promoting neoplasia was recently demonstrated in a landmark study using an original mouse model for hepatocellular carcinoma.10 DLC1 knock-down cooperated with c-myc overexpression to induce hepatocellular carcinomas in mice.10 The loss of DLC1 conferred dependency on the RhoA pathway, as suppression of RhoA inhibited the growth of DLC1-negative hepatoma cells.10 Such events are recapitulated in pathogenesis of MM. Amplification or complex chromosomal translocations deregulate myc gene during progression of MM and as shown here reintroduction of DLC1 decreased RhoA activity and suppressed myeloma cells migration and invasion.2,7,42 RhoA can be activated in response to B cell receptor (BCR) signaling and regulates BCR-dependent cell proliferation.43 Conceivably, ectopic restoration of DLC1 expression may interfere with BCR signaling resulting in reduction of RhoA activity and inhibition of myeloma cell proliferation.
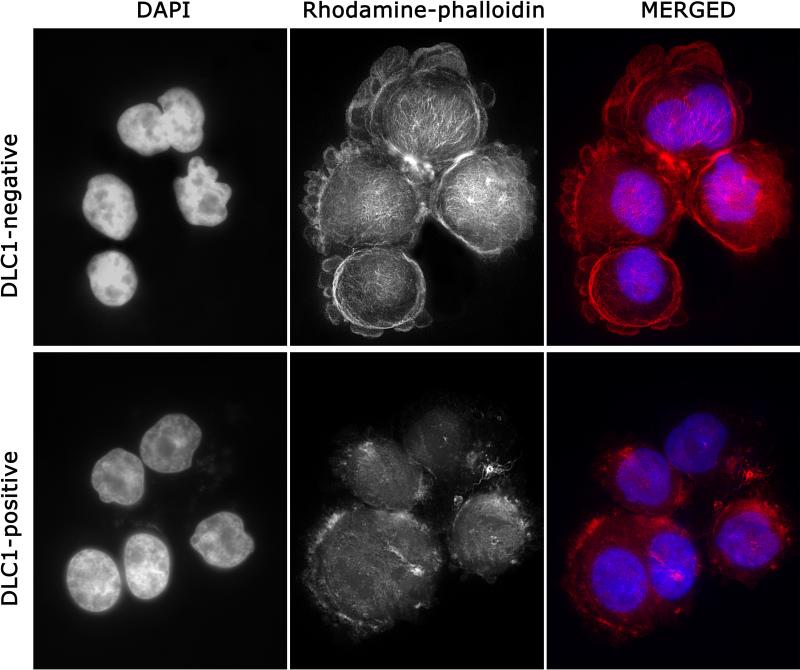
Since Rho proteins play an important role in the control of actin cytoskeleton organization and focal adhesion assembly we examined the effect of DLC1 expression on cytoskeletal organization in MM cells. 44, 45 DLC1-negative Delta47 cells showed a dense web of F-actin with strong presence on the cell periphery and cytoplasmic protrusions, and detectable fibers throughout the cytoplasm (Figure 6, top). Cells in which DLC1 expression was restored by adenoviral transduction (Figure 6, bottom) showed an overall reduction in F-actin, with sporadic accumulations on the perimeter, and almost no intracytoplasmic fibers. Although relatively little data are available on the characteristics of the MM cell cytoskeleton, our results appear to be consistent with observations that the amount of detectable F-actin and its intracellular distribution in MM cells fluctuates relative to cell motility; 46 motile cells show an increased overall density of F-actin in both the cortical band and cytoplasm in contrast to immotile cells, which contain fewer actin filaments, localized almost exclusively to the cell perimeter.
Figure 6.
Combined DAPI and Rhodamine-phalloidin staining of DLC1-negative and DLC1-positive Delta47 myeloma cells shows changes in the amount and organization of F-actin associated with restored expression of the DLC1 gene
Treatment of MM cells with Wnt or IGF-1 has been shown to cause rearrangements of the actin that are associated with activation of RhoA.19, 24, 47 The levels of F-actin and changes in its intracellular distribution in DLC1-transduced MM cells seem to be a consequence of the DLC1-mediated reduction of RhoA activity and is consistent with observations on cytoskeleton changes in DLC1-transduced prostate, liver and lung tumor cells.21, 39, 48 It has been shown that Wnt-mediated migration of MM cells requires activation of RhoA GTPase and DLC-1, a potent GAP for RhoA, may thus play a critical role in myeloma cell migration and invasion.19
In conclusion, our results show for the first time that DLC1 has oncosuppressive activity in a hematological cancer. Signaling pathways modulated by DLC1 may have wide therapeutic applications. 10, 32,49 Current observations show that DLC1 suppression of myeloma cell motility involves RhoA pathway that should be considered as one of the potential molecular targets for therapeutic interventions.
Acknowledgments
This research was supported by the Intramural Research Program of the National Cancer Institute, NIH. We thank Dr. Michael Kuehl (Center for Cancer Research, National Cancer Institute, Bethesda MD) for providing DNA and RNA samples isolated from multiple myeloma cell lines, critical reading of the manuscript and most helpful suggestions. We also thank to Dr. Sheng Chen for kindly providing plasma cells samples from healthy donors.
REFERENCES
- 1.Ries LAG, et al., editors. SEER Cancer Statistics Review, 1975–2004. National Cancer Institute; Bethesda, USA: 2007. [Google Scholar]
- 2.Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7:585–598. doi: 10.1038/nrc2189. [DOI] [PubMed] [Google Scholar]
- 3.Kyle RA, Rajkumar SV. Multiple myeloma. Blood. 2008;111:2962–2972. doi: 10.1182/blood-2007-10-078022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent clasification of multiple myeloma. J. Clinical Oncology. 2005;23:6333–6338. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 5.Spink CF, Gray LC, Davies FE, Morgan GJ, Bidwell JL. Haplotypic structure across the I kappa B alpha gene (NFKBIA) and association with multiple myeloma. Cancer Lett. 2007;246:92–99. doi: 10.1016/j.canlet.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9:313–325. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 7.Kuehl WM, Bergsagel PL. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer. 2002;2:175–187. doi: 10.1038/nrc746. [DOI] [PubMed] [Google Scholar]
- 8.Yuan BZ, Miller MJ, Keck CL, Zimonjic D, Thorgeirsson SS. Cloning, characterization and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homogous to rat RhoGAP. Cancer Res. 1998;58:2196–2199. [PubMed] [Google Scholar]
- 9.Durkin ME, Yuan BZ, Zhou X, Zimonjic DB, Lowy DR, Thorgeirsson SS, et al. DLC-1: a Rho GTPase-activating protein and tumour suppressor. J Cell Mol Med. 2007;11:1185–1207. doi: 10.1111/j.1582-4934.2007.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue W, Krasnitz A, Lucito R, Sordella R, VanAelst L, Cordon-Cardo C, et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22:1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi H, Guo J, Duff DJ, Rahmatpanah F, Chitima-Matsiga R, Al-Kuhlani M, et al. Discovery of novel epigenetic markers in non-Hodgkin's lymphoma. Carcinogenesis. 2007;28:60–70. doi: 10.1093/carcin/bgl092. [DOI] [PubMed] [Google Scholar]
- 12.Taylor KH, Pena-Hernandez KE, Davis JW, Arthur GL, Duff DJ, Shi H, et al. Large-scale CpG methylation analysis identifies novel candidate genes and reveals methylation hotspots in acute lymphoblastic leukemia. Cancer Res. 2007;67:2617–2625. doi: 10.1158/0008-5472.CAN-06-3993. [DOI] [PubMed] [Google Scholar]
- 13.Pike BL, Greiner TC, Wang X, Weisenburger DD, Hsu YH, Renaud G, et al. DNA methylation profiles in diffuse large B-cell lymphoma and their relationship to gene expression status. Leukemia. 2008;22:1035–1043. doi: 10.1038/leu.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ying J, Li H, Murray P, Gao Z, Chen YW, Wang Y, et al. Tumor-specific methylation of the 8p22 tumor suppressor gene DLC1 is an epigenetic biomarker for Hodgkin, nasal NK/T-cell and other types of lymphomas. Epigenetics. 2007;2:15–21. doi: 10.4161/epi.2.1.3883. [DOI] [PubMed] [Google Scholar]
- 15.Song YF, Xu R, Zhang XH, Chen BB, Chen Q, Chen YM, et al. High-frequency promoter hypermethylation of the deleted in liver cancer-1 gene in multiple myeloma. J Clin Pathol. 2006;59:947–951. doi: 10.1136/jcp.2005.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu L, Hatjiharissi E, Ciccareli BT, Roccaro AM, Adamia S, Sacco A, Hunter ZR, et al. Expression of the deleted in liver cancer-1 gene is regulated by DNAmethylation and is a target for therapy in Waldenstrom's macroglobulinemia. J Clin Oncol. 2008:26. abstr 19505. [Google Scholar]
- 17.Strutt DI, Weber U, Mlodzik M. The role of RhoA in tissue polarity and Frizzled signalling. Nature. 1997;387:292–295. doi: 10.1038/387292a0. [DOI] [PubMed] [Google Scholar]
- 18.Thompson MD, Monga SP. WNT/beta-catenin signaling in liver health and disease. Hepatology. 2007;45:1298–1305. doi: 10.1002/hep.21651. [DOI] [PubMed] [Google Scholar]
- 19.Qiang YW, Walsh K, Yao L, Kedei N, Blumberg PM, Rubin JS, et al. Wnts induce migration and invasion of myeloma plasma cells. Blood. 2005;106:1786–1793. doi: 10.1182/blood-2005-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan M, Tripathi V, Zhou X, Popescu NC. Adenovirus-mediated restoration of the expression of the tumor suppressor gene DLC1 inhibits the proliferation and tumorigenicity of aggressive, androgen-independent human prostate cancer cell lines: prospects for gene therapy. Cancer Gene Therapy. 2008;15:371–378. doi: 10.1038/cgt.2008.13. [DOI] [PubMed] [Google Scholar]
- 21.Zhou X, Zimonjic DB, Park S-W, Yang X-Y, Durkin ME, Popescu NC. DLC1 suppresses distant dissemination of human hepatocellular carcinoma cells in nude mice through reduction of RhoA GTPase activity, actin cytoskeletal disruption and down regulation of genes involved in metastasis. Int J Oncology. 2008;32:1285–1291. doi: 10.3892/ijo_32_6_1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan M, Zhou X Soulitzis N, Spandidos DA, Popescu NC. Abberant Methylation and Deacetylation of deleted in Liver Cancer-1 gene in Prostate Cancer: Potential Clinical Applications. Clin Cancer Res. 2006;12:1412–1419. doi: 10.1158/1078-0432.CCR-05-1906. [DOI] [PubMed] [Google Scholar]
- 23.Chen S, Sims GP, Chen XX, Gu YY, Chen S, Lipsky PE. Modulatory effects of 1, 25-dihydroxyvitamin D3 on human B cell differentiation. J Immunol. 2007;179:1634–1647. doi: 10.4049/jimmunol.179.3.1634. [DOI] [PubMed] [Google Scholar]
- 24.Qiang YW, Yao L, Tosato G, Rudikoff S. Insulin-like growth factor I induces migration and invasion of human multiple myeloma cells. Blood. 2004;103:301–308. doi: 10.1182/blood-2003-06-2066. [DOI] [PubMed] [Google Scholar]
- 25.Durkin ME, Avner MR, Huh CG, Yuan BZ, Thorgeirsson SS, Popescu NC. DLC-1, a Rho GTPase-activating protein with tumor suppressor function, is essential for embryonic development. FEBS Lett. 2005;579:1191–1196. doi: 10.1016/j.febslet.2004.12.090. [DOI] [PubMed] [Google Scholar]
- 26.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2003;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 27.Herman JG, Baylin SB. Mechanisms of disease: Gene silencing in cancer in association with promoter hypermethylation. New England Journal Of Medicine. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 28.Galm O, Wilop S, Reichelt J, Jost E, Gehbauer G, Herman JG, et al. DNA methylation changes in multiple myeloma. Leukemia. 2004;18:1687–1692. doi: 10.1038/sj.leu.2403434. [DOI] [PubMed] [Google Scholar]
- 29.Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66:6361–6369. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 30.Cheng JC, Yoo CB, Weisenberger DJ, Chuang J, Wozniak C, Liang G, et al. Preferential response of cancer cells to zebularine. Cancer Cell. 2004;6:151–158. doi: 10.1016/j.ccr.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Manero G, Yang H, Bueso-Ramos C, Ferrajoli A, Cortes J, Wierda WG, et al. Phase 1 study of the histone deacetylase inhibitor vorinostat (suberoylanilide hydroxamic acid (SAHA)) in patients with advanced leukemias and myelodysplastic syndromes. Blood. 2008;111:1060–1066. doi: 10.1182/blood-2007-06-098061. [DOI] [PubMed] [Google Scholar]
- 32.Lahoz A, Hall A. DLC1:a significant GAP in the cancer genome. Genes Dev. 2008;22:1724–1730. doi: 10.1101/gad.1691408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stafford LJ, Vaidya KS, Welch DR. Metastasis suppressor genes in cancer. The Int J Biochem Cell Biol. 2008;40:874–891. doi: 10.1016/j.biocel.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 34.Goodison S, Yuan J, Sloan D, Kim R, Li C, Popescu NC, et al. The RhoGAP protein DLC-1 functions as a metastasis suppressor in breast cancer cells. Cancer Res. 2005;65:6042–6053. doi: 10.1158/0008-5472.CAN-04-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santos E, Tronick SR, Aaronson SA, Pulciani S, Barbacid M. T24 human bladder carcinoma oncogene is an activated form of the normal human homologue of BALB-and Harvey-MSV transforming genes. Nature. 1982;298:343–347. doi: 10.1038/298343a0. [DOI] [PubMed] [Google Scholar]
- 36.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 37.Sahai EM, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 38.Gomez del Pulgar T, Benitah SA, Valeron PF, Espina C, Lacal JC. Rho GTPase expression in tumourigenesis: evidence for a significant link. Bioessays. 2005;27:602–613. doi: 10.1002/bies.20238. [DOI] [PubMed] [Google Scholar]
- 39.Wong CM, Lee JM, Ching YP, Jin DY, Ng IO. Genetic and epigenetic alterations of DLC-1 gene in hepatocellular carcinoma. Cancer Res. 2003;63:7646–7651. [PubMed] [Google Scholar]
- 40.Qian X, Li G, Asmussen HK, Asnaghi L, Vass WC, Braverman R, et al. Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities. Proc Natl Acad Sci U S A. 2007;104:9012–9017. doi: 10.1073/pnas.0703033104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Healy KD, Hodgson L, Kim TY, Shutes A, Maddileti S, Juliano RL, et al. DLC-1 suppresses non-small cell lung cancer growth and invasion by RhoGAP-dependent and independent mechanisms. Mol Carcinog. 2007;47:326–337. doi: 10.1002/mc.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gabrea A, Martelli ML, Qi Y, Roschke A, Barlogie B, Shaughnessy JD, et al. Secondary genomic rearrangements involving immunoglobulin or MYC loci show similar prevalences in hyperdiploid and nonhyperdiploid myeloma tumors. Genes Chromosomes Cancer. 2008;47:573–590. doi: 10.1002/gcc.20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saci A, Carpenter CL. RhoA GTPase regulates B cell receptor signaling. Mol Cell. 2005;17:205–214. doi: 10.1016/j.molcel.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 44.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 45.Wheeler AP, Ridley AJ. Why three Rho proteins? RhoA, RhoB, RhoC, and cell motility. Exp Cell Res. 2004;301:43–49. doi: 10.1016/j.yexcr.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 46.Menu E, Braet F, Timmers M, Van Rieta I, Van Camp B, Vanderkerken K. The F-actin content of multiple myeloma cells as a measure of their migration. Ann. N. Y. Acad. Sci. 2002;973:124–134. doi: 10.1111/j.1749-6632.2002.tb04620.x. [DOI] [PubMed] [Google Scholar]
- 47.Qiang YW, Endo Y, Rubin JS, Rudikoff S. Wnt signaling in B-cell neoplasia. Oncogene. 2003;22:1536–1545. doi: 10.1038/sj.onc.1206239. [DOI] [PubMed] [Google Scholar]
- 48.Yuan BZ, Jefferson AM, Millecchia L, Popescu NC, Reynolds SH. Morphological changes and nuclear translocation of DLC1 tumor suppressor protein precede apoptosis in human non-small cell lung carcinoma cells. Exp Cell Res. 2007;313:3868–3880. doi: 10.1016/j.yexcr.2007.08.009. 2007. [DOI] [PubMed] [Google Scholar]
- 49.Wong CC, Wong CM, Ko FC, Chan LK, Ching YP, Yam JW, et al. Deleted in liver cancer 1 (DLC1) negatively regulates Rho/ROCK/MLC pathway in hepatocellular Carcinoma. PLoS ONE. 2008;3:e2779. doi: 10.1371/journal.pone.0002779. doi: 10.1371/journal.pone.0002779. [DOI] [PMC free article] [PubMed] [Google Scholar]