Abstract
Two hundred twenty-two individuals of the southern plains woodrat (Neotoma micropus) were captured from 198 excavated middens at 10 discrete collecting sites from a single population in south-central Texas. Field data, mitochondrial D-loop haplotypes, and polymorphic microsatellite loci (5–7) were used to determine genetic patterns in parentage, relatedness, and mating strategy. Microsatellite loci were highly polymorphic (average observed heterozygosity = 0.859) and were used to construct genotypes that were unique for each individual (probability of identical genotypes: 1 in 2,104,567). Results indicated a high frequency of multiple paternity (6 of 9 litters), evidence of repeat mating between the same 2 individuals, and no indication of male dominance at any collection site. Examination of these data suggested a promiscuous mating system. Within a site, average relatedness between adult females was similar to that between adult males. A higher level of cohabitation from that previously documented was recorded and finer-scale analyses revealed high levels of relatedness between most cohabiting individuals. Taken with results from other studies of mating behaviors of N. micropus, our results suggest that mating and social behavior of this species are likely influenced by population density.
Keywords: cohabitation, mating system, microsatellites, multiple paternity, relatedness
Several factors (e.g., demography, density, resource competition and availability, life-history characteristics, spatial distribution, and dispersal of individuals) influence mating systems of a given species. Consequently, mating strategies may differ across the geographic range of a species (Clutton-Brock 1989; Jones et al. 2001; Nievergelt et al. 2002), especially if aforementioned factors vary. In recent years, genetic studies have been used to corroborate field data studies of mating systems (Avise 1994; Queller et al. 1993). Examples of the utility of genetic data include examination of relatedness between individuals (Fournier et al. 2002; Onorato et al. 2004; Painter et al. 2000; Taylor et al. 1997; Wilmer et al. 2000; Yu et al. 2001), maternal and paternal lineage assessment (Haynie et al. 2003; Jones and Avise 1997; Kays et al. 2000; Onorato et al. 2004), occurrence of multiple mating events (Baker et al. 1999; Berteaux et al. 1999; Burton 2002; Carling et al. 2003; Valenzuela 2000), and assessment of reproductive success (Blanchfield et al. 2003; Coltman et al. 1999; Fabiani et al. 2004; Matocq 2004; Scribner et al. 1993; Topping and Millar 1998).
Although genetic information has been increasingly applied to studies of mating strategies for a variety of mammalian taxa, rodents remain a primary model. Advantageous characteristics of this group include well-documented natural history attributes for many species; short gestation periods with >1 offspring per litter and several litters per year (Topping and Millar 1998); distributions encompassing a variety of habitats; and occurrence of several mating systems, including monogamy (Ribble 1991), polygyny (Rusu and Krackow 2004), and promiscuity (Heske and Ostfeld 1990; Matocq 2004; Schulte-Hostedde et al. 2004).
The southern plains woodrat (Neotoma micropus) has been characterized as solitary, asocial (Conditt and Ribble 1997; Johnson 1952; Raun 1966), and promiscuous, although it was noted that the mating system could range from polygyny to promiscuity (Conditt and Ribble 1997). Woodrats are known for their conspicuous aboveground nests or middens (Birney 1973; Raun 1966) and long-term occupation of these sites (Matocq and Lacey 2004). Middens can be located easily, providing an efficient method for capturing individuals. Additionally, numerous studies documenting natural history attributes of N. micropus are available (Birney 1973; Conditt and Ribble 1997; Finley 1958; Henke and Smith 2000; Johnson 1952; Merkelz and Kerr 2002; Raun 1966; Suchecki et al. 2004; Thies and Caire 1990, 1991), as well as studies regarding population genetic data (Mendez-Harclerode et al. 2005, 2007). These characteristics make the southern plains woodrat an excellent model for studies of mating strategies and social structure.
We used field and molecular genetics methods to determine genetic patterns in parentage, relatedness, and mating strategy of the southern plains woodrat. Data from 10 collection sites in south-central Texas were examined individually and together to investigate social parameters for this population, and to assess the utility of methods appropriate to examine discrete point-in-time data in mating structure studies.
MATERIALS AND METHODS
Collection site selection
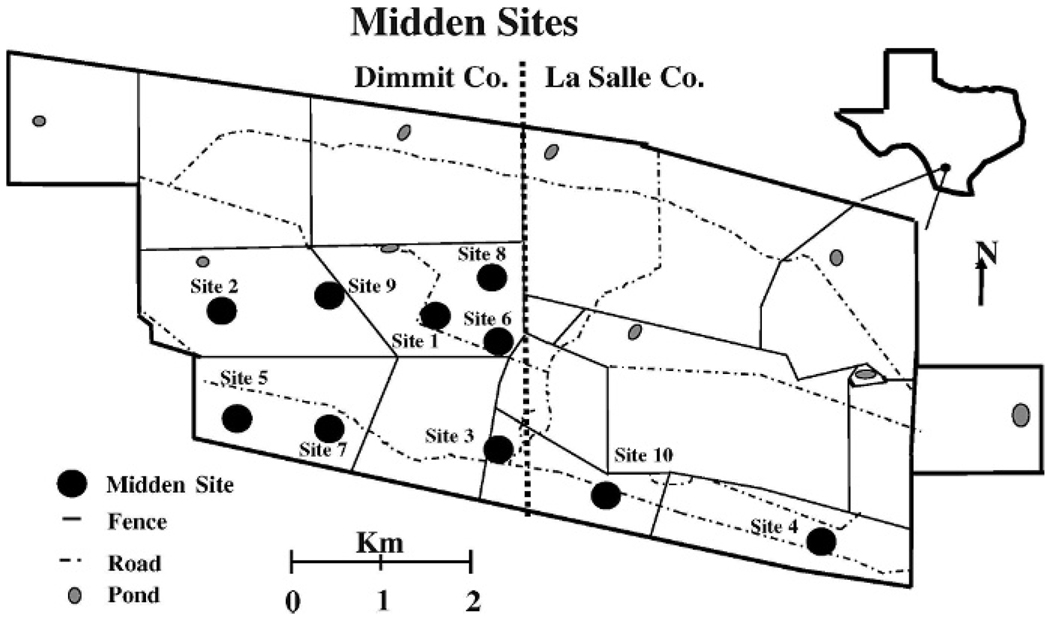
All collection sites were distributed across the southernmost portions of Chaparral Wildlife Management Area in Dimmit and La Salle counties, Texas. Collection sites were selected from areas possessing a high density of woodrats to maximize the number of woodrats available for study. Protocol for site selection was outlined in Suchecki et al. (2004), who previously defined sites as a circular area (25-m radius), >500 m from other collection sites, and possessing a minimum of 10 middens. Location and spatial distribution of collection sites are depicted in Fig. 1.
FIG. 1.
Map of the Chaparral Wildlife Management Area, Texas, showing the locations of the 10 midden sites.
Collection and processing
Woodrats were captured by hand during excavation of the subterranean tunnel systems associated with middens (see Suchecki et al. [2004] for a detailed description). Middens were excavated at new sites 4 times per year (January, March, June, and October) from March 2001 to June 2003 and exact locations were designated by Universal Transverse Mercator coordinates obtained from a handheld global positioning system device. Ten separate collection sites total were examined (1 per collecting trip) and all middens within a site were excavated during a single collection trip to ensure no migration between sites. All excavation occurred during daylight hours, when animals were least active, to maximize the number of captured individuals and increase the likelihood that individuals were in their home midden. Captured individuals were weighed, measured, and demographics associated with midden occupancy were recorded. These include number, age, sex, and field maternity assessment where applicable. Individuals were assigned to 1 of 4 age categories: adult, subadult, juvenile, or pup based on field assessment of molting patterns (Suchecki et al. 2004), mean nose-to-rump length, and mass. Pups were defined as individuals that were in utero at time of collection, attached to mammae of an adult female, or were incapable of independent movement outside the midden.
With the exception of pregnant females, animals were euthanized at the field site following methods (ketamine–xylazine overdose followed by cardiothoracic compression) approved by the American Society of Mammalogists Animal Care and Use Committee (Gannon et al. 2007) and were given an individual museum identification number (TK). Animals were prepared as voucher specimens with a standard set of tissues (liver, muscle, lung, spleen, kidney, and heart) collected from each individual and deposited in the Museum of Texas Tech University. Occasionally, epithelial tissues (toe or ear clips) were collected.
Assessment of parentage in the field
Assessments of maternity for all putative offspring were recorded in the field. Maternity was definitively assigned to offspring from females found pregnant at time of collection. Pregnant females were transported to the University of Texas Medical Branch and offspring were born in captivity; otherwise, embryos were collected upon euthanization of the female. Cohabiting adult females and pups were considered probable mother–offspring pairs, and females were considered prospective mothers of cohabiting juvenile and subadult individuals. However, individuals of these age classes were only included in parentage assessment if there were ≥2 individuals of that age classification collected with the female, or in cases in which offspring of different ages (multiple generations) were collected from a single midden.
No field assessment of paternity was possible; therefore, all adult males from a collection site were considered as potential sires of offspring from that site. Because males were euthanized upon capture and collection sites were >500 m apart, with consecutively sampled sites separated by much greater distances than 500 m (Fig. 1), it was highly unlikely that a male sired offspring at >1 site.
DNA isolation and D-loop sequencing
Genomic DNA was isolated from liver, kidney, and epithelial tissues using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minnesota) and methods outlined by Mendez-Harclerode et al. (2005). Isolates were used as a DNA source for amplification of both D-loop and microsatellite amplicons.
Approximately 963 base pairs of the D-loop region were amplified using reverse primers (2350-5—Castro-Campillo et al. 1999) and forward primers (N-mic 5′—Mendez-Harclerode et al. 2005). The following polymerase chain reaction protocol was used for amplification: 25 µl of 2× FailSafe PCR Premix K (Epicentre Biotechnologies, Madison, Wisconsin), 2.5 µl of 3′ and 5′ primers (20 mM), 0.5 µl of Taq, 18.5 µl of double-distilled H2O, and 1 µl of genomic DNA, and the thermal profile: 93.5°C for 1 min, 33 cycles at 93.5°C for 40 s, 49°C for 40 s, 72°C for 2 min, and a final extension at 72°C for 2 min. Polymerase chain reaction amplicons were purified using a QIAquick kit (Qiagen Inc., Chatsworth, California). Polymerase chain reaction primers and 2 internal primers (1115 Reverse and 500 Forward—Mendez-Harclerode et al. 2005) were used for sequencing. Samples were sequenced with a 3100-Avant Genetic Analyzer (Applied Biosystems, Foster City, California) and proofed and aligned using Sequencher 3.1 software (Gene Codes, Ann Arbor, Michigan). Sequences were deposited into GenBank (AY338507–AY338525, AY338529–AY338549, AY338551–AY338585, and DQ00012–DQ000153).
Haplotypes were assigned using Arlequin 2.0 software (Schneider et al. 2000). Novel haplotypes were defined by at least 1 base-pair difference between sequences (Bickham et al. 1996, 1998a, 1998b; Mendez-Harclerode et al. 2005, 2007; Trujillo et al. 2004), with sequences obtained from both forward and reverse strands. Haplotypes differing at a single site were reassessed following methods described in Mendez-Harclerode et al. (2005, 2007). Chromatograms for unique haplotypes were reexamined for verification or, if necessary, the polymerase chain reaction product was resequenced.
Microsatellite polymerase chain reaction and scoring
Microsatellite primers specific to Neotoma (Nma01, Nma04, Nma06, Nma10, and Nma11—Castleberry et al. 2000) were used to amplify 5 polymorphic loci for each individual. Two additional loci (Nma14 and Nma15—Castleberry et al. 2000) were amplified for dams, offspring, and potential sires to provide greater resolution in parentage analyses. The following polymerase chain reaction protocol, modified from Mendez-Harclerode et al. (2007), was used for amplification: 2.5 µl of 10× buffer, 1.5 µl of 25 mM MgCl2, 1 µl of 10 mM deoxynucleoside triphosphates (2.5 mM), 0.75 µl of 3′ and 5′ primer (10 mM), 0.25 µl of Taq, 17.75 µl of double-distilled H2O, and 0.4 µl of genomic DNA, and the thermal profile: 94°C for 2 min, 35 cycles at 94°C for 30 s, 57°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 10 min. Amplicons were analyzed using an ABI 3100-Avant Genetic Analyzer with GeneScan Analysis software version 3 (Applied Biosystems). Alleles were visualized and scored using GeneMapper software version 3 (Applied Biosystems).
Composite genotypes from 5–7 microsatellite loci were analyzed for each individual using Cervus 2.0 software (Marshall et al. 1998) to estimate allele frequencies, null allele frequencies, observed and expected heterozygosity values, parental exclusionary power, and values of polymorphic information content. Consistency with Hardy–Weinberg equilibrium was estimated using the FSTAT program (Goudet 1995). Probability of identity, likelihood of 2 randomly selected individuals possessing identical genotypes, was estimated using IDENTITY software (Wagner and Sefc 1999). Genotyping error rates were estimated per allele and per locus by methods described in Hoffman and Amos (2005) using data from unintentionally resampled individuals as well as intentional repeat genotyping of ~10% of individuals for each locus. Error rates also were assessed by examining genotypic mismatches of known mother–offspring pairs.
Relatedness
Relatedness coefficients (r-values) were estimated from 5 microsatellite loci (Nma01, Nma04, Nma06, Nma10, and Nma11) using Relatedness 4.2 software (Queller and Goodnight 1989), which generates estimates using an identity-by-decent approach. Relatedness values range from −1.000 to 1.000, with negative values indicating that relatedness between dyads is less than that expected between dyads chosen at random, whereas positive values indicate some degree of relatedness. All individuals were included in estimation of allele frequencies, providing more accurate values for collection sites with known clusters of highly related individuals (Queller and Goodnight 1989). Standard errors were estimated by jackknifing across all loci. Pairwise r-values were estimated for all pairs of individuals (dyads). Average r-values within sites were estimated for all sites including all individuals and for adults of both sexes of each site. Although Relatedness 4.2 software is useful in estimating average relatedness within and among groups, pairwise estimations of relatedness can be variable, thus estimates are more accurate when used in correlation with other variables (Queller and Goodnight 1989). Therefore, potential familial relationships were evaluated within sites using a combination of field data (cohabitation and spatial), molecular data (haplotypes and genotypes), and relatedness values with associated likelihood ratios (significance values P = 0.05, 0.01, and 0.001), estimated by Kinship 1.2 software (Goodnight 2000). Significance values (estimated using 100,000 iterations) were associated with critical likelihood values for each possible relationship (maternity, paternity, and sibship).
To determine intrasexual differences in relatedness between adults within a site, pairwise r-values between adult females were compared to values for adult males using the 2-sample Kolmogorov–Smirnov test (Matlab software; The MathWorks, Inc. 1994–2006, Natick, Massachusetts). This approach provided an estimate of the maximum difference between cumulative distributions of r-values between sexes. A Kolmogorov–Smirnov statistic (D), P-value, and power analysis were estimated using 1,000 iterations per test and an alpha value of 0.05.
Analysis of parentage
Assessments of maternity for possible mother–offspring relationships were evaluated by comparing D-loop haplotypes between the putative dam and offspring. Microsatellite genotypes of possible mother–offspring pairs initially were examined in a pairwise manner and later using Cervus 2.0 software (Marshall et al. 1998) to confirm the presence of maternal alleles at all loci for each offspring. Individuals of known or probable maternity for which >1 genotype mismatch occurred were excluded from further analyses. All pups for which maternity was confirmed were deemed candidates for inclusion in paternity assignment and assessment of multiple paternity.
Paternity was assessed using 2 methods. The 1st used exclusionary methods and male genotypes. Paternal alleles were identified by examination of maternal and offspring genotypes using the method described in Burton (2002). If mother and offspring possessed identical heterozygous genotypes at a given locus, both alleles were considered to be potentially paternal in origin. All paternal alleles were recorded for each locus, resulting in a paternal genotypic profile for each litter and individual offspring (Shurtliff et al. 2005). For family groups composed of 1 female and 2 generations of offspring, each litter was considered independently and in combination with the other litter as a family unit. Males were considered as candidate sires only for offspring collected from the same sampling site. The genotype of each candidate male was compared with each paternal genotypic profile both for litters and individual offspring. Males with no genotypic mismatches and whose genotype could account for all paternal alleles in a litter were considered the sire of the litter. Paternity was evaluated and assigned to individual offspring following the same procedure. Because of possible scoring errors in microsatellites, males with ≤1 mismatch were noted as putative sires to be evaluated further. Genotypic data for putative sires and offspring were reevaluated to ensure that discrepancies were not due to scoring error. Sires were not assigned to litters or individuals if >1 mismatch existed between the paternal genotypic profile and candidate males.
The 2nd method used relatedness values estimated with Relatedness 4.2 (Queller and Goodnight 1989) and Kinship 1.2 software (Goodnight 2000). Relatedness values (r-values) and evaluation of pedigree relationships were used for only those individuals for which paternity or putative paternity was assigned according to genotypic comparisons. Pairwise r-values were examined between males and offspring and between males and dam of the offspring. Theoretically, an r-value of 0.5 indicated a 1st-order relationship (Queller and Goodnight 1989), which was defined as those between parent and offspring and between full siblings. However, given that pairwise estimations of relatedness can be variable (Queller and Goodnight 1989), r-values from known mother–offspring pairs were used to determine the minimum r-value to be considered as indicating a 1st-order relationship. If r-values between sire and offspring indicated a 1st-order relationship and r-values between sire and dam indicated no potential relatedness (r = 0.0), males were considered as the sire of the litter or individual. However, if a 1st-order or 2nd-order relationship, such as that between half siblings (theoretical r = 0.25), was indicated between sire and dam, the potential sire could be a sibling or other close relative to the offspring. In this case, the male was not considered as the sire of the litter or individual. Criteria in this method excluded parentage resulting from mating events between closely related individuals, as incorrect parentage assignment based on this assumption would introduce type II error.
Multiple paternity and repeat mating
Litters with ≥2 offspring for which maternity was confirmed from field and genetic data were included in multiple paternity assessments. Multiple paternity was assessed in 2 ways. First, litters containing ≥3 offspring were considered multiply sired if ≥3 paternal alleles were detected at ≥2 loci (Burton 2002; Fitzsimmons 1998; Shurtliff et al. 2005; Valenzuela 2000). Second, using final paternity assignments, litters for which >1 male was assigned paternity, or for which paternity was assigned for ≥1 but not all individuals in a litter, were considered as multiply sired. In family units with 2 generations of offspring, females were considered to have engaged in repeat mating with the same male if paternity was assigned to the given male for at least 1 offspring from each of the 2 litters.
RESULTS
One hundred ninety-eight middens were excavated at 10 sites (Fig. 1) and 222 woodrats were captured. No significant differences in number of individuals were found between sexes; however, age structure differed because juveniles and pups were underrepresented in the study (28% of captures; Appendix I).
APPENDIX I.
Comparison of age and sex of plains woodrats (Neotoma micropus) collected across the 10 collection sites (Chaparral Wildlife Management Area) by collecting date (2001–2004). Roman numerals refer to midden sites. Numbers of individual woodrats for each age and sex are given in parentheses (males, females, unknown).
| Age class | ||||||
|---|---|---|---|---|---|---|
| Site | Date | No. total | Adult | Subadult | Juvenile | Pup |
| I | March 2001 | 33 (15, 16, 2) | 10 (4, 6) | 0 (0, 0) | 6 (4, 2) | 17 (7, 8, 2) |
| II | June 2001 | 25 (13, 12) | 13 (6, 7) | 12 (7, 5) | 0 (0, 0) | 0 (0, 0, 0) |
| III | October 2001 | 23 (10, 13) | 13 (7, 6) | 5 (1, 4) | 5 (2, 3) | 0 (0, 0, 0) |
| IV | January 2002 | 19 (9, 9, 1) | 14 (6, 8) | 3 (2, 1) | 0 (0, 0) | 2 (1, 0, 1) |
| V | March 2002 | 27 (11, 16) | 13 (6, 7) | 1 (1, 0) | 2 (1, 1) | 11 (3, 8, 0) |
| VI | June 2002 | 20 (10, 10) | 13 (6, 7) | 3 (2, 1) | 4 (2, 2) | 0 (0, 0, 0) |
| VII | October 2002 | 19 (11, 8) | 12 (6, 6) | 4 (4, 0) | 3 (1, 2) | 0 (0, 0, 0) |
| VIII | January 2003 | 19 (13, 6) | 16 (10, 6) | 2 (2, 0) | 1 (1, 0) | 0 (0, 0, 0) |
| IX | March 2003 | 19 (10, 7, 2) | 9 (4, 5) | 2 (1, 1) | 2 (2, 0) | 6 (3, 1, 2) |
| X | June 2003 | 18 (8, 10) | 11 (5, 6) | 4 (2, 2) | 3 (1, 2) | 0 (0, 0, 0) |
| Total | 222 (110, 107, 5) | 124 (60, 64) | 36 (22, 14) | 26 (14, 12) | 36 (14, 17, 5) | |
Forty-four haplotypes were present in 216 individuals (no haplotype data for 6 individuals) with an average of 5.8 haplotypes per collection site. Nine haplotypes (20.5%) occurred at multiple sites and the remaining 35 (79.5%) were unique to 1 of the 10 sites.
All microsatellite loci had moderate to high values for observed heterozygosity with values ranging from 0.717 to 0.924 (X̄ = 0.859), resulting in high parental exclusionary probabilities (Table 1). Additionally, there were moderate to high levels of variability within loci (polymorphic information content values), and no repetition of genotypes between individuals. Each individual possessed a unique genotype and the probability of repeat genotypes was 4.751571 × 10−7 (~1 individual in 2,104,567). The error rate estimated from mother–offspring mismatch data was 0.025, whereas the error rate estimated from repeat genotyping at all loci was 0.042. Six loci (Nma01, Nma04, Nma06, Nma10, Nma11, and Nma15) were in Hardy–Weinberg equilibrium based on an adjusted 95% significance level (P = 0.007, Bonferroni correction) and 7,000 permutations. Significant deviations from Hardy–Weinberg equilibrium were detected in locus Nma14 and across all loci collectively.
TABLE 1.
Microsatellite data (Cervus 2.0) for 222 individual plains woodrats (Neotoma micropus) at 5 microsatellite loci. Number of alleles (k), number of individuals typed for at each locus (N), observed (HO) and expected (HE) heterozygosity, polymorphic information content (PIC), total exclusionary power with 1 parent (Excl(1)) and 2 parents (Excl(2)), and null allele frequency (NF) are reported per locus and as a mean over all loci.
| Locus | k | N | HO | HE | PIC | Excl(1) | Excl(2) | NF |
|---|---|---|---|---|---|---|---|---|
| Nma1 | 18 | 219 | 0.840 | 0.901 | 0.891 | 0.664 | 0.799 | +0.0335 |
| Nma4 | 23 | 219 | 0.776 | 0.809 | 0.795 | 0.489 | 0.663 | +0.0173 |
| Nma6 | 15 | 219 | 0.804 | 0.836 | 0.816 | 0.513 | 0.681 | +0.0173 |
| Nma10 | 38 | 219 | 0.895 | 0.951 | 0.946 | 0.813 | 0.896 | +0.0280 |
| Nma11 | 14 | 219 | 0.689 | 0.717 | 0.690 | 0.339 | 0.525 | +0.0144 |
| Nma14 | 13 | 162 | 0.741 | 0.890 | 0.877 | 0.628 | 0.773 | +0.0902 |
| Nma15 | 15 | 158 | 0.924 | 0.910 | 0.899 | 0.681 | 0.811 | −0.0098 |
| X̄ | 19.43 | 246 | 0.859 | 0.845 | 0.99878 | 0.99995 |
Average relatedness within sites (r = 0.122) was greater than among sites (r = −0.005). Relatedness values for known mother–offspring pairs ranged from r = 0.315 to 0.704, with a mean r-value of 0.515.
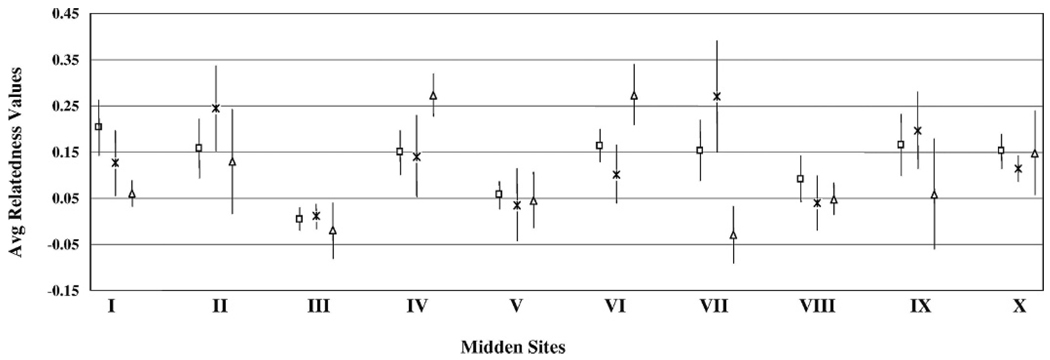
Average r-values for adults of both sexes (per site) varied across sites (Fig. 2). In 4 cases, the average r-value was >0.2 for either sex (males—sites IV and VI; females—sites II and VII). Differences in relatedness between adult females compared to differences between adult males within a site were significant at site VII (2-sample Kolmogorov–Smirnov test; D = 0.6, P = 0.018 at α = 0.05 and power = 0.879). Two cases of negative average relatedness occurred in males (sites III and VII); however, average relatedness values for site III, both overall and for adult females, was only slightly >0.
FIG. 2.
Range of r-values, with the centerpoint defining the average r-value at each site for all individual plains woodrats (Neotoma micropus; open squares), for adult females (times symbols), and for adult males (open triangles). Vertical bars represent SE.
Cohabitation
Composition of cohabitants varied and included adult males and females, adult males and females with offspring, only adult females, adult females with offspring, adult males with nonadults, and nonadult males and females (Table 2).
TABLE 2.
Records of cohabitation of plains woodrats (Neotoma micropus) by age and sex. S.M# indicates site and midden number. Age and sex of individuals are indicated as follows: adults (A), subadults (Sub), males (m), females (f), and offspring (O). Haplotypes (H) are indicated as shared (✓) or not shared (×). Relatedness values in middens with 2 occupants are indicated in the r1 column; values in columns r2−r5 report r-values between adult(s) and other occupants.
| Cohabitants | S.M# | H | r1 | r2 | r3 | r4 | r5 |
|---|---|---|---|---|---|---|---|
| Am/Af | 3.57 | ✓ | 0.64 | — | — | — | — |
| 4.84 | ✓ | 0.512 | — | — | — | — | |
| 5.99 | ✓ | 0.487 | — | — | — | — | |
| 6.127 | ✓ | 0.35 | — | — | — | — | |
| 7.143 | × | 0.296 | — | — | — | — | |
| 8.152 | ✓ | −0.022 | — | — | — | — | |
| 8.166 | ✓ | 0.334 | — | — | — | — | |
| 10.191 | × | 0.07 | — | — | — | — | |
| Am/Am/Af | 4.7 | ✓ | 0.731 | — | — | — | — |
| 0.522 | — | — | — | — | |||
| 0.597 | — | — | — | — | |||
| Am/Af/O | 1.19 | ✓ | 0.457 | ||||
| m | ✓ | 0.175 | 0.472 | 0.441 | 0.127 | ||
| f | ✓ | 0.449 | 0.387 | 0.483 | 0.329 | ||
| 4.79 | ✓ | 0.454 | |||||
| m | ✓ | 0.022 | — | — | — | ||
| f | ✓ | 0.355 | — | — | — | ||
| 4.82 | × | 0.188 | |||||
| m | ✓ | 0.185 | 0.242 | — | — | ||
| f | × | 0.616 | 0.38 | — | — | ||
| 7.131 | ✓ | 0.524 | |||||
| m | ✓ | 0.524 | 0.371 | 0.266 | — | ||
| f | ✓ | 0.664 | 0.352 | 0.375 | — | ||
| Af/Af | 6.122 | × | 0.07 | — | — | — | — |
| Af/Af/Subm/f | 2.3 | ✓ | 0.437 | ||||
| f | ✓ | 0.508 | 0.095 | 0.489 | — | ||
| f | ✓ | 0.432 | 0.32 | 0.057 | — | ||
| Af/Subm | 2.39 | ✓ | 0.236 | — | — | — | — |
| 2.42 | × | 0.045 | — | — | — | — | |
| 2.45 | ✓ | 0.2 | — | — | — | — | |
| 6.109 | ✓ | 0.053 | — | — | — | — | |
| 10.109 | ✓ | 0.582 | — | — | — | — | |
| Af/Subf | 10.185 | ✓ | 0.655 | — | — | — | — |
| Af/Subm/f | 2.43 | ✓ | 0.619 | 0.467 | — | — | |
| 3.52 | ✓ | — | 0.524 | 0.443 | — | ||
| 3.56 | ✓ | 0.729 | 0.298 | — | — | ||
| 10.193 | ✓ | 0.523 | 0.611 | — | — | ||
| Am/O | 6.126 | ✓ | 0.515 | — | — | — | — |
| 8.147 | ✓ | 0.392 | — | — | — | — | |
| Subm/f | 2.34 | ✓ | 0.333 | — | — | — | — |
| 0.643 | — | — | — | — | |||
| 0.621 | — | — | — | — |
Cohabitation between adult males and females was recorded in 13 middens at 8 of 10 collection sites, during all seasons. Of the 13 cases of adult males and females cohabiting, 8 involved an adult male and female as a solitary pair, the remaining 5 involved additional individuals. In 4 of the 5 cases involving additional individuals, adult male and female cohabitants were collected with pups, juveniles, or subadults, whereas in the remaining case, 2 adult males cohabited with an adult female. Adult females were found cohabiting twice, once as a pair, and once with 2 subadult males and 1 subadult female. Eleven cases involved cohabitation of an adult female with 1 or more subadults.
Eighteen cases of an adult female cohabitating with pups or juveniles were recorded. All were determined to be mother–offspring units and were evaluated in paternity assessment. Two cases of an adult male cohabitating with a juvenile and subadults were recorded. Additionally, sub–adults cohabited in the absence of an adult only once.
Parentage
Maternity was confirmed or assigned for 53 individuals, from 22 middens, resulting in 27 candidate litters, including 5 family units composed of multiple generations (Appendix II). Each litter in a family unit of multiple generations was considered independently and concurrently with the other litter. Haplotypes corresponded between all mother–offspring pairs, in positive, probable, and prospective categories of maternal assessment based on field data.
APPENDIX II.
Results of paternity assessments per litter and individual for each method and overall. Litter composition per site and midden number (S.M#) and age are listed in columns 1–3. Offspring are listed according to individual identification number (TK) and age is indicated as pup (p), pup in utero (p′), juvenile (j), or subadult (s). Results of paternity assessment for each individual for each method are indicated. Nonassignment is indicated in the same manner for all methods (×). Assignment of paternity is indicated in method I (exclusion) with individual identification (TK) of sire as well as number of mismatches in parentheses. Method II (relatedness) evaluations are reported as check marks (✓) when r-values for both offspring–sire and dam–sire pairs fell within range stipulated in the text. Significance values (P-values) for 1st-order offspring–sire relationships as estimated using Kinship 1.2 are indicated in superscript. Results per litter or family unit indicate nonassignment (none), ≥1 sire assigned (>1), the TK number of the sire assigned, or paternity unresolved (?). Overall paternity assignments are indicated in the same manner.
| S.M# | Offspring | Age | Exclusion | Relatedness | Overall |
|---|---|---|---|---|---|
| 1.17 | None | None | None | ||
| 98051 | p | × | × | × | |
| 98052 | p | × | × | × | |
| 1.19 | >1 | None | None | ||
| 98064 | p | × | × | × | |
| 98065 | p | × | × | × | |
| 98066 | j | 98082 (1) | × | × | |
| 98067 | j | × | × | × | |
| 1.21 | >1 | >1 | >1 | >1 | |
| 98070 | p | × | × | × | |
| 98071 | p | 98048 (1) | ✓ 0.001 | 98048 | |
| 98072 | p | × | × | × | |
| 1.22 | >1 | >1 | None | ||
| 98074 | p | 98048 (0) | × | × | |
| 98075 | p | × | × | × | |
| 98076 | p | × | × | × | |
| 98077 | j | 98048 (1) | ✓× | × | |
| 98082 (1) | × | ||||
| 1.23 | None | None | None | ||
| 98184 | p′ | × | × | × | |
| 98185 | p′ | × | × | × | |
| 98080 | j | × | × | × | |
| 98081 | j | × | × | × | |
| 2.43 | None | None | None | ||
| 98134 | s | × | × | × | |
| 98135 | s | × | × | × | |
| 3.49 | None | None | None | ||
| 98144 | j | × | × | × | |
| 98145 | j | × | × | × | |
| 3.52 | >1 | >1 | >1 | ||
| 98149 | s | × | × | × | |
| 98150 | s | 98147 (0) | ✓ 0.001 | 98147 | |
| 98163 (1) | × | ||||
| 3.56 | >1 | >1 | >1 | ||
| 98165 | s | × | × | × | |
| 98166 | s | 98147 (1) | ✓ 0.001 | 98147 | |
| 4.67 | None | None | None | ||
| 98357 | p′ | × | × | × | |
| 5.89 | >1 | >1 | >1 | ||
| 98351 | p′ | 98255 (0) | × 0.001 | ? | |
| 98352 | p′ | 98275 (0) | ✓ 0.001 | 98275 | |
| 5.92 | None | None | None | ||
| 98259 | p | × | × | × | |
| 98260 | p | × | × | × | |
| 98260 | p | × | × | × | |
| 5.96 | 98257 | >1 | 98257 | ||
| 98267 | j | 98257 (0) | ✓ 0.001 | 98257 | |
| 98255 (1) | ✓ 0.001 | ||||
| 98268 | j | 98257 (0) | ✓ 0.001 | 98257 | |
| 5.99 | None | None | None | ||
| 98353 | p′ | × | × | × | |
| 5.103 | >1 | None | None | ||
| 98355 | p′ | 98255 (1) | × | × | |
| 98356 | p′ | × | × | × | |
| 6.118 | None | None | None | ||
| 98369 | j | × | × | × | |
| 98370 | j | × | × | × | |
| 7.131 | >1 | >1 | >1 | ||
| 98418 | j | × | × | × | |
| 98419 | j | 98437 (1) | ✓ 0.001 | 98437 | |
| 98420 | s | 98437 (0) | ✓ 0.001 | 98437 | |
| 9.175 | >1 | >1 | >1 | ||
| 112102 | p′ | 112056 (1) | × ✓ | ? | |
| 112057 (1) | × ✓ | ||||
| 112064 | j | 112056 (0) | ✓ 0.001 | Either | |
| 112057 (0) | ✓ 0.001 | ||||
| 112065 | j | × | × | × | |
| 9.177 | >1 | 112062 | 112062 | ||
| 112068 | p | 112062 (0) | ✓ 0.001 | 112062 | |
| 112069 | p | 112056 (1) | × | 112062 | |
| 112062 (1) | ✓ 0.01 | ||||
| 9.179 | 112060 | 112060 | 112060 | ||
| 112077 | p | 112060 (1) | ✓ 0.001 | 112060 | |
| 112078 | p | 112060 (1) | ✓ 0.001 | 112060 | |
| 10.193 | None | None | None | ||
| 112136 | s | × | × | × | |
| 112137 | s | × | × | × | |
| 10.197 | None | None | None | ||
| 112141 | j | × | × | × | |
| 112142 | j | × | × | × | |
| 112143 | j | × | × | × |
Final (overall) conclusions of paternity were based on combined support from the 2 methods (Appendix II). Four litters (5.96, 7.131 [subadult], 9.177, and 9.179) were each determined to be sired by 1 male/litter. Six litters were determined to have >1 sire (1.21, 3.52, 3.56, 5.89, 7.131, and 9.175) and 1 litter had unresolved paternity (9.175-p′) between 2 sires. For 16 litters, no sire was assigned. Individually, paternity was assigned for 12 individuals, and was considered to be unresolved for 3 individuals. Paternity was not assigned for the remaining 38 individuals.
Twenty-one litters were included in the assessment of multiple paternity according to genotypic profiles. Of the 4 litters having ≥3 offspring, 1 (10.197) had ≥3 paternal alleles at 2 loci, and was considered multiply sired. Following final paternity assignments of the 21 eligible litters, evidence of multiple paternity was found in 5 litters, and 3 litters were determined to be singly sired. Following both methods of assessment of multiple paternity, 6 litters were determined to be multiply sired.
There were 5 family groups composed of 1 female and 2 generations of offspring (Appendix II). Two of these family groups (7.131 and 9.175) had litters in which multiple paternity occurred in 1 litter. Repeat mate choice was observed in 1 family unit (7.131), that is 1 sire was assigned to individuals in separate litters of 1 family unit. Although paternity could not be resolved for the unborn pup in the other family group (9.175), the likely candidate sires correspond with those of the juvenile sibling. Paternity was not assigned for individuals in the remaining 3 family units.
DISCUSSION
Values of haplotype and nucleotide diversity indicated a high number of closely related haplotypes with low levels of population substructure due to maternal lineages. Haplotypes were moderately to highly variable and were useful in assessing maternity and possible associations of individuals within a sampling area with a high density of individuals. These results concurred with those of Mendez-Harclerode et al. (2005, 2007) that were obtained during the same collection periods from nearby localities.
Seven microsatellite loci were used in parentage analyses and 5 loci were used for relatedness and kinship estimations. Error rates associated with an increase in number of microsatellite loci can greatly affect accuracy of parentage, relatedness, and kinship assignments and estimations, due to the fact that associated methods of data analysis use data on allele frequency and many do not make allowances for error (Blouin et al. 1996). In addition, when using exclusion as a method of parentage analysis, error in microsatellite data could result in either wrongful exclusion or inclusion of an individual as a potential parent, although the former is more likely except in cases in which potential parents are highly related. Instances of this type of error did occur, resulting in unresolved paternity assignments.
Data on relatedness revealed several possible patterns. Average relatedness values within sites were greater than the overall observed average and were suggestive of higher average relatedness values than would be expected in a panmictic population. In addition, within a site there was no significant difference between sexes in relatedness, with the exception of site VII. These patterns could be the result of a lack of long-range dispersal from the natal nest, and the latter could indicate a lack of sex-biased kin clustering.
To discriminate accurately among specific relationships (maternity, paternity, and full sibship) using only r-values, a larger number of highly polymorphic microsatellite loci than was used in this study would be needed (Blouin et al. 1996). Therefore, any results from an analysis using pairwise r-values (Relatedness 4.2) were used only with corresponding field data and genotypic profiles of individuals involved. Additionally, known mother–offspring pairs were used as a reference for assessing possible 1st-order relationships and supporting the accuracy of these values.
By using a maternally inherited marker and 5–7 microsatellite loci in conjunction with field data, maternal relationships were confidently assigned and confirmed. In turn, knowledge of those relationships was used as a scale to increase confidence with which other associations between individuals were evaluated.
Paternity assessments offered additional data leading to a greater understanding of social dynamics in areas of high density, including evidence of a high frequency of multiple paternity, occurrence of repeat mating events between the same 2 individuals, and no indication of male dominance at any collection site. Additionally, there was more evidence for promiscuity in females than in males, but this likely was an artifact of the low number of collected sires.
Overall, sires were assigned for only 12 of 53 offspring, although 2 methods were used to deduce paternity. However, given that the exclusionary power to identify the 2nd parent of an offspring if 1 was known was 99.9%, and the probability of identity was high (~1 individual in 2,104,567), it is highly unlikely that the low number of paternities assigned was due to methods, but rather suggests that females were predominantly mating with males outside the immediate sampling area, with sires for other offspring considered as occurring outside the sampling area. Recorded areas of home ranges of this species are highly variable (Conditt and Ribble 1997; Henke and Smith 2000; Johnson 1952; Raun 1966), and may be related to population density, that is, low densities are associated with larger home ranges, presumably for mating purposes. Given that our sampling area (1,964 m2) was greater than the largest reported mean home range for this species (1,899 m2—Conditt and Ribble 1997), it seemed probable that home ranges of males for this population would fall within the range of the sampling area.
In a few cases, 1st-order relatedness between dam and potential sire, or between 2 potential sires, resulted in unresolved paternity assignments, or could have potentially resulted in false paternity assignment. Additionally, because we assumed that dams do not mate with a higher-order relative, type II error was potentially introduced. Because there was evidence both for individuals of 1st- and 2nd-order relatedness living in close proximity, and that females mate with males outside of their immediate area, a type II error was less likely.
Initially, a widely used likelihood approach using Cervus 2.0 software was considered as a paternity assessment tool. However, this program is sensitive to discrepancies in certain variables, namely, accuracy of the estimate and proportion of candidate parents sampled (see Marshall et al. 1998). As was the case with this data set, without knowledge of the proportion of candidate parents (males in this case), or life-history information needed to accurately estimate this value, the accuracy of resultant assignments can be greatly skewed. Therefore, Cervus 2.0 proved to be inappropriate for this data set, but its assessment as a paternity assignment tool provided useful information and precautionary perspective on choice of parentage analysis programs.
Few studies have reported cohabitation data for N. micropus (Raymond et al. 2003; Suchecki et al. 2004) and none have documented a high frequency of cohabitation in areas of high densities of woodrats. Of the studies that have documented this behavior, cohabitation events have been associated solely with mate pairing. Using cohabitation records coupled with genetic data, it is apparent that mate pairing may not be the principal purpose for cohabitation in our population.
In cases of adult females and males collected as a solitary pair or with offspring, support for a higher-order relationship between the female and male was the dominant pattern observed. In half of the cases in which offspring were present (n = 4), the adult male was highly related to ≥1 of the resident offspring. These results indicate that cohabitation between adult females and males in this population is principally either between nondispersed siblings or an adult female with an older, nondispersed male offspring, or mate-pairing is occurring prevalently between highly related individuals. Varying levels of relatedness were found between adult females in cohabitation with subadult males (n = 5), with both higher-order and nonrelated dyads represented. Cases of unrelated adult males and females found in the same midden, both adults and subadults, could represent mate pairing, the attempt of 1 individual to displace the current resident, or cohabitation, given the energy required to excavate and maintain a midden.
Only 2 cases of cohabitation of adult females were recorded. These cases likely represent 2 newly dispersed females that were recently displaced from their natal nest, based on season of collection and the low observed frequency of adult females cohabiting, and 2 nondispersed siblings that remained in cohabitation even subsequent to breeding. Alternately, the latter pair may be the single example detected of cohabitation between adult females. Cases of an adult female cohabiting either with a subadult female or with multiple subadults likely represented a female and nondispersed offspring.
Previous studies conducted in densely populated areas (Raymond et al. 2003; Suchecki et al. 2004; Thies and Caire 1991) recorded cohabitation of adult males and females within middens, although Raymond et al. (2003) and Thies and Caire (1991) considered cohabitation to be the result of mate pairing. In contrast, the density (110.6 woodrats/ha) reported by Suchecki et al. (2004) was higher than previous studies, suggesting that density might influence cohabitation. The population studied by Conditt and Ribble (1997) had low densities (2.5–5.8 woodrats/ha), with only 13 individuals captured, making it difficult to discern between mating systems and density as being responsible for cohabitation. Additionally, because cohabitation was observed during all seasons, and pups or embryos were documented predominantly during the normal breeding seasons, it is unlikely that cohabitation occurred solely for mate pairing.
Relatedness assessments also support the conclusions that cohabitation between adult males and females was likely due to high population density and potentially to nondispersal of male offspring instead of mate pairing. Arguments for this include high frequencies of 1st-order relationships among adult males and females found as solitary pairs, and high relatedness of dyads found in cohabitation with ≥1 or more generations of offspring. If cohabitation between adult males and females was the result of mate pairing, there would be the possibility of high frequencies of inbreeding and either paternal participation in care of offspring or mate guarding.
Although this study cannot discriminate among these possibilities, presence of 1st-order relatives of both sexes within sites, moderate average relatedness within sites, evidence from paternity assignments (females likely do not mate as frequently with proximal males), and availability of nonrelated or less-related mates due to high population density, inbreeding at high frequencies is unlikely. Cohabitation between pairs of adult males and females in comparison with low observed frequency of female–female cohabitation could be evidence for sex-biased dispersal from the natal nest, or alternatively, natal philopatry of male offspring.
Although the sampling scheme employed in this study was not designed to examine social behavior or mating strategies, we were able to draw several conclusions from our results. First, paternity assignments indicated evidence of a high frequency of multiple paternity, occurrence of repeat mating events between the same 2 individuals, and no indication of male dominance at collection sites. Second, data were consistent with a promiscuous mating system, as are data from other studies of southern plains woodrats (Conditt and Ribble 1997; Merkelz and Kerr 2002; Thies and Caire 1991). Third, cohabitation appeared to be more frequent than reported in previous studies (Raymond et al. 2003; Suchecki et al. 2004; Thies and Caire 1991), and occurred for reasons other than mate pairing, perhaps related to the high density of woodrats at the site. Additionally, we were able to assess the appropriateness of methods commonly used for assessing parentage and mating systems for a point-in-time sampling scheme, a research design not conventionally used for this type of study.
RESUMEN
Doscientos veintidós individuos pertenecientes a la especie de rata magueyera (Neotoma micropus) fueron capturados en 198 nidos escavados en 10 sitios de colecta separados pertenecientes a una sola población en el sur central de Tejas, USA. Información de campo, haplotipos de D-loop de mitocondria y 5 a 7 loci de microsatélites polimórficos fueron usados para determinar patrones de paternidad genéticos, grado de relación y estrategia de apareamiento. Los loci de microsatélites resultaron altamente polimórficos (promedio de heterocigosis observado = 0.859) y fueron usados para construir genotipos únicos para cada individuo (probabilidad de genotipos idénticos = 1 en 2, 104,567) Resultados indicaron alta frecuencia de paternidad múltiple (6 de 9 camadas), evidencia de apareamiento repetido entre la misma pareja de individuos, y ausencia de indicación de dominación masculina en ninguno de los sitios de colecta. Estos datos sugieren un sistema de apareamiento promiscuo. Dentro de cada sitio de colecta, el promedio de grado de relación entre hembras adultas fue similar al de machos adultos. Un alto grado de cohabitación en comparación a aquellos reportados con anterioridad fue registrado, y análisis de escala más fina revelaron altos niveles de grado de relación entre la mayoría de los individuos cohabitantes. Nuestros resultados, cuando son tomados en cuenta justo con los resultados de otros estudios del comportamiento de apareamiento de N. micropus, sugieren que es probable que la densidad de población influya el comportamiento social y de apareamiento en esta especie.
ACKNOWLEDGMENTS
We thank R. K. Chesser, R. E. Strauss, R. Van Den Bussche, and M. L. Haynie for valuable help with the data analysis and comments on the manuscript. B. R. Amman, J. G. Brant, D. S. Carroll, M. B. Cajimat, N. D. Durish, J. D. Hanson, M. L. Haynie, M. Kageyama, L. K. Longhofer, R. McAliley, C. Milazzo, Jr., M. L. Milazzo, S. A. Reeder, J. R. Suchecki, and A. Vestal provided valuable help in the field. D. C. Ruthven III and D. R. Synatzske facilitated fieldwork. This research was financially supported by National Institutes of Health grant AI-41435, entitled “Ecology and Epidemiology of Emerging Arenaviruses in the Southwestern United States.”
Contributor Information
B. Dnate’ Baxter, Department of Biology, Texas Tech University, Lubbock, TX 79409-3191, USA.
Francisca M. Mendez-Harclerode, Department of Biology, Bethel College, North Newton, KS 67117-0531, USA
Charles F. Fulhorst, Department of Pathology, University of Texas Medical Branch, Galveston, TX 77555-0609, USA
Robert D. Bradley, Department of Biology, Texas Tech University, Lubbock, TX 79409-3191, USA Natural Science Research Laboratory, Museum of Texas Tech University, Lubbock, TX 79409-3191, USA.
LITERATURE CITED
- Avise JC. Molecular markers, natural history and evolution. New York: Chapman and Hall; 1994. [Google Scholar]
- Baker RJ, Makova KD, Chesser RK. Microsatellites indicate a high frequency of multiple paternity in Apodemus (Rodentia) Molecular Ecology. 1999;8:107–111. doi: 10.1046/j.1365-294x.1999.00541.x. [DOI] [PubMed] [Google Scholar]
- Berteaux D, Beaty J, Rengifo E, Bergeron J-M. Multiple paternity in meadow voles (Microtus pennsylvanicus): investigating the role of the female. Behavioral Ecology Sociobiology. 1999;45:283–291. [Google Scholar]
- Bickham JW, Loughlin TR, Calkins DG, Wickliffe JK, Patton JC. Genetic variability and population decline in Steller sea lions from the Gulf of Alaska. Journal of Mammalogy. 1998a;79:1390–1395. [Google Scholar]
- Bickham JW, Loughlin TR, Wickliffe JK, Burkanov VN. Genetic variation in the mitochondrial DNA of Steller sea lions: haplotype diversity and endemism in the Kuril Islands. Biosphere Conservation. 1998b;1:107–117. [Google Scholar]
- Bickham JW, Patton JC, Loughlin TR. High variability for control region sequences in a marine mammal: implications for conservation and biogeography of Steller sea lions (Eumetopias jubatus) Journal of Mammalogy. 1996;77:95–108. [Google Scholar]
- Birney EC. Systematics of three species of woodrats (genus Neotoma) in central North America. Miscellaneous Publications, Museum of Natural History, University of Kansas. 1973;58:1–173. [Google Scholar]
- Blanchfield PJ, Ridgway MS, Wilson CC. Breeding success of male brook trout (Salvelinus fontinalis) in the wild. Molecular Ecology. 2003;12:2417–2428. doi: 10.1046/j.1365-294x.2003.01917.x. [DOI] [PubMed] [Google Scholar]
- Blouin MS, Parsons M, Lacaille V, Lotz S. Use of microsatellite loci to classify individuals by relatedness. Molecular Ecology. 1996;5:393–401. doi: 10.1111/j.1365-294x.1996.tb00329.x. [DOI] [PubMed] [Google Scholar]
- Burton C. Microsatellite analysis of multiple paternity and male reproductive success in the promiscuous snowshoe hare. Canadian Journal of Zoology. 2002;80:1948–1956. [Google Scholar]
- Carling MD, Wiseman PA, Byers JA. Microsatellite analysis reveals multiple paternity in a population of wild pronghorn antelopes (Antilocapra americana) Journal of Mammalogy. 2003;84:1237–1243. [Google Scholar]
- Castleberry SB, King TL, Wood PB, Ford WM. Microsatellite DNA markers for the study of Allegheny woodrat (Neotoma magister) populations and cross-species amplifications in the genus Neotoma. Molecular Ecology. 2000;9:824–826. doi: 10.1046/j.1365-294x.2000.00915-4.x. [DOI] [PubMed] [Google Scholar]
- Castro-Campillo A, Roberts HR, Schmidly DJ, Bradley RD. Systematic status of Peromyscus boylii ambiguous based on morphologic and molecular data. Journal of Mammalogy. 1999;80:1214–1231. [Google Scholar]
- Clutton-Brock TH. Mammalian mating systems. Proceedings of the Royal Society of London, B. Biological Sciences. 1989;236:339–372. doi: 10.1098/rspb.1989.0027. [DOI] [PubMed] [Google Scholar]
- Coltman DW, Bancroft DR, Robertson A, Smith JA, Clutton-Brock TH, Pemberton JM. Male reproductive success in a promiscuous mammal: behavioural estimates compared with genetic paternity. Molecular Ecology. 1999;8:1199–1209. doi: 10.1046/j.1365-294x.1999.00683.x. [DOI] [PubMed] [Google Scholar]
- Conditt SA, Ribble DO. Social organization of Neotoma micropus, the southern plains woodrat. American Midland Naturalist. 1997;137:290–297. [Google Scholar]
- Fabiani A, Galimberti F, Sanvito S, Hoelzel AR. Extreme polygyny among southern elephant seals on Sea Lion Island, Falkland Islands. Behavioral Ecology. 2004;15:961–969. [Google Scholar]
- Finley RB., Jr The woodrats of Colorado: distribution and ecology. University of Kansas Publications, Museum of Natural History. 1958;10:213–552. [Google Scholar]
- Fitzsimmons NN. Single paternity of clutches and sperm storage in promiscuous green turtle (Chelonia mydas) Molecular Ecology. 1998;7:575–584. doi: 10.1046/j.1365-294x.1998.00355.x. [DOI] [PubMed] [Google Scholar]
- Fournier D, Aron S, Milinkovitch MC. Investigation of the population genetic structure and mating system in the ant Pheidole pallidula. Molecular Ecology. 2002;11:1805–1814. doi: 10.1046/j.1365-294x.2002.01573.x. [DOI] [PubMed] [Google Scholar]
- Gannon WL, Sikes RS The Animal Care and Use Committee of the American Society of Mammalogists. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. Journal of Mammalogy. 2007;88:809–823. doi: 10.1093/jmammal/gyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnight KF. [Accessed June 2005];Kinship (version 1.2) 2000 http://gsoft.smu.edu/GSoft.html.
- Goudet J. FSTAT v1.2. A computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Haynie ML, Van Den Bussche RA, Hoogland JL, Gilbert DA. Parentage, multiple paternity, and breeding success in Gunnison’s and Utah prairie dogs. Journal of Mammalogy. 2003;84:1244–1253. [Google Scholar]
- Henke SE, Smith SA. Use of aluminum balls to determine home ranges of woodrats. Southwestern Naturalist. 2000;45:352–355. [Google Scholar]
- Heske EJ, Ostfeld RS. Sexual dimorphism in size, relative size of testes, and mating systems in North American voles. Journal of Mammalogy. 1990;71:510–519. [Google Scholar]
- Hoffman JI, Amos W. Microsatellite genotyping errors: detection approaches, common sources and consequences for paternal exclusion. Molecular Ecology. 2005;14:599–612. doi: 10.1111/j.1365-294X.2004.02419.x. [DOI] [PubMed] [Google Scholar]
- Johnson CW. M.S. thesis. Austin: University of Texas; 1952. The ecological life history of the packrat, Neotoma micropus, in the brushlands of southwest Texas. [Google Scholar]
- Jones AG, Avise JC. Microsatellite analysis of maternity and the mating system in the Gulf pipefish Syngnathus scovelli, a species with male pregnancy and sex-role reversal. Molecular Ecology. 1997;6:203–213. doi: 10.1046/j.1365-294x.1997.00173.x. [DOI] [PubMed] [Google Scholar]
- Jones AG, Walker D, Lindstrom K, Kvarnemos C, Avise JC. Surprising similarity of sneaking rates and genetic mating patterns in two populations of sand goby experiencing disparate sexual selection regimes. Molecular Ecology. 2001;10:461–469. doi: 10.1046/j.1365-294x.2001.01193.x. [DOI] [PubMed] [Google Scholar]
- Kays RW, Gittleman JL, Wayne RK. Microsatellite analysis of kinkajou social organization. Molecular Ecology. 2000;9:743–751. doi: 10.1046/j.1365-294x.2000.00921.x. [DOI] [PubMed] [Google Scholar]
- Marshall TC, Slate J, Kruuk LEB, Pemberton JM. Statistical confidence for likelihood-based paternity inference in natural populations. Molecular Ecology. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- Matocq MD. Reproductive success and effective population size in woodrats (Neotoma macrotis) Molecular Ecology. 2004;13:1635–1642. doi: 10.1111/j.1365-294X.2004.02173.x. [DOI] [PubMed] [Google Scholar]
- Matocq MD, Lacey EA. Philopatry, kin clusters, and genetic relatedness in a population of woodrats (Neotoma macrotis) Behavioral Ecology. 2004;15:647–653. [Google Scholar]
- Mendez-Harclerode FM, Fulhorst CF, Milazzo ML, Ruthven DC, III, Bradley RD. Genetic diversity within the southern plains woodrat (Neotoma micropus) in southern Texas. Journal of Mammalogy. 2005;86:180–190. doi: 10.1644/1545-1542(2005)086<0180:gdwtsp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Harclerode FM, Strauss RE, Fulhorst CF, Milazzo ML, Ruthven DC, III, Bradley RD. Microsatellite and DNA sequence data indicate high levels of intrapopulation genetic diversity in woodrats (Neotoma micropus) Journal of Mammalogy. 2007;88:360–370. doi: 10.1644/05-MAMM-A-377R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkelz RM, Kerr SF. Demographics, den use, movements, and absence of Leishmania mexicana in southern plains woodrats (Neotoma micropus) Southwestern Naturalist. 2002;47:70–77. [Google Scholar]
- Nievergelt CM, Mutschler T, Feistner ATC, Woodruff DS. Social system of the Alaotran gentle lemur (Hapalemur griseus alaotrensis): genetic characterization of group composition and mating system. American Journal of Primatology. 2002;57:157–176. doi: 10.1002/ajp.10046. [DOI] [PubMed] [Google Scholar]
- Onorato DP, Hellgren EC, Van Den Bussche RA, Skiles JR., Jr Paternity and relatedness of American black bears recolonizing a desert montane island. Canadian Journal of Zoology. 2004;82:1201–1210. [Google Scholar]
- Painter JN, Crozier RH, Poiani A, Robertson RJ, Clarke MF. Complex social organization reflects genetic structure and relatedness in the cooperatively breeding bell miner, Manorina melanophrys. Molecular Ecology. 2000;9:1339–1347. doi: 10.1046/j.1365-294x.2000.01012.x. [DOI] [PubMed] [Google Scholar]
- Queller DC, Goodnight KF. Estimating relatedness using genetic markers. Evolution. 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. [DOI] [PubMed] [Google Scholar]
- Queller DC, Strassmann JE, Hughes CR. Microsatellites and kinship. Trends in Ecology & Evolution. 1993;8:285–288. doi: 10.1016/0169-5347(93)90256-O. [DOI] [PubMed] [Google Scholar]
- Raun GG. A population of woodrats (Neotoma micropus) in southern Texas. Bulletin of Texas Memorial Museum. 1966;11:1–62. [Google Scholar]
- Raymond RW, Mchugh CP, Witt LR, Kerr SF. Temporal and spatial distribution of Leishmania mexicana infections in a population of Neotoma micropus. Memorias do Instituto Oswaldo Cruz Rio de Janerio. 2003;98:171–180. doi: 10.1590/s0074-02762003000200002. [DOI] [PubMed] [Google Scholar]
- Ribble DO. The monogamous mating system of Peromyscus californicus as revealed by DNA fingerprinting. Behavioral Ecology and Sociobiology. 1991;29:161–166. [Google Scholar]
- Rusu AS, Krackow S. Kin-preferential cooperation, dominance-dependent reproductive skew, and competition for mates in communially nesting female house mice. Behavioral Ecology and Sociobiology. 2004;56:298–305. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Genetics and Biometry Laboratory. Geneva, Switzerland: University of Geneva; 2000. Arlequin v.2.000: a software for population genetics data analysis. [Google Scholar]
- Schulte-Hostedde AI, Millar JS, Gibbs HL. Sexual selection and mating patterns in a mammal with female-biased sexual size dimorphism. Behavioral Ecology. 2004;15:351–356. [Google Scholar]
- Scribner KT, Congdon J, Smith MH, Chesser RK. Annual differences in female reproductive success affect spatial and cohort-specific genotypic heterogeneity in painted turtles. Evolution. 1993;47:1360–1373. doi: 10.1111/j.1558-5646.1993.tb02160.x. [DOI] [PubMed] [Google Scholar]
- Shurtliff QR, Pearse DE, Rogers DS. Parentage analysis of the canyon mouse (Peromyscus crinitus): evidence for multiple paternity. Journal of Mammalogy. 2005;86:531–540. [Google Scholar]
- Suchecki JR, Ruthaven DC, III, Fulhorst CF, Bradley RD. Natural history of the southern plains woodrat Neotoma micropus (Rodentia: Muridae) from southern Texas. Texas Journal of Science. 2004;56:131–140. [Google Scholar]
- Taylor AC, Horsup A, Johnson CN, Sunnucks P, Sherwin B. Relatedness structure detected by microsatellite analysis and attempted pedigree reconstruction in an endangered marsupial, the northern hairy-nosed wombat, Lasiorhinus krefftii. Molecular Ecology. 1997;6:9–19. doi: 10.1046/j.1365-294x.1997.00146.x. [DOI] [PubMed] [Google Scholar]
- Thies M, Caire W. Association of Neotoma micropus nests with various plant species in southwestern Oklahoma. Southwestern Naturalist. 1990;35:80–102. [Google Scholar]
- Thies M, Caire W. Nearest-neighbor analysis of the spatial distribution of houses of Neotoma micropus in southwestern Oklahoma. Southwestern Naturalist. 1991;36:233–237. [Google Scholar]
- Topping MG, Millar JS. Mating patterns and reproductive success in the bushy-tailed woodrat (Neotoma cinerea), as revealed by DNA fingerprinting. Behavioral Ecology and Sociobiology. 1998;43:115–124. [Google Scholar]
- Trujillo RG, Loughlin TR, Gemmell NJ, Patton JC, Bickham JW. Variation in microsatellites and mtDNA across the range of the Steller sea lion, Eumetopias jubatus. Journal of Mammalogy. 2004;85:338–346. [Google Scholar]
- Valenzuela N. Multiple paternity in side-neck turtles Podocnemis expansa: evidence form microsatellite data. Molecular Ecology. 2000;9:99–105. doi: 10.1046/j.1365-294x.2000.00806.x. [DOI] [PubMed] [Google Scholar]
- Wagner HW, Sefc KM. IDENTITY 1.0. Centre for Applied Genetics. Vienna, Austria: University of Agricultural Sciences; 1999. [Google Scholar]
- Wilmer JW, Overall AJ, Pomeroy PP, Twiss SD, Amos W. Patterns of paternal relatedness in British grey seal colonies. Molecular Ecology. 2000;9:283–292. doi: 10.1046/j.1365-294x.2000.00872.x. [DOI] [PubMed] [Google Scholar]
- Yu H-T, Liao Y-Y, Kao C-H. Relatedness structure and individual identification in a semi-fossorial shrew (Soricidae: Anourosorex squamipes)—an application of microsatellite DNA. Zoological Studies. 2001;40:226–232. [Google Scholar]