Abstract
Nitric oxide (NO•) is a toxin, and various life forms appear to have evolved strategies for its detoxification. NO•-resistant mutants of Escherichia coli were isolated that rapidly consumed NO•. An NO•-converting activity was reconstituted in extracts that required NADPH, FAD, and O2, was cyanide-sensitive, and produced NO3−. This nitric oxide dioxygenase (NOD) contained 19 of 20 N-terminal amino acids identical to those of the E. coli flavohemoglobin. Furthermore, NOD activity was produced by the flavohemoglobin gene and was inducible by NO•. Flavohemoglobin/NOD-deficient mutants were also sensitive to growth inhibition by gaseous NO•. The results identify a function for the evolutionarily conserved flavohemoglobins and, moreover, suggest that NO• detoxification may be a more ancient function for the widely distributed hemoglobins, and associated methemoglobin reductases, than dioxygen transport and storage.
Nitric oxide (NO•) is a free radical with multiple and diverse biological functions. It is a common product of enzymic and nonenzymic oxidations of reduced nitrogen compounds and of reductions of nitrogen oxides. In bacteria, the metabolic reduction of inorganic nitrate (NO3−) or nitrite (NO2−) is an important source of toxic NO• (1). In mammals, NO• is produced by arginine-oxidizing NO• synthases (2). At low levels, NO• functions as a membrane-permeable signaling molecule controlling blood pressure and memory (3, 4). During responses to infection, foreign bodies, or tissue injury, toxic amounts of NO• are produced by an inducible NO• synthase isoform presumably to help kill or inhibit the growth of invading microorganisms or neoplastic tissue (5).
Given the diverse and abundant sources of NO•, and the prevalence of NO•-sensitive targets (6), cells would be expected to benefit from defenses that limit damage by NO• and NO•-derived reactive nitrogen species. In principle, these defenses would be analogous to the well-characterized antioxidant systems which protect aerobic organisms from reactive oxygen species. Indeed, several cellular adaptations, and cellular reactions of NO• or NO•-derived species, have been proposed to participate in the overall protection against NO• toxicity in bacteria (5, 7) and mammalian cells (8). In addition, NO• reductases have been shown to function in NO• detoxification; however, their expression appears to occur during the anaerobic assimilation and dissimilation of nitrogen from nitrite and nitrate (1, 9, 10).
Previously, we demonstrated an NO• sensitivity of the Escherichia coli and the mitochondrial aconitases in cells and in vitro (11). During the course of investigation, we observed an O2-dependent and cyanide-sensitive mechanism for aconitase protection which correlated with the rate of NO• consumption by E. coli (12). In E. coli, aconitase protection and NO• consumption was induced in response to NO• exposure, and this increase was dependent upon protein synthesis. Moreover, this aerobic cyanide-sensitive NO• consumption activity appeared to constitute part of an adaptive defense against NO• toxicity. Thus, we supposed that E. coli mutants containing elevated activity might be selected for resistance to NO•-mediated killing, and furthermore, that they could be used to facilitate the biochemical and genetic characterization of the NO• scavenging activity.
Here, we describe the isolation and characterization of an O2-dependent and cyanide-sensitive nitric oxide dioxygenase (NOD) from NO•-resistant E. coli, which is a flavohemoglobin. The identification of NOD with flavohemoglobin is significant because the cellular function of (flavo)hemoglobins in E. coli, yeast, and other organisms has long remained obscure whereas physical, biochemical, and genetic information has been bountiful. But, perhaps more significantly, the remarkable amino acid homology of the respective hemoglobin and flavin domains of flavohemoglobins with hemoglobins and methemoglobin reductases from distantly related animals, plants, protists, helminths, and bacteria (13–16) suggests that the more primitive role of the well-characterized and ubiquitous O2-binding hemoglobins is NO• detoxification.
MATERIALS AND METHODS
Bacteria Strains, Mutagenesis, and Plasmid Constructs.
E. coli strain RB9060 (Δhmp) and the parent strain YCM10 (17) were generously provided by Alex Ninfa (Univ. of Michigan). Strain AB1157 was from Bruce Demple (Harvard Univ.) and was mutagenized with N-methyl-N′-nitro-N-nitrosoguanidine as described (18). The hmp gene was cloned by using the respective 5′ and 3′ PCR oligonucleotide primers 5′-CCGAATTCTTGTGCGATAACAGGTCTTGAC-3′ and 5′-AGGGATCCACGCGGCAATTTAAACCGCGTC-3′, which contain homology to sequences flanking the hmp coding sequence and operator–promoter region (19). PCR products were gel purified, digested with EcoRI and BamHI, and ligated in the plasmid vector pAlter (Promega), and plasmids were transformed into JM109 cells.
Bacterial Growth, Harvest and Extract Preparation.
Culture growth, gas exposures, and medium preparation was as described (11). Medium contained 12 μg/ml tetracycline for cells containing plasmids. Cells were harvested, cell-free extracts were prepared in 50 mM potassium phosphate buffer (pH 7.8) containing 0.1 mM EDTA, and extract protein was assayed as described (11).
NO• Consumption and NOD Assays.
NO• consumption was followed polarographically by using an NO• microelectrode (Diamond General, Ann Arbor, MI). Cellular NO• consumption was determined from rates of NO• removal in a 2 ml assay containing minimal glucose medium and 1 μM NO• incubated at 37°C. NOD activity was measured at 37°C in a reaction mix containing 50 mM potassium phosphate buffer (pH 7.8), 0.1 mM EDTA, 2.5 mM glucose 6-phosphate (G6P), 0.5 unit/ml glucose-6-phosphate dehydrogenase (G6PD), 0.2 mM NADP+, 1 μM FAD, and 1 μM NO•. O2 was depleted from the reaction mixture by preincubating the mixture for 5 min with Aspergillus glucose oxidase (2 units/ml) (Sigma), glucose (10 mM), and bovine liver catalase (130 units/ml) (Boehringer Mannheim), and O2 depletion was verified with an O2 electrode. Rates were corrected for extract or cell-independent NO• decomposition.
Nitrite and Nitrate Assays.
NO2− was measured with the Griess reagent, and NO3− was determined from the nitrate reductase (Boehringer Mannheim) catalyzed formation NO2− (20).
NOD Isolation and Sequencing.
Cell-free lysates were prepared from a culture of the E. coli mutant PG118 grown with vigorous aeration in 1 liter of phosphate-buffered Luria–Bertani medium. Cells were harvested by centrifugation and lysed in 10 ml of a 50 mM potassium phosphate buffer (pH 7.8) containing 0.1 mM EDTA and 1 mM DTT (buffer A) plus ≈0.5 mg of DNase. Extracts were resuspended at 12 mg/ml protein, and proteins were precipitated with 33% ammonium sulfate on ice and were removed by centrifugation. NOD activity was precipitated from the supernatant with 55% ammonium sulfate, and the ammonium sulfate precipitant was resuspended in buffer A and dialyzed extensively against buffer A. Dialysates (≈100 mg protein) were then diluted in buffer A to 2 mg/ml and mixed by gentle inversion with 5 ml of FAD-agarose (Sigma) at 4°C overnight. FAD-agarose was loaded on a column and washed extensively with buffer A. FAD binding proteins were eluted with 20 mM FAD in 25 mM potassium phosphate buffer (pH 7.8) containing 50 μM EDTA and 1 mM DTT, and the eluate was concentrated with a Millipore ultrafree concentrator. FAD-agarose eluates were fractionated on a Superdex 200 column (Pharmacia/LKB) in buffer A containing 4 mM 2-mercaptoethanol in place of DTT, and fractions containing activity were pooled and concentrated, and then loaded on a HiTrap Mono Q column (Pharmacia/LKB), washed with buffer A, eluted with 0.2 M NaCl in buffer A, and concentrated. N-terminal amino acid sequencing by automated Edman degradation was done under contract by Commonwealth Biotechnologies (Norfolk, VA).
Determination of Heme and Flavin.
Heme content was assayed by the alkaline pyridine method by using bovine hemin as the standard (21), and FAD was determined from the fluorescence of boiled flavohemoglobin/NOD monitored at 520 nm and excited at 460 nm with an FAD standard (22).
RESULTS
Selection of NO•-Resistant Mutants.
E. coli strain AB1157 was mutagenized with N-methyl-N′-nitro-N-nitrosoguanidine and NO•-resistant mutants were selected by plating 2.5 million log phase bacteria on 10-cm Petri dishes containing phosphate-buffered LB agar medium and 50 μM phenazine methosulfate and by incubating culture dishes under a constantly replenished atmosphere containing 10% air and 960 ppm NO• balanced with N2 for 16 h at 37°C. Plating density and the redox-cycling agent phenazine methosulfate appeared important for achieving uniform and maximal NO•-dependent killing of E. coli on the agar possibly because of the enhancement of NO•-mediated killing by O2⨪/ONOO−, or H2O2.
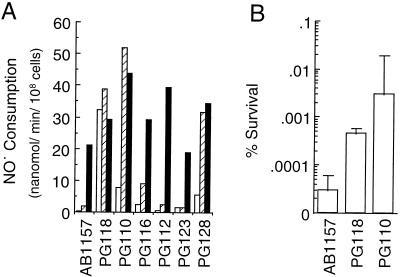
Thirty putative NO•-resistant mutants were isolated from several dishes. Mutants were grown in semi-aerated cultures, and their rates of NO• consumption were measured. Six mutants with an elevated basal NO• consumption rate were identified and characterized for their level of expression of the NO• consumption activity during vigorous aeration and following exposure to relatively high NO• levels (Fig. 1A). Three of these mutants, PG110, PG118, and PG128, expressed high constitutive levels of NO• consumption activity, with PG110 expressing the highest level with NO• exposure. Elevated NO• consumption by PG118 and PG110 contributed to respective and approximate 25- and 150-fold increases in resistance to NO•-mediated killing (Fig. 1B).
Figure 1.
Elevated NO• consumption by NO•-resistant mutants of E. coli. (A) Cultures were grown in phosphate-buffered LB medium with minimal aeration (open bars), with vigorous shaking under air (hatched bars), or under an atmosphere containing 960 ppm NO• in 21% O2 balanced with N2 for 60 min (filled bars), and cells were harvested and washed and NO• consumption was measured. (B) A total of 2.5 million cells from logarithmic cultures of AB1157, PG110, and PG118 were plated on phosphate-buffered LB agar medium containing phanazine methosulfate (50 μM) and were exposed to 960 ppm NO• under an atmosphere of 10% air balanced with N2 or to air at 37°C for 16 h. Cultures were then incubated at 37°C under a normal air atmosphere for 24 h to allow the outgrowth of bacterial colonies. Colonies were counted and percent survival was calculated relative to air exposed controls (n = 2; ±SD).
Assay of the NO• Converting Activity in Cell-Free Extracts.
Cell pellets and extracts of mutants exhibited a yellowish-brown hue which correlated with the level of NO• consumption activity. Soluble cell extracts prepared from the overexpressor PG118 also displayed a greater absorption at 427 nm and at ≈550 nm than extracts prepared from the parental strain AB1157 (data not shown). The absorption peak at 427 nm was reminiscent of the heme Soret band, and the 550 nm absorption was in the region of the heme α/β bands. Thus, PG118 appeared to express more heme or hemoproteins than AB1157. Importantly, the striking yellowish-brown hue and the absorption spectra of mutant extracts suggested that heme, flavin or cofactors with similar spectral properties may be associated with the activity.
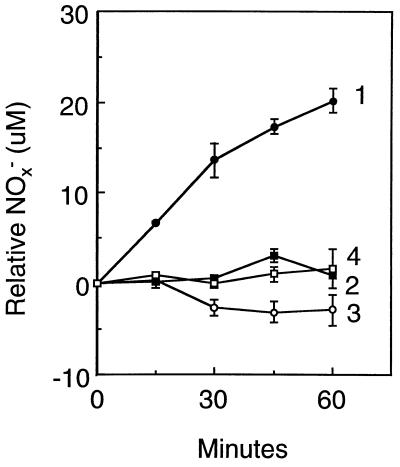
Extracts displayed no measureable NO• converting activity when tested alone and thus appeared to require added cofactors. The requirement of O2 for NO• consumption by cells (12) suggested the involvement of an oxygenase activity rather than an anaerobic NO• reductase activity (9, 10). In addition, several flavin and heme-containing enzymes oxygenate nitrogenous compounds, including organic nitroxides in reactions requiring NAD(P)H, O2, and flavins (23). Indeed, an NO• consumption activity was observed in a reaction containing FAD and an NADPH regenerating system (Table 1). This activity was elevated 58-fold in extracts of PG118 when compared with those from AB1157. This elevation was comparable to the 20-fold increase in NO• consumption observed in the mutant cells (Fig. 1A). FAD, O2, and NADPH were all essential for the activity, and moreover, the activity was cyanide-sensitive as observed in whole cells (12). In a reaction catalyzed by the activity in PG118 extracts, NO3−, but not NO2−, was produced (Fig. 2). These results support a role for an enzymic NOD activity in the mechanism of NO• conversion.
Table 1.
Reconstitution of an O2-dependent and cyanide-sensitive NO• consumption activity in cell-free extracts
| Condition | Activity
|
|
|---|---|---|
| nmol/min/mg | % | |
| AB1157 | ||
| Complete | 15 | 100 |
| PG118 | ||
| Complete | 865 | 100 |
| −FAD | 0 | 0 |
| −NADP+/G6P/G6PD | 0 | 0 |
| −O2 | 0 | 0 |
| +25 μM NaCN | 393 | 45 |
| +250 μM NaCN | 19 | 2 |
Cell-free extracts were prepared from log phase aerobic cultures grown in phosphate-buffered LB medium and were assayed with and without additions under standard conditions as described.
Figure 2.
Formation of NO3− by mutant extracts. NO3− formation in NO• consumption reactions catalyzed by 150 μg of PG118 (line 1) or AB1157 (line 2) extract protein was measured in a 10 ml reaction mixture containing 50 mM potassium phosphate buffer (pH 7.8), 0.1 mM EDTA, 0.2 mM NADP+, 2.5 mM glucose-6-phosphate, 0.5 unit/ml glucose-6-phosphate dehydrogenase, 1 μM FAD, and 10 μg/ml bovine erythrocyte copper and zinc-containing superoxide dismutase. Superoxide dismutase was included to inhibit NO2− and NO3− formation by the O2⨪-dependent oxidation of NO•. The reaction was equilibrated with an atmosphere containing 480 ppm NO• in 10% air balanced with N2 by vigorous shaking in a gyrorotatory water bath at 37°C. NO2− formation with PG118 (line 3) and AB1157 (line 4) extracts was also assayed. Extract-catalyzed NO3− and NO2− formation were calculated by subtracting the amount of extract-independent NO3− and NO2− formation. The amount of PG118 and AB1157 extract added was sufficient to catalyze the conversion of 47 and 0.3 μM NO• per min in the standard NO• consumption assay, respectively. Error bars represent SD where n = 3.
Isolation and Identification of the NOD with Flavohemoglobin.
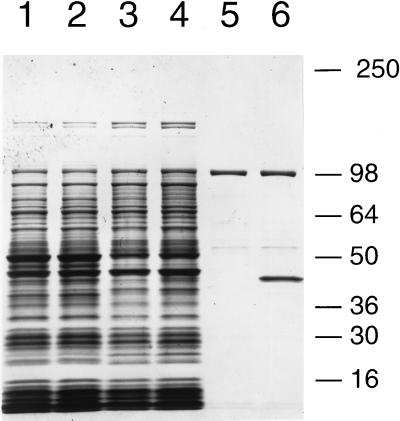
The NOD required 50–100 nM FAD for half-maximal activity and was fully stimulated at 1 μM (data not shown), suggesting an affinity of the enzyme for FAD. We utilized this property to absorb FAD binding proteins from extracts with FAD-agarose, and compared proteins from the overexpressing mutant PG118 with those from the parental strain AB1157. As shown by the Coomassie blue-stained SDS/PAGE gel in Fig. 3, PG118 expressed a unique ≈43-kDa protein (compare lanes 5 and 6). This ≈43-kDa protein was also more abundant in crude extracts (compare lanes 1 and 2) and in a dialyzed ammonium sulfate fraction (compare lanes 3 and 4) prepared from PG118 when compared with extracts prepared from strain AB1157.
Figure 3.
Identification of a unique FAD-binding protein in cell-free extracts of the NO•-resistant mutant PG118. Cell-free extracts at 25 μg (lanes 1 and 2), dialyzed 33–55% ammonium sulfate fractions at 25 μg (lanes 3 and 4), and proteins eluted from FAD-agarose (lanes 5 and 6) were separated on 8–16% SDS/PAGE and stained with Coomassie blue. Samples from AB1157 were separated in lanes 1, 3, and 5, and the corresponding PG118 samples were in lanes 2, 4, and 6. FAD-agarose (0.1 ml) was boiled in SDS sample buffer following an overnight incubation with an equal volume of buffer A containing 2 mg protein from the ammonium sulfate fractions and following extensive washing. One-twentieth of the total amount eluted was loaded on the gel.
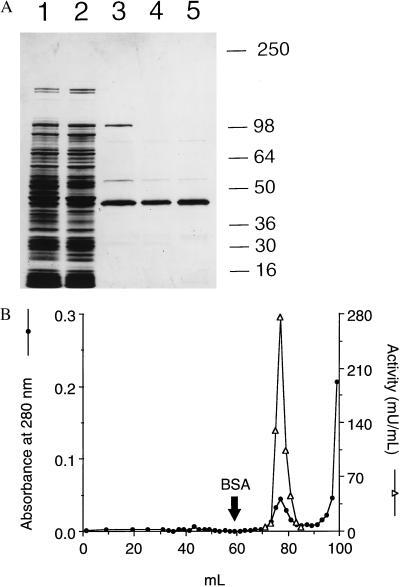
The ≈43-kDa FAD-binding protein cofractionated with the NOD activity in a scheme that included affinity elution from FAD-agarose (Table 2). Separation of the activity on Superdex 200 and on Mono Q anion exchange resin produced a near homogeneous protein as revealed by SDS/PAGE gel analysis (Fig. 4A). The NOD activity eluted with the major protein peak on Superdex 200 gel in fractions significantly beyond the ≈66-kDa molecular size marker BSA (Fig. 4B), indicating that NOD exists as a monomer under these conditions. It should be noted that there was a progressive loss of NOD activity during the fractionation. Nevertheless, the ≈43-kDa FAD-binding protein cofractionated with NOD activity, thus suggesting a loss of activity of the ≈43-kDa protein rather than the separation from a contaminating activity.
Table 2.
Fractionation of NOD
| Fraction | Volume, ml | Protein, mg | Activity, units | Specific activity, units/mg | Recovery, % |
|---|---|---|---|---|---|
| Cell-free lysate | 9.5 | 195 | 431 | 2.2 | 100 |
| Ammonium sulfate | 3.8 | 95 | 319 | 3.4 | 81 |
| FAD-agarose | 0.7 | 0.35 | 23 | 65.7 | 5 |
| Superdex 200 | 0.30 | 0.074 | 0.5 | 6.8 | 0.11 |
| Mono Q | 0.10 | 0.057 | 0.3 | 5.3 | 0.07 |
Figure 4.
Fractionation of NOD from a mutant overexpressor. (A) Fractions were separated on a 8–16% SDS/PAGE gel and stained with Coomassie blue. Lanes: 1, cell-free lysate, 25 μg; 2, 33–55% ammonium sulfate dialysate, 25 μg; 3, FAD-agarose eluate concentrate, 2 μg; 4, Superdex 200 pooled concentrate, 1.2 μg; and 5, Mono Q 0.2 M NaCl eluate concentrate, 2.9 μg. (B) Superdex 200 elution profile of the NOD activity and protein. The arrow points to the elution point of the marker BSA.
The Mono Q fraction, containing the purified ≈43-kDa FAD-binding protein/NOD, was subjected to N-terminal amino acid sequence analysis. The first 20 cycles of N-terminal sequencing by automated Edman degradation gave the sequence MLDAQTIATVLATIPLLVET. This sequence was used to search for identities to known E. coli proteins and to all translated ORFs in the complete E. coli genome by using the blast sequence analysis program provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). Nineteen of the 20 N-terminal amino acids determined showed sequence identity with the N terminus of flavohemoglobin (19) and shared no significant homologies with any other known proteins or translated ORFs. Only the eleventh amino acid disagreed with the flavohemoglobin sequence in which leucine (L) was assigned in place of lysine. This difference may be attributed to an ambiguity in the gas chromatographs for this position.
The isolated E. coli flavohemoglobin is a 43.8-kDa monomer that reportedly contains 0.27–0.42 mol FAD and 0.16–0.22 mol heme per mole protein (22). Importantly, it should be noted that there was significant deficiency of both heme and FAD in those preparations. We also observed a low heme Soret band absorption as well as a decrease in the yellowish-brown color of the flavohemoglobin during our isolation; we were unable to quantitate the heme or FAD content given the low yields. However, we were able to demonstrate an activity increase with added hemin. Hemin at 1 μM maximally stimulated the activity of the isolated NOD, giving 135 U per mg (data not shown). The hemin-stimulated activity, as well as the native activity, was partly inhibited by manganese-containing superoxide dismutase (15–150 μg/ml) indicating a role of O2−· in the catalytic mechanism. Moreover, we have since been able to stabilize NOD activity and increase the heme content of flavohemoglobin/NOD preparations by including the ferric heme ligand azide (10 mM) during gel filtration. Azide stabilized preparations contained 0.10 mol heme and 0.01 mol FAD per mole protein, and had a specific activity of 14.1 units/mg protein in the standard NOD assay containing added FAD. Thus, by the criteria of amino acid sequence identity, molecular weight, and cofactor requirements, the isolated NOD activity is flavohemoglobin.
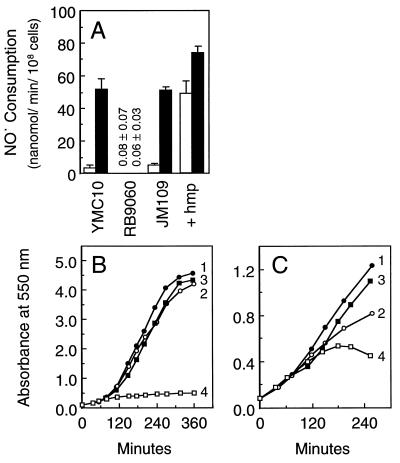
To verify that flavohemoglobin/NOD catalyzed NO• consumption by cells, and that NOD was an enzymic function of flavohemoglobin, we measured NO• consumption by cells, and NOD activity in cells, deficient in or overexpressing flavohemoglobin. In addition, we measured the effects of flavohemoglobin expression on steady-state O2⨪ levels in cells because elevated production of O2⨪ by flavohemoglobin in cells, as previously supposed (24), may provide a catalytic route for NO• decomposition given the extremely rapid reaction of free O2⨪ with NO•. The E. coli mutant RB9060 deleted in the hmp gene encoding flavohemoglobin (17) was essentially void of basal and induced catalytic NO• converting activity when compared with the parental strain YMC10 (Fig. 5A). In addition, JM109 cells expressing flavohemoglobin from a multi-copy plasmid bearing only the hmp gene catalyzed NO• consumption at a 10-fold higher rate than JM109 cells containing the plasmid vector pAlter only, and furthermore, exposure of cells to NO• elevated this expression ≈1.5-fold. Measurements of NOD activity in cell extracts provided similar results; basal and NO•-induced RB9060 cells contained only a very low noninducible “NO• converting activity,” whereas overexpressors contained elevated activity relative to controls. Measurements of steady-state O2⨪ levels, by using the sensitive aconitase method (25), revealed no measureable differences between control cells and cells overexpressing flavohemoglobin. JM109 cells containing vector only or vector plus hmp, grown under air as described in the legend to Fig. 5A, contained comparable aerobic aconitase activity levels and percent active. The respective aerobic and percent active aconitase for the control and the overexpressor were 165 ± 30 and 166 ± 26 mU/mg protein (n = 3; ±SD); and 88 ± 2% and 85 ± 5% (n = 3; ±SD). Thus, the mechanism of flavohemoglobin-catalyzed decomposition of NO• does not depend upon the oxidase activity of flavohemoglobin and free O2⨪.
Figure 5.
Effects of flavohemoglobin expression on cellular NO• consumption activity and NO• toxicity. (A) NO• consumption activities of the hmp deletion mutant (RB9060), its parent (YMC10), the flavohemoglobin overexpressor strain JM109 containing hmp in the multi-copy plasmid vector pAlter (+ hmp), and its control strain containing pAlter only (JM109) were measured during growth in air (open bars) or following a 60-min exposure to 960 ppm NO• balanced with N2 and 21% O2 (filled bars). Strains YMC10 (lines 1 and 2) and RB9060 (lines 3 and 4) were grown continuously under air (B) or N2 (C), or were exposed after 60 min (lines 2 and 4) to 960 ppm NO• balanced with N2 and 21% O2 (B) or to 240 ppm NO• balanced with N2 (C). Growth was monitored at 550 nm. Aerobic and anaerobic cultures were initiated in phosphate-buffered LB medium with 2% inocula from overnight aereobic cultures and with 4% inocula from static overnight anoxic cultures, respectively. Glucose (20 mM) was added for anaerobic growth. Cells were treated at an A550 = 0.3 for measurements of NO• consumption activity. Error bars represent the SD of three measurements.
Importantly, aerobic growth of flavohemoglobin-deficient RB9060 strain was more sensitive to NO• inhibition than the parental strain (Fig. 5B, compare lines 2 and 4), and the protection afforded by flavohemoglobin appeared greater during growth with O2 than in its absence (Fig. 5C, compare lines 2 and 4). Thus, cells containing flavohemoglobin conferred resistance to higher NO• levels aerobically than anaerobically. The results further demonstrate a defensive role of flavohemoglobin in the protection against NO• toxicity and support the proposed NOD function of flavohemoglobin. The ability of flavohemoglobin to protect anaerobic growth suggests additional protective mechanisms.
DISCUSSION
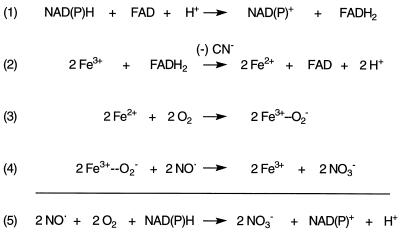
Scheme S1 delineates the proposed reaction sequence for flavohemoglobin-catalyzed NO• dioxygenation which incorporates the NADPH, FAD, and O2-dependence, the cyanide sensitivity (Table 1), the formation of NO3− (Fig. 2), and the extensive structural and biochemical information for the (flavo)hemoglobin family. FAD is proposed to function as the electron carrier from NAD(P)H to the ferric heme prosthetic group. In the homologous Alcaligenes flavohemoglobin the isoalloxazine ring of FAD and the heme propionic group are bridged by highly conserved amino acids and a water molecule that are thought to act as an electron conduit (26). Spectroscopic studies of the E. coli flavohemoglobin also support an electronic linkage of flavin and heme in which NAD(P)H reduces FAD to the leukoflavin (Eq. 1), and the FAD hydroquinone and semiquinone then univalently and sequentially reduce the heme–Fe3+ (Eq. 2) (27). Flavohemoglobin heme–Fe2+ binds O2 with high affinity to form heme–Fe2+–O2 (22, 27–31), which likely exists in a quasi equilibrium with heme–Fe3+–O2⨪ (Eq. 3) as previously suggested for the homologous hemoglobin and myoglobin (32). Indeed, the ability of oxygenated flavohemoglobin (24, 29–31), like oxyhemoglobin (33), to slowly release O2⨪ and of superoxide dismutase to favor deoxygenation (22, 33) supports this concept. The heme–Fe3+–O2⨪ intermediate would then be set to react rapidly with NO• to produce nitrate (NO3−) (Eq. 4) as previously demonstrated for the mammalian hemoglobin (34). The proposed reactions may also explain the cyanide sensitivity of NOD because the binding of cyanide to heme–Fe3+ would be expected to interfere with heme–Fe3+ reduction (Eq. 2) and subsequent high affinity O2 binding to the heme–Fe2+ as is well-known for hemoglobin. In summary, the reaction steps for NOD, as well as the overall stoichiometry (Eq. 5), resemble the well-documented reactions of the mammalian hemoglobin system.
Scheme 1.
.
The reaction of NO• with the heme–Fe3+–O2⨪ to form NO3− may bear similarity to the spontaneous reaction of NO• with free O2⨪ to form peroxynitrous acid (HOONO) and the subsequent nonenzymic decomposition of HOONO in free solution to form NO3− (35). The heme–Fe3+ may facilitate an oxygen bond rearrangement by participating in a iron-mediated oxygen bond shift analogous to the proton-mediated shift suggested for the nonenzymic mechanism for HOONO decomposition to NO3−. Importantly, however, the NOD-catalyzed mechanism would be more advantageous for NO• detoxification because its rate would not be dependent on high concentrations of toxic O2⨪ nor would NOD release the toxic intermediates HOONO and HO•.
Flavohemoglobins belong to a family of evolutionarily related proteins which includes the well-known O2-carrying red blood cell hemoglobins and associated flavin-containing methemoglobin reductases (13, 14). The bacterial Alcaligenes flavohemoglobin has a flavin and NAD(P)H-binding reductase domain and a hemin-binding hemoglobin domain with a high degree of structural homology to the mammalian hemoglobin and methemoglobin (cytochrome b5) reductase (26). Moreover, it has been repeatedly suggested that (flavo)hemoglobins may have a common ancestral function; however, that function has remained enigmatic. In this regard, Keilin (36) pointed out long ago that it was unlikely that the microbial hemoglobins participated in O2 transport or storage because there are no diffusion limits for O2 delivery to unicellular organisms nor are their levels of hemoglobin sufficient for significant O2 storage.
Numerous observations have since led to different hypotheses regarding the function of the bacterial and fungal (flavo)hemoglobins and plant (leg)hemoglobins. These proposals have included O2 sensing (28, 37) scavenging or delivery (15), ferrisiderophore reduction (38), heme sequestration (39), protection against reactive oxygen species (40), and NO• reduction (41). Here, we have demonstrated a function for the E. coli flavohemoglobin in the catalysis of NO• dioxygenation that may clarify some observations and provide new insights. For example, it may explain the recent demonstration of a sensitivity of flavohemoglobin (hmp) mutants of Salmonella typhimurium to nitrous acid or S-nitrosoglutathione during aerobic growth conditions (42) much as E. coli hmp mutants were protected from authentic NO• (Fig. 5B). The induction of transcription of the E. coli hmp gene (43), as well as the NOD activity (Fig. 5A and ref. 12), by NO•, NO2−, or NO3− may be explained by the defensive function of NOD. The induction of bacterial (flavo)hemoglobins by hypoxia (44–46) may be beneficial because of the advantage of increasing NOD activity when the substrate O2 becomes limiting for efficient NO• dioxygenation. The decreased virulence of the plant pathogen Erwinia chrysanthemi having mutations in flavohemoglobin (hmp) (47) is interesting because it suggests that NO• may also function in the immune defense of plants. On the other hand, nonsymbiotic and symbiotic plant hemoglobins (48) may protect plants from NO• produced via the oxidation of ammonia or the reduction of NO2− or NO3− either released by the N2-fixing rhizobia or present in the soil. An NO• detoxification function may also explain the ability of exogenously added oxyleghemoglobin to increase respiration in Rhizobia (49) because NO• is a potent inhibitor of terminal respiratory oxidases (6). The proposed NOD function of flavohemoglobins does not, however, explain the anaerobic protection afforded against NO• donors in S. typhimurium (42) or NO• gas in E. coli (Fig. 5C), or the function of elevated flavohemoglobin/NOD expression in anaerobic E. coli during NO3− or NO2− respiration (12, 43) because flavohemoglobin does not appear to effectively convert NO• in vitro or in cells in the absence of O2 (Table 1, unpublished results, and ref. 41). It is thus likely that flavohemoglobins serve additional function(s).
Estimates from amino acid divergence place the origin of the primeval hemoglobin molecule at ≈1.8 billion years ago (13, 14). This age coincides well with the estimated origin of the oxygenic blue-green algae (2.3 billion years) and free O2 in the atmosphere (≈1.8 billion years) (50). The evolution of hemoglobin/NOD with O2 makes sense because NOD requires O2 and because toxic NO• is produced via the O2-dependent oxidation or combustion of reduced nitrogen compounds. Finally, it is worth noting that the well-studied hemoglobins and myoglobins may not be the only O2 transport/storage proteins to have evolved from a dioxygenase function; the unrelated abalone “myoglobins” apparently formed from indolamine 2,3-dioxygenase (51).
Acknowledgments
We are grateful to Drs. Alex Ninfa and Bruce Demple for providing bacterial strains. We especially thank Drs. Irwin Fridovich and Dan Nebert for their comments on early drafts of the manuscript. This work was supported in part by an American Heart Association Scientist Development Grant (P.R.G.).
ABBREVIATIONS
- NOD
nitric oxide dioxygenase
- LB
Luria–Bertani
References
- 1.Goretski J, Zafiriou O C, Hollocher T C. J Biol Chem. 1990;265:11535–11538. [PubMed] [Google Scholar]
- 2.Marletta M A. J Biol Chem. 1993;268:12231–12234. [PubMed] [Google Scholar]
- 3.Kelm M, Schrader J. Circ Res. 1990;66:1561–1575. doi: 10.1161/01.res.66.6.1561. [DOI] [PubMed] [Google Scholar]
- 4.Snyder S H, Bredt D S. Sci Am. 1992;266:68–71. doi: 10.1038/scientificamerican0592-68. [DOI] [PubMed] [Google Scholar]
- 5.Fang F C. J Clin Invest. 1997;99:2818–2825. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry Y, Lepoirre M, Drapier J C, Ducrocq C, Boucher J L, Guissani A. FASEB J. 1993;7:1124–1134. doi: 10.1096/fasebj.7.12.8397130. [DOI] [PubMed] [Google Scholar]
- 7.Nunoshiba T, Derojas-Walker T, Tannenbaum S R, Demple B. Infect Immun. 1995;63:794–798. doi: 10.1128/iai.63.3.794-798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwarz M A, Lazo J S, Yalowich J C, Allen W P, Whitmore M, Bergonia H A, Tzeng E, Billiar T R, Robbins P D, Lancaster J R, Jr, Pitt B R. Proc Natl Acad Sci USA. 1995;92:4452–4456. doi: 10.1073/pnas.92.10.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakahara K, Tanimoto T, Hatano K, Usuda K, Shoun H. J Biol Chem. 1993;268:8350–8355. [PubMed] [Google Scholar]
- 10.Costa C, Macedo A, Moura I, Moura J J G, Le Gall J, Berlier Y, Liu M-Y, Payne W J. FEBS Lett. 1990;276:67–70. doi: 10.1016/0014-5793(90)80508-g. [DOI] [PubMed] [Google Scholar]
- 11.Gardner P R, Costantino G, Szabó C, Salzman A L. J Biol Chem. 1997;272:25071–25076. doi: 10.1074/jbc.272.40.25071. [DOI] [PubMed] [Google Scholar]
- 12.Gardner, P. R., Costantino, G. & Salzman, A. L. (1998) J. Biol. Chem. 273, in press. [DOI] [PubMed]
- 13.Hardison R C. Proc Natl Acad Sci USA. 1996;93:5675–5679. doi: 10.1073/pnas.93.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu H, Riggs A F. Proc Natl Acad Sci USA. 1992;89:5015–5019. doi: 10.1073/pnas.89.11.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wittenberg J B, Wittenberg B A. Annu Rev Biophys Chem. 1990;19:217–241. doi: 10.1146/annurev.bb.19.060190.001245. [DOI] [PubMed] [Google Scholar]
- 16.Membrillo-Hernandez J, Poole R K. FEMS Microbiol Lett. 1997;155:179–184. doi: 10.1016/s0378-1097(97)00384-4. [DOI] [PubMed] [Google Scholar]
- 17.Bueno R, Pahel G, Magasanik B. J Bacteriol. 1985;164:816–822. doi: 10.1128/jb.164.2.816-822.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg J T, Demple B. EMBO J. 1986;7:2611–2617. doi: 10.1002/j.1460-2075.1988.tb03111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasudevan S G, Armarego W L F, Shaw D C, Lilley P E, Dixon N E, Poole R K. Mol Gen Genet. 1991;226:49–58. doi: 10.1007/BF00273586. [DOI] [PubMed] [Google Scholar]
- 20.Green L C, Wagner D A, Glogowski J, Skipper P L, Wishnok J S, Tannenbaum S R. Anal Biochem. 1982;126:131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- 21.Appleby C A. Methods Enzymol. 1978;52:157–166. doi: 10.1016/s0076-6879(78)52018-1. [DOI] [PubMed] [Google Scholar]
- 22.Ioannidis N, Cooper C E, Poole R K. Biochem J. 1992;288:649–655. doi: 10.1042/bj2880649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Testa B. In: Biochemistry of Redox Reactions. Testa B, Caldwell J, editors. San Diego: Academic; 1995. pp. 164–202. [Google Scholar]
- 24.Membrillo-Hernández J, Ioannidis N, Poole R K. FEBS Lett. 1996;382:141–144. doi: 10.1016/0014-5793(96)00154-8. [DOI] [PubMed] [Google Scholar]
- 25.Gardner P R, Fridovich I. J Biol Chem. 1992;267:8757–8763. [PubMed] [Google Scholar]
- 26.Ermler U, Siddiqui R A, Cramm R, Friedrich B. EMBO J. 1995;14:6067–6077. doi: 10.1002/j.1460-2075.1995.tb00297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper C E, Ioannidis N, D’mello R, Poole R K. Biochem Soc Trans. 1994;22:709–713. doi: 10.1042/bst0220709. [DOI] [PubMed] [Google Scholar]
- 28.Poole R K, Ioannidis N, Orii Y. Proc R Soc London B. 1994;255:251–258. doi: 10.1098/rspb.1994.0036. [DOI] [PubMed] [Google Scholar]
- 29.Orii Y, Ioannidis N, Poole R K. Biochem Biophys Res Commun. 1992;187:94–100. doi: 10.1016/s0006-291x(05)81463-9. [DOI] [PubMed] [Google Scholar]
- 30.Poole R K, Rogers N J, D’mello R A, Hughes M N, Orii Y. Microbiology. 1997;143:1557–1565. doi: 10.1099/00221287-143-5-1557. [DOI] [PubMed] [Google Scholar]
- 31.Poole R K, Ioannidis N, Orii Y. Microbiology. 1996;142:1141–1148. doi: 10.1099/13500872-142-5-1141. [DOI] [PubMed] [Google Scholar]
- 32.Weiss J J. Nature (London) 1964;202:83–84. doi: 10.1038/202083b0. [DOI] [PubMed] [Google Scholar]
- 33.Misra H P, Fridovich I. J Biol Chem. 1972;247:6960–6962. [PubMed] [Google Scholar]
- 34.Doyle M P, Hoekstra J W. J Inorg Biochem. 1981;14:351–358. doi: 10.1016/s0162-0134(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 35.Pryor W A, Squadrito G L. Am J Physiol. 1995;268:L699–L722. doi: 10.1152/ajplung.1995.268.5.L699. [DOI] [PubMed] [Google Scholar]
- 36.Keilin D. Nature (London) 1953;172:390–393. doi: 10.1038/172390a0. [DOI] [PubMed] [Google Scholar]
- 37.Poole R K. Antonie Van Leeuwenhoek. 1994;65:289–310. doi: 10.1007/BF00872215. [DOI] [PubMed] [Google Scholar]
- 38.Andrews S C, Shipley D, Keen J N, Findlay J B C, Harrison P M, Guest J R. FEBS Lett. 1992;302:247–252. doi: 10.1016/0014-5793(92)80452-m. [DOI] [PubMed] [Google Scholar]
- 39.Vasudevan S G, Tang P, Dixon N E, Poole R K. FEMS Microbiol Lett. 1995;125:219–224. doi: 10.1111/j.1574-6968.1995.tb07361.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhao X-J, Raitt D, Burke P V, Clewell A S, Kwast K E, Poyton R O. J Biol Chem. 1996;271:25131–25138. doi: 10.1074/jbc.271.41.25131. [DOI] [PubMed] [Google Scholar]
- 41.Cramm R, Siddiqui R A, Friedrich B. J Biol Chem. 1994;269:7349–7354. [PubMed] [Google Scholar]
- 42.Crawford M J, Goldberg D E. J Biol Chem. 1998;273:12543–12547. doi: 10.1074/jbc.273.20.12543. [DOI] [PubMed] [Google Scholar]
- 43.Poole R K, Anjum M F, Membrillo-Hernandez J, Kim S O, Hughes M N, Stewart V. J Bacteriol. 1996;178:5487–5492. doi: 10.1128/jb.178.18.5487-5492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dikshit K L, Spaulding D, Braun A, Webster D A. J Gen Microbiol. 1989;135:2601–2609. doi: 10.1099/00221287-135-10-2601. [DOI] [PubMed] [Google Scholar]
- 45.Lacelle M, Kumano M, Kurita K, Yamane K, Zuber P, Nakano M M. J Bacteriol. 1996;178:3803–3808. doi: 10.1128/jb.178.13.3803-3808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Potts M, Angeloni S V, Ebel R E, Bassam D. Science. 1992;256:1690–1692. doi: 10.1126/science.256.5064.1690. [DOI] [PubMed] [Google Scholar]
- 47.Favey S, Labesse G, Vouille V, Boccara M. Microbiology. 1995;141:863–871. doi: 10.1099/13500872-141-4-863. [DOI] [PubMed] [Google Scholar]
- 48.Andersson C R, Ostergaard E, Llewellyn D J, Dennis E S, Peacock W J. Proc Natl Acad Sci USA. 1996;93:5682–5687. doi: 10.1073/pnas.93.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Appleby C A, Turner G L, Manicol P K. Biochim Biophys Acta. 1975;387:461–474. doi: 10.1016/0005-2728(75)90086-9. [DOI] [PubMed] [Google Scholar]
- 50.Giess A C. Cell Physiology. 5th Ed. Philadelphia: Saunders; 1979. pp. 43–44. [Google Scholar]
- 51.Suzuki T. J Protein Chem. 1994;13:9–13. doi: 10.1007/BF01891987. [DOI] [PubMed] [Google Scholar]